Graphical abstract
Keywords: Anther cuticle, Cell death, Spikelet development, Subtilisin like protease, Wax and cutin
Highlights
-
•
OsSUBSrP1 regulates anther cuticle biosynthesis.
-
•
OsSUBSrP1 is essential for apical spikelet development.
-
•
OsSUBSrP1 plays an important role in maintaining ROS-mediated programmed cell death.
Abstract
Introduction
Panicle abortion is a severe physiological defect and causes a reduction in grain yield.
Objectives
In this study, we aim to provide the characterization and functional analysis of a mutant apa1331 (apical panicle abortion1331).
Methods
The isolated mutant from an EMS-mutagenized population was subjected to SSR analysis and Mutmap assay for candidate gene mapping. We performed phenotypic analysis, anthers cross-sections morphology, wax and cutin profiling, biochemical assays and phylogenetic analysis for characterization and evaluation of apa1331. We used CRISPR/Cas9 disruption for functional validation of its candidate gene. Furthermore, comparative RNA-seq and relative expression analysis were performed to get further insights into mechanistic role of the candidate gene.
Results
The anthers from the apical spikelets of apa1331 were degenerated, pollen-less and showed defects in cuticle formation. Transverse sections of apa1331 anthers showed defects in post-meiotic microspore development at stage 8–9. Gas Chromatography showed a significant reduction of wax and cutin in anthers of apa1331 compared to Wildtype (WT). Quantification of H2O2 and MDA has indicated the excessive ROS (reactive oxygen species) in apa1331. Trypan blue staining and TUNEL assay revealed cell death and excessive DNA fragmentation in apa1331. Map-based cloning and Mutmap analysis revealed that LOC_Os04g40720, encoding a putative SUBTILISIN-LIKE SERINE PROTEASE (OsSUBSrP1), harbored an SNP (A > G) in apa1331. Phenotypic defects were only seen in apical spikelets due to highest expression of OsSUBSrP1 in upper panicle portion. CRISPR-mediated knock-out lines of OsSUBSrP1 displayed spikelet abortion comparable to apa1331. Global gene expression analysis revealed a significant downregulation of wax and cutin biosynthesis genes.
Conclusions
Our study reports the novel role of SUBSrP1 in anther cuticle biosynthesis by ROS-mediated programmed cell death in rice.
Introduction
The panicle is a reproductive organ in rice, which directly determines the grain yield [1]. Various genetic and environmental factors regulate the spatiotemporal arrangement of a panicle by affecting its reproductive success and developmental decisions [2]. Abortion of spikelets, also called panicle degeneration, is highly detrimental to plants and causes a significant reduction in yield [3], [4], [5]. Although, several genetic factors have been reported that control the apical panicle development in rice. Nevertheless, the molecular basis of mechanisms and why abortion remains in the apical portion still needs a comprehensive understanding.
Development of the panicle begins with the transition of juvenile to the reproductive phase. Various changes at these transitions including shoot, inflorescence and branch meristems could bring severe defects to panicle development [6]. SMALL PANICLE1 (SP1) controls the length of the panicle by encoding a nitrate transporter of a PEPTIDE TRANSPORTER (PTR) family [7]. Similarly, ERRECT PANICLE 2 (EP), LAX PANICLE1 (LAX1) and LAX2 are reported to involve in the development of axillary and branch meristems formation [8], [9], [10]. ABERRANT PANICLE ORGANIZATION 1 (APO1) and APO2 work in the temporal regulation of meristems, which ultimately affect the inflorescence development [11], [12]. SQUAMOSA PROMOTER BINDING PROTEIN LIKE6 (SPL6) regulates the signaling outputs by repressing an active transducer INOSITOL-REQUIRING ENZYME 1 (IRE1) of cell death and control panicle development. SPL6 deficient mutant plants revealed hyperactivation of IRE1 and displayed apical panicle abortion [13]. ALUMINUM ACTIVATED MALATE TRANSPORTER (OsALMT7) controls the panicle development by transporting malate to the vascular bundles [3]. A mutation in CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE 31 (OsCIPK31) also displayed apical spikelet abortion phenotype [14]. Mutation in TUTOU1 that encodes a SUPPRESSOR OF CAMP RECEPTOR (SCAR) like protein also exhibited the apical degeneration phenotype due to disorganization of actin [15]. OsC6 encodes a LIPID TRANSFER PROTEIN (LTP) and is involved in the post-meiotic development of anthers. Plants silenced for OsC6 displayed the defective development of pollen exine and tapetum [16]. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) played an important role in anther cuticle development, and dps1 plants showed an accumulation of ROS (reactive oxygen species) in apical spikelets [5].
During panicle development, abortion of spikelets frequently occurs either at basal or apical portions of the panicle due to unfavorable conditions [17]. Extreme temperature, malnutrition and drought stress have been identified as potential factors promoting panicle degeneration [3], [17], [18], [19], [20]. ROS are oxidizing agents and cause dynamic injury to various biological events [21]. Impairments in the panicle, heading date, plant height and number of grains are associated with ROS accumulation that causes damage to cellular tissues and machinery [14], [15], [22]. Unbalanced ROS homeostasis also leads to defects in meiosis and microspore development [23]. Programmed cell death (PCD) is a controlled process involved in rupturing the nuclear membrane, cytoplasmic shrinkage and swelling of the endoplasmic reticulum [24].
Anthers are the male reproductive organs present inside the spikelet and produce pollen grains via meiosis and mitosis [25]. Anthers have four layers; epidermis, endothecium, middle layer and tapetum. Tapetum is the innermost layer, which serves as a source of nutrients and enzymes during microspore development [26]. Tapetum plays a key role in pollen development and seed setting [27]. Normal tapetum development is often associated with a well-developed cuticle, and it protects anthers from external damages. The cuticle is mainly composed of wax and cutin monomers. Any abnormality in cuticle development leads to the impaired tapetum that ultimately causes the failure in the seed setting [28], [29], [30]. Several factors affect cuticle development, such as high temperature, excessive accumulation of ROS and imbalance of certain hormones [31], [32], [33]. These factors are either controlled by genetic elements or environmental cues. Anther cuticle and walls share several phenolic and lipidic precursors. Wax and cutin play an essential role in pollen mother cell (PMC) development. C16 and C18 and their derivatives are called cutin, constitute the cuticle of anthers [34]. Several genes controlling cuticle and tapetum development have been identified, such as WAX-DEFICIENT ANTHER1 (WDA1), DPS1, POST-MEIOTIC DEFICIENT ANTHER1 (PDA1), ATP BINDING CASSETTE TRANSPORTER G26 (OsABCG26), NO POLLEN1 (NP1), IRREGULAR POLLEN EXINE1 (IPE1), DEFECTIVE POLLEN WALL2 (DPW2), and DPW3 have been reported to play their role in anther development by regulating wax and cutin pathway [5], [35], [36], [37], [38], [39], [40], [41]. Similarly, MALE STERILE2 (MS2) and DPW2 play their part in developing pollens by regulating the expression of FATTY ACYL TRANSFERASE [40], [42].
The protein family of SUBTILISIN-LIKE SERINE PROTEASE (Subtilisin) is not well studied in plants. However, their role as CASPASES in animals has been widely reported. Subtilisins display a cleavage specificity and have a particular function in PCD [43], [44]. Plant subtilisins have been reported to play their role in the processing of peptide growth factors [45], fruit ripening [46], xylem differentiation [47], plant-pathogen recognition [48] and determination of silique number [49]. A few plant studies have also indicated their role in reproductive organs, and a meiotic protein was reported to involve in events of late microsporogenesis [50], [51].
The current study is a genetic analysis and functional validation of our previous preliminary study of a gene mapped on chromosome 4 controlling panicle abortion phenotype [52]. We presented a novel function of a putative OsSUBSrP1, which is essential for anther cuticle formation and panicle development. An ethyl methane-sulfonate (EMS) generated mutant apa1331 and CRISPR/Cas9 mediated knock-out lines displayed a significant reduction in seed setting rate due to loss of function of OsSUBSrP1. Thus, we propose that a mutation in OsSUBSrP1 produces defects in anther cuticle biosynthesis and triggers ROS accumulation. Our data present the first insight into the novel role of OsSUBSrP1 in seed setting rate by regulating wax and cutin pathway.
Materials and methods
Experimental materials
The panicle mutant apa1331 was derived from an indica maintainer line Yixiang 1B (WT) by EMS mutagenesis. Apa1331 was screened from an M2 population, and the trait of apical abortion was stably inherited. Yixiang 1B is a backbone maintainer line of indica hybrid rice, bred from the Institute of Agricultural Sciences, Yibin, Sichuan, China. More than 50 hybrids have been released using its corresponding CMS Yixiang 1A as the maternal parent. The apa1331 was used as a female parent and crossed with the WT and 02428 (japonica) cultivars to construct two F2 mapping populations. Plants were grown under natural conditions in the experimental fields at Rice Research Institute, Sichuan Agricultural University, Chengdu (N30.67°, E104.06°), or alternatively at Lingshui (N18.47°, E110.04°), Hainan Province, China.
Scanning electron microscopy
Scanning electron microscopy was performed as described by Chun et al. [53].
DAB staining and quantification of ROS
3,3′-Diaminobenzidine (DAB) staining of the spikelets of WT and apa1331 was done according to a previously described method [54]. ROS was quantitatively measured in the form of H2O2 from 6 cm fresh panicles of WT and apa1331 using a ROS assay Kit (Beyotime, Shanghai, China) by following a method of Zafar et al. [5].
Trypan blue staining and quantification of malondialdehyde (MDA) contents
Trypan blue staining of WT and apa1331 spikelets was performed according to Zafar et al. [55]. MDA contents were quantified from 6 cm fresh panicles of WT and apa1331 using an MDA assay kit purchased from Nanjing Jiancheng Bioengineering Institute according to the manufacturer’s instruction.
Transverse sectioning of anthers
Different stages of anther development were selected from WT and apa1331 panicles as previously described by Wilson et al. [56]. Paraffin-embedded anthers were cut into 4μ thin slides using a microtome (LEICA RM2255).
TUNEL assay
The samples were selected at the 13 cm stage of panicle development from WT and apa1331 prepared according to protocol mentioned in the study of Zafar et al. [5].
Quantitative relative expression analysis
RT-qPCR data of genes were determined from the 6 cm panicle tissues taken only from apical spikelets of WT (normal tissues) and apa1331 (degenerated tissues). PCR amplification and quantitative relative expression were performed according to the previous study of Wu et al. [57].
Genetic analysis of apa1331
Map-based cloning was performed with more than 700 SSR markers using 300 mutant individuals selected from the F2 population. Polymorphic bands were screened from all the chromosomes. For the MutMap assay, F2 plants were backcrossed with apa1331 to generate a BC1F2 population. 25 individuals from BC1F2 population with apical abortion phenotype were selected, and MutMap analysis was performed according to Abe et al. [58].
Development of knock-out lines using CRISPR/cas9 system
A CRISPR/Cas9 vector (BGK03), carrying rice U6 promoter and a single-guide RNA (sgRNA), was constructed for targeting OsSUBSrP1 to develop its knock-out lines. A target sequence (3′- CCATGCGCGACCTCAAGTGT-5′) with protospacer adjacent motif (PAM) sequence “CCA” was identified in the first exon of OsSUBSrP1. Two oligos were designed using an online Biogle tool (http://biogle.cn/index/excrispr). Knock-out constructs used in the current study were developed using Biogle Biotech Kit (BGK03) purchased from Hongzhou Biogle Biotechnology Co., Ltd. The possible off-target effects were prevented by BLAST search using target sequence in Gramene (www.gramene.org) database. Oligo dimers were synthesized according to the manufacturer’s guidelines. Briefly, synthesized oligos were dissolved in water to 10 µM. A reaction mixture (18 µl aneal buffer, 1 µl forward oligo and 1 µl reverse oligo) was heated (95 °C for 3 min) in PCR, and then slowly reduced to 20 °C at a rate of 0.2 °C/s. Oligo dimers were constructed into CRISPR/Cas9 vector by mixing the following components (2 µl CRISPR/Cas9 vector, 1 µl oligo dimer, 1 µl enzyme mix and H2O was added to 10 µl), and kept at a temperature of 20 °C for 1 h. The 10 µl of a recombinant mixture containing the CRISPR/Cas9 vector was transformed into an E. coli strain Trans-DH5α (100 µl) by following the guidelines of Ma et al. [59]. The mixture was added into preheated liquid LB (700ul) and cultured at 37 °C/200 rpm for 1 h. The reaction mixture was plated on the solid LB media having kanamycin at 37 °C overnight. Positive clones were confirmed by sequencing from Qingke Biology Co., Ltd. The constructed vector was sent to Boyun Biotechnology Co., Ltd for genetic transformation into the calli of a rice cultivar Zhonghua11 (ZH11) using a method of Toki et al. [60]. The homozygous T3 plants were used for phenotypic analysis.
Phylogenetic analysis
Sequences of characterized and putative subtilisins were downloaded from NCBI and Phytozome databases and analyzed according to Tripathy et al. [61]. A phylogenetic tree was constructed using a neighbor-joining method at 1000 bootstrap replicate in software MEGA-X [62].
Transcriptome profiling
Transcriptome analysis was performed using three replicates from the panicles of WT and apa1331. Total RNA was extracted from 6 cm panicles when the signs of programmed cell death became visible in the apical spikelet of apa1331 using the manufacturer’s protocol (mirVana miRNA Isolation Kit, Ambion). A total of 6 samples were sent to OE Biotechnology (Shanghai, China) for complete mRNA transcriptome analysis by following a method reported by Li et al. [63]. Samples integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA integrity number (RIN) ≥ 7 were subjected to the subsequent analysis. According to the manufacturer’s instructions, the libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA). Then, these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten), and 125 bp/150 bp paired-end reads were generated. The functional annotation of DEGs was performed according to significantly enriched biological processes according to a GO Consortium [64].
Analysis of anther wax and cutin
The fatty acids contents were determined using 25 mg of fresh anthers from WT and apa1331 according to the instructions of Jung et al. [35] through GC–MS (Gas Chromatograph-2010 Plus SHIMADZU). Briefly, the weighed anthers samples were crushed into powder with liquid nitrogen. A lipid extraction buffer containing chloroform, methanol and water (8:4:3 vol) was added to the sample and vortexed for 1 min. The reaction mixture was sonicated in ice for 5 min, and the process was repeated 3–5 times and kept standing for 2 h. The mixture was centrifuged at 3000 rpm for 15 min. The chloroform residue was transferred into a 10 ml tube. The residue was washed twice by adding 3 ml of extraction buffer, and obtained chloroform was transferred to the previously mentioned 10 ml tube. The tube was placed in a vacuum drier to extract plant triglycerides. For saponification, 2 ml of a solution (KOH-CH3OH, 0.5 mol/L) was added to the reaction tube, sealed and vortexed for 1 min. The reaction tube was placed in boiling water until the oil droplet completely disappeared from the surface. The reaction was mixed several times during saponification to get all free fatty acids. For methyl esterification, the reaction mixture was cooled for 3 min, and 2 ml of BF3-CH3OH was added, vortexed (10 s) and placed in a water bath (80 °C). For extraction, 1 ml of n-hexane and 5 ml of saturated NaCl solution were added to the mixture, vortexed and centrifuged at 2000 rpm for 5 min. The n-hexane layer (not less than 500 µl) was inserted into the Gas Chromatograph. The composition of fatty acids was presented in the form of relative percentage as performed by Liu et al. [65]
Results
apa1331 displayed aborted apical spikelets
To understand the genetic regulation of panicle development, we identified a mutant named apical panicle abortion1331 (apa1331), which shows a stable phenotype of panicle abortion. This mutant was screened through an EMS mutagenized population of a maintainer line (Yixiang 1B), taken as WT in the current study.
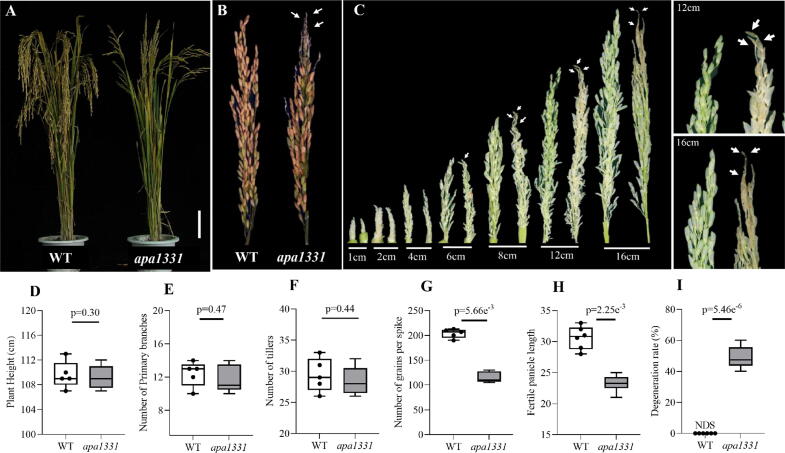
At maturity, apa1331 showed abortion of apical spikelets and had significantly lower seed yield than WT (Fig. 1, A). The panicle of apa1331 started to develop aborted spikelets at apical portions, while WT did not reveal any abortion (Fig. 1, B). To differentiate, when apa1331 started to show abortion, we carefully observed different stages of panicle development and compared them with WT (Fig. 1C). Until the 5 cm length of the panicle development, the developmental course of apa1331 was normal like that of WT and did not reveal any visible abortion signs. At 6 cm length of panicle development, spikelet abortion became visible in the apical spikelets of apa1331. Apical spikelets began to degenerate when panicle was still enclosed in the flag leaf. As the panicle grows in its length, apical spikelets abortion became severe. The ∼6 cm length of panicle development was regarded as the stage of divergence between apa1331 and WT. Various agronomic traits, e.g., plant height, number of primary branches, number of tillers, and 1000-grain weight did not show any significant differences in apa1331 compared to WT (Fig. 1D-F). However, the number of grains per spike and fertile panicle length were significantly decreased in apa1331 (Fig. 1G-H). Degeneration rate, which is the percentage of degenerated/aborted spikelets to the total number of spikelets in a panicle, in apa1331 was up to 53% (Fig. 1I). At the same time, WT did not show abortion of spikelets. The seed setting rate in apa1331 was decreased up to 40–53% compared to WT. Thus, apa1331 is an apical panicle abortion mutant and significantly reduces the number of fertile spikelets and causes a massive loss in grain yield.
Fig. 1.
Phenotypic observation and agronomic traits of apical panicle abortion1331 (apa1331), (A) Plant morphology of a Wild Type (WT) plant showing normal panicle development, while an apa1331 plant shows apical spikelet abortion grown under field conditions. (B) Mature panicle of WT and apa1331.(C) Different stages of WT and apa1331 panicle at a length of 1 cm, 2 cm, 4 cm, 6 cm, 8 cm, 12 cm and 16 cm. White arrows show the aborted spikelets in apa1331. Panels to the right of C are enlarged views of 12 cm and 16 cm panicles of WT and apa1331. Box and whisker plots (D-I) show the comparison of plant height (D), number of primary branches (E), number of tillers (F), number of grains per spike (G), fertile panicle length (H) and degeneration rate between WT and apa1331(I), respectively. The student’s t-test was used to calculate the significance of the data. Whereas, NDS: no degenerated spikelets. Bars are equal to 10 cm in (A).
The apical spikelets of apa1331 are pollen-less and defective in anther cuticle formation
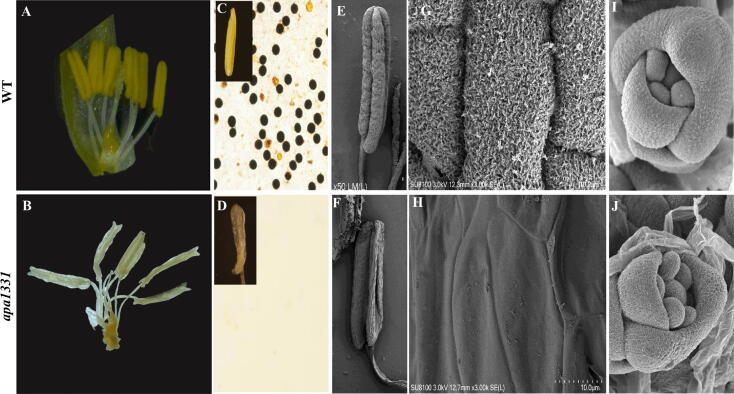
To know the basis of the low seed setting rate of apa1331, we studied the detailed structure of the apical spikelet using stereo and electron microscopy. The anthers of apa1331 were degenerated, while WT anthers were normal and yellowish (Fig. 2A-B). WT anthers had normal and viable pollens that were darkly stained in potassium iodide (KI) staining, while apa1331 anthers were entirely pollen-less (Fig. 2C-D). It is worth mentioning that the basal spikelets of apa1331 were normal and showed fertile spikelets comparable to WT. However, the only apical spikelets were found sterile. Previous studies have revealed the defective development of anthers cuticle in sterile mutants [29], [66]. Accordingly, the scanning electron microscopic (SEM) observation revealed a presence of three-dimensional nano-ridges of cuticle on the surface of WT anthers, while apa1331 showed a complete lack of cuticle (Fig. 2E-H). The lack of cuticle formation on the surface of apa1331 anthers encouraged a further deep look into microsporogenesis and meristem development. SEM of apa1331 did not show any observable aberration in meristem formation at ∼1.5 cm length of panicle (Fig. 2I-J). This data suggests that apa1331 showed defective anther cuticle formation and did not show any abnormality at meristem development.
Fig. 2.
Microscopic examination and anther cuticle formation of Wild type (WT) and apical panicle abortion1331 (apa1331). (A-B) Comparative morphology of WT and apa1331 spikelet obtained by stereomicroscope. (C-D) Potassium iodide (KI) staining of pollens and their enlarged view of WT (C) and apa1331 (D).(E-F) Scanning electron micrographs (SEM) of anthers of WT (E) and apa1331 (F).(G-H) Zoomed view of anther cuticle surface of WT (G) and apa1331 (H). (I-J) Inflorescence meristem development of WT (I) and apa1331 (J). Bars are equal to 1.5 mm in A-B, 0.5 mm in C-D, 50 µm in G-H and 500 µm in I-J.
Post-meiotic microspore development was defective in apa1331
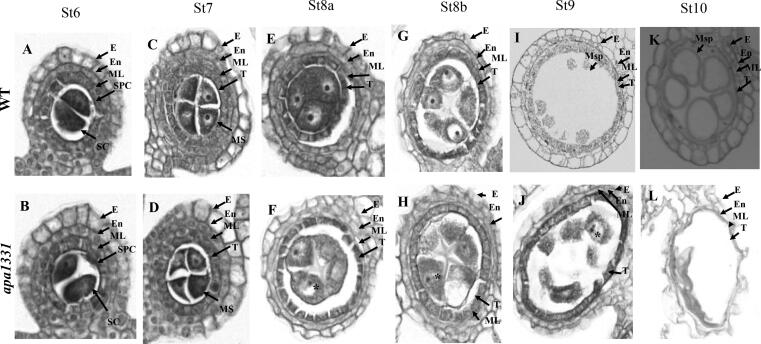
To reveal any cytological defects in microspore development, cross-sections of different stages of anthers were analyzed as discussed by Wilson et al. [56]. Transverse sections of WT and apa1331 anthers did not reveal any observable difference until stage 7, and both showed a regular structure of all layers e.g., epidermis, endothecium, middle layer and tapetum (Fig. 3A-D). During the early stage 8, the PMCs of WT went under normal meiosis; however, the PMCs nuclei of apa1331 were neither round (asterisk) nor enclosed by a well-developed callose wall (Fig. 3E-F). Moreover, the cytoplasm of apa1331 was trapped within a cloud (Fig. 3F). During late stage 8, in apa1331, the cloud around the cytoplasm became more evident, and PMC nuclei were irregular (asterisk) and degenerative (Fig. 3G-H). In contrast, PMC nuclei of WT were darkly stained and showed a regular (round) shape (Fig. 3G). During stage 9, WT tapetum cells were normal, but that of apa1331 were swollen and significantly degenerated microspore cells and nucleus were seen (Fig. 3I-J). Microspore development of apa1331 was seriously affected after stage 9 and showed severe degeneration of microspores (Fig. 3K-L). During stage 10, the tapetum of WT started to degenerate, and microsporocytes were found vacuolated. However, the microspores of apa1331 were collapsed and remnants of degenerated microspores were present in the form of cellular debris (Fig. 3L). Due to severe degeneration in apa1331 anthers, we were unable to differentiate later stages of development. The results of cross-sections morphology suggest that microspore development after meiosis was strongly affected in apa1331.
Fig. 3.
Transverse sections of anthers showing defects of microspore development in apical panicle abortion1331 (apa1331). (A-L) Transverse micrographs of Wild type (WT) and apa1331 anthers development at (A-B) St6: showing normal thickening of anthers walls, (C-D) St7: showing the normal development of all anther layers in apa1331, (E-F) St8a: showing the trapped cytoplasm in a cloud, and degeneration of pollen mother cell (PMC) nucleus in apa1331. At the same time, the PMC of WT was regular and round. Asterisk (*) indicates the position of degenerating PMC nucleus. (G-H) St8b: Trapped cytoplasm and degeneration of PMC nucleus of apa1331 become more evident, (I-J) St9: Swollen tapetum and degeneration of microspore of apa1331,(K-L) St10: At late microspore mother cell development, apa1331 showed complete degeneration of microsprocyte in the form of a degenerated tissue. Whereas, E; epidermis, En; endothecium, ML: middle layer, SPC; secondary parietal cells. SC; sporocytes, T; tapetum, and Msp; microsporocyte. Bars in A-L are 25 µm.
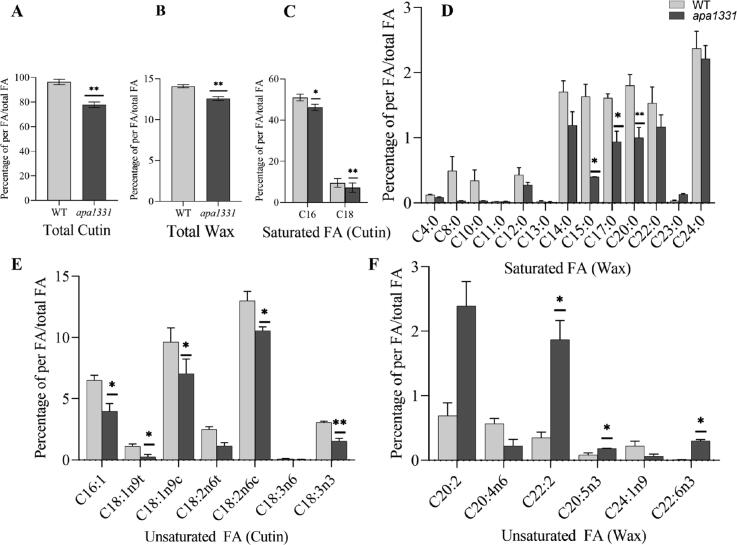
Reduction of wax and cutin contents in apa1331 anthers
SEM of anthers surface revealed a pronounced lack of cuticle in apa1331. To confirm the differences of wax and cutin, which are important components of anthers cuticle, we measured the fatty acid profiles of apa1331 and WT anthers by Gas Chromatography. Composition data of wax and cutin were presented in percentages as described by Liu et al. [65]. Results revealed a significant (P < 0.01) decrease in total cutin and wax contents in apa1331 as compared to WT (Fig. 4A-B). Among saturated fatty acids, palmitic acid (C16:0) and steric acid (C18:0) constitute the cutin monomers and were significantly (P < 0.01) decreased in apa1331 as compared to WT (Fig. 4C). Similarly, the percentage of saturated wax, especially C15:0, C17:0 and C20:0 were also significantly reduced in apa1331 (Fig. 4D). Similarly, unsaturated cutin e.g., C16:1, C18:1nt, C18:1nc and C18:1n3 were also significantly decreased in apa1331 (Fig. 4E). However, contrary to other fatty acids, unsaturated wax contents e.g., C22:2, C20:5n3, and C22:6n3 were significantly (P < 0.05) increased in apa1331 compared to WT (Fig. 4F). Consistent with our previous observations, chemical composition analysis of wax and cutin revealed that the candidate gene of apa1331 regulates the wax and cutin contents and anther cuticle formation.
Fig. 4.
Composition of wax and cutin contents in anthers of Wild type (WT) and apical panicle abortion1331 (apa1331), (A) Total amount of cutin in anthers. (B) Total amount of wax in anthers. (C) Amount of saturated fatty acids (cutin) in anthers. (D) Amount of saturated fatty acids (wax) in anthers. E) Amount of unsaturated fatty acids (cutin) in anthers. (F) Amount of unsaturated fatty acids (wax) in anthers of WT and apa1331. The values represent the average of three individual repeats. Data in (A-F) are presented in percentages of per fatty acid to total fatty acids. The student’s t-test was used to calculate the significance of data, where *p < 0.05 and **p < 0.01.
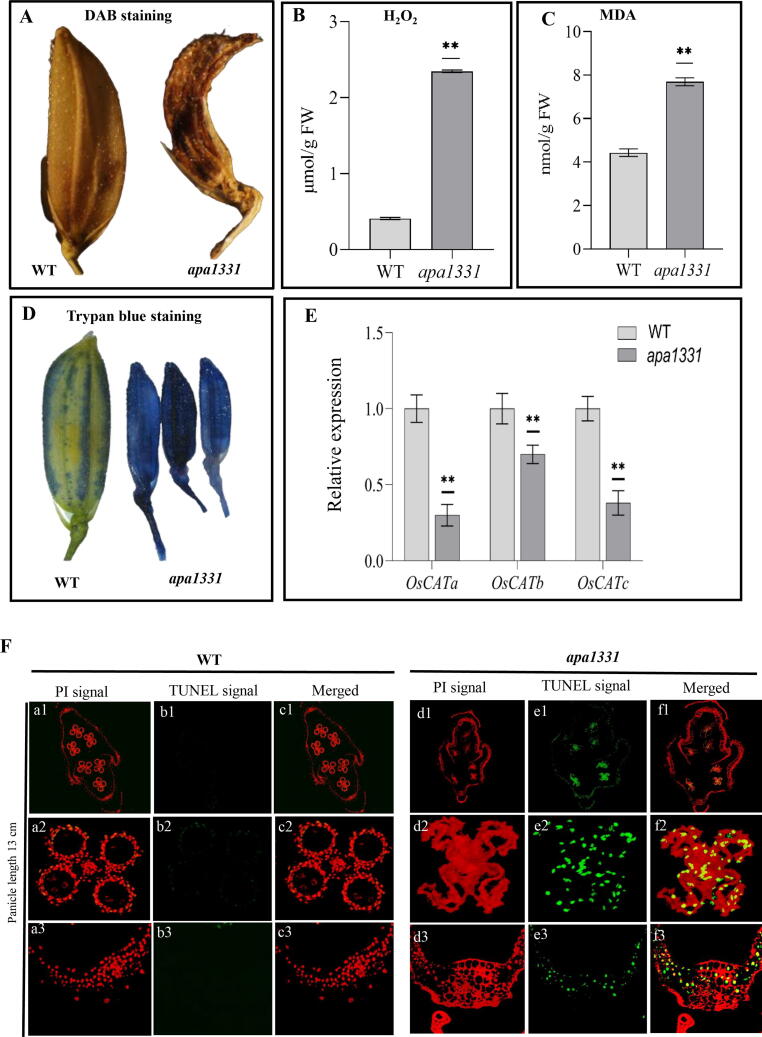
Apical spikelets of apa1331 had higher level of ROS
Excessive ROS accumulation could happen either due to genetic factors or in response to biotic and abiotic stresses [67]. These ROS molecules could cause damage to the cell wall structure, macromolecules, DNA and proteins, leading to the degenerated tissues [68]. To further investigate the cause of abortion of spikelets of apa1331, we detected the cellular ROS by DAB staining (Fig. 5A). The quantitative measurements revealed that H2O2 was significantly (P < 0.01) increased in apical spikelets of apa1331 than that of WT (Fig. 5B). Malondialdehyde (MDA) is an indicator of local ROS production and is involved in the peroxidation of fatty acids [69]. Consistently, MDA was also significantly increased in apa1331 spikelets (Fig. 5C). Trypan blue staining is used to test the viability of cells [70]. Dark staining of the apical spikelet of apa1331 showed cells viability was compromised, while light staining of WT showed spikelets cells were viable (Fig. 5D). CATALASE (OsCATa), OsCATb, and OsCATc are antioxidant enzymes coding genes and are involved in the scavenging activities of ROS [71]. Relative expressions of OsCATa, OsCATb, and OsCATc were significantly decreased in apa1331 spikelets compared to WT (Fig. 5E, Table S1). Together, DAB staining, measurement of H2O2 and relative expression data revealed the excessive accumulation of ROS in apa1331 caused the abortion and damage to apical spikelets. At the same time, trypan blue staining has indicated the death of apical spikelets cells.
Fig. 5.
Analysis of reactive oxygen species (ROS), cell viability, cell death and DNA fragmentation in apical panicle abortion1331 (apa1331), (A) Dark DAB (3,3–diaminobenzidine) staining of the spikelet of apa1331 shows the excessive accumulation of hydrogen peroxide (H2O2) compared to WT. (B) Quantification of H2O2 was significantly increased compared to WT. (C) Quantification of malondialdehyde (MDA) was significantly increased compared to WT. (D) Trypan blue staining revealed the compromised cell viability in apical spikelet apa1331. (E) Significant downregulation of CATALASES, e.g., OsCATa, OsCATb and OsCATc in the panicle of apa1331 than WT. (F) TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay showing DNA fragmentation in spikelet of apa1331. Tissues were taken from the whole spikelet (a1 and d1), anther (a2 and d2) and glume (a3 and b3). Propidium iodide (PI) and iso-thiocyanate produce red and green (b and e) fluorescence, respectively. Yellow (c and f) fluorescence is the combined signals of PI and iso-thiocyanate. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Apical spikelets and anthers of apa1331 showed increased cell death and DNA fragmentation
Trypan blue staining indicated the presence of cell death in apical spikelets of apa1331. We hypothesized that apical spikelets of apa1331 would have undergone an abnormal PCD and DNA fragmentation. To test this hypothesis, we performed a TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling) assay to detect apoptosis. Spikelet and anthers of 13 cm panicle length were used as representatives (Fig. 5F). WT (Fig. 5Fa-c) showed the absence of TUNEL signal in all parts of spikelets (c1) including anther (c2) and glume (c3). However, apa1331 (Fig. 5Fd-f) showed positive TUNEL signals in all parts of the spikelet (e1) including anther (e2) and glume (e3). These findings suggest that DNA fragmentation has occurred in apa1331 at the single-cell level that causes cell death in spikelets and anthers.
OsSUBSrP1 encodes a putative protein of a subtilisin family
To find the candidate gene of the characterized mutant phenotype, we developed two F2 mapping populations. These mapping populations were derived by crossing apa1331 with an indica cv. Yixiang 1B and a japonica cv. 02428, respectively. In F1 generations of both populations, all plants did not show an abortion phenotype. Genetic analysis of these populations revealed that a single recessive gene controls the phenotype of panicle abortion in apa1331 [52].
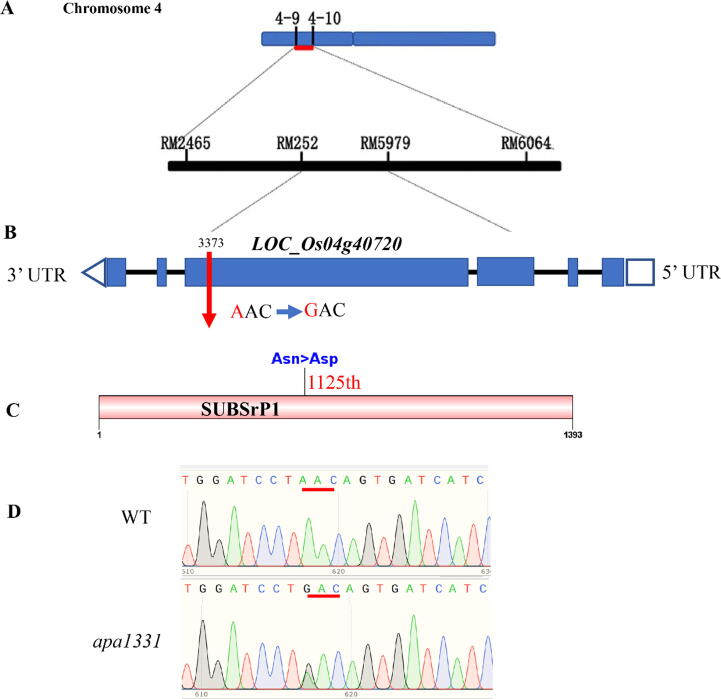
Primary mapping of apa1331 revealed that the candidate gene resides between the SSR marker 4–9 and 4–10 on the short arm of chromosome 4 (Fig. 6A). The gene was narrowed down between RM252 and RM5979 in a distance of 16.4 cM [52]. Due to the further unavailability of InDel markers in this region, we performed a MutMap assay to find any mutation in the primary mapped region by following a method of Abe et al. [58]. MutMap whole genome sequencing (Illumina HiSeq 2500 platform) revealed an SNP index of 1 due to the presence of a single nucleotide mutation in (Os04g0483400) LOC_Os04g40720 [52]. Consistent with the primary mapping by SSR markers, Mutmap results showed an SNP (A > G) located between the previously mapped region of SSR markers RM252 and RM5979. According to MutMap, the candidate gene of apa1331 phenotype was LOC_Os04g40720, and it has six exons, present in reverse order in the MSU database. The mutation was located in the 4th exon of LOC_Os04g40720, changing the 3373th codon from AAC to GAC (Fig. 6B). This SNP in apa1331 has ultimately substituted an amino acid (asparagine into aspartic acid). According to Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), LOC_Os04g40720 annotates a putative protein of SUBTILISIN LIKE SERINE PROTEASE [72]. Candidate protein has a total residue length of 1393 amino acids, and mutation (Asn > Asp) was occurred in 1125th amino acid (Fig. 6C). To further verify the MutMap candidate gene analysis, we cloned and sequenced the candidate gene “LOC_Os04g40720” in apa1331 and WT. The comparative chromatograms of WT and apa1331 validated the presence of an SNP at the same position consistent with MutMap analysis (Fig. 6D).
Fig. 6.
Fine mapping and genetic analysis of SUBTILISIN-LIKE SERINE PROTEASE 1 (OsSUBSrP1). (A) Primary mapping of the OsSUBSrP1 controlling apical panicle abortion1331 (apa1331) phenotype on chromosome 4 between SSR markers 4–9 and 4–10. (B) Genomic structure of OsSUBSrP1 (LOC_Os04g40720) and position of an SNP, where blue boxes are representing exons, blue lined boxes are up and downstream sequences and black lines are indicating introns. The red arrow shows the position of an SNP that was harbored in the 4th exon of OsSUBSrP1.(C) SUBSrP1 was consist of 1393 amino acids, and black line show the position of a non-synonymous substitution (Asn > Asp) of 605th amino acid. (D) The comparative chromatograms cloned and sequenced from WT (up) and apa1331 (down), where the red lines show the substituted sequence (AAC > GAC) of the codon. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Phylogenetic and annotation analysis of the candidate gene of apa1331 shows that it encodes a novel protein and has not been characterized before. However, its orthologs in Brachypodium (Bradi5g13740), maize (GRMZM2G063163) and sorghum (Sb06g0200570) encode putative subtilisin-like serine proteases. According to MEROPS database, there are more than 50 families of serine protease [73]. A study has identified 63 and 56 subtilases in rice and Arabidopsis, respectively [74], [75]. These two plant species also appear to encode a similar number (206 and 222) of putative subtilises in Arabidopsis and rice, respectively [61]. Although the genome size of rice (389 MB) is much higher than that of Arabidopsis (125 MB). Another genomic evolution study has categorized serine protease into 9 families, among them subtilisins are the second largest family having a catalytic triad of Asp (D), His (H) and Ser (S) residues with no other structural similarity [76], [77]. Accordingly, the candidate protein of apa1331 has also no known structural similarity with other characterized proteins. BLAST of referenced protein showed only a 37–45% sequence similarity with most of the uncharacterized proteins of species other than rice (Figure S1). None of the members of its orthologs genes have been functionally characterized before. Amino acid sequence alignment of blasted proteins showed conserved residues of catalytic triads of D, H and S (Figure S2 A). A phylogenetic tree was constructed, revealing rice subtilisins can be divided into three major clades (Figure S2 B). The largest clade shared 26 genes that were further divided into two subclades. While both second and third clades shared two genes each. The reference gene (Os04g0483400) was placed into a seprate clade with another gene Os07g0651400.
Genetic analysis revealed that a-synonymous subtitution (Asn-Asp) of an amino acid in apa1331 has not occurred in the catalytic triad of subtilisin. It suggests the additional and criticle role of another amino acid (Asn) combined with other catalytic traids for proper functioning. The presence of a catalytic triad of D, H and S and functional homology with subtilisin orthologs suggest that the candidate protein of apa1331 encodes a subtilisin. Hence, we tentatively named the reference protein of apa1331 as putative SUBSrP1.
SUBSrP1 is preferentially expressed in the upper part of a young panicle
Members of the subtilisin protease family showed variation in spatio-temporal expression under different conditions [78]. According to Rice anther expression (https://www.cpib.ac.uk/anther/riceindex.html) plots [79] the expression of OsSUBSrP1 during anther development was also found variable (Figure S3 A). According to BAR ePlant Browser [80] database (bar.utoronto.ca/eplant) relative expression spectrum of OsSUBSrP1 showed the highest expression in a young panicle at stage P1 and P2 (Figure S3 B). To answer an important question, e.g., why only apical spikelets of apa1331 have degenerated? We determined the relative expression spectrum of OsSUBSrP1 in the root, leaf, stem and different stages of panicle development in WT (Figure S4). Relative expression of OsSUBSrP1 in 5 cm panicle was higher than that of 2 cm and 12 cm panicle length. We further divided the 5 cm panicle into the upper, middle and lower part and quantified the relative expression of OsSUBSrP1. Result revealed that OsSUBSrP1 was preferentially expressed in different parts, where upper part showed highest expression followed by middle part, and minimum in lower part of WT panicle. Variation in the expression of OsSUBSrP1 redundancy between different parts of the panicle disentangled its phenotypic defect from the lower and middle part of the panicle. Hence, the phenotypic defect of the loss of function of OsSUBSrP1 was majorly seen in the apical spikelets only.
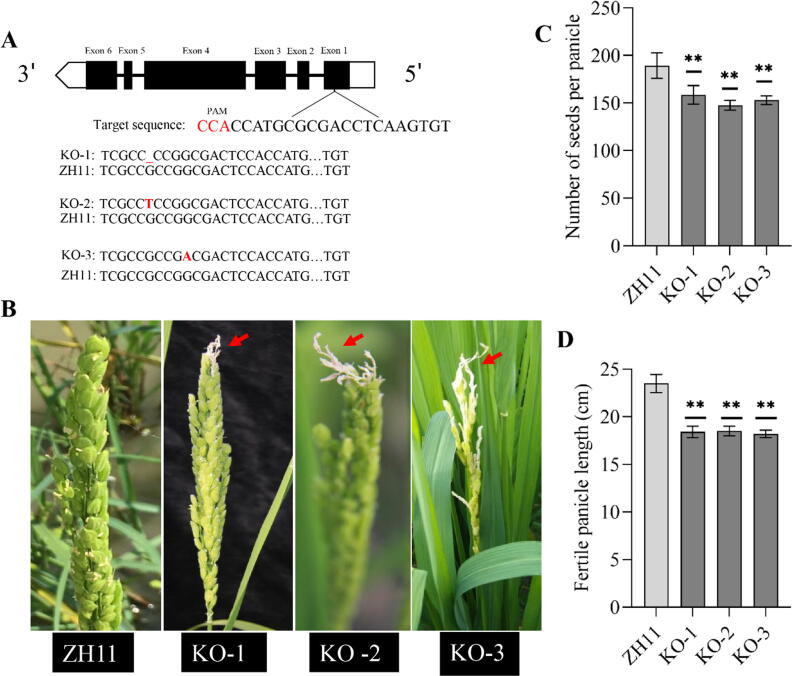
CRISPR-Cas9 targeted knock-out lines showed the phenotype of apical spikelet abortion
To validate the function of OsSUBSrP1, a CRISPR-Cas9 vector containing a sgRNA was constructed and transformed into calli of ZH11 to develop knock-out (KO) lines. The target sequene sequence (3′- CCATGCGCGACCTCAAGTGT-5′) was located in first exon of OsSUBSrP1 (Fig. 7A). Three independent knock-out (KO) lines showed the abortion of apical spikelet. Sequencing revealed the one bp (G) deletion, one bp substitution (G to T) and one bp substitution (G to A) in KO-1, KO-2, and KO-3, respectively. KO-1, KO-2 and KO-3 showed the abortion of apical spikelets, while the ZH11 did not show any abortion phenotype at the heading stage (Fig. 7B). Similar to apa1331, the number of seeds per panicle and fertile panicle length was significantly (P < 0.01) decreased in KO-1, KO-2, and KO-3 compared to ZH11 (Fig. 7C-D). Overall, genetic analysis of apa1331 and CRISPR/Cas9-targeted mutagenesis of KO lines revealed that mutation in OsSUBSrP1 causes a significant decrease in seed setting due to abortion of apical spikelets. It validates the function of OsSUBSrP1 in apical spikelet development in rice.
Fig. 7.
CRISPR/Cas9 mediated knock-out (KO) lines of SUBTILISIN-LIKE SERINE PROTEASE 1 (OsSUBSrP1) showed the phenotype of apical abortion. (A) The position of target sequence (PAM and gRNA) in gene structure of OsSUBSrP1. Sequence comparison reveals a one bp (-) deletion in KO-1, substitution of G to T in KO-2 and substitution of G to A in KO-3 (B) Panicle of KO-1, KO-2, and KO-3 show the phenotype of apical abortion, while the Zhonghua11 (ZH11) did not show abortion. (C) Number of seeds per panicle was significantly (P < 0.01) decreased in KO-1, KO-2, and KO-3. (D) Fertile length of panicle was significantly decreased due to apical panicle abortion in KO-1, KO-2, and KO-3. The student’s t-test was used to calculate the significance of data, whereas **p < 0.01.
Global gene expression analysis revealed OsSUBSrP1 mediated downregulation of wax and cutin biosynthesis pathway genes
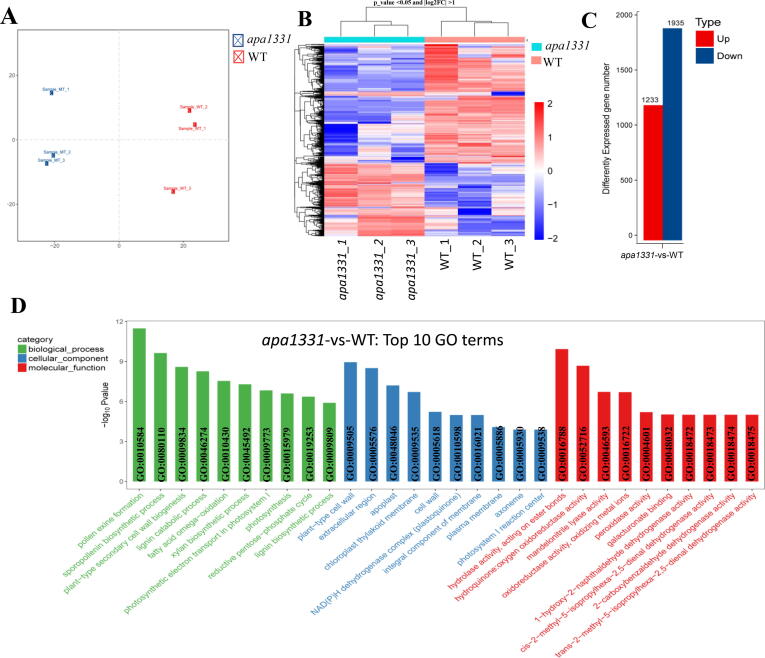
Abnormal PCD and elevated ROS levels can affect the expression of many wax and cutin pathway genes [5], [39]. To elucidate the transcriptional changes associated with functional pathways due to mutation in OsSUBSrP1, a comparative transcriptome profiling of WT and apa1331 was performed using RNA sequencing Illumina Platform (HiSeqTM 2500). Three individual sampling repeats were selected from WT and apa1331 apical spikelets at the 6 cm stage (point of divergence between WT and apa1331). High-throughput RNA sequencing generated high-quality data with an adequate sequencing depth. Principal component analysis (PCA) revealed 76% (PC1) and 19% (PC2) variance between WT and apa1331 samples (Fig. 8A). The heat map showed a sum of 3168 differentially expressed genes (DEGs) between apa1331 and WT (Fig. 8B). Among 3168 DEGs, 1935 and 1233 DEGs were found down-regulated and up-regulated in apa1331 compared to WT, respectively (Fig. 8C). We further subjected the significantly [fold change (FC) > 2 and a p < 0.05] DEGs to gene ontology (GO) enrichment analysis and Kyoto encyclopedia of genes and genomes (KEGG) pathway functional classification.
Fig. 8.
Global gene expression analysis of Wild type and apical panicle abortion1331, (A) Principal component analysis (PCA) shows 76% and 19% variations in the samples contributed by PC1 and PC2, respectively. (B) Heat map showing the differentially expressed genes (DEGs) with log2FC > 1 at p-value < 0.05 in apa1331 compared to WT. (C) Graph showing statistics of DEGs between WT and apa1331, where red and blue color are showing the number of up and down-regulated genes, respectively. (D) Comparison of top 30 downregulated gene ontology (GO) terms. The x-axis shows the GO terms of biological processes, cellular components and molecular functions in green, blue and red color, respectively, while the y-axis shows the -log10 (p-value) of differentially enriched GO terms between WT and apa1331. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
GO enrichment analysis of significantly enriched DEGs between apa1331 and WT was categorized into biological processes, cellular components and molecular functions. Among the top 30 down-regulated DEGs, pollen exine function (GO:0010584) was most significantly down-regulated biological process in apa1331 (Fig. 8D). While other important biological processes associated with these downregulated DEGs were sporopollenin biosynthetic process (GO:0080110), fatty acid omega oxidation (GO:0010430) and lignin biosynthetic process (GO:000980). While most significantly downregulated cellular components and molecular functions associated with these DEGs were plant-type cell wall (GO:0009505) and hydrolase activity, acting on ester bonds (GO:00016788), respectively. GO classification indicates the downregulation of pollen exine, sporopollenin, fatty acid omega-oxidation related biological processes in apa1331 panicle.
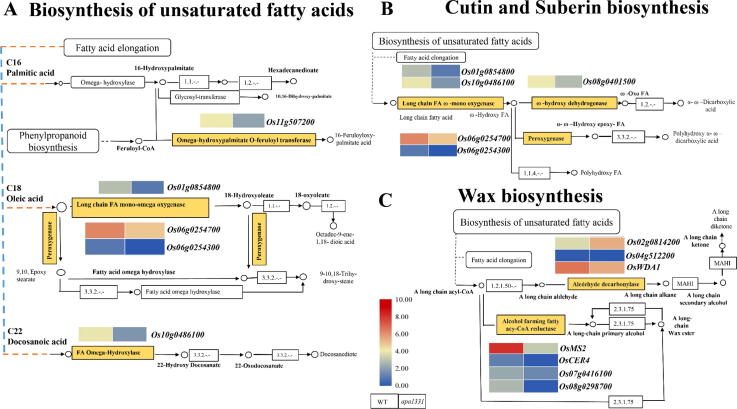
We mapped all DEGs to their respective KEGG enrichment analysis to get further insights into the specific pathways, which regulate the above-mentioned biological processes. Among them, the cutin, suberin, and wax biosynthesis pathway (K00073) was the most relevant KEGG pathway with the highest (15.3%) number of DEGs (Fig. 9). 13 DEGs were involved in the wax and cutin biosynthesis pathway. Among the 13 genes, 12 genes (Os11g0507200, Os01g0854800, Os06g0254700, Os06g0254300, Os10g0486100, Os08g0401500, Os04g0512200, OsWDA1, OsMS2 (MALE STERILITY2), OsCER4, Os07g0416100 and Os08g0298700) were significantly down-regulated. While only one gene Os02g0814200 annotated for aldehyde decarbonylase (EC: 4.1.99.5) was up-regulated (FC = 3.341) in apa1331 compared to WT. The heat map of these 12 down-regulated and one up-regulated genes represents significant changes in their expression in apa1331 compared to WT. Among the down-regulated genes e.g., Os11g0507200 encodes a transferase family protein and is involved in the biosynthesis of unsaturated fatty acids, especially cutin and suberin. Os01g0854800 and Os10g0486100 encode a cytochrome P450 protein and are involved in the long-chain fatty acid mono-oxygenase activity (EC:1.14.14.80). Os06g0254300 encodes a peroxygenase and is involved in the cutin, suberin and wax biosynthesis pathway. OsWDA1, OsMS2, and OsCER4 have been reported to regulate the wax and cutin biosynthesis and pollen development in rice [30], [35], [81].
Fig. 9.
SUBTILISIN-LIKE SERINE PROTEASE 1 (OsSUBSrP1) regulates the expression of wax, suberin, and cutin biosynthesis pathway genes, (A) Comparative transcriptome analysis of Wild type (WT) and apical panicle abortion1331 (apa1331) revealed the downregulation of genes involved in the biosynthesis of the unsaturated fatty acids. (B) Comparative transcriptome analysis of WT and apa1331 revealed the downregulation of genes involved in the biosynthesis of cutin and suberin biosynthesis. (C) Comparative transcriptome analysis of WT and apa1331 revealed the up and downregulation of genes involved in the wax biosynthesis. The heat maps showed the normalized expression value of fragments per kilobase of transcript per million (FPKM) for a specific gene (brown rectangle). Figure legend shows the normalized expression value key scale, in which dark red and blue show the highest and lowest expression, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To validate the expression of DEGs obtained in RNA-seq data, we randomly selected five genes among up-regulated and down-regulated DEGs validated their relative expression by RT-qPCR (Figure S5, Table S1). The level of ENT-COPALYL DIPHOSPHATE SYNTHASE 4 (OsCPS4), CXE CARBOXYLASE, POLYAMINE OXIDASE, DIISOPROPYL-PHOSPHO-FLUORIDATE (DFP) and GLUCSOYL HYDROLASE were significantly up-regulated in apa1331 compared to WT. LTPL72, PURPLE ACID PRECURSOR, CYTOCHROME P450 FAMILY, 3-OXOACYLE-REDUCTASE and OsC6 were significantly down-regulated in apa1331 compared to WT. The relative expression of genes analyzed by qRT-PCR was consistent with that RNA-seq profiling. These results support that mutation in OsSUBSrP1 causes a significant decrease in wax and cutin contents due to downregulation of cutin, suberin and wax biosynthesis pathway genes.
Discussion
Successful plant reproduction is a sophisticated process involving proper development of male and female reproductive organs. This process is controlled by a set of different genetic factors working together in different pathways. Identifying the role of these genetic factors is crucial to understand the mechanism of reproduction, and the knowledge could be applied in hybrid-breeding programs to accelerate the crop yields. In this study, we have identified the role of a novel genetic factor called OsSUBP1 in rice male reproductive development using mutation breeding, transcriptomic and genome editing approaches. The OsSUBP1 was cloned from a mapping population using map-based cloning based on a panicle abortion mutant called apa1331. The function was confirmed by knocking-out the target gene using CRISPR/Cas9, a highly sensitive genome editing approach [82].
OsSUBSrP1 is essential for normal apical spikelet development
Genetic analysis of apa1331 has revealed that it bears an SNP (A > G) in the 4th exon that causes the dysregulation in OsSUBSrP1 (Fig. 6). OsSUBSrP1 encodes a putative subtilisin and its role in the plants was not reported before. A study has indicated the secretion of a serine proteinase at late microsporogenesis and primary expression in tapetal cells [51]. It was also stated that the serine protease is a preproprotein, which acquires glycans during movement from the Golgi body and mitochondria to other organelles. Another study revealed the preferential expression of a preproprotein RICE SUBTILISIN-LIKE SERINE PROTEASE 1 (RSP1) in ovaries and pistils [50]. Few family members have also been differentially expressed in tomatoes [83]. Martinez et al. reported SENESCENCE-ASSOCIATED SUBTILISIN LIKE PROTEASE (SASP) in Arabidopsis, and its loss of function mutant showed increased inflorescence branches at the reproductive stage [49]. They also reported that the subtilisin-like SASP function is conserved at-least between rice and Arabidopsis. However, functional characterization of any individual plant serine protease has not been reported so far. Nonetheless, some findings have indicated the presence of SUBSrP in rice and preferential expression in flower development [49], [50]. Our study presents that loss of function of OsSUBSrP1 produced apical spikelet abortion. Knock-out of OsSUBSrP1 also produces apical abortion, which verifies its indispensability for normal apical spikelet development in rice.
OsSUBP1 is involved in anther cuticle formation and regulates wax and cutin biosynthesis pathway
Fatty acids are essential molecules required during anther, especially tapetum development [29]. Anthers of the apical spikelets of apa1331 were pollen-less and sterile. Their SEM also revealed defects in anther cuticle formation (Fig. 2E-G). Transverse sections of apa1331 revealed the abnormal swelling of tapetum and degeneration of the PMC nucleus at stage 9 (Fig. 3K-L). Anther cuticle is composed of cutin and wax, which are long-chain fatty acids, alcohols and alkanes [84]. Comparative fatty acid profiling revealed a substantial decrease in wax and cutin in apa1331 (Fig. 4). Previous studies showed that reduction of wax and cutin contents causes defects in pollen development [16], [29], [42], [66]. Decrease in wax and cutin contents also suggests the substantial loss of water and alters cuticle properties [85]. Transcriptome profiling of apical spikelets also revealed the significant downregulation of wax and cutin pathway genes in apa1331. Downregulation of wax and cutin genes supports the observation of decreased anther cuticle formation (Fig. 9). Global gene expression analysis revealed that 13 genes were directly involved in the metabolism of fatty acids. OsWDA1, OsMS2 and OsCER4 have already been reported to regulate wax biosynthesis, pollen exine and anther walls formation [35], [42], [65], [81]. Os01g0854800, Os06g0254300 and Os10g0486100 also encode different enzymes involved in regulating cutin, suberin and wax biosynthesis pathway. A considerable decrease in saturated and unsaturated wax and cutin in apa1331 was likely due to the down-regulation of genes mentioned above. These findings support that OsSUBSrP1 is essential for normal development of PMC and anther cuticle formation. Our data suggest OsSUBSrP1 plays an essential role in anther cuticle formation by regulating the expression of wax and cutin biosynthesis genes.
OsSUBSrP1 regulates ROS-mediated cell death
DAB staining and measurement of H2O2 revealed an excessive accumulation of ROS in apical spikelets of apa1331 (Fig. 5). Enhanced level of MDA and decreased relative expression level of OsCATa, OsCATb and OsCATc revealed the excessive peroxidation of fatty acids and ROS imbalance in apa1331 (Fig. 5E). Dark staining of trypan blue and strong positive TUNEL signals revealed DNA fragmentation and cell death in apa1331 (Fig. 5F). Previous studies have reported that the excessive occurrence of ROS causes cell death in the apical spikelets [3], [14]. Enhanced ROS levels and increased PCD in the apical spikelets indicate their correlation with panicle abortion. Excessive PCD has been reported to play its role in panicle development and spikelet abortion [16], [35]. Enhanced PCD is usually accompanied due to disturbance in the ROS homeostasis or external stimuli [86]. Aborted pollen grains, failure of normal microspore development and abnormal anther development have been reported due to excessive accumulation of ROS [5], [67]. ROS are important signaling molecules involved in the rupture of tapetum for pollen grain development. Still, its accumulation beyond the threshold would produce oxidative stress to cellular components [87]. Hence, it is logical to refer that apical sterility in apa1331 was caused due to excessive bursts of ROS and abnormal cell death occurrence. Our findings provide the basis of the mechanism by which homeostasis in ROS-mediated PCD was possibly dysregulated due to the mutation in OsSUBSrP1. Our data support that apical spikelet abortion onset was started due to mutation in OsSUBSrP1 and validate the potential correlation between cell death and ROS homeostasis for panicle development.
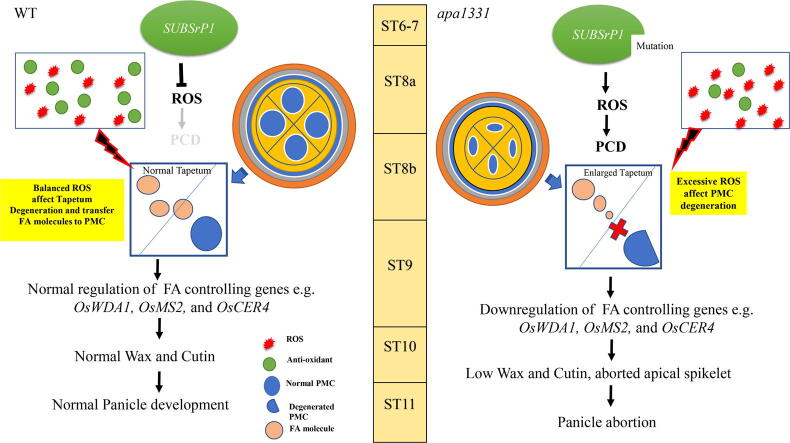
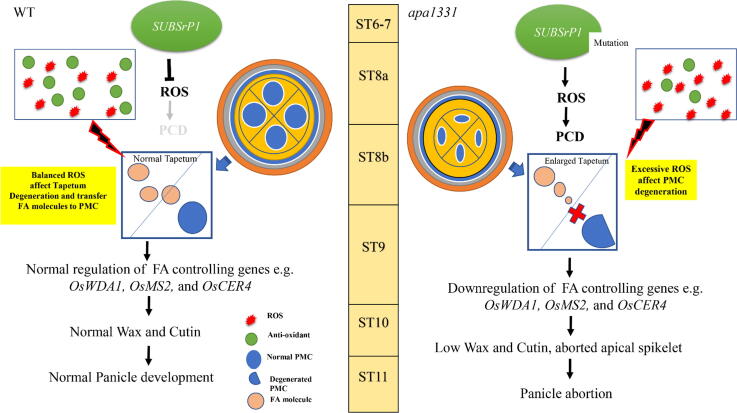
In summary, our results revealed that SUBSrP1 is essential to control the excessive outburst of ROS and programmed cell death (Fig. 10). A mutation in SUBSrP1causes dysregulation in its function and induces abnormal cell death to apical spikelet of apa1331. In WT, the balanced homeostasis of ROS and antioxidants produce a normal degeneration of tapetum that is essential for normal microspore development. However, excessive ROS and abnormal cell death cause the faulty development of PMCs and defective cuticle formation in apa1331. Mutation in OsSUBSrP1 results in the impairment in transferring fatty acid molecules to developing pollen that causes sterility. Defective cuticle formation and a significant decrease in wax and cutin contents cause the excessive water loss that produced aborted apical spikelet in apa1331. However, in WT, normal development of anther cuticle and wax saves enough water for cell viability and provides fatty acid to develop regular pollen grains. Our hypothesized model supports that SUBSrP1 is essential for maintaining ROS homeostasis by regulating wax and cutin development in rice.
Fig. 10.
An Illustration showing the function of SUBTILISIN-LIKE SERINE PROTEASE 1 (OsSUBSrP1) in homeostasis of Reactive oxygen species (ROS)-mediated programmed cell death (PCD) for anther cuticle development and seed setting rate,OsSUBSrP1 is essential for maintaining a balance of ROS-mediated PCD and plays a key role in developing apical spikelets by regulating the expression of wax and cutin pathway genes. A mutation in OsSUBSrP1 causes dysregulation in its protein and produces a defect in anther cuticle formation in apical panicle abortion1331 (apa1331). Significant decrease of wax and cutin contents in anthers of apa1331 causes excessive water loss from the surface of anthers that ultimately produce aborted spikelets.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Asif Ali: Methodology, Software, Formal analysis, Data curation, Validation, Investigation, Writing – original draft. Tingkai Wu: Formal analysis, Methodology. Hongyu Zhang: Resources. Peizhou Xu: Validation, Data curation. Syed Adeel Zafar: Conceptualization, Validation, Writing – review & editing. Yongxiang Liao: Resources. Xiaoqiong Chen: Data curation. Hao Zhou: Software. Yutong Liu: Resources. Wenming Wang: Supervision, Writing – review & editing. Xianjun Wu: Project administration, Funding acquisition, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge grants-in-aid from Department of Science and Technology of Sichuan Province (2021YFYZ0020, 2018JY0144), NSFC (National Natural Science Foundation of China) Grant No. 31771763 and a Breeding Research Project (21ZDYF2186). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. Authors are thankful to Dr. Hongyu Zhang for providing mutant for follow up functional study. Authors are thankful to Wenfei Zhang (OE Biotechnology Co., Ltd, Shanghai, China) for support in sequencing.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.01.003.
Contributor Information
Wenming Wang, Email: j316wenmingwang@163.com.
Xianjun Wu, Email: wuxjsau@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Zhu Z., Tan L., Fu Y., Liu F., Cai H., Xie D., et al. Genetic control of inflorescence architecture during rice domestication. Nat Commun. 2013;4(1) doi: 10.1038/ncomms3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teo Z.W.N., Song S., Wang Y.-Q., Liu J., Yu H. New insights into the regulation of inflorescence architecture. Trends Plant Sci. 2014;19(3):158–165. doi: 10.1016/j.tplants.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Heng Y., Wu C., Long Y.u., Luo S., Ma J., Chen J., et al. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell. 2018;30(4):889–906. doi: 10.1105/tpc.17.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu H., Dai Z., Li L., Wang J., Miao X., Shi Z. OsRAMOSA2 shapes panicle architecture through regulating pedicel length. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zafar S.A., Patil S.B., Uzair M., Fang J., Zhao J., Guo T., et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS 1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020;225(1):356–375. doi: 10.1111/nph.16133. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka W., Pautler M., Jackson D., Hirano H. Grass meristems II – Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 2013;54(3):313–324. doi: 10.1093/pcp/pct016. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Qian Q., Fu Z., Zeng D., Meng X., Kyozuka J., et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009;58(4):592–605. doi: 10.1111/j.1365-313X.2009.03799.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhu K., Tang D., Yan C., Chi Z., Yu H., Chen J., et al. ERECT PANICLE2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics. 2010;184(2):343–350. doi: 10.1534/genetics.109.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikawa T., Kyozuka J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell. 2009;21(4):1095–1108. doi: 10.1105/tpc.108.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabuchi H., Zhang Y.u., Hattori S., Omae M., Shimizu-Sato S., Oikawa T., et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell. 2011;23(9):3276–3287. doi: 10.1105/tpc.111.088765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda K., Nagasawa N., Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol. 2005;282(2):349–360. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Ikedakawakatsu K., Maekawa M., Izawa T., Itoh J.I., Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012;69(1):168–180. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q.-L., Sun A.-Z., Chen S.-T., Chen L.-S., Guo F.-Q. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat Plants. 2018;4(5):280–288. doi: 10.1038/s41477-018-0131-z. [DOI] [PubMed] [Google Scholar]
- 14.Peng Y., Hou F., Bai Q., Xu P., Liao Y., Zhang H., et al. Rice calcineurin B-like protein-interacting protein kinase 31 (OsCIPK31) is involved in the development of panicle apical spikelets. Front Plant Sci. 2018;9(1661) doi: 10.3389/fpls.2018.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai J., Zhu X., Wang Q., Zhang J., Chen H., Dong G., et al. Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 2015;169(2):1179–1191. doi: 10.1104/pp.15.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010;154(1):149–162. doi: 10.1104/pp.110.158865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y., Yamamoto Y., Yoshida T., Nitta Y., Miyazaki A. Response of differentiated and degenerated spikelets to top-dressing, shading and day/night temperature treatments in rice cultivars with large panicles. Soil Sci Plant Nutrit. 2000;46(3):631–641. [Google Scholar]
- 18.Itoh J.-I., Nonomura K.-I., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46(1):23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- 19.Smith A.M., Stitt M. Coordination of carbon supply and plant growth. Plant, Cell Environ. 2007;30(9):1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 20.Ali A., Xu P., Riaz A., Wu X. Current advances in molecular mechanisms and physiological basis of panicle degeneration in rice. Int J Mol Sci. 2019;20(7):1613. doi: 10.3390/ijms20071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D., Yuan Z., An G., Dreni L., Hu J., Kater M.M. In: Genetics and genomics of rice. Zhang Q., Wing R.A., editors. Springer; New York, NY: 2013. Panicle development; pp. 279–295. [Google Scholar]
- 22.Weng X., Wang L., Wang J., Hu Y., Du H., Xu C., et al. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014;164(2):735–747. doi: 10.1104/pp.113.231308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi J., Moon S., Lee Y.-S., Zhu L.u., Liang W., Zhang D., et al. Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 2016;170(3):1611–1623. doi: 10.1104/pp.15.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papini A., Mosti S., Brighigna L. Programmed-cell-death events during tapetum development of angiosperms. Protoplasma. 1999;207(3-4):213–221. [Google Scholar]
- 25.Ling S., Chen C., Wang Y., Sun X., Lu Z., Ouyang Y., et al. The mature anther-preferentially expressed genes are associated with pollen fertility, pollen germination and anther dehiscence in rice. BMC Genomics. 2015;16(1):1–17. doi: 10.1186/s12864-015-1305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedinger P. The remarkable biology of pollen. Plant Cell. 1992;4(8):879. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzair M., Xu D., Schreiber L., Shi J., Liang W., Jung K.-H., et al. PERSISTENT TAPETAL CELL2 is required for normal tapetal programmed cell death and pollen wall patterning. Plant Physiol. 2020;182(2):962–976. doi: 10.1104/pp.19.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N.a., Zhang D.-S., Liu H.-S., Yin C.-S., Li X.-X., Liang W.-q., et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18(11):2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z.e., Lin S., Shi J., Yu J., Zhu L.u., Yang X., et al. Rice No Pollen 1 (NP 1) is required for anther cuticle formation and pollen exine patterning. Plant J. 2017;91(2):263–277. doi: 10.1111/tpj.13561. [DOI] [PubMed] [Google Scholar]
- 30.Shi J., Tan H., Yu X.-H., Liu Y., Liang W., Ranathunge K., et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell. 2011;23(6):2225–2246. doi: 10.1105/tpc.111.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y., Bai Q., Xu P., Wu T., Guo D., Peng Y., et al. Mutation in rice abscisic acid2 results in cell death, enhanced disease-resistance, altered seed dormancy and development. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rang Z.W., Jagadish S.V.K., Zhou Q.M., Craufurd P.Q., Heuer S. Effect of high temperature and water stress on pollen germination and spikelet fertility in rice. Environ Exp Bot. 2011;70(1):58–65. [Google Scholar]
- 33.Zhang C.X., Feng B.H., Chen T.T., Zhang X.F., Tao L.X., Fu G.F. Sugars, antioxidant enzymes and IAA mediate salicylic acid to prevent rice spikelet degeneration caused by heat stress. Plant Growth Regul. 2017;83(2):313–323. [Google Scholar]
- 34.Nawrath C. Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol. 2006;9(3):281–287. doi: 10.1016/j.pbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Jung K.-H., Han M.-J., Lee D.-Y., Lee Y.-S., Schreiber L., Franke R., et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. 2006;18(11):3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L.u., Shi J., Zhao G., Zhang D., Liang W. Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J Plant Biol. 2013;56(1):59–68. [Google Scholar]
- 37.Chang Z., Chen Z., Yan W., Xie G., Lu J., Wang N.a., et al. An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 2016;253:21–30. doi: 10.1016/j.plantsci.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y., Wang Y., Mi X., Shan J., Li X., Xu J., et al. The QTL GNP1 encodes GA20ox1, Which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLOS Genetics. 2016;12(10) doi: 10.1371/journal.pgen.1006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Zhang H., Sun H., Luo H., Zhao L.i., Dong Z., et al. IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 2017;173(1):307–325. doi: 10.1104/pp.16.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu D., Shi J., Rautengarten C., Yang L.i., Qian X., Uzair M., et al. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol. 2017;173(1):240–255. doi: 10.1104/pp.16.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondol P.C., Xu D., Duan L., Shi J., Wang C., Chen X., et al. Defective Pollen Wall 3 (DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice. New Phytol. 2020;225(2):807–822. doi: 10.1111/nph.16161. [DOI] [PubMed] [Google Scholar]
- 42.Chen W., Yu X.-H., Zhang K., Shi J., De Oliveira S., Schreiber L., et al. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011;157(2):842–853. doi: 10.1104/pp.111.181693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vartapetian A.B., Tuzhikov A.I., Chichkova N.V., Taliansky M., Wolpert T.J. A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ. 2011;18(8):1289–1297. doi: 10.1038/cdd.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffeen W.C., Wolpert T.J. Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell. 2004;16(4):857–873. doi: 10.1105/tpc.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava R., Liu J.X., Howell S.H. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 2008;56(2):219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Othman R., Nuraziyan A. Fruit-specific expression of papaya subtilase gene. J Plant Physiol. 2010;167(2):131–137. doi: 10.1016/j.jplph.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 47.Zhao C., Johnson B.J., Kositsup B., Beers E.P. Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 2000;123(3):1185–1196. doi: 10.1104/pp.123.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueiredo A., Monteiro F., Sebastiana M. Subtilisin-like proteases in plant–pathogen recognition and immune priming: a perspective. Front Plant Sci. 2014;5:739. doi: 10.3389/fpls.2014.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez D.E., Borniego M.L., Battchikova N., Aro E.-M., Tyystjärvi E., Guiamét J.J. SASP, a Senescence-Associated Subtilisin Protease, is involved in reproductive development and determination of silique number in Arabidopsis. J Exp Bot. 2015;66(1):161–174. doi: 10.1093/jxb/eru409. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida K.T., Kuboyama T. A subtilisin-like serine protease specifically expressed in reproductive organs in rice. Sex Plant Reprod. 2001;13(4):193–199. [Google Scholar]
- 51.Taylor A.A., Horsch A., Rzepczyk A., Hasenkampf C.A., Riggs C.D. Maturation and secretion of a serine proteinase is associated with events of late microsporogenesis. Plant J. 1997;12(6):1261–1271. doi: 10.1046/j.1365-313x.1997.12061261.x. [DOI] [PubMed] [Google Scholar]
- 52.Hou F., Peng Y., Han X., Bai Q., Gu C., Sun C., et al. Identification and gene mapping of a panicle apical abortion mutant (paa1331) in rice (in Chinese) Chin Sci Bull. 2018;63:3192–3203. [Google Scholar]
- 53.Chun Y., Fang J., Zafar S.A., Shang J., Zhao J., Yuan S., et al. MINI SEED 2 (MIS2) encodes a receptor-like kinase that controls grain size and shape in rice. Rice. 2020;13(1):1–17. doi: 10.1186/s12284-020-0368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J., Yang R., Yang Z., Yao S., Zhao S., Wang Y.u., et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants. 2017;3(1) doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- 55.Adeel Zafar S., Uzair M., Ramzan Khan M., Patil S.B., Fang J., Zhao J., et al. DPS1 regulates cuticle development and leaf senescence in rice. Food Energy Secur. 2021;10(1) doi: 10.1002/fes3.v10.1. [DOI] [Google Scholar]
- 56.Wilson Z.A., Zhang D. From Arabidopsis to rice: pathways in pollen development. J Exp Bot. 2009;60(5):1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- 57.Wu T., Ali A., Wang J., Song J., Fang Y., Zhou T., et al. A homologous gene of OsREL2/ASP1, ASP-LSL regulates pleiotropic phenotype including long sterile lemma in rice. BMC Plant Biol. 2021;21(1) doi: 10.1186/s12870-021-03163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30(2):174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 59.Ma X., Zhu Q., Chen Y., Liu Y.-G. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Molecular Plant. 2016;9(7):961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47(6):969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 61.Tripathi L.P., Sowdhamini R. Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics. 2006;7(1):1–31. doi: 10.1186/1471-2164-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Battistuzzi F.U. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Consortium G.O. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J., Zhu L., Wang B., Wang H., khan I., Zhang S., et al. BnA1. CER4 and BnC1. CER4 are redundantly involved in branched primary alcohols in the cuticle wax of Brassica napus. Theor Appl Genet. 2021;134(9):3051–3067. doi: 10.1007/s00122-021-03879-y. [DOI] [PubMed] [Google Scholar]
- 66.Men X., Shi J., Liang W., Zhang Q., Lian G., Quan S., et al. Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J Exp Bot. 2017;68(3):513–526. doi: 10.1093/jxb/erw445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu L., Liang W., Yin C., Cui X., Zong J., Wang X., et al. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell. 2011;23(2):515–533. doi: 10.1105/tpc.110.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomez J.M., Jimenez A.I., Olmos E., Sevilla F. Location and effects of long-term NaCl stress on superoxide dismutase and ascorbate peroxidase isoenzymes of pea (Pisum sativum cv. Puget) chloroplasts. J Exp Bot. 2003;55(394):119–130. doi: 10.1093/jxb/erh013. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt A., Kunert K.J. Lipid peroxidation in higher plants: the role of glutathione reductase. Plant Physiol. 1986;82(3):700–702. doi: 10.1104/pp.82.3.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu X., Yin J., Liang S., Liang R., Zhou X., Chen Z., et al. The multivesicular bodies (MVBs)-localized AAA ATPase LRD6-6 inhibits immunity and cell death likely through regulating MVBs-mediated vesicular trafficking in rice. PLOS Genetics. 2016;12(9):e1006311. doi: 10.1371/journal.pgen.1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wutipraditkul N., Boonkomrat S., Buaboocha T. Cloning and characterization of catalases from rice, Oryza sativa L. Biosci Biotechnol Biochem. 2011;75(10):1900–1906. doi: 10.1271/bbb.110214. [DOI] [PubMed] [Google Scholar]
- 72.Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35(Database):D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38(suppl_1):D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rautengarten C., Steinhauser D., Büssis D., Stintzi A., Schaller A., Kopka J., et al. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol. 2005;1(4):e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beers E.P., Jones A.M., Dickerman A.W. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65(1):43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23(9):347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 77.Siezen R.J., Leunissen J.A.M. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6(3):501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itzhaki H., Naveh L., Lindahl M., Cook M., Adam Z. Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J Biol Chem. 1998;273(12):7094–7098. doi: 10.1074/jbc.273.12.7094. [DOI] [PubMed] [Google Scholar]
- 79.Lin H., Yu J., Pearce S.P., Zhang D., Wilson Z.A. RiceAntherNet: a gene co-expression network for identifying anther and pollen development genes. Plant J. 2017;92(6):1076–1091. doi: 10.1111/tpj.13744. [DOI] [PubMed] [Google Scholar]
- 80.Waese J., Fan J., Pasha A., Yu H., Fucile G., Shi R., et al. ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell. 2017;29(8):1806–1821. doi: 10.1105/tpc.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X., Guan Y., Zhang D.u., Dong X., Tian L., Qu L.Q. A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 2017;173(2):944–955. doi: 10.1104/pp.16.01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ibrahim S., Saleem B., Rehman N., Zafar S.A., Naeem M.K., Khan M.R. CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J Adv Res. 2021 doi: 10.1016/j.jare.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jordá L., Coego A., Conejero V., Vera P. A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem. 1999;274(4):2360–2365. doi: 10.1074/jbc.274.4.2360. [DOI] [PubMed] [Google Scholar]
- 84.Samuels L., Kunst L., Jetter R. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59(1):683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 85.Aharoni A., Dixit S., Jetter R., Thoenes E., Van Arkel G., Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mittler R., Del Pozo O., Meisel L., Lam E. Pathogen-induced programmed cell death in plants, a possible defense mechanism. Dev Genet. 1997;21(4):279–289. doi: 10.1002/(SICI)1520-6408(1997)21:4<279::AID-DVG5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 87.Duan Q., Kita D., Johnson E.A., Aggarwal M., Gates L., Wu H.-M., et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014;5(1):1–10. doi: 10.1038/ncomms4129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.