Background:
This study investigated bacterial colonization of the foam eluate after negative-pressure wound therapy (NPWT) with instillation and dwell time (NPWTi-d) to obtain an indication of possible recontamination of the wound during NPWTi-d. To detect bacterial colonization and the extent of planktonic and nonplanktonic bioburden as comprehensively as possible, routine culture and molecular biology methods were used.
Methods:
Before (time point 1) and after (median 3.0 days; time point 2) NPWT (n = 15) and NPWTi-d with antiseptic installation (n = 15), wound bed [22 acute, eight chronic wounds; median age, 51 years (range, 24 to 91); 26 men], foam, and eluate were examined by routine culture methods and fluorescence in situ hybridization (FISH), polymerase chain reaction, and FISH sequencing (FISHseq).
Results:
At time point 2, 94.9% (37 of 39) of the pathogens identifiable in the eluate were also detected in the wound bed. Foam and eluate were always bacterially contaminated. NPWTi-d resulted in a significant reduction in the number of pathogen species compared with NPWT (NPWTi-d, time point 1 versus time point 2: P = 0.026; NPWT, time point 1 versus time point 2: not significant). Routine culture of wound bed samples at time point 2 identified only 28 of 52 (53.8%) of the pathogens, whereas examination of wound bed, foam, and eluate and additional FISHseq use detected 50 of 52 (96.2%) of the bacterial species. FISHseq identified biofilm in one and microcolonies in 10 wounds (time point 2).
Conclusions:
The bacterial load of the foam is flushed back into the wound during NPWTi-d. FISHseq should be used in addition to the routine culture method when pathogen identification and detection of nonplanktonic bacterial growth is particularly important for the patient’s therapy.
Clinical question/level of evidence:
Therapeutic, V.
Negative-pressure wound therapy (NPWT) has dramatically changed the care of complex wounds.1,2 Another important pillar in wound treatment is wound irrigation, which usually concludes the operation and is routinely performed during dressing changes. The combination of these two wound treatment options has been implemented technically since 1996 for retrograde instillation of a topical solution with removal using alternating negative pressure cycles as an important evolution of the NPWT concept.3,4 Modern NPWT with instillation and dwell time (NPWTi-d) permits a constantly computer-controlled programmable instillation and allows the wound solution to dwell in the wound bed for a user-selected period.5
Improved clinical outcomes have been reported consistently with the use of NPWTi-d combined with saline instillation or antiseptic solutions versus conventional NPWT.6–10 The clinical benefit, the added comfort, and process simplification of solution delivery in tandem with NPWT have prompted use of NPWTi-d as first-line therapy in a wider subset of complex wounds.11 NPWTi-d also was shown to reduce both free-living (planktonic) bacteria and biofilm-associated (nonplanktonic) bacteria significantly, whereas wounds treated with conventional NPWT did not show this clear effect.7,12,13 So it seems understandable that the most recent international guideline recommends the use of NPWTi-d, especially in contaminated wounds, using saline or antiseptic solution as instillation fluid in wounds with heavy bioburden.14
The recent literature does not, however, take into account the fact that the wound filler is usually colonized by bacteria after irrigation at the time of the foam change.15–17 Because the foam is rinsed retrogradely several times a day when using NPWTi-d, it must be assumed that bacteria are being repeatedly flushed back into the wound from this foam. Against this background, the extent of the contamination of the typically used polyurethane foam and the severity of possible bacterial recontamination of the wound bed are of great interest. There are no published studies that evaluate the eluate bioburden to prove whether pathogens from the foam are flushed back into the wound bed retrogradely during NPWTi-d. In addition, previous investigations of bacterial foam colonization were limited to conventional microbiological methods of detecting routinely culturable bacteria.17
In this study, we sought to investigate the extent and spatial distribution of the planktonic and biofilm-associated microbial load of the eluate after retrograde irrigation of the NPWT foam and wound tissue samples as comprehensively as possible. Bacterial colonization was analyzed using standard routine cultures and molecular analyses.
PATIENTS AND METHODS
Study Design and Patient Population
This prospective cohort study was performed in a center for septic defect wounds. We included 30 consecutive hospitalized patients (age 18 years or older) with acute or chronic soft tissue and bone infections receiving NPWT (n = 15) or NPWTi-d (n = 15). The manner in which patients were assigned to the groups (NPWT or NPWTi-d) was at the discretion of the respective surgeon. The study was approved by the Ethics Committee Berlin, Germany (EA1/055/15). All patients gave written informed consent to participate in this study.
Sample Collection
Tissue samples were taken from the wound bed of open wounds before any contact with a disinfectant in primarily closed infection situations after incision and exploration of the infection area (time point 1). Skin disinfection was performed in the operating theater immediately before surgery (Skinsept G; Ecolab, Monheim am Rhein, Germany). After radical débridement, the wound was irrigated extensively with antiseptic solutions and then closed using NPWT or NPWTi-d. After removal of the NPWT dressing (time point 2), foam and wound bed samples were taken. In addition, in both groups, the foam was rinsed retrogradely with 20 mL NaCl through the connecting tube under sterile conditions, simulating the retrograde instillation phase of NPWTi-d.
NPWT and NPWTi-d Application
Patients were assigned to either NPWT or NPWTi-d (V.A.C. Granufoam Dressing or V.A.C. Veraflo Dressing, 3M, St. Paul, Minn.) with antiseptic instillation solution [sodium hypochlorite/hypochlorous acid (HOCl); Granudacyn NPWT wound irrigation solution, Mölnlycke, Gothenburg, Sweden] immediately after surgical débridement. HOCl is the only antiseptic fluid that shows no toxic effects toward cartilage and nerve tissue.18 A continuous negative pressure of 100 or 125 mmHg was set. For NPWTi-d, HOCl instillation with dwell time was performed for 20 minutes every 6 hours.
Microbiologic Analysis
Standard Culture Methods
The processing of culture diagnostics was carried out under the usual conditions in microbiology using commercially available solid culture media (Schaedler agar, Columbia boiled blood agar, Columbia sheep blood agar, MacConkey agar, Candida selective agar) and various liquid enrichment media (brain-heart dextrose broth, thioglycolate broth) with incubation times of 2 to 7 days. A semiquantitative algorithm was used to assess growth on the solid media. Simple manual procedures as well as the BioMerieux Vitek 2 system, which was also used for antibiotic susceptibility testing (determination of minimum inhibitory concentration), were available for identification of the cultured microorganisms. As an alternative, the inhibition zone diameter was determined in the agar diffusion test originally according to Bauer et al.,19 now being performed following the standards of the European Committee on Antimicrobial Susceptibility Testing and the German National Antibiotics Committee.
Molecular Analysis Using Fluorescence In Situ Hybridization Sequencing
Fluorescence in situ hybridization (FISH) sequencing (FISHseq) describes the combination of molecular imaging of microorganisms using FISH with 16S rRNA gene polymerase chain reaction and subsequent sequencing, both out of consecutive methacrylate-embedded tissue sections.
Sample Embedding and FISH
Tissue samples were fixed in FISHopt fixation solution (MoKi Analytics, Berlin, Germany) and embedded utilizing cold polymerizing resin (Technovit 8100; Kulzer, Wehrheim, Germany) and sectioned as described previously.20 FISH analysis was performed according to Schoenrath et al.21 Sections were hybridized with pan-bacterial probe EUB338Cy322 to visualize the entire bacterial population. A nonsense probe NON EUB338Cy523 was used to exclude nonspecific probe binding. For visualization of nucleic acids in host cell nuclei and bacteria, 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) was included. In cases where a positive EUB338Cy3 signal was found, specific probes related to culture or sequencing results were used to confirm microbial findings by FISH. For microscopy, an epifluorescence microscope (Axioplan 2 and AxioImager Z2; Carl Zeiss, Jena, Germany) equipped with narrow band filter sets (AHF Analysentechnik, Tübingen, Germany) was used. Digital images were generated using the ZEN and the AxioVision software from Zeiss. Microorganisms detected were classified empirically as planktonic (single bacteria), microcolonies (clusters of up to 30 microorganisms), or biofilms (communities of more than three layers of adjacent bacteria over a length of more than 20 µm).
DNA Extraction, 16S rRNA Gene Polymerase Chain Reaction Amplification, and Sequencing
DNA was extracted from consecutive sections of the embedded samples and polymerase chain reaction was performed using the pan-bacterial primer set time point U1 and RTU324 for the amplification of the 16S rRNA gene as described.20 Subsequent sequencing of amplicons was performed using a commercial sequencing service (LGC Genomics, Berlin, Germany) and analyzed with the commercial analysis pipeline SmartGene (Lausanne, Switzerland) as described previously.25
Statistics
The data were analyzed and compiled in tables and graphs (Microsoft Excel 2010; Microsoft Corp., Redmond, WA; GraphPad Prism 9; GraphPad Software, Inc., San Diego, CA; biorender.com, Toronto, Canada). Statistical analyses were performed using SPSS 25 for Windows (IBM Corporation, Chicago, IL). Data to be presented descriptively were calculated as frequency, median with 25th and 75th percentile, or minimum and maximum. To test for normality distribution, the Kolmogorov-Smirnov test was used. Parametric data were analyzed with the t test and nonparametric data with the Mann-Whitney U test. Analysis of linked nonparametric data was performed with the Wilcoxon test. Statistical significance was defined as P ≤ 0.05.
RESULTS
Demographics, Antibiotic Treatment, NPWT Duration, and Specimen Collection
The median age of the patients (26 male) was 51 years (24 to 91). Nineteen patients had already received antibiotic therapy before the first biopsy [n = 16; median 2.5 days (0 to 50)]. In two patients, antibiotic therapy started on the day of surgery; in one patient, immediately before biopsy; and in 11 patients, after specimen collection. At time point 2, all patients received antibiotics. There were 22 acute and eight chronic wounds (Table 1). There are no significant demographic differences between the NPWT and NPWTi-d groups (Table 2). At time point 1, white blood cell and C-reactive protein levels in patients with acute wounds were 12.0 (median 25th to 75th percentile, 9.55 to 15.05) and 73.7 (median 25th to 75th percentile, 44.93 to 215.28), respectively, and in patients with chronic wounds, 8.3 [median 25th to 75th percentile, 7.85 to 9.63 (P = 0.027 versus white blood cells in acute wounds)] and 12.1 [median 25th to 75th percentile, 9.13 to 49.1 (P = 0.097 versus C-reactive protein in acute wounds)]. In the NPWT group, the median interval until foam change was 3 days (range, 1 to 5 days); in the NPWTi-d group, a median of 2 days (range, 1 to 5 days). At time point 1, a median of 2.8 wound bed tissue samples (range, one to four) were analyzed using standard culture methods and a median of 1.3 samples (range, one to three) were examined using molecular biology methods. At time point 2, a median of 2.0 wound bed tissue samples (range, one to four) were analyzed using standard culture methods and a median of 1.0 samples (range, one to two) were examined using molecular biology methods. In addition, at least one foam sample and the eluate were analyzed with both microbiological methods. In total, 235 samples were examined with routine microbiological methods and 145 samples with FISHseq.
Table 1.
Demographic Data and Wound Conditions in Both Groupsa
| Therapy/Sex/Age, yrs | Diagnosis and Wound Conditions | Antibiotic Therapy Duration before Time Point 1,b days | NPWT Interval between Time Points 1 and 2,b days |
|---|---|---|---|
| NPWT/M/40 | Right lower leg phlegmone; redness, swelling, closed; acute wound | 1 | 2 |
| NPWT/M/53 | SSI after ORIF; redness, overheating, putrid secretion, open; acute wound | 1 | 4 |
| NPWT/F/73 | Bursitis olecranii; putrid, redness, swelling, overheating, closed; acute wound | Unknown | 2 |
| NPWT/M/56 | Ventral lower leg abscess; redness, overheating, putrid secretion, open; acute wound | Perioperative (single shot after sample collection) | 2 |
| NPWT/M/68 | DFS; necrosis plate, wet infected margin, open; chronic wound | Perioperative (single shot before sample collection) | 3 |
| NPWT/M/66 | Sepsis in infected left leg ulcer; odorous, moist, necrotic, cut through stocking to bone, open; chronic wound | Preoperative in ED before surgery | 3 |
| NPWT/M/54 | Soft-tissue infection after fibula head fracture; conservative therapy, redness, swelling, overheating, open; chronic wound | 50 | 5 |
| NPWT/M/33 | Infected wound margin necrosis in skin of right knee joint with osteomyelitis; 7×5 cm serous secreting wound, fibrin threads, suture material enclosed, open; acute wound | Postoperative (time point 2; 5 days after first surgery) | 5 |
| NPWT/F/91 | Left lower leg ulcus cruris; covered ulcer, 12×8 cm, redness, overheating, open; chronic wound | 3 | 2 |
| NPWT/M/46 | Right lower leg injection abscess; two abscesses, 6×3 and 5×3 cm, putrid, secreting, porus, open; acute wound | 1 | 3 |
| NPWT/M/43 | Left phlegmonous bursitis olecranii; massive swelling, redness, overheating, closed; acute wound | Perioperative (single shot after sample collection) | 1 |
| NPWT/M/27 | Infectious pseudarthrosis of the right tibia in a gunshot fracture; pretibial swelling, reddened scar, hemorrhagic crusts, closed; chronic wound | Perioperative (single shot after sample collection) | 4 |
| NPWT/M/31 | Right foot shattershot fracture; swelling of the forefoot, nonirritant bullet wounds, closed; acute wound | 4 | 4 |
| NPWT/M/74 | Right ankle joint empyema; redness, swelling, open ulcer with greenish coating, open; acute wound | 1 | 3 |
| NPWT/M/32 | Right lower leg abscess; redness, swelling, closed; acute wound | Preoperative in ED before surgery | 3 |
| NPWTi-d/M/29 | Right elbow fistulous osteitis; necrosis plate, serous secreting fistula, open; chronic wound | 16 | 5 |
| NPWTi-d/M/24 | Chronic osteomyelitis of the ulna with fistula on the right forearm; putrid secretion, redness, swelling, open; chronic wound | 1 | 2 |
| NPWTi-d/M/74 | Right lower leg phlegmon; redness, swelling, overheating, closed; acute wound | 4 | 2 |
| NPWTi-d/M/62 | Right gluteal region abscess; redness, swelling, overheating, putrid secretion, open; acute wound | 2 | 2 |
| NPWTi-d/M/57 | Infected ulcer with erysipelas in left foot stump osteomyelitis; redness, pain, closed; acute wound | 4 | 4 |
| NPWTi-d/M/65 | Chronic wound-healing disorder after LL amputation with PAD; moist necrotic plate, open; chronic wound | Postoperative (first postoperative day) | 2 |
| NPWTi-d/F/47 | Bone marrow phlegmon radius in right forearm abscess; redness, swelling, overheating, putrid secretion, open; acute wound | 1 | 2 |
| NPWTi-d/M/35 | Left foot abscess; redness, swelling, overheating, putrid secretion, open; acute wound | Perioperative (single shot after sample collection) | 2 |
| NPWTi-d/M/33 | Left humerus bullet fracture; shotgun channel open, no inflammation; acute wound | Perioperative (single shot after sample collection) | 3 |
| NPWTi-d/M/49 | Subcutaneous abscess hollow of the knee on both sides; redness, swelling, overheating, putrid secretion, open; acute wound | Perioperative (single shot after sample collection) | 3 |
| NPWTi-d/M/78 | Postoperative hip joint abscess after periprosthetic fracture treatment; redness, swelling, overheating, closed; acute wound | Perioperative (single shot after sample collection) | 1 |
| NPWTi-d/M/45 | Right thigh phlegmone; acute wound | 4 | 4 |
| NPWTi-d/F/78 | Right buttocks postinterventional phlegmon after injection; swelling, closed; acute wound | 2 | 4 |
| NPWTi-d/M/28 | SSI after claviulc ORIF; redness, swelling, overheating, putrid secretion, open; acute wound | Perioperative (single shot after sample collection) | 3 |
| NPWTi-d/M/61 | Right lower leg abscess, redness, swelling, wet necrosis, open; acute wound | Perioperative (single shot after sample collection) | 2 |
DFS, diabetic foot syndrome; ED, emergency department; LL, lower leg; ORIF, open reduction and internal fixation; PAD, peripheral artery disease; SSI, surgical-site infection.
NPWT, n = 15; NPWTi-d, n = 15.
Time point 1 involves débridement and the beginning of NPWT; time point 2, first dressing (foam) change.
Table 2.
Demographic and Treatment Details of Both Groups
| Demographic and Treatment Details | NPWT (n = 15) | NPWTi-d (n = 15) |
|---|---|---|
| Sex, M/F | 13/2 | 13/2 |
| Age, yrs, mean (range) | 52 (27–91) | 51 (24–78) |
| Wound site | ||
| Upper extremity | 2 | 4 |
| Lower extremity | 13 | 8 |
| Buttock or trunk | 0 | 3 |
| Type of infected wound situation | ||
| Acute | 10 | 12 |
| Chronic | 5 | 3 |
| Open/closed | 9/6 | 10/5 |
| Comorbidities | ||
| HIV/hepatitis C/drug abuse | 3/15 | 3/15 |
| Diabetes/peripheral artery disease | 3/15 | 5/15 |
| NPWT treatment, days, median (range) | 3 (1–5) | 2 (1–5) |
| Start of antibiotic application | ||
| Days before operation; median (range) | 8; 2 (1–50) | 8; 3 (1–16) |
| On the day of operation before probe biopsy | 3 | 0 |
| After probe biopsy | 4 | 7 |
| Reduction of bacterial species, wound bed (time point 1) versus wound bed (time point 2),a P | 0.317 | 0.026 |
Time point 1 involves débridement and the beginning of NPWT; time point 2, first dressing (foam) change.
Bacterial Spectrum of Wound Beds at Time Points 1 and 2
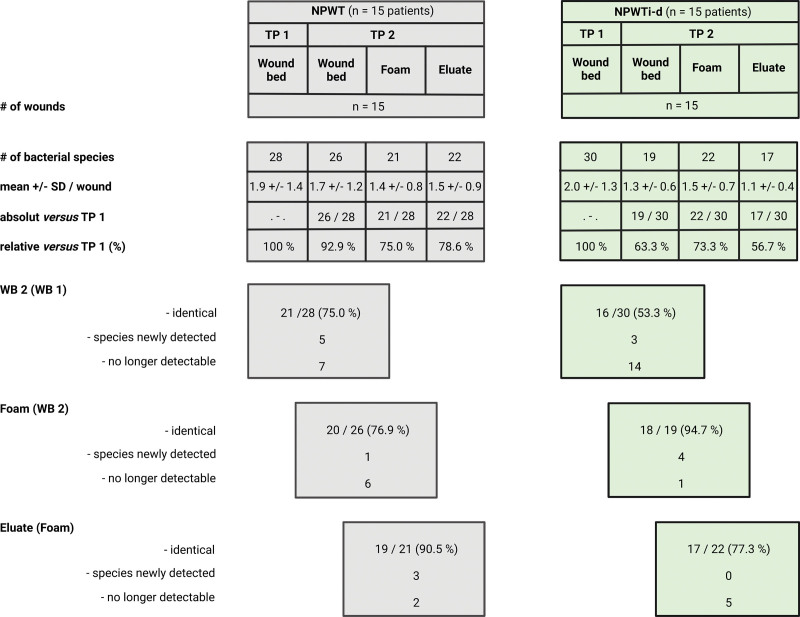
The additional use of molecular biological techniques enabled the identification of a markedly higher proportion of pathogens. In time point 1, the presence of 58 bacterial colonizations was detected in all 30 wound beds (monomicrobial, n = 16 wounds; polymicrobial, n = 14 wounds). Of these 58, 43 (74.1%) could be detected using the routine microbiological culture–based detection method and 53 (91.4%; the bacterial species could only be identified in 53 cases) with FISHseq. In time point 2, the identification of bacteria in wound bed samples increased from 28 of 52 (53.8%) to 36 of 52 (69.2%) because of the addition of FISHseq to the microbiological diagnostics (52 bacteria were detected in time point 2). At time point 2, in no case was the wound bed free from colonization. In 63.8% (37 of 58), the pathogens detected at time point 1 could also be detected in the wound bed at time point 2 [NPWT group, 21/28 (75%); NPWTi-d group, 16 of 30 (53.3%); Figs. 1 and 2].
Fig. 1.
Consistency of detection of identified bacterial species in wound bed, foam, and eluate before and after NPWT or NPWTi-d application in all 30 patients. The numbers given in the circles, separated according to form of therapy, indicate the frequency of the detection of the respective species. A total of 39 different bacterial species were identified at the two time points (time points 1 and 2) and in all sample materials (wound bed, foam, eluate). Gray circle: NPWT; green circle: NPWTi-d.
Fig. 2.
Number of bacterial species as detected by all diagnostic methods in both groups (NPWT and NPWTi-d). Shown is the number in different samples [wound bed at time point 1 (WB1), wound bed at time point 2 (WB2), foam, and eluate]. Created with BioRender.com.
Foam as Pathogen Reservoir and Possible Pathogen Backwash
Of the bacteria identified in the wound bed at time point 2, 84.4% (38 of 45) were also detectable in the foam (NPWT group, 76.9%; NPWTi-d group, 94.7%; Fig. 2). A total of 90.7% (39 of 43) of the pathogens identifiable in the foam could also be detected in the wound bed. In no case was the foam free of pathogens. Five pathogens that were not detectable at time point 2 in the wound bed were newly detected in the foam analysis [one in NPWT (Clostridium perfringens) and four in NPWTi-d (Staphylococcus warneri, Pseudomonas sp., Proteus mirabilis, Acinetobacter sp.)]. Regardless of the type of NPWT, no eluate was free from bacteria. Of the pathogens identifiable in the foam, 83.7% (in both groups; 36 of 43) could also be detected in the eluate. A total of 92.3% (36 of 39) of the pathogens identifiable in the eluate could also be detected in the foam and 94.9% (37 of 39) in the wound bed.
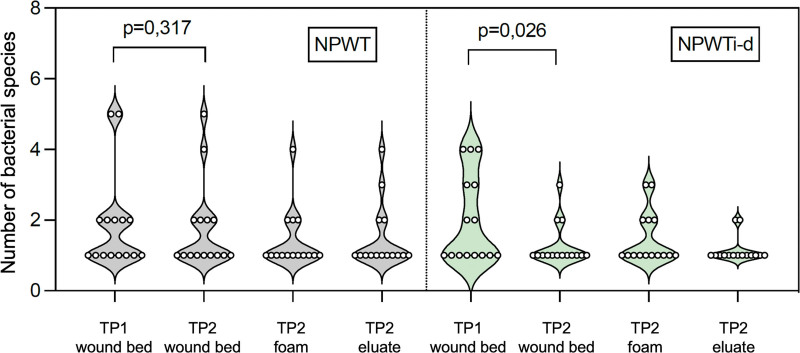
Effect of Antiseptic NPWTi-d on Bacterial Wound Contamination
The microbiological and molecular biological analysis of the tissue samples of both therapy forms showed a significantly reduced number of pathogen species from time point 1 to time point 2 (P = 0.012). In the NPWT group, 75% (21 of 28) of the pathogens identified at time point 1 were also detected at time point 2 in the wound bed, compared with only 53.3% (16 of 30) in the NPWTi-d group. The application of NPWTi-d led to a statistically significant reduction in the number of pathogen species detected compared with NPWT (NPWTi-d, wound bed at time point 1 versus wound bed at time point 2: P = 0.026; NPWT, time point 1 versus time point 2: P = 0.317; Fig. 3). Five pathogens were newly detected at time point 2 in the NPWT group and three in the NPWTi-d group (Figs. 1 and 2).
Fig. 3.
Number of bacterial species identified in each individual patient in the wound bed (time points 1 and 2), foam, and eluate at time point 2 in the NPWT group (n = 15 patients) and NPWTi-d group (n = 15 patients). Created with GraphPad Prism 9.
Biofilm in Tissue Samples and NPWT Foam
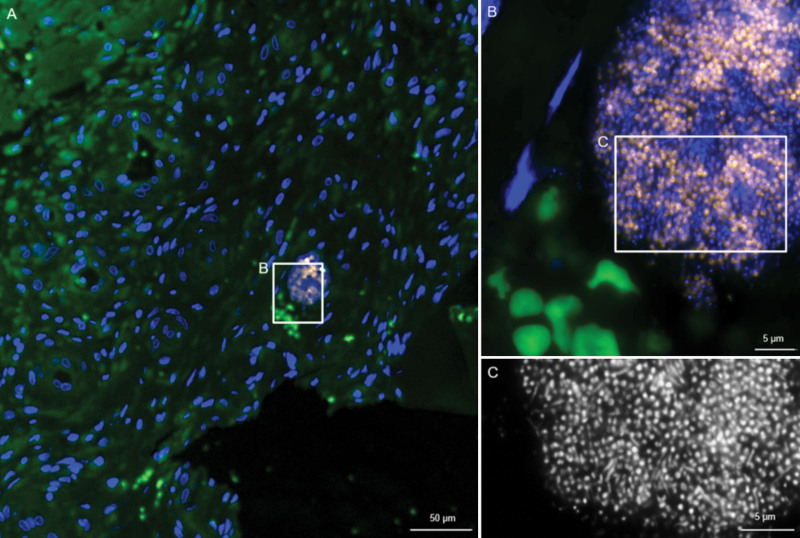
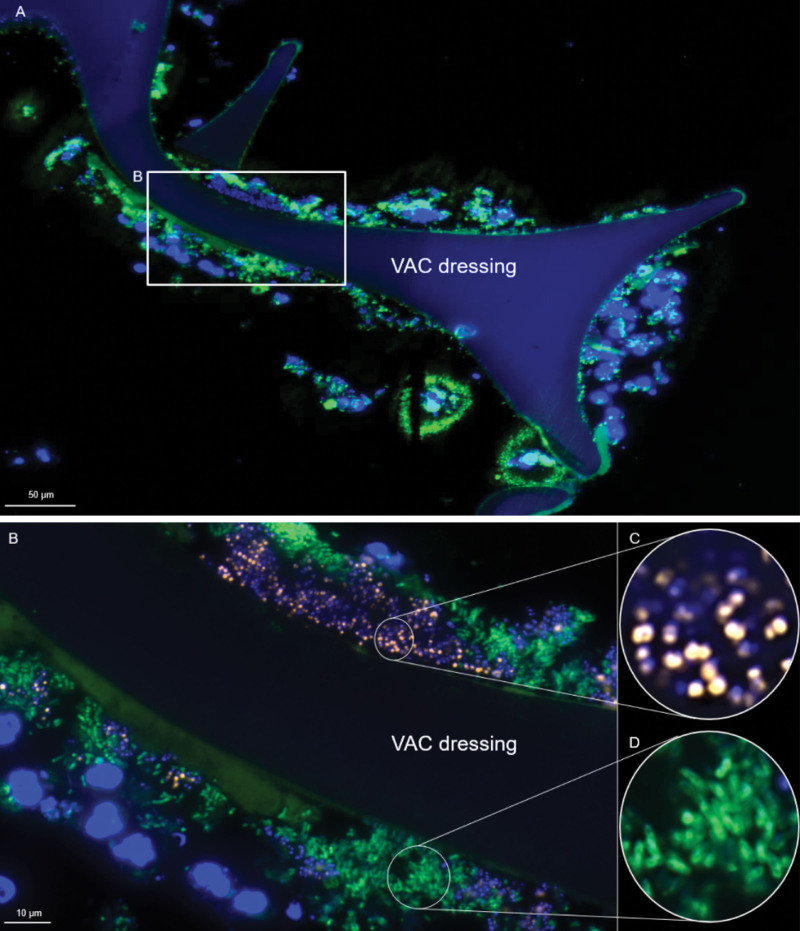
In all 30 patients, growth behavior of 35 bacteria in the wound bed (time point 1 to time point 2) and of 32 bacteria in the wound bed at both time points and in the foam was investigated. At time point 1, planktonic bacteria could be detected in 16 wounds (NPWT, nine; NPWTi-d, seven), microcolonies in eight wounds (NPWT, two; NPWTi-d, six), and biofilm in six wounds (NPWT, four; NPWTi-d, two) (Fig. 4). The analysis of the foam (time point 2) in all 30 patients revealed the presence of planktonic growth form in 24 patients (NPWT, 14 of 15; NPWTi-d, 10 of 15), microcolonies in four patients (NPWT, 0 of 15; NPWTi-d, four of 15), and biofilm in two patients (NPWT, one of 15; NPWTi-d, one of 15) (Fig. 5). In these two biofilm cases, the bacterial species could be detected in the foam and eluate by routine microbiological culture as well.
Fig. 4.
Fluorescence in situ hybridization of wound tissue in a patient before NPWT. Culture was positive for Streptococcus sp. and Enterobacteriaceae. (Left) Overview shows the tissue in green and host cell nuclei in blue. Inset marks a region where bacteria detected by the panbacterial FISH probe are visible within a small biofilm. At higher magnification (above, right), the Streptococcus genus-specific FISH probe (orange)26 shows a strong fluorescence signal, indicating active bacteria. In DAPI (below, right), additional rods are visible, in line with Enterobacteriaceae detected by culture.
Fig. 5.
Fluorescence in situ hybridization of a NPWT foam that was culture-positive for Pseudomonas aeruginosa and Staphylococcus aureus. The section was hybridized with the Staphylococcus genus–specific probe STAPHYCY327 and the Pseudomonas genus-specific probe PSMGFITC.28 DAPI was used for visualization of nucleic acids in host cell nuclei and bacteria. (Above) Overview shows negative pressure wound therapy foam colonized with host cell nuclei and bacterial biofilms. Inset marks a region were FISH-positive bacteria are visible within biofilms. At higher magnification (below), the Staphylococcus genus–specific FISH probe (first round inset, orange) and the Pseudomonas genus–specific FISH (second round inset, green) shows a strong fluorescence signal, indicating active bacteria of both species.
DISCUSSION
This study investigates the bacterial load of the eluate after retrograde rinsing of the NPWT foam to clarify whether the use of NPWTi-d leads to recontamination of the wound with pathogens. To our knowledge, our study is the first to analyze the bacterial load and growth behavior of organisms in NPWT foams, their eluate, and in the wound bed using both standard cultural microbiological and molecular biological methods such as FISHseq. The most important finding is that without exception, all NPWT foams and eluates showed a high bioburden. Even though the number of different bacterial species per wound was significantly lower in the NPWTi-d group, antiseptic instillation fluid was not able to produce a pathogen-free condition in the eluate, the foam, or the wound bed.
Other studies have shown that the foam presents a high bacterial load in the context of NPWT. Anagnostakos and Mosser15 found at least one type of microorganism in 39 out of 101 foams (39%) and polymicrobial contamination in 26%. Yusuf et al.17 found that 97% of foams were colonized, 54% of which were polymicrobial. Our results showed that 100% of foams were colonized, 30% of which were polymicrobial. Just as the major difference between the two studies cited can be explained by the use of sonication by Yusuf et al.,17 the results presented here show that the use of additional FISHseq can more sensitively detect bacterial colonization by fastidious or resting bacteria that would otherwise have remained unidentifiable.
Yusuf et al.17 demonstrated a high bacterial count in the foam after NPWT but did not take tissue samples from the wound. Based on this work, it was hoped that pathogens would collect in the NPWT foam, leaving the wound clean. However, other research groups have shown the tendency of pathogens to persist in the wound during NPWT therapy.29–31 The results of our study confirm not only the persistence of pathogens in the wound and in the foam but also that with NPWTi-d the pathogens can be flushed back and forth with each instillation cycle and thus may repeatedly recontaminate the wound.
According to the recent literature on NPWTi-d, there is a trend toward reduced bioburden with antiseptic solutions compared with normal saline.32,33 Our decision to instill HOCl was primarily based on the results of previously published studies describing a broad spectrum of microbiocidal activity and noncytotoxic effects to cartilage and nervous structures.18,34–36 The results of bacterial load reduction may vary depending on the antiseptic solution used in NPWTi-d. Although there was a significant reduction in bacterial species in the wound bed in the NPWTi-d group compared with the NPWT group, no wound bed became bacteria-free. Similar results were observed in previous studies in which complete reduction of bacterial contamination was not observed after NPWTi-d.14,37,38
The bacteriological profiling of infected wounds could be improved significantly in our study by supplementing the standard cultivation method with molecular biological diagnostic techniques such as FISHseq. With this method, the detection rate of bacterial pathogens in the wound bed could be increased from just over 70% using the standard cultivation method to almost 95% using FISHseq at time point 1 and from 50% using the standard cultivation method to 70% using FISHseq at time point 2, confirming infection by visualization of the microorganism within the tissue. Bacteria embedded in biofilms are difficult to detect and to culture,17,39 a fact that may also contribute to the underestimation of bioburden.
These study results are limited by several factors, including the way patients were assigned to the two groups (NPWT or NPWTi-d). There was no randomization procedure, and allocation was at the discretion of the individual surgeons, so selection bias cannot be excluded, although there were no relevant differences in group characteristics. Although several surgeons were involved, all patients were treated uniformly according to the established standards of a military wound center for septic defects. The heterogeneity in antibiotic administration should be noted. In particular, antibiotic administration, which in some cases was already started preoperatively, influenced the detection of bacteria in intraoperative microbiological samples, because preoperative antibiotic administration can lead to false-negative results.40 On the other hand, the fact that antibiotic administration was often started before surgery in our study corresponds with the realities of patient care, as unsuccessfully treated cases or patients with complications are usually transferred to our treatment center with antibiotic therapy already in progress. However, our observations on contamination of the eluate or foam and the wound bed as well as on the consistency of the bioburden remain valid. Thus, the current study adds important new insights to the literature and provides a basis for exploring further clinical trial options to understand and demonstrate the effect of NPWTi-d on wound healing, particularly in wounds with high bacterial bioburden. It would be interesting to conduct further clinical studies to investigate the influence of antiseptic foams, such as silver foams, on reducing bacterial load when using NPWT or NPWTi-d.
In summary, all wounds and foams remained colonized by bacteria after NPWT. In most cases, the original pathogens were still in the wound and foam after NPWT. The eluate of NPWT foams has a bacterial load and this load is at risk to be washed back into the wound as part of NPWTi-d. Nevertheless, the number of bacterial species in the wound is reduced after NPWT with antiseptic instillation compared with conventional NPWT. Therefore, NPWTi-d should be used for contaminated wounds. The analysis of nonplanktonic bacterial life forms during NPWT shows that there is a tendency to detect fewer microcolonies and biofilms in the wound bed and foam even after a short NPWT period. Our observations underline the question of the significance of the quantity of the pathogens in wounds, which has not yet been adequately answered. Through the additional use of molecular biological examination techniques (FISHseq), a significantly higher proportion of bacterial species can be identified. This technique is costly and not necessary for every wound, but it should be recommended for therapy-resistant infection situations without previous microbiological pathogen detection.
CONCLUSIONS
We investigated the foam and eluate of NPWT for the first time and found that NPWT poses a risk of backwashing bacteria into the wound. NPWT cannot achieve complete decontamination of wounds after one cycle, but by using NPWTi-d, the bacterial load in contaminated wounds can be significantly reduced compared with conventional NPWT. The use of additional molecular biological methods, such as FISHseq, can significantly improve pathogen detection in difficult therapy-resistant wounds and offer further therapeutic options.
ACKNOWLEDGMENTS
The authors thank the medical colleagues of the Department of Trauma Surgery and Orthopaedics and the surgical assistants of their hospital who helped obtain samples from the study participants; Dr. Dennis Vogt for contributions and suggestions in the interpretation of the study results; Drs. Daniel Köppen and Nadine Schäfer for support in the statistical analysis; and the biofilm center team for technical support during fluorescence in situ hybridization sequencing analysis. This study was funded by the German Armed Forces as a special research project (SoFo 15K4-S-121517) of the Office of Defence Medical Research and Development. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure: The authors have no financial interest to declare in relation to the content of this article.
REFERENCES
- 1.Téot L, Guillot-Masanovic M, Miquel P, et al. Clinical impact of negative-pressure wound therapy: a 1,126-patient observational prospective study. Wound Repair Regen. 2014;22:341–350. [DOI] [PubMed] [Google Scholar]
- 2.Apelqvist J, Willy C, Fagerdahl AM, et al. EWMA document: negative pressure wound therapy. J Wound Care. 2017;26(Suppl 3):S1–S154. [DOI] [PubMed] [Google Scholar]
- 3.Moch D, Fleischmann W, Westhauser A. [Instillation vacuum sealing: report of initial experiences.] Langenbecks Arch Chir Suppl Kongressbd. 1998;115:1197–1199. [PubMed] [Google Scholar]
- 4.Timmers MS, Graafland N, Bernards AT, Nelissen RG, van Dissel JT, Jukema GN. Negative pressure wound treatment with polyvinyl alcohol foam and polyhexanide antiseptic solution instillation in posttraumatic osteomyelitis. Wound Repair Regen. 2009;17:278–286. [DOI] [PubMed] [Google Scholar]
- 5.Ludolph I, Fried FW, Kneppe K, Arkudas A, Schmitz M, Horch RE. Negative pressure wound treatment with computer-controlled irrigation/instillation decreases bacterial load in contaminated wounds and facilitates wound closure. Int Wound J. 2018;15:978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lessing C, Slack P, Hong KZ, Kilpadi D, McNulty A. Negative pressure wound therapy with controlled saline instillation (NPWTi): dressing properties and granulation response in vivo. Wounds. 2011;23:309–319. [PubMed] [Google Scholar]
- 7.Goss SG, Schwartz JA, Facchin F, Avdagic E, Gendics C, Lantis JC, 2nd. Negative pressure wound therapy with instillation (NPWTi) better reduces post-debridement bioburden in chronically infected lower extremity wounds than NPWT alone. J Am Coll Clin Wound Spec. 2012;4:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim PJ, Attinger CE, Steinberg JS, et al. The impact of negative-pressure wound therapy with instillation compared with standard negative-pressure wound therapy: a retrospective, historical, cohort, controlled study. Plast Reconstr Surg. 2014;133:709–716. [DOI] [PubMed] [Google Scholar]
- 9.Omar M, Gathen M, Liodakis E, et al. A comparative study of negative pressure wound therapy with and without instillation of saline on wound healing. J Wound Care. 2016;25:475–478. [DOI] [PubMed] [Google Scholar]
- 10.Qiu Y, Li Y, Gao B, et al. Therapeutic efficacy of vacuum sealing drainage-assisted irrigation in patients with severe multiple-space infections in the oral, maxillofacial, and cervical regions. J Craniomaxillofac Surg. 2019;47:837–841. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Gabriel A, Lantis J, Téot L. Clinical recommendations and practical guide for negative pressure wound therapy with instillation. Int Wound J. 2016;13:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C, Goss SG, Alcantara S, Schultz G, Lantis Ii JC. Effect of negative pressure wound therapy with instillation on bioburden in chronically infected wounds. Wounds. 2017;29:240–246. [PubMed] [Google Scholar]
- 13.Hall-Stoodley L, Stoodley P, Kathju S, et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol. 2012;65:127–145. [DOI] [PubMed] [Google Scholar]
- 14.Kim PJ, Attinger CE, Constantine T, et al. Negative pressure wound therapy with instillation: international consensus guidelines update. Int Wound J. 2020;17:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anagnostakos K, Mosser P. Bacteria identification on NPWT foams: clinical relevance or contamination? J Wound Care. 2012;21:333–334, 336–339. [DOI] [PubMed] [Google Scholar]
- 16.Scherrer AU, Bloemberg G, Zbinden R, et al. ; VASGRA Cohort. Prosthetic vascular graft infections: bacterial cultures from negative-pressure-wound-therapy foams do not improve diagnostics. J Clin Microbiol. 2016;54:2190–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusuf E, Jordan X, Clauss M, Borens O, Mäder M, Trampuz A. High bacterial load in negative pressure wound therapy (NPWT) foams used in the treatment of chronic wounds. Wound Repair Regen. 2013;21:677–681. [DOI] [PubMed] [Google Scholar]
- 18.Kramer A, Dissemond J, Kim S, et al. Consensus on wound antisepsis: update 2018. Skin Pharmacol Physiol. 2018;31:28–58. [DOI] [PubMed] [Google Scholar]
- 19.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Tech Bull Regist Med Technol. 1966;36:49–52. [PubMed] [Google Scholar]
- 20.Moter A, Leist G, Rudolph R, et al. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology (Reading). 1998;144(Pt 9):2459–2467. [DOI] [PubMed] [Google Scholar]
- 21.Schoenrath F, Kursawe L, Nersesian G, et al. Fluorescence in situ hybridization and polymerase chain reaction to detect infections in patients with left ventricular assist devices. ASAIO J. 2021;67:536–545. [DOI] [PubMed] [Google Scholar]
- 22.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. [DOI] [PubMed] [Google Scholar]
- 24.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas P, Petrich A, Schulze J, et al. Distribution and phylogeny of Brachyspira spp. in human intestinal spirochetosis revealed by FISH and 16S rRNA-gene analysis. Anaerobe. 2017;47:25–32. [DOI] [PubMed] [Google Scholar]
- 26.Gescher DM, Kovacevic D, Schmiedel D, et al. Fluorescence in situ hybridisation (FISH) accelerates identification of Gram-positive cocci in positive blood cultures. Int J Antimicrob Agents. 2008;32(Suppl 1):S51–S59. [DOI] [PubMed] [Google Scholar]
- 27.Trebesius K, Leitritz L, Adler K, Schubert S, Autenrieth IB, Heesemann J. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med Microbiol Immunol. 2000;188:169–175. [DOI] [PubMed] [Google Scholar]
- 28.Pawar V, Komor U, Kasnitz N, et al. In vivo efficacy of antimicrobials against biofilm-producing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;59:4974–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braakenburg A, Obdeijn MC, Feitz R, van Rooij IA, van Griethuysen AJ, Klinkenbijl JH. The clinical efficacy and cost effectiveness of the vacuum-assisted closure technique in the management of acute and chronic wounds: a randomized controlled trial. Plast Reconstr Surg. 2006;118:390–397; discussion 398. [DOI] [PubMed] [Google Scholar]
- 30.Lalliss SJ, Stinner DJ, Waterman SM, Branstetter JG, Masini BD, Wenke JC. Negative pressure wound therapy reduces pseudomonas wound contamination more than Staphylococcus aureus. J Orthop Trauma. 2010;24:598–602. [DOI] [PubMed] [Google Scholar]
- 31.Mouës CM, Vos MC, van den Bemd GJ, Stijnen T, Hovius SE. Bacterial load in relation to vacuum-assisted closure wound therapy: a prospective randomized trial. Wound Repair Regen. 2004;12:11–17. [DOI] [PubMed] [Google Scholar]
- 32.Diehm YF, Fischer S, Wirth GA, et al. Management of acute and traumatic wounds with negative-pressure wound therapy with instillation and dwell time. Plast Reconstr Surg. 2021;147:43S–53S. [DOI] [PubMed] [Google Scholar]
- 33.Kim PJ, Attinger CE, Steinberg JS, et al. Negative-pressure wound therapy with instillation: International consensus guidelines. Plast Reconstr Surg. 2013;132:1569–1579. [DOI] [PubMed] [Google Scholar]
- 34.Aragón-Sánchez J, Lázaro-Martínez JL, Quintana-Marrero Y, Sanz-Corbalán I, Hernández-Herrero MJ, Cabrera-Galván JJ. Super-oxidized solution (Dermacyn Wound Care) as adjuvant treatment in the postoperative management of complicated diabetic foot osteomyelitis: preliminary experience in a specialized department. Int J Low Extrem Wounds. 2013;12:130–137. [DOI] [PubMed] [Google Scholar]
- 35.Tata MD, Kwan KC, Abdul-Razak MR, Paramalingam S, Yeen WC. Adjunctive use of superoxidized solution in chest wall necrotizing soft tissue infection. Ann Thorac Surg. 2009;87:1613–1614. [DOI] [PubMed] [Google Scholar]
- 36.Wolvos TA. Negative pressure wound therapy with instillation: the current state of the art. Surg Technol Int. 2014;24:53–62. [PubMed] [Google Scholar]
- 37.Chowdhry SA, Wilhelmi BJ. Comparing negative pressure wound therapy with instillation and conventional dressings for sternal wound reconstructions. Plast Reconstr Surg Glob Open. 2019;7:e2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis K, Bills J, Barker J, Kim P, Lavery L. Simultaneous irrigation and negative pressure wound therapy enhances wound healing and reduces wound bioburden in a porcine model. Wound Repair Regen. 2013;21:869–875. [DOI] [PubMed] [Google Scholar]
- 39.Kirketerp-Møller K, Jensen PØ, Fazli M, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46:2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Mayahi M, Cian A, Lipsky BA, et al. Administration of antibiotic agents before intraoperative sampling in orthopedic infections alters culture results. J Infect. 2015;71:518–525. [DOI] [PubMed] [Google Scholar]