Graphical abstract
Keywords: Fragaria x ananassa, Holobiont, Phyllosphere, Plant growth-promoting bacteria, Pseudomonas fluorescens, Rhizophagus irregularis
Highlights
-
•
NGS was used to precisely describe the microbiome of three strawberry cultivars.
-
•
Plant organs and genotype shape the composition of the microbiomes.
-
•
Strawberry microbiome delivers several beneficial functions to the plant.
-
•
Functional microbiome diversity correlates with cultivars’ phenotypical differences.
-
•
Microbiome manipulation may enhance agricultural performances.
Abstract
Introduction
Specific microbial communities are associated to host plants, influencing their phenotype and fitness. Despite the rising interest in plant microbiome, the role of microbial communities associated with perennial fruit plants remains overlooked.
Objectives
This work provides the first comprehensive description of the taxonomical and functional bacterial and fungal microbiota of below- and above-ground organs of three commercially important strawberry genotypes under cultural conditions.
Methods
Strawberry-associated fungal and bacterial microbiomes were characterised by Next-Generation Sequencing and the potential functions expressed by the bacterial microbiome were analysed by both in silico and in vitro characterisation of plant growth-promoting abilities of native bacteria. Additionally, the association between the strawberry microbiome, plant disease tolerance, plant mineral nutrient content, and fruit quality was investigated.
Results
Results showed that the strawberry core microbiome included 24 bacteria and 15 fungal operational taxonomic units (OTUs). However, plant organ and genotype had a significant role in determining the taxonomical and functional composition of microbial communities. Interestingly, the cultivar with the highest tolerance against powdery mildew and leaf spot and the highest fruit productivity was the only one able to ubiquitously recruit the beneficial bacterium, Pseudomonas fluorescens, and to establish a mutualistic symbiosis with the arbuscular mycorrhiza Rhizophagus irregularis.
Conclusion
This work sheds light on the interaction of cultivated strawberry genotypes with a variety of microbes and highlights the importance of their applications to increase the sustainability of fruit crop production.
Introduction
Crop plants are associated with wide diversity of microorganisms, which differently colonise plant compartments [1]. Such microbial biocoenosis influences plant phenotype, fitness, growth, fruit production, and quality, by contributing to plant nutrition, tolerance to abiotic stresses, and control of pathogenic or opportunistic species [2]. In this view, the individual plant can be considered as a holobiont, i.e. the superorganism encompassing the host and its associated microbial community [3], [4]. The association between terrestrial plants and microbes developed at least 460 million years ago, as suggested by the fossil evidence of arbuscular mycorrhizae on some of the earliest land plants [4]. To date, many questions regarding plant–microbe associations remain unanswered, including the factors determining the community assemblage and diversity of the plant microbiome [2]. Increasing evidence suggests that plants can actively recruit beneficial microflora to facilitate their adaptation to environmental conditions and changes [2]. However, further studies are needed to generalize this hypothesis, and enable practical applications, especially for perennial crops such as fruit trees [5]. To date, most experiments on plant microbiome have focused either on model plants (i.e. Arabidopsis thaliana) or economically important, annual herbaceous monocotyledons, such as corn and rice [6]. Perennial plants, on the other hand, are exposed to radically changing environmental conditions (including freezing winter temperatures, dry seasons, periodic flooding) [7]. Therefore, the microbial community associated with such plants has evolved to last for more than a growing season, thus suggesting a metabolic selectivity and intimate connection with the host, allowing its stability despite environmental stresses. Furthermore, perennial crops may promote microbial ecological complexity, increasing the richness of bacterial and fungal beneficial communities, due to their extensive root networks and allocation of carbon to the rhizosphere [8]. In addition, microbiome research has so far primarily taken into consideration the rhizosphere, while other plant-associated niches have been relatively neglected [9]. The study of bacterial and fungal microbiota colonising different plant compartments under agronomic conditions provides key information to unfold agricultural constraints and achieve a successful microbial manipulation in farmlands [10].
The cultivated strawberry (Fragaria × ananassa Duch., fam. Rosaceae) is an important fruit crop, originated approximately 300 years ago from the hybridisation between ecotypes of wild octoploid species: Fragaria chiloensis subsp. chiloensis from South America and Fragaria virginiana subsp. virginiana from North America [11]. In the last decade (2008–2018), the global strawberry cultivation area has increased by 14% [12]. In 2016, the global strawberry gross production was estimated at 17 billion US$ with China having the largest market share (6.48 bn US$), followed by Europe (3.49 bn US$) and the United States (3.47 bn US$) [12]. The high adaptability of strawberry to different conditions allows its cultivation under a wide range of environments and agronomical managements (from the Mediterranean to the Nordic climates) making the fruit available on the market, almost independently of the season [13]. For this reason, strawberry fruit represents an important and valuable portion of daily fresh food consumption [14]. Strawberry is greatly appreciated for its aroma and nutraceutical properties. Among others, strawberry fruit contains phytochemicals, such as anthocyanins and ellagitannins which may prevent human health issues induced by oxidative stress [14]. While strawberry productivity and quality can be positively improved by beneficial microorganisms [15], the cultivation is challenged by a large variety of pathogens, which cause substantial economic losses and require the frequent application of pesticides. Among these diseases, red stele (Phytophthora fragariae), powdery mildew (Podosphaera aphanis), and leaf spot are the most serious concerns worldwide [16]. Powdery mildew mainly affects the photosynthetic ability of strawberries cultivated in humid environments [17], which leads to a strong reduction of growth and productivity with major yield losses [18]. Leaf spot diseases, which in severe conditions may lead to plant death, are caused by different pathogens, including bacteria (Xanthomonas fragariae) and fungi (Colletotrichum gloeosporiodes, Mycosphaerella fragariae, Cercospora fragariae, Mycosphaerella louisianae, Septoria fragariae, S. aciculosa, S. fragariaecola, etc.). For most of these pathogens and/or diseases, genetic resistance traits are not available among commercial strawberry cultivars (i.e. human-selected clonal genotypes) [19]. In addition, several cultivars present at the same time, flowers, fruits, and leaves, and therefore disease control practices may lead to pesticide residue accumulation on berries [13]. Biological control is a promising and safer alternative to the use of xenobiotic pesticides. Some commercially available, beneficial microorganisms (i.e. Ampelomyces quisqualis, Bacillus subtilis, Trichoderma harzianum, Glomus spp.) have been tested for disease control in strawberry, yet none of them has demonstrated sufficient reliability, persistence, and/or cost-effectiveness to replace chemical pesticides [16]. The unsatisfactory degree of disease control and the high variability of results obtained in different locations and seasons with commercial beneficial microorganisms can be explained by the fact that those microbes are in most cases non-native to the strawberry plant microbiome. Several studies suggest that biological control agents isolated from the host plant microbiome have superior efficacy in comparison to non-indigenous microbial inoculants [20].
Traditionally, microbiome studies focused on the taxonomy of plant-associated microbial communities. However, functional diversity should receive reasonable consideration in the context of sustainable agriculture as it has been recognized as the best predictor of ecosystem processes and properties [21]. Indeed, metabolic functionalities provided by the microbiome are essential for supporting plants in coping with adverse environmental conditions [3], [5], and are adapted to specific plant niches [22]. For the above reasons, the characterisation of the plant native microbiome both from a taxonomical and functional point of view is a key step for the successful selection of beneficial microorganisms [1]. Therefore, this study aimed to provide a complete description of the strawberry holobiome, including both fungal and bacterial populations, and to identify the core microbiome, from soil, plant-soil interface (rhizosphere), and plant compartments (roots and above-ground organs) using Next Generation Sequencing (NGS). For this purpose, three commercially important strawberry genotypes ('Elsanta', 'Darselect', and 'Monterey') were used. Additionally, Plant Growth Promoting (PGP) functions potentially expressed by the bacterial microbiota were analysed both in silico and in vitro. Furthermore, the effects of strawberry genotypes, soil and plant compartments on the richness and microbial community composition were studied, with a focus on pathogenic and beneficial microbes. Finally, the links between strawberry microbiomes, plant mineral nutrient content, and fruit quality traits were investigated. This study provides the first in-depth and comprehensive view of the horticultural crop microbiome in relation to plant genotype, health, nutritional status, and fruit quality parameters, prompting the selection of the most effective indigenous microorganisms for future field application.
Materials and Methods
Strawberry cultivation, phenotypic characterisation, and disease severity ranking
Three Fragaria × ananassa cultivars (genotypes) were used: the everbearing genotypes 'Elsanta' and 'Darselect' (widely cultivated in Northern Italy), and the day-neutral variety 'Monterey'. Bareroot strawberry plants were bought from CREA (Forlì, Italy), COVIRO (Ravenna, Italy) and SANTORSOLA (Trento, Italy) for 'Darselect', 'Monterey', and 'Elsanta' genotype, respectively. Plants were transplanted in early June-July 2017 into 48.5 × 22 × 11 cm white plastic pots, filled with a commercial blond sphagnum peat moss soil (pH 5.2–5.8) (Vigorplant s.r.l, Lodi, Italy). Each pot contained 6 plants with a distance of 16.7 cm between each other. These pots were maintained at 1.2 m above ground under a rainproof tunnel (18 m × 3.50 m × 5.60 m) located in-field at the experimental station of Pergine Valsugana (Trento, Italy; 46°07′N, 11°22′E, 450 a.s.l.). Plants were fertigated using a drip system (Table S1). Over the growing season (25, 35, and 50 days after transplant) root and leaf apparatus, as well as fruits, of 50 plants were weighted. Moreover, 100 additional plants of each genotype grown in the same conditions were weekly monitored for powdery mildew and leaf spot symptoms. Disease index on leaves was visually ranked using a 0–5 scale (0 = no symptoms; 5 = severe symptoms) (Table S2).
Sampling
At the end of the production cycle, for each genotype, four asymptomatic plant replicates from different pots distributed in the field area were collected. Plant-soil compartments were defined as described in [23]. The plants were immediately brought to the laboratory and processed. Briefly, bulk soil was collected from the growing pots and suspended in sterile 10 mM MgSO4 solution. The use of MgSO4 is required for avoiding osmotic potential imbalances of cells and it is commonly used for microbial suspension preparation, serial dilutions, and washing of plant organs [1]. Plants were divided into above-ground tissues (leaves, stems, crown) and roots. Roots were shaken to release loosely-associated soil, then washed in sterile 10 mM MgSO4 solution under vigorous shaking to collect the rhizospheric soil. Above-ground tissues and root samples were ground with mortar and pestle, and suspended in sterile 10 mM MgSO4 solution. For each sample, one aliquot of MgSO4, in which the washing of the organs was carried out, was used to perform serial dilutions. Dilutions were plated on Lysogenic Broth (LB) agar medium (Sigma Aldrich, St. Louis, MO, USA) amended with cycloheximide (100 μg ml−1) to prevent fungal growth. The remaining washing volume was stored at −20 °C until DNA extraction.
Bacterial isolation and functional characterisation
LB agar plates, prepared as described above, were incubated at 27 °C for 24 h. Colonies were phenotypically characterised and for each phenotype in a repetition, a single colony was randomly collected from the plates at the highest dilution. After purification, isolates were stored at −80 °C in LB broth supplemented with 20% v/v glycerol [24]. DNA was extracted from each bacterial isolate using GenElute Bacterial Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA), following the manufacturer’s instructions. REP-PCR was performed using BOXA1R and genomic DNA patterns obtained were visualized on a polyacrylamide gel (Fig. S1), as described in [25]. All primers’ references used in this work are shown in supplementary information (Table S3). Bacterial isolates were identified by 16S rRNA sequencing as described in [26]. Bacterial isolates (19, 14, and 10 for 'Monterey', 'Elsanta' and 'Darselect', respectively) were functionally characterised for acetoin, IAA, ammonia, and siderophores production, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity, X. fragariae, and B. cinerea inhibition as described in [26].
Analysis of plant mineral composition and fruit quality traits
General procedures were performed as previously reported [27]. Elemental analysis (%C, H, and N) was performed in duplicate on Thermo ScientificTM FLASH 2000 elemental analyser (ThermoFisher Scientific, Waltham, MA, USA). Measurements of the Mg, P, K, Ca, Fe, Mn, Co, Ni, Zn. Sr, Ba, and Pb were performed by Thermo ScientificTM XSeries-II ICP/qMS analyser equipped with Peltier cooled (3 °C) spray chamber and Teledyne CETAC ASX 520 autosampler (Teledyne CETAC, Omaha, NE, USA). The instrument tuning was performed daily, HNO3 4% Indium solution (100 ppb) was used as an internal standard and ICP-multi-element solution (IV-ICP-MS-71A, Inorganic Ventures, Christiansburg, VA, USA) was used for quantitation. 0.4 g of each sample were weighted in the microwave quartz vessels and added of 1.5 mL sub-boiling HNO3 and 3.5 mL H2O. Digestion was performed by a microwave-assisted Ultrawave FKV autoclave (FKV s.r.l., Bergamo, Italy). The resulting solutions were diluted up to 15 g with Milli-Q water and microfiltered (Ø 0.22 mm) before analysis. Results are given as mean value ± standard deviation (3 independent measurements, 3 repetitions each) (Table S7).
Strawberry fruits for each cultivar were weighted. Fruit firmness was measured by a texture analyser (Zwick Roell, Genova, Italy) using the penetration test methodology that was previously developed for raspberry [28]. In this study, only the maximum force value (N) was considered, since this parameter is usually highly related to berry firmness [28]. Total soluble sugar content (SSC) was measured on strawberry fruit juice with a hand-held Atago digital refractometer (Optolab, Modena, Italy). Titratable acidity (TA) was determined on strawberry juice diluted (1:2) in distilled water by titration with NaOH to pH 8.1 and expressed as citric acid equivalents.
DNA extraction and Illumina sequencing
DNA was extracted from 250 mg of each homogenized bulk soil, rhizosphere, root, and above-ground organs using the MoBio PowerSoil kit (MO BIO Laboratories, Carlsbad, CA, USA), following the manufacturer’s instructions. DNA quality and quantity were measured by spectrophotometric quantification with a NanoDrop ND-8000 V1.1.1 spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). DNA extracts were then stored at –20 °C before further analysis. The extracted DNA samples were sent to RTL Genomics (Lubbock, TX, USA) for Paired-end Illumina MiSeq sequencing. The V5, V6, and V7 regions of the 16S rRNA gene and ITS2 regions of the nuclear ribosomal internal transcribed spacer (ITS) rRNA gene were targeted for bacteria and fungi respectively. DNA extracts were amplified for sequencing in a two-step process: the forward primer was constructed with the Illumina i5 sequencing primer (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3′), and the 799F (5′-AACMGGATTAGATACCCKG-3′) for bacteria and the fITS7 primer (5′- GTGARTCATCGAATCTTTG-3′) for fungi. The reverse primer was constructed with the Illumina i7 sequencing primer (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3′) and the 1193r (5′-ACGTCATCCCCACCTTCC-3′) for bacteria or the ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primer for fungi (Table S3). The selected primer set for bacteria (799F and 1193r) can avoid contamination from plastid DNA. Reaction for the amplification of bacteria was performed as described in [29]. For fungi, the amplification cycle was as follows: 95 °C for 15 min, then 35 cycles of 94 °C for 30 sec, 54 °C for 40 sec, 72 °C for 1 min, followed by one cycle of 72 °C for 10 min and 4 °C hold. Products from the first stage amplification were added to a second PCR aimed to qualitatively determine concentrations. The second PCR reaction was performed using Nextera primers i5 index and i7 index (Illumina Inc., San Diego, CA, USA) (Table S3). The second stage amplification was run the same as the first stage except for 10 additional cycles. Amplified products were visualized with eGels (Life Technologies, Grand Island, NY, USA). The products were then pooled equimolar, and each pool was size selected in two rounds using Agencourt AMPure XP (BeckmanCoulter, Indianapolis, IN, USA) in a 0.75 ratio for both rounds. The size selected pools were then quantified using the Quibit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA) and loaded on an Illumina MiSeq (Illumina, Inc, San Diego, CA, USA) 2x300 flow cell at 10 pM.
nifH gene and Pseudomonas fluorescens detection
To verify the nitrogen-fixing activity and the presence of putative PGP species that were found to be discriminant genotypes and/or compartments, specific detection protocols were applied. nifH gene was used as a target to detect N-fixing activity, being a highly conserved gene [30]. The presence of the nifH gene in samples was verified by PCR using nifH gene-specific primers, as previously described (Table S3). Pseudomonas fluorescens detection in 'Monterey' genotype was performed as follows: DNA from bulk soil and rhizosphere samples was extracted as above; roots and above-ground parts of strawberry plants were surface-sterilized two times with deionized water and 70% ethanol and washed 3 times sterile water, organs were let 3 h in sterile water. Then, DNA of strawberry roots and above-ground part was extracted as above and amplified as described elsewhere (Table S3). For both nifH and Ps. fluorescens detection, amplification products were visualized through agarose gel 1.5% electrophoresis.
Bioinformatics
High-quality reads from the paired-end sequences generated by the Illumina MiSeq sequencing platform were extracted using Mothur [31] and OBI Tools [32] software suits. PANDAseq was used to merge forward and reverse raw reads from the same sample by using the simple-bayesian algorithm with a minimum overlap of 80 and 20 nucleotides for bacteria and fungi, respectively. All the merged reads were then trimmed with the following parameters: (i) minimum length of 350 (bacteria) and 120 (fungi), (ii) minimum average Phred score of 25 on the trimmed length, (iii) no ambiguities in the sequence length, and (iv) maximum length of 20 homopolymers in the sequence. The reads were then pre-clustered using CD-HIT-EST, allowing a maximum of 1% of dissimilarity and with only one base allowed per indel, to merge those reads arising likely from sequencing errors. Chimeric sequences were detected using the Uchime algorithm [33] as implemented in Mothur and removed. Reads from each sample were pooled together and were dereplicated into unique sequences and sorted by decreasing abundance. The resulting reads were then clustered into operational taxonomic units (OTUs) using the CD-HIT-EST algorithm [34] at a threshold of 97% sequence similarity. The OTU representative sequences (defined as the most abundant sequence in each OTU) were taxonomically assigned against the reference sequences from the SILVA database v132 for prokaryote 16S [35] and from the Unite database (version unite.v7) [36] for fungal ITS using the naive Bayesian classifier [37] as implemented in Mothur using the default parameters. All the sequences identified as non-target organisms were removed from bacterial and fungal datasets. Rare OTUs (singletons), which potentially might represent artificial sequences were removed. The read counts were rarefied to the smallest read number per sample (10,930 and 8,077 reads for bacteria and fungi, respectively). Ecological functions were determined for each OTU using FAPROTAX for bacteria [38], and FUNGuild [39] for fungi. The ecological functions of bacteria obtained by FAPROTAX were also manually checked against other references for their presence in the terrestrial system [40]. Arbuscular mycorrhizae, ectomycorrhizae, ericoid mycorrhizae, endophytes, dark septate endophytes, and mycoparasites were grouped as potential beneficial fungi. All fungal plant pathogens were checked again for their taxonomic identifications and their DNA-based Species Hypotheses (SH) are presented in Supplementary Table S4. Potential beneficial bacteria (N fixing, plant-growth-promoting, and biological control agents) were manually assigned using all available references (Table S5). Prediction of the bacterial functions according to the KEGG database was also performed. Tax4fun package [41] was used to perform the functional prediction, and the results were analysed following MicrobiomeAnalyst online pipeline ((https://www.microbiomeanalyst. ca/) [42]. KEGG orthologs and modules involved in PGP mechanisms were chosen using the KEGG database and several reviews. The Illumina sequencing of all bacterial and fungal datasets are deposited in The National Center for Biotechnology Information (NCBI) database under BioProject: PRJNA556362 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA556362).
Statistical analysis
To assess the coverage of the sequencing depth, individual rarefaction analysis for each sample using the function ‘diversity’ in Past was performed. At the analysed sequencing depth, all individual rarefactions showed to be sufficient to infer bacterial and fungal community composition and richness in the samples (Fig. S2). The core microbiome was defined as the bacterial and fungal communities that are comprised of OTUs that were detected in all strawberry genotypes and present in more than 75% of the samples. The effects of strawberry genotype, soil, and plant compartments (bulk soil, rhizosphere, root, and above-ground organs) on bacterial and fungal OTUs richness were analysed using two-way analysis of variance (ANOVA), incorporating the Jarque-Bera JB test for normality. The effects of strawberry genotype, soil, and plant compartment on bacterial and fungal community compositions were visualized using Non-metric MultiDimensional Scaling (NMDS) based on the presence-absence data and Jaccard distance measure. Colored ellipses in NMDS ordinations are 95% confidence intervals of species centroids for each treatment level. The significant effect of the strawberry genotype, soil, and plant compartment on bacterial and fungal community compositions was determined using two-way Analysis of Similarity (ANOSIM) and two-way Permutational multivariate analysis of variance (PERMANOVA) based on the presence-absence data and Jaccard distance measure over 999 permutations. Since relative abundance data from NGS may not be fully used quantitatively, the microbial community composition was analysed using both presence/absence and relative abundance data sets. The results from presence/absence data are presented in the main text and the corresponding results using relative abundance data (with Bray–Curtis distance measure) are presented in Supplementary Information (Table S6). NMDS ordination based on presence-absence data and the Jaccard dissimilarity measure coupled with the envfit function of the vegan package in R version 3.2.2 were used to investigate the links between each bacterial and fungal community composition (bulk soil, rhizosphere, root, and above-ground organs) and soil nutrient parameters, strawberry genotypes, fruit quality parameters (SSC and TA). NMDS stress values were between 0.06 and 0.13. Differences in phenological characteristics, PGP KEGG modules, Kegg Orthologies (KOs), and chemical composition of roots and above-ground strawberry of the three genotypes were tested by One-way ANOVA, followed by multiple comparisons by Tukey’s test, performed using Past version 2.17.
Results
Phenotypical differences of strawberry cultivars
Flower differentiation, fruit production, plant architecture, and growth rate were observed during the growing season. Leaf and root apparatus of 'Elsanta' were significantly heavier in respect to the other genotypes, whereas 'Monterey' showed the highest productivity and the largest fruit size (Table 1).
Table 1.
Phenological characteristics of different strawberry genotypes. Data are expressed as mean ± SE. Different letters indicate significant differences between genotypes according to One-way ANOVA followed by multiple comparisons by Tukey’s test.
| Cultivar | Leaves g/plant at 50 days after transplant | Root g/plant at 50 days after transplant | Harvested red fruit/week/plant (g) | Single fruit weight (g) |
|---|---|---|---|---|
| Monterey | 7.76 ± 0.90b | 30.04 ± 3.29 ab | 53.87 ± 5.48 a | 16.24 ± 0.76 a |
| Elsanta | 15.32 ± 1.54 a | 39.54 ± 2.74 a | 34.24 ± 5.81 ab | 10.24 ± 0.55b |
| Darselect | 9.44 ± 1.39b | 20.25 ± 3.64b | 30.87 ± 6.73b | 9.43 ± 1.07b |
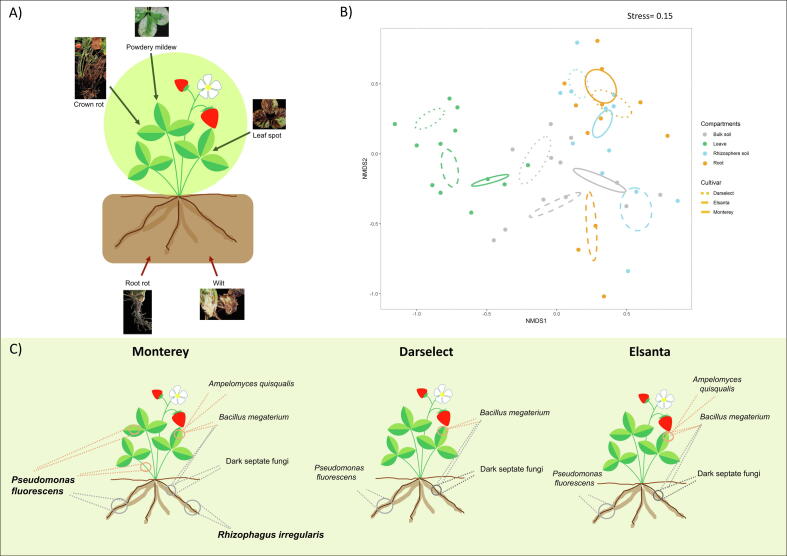
The susceptibility of the different genotypes to the main strawberry diseases was also evaluated (Table S2). Plants of 'Monterey' showed the highest tolerance to leaf spot and powdery mildew in comparison to the other genotypes. On the other hand, 'Elsanta' showed to be less sensitive than 'Darselect' to leaf spot, but highly susceptible to powdery mildew.
Composition of strawberry microbiomes
Bacterial 16S rRNA and ITS gene communities were profiled in bulk soil, rhizosphere, root, and above-ground organ samples in the three strawberry genotypes (Fig. 1a). In roots and above-ground organs, epiphytic and endophytic microorganisms were targeted jointly. In total, 1,531,637 (average of 31,909 reads per sample) and 739,458 (average of 15,405 reads per sample) high-quality reads were generated for bacteria and fungi, respectively, excluding chimeric sequences. Singletons that may come from sequencing errors were removed and all datasets were normalized to 10,930 sequences for bacteria and 8,077 sequences for fungi. Rarefaction curves showed sufficient sequencing effort for most of the samples (Fig. S2a, b). Observed richness was used directly as a diversity measure for both bacteria and fungi (Fig. 1b,c). In total, 26,434 bacterial and 1,716 fungal OTUs were detected. Among the three strawberry genotypes, 'Darselect' displayed the lowest bacterial richness in all compartments (Fig. 1d). In general, above-ground organs had a lower bacterial richness compared to the other compartments. In contrast, fungal microbiome diversity was fairly homogenous among different compartments (Fig. 1e).
Fig. 1.
Composition of strawberry microbiomes. (A) Overall community composition of bacteria and fungi in different soil and plant compartments and genotypes, estimated using presence-absence data. (B) Bacterial and (C) fungal OTU richness in different soil and plant compartments and genotypes. Bacterial (D) and (E) fungal composition similarity among different soil and plant compartments and genotypes, shown by nonmetric multidimensional scaling (NMDS) plot.
The total bacterial and fungal community assemblages were compared using two-way PERMANOVA to identify the main drivers of the microbiome composition (Table 2). Microbial compositions were strongly dependent on the analysed genotype (bacteria F = 1.87, P = 0.002; fungi F = 2.93, P = 0.001), on the compartment (bacteria F = 4.27, P = 0.001; fungi F = 3.56, P = 0.001), and on genotype × compartment interaction (bacteria F = 1.44, P = 0.001; fungi F = 1.51, P = 0.001; Table 2). In particular, NMDS highlighted that both bacterial and fungal above-ground microbiome compositions strongly differed from below-ground ones (Fig. 1d, e). Similar results were obtained by means of two-way ANOSIM analysis (Table 2) and by using relative abundance data, instead of presence-absence (Table S6).
Table 2.
Effect of genotype, soil, and plant compartment on the richness and community composition of the strawberry microbiome. Nd = not determined; Significant P values are highlighted in bold.
| Microorganisms/Factors |
Richness (Two way ANOVA) |
Community composition (Two-way ANOSIM) |
Community composition (Two-way PERMANOVA) |
|||
|---|---|---|---|---|---|---|
| F | P | R | P | PseudoF | P | |
| Total bacteria | ||||||
| Genotype | 12.15 | 0.000 | 0.65 | 0.001 | 1.87 | 0.002 |
| Compartment | 32.47 | 0.000 | 0.83 | 0.001 | 4.27 | 0.001 |
| Genotype × compartment | 2.55 | 0.037 | nd | nd | 1.44 | 0.001 |
| Potential beneficial bacteria | ||||||
| Genotype | 4.92 | 0.013 | 0.34 | 0.001 | 1.61 | 0.001 |
| Compartment | 20.86 | 0.000 | 0.48 | 0.001 | 2.87 | 0.001 |
| Genotype × compartment | 1.81 | 0.125 | 1.35 | 0.001 | ||
| Fungi | ||||||
| Genotype | 1.74 | 0.191 | 0.78 | 0.001 | 2.93 | 0.001 |
| Compartment | 19.00 | 0.000 | 0.76 | 0.001 | 3.56 | 0.001 |
| Genotype × compartment | 2.00 | 0.092 | nd | nd | 1.51 | 0.001 |
| Potential beneficial fungi | ||||||
| Genotype | 9.23 | 0.001 | 0.22 | 0.001 | 2.05 | 0.004 |
| Compartment | 13.13 | 0.000 | 0.47 | 0.001 | 4.05 | 0.001 |
| Genotype × compartment | 3.46 | 0.008 | nd | nd | 1.33 | 0.033 |
| Plant pathogenic fungi | ||||||
| Genotype | 3.65 | 0.036 | 0.43 | 0.001 | 3.34 | 0.001 |
| Compartment | 4.30 | 0.011 | 0.51 | 0.001 | 3.90 | 0.001 |
| Genotype × compartment | 2.92 | 0.020 | nd | nd | 1.58 | 0.002 |
Community composition based on presence-absence data indicates that Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Bacteroidia are the bacterial groups representing the backbone of the strawberry bacterial microbiome in all plant and soil compartments, accounting on average for 80% of the total detected OTUs (Fig. 1a). Above-ground organs of ‘Darselect’ and ‘Elsanta’ were dominated by Actinobacteria (54 and 53% respectively), whereas ‘Monterey’ was mostly colonised by Gammaproteobacteria (Fig. 1a). Below-ground compartments of the three cultivars were dominated by Alphaproteobacteria (Fig. 1a).
Regarding the strawberry mycobiome, Sordariomycetes, Dothideomycetes, Leotiomycetes, and Agaricomycetes were the most represented fungal classes in all plant and soil compartments accounting for 64% of total OTUs based on presence-absence data (Fig. 1a), but their percentages varied depending on cultivar and compartment. Dothideomycetes were predominant in leaves of the three F. × ananassa genotypes (34% 'Elsanta', 28% 'Darselect', 27% 'Monterey'), whereas below-ground compartments of all genotypes were mostly dominated by Sordariomycetes (Fig. 1a).
Overall bacterial and fungal microbial community composition was also analysed based on relative-abundance data (Fig. S3). In this view, Actinobacteria was the predominant bacterial group in all genotypes and compartments. The lowest percentages were found in the rhizosphere and bulk soil of 'Elsanta' (31 and 33%, respectively), whereas 'Elsanta' and 'Darselect' above-ground compartments showed a high group homogeneity, being dominated by Actinobacteria for 98 and 99%, respectively (Fig. S3). Alphaproteobacteria were homogenously represented in plant and soil compartments of the three genotypes. Gammaproteobacteria were almost absent in the above-ground compartments of 'Elsanta' and 'Darselect', while they were the second most represented group in 'Monterey' (26%) (Fig. S3). The fungal community composition, based on relative abundance data, showed a good level of homogeneity between different genotypes. Dothideomycetes were the predominant group in the above-ground organs of the three genotypes (Fig. S3), followed by Tremellomycetes, Leotiomycetes, and Sordariomycetes. whereas below-ground organs of the three cultivars were dominated by Sordariomycetes (Fig. S3).
Commonalities in the microbiomes of the three cultivars in the different plant compartments are shown (Fig. 2). Alphaproteobacteria, Gammaproteobacteria and Actinobacteria (plus Deltaproteobacteria and Bacteroidia, except for above-ground organs) are common to the three genotypes in all plant compartments (Fig. 2). Regarding the fungal microbiome, Sordariomycetes, Dothideomycetes, Leotiomycetes, Eurotiomycetes, and Agaricomycetes were found in the below-ground organs of the three genotypes. The above-ground core was similar except for the absence of Eurotiomycetes and Agaricomycetes (Fig. 2).
Fig. 2.
Identification of bacterial and fungal core microbiomes. Core microbiome taxonomic compositions, both concerning the whole plant and for each organ, are described. Quantity of bacterial and fungal OTU unique for each strawberry genotype, as well as overlaps, are reported. M= 'Monterey', E= 'Elsanta', D= 'Darselect'.
Ubiquitous microbes found from the soil to the above-ground plant organs of all three strawberry genotypes were identified. Among these core microbes, 24 OTUs were bacteria (mainly Micrococcales) and 15 were fungi (mainly Ascomycota) (Table S8).
Functions potentially expressed by plant-associated bacteria and fungi
The plant-associated microbial community is pivotal in delivering multiple functions to the plant, such as plant growth promotion and increase of abiotic/biotic stress resilience. In this work, the functional diversity of bacterial and fungal microbiomes was studied following several approaches.
In silico prediction of the microbial functions was first performed using FAPROTAX and FUNGuild. These are functional prediction tools providing guild characteristics of bacterial and fungal taxa, respectively. 3,845 bacterial (15% of all detected bacteria) and 706 fungal (41% of all detected fungi) OTUs were assigned to a putative functional group. Twenty bacterial and sixteen fungal functional groups colonised different soil and plant compartments of strawberry plants (Table S9). Chemoheterotrophy, methanol oxidation, intracellular parasitism, predation/exoparasitism were the dominant bacterial functions, while saprophytism, plant pathogenic, and endophytic colonisation were dominant among the fungal functions.
Within the bacterial OTUs, specific functions relevant to plant health, fitness, and growth were further explored (Table S5). 285 OTUs were assigned to 16 potential N-fixing genera, and 129 OTUs as species are known for their activity as biological control agents (BCA) and/or plant growth promoting bacteria (PGPB). Both compartment and genotype had a significant role in defining plant-associated beneficial bacterial community, according to ANOSIM and PERMANOVA (P < 0.001) (Fig. S4a; Table 2; Table S6). In below-ground organs, commonalities of N-fixing bacteria between the three strawberry genotypes consisted of Rhizobiaceae, Devosiaceae, Xanthobacteraceae, and Burkholderiaceae, whereas in above-ground organs no overlap was identified (Fig. 2). Besides the identification of commonalities of the three genotypes in different organs, the distribution of OTUs among organs of the same genotype was further highlighted (Fig. 3). N-fixing bacteria were found to be widely distributed both in below- and above-ground compartments. However, above-ground organs were colonised by fewer N-fixing bacteria in respect to below-ground compartments. Above-ground organs of 'Monterey' and 'Darselect' were characterised by the presence of Methylobacterium spp., which was found only in this compartment. Aminobacter spp. was uniquely found in the root and rhizosphere of 'Darselect', whereas Phyllobacterium spp. was characteristic of 'Monterey' (Fig. 3a).
Fig. 3.
Genotype-specific functional microorganisms. Venn diagram showing (A) Potential Nitrogen-fixing genera, (B) Potential Plant Growth Promoting Bacteria and Biological Control Agents species, (C) Potential Fungal pathogens species, and (D) Potential Fungal Beneficial species present in each compartment (B = bulk soil; RH = rhizosphere; R = Roots, AGO = Above-ground organs) of the three strawberry cultivars. Intersections indicate microbes simultaneously present in more than one organ are also presented (genera abbreviations Ps. = Pseudomonas; B. = Bacillus).
Regarding the bacterial beneficial microbiome, in 'Monterey', 19% of beneficial OTUs were able to simultaneously colonise below- and above-ground organs, whereas in 'Elsanta' and 'Darselect' only one OTU (identified as B. megaterium) was found to colonise both underground and above-ground organs. Ps. fluorescens is known for its PGP activity and the ability to control several plant diseases [43]. NGS analysis revealed that Monterey was the only genotype colonised by Ps. fluorescens both in below- and above-ground organs (Fig. 3b). Its ability to colonise different plant compartments was further confirmed by PCR analysis. Indeed, populations of Ps. fluorescens could be detected in the soil and tissues (both internal and external) of 'Monterey' (Fig. S5).
Nitrogen (N) is one of the essential nutrients for ensuring plant growth and productivity. In agriculture, it might become a limiting factor and therefore external application is generally needed. N-fixing microorganisms, able to convert the unavailable atmospheric N2 into an accessible form, belong to Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Firmicutes, and Cyanobacteria phyla [44]. nifH is a highly conserved gene, generally used as a marker for nitrogen fixation processes in natural habitats. In this work, PCR analysis assessed the presence of the nifH gene in bulk soil, rhizosphere, and root samples of the three strawberry genotypes.
Besides investigating the potential functionalities of the bacterial microbiome using FAPROTAX and databases research, biochemical functions were analysed using Tax4fun [41]. Tax4Fun allows the prediction of functional traits comparing bacterial 16S rRNA genes with information available on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. According to Two-way PERMANOVA, the compartments had a significant role in shaping the functionalities expressed by the microbiome (Table 3). Modules and KOs related to PGP traits were strongly linked both to the organ and the genotype. On the whole, 26 modules and KOs were found to be significantly different (Table 4). Interestingly, in the above-ground organs of 'Monterey', genes linked to jasmonic acid (JA) biosynthesis, isoprenoid biosynthesis, and H2S production were found to be significantly more abundant. On the other hand, potential beneficial traits associated with below-ground organs were homogeneously found across the three genotypes.
Table 3.
Effect of genotype and soil/plant compartment on overall KEGG modules and KO and Plant Growth Promoting KEGG modules and KO composition throughout strawberry microbiome, according to Two-way PERMANOVA analysis. Significant P values are highlighted in bold.
| Feature/Factors |
Two-way PERMANOVA |
|
|---|---|---|
| Pseudo F | P | |
| KEGG Modules | ||
| Organ | 22.46 | 0.0001 |
| Cultivar | 1.76 | 0.12 |
| Organ × Cultivar | 1.43 | 0.15 |
| KEGG KO | ||
| Organ | 39.007 | 0.0001 |
| Cultivar | 1.95 | 0.1111 |
| Organ × Cultivar | 1.6 | 0.1088 |
| PGPB Kegg Modules | ||
| Organ | 18.74 | 0.0001 |
| Cultivar | 2.09 | 0.0564 |
| Organ × Cultivar | 1.42 | 0.1182 |
| PGPB Kegg KO | ||
| Organ | 24.04 | 0.0001 |
| Cultivar | 2.72 | 0.0361 |
| Organ × Cultivar | 1.39 | 0.1904 |
Table 4.
Significantly different Plant Growth Promoting KEGG modules and KOs in soil/plant compartments. Data are expressed as mean ± SE. Different letters indicate significant differences between genotypes according to One-way ANOVA followed by multiple comparisons by Tukey’s test.
| Organ | KEGG PGP MODULE or GENE | Monterey | Elsanta | Darselect |
|---|---|---|---|---|
| BULK SOIL | Pectin degradation | 42.25 ± 0.94b | 67 ± 5.93 a | 34 ± 2.20c |
| Polyamine biosynthesis, arginine => agmatine => putrescine => spermidine | 1315.75 ± 27.67 ab | 1389.25 ± 36.81 a | 1199.75 ± 33.61b | |
| C10-C20 isoprenoid biosynthesis, bacteria | 650.5 ± 11.13b | 704 ± 7.22 a | 643.75 ± 7.92b | |
| C10-C20 isoprenoid biosynthesis, archaea | 215.5 ± 5.5b | 277.5 ± 15.22a | 211 ± 10.98b | |
| Dissimilatory sulfate reduction, sulfate => H2S | 434 ± 10.36b | 506 ± 18.41 a | 442.75 ± 12.26b | |
| 1-aminocyclopropane-1-carboxylate deaminase | 42 ± 2.97b | 56 ± 2.55 a | 42.75 ± 1.31b | |
| Pyocyanine biosynthesis, chorismate => pyocyanine | 5.75 ± 0.63b | 19.25 ± 2.59 a | 8,75 ± 2.02b | |
| Abscisic acid biosynthesis, beta-carotene => abscisic acid | 3.5 ± 0.29b | 5.5 ± 0.29 a | 2.75 ± 0.25b | |
| RHIZOSPHERE | Assimilatory sulfate reduction, sulfate => H2S | 2578.5 ± 26.43c | 2847.75 ± 54.76 a | 2774.5 ± 45.10b |
| Betaine biosynthesis, choline => betaine | 1265.5 ± 15.78b | 1486 ± 77.35 a | 1429.5 ± 40.70 ab | |
| Butanoate metabolism | 4654.5 ± 26.36b | 5321 ± 217.89 a | 5044.25 ± 181.02 ab | |
| ROOT | GABA (gamma-Aminobutyrate) shunt | 1911.5 ± 45.42 ab | 2184.25 ± 92.87 a | 1833.75 ± 65.80b |
| C5 isoprenoid biosynthesis, non-mevalonate pathway | 3498.75 ± 31.54 ab | 3597.25 ± 83.17 a | 3277.25 ± 66.64b | |
| Ascorbate biosynthesis, plants, fructose-6P => ascorbate | 2918.75 ± 20.30b | 2970.25 ± 41.62 a | 2717 ± 72.87c | |
| Ascorbate biosynthesis, animals, glucose-1P => ascorbate | 1523.75 ± 5.65 ab | 1580.25 ± 5.12 a | 1477.25 ± 40.07b | |
| Polyamine biosynthesis, arginine => agmatine => putrescine => spermidine | 1231.25 ± 39.21b | 1044.5 ± 23.36c | 1287.75 ± 57.05 a | |
| GABA biosynthesis, eukaryotes, putrescine => GABA | 2914 ± 36.25 ab | 3086.75 ± 129.51 a | 2541 ± 178.62b | |
| Methanogenesis, methanol => methane | 150.5 ± 18.19 a | 72.75 ± 20.00b | 130.25 ± 15.43 ab | |
| Betaine biosynthesis, choline => betaine | 1458 ± 40.38 ab | 1599.25 ± 49.18 a | 1285.25 ± 48.17b | |
| methionine-gamma-lyase | 277.25 ± 13.47 a | 252.75 ± 10.94 a | 190 ± 13.23b | |
| IAA | 1006.75 ± 15.13 ab | 1059.25 ± 60.20 a | 827.25 ± 56.85b | |
| salycilic acid | 391.5 ± 10.32b | 458.5 ± 13.11 a | 319.5 ± 15.60c | |
| ABOVE-GROUND ORGANS | Jasmonic acid biosynthesis | 710.25 ± 45.82 a | 552 ± 44.68b | 624.5 ± 23.90 ab |
| C10-C20 isoprenoid biosynthesis, non-plant eukaryotes | 130.75 ± 8.08 a | 99.75 ± 9.09b | 116.5 ± 5.63 ab | |
| Purine degradation, xanthine => urea | 1378.5 ± 104.68b | 1775 ± 81.18 a | 1595 ± 98.93 ab | |
| Dissimilatory sulfate reduction, sulfate => H2S | 764.5 ± 22.26 a | 669.75 ± 15.14b | 710.75 ± 30.13 ab |
Using the FUNGuild predictive tool, fungi were divided into beneficial or pathogenic (Table S9). As for bacteria, beneficial fungal community showed to be strongly correlated to both genotype (F = 2.05, P = 0.004) and compartment (F = 4.05, P = 0.001) (PERMANOVA values genotype × compartment F = 1.33, P = 0.033; Fig. S4b; Table 2; Table S6). Cladosporium sphaerospermum was the only species common to the above-ground organs and rhizosphere of the three genotypes (Fig. 2). While most of the potentially beneficial fungal groups were similarly represented in the three strawberry genotypes, the arbuscular mycorrhizal fungus Rhizophagus irregularis showed a high frequency only in 'Monterey', while being completely absent in 'Elsanta' and 'Darselect' (Fig. 3d). In addition to the beneficial fungi recognized with FUNGuild, other species previously documented as beneficial to plants were also highlighted (Table S10).
Both genotype (F = 3.34, P = 0.001) and plant compartment (F = 3.90, P = 0.001), as well as their interaction (PERMANOVA values genotype × compartment F = 1.58; P = 0.002; Fig. 3c; Fig. 4b; Table 2; Table S6) play a key role in the abundance of pathogens in the fungal community associated to strawberry. Regarding commonalities of fungal pathogens in the three strawberry cultivars in different organs, Powellomycetaceae and Taphrinaceae were only found in bulk soil and above-ground organs, respectively (Fig. 2) whereas Podosphaera spp. was present in both below- and above-ground compartments of the three genotypes (Fig. 3c).
Fig. 4.
Distinctive features of populations of pathogens and disease-protecting microbes in different strawberry cultivars. (A) Fungal and bacterial pathogens affecting strawberry plant cultivation. (B) Nonmetric multidimensional scaling (NMDS) plot showing fungal pathogens composition similarity among different soil and plant compartments and genotypes. (C) Bacterial and fungal beneficial species colonising the different strawberry genotypes.
Isolation and functional characterisation of bacteria isolates
Viable bacterial counts were on average 5.02 × 107, 4.65 × 108, 4.29 × 107, 1.12 × 108 CFU/ml for leaves, rhizosphere, root, and soil, respectively (Fig. S6). No significant differences were found among population sizes detected on the same organ of different genotypes. A total of 70 colonies were selected from the highest countable dilution.
Ten genera of potential PGPB were uniquely isolated from this variety, whereas one and three genera were unique to 'Elsanta' and 'Darselect', respectively. Pseudomonas and Vagococcus genera were common to the three genotypes (Table S11), being the former isolated only from below-ground compartments. 43 isolates were tested in vitro for their plant growth-promoting activities. PGP traits were qualitatively screened (0 = no activity, 3 = highest activity). Bacterial isolates from Monterey showed the highest plant-growth-promoting potentiality both in the above- and below-ground organs (Table 5). Interestingly, in above-ground organs, 'Monterey' showed the highest number of indoleacetic acid (IAA)- and NH4+-producing bacteria, as well as more Xanthomonas fragariae antagonists when compared to 'Elsanta' and 'Darselect'. ACC deaminase-producing bacteria were not found in above-ground organs. Potential plant-growth promoting functions were more homogeneously spread in below-ground than above-ground organs across the three cultivars.
Table 5.
Plant Growth Promoting in vitro activities of bacteria isolated either in above-ground or below-ground organs of the three strawberry genotypes. Plant Growth Promoting traits were qualitatively screened (0 = no activity, 3 = highest activity) for each isolate (Table S11). For each trait, the indicated values represent the sum of activity levels of single isolates from each genotype.
| Monterey | Elsanta | Darselect | ||
|---|---|---|---|---|
| Above-Ground Organs | Acetoin Production | 2 | 0 | 1 |
| IAA production | 6 | 0 | 0 | |
| Siderophores | 0 | 0 | 0 | |
| Botrytis cinerea antagonism | 0 | 0 | 1 | |
| Xanthomonas fragariae antagonism | 5 | 0 | 2 | |
| NH4 + production | 3 | 1 | 0 | |
| ACC deaminase activity | 0 | 0 | 0 | |
| Below-Ground Organs | Acetoin Production | 6 | 2 | 6 |
| IAA production | 0 | 3 | 3 | |
| Siderophores | 6 | 3 | 4 | |
| Botrytis cinerea antagonism | 2 | 0 | 0 | |
| Xanthomonas fragariae antagonism | 6 | 1 | 2 | |
| NH4 + production | 5 | 7 | 2 | |
| ACC deaminase activity | 0 | 1 | 4 |
Effects of the strawberry microbiome on plant mineral composition and fruit quality
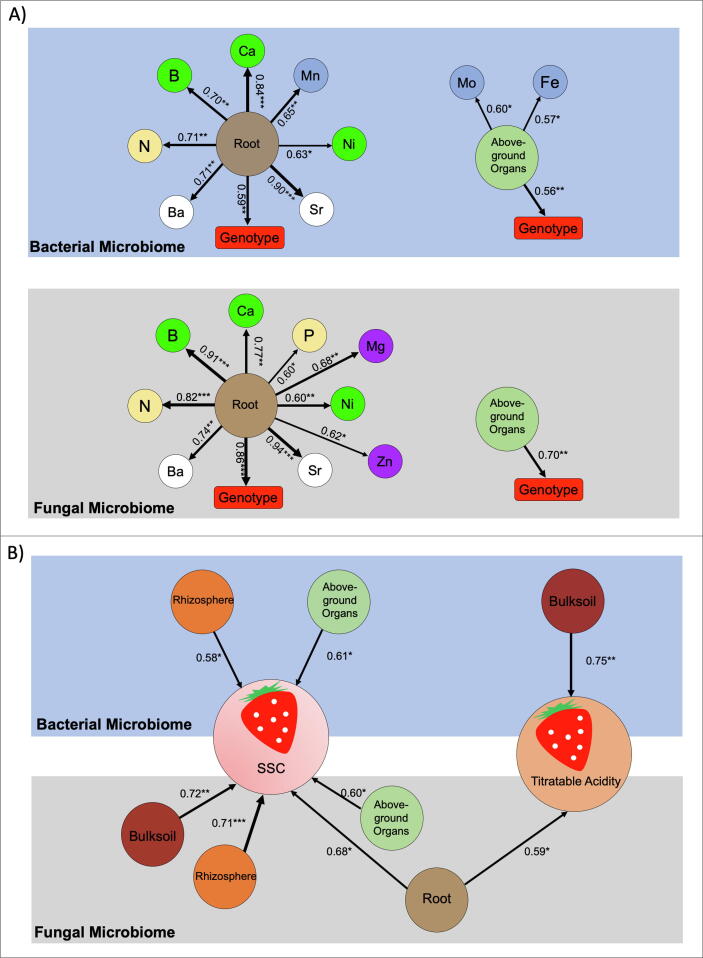
The contribution of bacterial and fungal microbiomes to the plant mineral composition (Fig. 5a) and fruit quality (Fig. 5b) was investigated. Significant correlations were observed between the mineral composition of the plant organs and the microbial community assemblage across different soil and plant compartments (Fig. 5a). In particular, the fungal microbiome colonising the root of strawberry was strongly determined by the genotype and was associated with the availability of B, Sr, and N. On the other hand, the fungal community associated with above-ground organs was not correlated to plant mineral composition. Compared to the fungal microbiome, the correlation of the bacterial community with the mineral elements was weak. However, below-ground bacterial microbiomes strongly correlated with Sr and Ca. In addition, microbes, and particularly those associated with soil and roots, contributed substantially, although indirectly, to fruit quality (Table S12). In detail, the fungal microbiome associated with the bulk soil and rhizosphere contributed the most to the development of SSC and TA of fruits (Fig. 5b). Rhizospheric and above-ground bacterial microbiome also contributed to SSC of fruits, whereas TA was related only to bacterial bulk soil microbiome.
Fig. 5.
Interactions between the strawberry microbiome, plant mineral composition, and the effect of the microbiome on fruit quality. (A) Correlations between bacterial and fungal microbiomes colonising roots and above-ground compartments with genotype and mineral nutrient content of the respective organ. Nutrients are indicated as essential macro-nutrients (yellow), universally essential cations (purple), elements playing an important ecological role both for plants and microbes (light blue), elements with specialized purposes in microbes (green), micro-nutrients (white); (B) Correlation between bacterial and fungal microbiomes colonising different plant compartments and total soluble solid content (SSC) and titratable acidity of fruits. Arrows thickness indicate strength of the correlation, R2 and p-values (* = 0.05; ** = 0.01; *** = 0.001) are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
Microbiome composition
In agreement with previous studies [4], above-ground organs displayed a lower bacterial and fungal alpha-diversity than below-ground ones. Such diversity in microbial community composition (Fig. 1a; Fig. S2a, b) and richness could be explained by the differences in the physical and chemical properties of the two environments. In fact, above-ground organs are subjected to oligotrophic and unstable conditions, with daily and seasonal fluctuations in temperature, humidity, and UV light [45], whereas the soil compartment is relatively more protected, stable, and nutrient-rich [46]. Despite the influence of plant genotype in the assembly of the cultivar-specific microbiota, the existence of a core microbiome, common to the three strawberry plant genotypes was reported (Fig. 2). The identification of ubiquitous microbes suggests that they are either able to colonise all soil and plant compartments, or they can move across the soil and the plant organs with a passive or active translocation from roots to the above-ground organs (e.g. leaves, runners).
Interestingly, several strawberry pathogens colonised the plants (Fig. 3c), and some of them were found among the core fungal microbiome, although no evident disease symptoms were observed. Such organisms include Plectosphaerella cucumerina (responsible for fruit root and collar rot), Botrytis caroliniana (gray mold), and Alternaria alternata (black leaf spot) (Table S8). The integration of different populations of potential pathogens (pathobiome) into a complex biotic environment [47], where they are involved in antagonistic and mutualistic interactions occurring among microbes and with the host plant, is likely to contribute to the control of such pathogens population, making the mere organ colonisation not sufficient, per se, for a successful infection causing disease symptoms [48]. On the whole, these observations suggest that abiotic (e.g. temperature, humidity, nutrient availability) [49] and biotic factors (e.g. plant-associated microbial consortia, plant resistance) [17] may have contributed to determine the fate of plant-pathogen interactions.
Plant growth-promoting functions of the microbiota and their contribution to plant phenotype
Domestication of crop plants has been suggested to have determined a reduction in the biodiversity of the associated microflora, in particular for functions regarding nutrition and stress tolerance [50]. On the other hand, cultivated plants might recruit microbes specifically exerting beneficial functions under cultural conditions. In this view, the ability to interact with such microbes may be regarded as a trait selected by domestication [51]. In this work, it was found that even after centuries of domestication and complex hybridisation [11], cultivated strawberry plants are associated with 16 nitrogen-fixing bacterial genera (Fig. 3a), which is more than what was reported in wild strawberry plant relatives (F. chiloensis, F. virginiana ssp. platypetala, F. × ananassa ssp. cuneifolia) (7 genera) [52] and comparable to the number of nitrogen-fixing genera (18) able to establish a nitrogen-fixing symbiosis with legumes (Table S13). Although bacterial taxa known to have N-fixing potential were surprisingly found in the above-ground habitat (Fig. 3a), nifH gene was not detected in this compartment by specific PCR. Considering that nitrogenase is inactivated by oxygen [53], the ability of these bacteria to interact with plant hosts is not necessarily related to their ability to fix nitrogen.
Tax4Fun predictive functional analysis of the bacterial microbiome showed that both the plant genotype and the organ significantly contributed to the overall assembly of metabolic functionalities (Table 3), which is in accordance with previous findings [54]. Modules and KOs specifically related to PGP traits were also investigated and the organ was found to significantly contribute to the assembly of metabolic functionalities of the microbiota. Metabolites or signals differently emitted by plant organs may explain organ colonisation by microorganisms potentially delivering different functions [54]. Additionally, both Tax4fun predictive analysis and in vitro screening of PGP traits showed that several PGP functions were different among cultivars (Table 4). However, in the case of below-ground organs, differences in PGP traits of the three genotypes were less marked. Similarly, studies about functional microbiomes of grapevine rootstocks of different genotypes showed that the potential ecological services were maintained across below-ground organs of different genotypes [55].
PGP functions might result in the production of specific metabolites, signals, or activation of pathogen defence mechanisms that may explain differences observed in the phenotype of different strawberry cultivars. ACC deaminase-producing bacteria have not been isolated from 'Monterey'. On the other hand, 'Elsanta' and 'Darselect' hosted ACC deaminase-producing bacteria. Thus, it might be speculated that these plants, less fit than 'Monterey' to the growing conditions, actively recruited bacteria expressing ACC deaminase activity to cope with stress. Indeed, this enzyme cleaves the ethylene precursor (ACC) into ammonia and alpha-ketobutyrate, preventing the accumulation of ethylene to detrimental levels [56]. 'Elsanta' plants showed significantly heavier root and leaf apparatuses, which might relate to functions linked to IAA and betaine production expressed by below-ground microflora. Indeed, root growth can be simulated through secretion of IAA by a variety of microbial species, including PGP, stress tolerance-inducing, as well as pathogenic ones [57]. Betaine is synthesized by plants in response to several environmental stresses, such as drought, salinity, and low temperatures [58], but betaine-producing bacteria (including B. subtilis and Arthtobacter globiformis [59] may also contribute to plant stress tolerance.
The roles of beneficial fungi on disease tolerance are almost unexplored in strawberry. This current work revealed the vast diversity of fungal partners of strawberry, which have not been thoroughly investigated so far, and include ectomycorrhizae, arbuscular mycorrhizae, ericoid mycorrhizae, endophytes, dark septate endophytes, and mycoparasites (Fig. 2, Fig. 3c,d). Future study should focus on isolation of these beneficial fungi and test for their contributions and mechanisms on plant disease tolerance.
Effect of the microbiota on disease resistance
The environmental factors, soil conditions, and pool of natural microbial inoculum were assumed to be comparable for all three strawberry genotypes, as plants were grown in the same cultural and environmental conditions. Therefore, the observed differences in associated bacterial and fungal communities (Fig. 2, Fig. 3) can be explained by the ability of the plant to adjust the composition of the associated microflora [60]. In this view, the lower susceptibility to powdery mildew and leaf spot observed in 'Monterey' over the season (Table S2), which is in agreement with existing literature [61], may be at least partly due to its ability to establish beneficial microbial relationships (Fig. 4a, c). At the same time, 'Elsanta' showed to be less sensitive than 'Darselect' to leaf spot, but highly susceptible to powdery mildew [62], [63]. Finding microbial patterns unique to a tolerant genotype suggests an important contribution of the microbiota to the defence strategy of strawberry plants against biotic stresses.
Indeed, 'Monterey' was characterised by the presence of Rhizophagus irregularis and above-ground Ps. fluorescens populations (Fig. 4). In several crop plants, the colonisation of the root systems by R. irregularis has been demonstrated to confer plant resistance to broad spectrum of pathogens by induced systemic resistance (ISR) and mycorrhizal-induced resistance (MIR) [64], [65]. Many Ps. fluorescens strains promote plant growth or protection, by mechanisms such as phosphorus solubilization, phytohormone production, competition against phytopathogens, elicitation of ISR, or production of antimicrobial compounds, such as cyanide or phenolics [43]. Ps. fluorescens strains isolated from raspberry fruit produce high levels of siderophores, which provide a competitive advantage against plant pathogens [26]. Furthermore, the same strains showed a strong ACC-deaminase activity that may regulate plant ethylene and, in turn, influence induced systemic resistance. Non-indigenous Ps. fluorescens strains have been already applied to strawberry plants, allowing them to anticipate flowering and fruiting, increase fruit yield and vitamin content [66], and control crown rot (Phytophthora cactorum) [67]. Notably, the inoculation of rice seed with a Ps. fluorescens strain for rice blast control resulted in the colonisation of roots, stems, and leaves [68], supporting that this species does not have strict organ preferences. In this work, three different Ps. fluorescens strains were isolated from the three strawberry genotypes. Remarkably, only the strains isolated from 'Monterey' often showed to have in vitro antagonistic activity against the pathogen Xanthomonas fragariae (Table 5 and Table S11).
Interestingly, the combined action of Pseudomonas spp. and Rhizophagus spp. has been explored in several crop species [65], [69]. In particular, a mixture of arbuscular mycorrhizae, which included Rhizophagus spp., and Pseudomonas fluorescens was successfully applied to strawberry, resulting in increased fruit production and quality [66]. The combination of Rhizophagus spp. and Ps. fluorescens has been proven to elicit plant systemic defence system in tomato via the activation of ethylene response to pathogen attack [65].
B. megaterium was the only OTU able to ubiquitously colonise 'Elsanta' and 'Darselect'. This bacterium has attracted considerable attention as a functional microbe in several crop species, including strawberry since it can solubilize phosphate and produce phytohormones [70]. Furthermore, it has been proven to be effective for the control of B. cinerea [71].
In above-ground organs, the KEGG modules responsible for JA, isoprenoid biosynthesis and H2S formation were higher in 'Monterey' than in the other genotypes. JA and its derivatives are responsible for many essential processes involved in plant growth and development, such as the immune response against necrotrophic pathogens and herbivorous insects [72] and regulation of ISR [73]. The production of JA from plant-associated bacteria is scarcely documented [74], although oxylipins have been reported to act as quorum sensing messengers in bacterial communication [75]. Volatile isoprenoids and H2S may exert a direct antimicrobial action, and/or enhance plant defences against pests and pathogens [76].
A further indication of the role of the microbiota in disease protection of 'Monterey' is offered by the high number of native X. fragariae antagonists isolated from this cultivar, in agreement with its low susceptibility to leaf spot disease.
For these reasons, disease tolerance in 'Monterey' may be partially explained by a contribution of phyllospheric microflora in plant signaling and biochemical functions, and by the ability of the plant to recruit microbiota components with protective action.
Interactions between the strawberry microbiome, plant mineral composition, and fruit production and quality
Bacterial and fungal microbiomes were correlated to plant mineral composition (Fig. 5a). Indeed, plant-associated microbiomes play a key role in improving plant nutrition by promoting both nutrient acquisition and nutrient use efficiency [77]. On the other hand, the host plant and its nutrient preferences impact its microbiome recruitment [78]. Significant correlations between fruit SSC and TA and bacterial and fungal microbiomes emerged from this work (Fig. 5b), suggesting a role of plant-associated microflora in fruit development. Such a role has been previously observed in strawberry [15] as well as in other species, e.g. for Bacillus spp. on flowers and leaves of sour cherry [79], and is not limited to interactions with above-ground organs. In fact, fruit development and ripening are finely regulated by phytohormones, in particular by ethylene, auxin, and gibberellins that can be produced by both fungi and bacteria. Ethylene is produced by a wide range of microbes starting from two alternative precursors, 2-keto-4-methyl-thiobutyric acid (KMBA) or 2-oxoglutarate [80]. Furthermore, several ACC deaminase-expressing bacterial species has been observed in this study (i.e. Methylobacterium sp., Pantoea sp., Erwinia sp., Pseudomonas sp.), whose ACC deaminase activity has been previously found on other berry fruits [26].
A high plant productivity and fruit weight, as observed in 'Monterey', may be partially explained by the hormonal and signaling effects of the associated microflora [25]. Indeed, this cultivar was rich in OTUs predicted to produce JA and isoprenoids (Table 4), as well as in IAA-producing isolates (Table 5). In strawberry, JA and derived compounds stimulate the development and ripening of fruits [81]. In addition to volatile compounds, responsible for fruit aroma, the isoprenoid family includes abscisic acid, brassinosteroids, cytokinins, gibberellins, phytoecdysteroids, and strigolactones, all with key roles in plant growth regulation [82]. IAA regulates fruit size and ripening at the early stages of fruit development [83]. The relative importance of these signals and the microbes that release them, on fruit production and quality remains to be assessed.
Arbuscular mycorrhizae have been proven to affect plant hormonal balance and metabolism. Indeed, their beneficial effect has been observed both in below- and above-ground organs [66]. Besides arbuscular mycorrhizae, PGPB are also able to affect fruit quality, mainly by modulating the interplay between ethylene and auxin metabolisms and providing essential nutrients [5], [26]. On the whole, these correlations suggest that bacteria and fungi can contribute to the host’s adaptation to growing conditions and, consequently, to fruit development.
Conclusions
Together with the characterisation of the taxonomical composition, the prediction of plant microbiota functional properties facilitates the study and exploitation of metabolic potentialities of the microbiome [3], [41]. Thus, several aspects, such as the plant performance, resistance, and resilience to stresses might be influenced by functional, rather than taxonomic diversity [21].
Although it may be difficult to dissect the plant properties uniquely deriving from its genotype, the contribution of its microflora, and the interactive effects of plant genotype with the specifically associated microbiota, it is important to note that the culture-dependent, biochemical characterisation of the bacterial isolates led to results similar to those obtained by in silico prediction of metabolic functionalities (viz. a culture-independent approach). These data offer two independent lines of confirmation of phenotypic observations, thus strengthening the results.
In conclusion, this work highlighted the interaction of cultivated strawberry genotypes with a variety of microbial species. Such interactions are specific to genotypes and compartments. The microbiome plays a key role in the plant’s ability to cope with biotic stress and in modulating fruit quality. These findings suggest that a comprehensive picture of plant holobiome is needed in order to shed light on the influence of microbial communities and key microbes on plant phenotype and performances. Further studies on microbiomes of crop plants can contribute to the advancement of plant production science, by providing a deeper insight into the interactions between crops and the microflora and evidencing applicative tools and strategies for an efficient and environmentally sustainable horticultural practice. However, the complexity and specificity of the patterns described in this work suggest that the idea to replace agrochemicals with a few universal beneficial microorganisms is not realistic. Therefore, breeding programs should aim at the selection of high-quality, climate-change resilient horticultural varieties with a remarkable capacity to establish symbiotic relationships with useful microorganisms [5]. The inclusion of microbial markers in marker-assisted selection will represent a paradigm shift in plant breeding.
Availability of data and materials
The datasets Illumina sequencing of all bacterial and fungal datasets generated during the current study are available in The National Center for Biotechnology Information (NCBI) database under BioProject: PRJNA556362 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA556362).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors are grateful to L. Giongo for providing accession to the fields and to P. Martinatti for plants management. S.F.M. Wahdan is financially supported by the Egyptian Scholarship (Ministry of Higher Education, external missions 2016/2017 call).
Funding
This work has been partially funded by the internal research budget of Dr.Witoon Purahong, Department of Soil Ecology, UFZ-Helmholtz Centre for Environmental Research.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.02.009.
Contributor Information
Francesco Spinelli, Email: francesco.spinelli3@unibo.it.
Witoon Purahong, Email: witoon.purahong@ufz.de.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Purahong W., Orrù L., Donati I., Perpetuini G., Cellini A., Lamontanara A., et al. Plant microbiome and its link to plant health: Host species, organs, and Pseudomonas syringae pv. actinidiae infection shaping bacterial phyllosphere communities of kiwifruit plants. Front Plant Sci. 2018;9:1563. doi: 10.3389/fpls.2018.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busby P.E., Newcombe G., Dirzo R., Whitham T.G., Thrall P. Differentiating genetic and environmental drivers of plant–pathogen community interactions. J Ecol. 2014;102(5):1300–1309. doi: 10.1111/1365-2745.12270. [DOI] [Google Scholar]
- 3.Lemanceau P., Blouin M., Muller D., Moënne-Loccoz Y. Let the core microbiota be functional. Trends Plant Sci. 2017;22(7):583–595. doi: 10.1016/j.tplants.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Cregger M.A., Veach A.M., Yang Z.K., Crouch M.J., Vilgalys R., Tuskan G.A., et al. The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome. 2018;6(1) doi: 10.1186/s40168-018-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sangiorgio D., Cellini A., Donati I., Pastore C., Onofrietti C., Spinelli F. Facing climate change: Application of microbial biostimulants to mitigate stress in horticultural crops. Agron. 2020;10:794. doi: 10.3390/agronomy10060794. [DOI] [Google Scholar]
- 6.Busby P.E., Soman C., Wagner M.R., Friesen M.L., Kremer J., Bennett A., et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017;15(3):e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutschick V.P., BassiriRad H. Extreme events as shaping physiology, ecology, and evolution of plants: toward a unified definition and evaluation of their consequences. New Phytol. 2003;160(1):21–42. doi: 10.1046/j.1469-8137.2003.00866.x. [DOI] [PubMed] [Google Scholar]
- 8.Hargreaves S.K., Hofmockel K.S. Physiological shifts in the microbial community drive changes in enzyme activity in a perennial agroecosystem. Biogeochem. 2014;117(1):67–79. doi: 10.1007/s10533-013-9893-6. [DOI] [Google Scholar]
- 9.Grady K.L., Sorensen J.W., Stopnisek N., Guittar J., Shade A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat Commun. 2019;10:4135. doi: 10.1038/s41467-019-11974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Z., Egidi E., Liu H., Kaur S., Singh B.K. New frontiers in agriculture productivity: Optimised microbial inoculants and in situ microbiome engineering. Biotechol Adv. 2019;37(6):107371. doi: 10.1016/j.biotechadv.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Edger P.P., Poorten T.J., VanBuren R., Hardigan M.A., Colle M., McKain M.R., et al. Origin and evolution of the octoploid strawberry genome. Nat Genet. 2019;51(3):541–547. doi: 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Food and Agriculture Organization of the United Nations. (1997). FAOSTAT statistical database. [Rome] :FAO
- 13.Mezzetti B., Giampieri F., Zhang Y.-T., Zhong C.-F. Status of strawberry breeding programs and cultivation systems in Europe and the rest of the world. J Berry Res. 2018;8(3):205–221. doi: 10.3233/JBR-180314. [DOI] [Google Scholar]
- 14.Battino M, Forbes-Hernandez TY, Gasparrini M, Afrin S, Mezzetti B, Giampieri F. The effects of strawberry bioactive compounds on human health. In: VIII International Strawberry Symposium 1156 (2018). 355-362. 10.17660/ActaHortic.2017.1156.54
- 15.Todeschini V., AitLahmidi N., Mazzucco E., Marsano F., Gosetti F., Robotti E., et al. Impact of beneficial microorganisms on strawberry growth, fruit production, nutritional quality, and volatilome. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husaini A.M., Neri D., editors. Strawberry: growth, development and diseases. CABI; Wallingford: 2016. [Google Scholar]
- 17.Amsalem L., Freeman S., Rav-David D., Nitzani Y., Sztejnberg A., Pertot I., et al. Effect of climatic factors on powdery mildew caused by Sphaerotheca macularis f. sp. fragariae on strawberry. Eur J Plant Pathol. 2006;114(3):283–292. doi: 10.1007/s10658-005-5804-6. [DOI] [Google Scholar]
- 18.Sargent D.J., Buti M., Šurbanovski N., Brurberg M.B., Alsheikh M., Kent M.P., et al. Identification of QTLs for powdery mildew (Podosphaera aphanis; syn. Sphaerotheca macularis f. sp. fragariae) susceptibility in cultivated strawberry (Fragaria× ananassa) PLoS ONE. 2019;14(9):e0222829. doi: 10.1371/journal.pone.0222829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitaker V.M., Knapp S.J., Hardigan M.A., Edger P.P., Slovin J.P., Bassil N.V., et al. A roadmap for research in octoploid strawberry. Hortic Res. 2020;7(1) doi: 10.1038/s41438-020-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haney C.H., Samuel B.S., Bush J., Ausubel F.M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants. 2015;1(6) doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson D.R., Pomati F. A brief guide for the measurement and interpretation of microbial functional diversity. Environ. microbial. 2020;22(8):3039–3048. doi: 10.1111/1462-2920.15147. [DOI] [PubMed] [Google Scholar]
- 22.Bai Y., Müller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nat. 2015;528(7582):364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 23.Wahdan S.F.M., Tanunchai B., Wu Y.-T., Sansupa C., Schädler M., Dawoud T.M., et al. Deciphering Trifolium pratense L. holobiont reveals a microbiome resilient to future climate changes. Microb. Open. 2021;10(4) doi: 10.1002/mbo3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feltham R.K.A., Power A.K., Pell P.A., Sneath P.H.A. A simple method for storage of bacteria at—76 C. J Appl. Bacteriol. 1978;44(2):313–316. doi: 10.1111/j.1365-2672.1978.tb00804.x. [DOI] [PubMed] [Google Scholar]
- 25.Sangiorgio D., Cellini A., Spinelli F., Farneti B., Khomenko I., Muzzi E., et al. Does organic farming increase raspberry quality, aroma and beneficial bacterial biodiversity. Microorganisms. 2021;9(8):1617. doi: 10.3390/microorganisms9081617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perpetuini G., Donati I., Cellini A., Orrú L., Giongo L., Farneti B., et al. Genetic and functional characterization of the bacterial community on fruit of three raspberry (Rubus idaeus) cultivars. J Berry Res. 2019;9(2):227–247. doi: 10.3233/JBR-180340. [DOI] [Google Scholar]
- 27.Durante C., Bertacchini L., Bontempo L., Camin F., Manzini D., Lambertini P., et al. From soil to grape and wine: Variation of light and heavy elements isotope ratios. Food Chem. 2016;210:648–659. doi: 10.1016/j.foodchem.2016.04.108. [DOI] [PubMed] [Google Scholar]
- 28.Giongo L., Ajelli M., Poncetta P., Ramos-García M., Sambo P., Farneti B. Raspberry texture mechanical profiling during fruit ripening and storage. Postharvest Biol. Tec. 2019;149:177–186. doi: 10.1016/j.postharvbio.2018.11.021. [DOI] [Google Scholar]
- 29.Bodenhausen N, Horton M.W., Bergelson J., Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana, PLoS ONE 2013; 8(2):e56329. 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed]
- 30.Boyd E.S., Peters J.W. New insights into the evolutionary history of biological nitrogen fixation. Front Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothurMothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer F., Mercier C., Bonin A., Le Bras Y., Taberlet P., Coissac E. Obitools: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour. 2016;16:176–182. doi: 10.1111/1755-0998.12428. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinforma Oxf Engl. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinforma Oxf Engl. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42(D1):D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kõljalg U., Larsson K.-H., Abarenkov K., Nilsson R.H., Alexander I.J., Eberhardt U., et al. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005;166(3):1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 37.Bai H.F., Li M.W., Wang D.S. Bayesian classifier based service-aware mechanism in 10G-EPON for smart power grid. Acta Photonica Sinica. 2013;42:668–673. doi: 10.3788/gzxb20134206.0668. [DOI] [Google Scholar]
- 38.Louca S., Parfrey L.W., Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Sci. 2016;353:1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen N.H., Song Z., Bates S.T., Branco S., Tedersoo L., Menke J., et al. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–248. doi: 10.1016/j.funeco.2015.06.006. [DOI] [Google Scholar]
- 40.Sansupa C., Wahdan S.F.M., Hossen S., Disayathanoowat T., Wubet T., Purahong W. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl Sci. 2021;11:688. doi: 10.3390/app11020688. [DOI] [Google Scholar]
- 41.Aßhauer K.P., Wemheuer B., Daniel R., Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinform. 2015;31:2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Prot. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 43.David B.V., Chandrasehar G., Selvam P.N. Pseudomonas fluorescens: a plant-growth-promoting rhizobacterium (PGPR) with potential role in biocontrol of pests of crops, Crop improvement through microbial. biotechnology. 2018:221–243. doi: 10.1016/B978-0-444-63987-5.00010-4. [DOI] [Google Scholar]
- 44.Pathania P., Rajta A., Singh P.C., Bhatia R. Bhatia Role of plant growth-promoting bacteria in sustainable agriculture, Biocatalysis and Agricultural. Biotechnology. 2020;30:101842. doi: 10.1016/j.bcab.2020.101842. [DOI] [Google Scholar]
- 45.Redford A.J., Fierer N. Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol. 2009;58(1):189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- 46.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69(4):1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vayssier-Taussat M., Albina E., Citti C., Cosson J.-F., Jacques M.-A., Lebrun M.-H., et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front. cell. infect. microbial. 2014;4 doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donati I., Cellini A., Sangiorgio D., Vanneste J.L., Scortichini M., Balestra G.M., et al. Pseudomonas syringae pv. actinidiae: Ecology, Infection Dynamics and Disease Epidemiology. Microb Ecol. 2020;80(1):81–102. doi: 10.1007/s00248-019-01459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandey P., Senthil-Kumar M. Plant-pathogen interaction in the presence of abiotic stress: What do we know about plant responses? Plant Physiol Rep. 2019;24(4):541–549. doi: 10.1007/s40502-019-00483-7. [DOI] [Google Scholar]
- 50.Pérez-Jaramillo J.E., Mendes R., Raaijmakers J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol. 2016;90(6):635–644. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez-Jaramillo J.E., Carrión V.J., de Hollander M., Raaijmakers J.M. The wild side of plant microbiomes. Microbiome. 2018;6:143. doi: 10.1186/s40168-018-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei N., Ashman T.L. The effects of host species and sexual dimorphism differ among root, leaf and flower microbiomes of wild strawberries in situ. Sci Rep. 2018;8:5195. doi: 10.1038/s41598-018-23518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallon J.R. The oxygen sensitivity of nitrogenase: a problem for biochemists and micro-organisms. Trends Biochem Sci. 1981;6:19–23. doi: 10.1016/0968-0004(81)90008-6. [DOI] [Google Scholar]
- 54.Zipfel C., Oldroyd E.D. Plant signalling in symbiosis and immunity. Nat. 2017;543(7645):328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 55.Marasco R., Rolli E., Fusi M., Michoud G., Daffonchio D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome. 2018;6:1–17. doi: 10.1186/s40168-017-0391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glick B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169(1):30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Spaepen S., Vanderleyden J., Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS microbio Rev. 2007;31(4):425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 58.Ashraf M., Foolad M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Env Exp Bot. 2007;59(2):206–216. doi: 10.1016/j.envexpbot.2005.12.006. [DOI] [Google Scholar]
- 59.Zou H., Chen N., Shi M., Xian M.o., Song Y., Liu J. The metabolism and biotechnological application of betaine in microorganism. Appl Microbiol Biotech. 2016;100(9):3865–3876. doi: 10.1007/s00253-016-7462-3. [DOI] [PubMed] [Google Scholar]
- 60.N.M. Morella, Multiple forces shape the phyllosphere microbiome: The importance of vertical transmission, environmental selection, and bacteriophages (Doctoral dissertation, UC Berkeley) (2019).
- 61.Masny A., Pruski K., Żurawicz E., Mądry W. Breeding value of selected dessert strawberry (Fragaria× ananassa Duch.) cultivars for ripening time, fruit yield and quality. Euphytica. 2016;207:225–243. doi: 10.1007/s10681-015-1480-6. [DOI] [Google Scholar]
- 62.Bestfleisch M., Luderer-Pflimpfl M., Höfer M., Schulte E., Wünsche J.N., Hanke M.-V., et al. Evaluation of strawberry (Fragaria L.) genetic resources for resistance to Botrytis cinerea. Plant Pathol. 2015;64(2):396–405. doi: 10.1111/ppa.12278. [DOI] [Google Scholar]
- 63.Bestfleisch M., Richter K., Wensing A., Wünsche J.N., Hanke M.-V., Höfer M., et al. Resistance and systemic dispersal of Xanthomonas fragariae in strawberry germplasm (Fragaria L.) Plant Pathol. 2015;64(1):71–80. doi: 10.1111/ppa.12232. [DOI] [Google Scholar]
- 64.Cameron D.D., Neal A.L., van Wees S.C.M., Ton J. Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci. 2013;18(10):539–545. doi: 10.1016/j.tplants.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Velivelli S.LS., Lojan P., Cranenbrouck S., de Boulois H.D., Suarez J.P., Declerck S., et al. The induction of Ethylene response factor 3 (ERF3) in potato as a result of co-inoculation with Pseudomonas sp. R41805 and Rhizophagus irregularis MUCL 41833 - a possible role in plant defense. Plant Signal Behav. 2015;10(2):e988076. doi: 10.4161/15592324.2014.988076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bona E., Lingua G., Manassero P., Cantamessa S., Marsano F., Todeschini V., et al. AM fungi and PGP pseudomonads increase flowering, fruit production, and vitamin content in strawberry grown at low nitrogen and phosphorus levels. Mycorrhiza. 2015;25(3):181–193. doi: 10.1007/s00572-014-0599-y. [DOI] [PubMed] [Google Scholar]
- 67.Agusti L., Bonaterra A., Moragrega C., Camps J., Montesinos E. Biocontrol of root rot of strawberry caused by Phytophthora cactorum with a combination of two Pseudomonas fluorescens strains. J. Plant Pathol. 2011:363–372. https://www.jstor.org/stable/41999007?seq=1#metadata_info_tab_contents [Google Scholar]
- 68.Vidhyasekaran P., Rabindran R., Muthamilan M., Nayar K., Rajappan K., Subramanian N., et al. Development of a powder formulation of Pseudomonas fluorescens for control of rice blast. Plant Pathol. 1997;46(3):291–297. doi: 10.1046/j.1365-3059.1997.d01-27.x. [DOI] [Google Scholar]
- 69.Bahmani M., Naghdi R., Kartoolinejad D. Milkweed seedlings tolerance against water stress: Comparison of inoculations with Rhizophagus irregularis and Pseudomonas putida. Environ. Technol. Innov. 2018;10:111–121. doi: 10.1016/j.eti.2018.01.001. [DOI] [Google Scholar]
- 70.Dias A.C.F., Costa F.E.C., Andreote F.D., Lacava P.T., Teixeira M.A., Assumpção L.C., et al. Isolation of micropropagated strawberry endophytic bacteria and assessment of their potential for plant growth promotion. World J Microbiol Biotechnol. 2009;25(2):189–195. doi: 10.1007/s11274-008-9878-0. [DOI] [Google Scholar]
- 71.Donmez M.F., Esitken A., Yildiz H., Ercisli S. Biocontrol of Botrytis cinerea on strawberry fruit by plant growth promoting bacteria. J. Anim. Plant Sci. 2011;21:758–763. [Google Scholar]
- 72.Ali M., Baek K.H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int J Mol Sci. 2020;21:621. doi: 10.3390/ijms21020621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pieterse C.M.J., Zamioudis C., Berendsen R.L., Weller D.M., Van Wees S.C.M., Bakker P.A.H.M. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52(1):347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 74.Piccoli P., Travaglia C., Cohen A., Sosa L., Cornejo P., Masuelli R., et al. An endophytic bacterium isolated from roots of the halophyte Prosopis strombulifera produces ABA, IAA, gibberellins A 1 and A 3 and jasmonic acid in chemically-defined culture medium. Plant Growth Regul. 2011;64(2):207–210. doi: 10.1007/s10725-010-9536-z. [DOI] [Google Scholar]
- 75.Martínez E., Cosnahan R.K., Wu M., Gadila S.K., Quick E.B., Mobley J.A., et al. Oxylipins mediate cell-to-cell communication in Pseudomonas aeruginosa. Comm. Biol. 2019;2:1–10. doi: 10.1038/s42003-019-0310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abbas M., Paul M., Huttner A. New and improved? A review of novel antibiotics for Gram-positive bacteria. Clin Microbiol Infect. 2017;23(10):697–703. doi: 10.1016/j.cmi.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 77.Trivedi P., Leach J.E., Tringe S.G., Sa T., Singh B.K. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18(11):607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 78.Cai F., Pang G., Miao Y., Li R., Li R., Shen Q., et al. The nutrient preference of plants influences their rhizosphere microbiome. Appl Soil Ecol. 2017;110:146–150. doi: 10.1016/j.apsoil.2016.11.006. [DOI] [Google Scholar]
- 79.Arikan Ş., Pirlak L. Effects of plant growth promoting rhizobacteria (PGPR) on growth, yield and fruit quality of sour cherry (Prunus cerasus L.) Erwerbs-obstbau. 2016;58:221–226. doi: 10.1007/s10341-016-0278-6. [DOI] [Google Scholar]
- 80.Nagahama K., Ogawa T., Fujii T., Fukuda H. Classification of ethylene-producing bacteria in terms of biosynthetic pathways to ethylene. J Ferment Bioeng. 1992;73(1):1–5. doi: 10.1016/0922-338X(92)90221-F. [DOI] [Google Scholar]
- 81.Han Y., Chen C., Yan Z., Li J., Wang Y. The methyl jasmonate accelerates the strawberry fruits ripening process. Sci. Hort. 2019;249:250–256. [Google Scholar]
- 82.Tarkowská D., Strnad M. Isoprenoid-derived plant signaling molecules: biosynthesis and biological importance. Planta. 2018;247(5):1051–1066. doi: 10.1007/s00425-018-2878-x. [DOI] [PubMed] [Google Scholar]
- 83.Symons G.M., Chua Y.-J., Ross J.J., Quittenden L.J., Davies N.W., Reid J.B. Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot. 2012;63(13):4741–4750. doi: 10.1093/jxb/ers147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets Illumina sequencing of all bacterial and fungal datasets generated during the current study are available in The National Center for Biotechnology Information (NCBI) database under BioProject: PRJNA556362 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA556362).