Abstract
Objective Our objective was to phenotype periodontal disease (PD) diagnoses from three different sections (diagnosis codes, clinical notes, and periodontal charting) of the electronic dental records (EDR) by developing two automated computer algorithms.
Methods We conducted a retrospective study using EDR data of patients ( n = 27,138) who received care at Temple University Maurice H. Kornberg School of Dentistry from January 1, 2017 to August 31, 2021. We determined the completeness of patient demographics, periodontal charting, and PD diagnoses information in the EDR. Next, we developed two automated computer algorithms to automatically diagnose patients' PD statuses from clinical notes and periodontal charting data. Last, we phenotyped PD diagnoses using automated computer algorithms and reported the improved completeness of diagnosis.
Results The completeness of PD diagnosis from the EDR was as follows: periodontal diagnosis codes 36% ( n = 9,834), diagnoses in clinical notes 18% ( n = 4,867), and charting information 80% ( n = 21,710). After phenotyping, the completeness of PD diagnoses improved to 100%. Eleven percent of patients had healthy periodontium, 43% were with gingivitis, 3% with stage I, 36% with stage II, and 7% with stage III/IV periodontitis.
Conclusions We successfully developed, tested, and deployed two automated algorithms on big EDR datasets to improve the completeness of PD diagnoses. After phenotyping, EDR provided 100% completeness of PD diagnoses of 27,138 unique patients for research purposes. This approach is recommended for use in other large databases for the evaluation of their EDR data quality and for phenotyping PD diagnoses and other relevant variables.
Keywords: periodontal disease, data quality, automated algorithms, electronic dental record, phenotype
Background and Significance
There is a significant increase in the utilization of electronic dental record (EDR) systems for patient care and reimbursement purposes. 1 2 It has been demonstrated that the patient care information documented in the EDR has invaluable utility in clinical research and for quality improvement purposes. 3 4 5 6 As a result, there has been a steep curve in using EDR data for research in the last decade. Researchers have developed advanced machine learning algorithms and statistical models to utilize EDR data to extract information, predict disease risk, and provide personalized treatment recommendations. 2 7 8 9 Despite this massive shift toward “big EDR data research,” the transition of the research results generated through EDR data to practice is limited and controversial. 10 11 There are challenges associated with questionable quality and reliability, missing information, and fragmented information in different sections of the EDR. Hence, it is critical to determine the quality of the EDR data before its intended use because poor data quality may lead to flawed outcomes. 6 11
EDR data also provide longitudinal patient care information for periodontal disease (PD) research. 12 13 PD is one of the most prevalent dental diseases which may cause tooth loss and poor quality of life if left untreated. 14 15 The prevalence of PD is high worldwide. For example, approximately 80% of adults in the United States have periodontal inflammation and 47% have destructive periodontitis. 14 16 17 Gingivitis is an inflammation of the gingiva (gums) surrounding teeth, while periodontitis is the inflammation and loss of the periodontal attachment and alveolar bone. 14 18 Further research is needed to advance our knowledge of these diseases, including information about their prevalence, incidence and progression in various populations, etiologic factors among vulnerable groups, and long-term efficacy of various treatment regimens and in developing prediction models to identify high-risk patients, with the ability to provide up-to-date real-world information. 13
EDR data are intended to support patient care and are not designed specifically for research. Hence, using EDR data for research presents challenges related to missing data, poor data completeness, and fragmented information. 6 10 11 19 For instance, patients' complete dental diagnosis information may not be available for all patients because dentists get reimbursed based on procedures performed, rather than diagnosis. 20 In addition, it is instrumental to assess the data quality of clinical variables. Data quality completeness implies the utilization of all sections of the EDR for relevant data, although the information could have been reported in multiple sections of the EDR. On the contrary, utilizing EDR data for research has several advantages such as providing patients' longer follow-up information that may be difficult to collect prospectively and providing “real-world” data at a significantly lower cost. 3 21
A few studies in dentistry have evaluated the quality of the EDR data. For example, Patel et al found that the cardiovascular disease information documented in the EDR may not be reliable because they are patient reported. 22 And at the same time, smoking information reported in the EDR may be a more reliable resource to obtain patients' detailed smoking information (smoking intensity and duration). 5 Thyvalikakath et al examined data quality of private dental practices through the National Dental PBRN Practices and found that patients' age and gender information were recorded for 100% of patients. 6 However, 8% of observations had incorrect data such as incorrect tooth number, tooth surface, primary teeth, supernumerary teeth, and tooth ranges, indicating multitooth procedures instead of posterior composite restorations or root canal treatment. Mullins et al have assessed the completeness of PD documentation in the EHR and found the feasibility of developing automated data extraction script using only structured data. 23 Despite this effort, as per our best knowledge, no study has attempted to improve the completeness of PD diagnoses by utilizing different sections of the EHR such as periodontal charting, clinical notes, and diagnoses reported by the clinicians.
The objectives of the study are two-fold: (1) to appraise data completeness and accuracy on the diagnosis of PD documented in a large EDR system at Temple University Kornberg School of Dentistry (TUKSoD) and (2) to develop and test automated computer algorithms by phenotyping PD diagnosis information from multiple sections of the EDR. The results of this study will allow us to determine the quality of PD-related clinical variables stored in the EDR, generating a dental-specific data quality framework and three automated Python programming algorithms to automatically diagnose PD based on the current disease classification. 24 In this study, we only considered chronic periodontitis classification system from the American Academy of Periodontology (AAP) and excluded classification of other types of periodontitis such as necrotizing, aggressive, and periodontitis as a manifestation of systemic diseases. 24
Methods
A retrospective study using EDR data from TUKSoD clinics was conducted. Patients' demographics, periodontal findings, clinical case notes, and PD diagnosis in the EDR were retrieved. The completeness of variables required to diagnose PD was determined and an automated computer algorithm to diagnose patients' PD statuses from periodontal charting findings was created. A natural language processing (NLP) program was also created to retrieve diagnoses recorded by clinicians in the clinical notes as free text. The two automated computer algorithms were developed to improve the completeness of PD diagnoses documented in the EDR. The performance of these programs was evaluated through manual review processes. Finally, data completeness before and after using the automated computer algorithms was assessed ( Fig. 1 ).
Fig. 1.
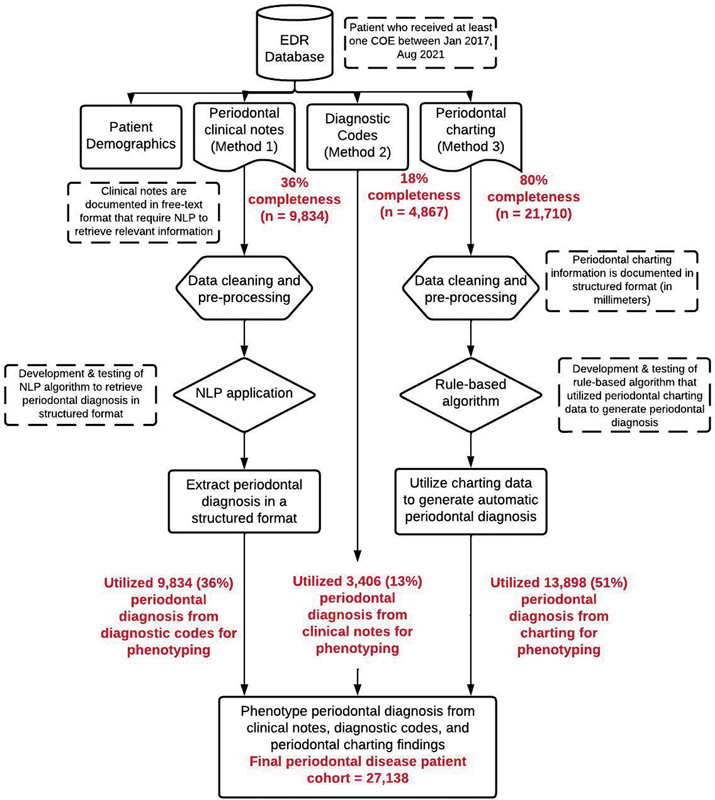
Overall workflow to phenotype periodontal disease information from three sections of the EDR. EDR, electronic dental records.
Data Retrieval and Patient Cohort
EDR (axiUm, Exan software, Las Vegas, Nevada, United States) data of patients who received at least one comprehensive oral examination (COE) at TUKSoD between January 1, 2017, and August 31, 2021 were used. There were 27,138 unique patients who received at least one COE during the study time. PD diagnosis documented during the patient's most recent visit was considered. For example, if the patient has received dental treatments in 2017, 2019, and 2021, then the charting updated during his/her 2021 visit was considered. The new patients during the study time period were excluded because the charting information may not be completed. To avoid including incomplete charting information, we only included those patients whose COE code indicated “complete” in the database. This demonstrates that the dental students have successfully completed documenting patients' charting information which was then reviewed and approved by the clinic faculty members. The dataset included patient demographics, diabetes history, PD diagnoses, periodontal charting, and smoking information (variables necessary to diagnose PD).
Periodontal Disease Information in Different Sections of the Electronic Dental Records
Patient's PD information was recorded in three different sections within the EDR.
Diagnosis section (Method [M] 1): A separate diagnosis section was provided in the EDR where clinicians were trained and instructed to document patients' detailed dental diagnoses, using the systemized nomenclature dental diagnostic system, including PD. This EDR section stores diagnosis information in a structured format as selected using a dropdown list.
Clinical notes section (M 2): Clinicians were provided a separate text box to write patients' clinical and prognosis information in periodontal evaluation forms. This section typically documents patients' gingival health, bone loss information, and PD diagnosis and stores this information in free text format.
Periodontal charting section (M 3): Clinicians documented periodontal findings such as clinical attachment loss (CAL), periodontal probing depth (PPD), bleeding on probing (BOP), and bone loss information. This information was stored in a structured format. For example, CAL and PPD information was documented in millimeters and BOP information was documented as Boolean (yes/no).
Completeness of Periodontal Disease Diagnosis Information
Completeness is the most assessed dimension of EDR data quality and refers to data availability or missing data. A similar model to that described by Weiskopf et al was used to calculate the completeness of the needed variables. 10 19 The proportions of present values by the total number of patients that received COE to examine completeness were determined. [Completeness = total reported observation per section (M1/M2/M3)/27,138)]. 10 19 However, as the diagnosis is typically not used for reimbursement purposes, this information may be recorded using a dropdown list but is often missing. Therefore, we assessed completeness of diagnosis documentation in different sections of the EDR. We then developed computer algorithms to provide automating diagnosis of patients' whose periodontal diagnosis is not recorded in any section of the EDR. The rationale is to improve the completeness of the diagnosis using all the possible sections of the EDR.
Development of an Automated Computer Algorithm (PerioDx Diagnoser) to Diagnose Patients' Periodontal Disease Status
The PerioDx Diagnoser in Python (open-source computer programming language) automatically classifies patients' PD status into healthy, gingivitis, or periodontitis using the criteria of the 2017 Classification of Periodontal Diseases ( Fig. 2 ). 24 Working with the clinical data extracted from the EDR, the PerioDx Diagnoser first identifies if patients' CAL, PPD, BOP, and bone loss information has been recorded, which are necessary variables to diagnose PD. For instance, if the patient has 40 BOP sites and have a total of 28 teeth present, then the BOP score would be [40/(28*6)168] 24%. Based on the BOP score, this patient would be classified as localized gingivitis. Similarly, it also automatically provides different grading and staging of periodontitis. For instance, if the patient has (1) bone loss of >33% (middle third), (2) CAL of 3 to 5 mm and PPD of >5 mm for more than 30% of dentition, and (3) >5 mobile teeth, then this patient is categorized as generalized stage IV periodontitis. To automatically diagnose patients' periodontitis status, utilization of periodontal bone loss information is critical. We obtained this information from the periodontal evaluation forms. At TUKSoD, the periodontal evaluation form has a dropdown list for clinicians to document patients' bone loss information. This includes (1) bone loss <15%, (2) bone loss between 15% and 33%, and (3) bone loss > 33%. In addition, we took one step further and determined the grade of periodontitis as well. We utilize patients' smoking and HbA1c levels to obtain periodontitis grading. For example, if the patient smokes >10 cigarettes a day and is diabetic with HbA1c ≥7, then this patient will be classified as grade C periodontitis case.
Fig. 2.
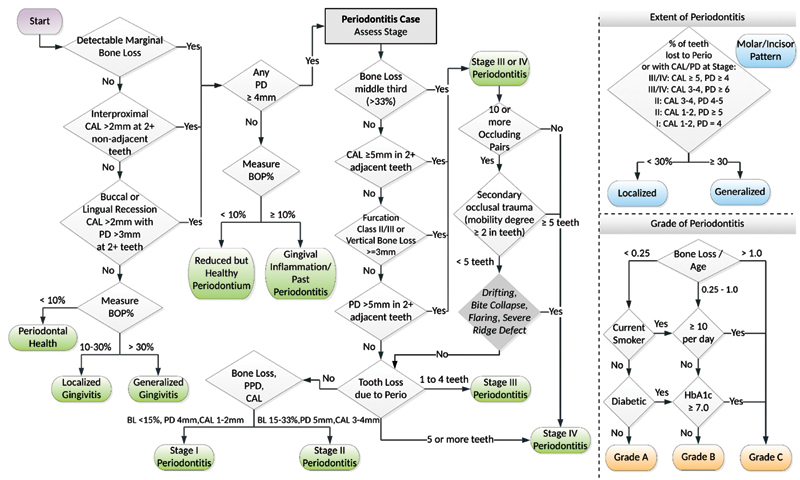
Process to diagnose patients' PD status automatically from periodontal findings based on the 2017 Classification of Periodontal and Peri-implant Diseases and Conditions. *BOP, bleeding on probing; *CAL, clinical attachment loss; *PD, pocket depth.
Development of a Natural Language Processing Algorithm (PerioDx Extractor) to Extract Patients' Periodontal Disease Diagnoses from Clinical Notes
As described in section 4.2, patients' PD diagnoses may also be available in the clinical notes section of the EDR within the free-text format. Unlike dropdown lists, free-text boxes allow clinicians to write their clinical findings without any limitations. However, free-text data are stored in an unstructured format and are difficult to mine for analysis. Extracting information out of free-text data needs experts' manual review of selected patient's clinical notes with further coding of structured/categorical variables to perform statistical analysis. 25 26 27 Therefore, an NLP algorithm (PerioDx Extractor) was developed to extract PD diagnoses automatically from the clinical notes. In this program, computer algorithms were trained to read and interpret lengthy clinical text documented by the clinicians, which were converted into structured/categorical format for further analysis. Below we describe detailed steps in developing and testing our NLP program.
We used a bottom-up approach to develop our NLP program. 20 First, we created manual annotation guidelines. Two domain experts manually reviewed 100 clinical notes that contained PD diagnosis, stage of periodontitis, grade of periodontitis, smoking histories, and their HbA1C level for diabetes diagnosis. We used “The extensible Human Oracle Suite of Tools” tool to annotate clinical notes. 28 29 For the PD diagnoses, we collected bag of words related to extent, stage, grade, and severity of PD as presented in the 2017 AAP classification. 24 For the diabetes status, we annotated HbA1C level <5 as normal, 5.7% ≤ HbA1C < 6.4% as prediabetes, and smoking status into nonsmoker (patient who never smoked), <10 cigarettes/day, and ≥10 cigarettes/day. We performed the manual annotation process with four iterations. After every iteration, we calculated the interrater agreement using Cohen's Kappa test. Disagreed concepts between the annotators were discussed and resolved through consensus.
We then developed a two-step NLP algorithm. First, we created a program to preprocess the data to remove special characters, capitalizations, removal of stop words, and created text chunks by tokenization. We performed this task using several Python libraries, such as the Natural Language Toolkit, Version 3.5, string-matching algorithm, regular expression, and pandas. 30 31 32 We then used a keyword approach from the gold standard dataset to only extract patients' clinical notes that had a mention of at least one PD diagnosis. This approach yielded a total of 4,867 clinical notes. The rationale for doing this task is to reduce false-positive error rate and to save processing time and computational power. In the second filter, we took one step further and identified keywords related to the staging and grading of periodontitis. In the second filter, we used two major functions including word stemming function and text similarity function in the Python library. We utilized “gensim” and “scikit-learn” functions in Python to provide the most closet word vector compared with the gold-standard word dictionary. We then merged both of the outputs (outputs from filtering step 1, and step 2) into one output to obtain final PD diagnoses. We also developed a Python program to automatically calculate average length of characters, sentences, and words of clinical text. 33 34 35 The performance of each of these filtering approaches was then tested through a manual review process as described below.
Evaluate the Performances of PerioDx Diagnoser and PerioDx Extractor
The performances of PerioDx Diagnoser and PerioDx Extractor were evaluated through a manual review process. Two domain experts manually reviewed 200 patients' randomly selected periodontal charting data and provided their PD diagnoses using the 2017 PD classification. The manually reviewed diagnoses were then compared against the diagnoses provided by PerioDx Diagnoser. Similarly, experts reviewed 400 randomly selected patients' clinical notes and annotated PD diagnoses documented by the clinicians. These diagnoses were then compared against the PD diagnoses extracted in a structured format by PerioDx Extractor. A confusion matrix containing true positive (TP), false positive (FP), true negative (TN), and false negative (FN) were created for both algorithms. Using this confusion matrix, we calculated precision (correctly predicted positive observations to the total predicted positive observations), recall (correctly predicted positive observations to all observations in actual class), and F-1 measure (weighted average of precision and recall) to assess performances. 36
Phenotype Periodontal Disease Diagnoses from three Electronic Dental Records Sections (M1 + M2 + M3)
Last, PD diagnoses were from three EDR sections: (1) diagnosis sections (M1), (2) clinical notes (M2), and (3) periodontal charting (M3). Automated diagnoses generated from M2 and M3, were merged with M1 to create the final PD patient cohort. During phenotyping, first, all available PD diagnoses from the diagnosis section documented by the clinicians (M1) were utilized, followed by the diagnoses extracted from clinical notes (M2), and then the diagnoses generated from periodontal charting data (M3). Finally, improved completeness of PD diagnoses was reported after phenotyping.
Results
Patient Demographics and Data Completeness
Our sample consisted of 27,138 unique dental patients who received at least one COE between January 1, 2017, and August 31, 2021. Our patients' most common age group was 58 to 67 years (19% [ n = 5,240]), followed by 48 to 57 years (18% [ n = 4,851]), and 28 to 37 years (17%[ n = 4,673]). More than half (57%) of our patients' race information was missing. Among the remaining 43% reporting race, African American was the most frequent race (28%), followed by white (12%). The majority of our patients were females (57%). Periodontal diagnosis codes were available for only 36% ( n = 9,834) of patients, and diagnoses in clinical notes were available for 18% ( n = 4,867). Complete periodontal charting data were available for 80% ( n = 21,710) of patients ( Fig. 1 ; mutually inclusive). After phenotyping (M1 + M2 + M3), the completeness of PD diagnoses improved for all patients ( n = 27,138) .
Periodontal Disease Patient Cohort after Phenotyping
We used a stepwise approach to create a final PD patient cohort from the three EDR sections. We first reported PD diagnoses that were available through the diagnosis codes section (M1; 36% of final cohort), followed by the clinical notes (M2; 18% of final cohort), and last, periodontal charting (M3; 46% of final cohort). Diagnoses generated out of 4,867 patients' clinical notes, 1,461 diagnoses were mutually inclusive with the M1 method (diagnosis codes). Therefore, during the final step, we utilized (M1; 36% of final cohort), followed by the clinical notes (M2; 13% of final cohort), and last, periodontal charting (M3; 51% of final cohort). The rationale for using this stepwise approach is that the automated diagnosis is not needed when the clinicians' diagnoses are available. Therefore, first, we reported diagnoses documented by the clinicians in M1 and M2 (mutually exclusive diagnoses), and the remaining missing diagnoses were obtained from the periodontal charting information. These diagnoses are the patient levels indicating the latest diagnosis per patient. Fig. 3 demonstrates the breakdown of PD diagnoses by each PD category and their data source (M1, M2, M3). We found consistent diagnosis categories across all data sources. In total, 11% of patients were healthy, 43% had gingivitis, 3% had stage I, 36% had stage II, and 7% had stage III/IV periodontitis. We also obtained patients' grading information with staging information by utilizing their medical history (diabetes) and social history (smoking) sections. For example, 2,629 patients had stage II grade A generalized periodontitis, 1,088 patients had stage II grade A localized periodontitis, and such as. Detailed information about detailed periodontitis grading and staging is described in Fig. 3 .
Fig. 3.
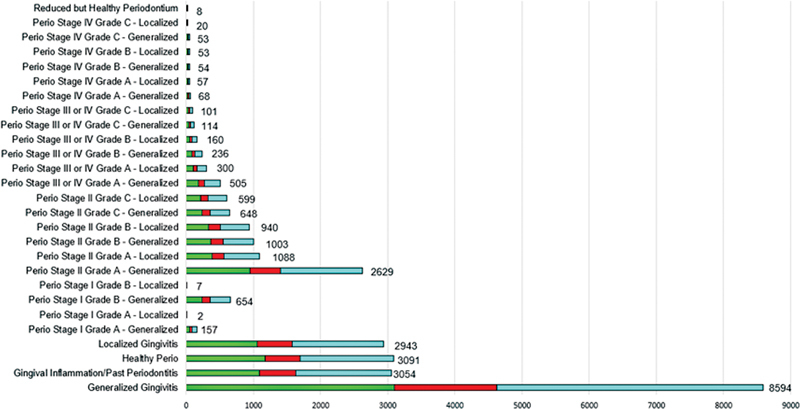
Phenotype periodontal diagnoses using diagnosis codes, clinical notes, and diagnosis generated from charting findings.
Average Length of Clinical Texts
The average length of sentences, words, and characters in clinical notes was 5.2, 72.0, and 521.26, respectively.
Performance of PerioDx Diagnoser and PerioDx Extractor
The PerioDx Diagnoser performed with 96% precision, 98% recall, and 97% of F-1 measure. Because periodontal charting data were present in a structured format, it was easier to compute and provide accurate diagnoses. However, the PerioDx Diagnoser could not provide diagnoses of the patients who had incomplete charting. Similarly, PerioDx Extractor with 91% precision, 87% recall, and 95% of F-1 measure to automatically extract patients' PD diagnoses from clinical and prognosis notes. Our expert manual reviewers found 224 TP, 154 TN, 22 FP, and 0 FN out of 400 manually reviewed clinical notes ( Table 1 ).
Table 1. Performance of PerioDx extractor.
| Total population: 400 | Predicted condition positive: 246 | Predicted condition negative: 154 | Informedness: 0.85 |
|---|---|---|---|
| Actual condition positive: 112 | True positive: 224 | False negative: 0 | True positive rate: 1 |
| Actual condition negative: 154 | False positive: 22 | True negative: 154 | False positive rate: 0.14 |
| Prevalence: 0.59 | Positive predictive Value: 0.91 | False omission Rate: 0 | Positive likelihood ratio: 7 |
| Accuracy: 0.94 | False discovery Rate: 0.089 | Negative predictive value: 1 | Markedness: 0.91 |
| Balanced accuracy: 0.93 | F1 Score: 0.95 | Fowlkes–Mallows index: 0.95 | Matthews correlation coefficient: 0.88 |
Upon error analysis of these, we identified a few reasons for these errors and these are described below.
There were many inconsistencies in the format of the text that the dental clinicians used to describe the patients' PD diagnoses in the clinical notes. For instance, in some cases, the text states negation concepts such as “not the presence of inflammation,” “not the presence of bone loss,” etc., the PerioDx Extractor falsely identified without consideration of negation concepts and falsely identified as positive cases.
In a few cases, the clinical notes contained acronyms or incomplete words to describe clinical findings of PD, such as “ging” for gingivitis, and “st. 2 perio” for stage II periodontitis. As a result, the algorithm was unable to identify those cases.
Discussion
This study developed advanced computational applications to phenotype PD diagnoses from multiple sections of the EDR. The completeness of data was poor (only 36%) when considering the diagnosis codes section where clinicians are supposed to diagnose PD diagnoses. However, the completeness improved to 100% when we utilized multiple sections of the EDR. We achieved this goal by developing two computational applications ( PerioDx Diagnoser and PerioDx Extractor ). As per our best knowledge, no other study has attempted to develop automated approaches to phenotype PD diagnoses from EDR. The results show that our algorithms that implemented the 2017 AAP classification system 24 were effective with 97% F-1 score in automatically diagnosing patients' detailed PD classification when these diagnoses were missing from the EDR.
Only few dental studies have evaluated the quality of EDR data. 5 6 8 12 20 21 22 23 For example, Patel et al. compared the self-reported cardiovascular disease (CVD) information with dental patients' medical records. They found low to no agreement and concluded that self-reported CVD information in the EDR may not be reliable research sources compared with electronic medical records. Similarly, the authors extracted dental patients' detailed smoking status for research. They used three machine learning models (support vector machines, random forest, and Naïve Bayes) to classify patients into light, intermediate, intermittent, past, or current smokers. Unlike the CVD study, they found that EDR provided more accurate information than electronic medical records. Thyvalikakath et al 6 examined data quality of private dental practices through the National Dental PBRN Practices and found that patients' age and gender information were recorded for 100% of patients. However, 8% of observations had incorrect data such as incorrect tooth number, tooth surface, primary teeth, supernumerary teeth, and tooth ranges, indicating multitooth procedures instead of posterior composite restorations or root canal treatment. Even though these studies provided meaningful insights on the quality of tooth-related variables, patient demographics, and medical and social histories, none assessed the data quality of PD diagnosis information. As per our best knowledge, there are no published studies that have evaluated the periodontal phenotype information from different sections of the EDR to improve the data quality and completeness.
The automated approaches generated in this study can be utilized to automatically document patients' PD diagnoses because of the fragmented reporting of diagnosis. Next, this phenotype approach could be utilized to first improve the completeness of the EDR data which then can be utilized to study PD. For example, this approach can be utilized to examine the long-term periodontal treatment outcomes and to develop prediction models.
One important takeaway from this study is that EDRs have a high potential to provide good quality patient information for research and quality improvement purposes. For example, this information can be used to develop prediction models for PD to assess future disease risk, which can be used to implement disease preventive approaches. However, it is critical to utilize all sections of the EDR to obtain the best quality data and to avoid biased outcomes, as demonstrated in this study. This study demonstrated the power of informatics methods to curate and mine the EDR data for research and quality improvement purposes. In this study, an interdisciplinary team of dentists, informaticists, and computer scientists developed computer applications that provided f-1 scores of 97 and 95% for obtaining automated PD diagnoses from periodontal charting and clinical notes, respectively. The domain experts manually reviewed and collected a bag of words for the PerioDx extractor program, and informatics researchers deployed them as an NLP algorithm in Python programming language.
This study has some limitations. The present results may not be generalizable because the writing patterns of clinical notes of PD diagnoses may vary by the school's culture, training, and experience of faculty members. However, our algorithm provides a foundation to develop personalized NLP pipelines as researchers would only have to update the bag of words in the NLP algorithm. Another limitation is that the study did not include data before 2017 because the current PD classification was introduced in 2017. Next, we did not consider negation concepts in our NLP program which resulted in a few FP cases. Finally, we had to rely on a substantial amount of manual review process which will be addressed in the future work by adding active learning component in our NLP algorithm.
Conclusions and Future Work
The EDR has a high potential to provide good quality PD diagnoses and charting information if phenotyping approaches are used, as demonstrated in this study. This study provided proof of the concept of evaluating the EDR data quality. This is important since data quality evaluation could minimize biased outcomes when the data are utilized for reporting or research. We successfully developed, tested, and implemented two automated algorithms (PerioDx Diagnoser and PerioDx Extractor) on large EDR datasets to improve the completeness of PD diagnoses in the EDR. Other investigators can use this approach to evaluate their EHR data quality and phenotype PD diagnoses and other relevant variables. Future work will determine the concordance between the PD diagnoses generated from PerioDx Diagnoser and clinician-documented diagnoses. We will also improve the performance of our NLP algorithm by adding more bag of words to our NLP dictionary. We will then use this information to develop data-driven prediction models to enhance disease prevention and examine long-term treatment outcomes.
Acknowledgment
We acknowledge dear reviewers and dear Dr. Thankam Paul Thyvalikakath for their critical feedback. Their feedback helped improve the scientific region of this research. J.P. would also like to thank Dr. Thankam Paul Thyvalikakath to teach the importance of data quality evaluation concept during his PhD education.
Conflict of Interest None declared.
Protection of Human Subjects and Animals in Research
This study was reviewed and approved by our institutional review board (IRB: 28321) granted authorization. In this retrospective study, we used deidentified patient datasets from the patients' EDR records; therefore, informed consents were not required to obtain.
References
- 1.Bruland P, Doods J, Storck M, Dugas M. What information does your EHR contain? Automatic generation of a Clinical Metadata Warehouse (CMDW) to support identification and data access within distributed clinical research networks. Stud Health Technol Inform. 2017;245:313–317. [PubMed] [Google Scholar]
- 2.Song M, Liu K, Abromitis R, Schleyer T L. Reusing electronic patient data for dental clinical research: a review of current status. J Dent. 2013;41(12):1148–1163. doi: 10.1016/j.jdent.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coorevits P, Sundgren M, Klein G O. Electronic health records: new opportunities for clinical research. J Intern Med. 2013;274(06):547–560. doi: 10.1111/joim.12119. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui Z, Wang Y, Patel J, Thyvalikakath T. Differences in medication usage of dental patients by age, gender, race/ethnicity and insurance status. Technol Heal Care. 2021;29(06):1099–1108. doi: 10.3233/THC-202171. [DOI] [PubMed] [Google Scholar]
- 5.Patel J, Siddiqui Z, Krishnan A, Thyvalikakath T P.Leveraging electronic dental record data to classify patients based on their smoking intensity Methods Inf Med 201857(5-06):253–260. [DOI] [PubMed] [Google Scholar]
- 6.National Dental PBRN Collaborative Group . Thyvalikakath T P, Duncan W D, Siddiqui Z. Leveraging electronic dental record data for clinical research in the National Dental PBRN Practices. Appl Clin Inform. 2020;11(02):305–314. doi: 10.1055/s-0040-1709506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson J I, Patel J S, Ramya M B. Longevity of crown margin repairs using glass ionomer cement: a retrospective study. Oper Dent. 2021;46(03):263–270. doi: 10.2341/20-062-C. [DOI] [PubMed] [Google Scholar]
- 8.Thyvalikakath T P, Padman R, Vyawahare K, Darade P, Paranjape R. Utilizing dental electronic health records data to predict risk for periodontal disease. Stud Health Technol Inform. 2015;216:1081. [PubMed] [Google Scholar]
- 9.Krois J, Ekert T, Meinhold L. Deep learning for the radiographic detection of periodontal bone loss. Sci Rep. 2019;9(01):8495. doi: 10.1038/s41598-019-44839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiskopf N G, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20(01):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S, Wagner J, Lupulescu-Mann N et al. Comparison of EHR-based diagnosis documentation locations to a gold standard for risk stratification in patients with multiple chronic conditions. Appl Clin Inform. 2017;8(03):794–809. doi: 10.4338/ACI-2016-12-RA-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel J, Zai A, Kumar K.Retrospective study of deriving periodontal disease diagnosis from periodontal findingsJ Dent Res 2020. Available at:https://iadr.abstractarchives.com/abstract/20iags-3323278/retrospective-study-of-deriving-periodontal-disease-diagnosis-from-periodontal-findings
- 13.Patel J, Zai A, Kumar K.Utilizing electronic dental record data to monitor periodontal disease progressionmobilize Comput Biomed Knowl; July 18–19 2019; Bethesda, Maryland; Abstract 18. Accessed September 22, 2022.
- 14.Eke P I, Thornton-Evans G O, Wei L, Borgnakke W S, Dye B A, Genco R J. Periodontitis in US adults: National Health and Nutrition Examination Survey 2009-2014. J Am Dent Assoc. 2018;149(07):576–5.88E8. doi: 10.1016/j.adaj.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramseier C A, Anerud A, Dulac M. Natural history of periodontitis: disease progression and tooth loss over 40 years. J Clin Periodontol. 2017;44(12):1182–1191. doi: 10.1111/jcpe.12782. [DOI] [PubMed] [Google Scholar]
- 16.Eke P I, Dye B A, Wei L. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86(05):611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albandar J M.Epidemiology and risk factors of periodontal diseases Dent Clin North Am 20054903517–532., v–vi [DOI] [PubMed] [Google Scholar]
- 18.Trombelli L, Farina R, Silva C O, Tatakis D N. Plaque-induced gingivitis: case definition and diagnostic considerations. J Clin Periodontol. 2018;45 20:S44–S67. doi: 10.1111/jcpe.12939. [DOI] [PubMed] [Google Scholar]
- 19.Weiskopf N G, Bakken S, Hripcsak G, Weng C. A data quality assessment guideline for electronic health record data reuse. EGEMS (Wash DC) 2017;5(01):14. doi: 10.5334/egems.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel J S.Utilizing Electronic Dental Record Data to Track Periodontal Disease Change 2020. Available at:https://scholarworks.iupui.edu/bitstream/handle/1805/23677/Patel_iupui_0104D_10455.pdf?sequence=1
- 21.Chan K S, Fowles J B, Weiner J P. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev. 2010;67(05):503–527. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 22.Patel J, Mowery D, Krishnan A, Thyvalikakath T. Assessing information congruence of documented cardiovascular disease between electronic dental and medical records. AMIA Annu Symp Proc. 2018;2018:1442–1450. [PMC free article] [PubMed] [Google Scholar]
- 23.Mullins J, Yansane A, Kumar S V. Assessing the completeness of periodontal disease documentation in the EHR: a first step in measuring the quality of care. BMC Oral Health. 2021;21(01):282. doi: 10.1186/s12903-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonetti M S, Greenwell H, Kornman K S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J Periodontol. 2018;89 01:S159–S172. doi: 10.1002/JPER.18-0006. [DOI] [PubMed] [Google Scholar]
- 25.Pathak J, Bailey K R, Beebe C E.Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium J Am Med Inform Assoc 201320(e2):e341–e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koleck T A, Dreisbach C, Bourne P E, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc. 2019;26(04):364–379. doi: 10.1093/jamia/ocy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalifa A, Meystre S.Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes J Biomed Inform 201558(Suppl):S128–S132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Li C, Xu J, Wu H. A patient-oriented clinical decision support system for CRC risk assessment and preventative care. BMC Med Inform Decis Mak. 2018;18 05:118. doi: 10.1186/s12911-018-0691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GitHub—chrisleng/ehost: Annotation Tool: The extensible Human Oracle Suite of Tools (eHOST)Accessed September 9, 2022 at:https://github.com/chrisleng/ehost
- 30.Ravanelli M, Parcollet T, Bengio Y.The Pytorch-kaldi Speech Recognition ToolkitICASSP—IEEE International Conference on Acoustics, Speech and Signal Processing;2019
- 31.Loper E, Bird S.NLTK: The Natural Language Toolkit. Proceedings of the ACL-02 Workshop on Effective Tools and Methodologies for Teaching Natural Language Processing and Computational LinguisticsPhiladelphia, PA: Association for Computational Linguistics. . doi:10.48550/arxiv.cs/0205028
- 32.Hammami L, Paglialonga A, Pruneri G. Automated classification of cancer morphology from Italian pathology reports using natural language processing techniques: a rule-based approach. J Biomed Inform. 2021;116:103712. doi: 10.1016/j.jbi.2021.103712. [DOI] [PubMed] [Google Scholar]
- 33.Geng W, Qin X, Wang Z, Kong Q, Tang Z, Jiang L.Model-based reasoning methods for diagnosis in integrative medicine based on electronic medical records and natural language processing medRxiv 2020:2020.07.12.20151746. 10.1101/2020.07.12.20151746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Dodoo N A, Wen T J, Ke L. Understanding Twitter conversations about artificial intelligence in advertising based on natural language processing. Int J Advert. 2021;41(04):685–702. [Google Scholar]
- 35.Yang F, Wang X, Ma H, Li J.Transformers-sklearn: a toolkit for medical language understanding with transformer-based models BMC Med Inform Decis Mak 202121(2, Suppl 2):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lalkhen A G, McCluskey A. Clinical tests: sensitivity and specificity. Contin Educ Anaesth Crit Care Pain. 2008;8(06):221–223. [Google Scholar]


