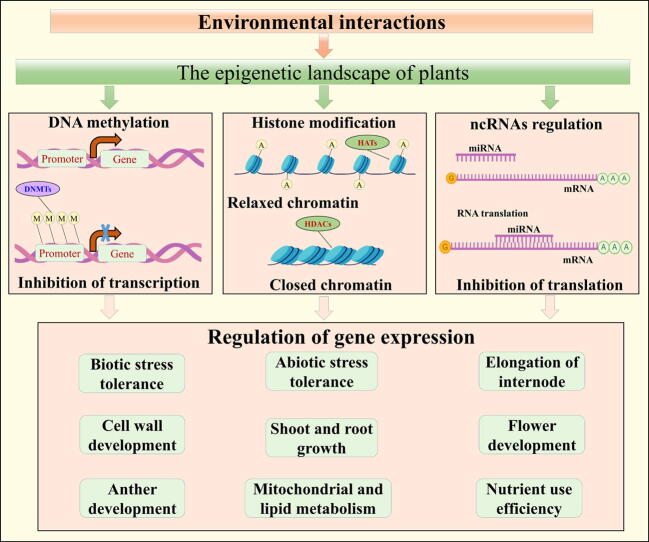
Graphical abstract
Keywords: Redox status, Oxidative stress, Epigenetic modifications, miRNA biogenesis, Chromatin rearrangement, Abiotic stress tolerance
Highlights
-
•
The redox status of the cell influences enzymes involved in plant epigenetic modifications.
-
•
The redox homeostasis in the cell is difficult to sustain because of unequal production of oxidant molecules.
-
•
Intracellular epigenetic modifications in plants mitigate damage response during stress.
-
•
High-throughput techniques have greatly advanced redox-mediated gene expression discovery.
-
•
The integrated view of the redox status and epigenetic changes in plants are still scarce.
-
•
We show opportunities for smart use of the redox status of the cell in development of the plant.
Abstract
Background
The oxidation-reduction (redox) status of the cell influences or regulates transcription factors and enzymes involved in epigenetic changes, such as DNA methylation, histone protein modifications, and chromatin structure and remodeling. These changes are crucial regulators of chromatin architecture, leading to differential gene expression in eukaryotes. But the cell’s redox homeostasis is difficult to sustain since the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) is not equal in plants at different developmental stages and under abiotic stress conditions. Exceeding optimum ROS and RNS levels leads to oxidative stress and thus alters the redox status of the cell. Consequently, this alteration modulates intracellular epigenetic modifications that either mitigate or mediate the plant growth and stress response.
Aim of review
Recent studies suggest that the altered redox status of the cell reform the cellular functions and epigenetic changes. Recent high-throughput techniques have also greatly advanced redox-mediated gene expression discovery, but the integrated view of the redox status, and its associations with epigenetic changes and subsequent gene expression in plants are still scarce. In this review, we accordingly focus on how the redox status of the cell affects epigenetic modifications in plants under abiotic stress conditions and during developmental processes. This is a first comprehensive review on the redox status of the cell covering the redox components and signaling, redox status alters the post-translational modification of proteins, intracellular epigenetic modifications, redox interplay during DNA methylation, redox regulation of histone acetylation and methylation, redox regulation of miRNA biogenesis, redox regulation of chromatin structure and remodeling and conclusion, future perspectives and biotechnological opportunities for the future development of the plants.
Key Scientific Concepts of Review
The interaction of redox mediators such as ROS, RNS and antioxidants regulates redox homeostasis and redox-mediated epigenetic changes. We discuss how redox mediators modulate epigenetic changes and show the opportunities for smart use of the redox status of the cell in plant development and abiotic stress adaptation. However, how a redox mediator triggers epigenetic modification without activating other redox mediators remains yet unknown.
Introduction
Gene expression is the part of the central dogma of cells supporting plant growth and development throughout the plant life cycle, but spatiotemporal gene expression depends on several factors: gene structure and organization and epigenetic processes. Epigenetic changes, such as DNA and histone proteins modifications, are crucial regulators of chromatin architecture, leading to differential gene expression. These changes are transient, thus playing a vital role during plant development and for abiotic stress adaptation [1]. Epigenetic-mediated gene expression undergoes dynamic changes such as switching stress and cell-specific genes on or off [2]. Epigenetically-induced phenotypic variations are generally inheritable without affecting the primary DNA sequence [3].
However, the internal cellular environment, particularly the oxidation-reduction (redox) status of the cell, influences enzymes involved in epigenetic changes, thereby controlling the activities of several transcription factors and other enzymes. For instance, enzymes involved in histone acetylation homeostasis require primary metabolites as substrates or cofactors whose levels are seriously influenced by the redox status of the cell [4]. The redox status also influences enzymes involved in methyl group donor and methyl group acceptor, which determine methylation efficiency. In recent times, redox-mediated gene expression discovery and its associations with epigenetic modifications is established in plants. Accruing evidence indicates that altered the redox status of the cell reform the cellular functions and epigenetic changes and regulates a wide range of functions in plants than previously understood. This occurs in all stages of plant growth and development, including adaptations to stresses [5]. However, despite the importance of the redox status of the cell, except for a few reviews [4], [5], [6], [7], there is a lack of organized literature on redox-related epigenetic changes in plants. Therefore, in this review, we attempt to highlight the role of cellular redox status in epigenetic changes in plant systems, especially in addressing abiotic stress tolerance, vernalization and stem cell development.
Redox components and signaling
The redox status is balanced by interactions of oxidant and antioxidant systems in the cell. The maintenance of the redox status of a living cell is dynamic because of the continuous production of oxidant molecules, reactive oxygen species (ROS) and reactive nitrogen species (RNS) (Supplementary Table 1). In plants, ROS production occurs in various cellular components (Fig. 1), such as in chloroplast during photosynthesis, in mitochondria during electron transport, in peroxisomes during photorespiration, and in the plasma membrane [8]. As with ROS, RNS generation is also ineluctable. RNS is generally derived from nitric oxide (NO). RNS is also generated within different cellular compartments by the action of enzymes such as NO reductase, nitrate reductase and nitric oxide synthase-like enzymes [9]. Both oxidant molecules regulate important cell signaling processes in cell differentiation, multiplication and migration and programmed cell death [10]. Though ROS and RNS are both maintained at optimum levels by scavenging systems (antioxidants), excessive oxidant production occurs when cells are stressed and derails the redox status, triggering an oxidative stress response; however, these species get reduced, counterbalancing oxidative processes to maintain the cell’s redox status [11]. During cellular processes, cellular redox status is an important indicator of a healthy plant system. Changes in the cell’s redox status influence or alter nuclear gene expression in several ways such as direct modification, transcription factor activation, epigenetic modification, etc. [4].
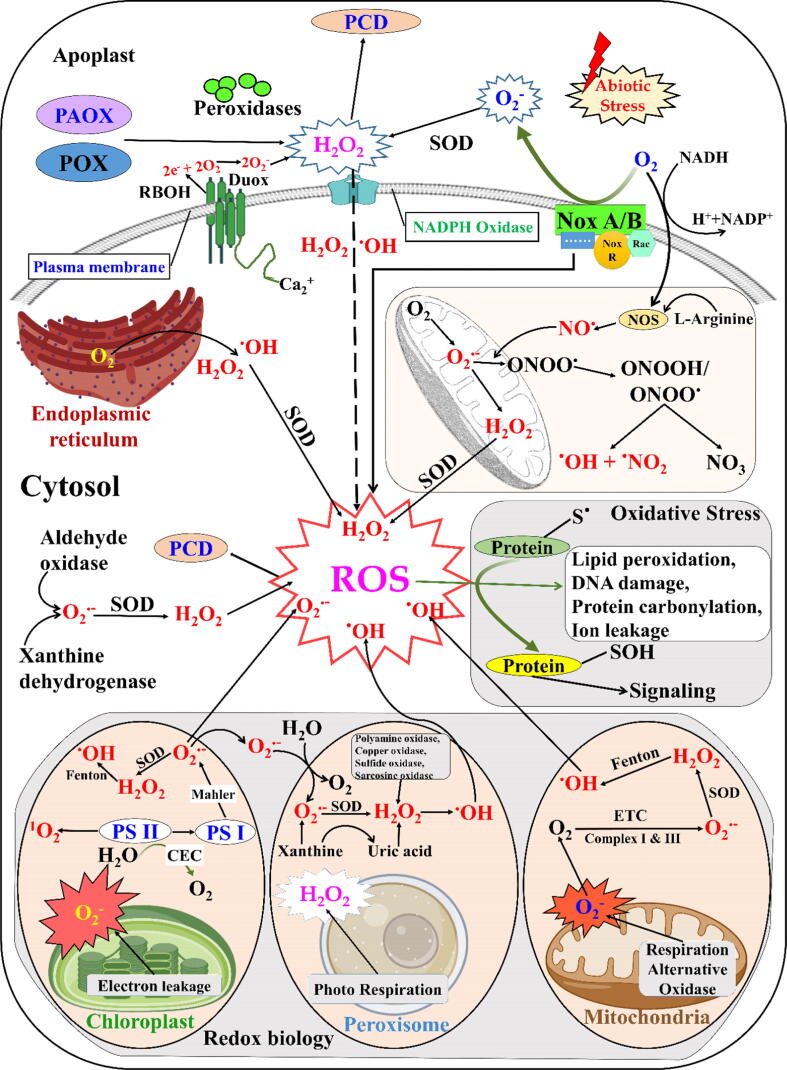
Fig. 1.
Redox process of cellular state. Different reactive oxygen species (ROS) and reactive nitrogen species (RNS) are produced in plants under abiotic stress to regulate gene expression for stress tolerance. ROS and RNS generation are activated during abiotic stress in the apoplastic space, chloroplasts, mitochondria, and peroxisomes. For instance, first, superoxide (O2•−) is generated by plasma membrane NADPH (nicotinamide adenine dinucleotide phosphate reduced) oxidases, and O2•− is immediately dismutated to hydrogen peroxide (H2O2) by superoxide dismutases (SOD) in the corresponding organelles. Peroxidases (POX) and polyamine oxidases (POAX) are other sources of apoplastic H2O2 generation [14]. These organelles also have mechanisms to detoxify the H2O; however, oxidative stress and programmed cell death (PCD) occur if ROS and RNS generation is beyond the homeostatic range.
Although oxidative stress due to the accumulation of excessive ROS and RNS is highly harmful to plant biological systems, optimum levels of ROS and RNS are necessary for regulating gene expression for stress tolerance. For instance, in Arabidopsis, mutants deficient in ROS production (knockout-NADPH oxidase D protein) were reported to be more sensitive to light stress [12]. Phytohormones like salicylic acid (SA) accomplish their activity by changing the redox status of the cell when the appropriate balance of reactive species is lost. For example, during salt stress, SA promotes the generation of endogenous ROS and NO and modulates their function [13].
Soluble redox carriers
During redox homeostasis, intracellular communication between various cellular compartments is performed by soluble molecules known as redox/electron carriers such as nicotinamide adenine dinucleotide (NADH), glutathione (GSH) and ascorbate (Asc) [15]. These molecules can quickly switch between reduced and oxidized states through the exchange of electrons (gain or loss of electrons). Thus, redox couples or redox guardians are essential for maintaining redox homeostasis and cellular metabolism. Redox pairs of these carrier molecules, such as NADH/NAD+, GSH/glutathione disulfide (GSSG) and Asc/dehydroascorbic acid (DHA), play a significant role at various plant growth stages and when plants are exposed to stress.
The imbalance of redox couples or preventing reactions of the redox pairs of these carrier molecules renders the cell more vulnerable to ROS. For example, Arabidopsis failed to develop past the embryonic stage without GSH whose function depends on the redox status of the cell [16]. The GSH:GSSG ratio is high in most cell organelles in plants growing under normal conditions but is altered by oxidative stress that increases the accumulation of GSSG to enhance stress tolerance. Thus, the GSH:GSSG ratio is related to the intracellular availability of H2O2. The glutathione reductase (gr1) mutant, encoding the peroxisome/cytosol enzymes, decreased the accumulation of GSSG in Arabidopsis leaf tissue, preventing survival [17]. Plants with NADPH deficiency also produced low GSH:GSSG ratios in the vacuole or endoplasmic reticulum, reviewed by Noctor et al. [18]. In tobacco lines, DHAR overexpression and knockdown respectively increased and decreased extractable foliar activity. The knockdown lines altered the Asc/DHA ratio in leaves under oxidative stress. Further, the GSH/GSSG ratio was higher in the DHAR overexpressed lines. A higher foliar level of Asc protected plants against oxidative stress [19]. Thus, redox carriers can act as markers of redox status and change the redox status of the cell.
Changes in the redox status of the cell induce several epigenetic modifications, including post-translational modifications (PTMs) in histone molecules [4], and either mitigate or mediate a response to damage during stress. The redox potential conditioned by the carrier molecules plays an important role in plants. For instance, an increase in NAD accumulation in a cytoplasmic male sterile II (CMSII) mitochondrial mutant of Nicotiana sylvestris was found to impart increased defense response and improved ozone tolerance [20]. By measuring the redox status using a redox-sensitive biosensor, namely reduction-oxidation sensitive green fluorescent protein (roGFP), Vivancos et al. [21] found that GSH plays a vital role in cell cycle progression in Arabidopsis. Similarly, an ascorbate deficient, vitamin C defective (vtc), mutant of Arabidopsis was reported to exhibit mild oxidative stress in the nucleus together with the arrest of cell cycle progression [22]. In the following sections, we summarize and discuss how the redox status of the cell affects epigenetic changes, regulation of miRNA biogenesis, chromatin remodeling and their collective role in abiotic stress tolerance.
Redox status alters the post-translational modification of proteins
The PTMs of the gene product determine the ultimate phenotype of any gene expression. PTMs introduced by oxidative stress mediate the epigenetic pathway, thereby leading to altered phenotypes [23]. For instance, in Arabidopsis, a comparative expression study between the wildtype and a redox compromised mutant stn7 (state transition 7) showed that many nuclear genes were not expressed in stn7 mutants in response to high-light treatments. Further, in wild-type plants, redox signals that arose from the chloroplast could activate two enzymes, nuclear histone acetyltransferase (HAT) and histone deacetylase (HDAC), to promote histone methylation/demethylation. Since methylation and demethylation are two interchangeable processes integral to epigenetic gene expression, the observations on the stn7 mutant are further evidence that redox signals alter gene expression through epigenetic and other regulatory pathways [23].
In Lablab purpureus, the induced elevation of O2•− and H2O2 after high-temperature treatment could be reversed by the application of SA and sodium nitroprusside (SNP), an NO donor. This reversal also showed a concomitant increase in the expression of antioxidant enzymes such as catalase (CAT), ascorbate peroxidase (APX), superoxide dismutase (SOD) and glutathione peroxidase (GPOX). Further, with the application of SA and SNP in high-temperature treated plants, the level of DNA methylation and demethylation was found to be higher than when SA and SNP were not applied [24].
Since PTMs are controlled by the redox status of the cell, protein conformations are adversely affected under oxidative stress. Oxidative stress-mediated PTMs can regulate cell cycle progression, and the maintenance of DNA methylation through cell division is a ubiquitin-dependent process. For instance, in wheat, exposure of seedlings to cadmium (Cd), methyl viologen and H2O2 induced oxidative stress and led to ubiquitylation of the cell cycle control protein, E2F/Rb related (RBR) transcription factors (TFs), leading, in turn, to the loss of protein function [25]. RBR/E2F association with chromatin factors (HDAC) plays a crucial role at every G1/S transition and participates in transcriptional regulatory complexes related to epigenetic changes [26], thus activating the cellular defence system. Further, the microenvironment and redox potential available to key amino acids determine cross-talk between protein and redox molecules. Generally, cysteine (Cys) and methionine (Met) residues are highly susceptible to reactive molecules. These findings indicate that redox changes affect epigenetic processes in plants and influence gene expression. The intrinsic mechanisms of redox changes during stress conditions in altering gene expression, particularly through epigenetic ways, are still not completely understood.
Intracellular epigenetic modifications
Intracellular modulations leading to epigenetic changes commonly occur in the nuclear matrix. PTMs introduced at the chromatin region constitute some of the major epigenetic focal points of the cell. Chromatins are organized nucleoprotein structures in the nucleus where the nucleosomes are arranged. Each nucleosome comprises histone proteins and DNA [27]. Each histone molecule ends with an N-terminal tail composed of various amino acids. These tails are where various PTMs occur. Various modifications were reported in histones, such as 13 types in H2A, 12 in H2B, and 21 each in H3 and H4 [28], [29], [30]. The most common histone-level PTMs are lysine acetylation, methylation, arginine methylation, citrullination, SUMOylation, carbonylation, phosphorylation, ubiquitylation, ADP ribosylation, proline isomerization and cytosine glutathionylation [28], [31], [32], [33], [34]. These PTMs are known to alter gene expression by modulating transcriptional regulatory protein binding and/or by influencing histone-DNA interactions. Among these PTMs, acetylation and methylation are well-characterized epigenetic events. Following heat treatment, Wang et al. [35] reported increased acetylation and decreased methylation in maize with increased ROS accumulation. The decreased methylation was concomitant with nucleolar enlargement, implying chromatin decondensation [36]. Wang et al. [35] found that demethylation among H3K9 histones increased superoxide scavenging by loosening DNA-histone contact points favoring gene expression.
Redox interplay during DNA methylation
Under stress conditions, changes in hormonal levels affect the redox status of the cellular system and are accompanied by changes in DNA and histone methylation. Maintenance of DNA methylation depends on (i) methyl source supplier, (ii) DNA methyltransferase (DNMT) activity, and (iii) DNA demethylase (DME) activity (Supplementary Fig. 1) [37]. In general, DNA methylation occurs at the promoter region and halts transcription. However, there are exceptions wherein methylation enhances gene transcription. This happens in the gene, repressor of silencing 1 (ROS1), wherein the methylation of the promoter improves transcription. ROS1 is an endonuclease III domain nuclear protein with bifunctional DNA glycosylase/lyase activity [38]. ROS1 acts only against methylated but not on unmethylated DNA, which establishes its DNA repair role. ROS1 requires a direct connection with the Fe-S assembly, which is highly influenced by the redox status of the cell [5].
Generally, increased accumulation of reactive species such as O2•- is reported to reduce DNA methylation. In tobacco cells, increased oxidative stress due to exposure to various inducers, such as aluminium, paraquat, salt and low temperature, activates the expression of glycerophosphodiesterase-like (NtGPDL) proteins. NtGPDL proteins are already demonstrated to be responsive to aluminium stress. Analyzing the methylation pattern through bisulfite methylation mapping at the genomic loci of the NtGPDL gene showed selective demethylation of the CG site in the coding region. Further, irrespective of the stress inducer, the promoter region was found to be unmethylated. Choi and Sano [39] proposed that plant responses to environmental stresses are partly regulated through epigenetic modifications such as DNA hypomethylation. Subsequent studies also confirm that increase in oxidative stress conditions could lead to DNA hypomethylation [40]. Treatment of tobacco suspension culture with a naphthoquinone allelopathic toxin, Juglone (5-hydroxy-1,4-naphthalenedione), was reported to induce overaccumulation of ROS with simultaneous DNA hypomethylation [41]. Similar accompaniment of DNA hypomethylation under oxidative stress induced by the treatment of Pisum sativum suspension culture with nicotinamide (precursor to NAD+) was reported by Berglund et al. [42]. Nicotinamide treatment was found to increase the level of GSH. Although epigenetic changes induced by increased GSH levels are well documented in animal and human cells [43], information regarding GSH-induced epigenetic alterations remains scarce in plants. Treatment of rice plants with a higher concentration of SNPs, NO donors, caused DNA hypomethylation [44]. Besides chemical stress inducers, physical factors, such as irradiation, were also found to induce demethylation. In Arabidopsis, irradiation of roots induced different methylation patterns on aerial plant parts, ranging from hemi-methylated to non-methylated, accompanied by ROS accumulation [45].
Epigenetic gene silencing in plants is established through the RNA-directed DNA methylation (RdDM) pathway involved with the production of small interfering RNAs (siRNA) [37]. Production of siRNA is mediated through the dicing activity of Dicer-like (DCL) proteins and the cleavage activity of RNase III-like (RTL) proteins. Both these proteins have conserved cysteine residue at the 230th position and are highly susceptible to oxidative stress conditions. In Arabidopsis, RTL2 regulates the expression of genes through the production of siRNA. In situ mutagenesis studies have shown that the cysteine 230 position is essential for cleavage leading to siRNA production. Incubation of RTL1 with GSH or GSSG was found to abolish RTL1 activity and prevented siRNA production by the glutathionylation of cysteine 230 under oxidative stress [46]. Similarly, regulation of DCL activity by SA and the redox status of the cell was established by Seta et al. [47], who found that DCL4 activity was enhanced by SA application through the accumulation of GSH. They concluded that the dicing activities of DCL3 and DCL4 were regulated by inorganic phosphate and the redox status of the cell, suggesting that homeostasis of gene silencing occurs according to the growing environment.
Regulation of SAM synthesis
The most common methyl source supplier in biological systems is S-adenyl methionine (SAM). SAM synthesis is established through (a) folate and (b) methionine cycles, as depicted in Fig. 2. The source of the methyl group for SAM production is the folate molecule. Folate enters through folate cycle pathways that involve bifunctional enzyme-like dihydrofolate reductase-thymidylate synthases (DHFR-TS) [4]. DHFR-TS helps in the conversion of dihydrofolate (DHF) to tetrahydrofolate (THF) by consuming the reducing equivalent from NADPH. Gorelova et al. [48] have shown that Arabidopsis DHFR mutants are incapable of assimilating folates. From folates, different derivatives are produced by different enzymes (Fig. 2). Homology-dependent gene silencing 1 (HOG1) is an enzyme that catalyzes the conversion of S-adenosyl homocysteine (SAH) to homocysteine (Hcy). During this process, SAM acts as a methyl donor for DNA methylation. In this manner, the Hcy/SAM cycle provides a constant supply of methyl groups for DNA methylation. The role of folate in driving this cycle is crucial, as observed in Arabidopsis, wherein impairment of folate production by sulfamethazine treatment was found to reduce DNA and histone methylation [49].
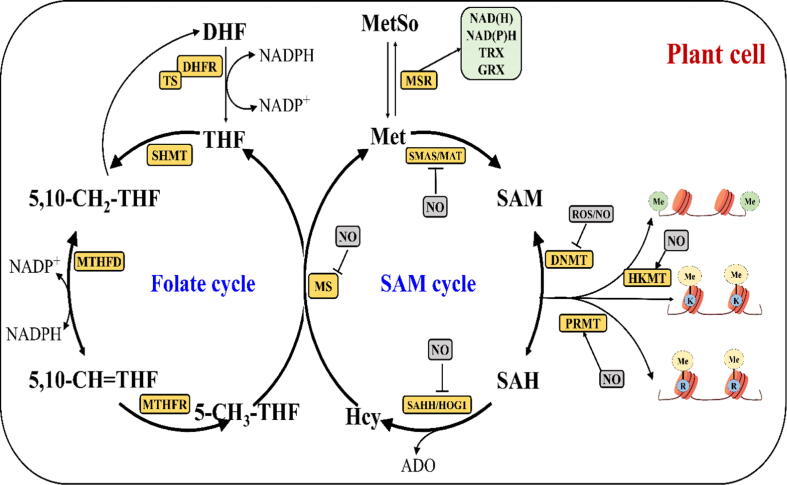
Fig. 2.
Redox components regulate S-adenyl methionine (SAM) synthesis through the folate cycle in plants. The folate cycle starts with the conversion of dihydrofolate (DHF) to tetrahydrofolate (THF) through dihydrofolate reductase-thymidylate synthase (DHFR-TS) by using reducing equivalents from NADPH (nicotinamide adenine dinucleotide phosphate). Methyl groups gained from THFs (5,10-CH2-THF, 5,10-CH = THF) are synthesized by serine hydroxymethyltransferase (SHMT) and methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase (MTHFD1). Methylenetetrahydrofolate reductase (MTHFR) reduces 5,10-CH = THF to 5-CH3-THF, from which the methyl group is transferred to homocysteine (Hcy) to synthesize methionine (Met) through the methionine synthase (MS) enzyme. Methionine synthesizes SAM through SAM synthase (SAMS), and SAM donates methyl groups to proteins or DNA through DNA methyltransferase (DNMT)/ histone lysine methyltransferase (HKMT)/ protein arginine N-methyltransferase (PRMT) and is converted to S-adenosyl homocysteine (SAH) which is further processed to Hcy through S-adenosyl homocysteine hydrolase/ homology-dependent gene silencing 1 (SAHH/HOG1). Redox components influence the following key enzymes: SAMS/methionine adenosyl transferases (MAT), DNMT/HKMT/PRMT, SAHH/HOG1, MS and methionine sulfoxide reductase (MSR). K: lysine; R: arginine; Me: methyl. ROS/NO is uncharacterized regulation. GRX, glutaredoxin; TRX, thioredoxin [4]. The schematic representation was adapted from Saravana Kumar et al. [4].
The folate cycle is the supplier of the methyl group to methionine and to the SAM cycle. It is found that enzymes involved in SAM synthesis through the folate cycle are regulated through the redox status of the cell or by using reducing equivalents (Table 1). Some of these enzymes, such as HOG1 [50] and MS [51] were found to undergo nitrosylation, the covalent modification of redox-sensitive cysteine residues by NO, suggesting that these enzymes are redox-regulated. Furthermore, during developmental processes and/or under stress, methionine undergoes oxidization to become methionine sulfoxide (MetSo), which, in turn, reverts to methionine by methionine sulfoxide reductase (MSR) activity, consuming NADPH in the process [52]. Evidence indicates that MSR enzyme activity is also redox-regulated in the cell. In Arabidopsis, GSH treatment was found to down-regulate MSR accumulation [53]. In vitro studies have shown that glutathionylation of MSR at the cysteine position plays a vital role in methionine regeneration. Lindermayr et al. [54] described nitrosylation of SAMS at the cysteine 113 position, following its incubation with nitrosoglutathione (GSNO), which abolished SAMS activity in Arabidopsis. They concluded that SAMS activity is influenced by the redox status of the cell. The redox status of the cell modulates DNA methylation by influencing one or more factors related to DNA hyper or hypomethylation. The fact that methylation efficiency depends on the reaction conditions between methyl group donors and acceptors, such as DNA or histones, suggests that the redox status of the cell can influence both donor and acceptor systems to induce epigenetic changes.
Table 1.
The list of enzymes involved in DNA methylation affected by the redox status of the cell.
| Epigenetic process and modifying enzyme | Redox status influence on enzymes | Described roles for the epigenetic process | References |
|---|---|---|---|
| Methionine sulfoxide reductase (MSR) | NADP increases the MSR transcript, and GSH helps regenerate MSR | MSR regenerates methionine from the oxidized methionine pool | [52], [55] |
| Homologous gene silencing 1 (HOG1) | Nitrosylation of HOG1 | Converts S-Adenosyl homocysteine to homocysteine | [50] |
| Detrahydrofolate reductase | NADPH is converted to NADP | Catalyzes the conversion of dihydrofolate to tetrahydrofolate | [48] |
| Methionine synthase (MS) | Nitrosylation | Converts homocysteine to methionine | [51] |
Note: NADP, nicotinamide adenine dinucleotide phosphate; GSH, glutathione.
Regulation of DNA methylation/demethylation
The redox status of the cell influences DNMT activity, inducing changes in methylation patterns. A methylation indicator, the presence of oxidative stress, has been shown to increase DNMT expression in plants. In in vitro grown birch plants, increased ROS activity in older calli elevated DNA methylation and was accompanied by an increased expression of DNMT genes such as domain rearranged methyltransferase (DRM), methyltransferase (MET) and chromomethylase (CMT) [56].
In demethylation, DNA demethylase activity is performed by DNA glycosylase by excising the entire methylated base instead of the methyl group alone. The removal of the methylated base from the methylated DNA region is accomplished through the base excision repair (BER) pathway. DNA glycosylase has an iron-sulfur (Fe-S) cluster assembly as a cofactor, which aids in gaining or losing electrons under oxidation. The assemblage of the Fe-S cluster with the apoprotein is mediated through the cytosolic Fe-S cluster assembly (CIA) pathway. The CIA pathway engages several scaffold and carrier proteins [57]. Demeter and ROS1 are 5-methylcytosine DNA glycosylases that play a role in fertilization [58]. This Fe-S cluster assembled DNA demethylase is activated during fertilization, following an oxidative burst in the central cell, leaving evidence that conditions similar to oxidative stress are required for demethylase activity.
Redox regulation of histone acetylation
Histone acetylation, another type of PTM, occurs in the nucleus. In contrast to methylation, acetylation of histones generally facilitates gene expression [59], although some histone methylation is also known to promote gene expression. In the cell, histone acetylation depends on the availability of the substrate, acetyl CoA, and is maintained through the antagonistic activity of two enzymes, HAT and HDAC. As with other PTMs, the activities of HAT/HADC enzymes, as well as acetyl CoA metabolism, are redox-regulated. Currently, several reports establish redox-mediated histone acetylation in plants and animals. Mengel et al. [60] report that, in Arabidopsis, treatment with NO or GSNO resulted in the inactivation of the HDAC complex, increasing histone acetylation. During heat-induced programmed cell death, increased accumulation of ROS was found to ensure hyperacetylation by overexpression of HAT in maize seedlings [35]. In sugar beet lines, Causevic et al. [61] have shown that different levels of ROS expression could direct different levels of histone acetylation.
Regulation of acetyl CoA
Acetyl CoA is a thioester between acetic acid and coenzyme A. The prime role of acetyl CoA is to act as an acetyl radical donor, a cofactor for several metabolic pathways in the cell, such as the Krebs cycle and the PTMs such as histone acetylation. Acetyl CoA is synthesized in plants in different cellular organelles such as mitochondria (Fig. 3), chloroplast, and peroxisomes. The oxidative stress in cells alters acetyl CoA metabolism, thereby significantly influencing histone acetylation [62]. In mitochondria, oxidative decarboxylation of pyruvate by the activity of the mitochondrial pyruvate dehydrogenase (mPDH) complex results in the generation of acetyl CoA. Oxidative decarboxylation of pyruvate is an integral step of the glycolysis pathway that occurs before the entry of acetyl CoA into the Krebs cycle. During this process, the PDH complex uses NAD+ as a cofactor for its catalytic activity, converts pyruvate to acetyl CoA and releases CO2 [63]. In Arabidopsis, it has been shown that altering the redox status (increased NADH/NAD + ) under in vitro conditions inhibits mPDH complex activity [64]. In a 2019 review, Dumont and Rivoal [65] point out that, in Arabidopsis, the PTMs of enzymes are involved in the tricarboxylic acid (TCA) cycle in a redox-dependent manner. The inhibition of the redox-dependent TCA cycle increases acetyl CoA and decreases ROS production by increasing citrate concentrations to increase histone acetylation. In roots and seedlings, pyruvate was shown to increase by oxidative stress. This increase impacts TCA cycle inhibition, suggesting that the redox status influences acetyl CoA synthesis.
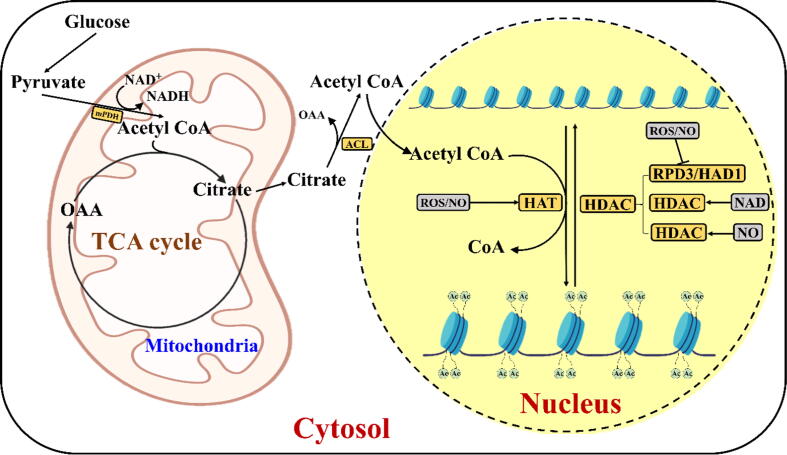
Fig. 3.
Acetyl CoA synthesis in the mitochondrial matrix and the regulation of histone acetylations by redox components. In the cytoplasm, the breakdown of glucose produces pyruvate, which enters the tricarboxylic acid (TCA) cycle that occurs in the mitochondria. Then, pyruvate is converted to acetyl-CoA through mitochondrial pyruvate dehydrogenase (mPDH) by reducing NAD +. Combining with acetyl-CoA, oxaloacetate (OAA) formed in the TCA cycle produces citrate. After entering the cytoplasm, citrate is converted back to OAA and acetyl-CoA through ATP-citrate lyase (ACL). Synthesized acetyl-CoA enters the nucleus and provides the acetyl group to the histone acetyltransferase (HAT) for histone acetylation. After receiving the acetyl group, HAT introduces acetylation marks (Ac) over the lysine residues of the histone tail, thereby loosening the nucleosome structure (DNA and histones) and facilitating gene expression. Histone deacetylase (HDAC) removes histone acetyl groups from the histone tail, thus leading to chromatin compaction. ROS, NO, and NAD+ influence different HAT and HDAC enzymes [4]. The schematic representation was adapted from Saravana Kumar et al. [4].
The other two mechanisms that generate acetyl CoA include fatty acid β-oxidation and amino acid catabolism. Amino acid catabolism involves amino acids such as leucine, lysine, phenylalanine, tryptophan and tyrosine. The catalytic conversion of acetyl CoA into fatty acids is regulated by the enzyme acetyl-CoA carboxylase 1 (ACC1) in animals and plants. The acc1 mutants of Arabidopsis that are defective in fatty acid synthesis are reported to accumulate a high level of acetyl CoA [66]. A high accumulation of acetyl CoA could induce increased histone acetylation [67].
Regulation of histone acetyltransferase
HAT enzymes mediate acetylation by combining with other factors to form a HAT complex, whose activity is affected by oxidative stress. The elongator protein, a component of the HAT complex, is known to mediate ABA response, anthocyanin biosynthesis and oxidative stress tolerance in Arabidopsis [68]. In maize, ROS generation occurs in seedlings exposed to elevated salinity conditions in the form of increased electrolyte leakage and H2O2 production, thus inducing several antioxidant pathway genes [69]. A collateral increase in HAT enzyme expressions, such as ZmHATB and ZmGCN5, was observed and was accompanied by the upregulation of cell wall synthesis genes, ZmEXPB2 (expansin-B2) and ZmXET (xyloglucan endotransglycosylase). Upregulation of cell wall synthesis genes was mediated by increased acetylation in promoter regions, evidence that HAT enzymes mediate the expression of various stress-responsive genes by modifying their acetylation status during stress [70].
Regulation of histone deacetylases
Available evidence shows that HDAC expression is primarily regulated by the redox status of the cell leading to histone deacetylation (Supplementary Table 2). Different PTMs mediated by the redox status of the cell also affect HDAC activity. HDACs are enzymes with deacetylation activity that are classified based on both mechanistic similarity and phylogenetic comparison [71]. There are generally two types, the predominant non-sirtuin types and the sirtuin types. Non-sirtuin HDACs have Zn or Fe ions embedded in their structure while sirtuins use NAD+ as a cofactor. A comprehensive phylogenetic analysis of HDAC-like sequences, across sequenced genomes, established that non-sirtuin HDAC enzymes are best classified into three distinct groups, Class I, II, and IV, while the sirtuin-like enzymes remained classified in Class III. Class I HDACs are HDA1, HDA2, HDA3 and HDAC8; Class II includes HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10; Class III includes sirtuin-2 like (SIRT) HDACs such as SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7; and Class IV contains HDAC11. Arabidopsis HDACs, for example, constitute 18 genes, grouped as RPD3 (reduced potassium dependency 3)/HDA1 (histone deacetylase 1) (RPD3/HDA1), SIR2 (silent information regulator 2), and HD2, the plant-specific histone deacetylase 2 [71].
Non-sirtuin HDACs
Non-sirtuins, the major group of HDACs, are predominantly involved in regulating histone acetylation in cells. Under oxidative stress, HDACs are found to be unrecruited from gene repression, driving increased acetylation and consequent gene expression. Ding et al. [72] report that, in rice, during the compatible host-pathogen interaction, the transcript level of HDT701 (histone 4 deacetylase 701), an HD2 subfamily gene, is increased, in contrast to its lowered expression during incompatible interaction. They found that transgenic overexpression of HDT701 led to a reduction in H4 acetylation leading to susceptibility to rice pathogens, salt, drought and osmotic stresses. Decreased H4 acetylation is associated with delayed seed germination [72], [73]. The pattern was found to be reversed with the silencing of HDT701, whereupon elevated H4 acetylation was observed along with an increased transcription of PRRs and defense genes. Additionally, treating with pathogen-associated molecular pattern (PAMP) elicitors, flg22 and chitin, increased ROS levels in transgenic plants. Increased transcription of defense genes was found to be linked to enhanced acetylation marks in their promoters.
Non-sirtuin HDAC members are redox-sensitive and are produced during different oxidative stress conditions. In plants, HDACs get inactivated under oxidative stress to increase histone acetylation which helps the expression of many abiotic stress resistance genes [74]. There are several reports of redox switch regulation of HDACs such as HDA1, HDA2, HDA3, HDAC4, HDAC5, HDAC6 and HDAC8 during oxidative stress conditions [74].
PTMs involving HDACs either make proteins inactive or loosen contact with the surrounding DNA. Among the deacetylases, HDA2 is among the most characterized. Ito et al. [75] demonstrated that S-nitrosylation of HDA2 leads to hyperacetylation. When H2O2 was administered, S-nitrosylation was induced on HDA2, driving histone acetylation. Similarly, S-nitrosylation of HDAC6, HDAC8 was also found to occur under oxidative stress [74]. Plants are also observed to possess HDA6, a homolog of human HDAC1 [76] and yeast Reduced potassium dependency 3 (RPD3) [77]. Plant HDACs work as a complex, and S-nitrosylation of either HDACs or their complexes could result in the inactivation of HDACs [78]. In general, HDACs lack DNA binding properties and, hence, form complexes with large multiprotein sub-units.
Like any other stress response genes, HDACs also undergo PTMs under stress, when the cysteine residue changes to alter protein development, functions and subsequent protein–protein interactions. Mengel et al. [60] have shown that the application of GSH to Arabidopsis seedlings grown in a liquid medium increased histone acetylation due to the reduction of HDAC activity. In vitro studies have revealed that GSSG mediates protein glutathionylation at the cysteine position. In Arabidopsis, the expression of genes related to SA biosynthesis and SA-mediated defense-associated pathogenesis-related (PR) proteins were repressed by an RPD3/HDA1 class deacetylase. In the case of HDA19, which deacetylates the promoter regions, oxidative stress conditions induced by pathogen attack abolish HDA19 activity to drive promoter acetylation leading to SA accumulation and PR protein expression inciting defense response [79]. Later, Liu et al. [80] demonstrated that increased ROS production, after the addition of SA and flg22 into Arabidopsis cell suspension cultures, led to the oxidation of two deacetylases, HDA9 and HDA19, improving the acetylation of stress-responsive genes.
Likewise, in Arabidopsis under cold conditions, the CULLIN4-based ubiquitin E3 ligase complex promotes HD2C degradation (increased acetylation levels of histone H3), which allows the activation of cold-responsive (COR) genes [81]. Further, HD2C interacts with other HDACs to either increase or decrease acetylation levels to regulate stress tolerance. For instance, in Arabidopsis, in double mutant plants (hda6, hd2c, and hda6/hd2c-1), HD2C directly interacted with HDA6, was bound to histone H3 and was associated with increased histone H3K9K14 acetylation and decreased histone H3K9 dimethylation, thus activating ABA-responsive genes [82]. In addition, HD2C interaction with the BRM-containing SWI/SNF chromatin remodeling complex regulates Arabidopsis heat stress response [83]. According to ROS status in the cell, plants require either low or high levels of acetylation to regulate stress tolerance. In Arabidopsis, the HDA19-MSI1 complex maintains low levels of acetylation of histone H3 at lysine 9 and fine-tunes ABA signaling [84]. HDA19 formation with other proteins is also important for brassinosteroid signaling to facilitate histone deacetylation of ABI3 chromatin [85], suggesting that the redox status of the cell regulates plant stress tolerance through histone modifications.
Sirtuin HDACs
Sirtuin HDACs use NADH, a redox carrier cofactor molecule known for its deacetylation activity. NADH is a small metabolite transporting reducing equivalents to different cell compartments [71]. NADH shuttles between oxidized (NAD+) and reduced states through electron exchange, mediating different metabolic reactions inside the cell. The transition of NADH between the two oxidation states depends on the redox status of the cell, qualifying it as an indicator of energy and the redox status of the cell. Although the redox status of the cell and its impact on NADH is not well documented in plants, it is widely accepted that, under oxidative stress conditions, the redox status of NAD+ shifts towards a more reduced state. Since the activity of sirtuin HDACs is NAD+ dependent, the cellular NAD+ level and the NAD+/NADH ratio are the major determinants of their deacetylation activity [86]. Generally, sirtuins are assemblers of heterochromatin (silent information) during gene expression. However, sirtuin type class III HDACs are involved in the deacetylation of histones as well as non-histone proteins.
Among seven sirtuin isoforms (SIRT1-7), the activity of the SIRT2 gene involves several PTMs that are redox-mediated. Under oxidative stress, various protein kinases, which introduce PTMs in the SIRT1, are activated. Common oxidative stress-induced protein kinases are casein kinase II (CKII) [87], cell division control 2 (Cdc2) [88] and 5′ adenosine monophosphate-activated protein kinase (AMPK) [89]. Phosphorylation of SIRT2 by protein kinases makes it dysfunctional, leading to increased acetylation. Although SRT1 expression is promoted under mild oxidative stress conditions, during severe oxidative stress, SRT1 gets inactivated through PTMs such as glutathionylation [90] and nitrosylation [91]. Additionally, NAD+ depletion under oxidative stress can also suppress SRT1 activity [92]. In plants, cellular redox status recruits SIRT genes to alter the expression of ROS responsive genes through NAD+ mediated activity. Unlike in animal systems, the number of sirtuins reported in plant systems is low [93]. Two SIRT genes each are reported from Arabidopsis (AtSRT1 and AtSRT2) and rice (OsSRT1 and OsSRT2) [94], and one from durum wheat (WhSRT2) [95].
Redox regulation of histone methylation
Unlike histone acetylation, methylation does not affect the net charge of histone residues but modifies the bulkiness instead. The bulkiness or hydrophobicity between the residues opens up new platforms for the binding of other regulatory proteins. Classified as mono-, di- and tri-methylation, based on the number of methyl groups attached over the histone molecules, different methylations have different effects on the expression of genes. For instance, trimethylation of the 27th lysine residue of H3 (H3K27me3), usually carried out by the polycomb group (PcG) protein, leads to the repression of genes, while trimethylation at the fourth lysine of H3 (H3K4me3), catalyzed by Arabidopsis Trithorax (ATX) group proteins, leads to active transcription [96]. Like other PTMs, histone methylation is also affected by various redox components in the cell.
As with DNA methylation, SAM is the source supplier of the methyl group (added to the lysine and arginine residues of histones) for histone methylation. In Andropogon virginicus, AvSAMS gene expression is induced by different metal toxicities and other oxidative stress conditions [97]. When Arabidopsis is transformed with the AvSAMS gene, the transgenic plants confirm increased tolerance to aluminum and other metal ions; during aluminum stress, the histone methylation level of the differentially expressed genes was significantly altered compared to the level in wild-type plants. Ezaki et al. [97] also concluded that SAMS expression under oxidative stress-regulated histone methylation influences the expression of stress-responsive genes. Since folate is necessary for SAM production and to carry out the methionine cycle, folate inhibitors can play a significant role in controlling methylation. Sulfamethazine is a potential antagonist to folate synthesis in the cell, affecting epigenetic modifications. When applied, sulfamethazine reduces the folate level in plants, thereby reducing DNA and histone methylation [49]. Although stress-mediated histone methylation is known, the details of interconnectivity between oxidative stress and histone methyltransferase/demethylase reactions are seldom detailed.
Regulation of histone methyltransferase
Histone methyltransferases (HMTs) catalyze the addition of one or more methyl groups to histone residues, particularly those of H3 and H4 histones. Using SAM as the methyl donor, histone-lysine N-methyltransferases (HKMT) and histone-arginine N-methyltransferases (HRMT) mediate methylation. Methyltransferase activity is influenced by stress-induced redox modification whereupon the methylome of stress-specific genes is controlled [98]. Depending upon the target residue, HMTs are of two types, lysine-specific HKMT and arginine-specific HRMT.
HKMT proteins possess conserved SET (Suppressor of variegation 3–9, Enhancer of Zeste, and Trithorax) domains for their catalytic activity. The HMTs of the SET domain-containing group (SDG) (SDG-HMTs) is found to be distributed across yeast, plants and animals [99]. SDG-HMTs are known to alter gene expression either at the transcriptional or post-transcriptional level. In mammalian cells, it was shown that ROS regulate the expression of mitochondrial and antioxidant genes by the action of the SET domain (SETD7) group of proteins [100]. Unlike in animal cells, direct evidence interlinking the redox status of the cell with SDG-mediated gene expression is lacking in plant cells. For instance, in Arabidopsis, it is well known that exposure to light and carbon alters the redox status of the cell. Li et al. [101] report that methylation patterns in light and carbon responsive genes are altered by SDG8 activity. Belonging to the SDG-HMT group, Arabidopsis Trithorax1 (ATX1) mediates H2O2 signaling. Kaurilind et al. [102] demonstrated that, when atx mutants were integrated with Arabidopsis catalase 2 (cat2) mutants, lesion development in the mutants got suppressed. The lesions in cat2 mutants are due to the excessive accumulation of H2O2, which leads to programmed cell death. Lesion suppression by the introduction of atx mutants implied that ATX1 mediates H2O2 signaling in the programmed cell death pathway.
Similarly, HRMT, also known as protein arginine N-methyltransferase (PRMT), regulates different developmental and stress responses in plants. A well-characterized PRMT in Arabidopsis, PRMT5, acts as a redox signaling mediator [103]. PRMT5 induces a di-methylation of the third arginine residue of the H4 histone (H4R3me2) and suppresses the expression of the flowering locus C (FLC) as well as of salt responsive genes [104], [105]. Under salinity stress, PRMT5 acts as a signal transducer of NO by methylating histone residues. The NO produced induces S-nitrosylation of PRMT5, thereby inducing its activity. The induced PRMT5 methylates the spliceosome of stress-response pre-mRNA. The change in the methylation status of the spliceosome affects protein splicing activities and leads to the development of alternative splice variants [105]. The production of these alternate variants increases the number of stress-specific gene products. Such post-transcriptional level variations are induced by NO through S-nitrosylation to regulate the production of stress-specific mRNA transcripts [103].
Regulation of histone demethylase reactions
Histone demethylases (HDMs) mediate the removal of the methyl group from histone molecules. HDMs are of two classes, the Jumonji C (JmjC) domain-containing histone demethylase (JHDM) and the lysine-specific demethylase (LSD) [106]. LSD belongs to a flavin adenosine dinucleotide (FAD)-dependent amine oxidase, while JHDM belongs to the ferrous and α-ketoglutarate-dependent dioxygenase family. Both enzymes follow different pathways to demethylate histones and use different cofactors for their activities [107]. The oxidative stress influence on these demethylase enzymes is directed through their cofactors, which, in turn, regulate enzyme activity and are influenced by the cellular environment.
Among the LSDs, LSD1 is the first characterized histone demethylase. It uses FAD as its cofactor [107]. Forneris et al. [108], [109] have shown that the demethylation reaction catalyzed by LSD1 is a flavin-dependent oxidative process in which the methylated lysine is oxidized. LSD1 essentially requires protonated nitrogen for its activity and is relatively active over mono- and di-methylated residues. In vitro studies have shown that the FAD cofactor in the enzyme donates electrons to the protonated nitrogen atom and becomes reduced with concomitant oxidation of C-N-methylamine. The resulting imine group reacts with a water molecule to form formaldehyde together with the removal of the methyl group. During this process, flavin undergoes redox switching and this property makes it a key player in transcription, cellular signaling and abiotic stress responses [110], [111]. Expression analysis has shown that LSD1 expression is directly proportional to the level of demethylation of mono-/di-methylated lysine residues in histone 3 (H3K4me1/2), leading to higher gene expression.
The second class of HDMs, the JHDMs, catalyzes the demethylation of mono-, di-, and tri-methylated histones in animals and plants [106]. JHDMs remove histone methylation marks while associating with wide functions in plant development [112], [113]. The JmjC domain protein of these HDMs harbors ferrous ions and α-ketoglutarate as cofactors for their activities. Oxidation of ferrous to ferric form leads to the inactivation of JmjC containing protein, promoting hypermethylation. Arabidopsis jmj16 mutants with aberrant JmjC domain-containing protein, Jmj16, accumulate a higher level of ROS accompanied by H3K4me3 hypermethylation [114]. Hypermethylation induces early leaf senescence by increasing the expression of senescence-associated genes such as WRKY53 (tryptophan (W) - arginine (R) - lysine (K) - tyrosine (Y) 53) and SAG201 (senescence-associated gene 201), indicating that Jmj16 is essential for maintaining the homeostatic level of ROS during oxidative stress and for subsequent methylation events.
Redox regulation of miRNA biogenesis
MicroRNAs (miRNAs) are yet another intermediary of gene regulation in plant systems. Most of their activities are epigenetic, such as heterochromatin formation, methylation, histone modification and gene silencing. In plants, evidence from several species indicates a strong interconnection between the redox status of the cell and miRNAs production. The type of oxidative stress was found to influence the expression of specific miRNAs, depending upon the plant species involved (Supplementary Table 3) [115]. In wheat, exposure to ozone was found to induce the expression of 21 ozone-specific miRNAs that are uninfluenced by H2O2 exposure [116]. Similarly, Jia et al. [117] have shown that the expression of miR156 was induced by ROS and GSH but not by H2O2. Apart from these, H2O2-responsive miRNAs were identified in rice [118], wheat [116] and Brachypodium [119]. When exposed to similar levels of H2O2, there were significant miRNA expression differences between species (Supplementary Table 3). Only two miRNAs (miR169 and miR528) were found to have similar expression levels between rice and Brachypodium under similar treatment conditions [118], [119]. In apple, the vegetative phase change is under the regulatory control of miR156, which is under the control of plastid-nucleus redox signals. When in vitro apple shoots were exposed to chemical inhibitors, the expression of miR156 was found to be concomitantly affected by GSH content and not by H2O2 level [117]. This shows that redox signaling works upstream of miRNA production.
Despite mounting evidence on the influence of redox elements on miRNA expression (Fig. 4), direct evidence remains scanty on how they influence the expression of different accessory proteins. Mutant plants with defective accessory proteins are compromised in miRNA biogenesis as seen among Arabidopsis mutants, dcl1, hen1 and hyl1 [120]. However, indirect evidence has shown that miRNA synthesis is inhibited by feedback mechanisms on which oxidative stress conditions have a significant influence. When oxidative stress affects miR162 and miR168, their feedback mechanisms also influence the proteins, DCL1 and AGO1, respectively [121], [122]. Although the DCL1 protein is involved in the production of miR162, it also binds to the stem-loop structure of miR162. In turn, in the feedback reaction, the redox-sensitive miR162 targets and cleaves DCL1 mRNA and pre-mRNAs [122]. In the case of dcl1 mutants, wherein the functional, active DCL1 protein is absent, miR162 biosynthesis is impaired. However, in wild-type plants, DCL1 production was restricted by the miR162 feedback, resulting in miR162 homeostasis. Similarly, AGO1 homeostasis is also under the feedback regulation of miR168 [121].
Fig. 4.
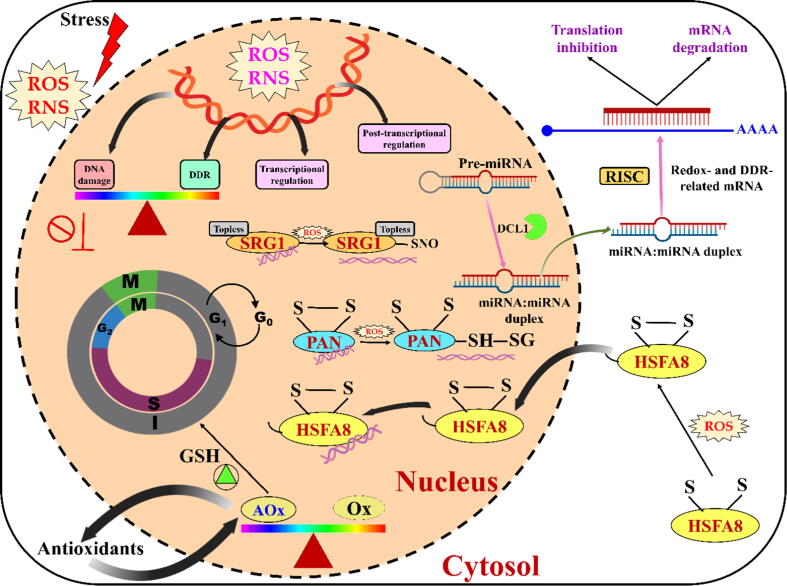
Cellular redox balance-DNA damage response (DDR)-miRNA triangle. Under various stress conditions, increased ROS production is usually present in different cell compartments, such as the nucleus, cytoplasm, etc., through which ROS and redox signals move to regulate gene expression. ROS accumulation in the nucleus can arrest the cell cycle by inducing DNA damage; however, the DDR reduces the negative effects of DNA damage by modulating the activity of microRNAs. To promote the cell cycle according to the redox environment, altered antioxidant and oxidant balance is required in the nucleus. In this circumstance, ROS and redox signals regulate gene expression at transcriptional and post-transcriptional levels. The picture illustrates redox-dependent transcriptional mechanisms involved in the regulation of redox-sensitive TF (SRG1, PAN, and HSFA8) expression to regulate gene expression affected by ROS/RNS. In fact, at the post-transcriptional level, miRNAs can also target mRNAs (redox- and DDR-related target mRNAs) to inhibit the translation of negative regulation of stress tolerance. miRNA, microRNA; ROS, reactive oxygen species; RNS, reactive nitrogen species; AOx, antioxidants; Ox, oxidants; DCL1, DICER-like1; PAN, perianthia; SRG1, SNO-regulated gene1; HSFA8, heat shock factor A8 [115]. The schematic representation was adapted from Cimini et al. [115].
Mediation of miRNA expression
Many oxidative stress conditions mediate the expression of miRNAs (Fig. 5) through the GSH-dependent redox signaling pathway. Cao et al. [116] showed induction of miRNA expression following H2O2 treatment in wheat. In Arabidopsis, Jagadeeswaran et al. [123] have revealed a redox signaling-dependent production of miR395, induced under sulfur deficiency. This miRNA is known to target a low-affinity transporter gene, Arabidopsis sulfate transporter 68 (AST68), and three ATP sulfurylases, APS1, APS3 and APS4. Similarly, miR395 production is found to be promoted under oxidative stress abetted by sulfur deprivation, the mechanism of which remains unknown. Analysis of miR395 expression within various genetic backgrounds suggests that miR395 biogenesis involves more pathways than oxidative stress. GSH dependency for miRNA production and expression was evident from the partial impairment of miR395 production in the GSH deficient cadmium sensitive mutant, cad2-1 [123]. In the case of an Arabidopsis triple mutant, carrying mutations in nitroreductase genes, ntra and ntrb, along with cad2, miR395 reduction is found to occur by up to 45%. Likewise, redox homeostatic or redox signaling components have been shown to play a vital role in miR395 biogenesis under different mutant backgrounds. Conversely, some mutants with impaired GSH synthesis were found to have miR395 production, indicating that miR395 expression could also be under the regulatory influence of pathways other than the oxidative stress pathway. Apart from this, the expression of miR398, which targets Cu/Zn superoxide dismutases, CSD1 and CSD2, was found to be regulated under oxidative stress conditions [123]. Sulfur deprivation-induced oxidative stress was found to leverage a high level of CSD1 accumulation, with concomitant down-regulation of miR398 expression. Further supporting this, no CSD1 accumulation was observed in the redox signaling mutants. With observations of specific patterns of miRNA expression, possibilities of occurrence of specific mediatory pathways under specific stress conditions cannot be ruled out and require additional experimental evidence.
Fig. 5.
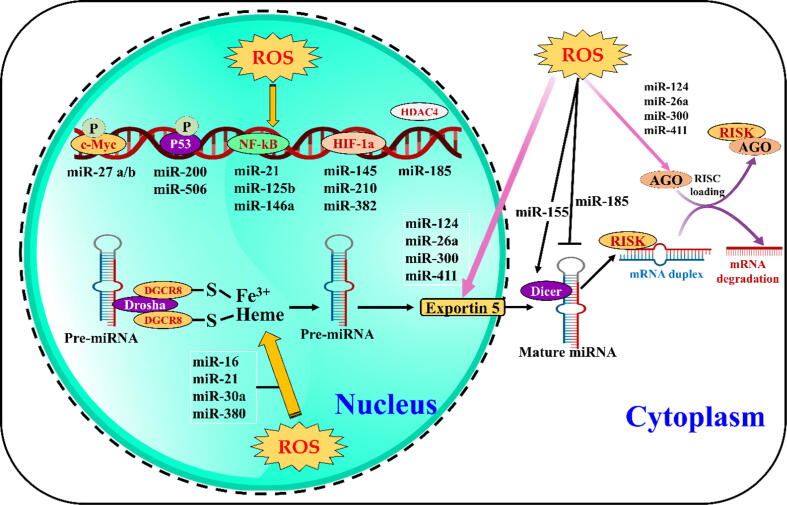
miRNA regulation under oxidative stress. ROS influence miRNA processes in the nucleus and cytoplasm and induce miRNA transcription by triggering transcription factors (TFs), such as c-Myc, p53, NF-κB, and HIF-1α. When TFs bind to the promoter, HDAC4 can suppress the TFs. ROS can regulate the expression of DGCR8, drosha, exportin 5 and dicer enzymes, thereby affecting miRNA biogenesis, translocation and maturation. Arrow and T arrow indicate activation and inhibition, respectively. miRNAs, microRNAs; ROS, reactive oxygen species; HDAC4, histone deacetylase 4; DGCR8, DiGeorge syndrome critical region 8; AGO, Argonaute; RISC, RNA-induced silencing complex [124]. The schematic representation was adapted from Carbonell and Gomes [124].
Redox regulation of chromatin structure and remodeling
As the carrier of genetic information, chromatin biology is all about the regulation of gene expression for cellular functions related to development and metabolism. Chromatin shuttles between compact and relaxed forms depending on the internal cellular conditions. Structural compaction of the chromatin generally halts transcription, while relaxation drives transcription and gene expression. For example, under low temperature, chromatin dynamics can regulate histone modifications, DNA methylation and ROS status, thereby activating cold-responsive genes [125] (Fig. 6) and flower induction during vernalization. Organizational modification of chromatin may be either covalent such as epigenetic (DNA methylation and histone post-translational modification), or non-covalent via chromatin modifications and remodeling [126]. As a result, the structural organization of chromatin is decided by histone status, histone variants and structure protein remodelers. ATP-dependent chromatin remodeler and poly (ADP-ribosyl) polymerase are two common remodeler proteins (PARP). Histone modifications include multiple changes in the amino acids of histone codes in the histone tail and interactions of regulatory proteins with modified histones are among the functions of histone modifications [127]. Modified histones tend to interact with chromo- and bromodomain proteins, which alter chromatin organization. Furthermore, interacting regulatory proteins have broader roles in DNA repair, recombination, and other plant developmental activities [128]. According to the level of arrangement and active involvement in transcription, Roudier et al. [129] and Wang et al. [130] have classified chromatin into four types: chromatin states 1 to 4 (CS1-CS4). CS1 represents the active state, CS2 the repressive state, CS3 the silent transposon and CS4 the intergenic regions.
Fig. 6.
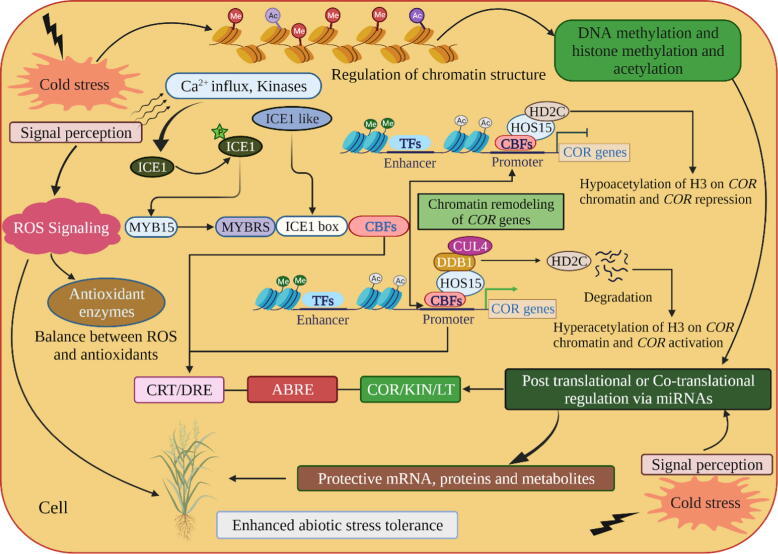
Schematic representation of epigenetic changes at low temperature. Increased levels of ROS and antioxidants under low temperature are associated with chromatin structure which triggers DNA and histone modifications, thereby activating COR genes. For example, in normal conditions, the HOS15 and HD2C complex represses COR gene expression due to hypoacetylation of H3 on COR chromatin. Under cold conditions, histone deacetylase 2C (HD2C) degradation triggers the hyperacetylation of H3 on COR chromatin and, thus, activates COR genes through CBF TFs. Using the high expression of the osmotically responsive gene 15 (HOS15) as a substrate receptor, the CULLIN4-based ubiquitin E3 ligase complex (CUL4) promotes HD2C degradation. The association of HOS15 with COR gene chromatin and HD2C degradation requires Powerdress (PWR), a HOS15-interacting protein. The HOS15/PWR complex and CBF TFs bind to the promoters of COR genes [81], [131]. Likewise, chromatin changes also activate miRNAs that influence COR gene expression through post-transcriptional modification. Cold stress also stimulates calcium influx which triggers protein kinases to activate ICE1. Activated ICE1 suppresses MYB15 and activates the expression of CBFs, thus regulating COR genes. A small star circle represents post-transcriptional modification, such as phosphorylation. ROS, reactive oxygen species; COR, cold-responsive genes; miRNAs, microRNAs; ICE1, an inducer of CBF expression 1; CBF, C-repeat binding factor; CRT, C-repeat elements; DRE, dehydration-responsive elements; ABRE, ABA-responsive element; KIN, cold-induced genes [125].
Regulation of ATP-dependent chromatin remodelers
ATP-dependent chromatin remodelling complexes (CRC) are nuclear enzymes that mediate the interaction between histones and DNA, to appreciate chromatin plasticity. They are multi-subunit protein complexes that are chromatin modifiers and remodelers and work in harmony to alter chromatin structure. Polycomb group (PcG) and trithorax group (TrxG) proteins constitute chromatin modifiers and interact with remodelers to alter chromatin organization [132]. Both groups of proteins work contrariwise in gene expression, wherein PcG maintains a repressed state, while TrxG maintains an active state. In Arabidopsis, PcG interacts with polycomb repressive complex 2 (PRC2) to promote the trimethylation of H3K27 to aid suppression of gene expression, while TrxG derepresses target genes and facilitates gene transcription [133], [134], [135]. These ATP-dependent chromatin remodelers utilize energy from ATP and mediate the alteration of histone-DNA modification. In a well-studied remodeler system, switch 2/sucrose non-fermenter 2 (SWI2/SNF2), the protein complex, comprising of protein subunits such as SNF5, SWP73 and SWI3, binds with the promoter region of the gene where different post-translational marks occur [136], [137]. Oxidative stress conditions influence the binding of the complex with the promoter and bring in regulatory changes [138], [139], [140], [141]. Similarly, gene expression demands are met at various developmental stages when histone modifiers and remodelers alter chromatin structural organization [128], [142]. In this review, we restrict the discussions to vernalization and stem cell development which are complex traits that depend on environmental variability.
Vernalization
Plants growing in temperate regions require vernalization, a procedure of flower initiation by exposure of seeds or plants to low temperatures (1–7 °C). Throughout vernalization, flower induction is facilitated through a series of gene expression events that involve chromatin rearrangement from the condensed state to the transcriptionally accessible state. In Arabidopsis and several other angiosperms, flowering is under the control of a floral repressor gene, which blocks a series of flowering-related genes to make the plants go into a non-flowering phase. One such gene is FLC, which encodes a MADS (minichromosome maintenance factor 1/ agamous (AG)/ deficiens/ serum response factor (SRF)-box TF) to suppress the floral integrator gene, thus upholding the vegetative phase [143]. FLC expression is regulated by different epigenetic pathways which are influenced by the redox status of the cell. During vernalization, FLC is attenuated by low-temperature exposure, thereby inducing flowering. Other proteins, such as vernalization 1 (VRN1), vernalization insensitive 3 (VIN3), and vernalization 5 (VRN5) /VIN3-like 1 (VIL1), are involved in vernalization [144], [145]. FLC attenuation occurs through chromatin rearrangement, wherein histone acetylation marks are erased [146], [147], [148]. FLC repression is mediated through VIN3 proteins, which bind upstream to the FLC region [146]. FLC repression requires trimethylation of H3K27 residue and deacetylation of H3 proteins; thus, VIN3 encodes for a plant homeodomain (PHD) protein interacting with the PRC2 to induce trimethylation and deacetylation of histone proteins, thereby leading to FLC attenuation [149], [150], [151]. In Arabidopsis, the redox regulation of VIN3 and PRC2 expression has been demonstrated and it was found that the expression of VIN3 is induced by nicotinamide treatment and, also, during hypoxic conditions. Likewise, chromatin-mediated gene expressions at the time of hypoxia are initiated [152] in which ROS and other oxidative stress conditions regulate the activity of PRC2. The PRC2 complex consists of four subunits, among which the VRN2-PRC2 subunit is responsible for substrate methylation. However, the expression of VRN2-PRC2 is oxygen level-dependent. So, any change in the O2 level will affect VRN2 activity [153]. NO-regulated protein arginine methyltransferase 5 (PRMT5) induces H4R3 demethylation in FLC gene promoter regions to reduce FLC expression to suppress flowering [154].
Stem cell development
During plant developmental processes, the redox regulation of chromatin plasticity is well accounted for in plants. The decision for a defined growth is met through stem cells present in the shoot apical meristem (SAM) and root apical meristem (RAM) regions [155], [156]. Meristematic activity and cell differentiation in the SAM are maintained by the wuschel (WUS) genes and in the RAM by wuschel-related homeobox 5 (WOX5) genes. The maintenance of stem cell activity and the differentiation process involve different forms of ROS [157], [158]. The accumulation of O2•− in the stem cell niche increases stem cell activity by activating WUS expression, while H2O2 accumulation in the periphery suppresses stem cell activity and allows for differentiation by inhibiting WUS activity. Both SAM and RAM developments in plants are under regulation by the redox status of the cell. In root cells, an increase in O2•− was found in the stem cell niche, while, in the periphery, H2O2 accumulation was found to be high. Thus, throughout stem cell development, redox modulation plays a significant role. Apart from WOX5 and WUS, there are other TFs involved in the stem cell process, such as plethora (PLT). Encodes for Apetala 2 (AP2) type TFs, PLT1 and PLT2, are involved in the maintenance of the root stem cell niche in Arabidopsis [159]. High-level PLT expression maintains stem cell activity, while low levels promote cell differentiation [160]. ABA-induced production of ROS is also known to affect the expression of PLT [161], [162]. In Arabidopsis, a pentatricopeptide repeat (PPR) protein, ABA Overly sensitive 8, (ABO8), maintains redox homeostasis. In the case of abo8 mutants, overaccumulation of ROS represses PLT1/2 expression [161]. The general control non-repressed 5 (GCN5) protein, belonging to the HAT family, is the first HAT that was found to associate histone acetylation to transcriptional regulation [163], [164]. GCN5, along with the adenosine deaminase 2 (ADA2b) transcriptional adapter (for alteration/deficiency in activation 2b), regulates the expression of PLT1/2 [165]. Among the several TrxG proteins reported in Arabidopsis, SDG2 regulates stem cell activity by maintaining H3K4me3 levels. It is observed that the chromatin assembly factor 1 (CAF1) works in a synergetic way along with SDG2 to regulate PLT1/2 expression [166]. The antagonistic activity of chromatin remodeling factor, Pickle (PKL), and the PcG protein, Curly leaf (CLF), decides root stem cell activity through the regulation of H3K27me3 [167].
Regulation of PARP1
Poly ADP- ribosylation (PAR) is a post-translational modification of histone and non-histone proteins carried out by the enzyme, poly (ADP-ribose) polymerase (PARP) [168]. In this process, NAD+ supplies ADP-ribose and releases nicotinamide as a by-product. Members of this protein possess a catalytic domain, the PARP signature, which catalyzes the addition of ADP-ribose to proteins [169]. Under stress conditions, the activity of PARP is essential to repair DNA [170]. Histone PAR makes the protein more anionic and causes the expulsion of histones from the chromatin structure and, thus, the chromatin will become relaxed [171]. Blocking PARP1 activity through chemical applications leads to a high level of NAD+ accumulation. Under stress, DNA breakage caused by the increased level of ROS accumulation is sensed by PARP which adds ADP-ribose units to the damaged DNA regions, recruiting other damage repair mechanisms [172], [173].
In Arabidopsis cell culture, a high level of PARP accumulation was found to be more likely to occur during the exponential growth stage than in the early and older stages of growth. A concomitant increase in NAD+ oxidation and glutathione production were reported when there was a high level of PARP accumulation [174]. Also, elevated PARP1 expression at the time of DNA damage is caused by oxidative stress conditions [175], [176], [177]. Thus, PARP is a major energy consumer during stress response [178], [179]. High PARP activity, however, depletes NAD+ content, which compels plants to replenish NAD+ content. For this, plants use other salvage pathways to synthesise NAD+. Thus, the total energy content of the plant system drops, leading to cell death. PARP mutant plants, as well as plants in which PARP1 activity is externally blocked, are shown to be resistant to oxidative stress where energy consumption in the form of NAD+ is protected [178], [180]. PARP inhibition enhances the growth of plants as PARP-mediated NAD+ consumption is prevented.
Conclusion, future perspectives and biotechnological opportunities
Plants adapt to stress conditions with their innate epigenetic rhythms such as DNA methylation, histone modification and chromatin remodeling. Since stress conditions involve redox homeostasis in plants, epigenetic components undergo associated changes, driving crucial regulations of gene expression.
Plant systems have an extensive battery of redox mediators such as ROS, RNS, antioxidants, etc. the interplay of which regulates redox homeostasis. Redox-regulated epigenetic marks are indirect indicators of the redox status of the cell. Thus, redox mediators are emerging key regulators of epigenetic rhythms that regulate the activities of various enzymes controlling various molecular processes involved in plant growth and development under different environments.
The understanding of the functional mechanisms of epigenetic modifications regulated by the redox status of the cell has significantly advanced in recent years; however more remains yet unknown, particularly the precise molecular mechanisms. For instance, how does one oxidant trigger epigenetic modification without activating other antioxidants as well as other redox-regulated epigenetic marks?
Also, epigenetic activities such as sumoylation of histones, phosphorylation, ADP-ribosylation, glycosylation, ubiquitinylation, and chromatin remodeling are relatively under-studied. Chromatin modifications, triggered by redox mediators under various stress conditions, require more investigation. Furthermore, the epigenetic role of enzymes, such as NADPH oxidase and antioxidant enzymes that help in ROS generation and scavenging, and GSH synthesis in the mitochondria, NO synthase-like enzymes and nitrate reductase are yet to be completely understood. Enzymes and transcription factors that are influenced the redox status of the cell require in-depth studies.
Since Epigenetic regulation is complicated since it involves several pathways. Thus, the crosstalk between various components of the epigenetic complex, including co-expression genes, transposable elements, non-coding RNAs, spliceosomes and proteomes, requires investigation, especially with respect to redox-mediated changes. Plant stress memory associated with the redox status of the cell also requires in-depth studies. N6-methyladenosine (6 mA), novel epigenetic modifications of DNA, are adequately understood in plants, but 6 mA associations with the redox status of the cell remain unidentified [181].
Current biotechnological resources and escalating developments offer compelling promise for solutions to the unsolved mysteries of epigenetic development in plants. Recent technological developments such as nanopore sequencing, ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) [182], epitranscriptomics [183], gene editing [184], proteomics, metabolomics, imaging of redox status components [185], etc. are driving discoveries of molecular mechanisms involved in redox-mediated epigenetic changes.
The broader understanding of the epigenetic development of plants will soon recast crop improvement strategies in economically and ecologically important crops and tree species. Therefore, it is necessary to comprehend the epigenetic rhythm of every target species, particularly when related to stress responses where redox-mediated regulations underpin the survival of the organism. This would lead us to grow crop cultivars with improved traits for food and energy security and climate resilience.
Compliance with Ethics Requirements
All the authors state that this article does not contain any studies with human or animal subjects performed by the any of the authors and have declared no conflict of interest.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (grant numbers 31870656 and 31470615) and the Zhejiang Provincial Natural Science Foundation of China (grant number LZ19C160001). MR is also supported by Metasequoia Faculty Research Start-up Funding (grant number 163100028) at Nanjing Forestry University. We would like to extend our sincere gratitude and appreciation to all reviewers for their valuable comments. Fig. 6 was created with BioRender.com.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.04.007.
Contributor Information
Muthusamy Ramakrishnan, Email: ramky@njfu.edu.cn.
Mingbing Zhou, Email: zhoumingbing@zafu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Ashapkin V.V., Kutueva L.I., Aleksandrushkina N.I., Vanyushin B.F. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int J Mol Sci. 2020;21:7457. doi: 10.3390/ijms21207457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekhawat K., Almeida-Trapp M., García-Ramírez G.X., Hirt H. Beat the heat: plant- and microbe-mediated strategies for crop thermotolerance. Trends Plant Sci. 2022 doi: 10.1016/j.tplants.2022.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Sharma M., Kumar P., Verma V., Sharma R., Bhargava B., Irfan M. Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol Biochem. 2022;179:10–24. doi: 10.1016/j.plaphy.2022.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Saravana Kumar R.M., Wang Y., Zhang X., Cheng H., Sun L., He S., et al. Redox components: key regulators of epigenetic modifications in plants. Int J Mol Sci. 2020;21:1419. doi: 10.3390/ijms21041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y., Issakidis-Bourguet E., Zhou D.X. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J Exp Bot. 2016;67:5291–5300. doi: 10.1093/jxb/erw310. [DOI] [PubMed] [Google Scholar]
- 6.Considine M.J., Foyer C.H. Redox regulation of plant development. Antioxid Redox Signal. 2014;21:1305–1326. doi: 10.1089/ars.2013.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locato V., Cimini S., De Gara L. ROS and redox balance as multifaceted players of cross-tolerance: epigenetic and retrograde control of gene expression. J Exp Bot. 2018;69:3373–3391. doi: 10.1093/jxb/ery168. [DOI] [PubMed] [Google Scholar]
- 8.You J., Chan Z. ROS regulation during abiotic stress responses in crop plants. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpas F.J., Río L.A.D., Palma J.M. Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes. Plants (Basel) 2019;8:37. doi: 10.3390/plants8020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foyer C.H., Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. [Google Scholar]
- 11.Kapoor D., Singh S., Kumar V., Romero R., Prasad R., Singh J. Antioxidant enzymes regulation in plants in reference to reactive oxygen species (ROS) and reactive nitrogen species (RNS) Plant Gene. 2019;19 [Google Scholar]
- 12.Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D.J., Coutu J., et al. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gémes K., Poór P., Horváth E., Kolbert Z., Szopkó D., Szepesi Á., et al. Cross-talk between salicylic acid and NaCl-generated reactive oxygen species and nitric oxide in tomato during acclimation to high salinity. Physiol Plant. 2011;142:179–192. doi: 10.1111/j.1399-3054.2011.01461.x. [DOI] [PubMed] [Google Scholar]
- 14.Hernández J.A., Gullner G., Clemente-Moreno M.J., Künstler A., Juhász C., Díaz-Vivancos P., et al. Oxidative stress and antioxidative responses in plant–virus interactions. Physiol Mol Plant Pathol. 2016;94:134–148. [Google Scholar]
- 15.Noctor G. Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ. 2006;29:409–425. doi: 10.1111/j.1365-3040.2005.01476.x. [DOI] [PubMed] [Google Scholar]
- 16.Cairns N.G., Pasternak M., Wachter A., Cobbett C.S., Meyer A.J. Maturation of arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006;141:446–455. doi: 10.1104/pp.106.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marty L., Siala W., Schwarzländer M., Fricker M.D., Wirtz M., Sweetlove L.J., et al. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:9109–9114. doi: 10.1073/pnas.0900206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noctor G., Mhamdi A., Chaouch S., Han Y.I., Neukermans J., Marquez-Garcia B., et al. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z., Gallie D.R. Increasing tolerance to ozone by elevating foliar ascorbic acid confers greater protection against ozone than increasing avoidance. Plant Physiol. 2005;138:1673–1689. doi: 10.1104/pp.105.062000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutilleul C., Garmier M., Noctor G., Mathieu C., Chétrit P., Foyer C.H., et al. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15:1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vivancos P.D., Dong Y., Ziegler K., Markovic J., Pallardó F.V., Pellny T.K., et al. Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 2010;64:825–838. doi: 10.1111/j.1365-313X.2010.04371.x. [DOI] [PubMed] [Google Scholar]
- 22.de Simone A., Hubbard R., de la Torre N.V., Velappan Y., Wilson M., Considine M.J., et al. Redox changes during the cell cycle in the embryonic root meristem of Arabidopsis thaliana. Antioxid Redox Signal. 2017;27:1505–1519. doi: 10.1089/ars.2016.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dietzel L., Gläßer C., Liebers M., Hiekel S., Courtois F., Czarnecki O., et al. Identification of early nuclear target genes of plastidial redox signals that trigger the long-term response of arabidopsis to light quality shifts. Mol Plant. 2015;8:1237–1252. doi: 10.1016/j.molp.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Rai K.K., Rai N., Rai S.P. Salicylic acid and nitric oxide alleviate high temperature induced oxidative damage in Lablab purpureus L plants by regulating bio-physical processes and DNA methylation. Plant Physiol Biochem. 2018;128:72–88. doi: 10.1016/j.plaphy.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 25.Pena L.B., Barcia R.A., Azpilicueta C.E., Méndez A.A.E., Gallego S.M. Oxidative post translational modifications of proteins related to cell cycle are involved in cadmium toxicity in wheat seedlings. Plant Sci. 2012;196:1–7. doi: 10.1016/j.plantsci.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Desvoyes B., Gutierrez C. Roles of plant retinoblastoma protein: cell cycle and beyond. EMBO J. 2020;39:e105802. doi: 10.15252/embj.2020105802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 28.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang D., Gallusci P., Lang Z. Fruit development and epigenetic modifications. New Phytol. 2020 doi: 10.1111/nph.16724. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q., Belden W.J. Molecular regulation of circadian chromatin. J Mol Biol. 2020;432:3466–3482. doi: 10.1016/j.jmb.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Ueda M., Seki M. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol. 2020;182:15. doi: 10.1104/pp.19.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L.-T., Wu K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav. 2010;5:1318–1320. doi: 10.4161/psb.5.10.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L.T., Luo M., Wang Y.Y., Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassan Y.I., Zempleni J. A novel, enigmatic histone modification: biotinylation of histones by holocarboxylase synthetase. Nutr Rev. 2008;66:721–725. doi: 10.1111/j.1753-4887.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang P., Zhao L., Hou H., Zhang H., Huang Y., Wang Y., et al. Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol. 2015;56:965–976. doi: 10.1093/pcp/pcv023. [DOI] [PubMed] [Google Scholar]
- 36.Xu L., Jiang H. Writing and Reading Histone H3 Lysine 9 Methylation in Arabidopsis. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H., Lang Z., Zhu J.K. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 38.Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 39.Choi C.S., Sano H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics. 2007;277:589–600. doi: 10.1007/s00438-007-0209-1. [DOI] [PubMed] [Google Scholar]
- 40.Nishida N., Kudo M. Oxidative Stress and Epigenetic Instability in Human Hepatocarcinogenesis. Dig Dis. 2013;31:447–453. doi: 10.1159/000355243. [DOI] [PubMed] [Google Scholar]
- 41.Poborilova Z., Ohlsson A.B., Berglund T., Vildova A., Provaznik I., Babula P. DNA hypomethylation concomitant with the overproduction of ROS induced by naphthoquinone juglone on tobacco BY-2 suspension cells. Environ Exp Bot. 2015;113:28–39. [Google Scholar]
- 42.Berglund T., Wallström A., Nguyen T.-V., Laurell C., Ohlsson A.B. Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol Biochem. 2017;118:551–560. doi: 10.1016/j.plaphy.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 43.García-Giménez J.L., Pallardó F.V. Maintenance of glutathione levels and its importance in epigenetic regulation. Front Pharmacol. 2014;5:88. doi: 10.3389/fphar.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou X., Zhuang T., Yin W., Miao Y., Wang B., Zhang Y., et al. DNA Methylation changes induced in rice by exposure to high concentrations of the nitric oxide modulator, sodium nitroprusside. Plant Mol Biol Rep. 2015;33:1428–1440. [Google Scholar]
- 45.Xu W., Wang T., Xu S., Xu S., Wu L., Wu Y., et al. Radiation-induced epigenetic bystander effects demonstrated in Arabidopsis thaliana. Radiat Res. 2015;183:511–524. doi: 10.1667/RR13909.1. [DOI] [PubMed] [Google Scholar]
- 46.Charbonnel C., Niazi A.K., Elvira-Matelot E., Nowak E., Zytnicki M., de Bures A., et al. The siRNA suppressor RTL1 is redox-regulated through glutathionylation of a conserved cysteine in the double-stranded-RNA-binding domain. Nucleic Acids Res. 2017;45:11891–11907. doi: 10.1093/nar/gkx820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seta A., Tabara M., Nishibori Y., Hiraguri A., Ohkama-Ohtsu N., Yokoyama T., et al. Post-translational regulation of the dicing activities of arabidopsis DICER-LIKE 3 and 4 by inorganic phosphate and the redox state. Plant Cell Physiol. 2017;58:485–495. doi: 10.1093/pcp/pcw226. [DOI] [PubMed] [Google Scholar]
- 48.Gorelova V., De Lepeleire J., Van Daele J., Pluim D., Meï C., Cuypers A., et al. Dihydrofolate reductase/thymidylate synthase fine-tunes the folate status and controls redox homeostasis in plants. Plant Cell. 2017;29:2831–2853. doi: 10.1105/tpc.17.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Deng X., Miki D., Cutler S., La H., Hou Y.J., et al. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell. 2012;24:1230–1241. doi: 10.1105/tpc.112.096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fares A., Rossignol M., Peltier J.B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Biophys Res Commun. 2011;416:331–336. doi: 10.1016/j.bbrc.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 51.Lozano-Juste J., Colom-Moreno R., León J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot. 2011;62:3501–3517. doi: 10.1093/jxb/err042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim J.C., You Z., Kim G., Levine R.L. Methionine sulfoxide reductase A is a stereospecific methionine oxidase. Proc Natl Acad Sci U S A. 2011;108:10472–10477. doi: 10.1073/pnas.1101275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Begara-Morales J.C., Sánchez-Calvo B., Luque F., Leyva-Pérez M.O., Leterrier M., Corpas F.J., et al. Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 2014;55:1080–1095. doi: 10.1093/pcp/pcu044. [DOI] [PubMed] [Google Scholar]
- 54.Lindermayr C., Saalbach G., Bahnweg G., Durner J. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem. 2006;281:4285–4291. doi: 10.1074/jbc.M511635200. [DOI] [PubMed] [Google Scholar]
- 55.Tarrago L., Laugier E., Rey P. Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Mol Plant. 2009;2:202–217. doi: 10.1093/mp/ssn067. [DOI] [PubMed] [Google Scholar]
- 56.Zeng F.-S., Sun F.-K., Liang N.-S., Zhao X.-T., Luo W., Zhan Y.-G. Dynamic change of DNA methylation and cell redox state at different micropropagation phases in birch. Trees. 2015;29:917–930. [Google Scholar]
- 57.Couturier J., Chibani K., Jacquot J.P., Rouhier N. Cysteine-based redox regulation and signaling in plants. Front Plant Sci. 2013;4:105. doi: 10.3389/fpls.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buzas D.M. Emerging links between iron-sulfur clusters and 5-methylcytosine base excision repair in plants. Genes Genet Syst. 2016;91:51–62. doi: 10.1266/ggs.16-00015. [DOI] [PubMed] [Google Scholar]
- 59.Ramazi S., Allahverdi A., Zahiri J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J Biosci. 2020;45:135. [PubMed] [Google Scholar]
- 60.Mengel A., Ageeva A., Georgii E., Bernhardt J., Wu K., Durner J., et al. Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 2017;173:1434–1452. doi: 10.1104/pp.16.01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Causevic A., Gentil M.-V., Delaunay A., El-Soud W.A., Garcia Z., Pannetier C., et al. Relationship between DNA methylation and histone acetylation levels, cell redox and cell differentiation states in sugarbeet lines. Planta. 2006;224:812–827. doi: 10.1007/s00425-006-0267-3. [DOI] [PubMed] [Google Scholar]
- 62.Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugden M.C., Holness M.J. The pyruvate carboxylase-pyruvate dehydrogenase axis in islet pyruvate metabolism: Going round in circles? Islets. 2011;3:302–319. doi: 10.4161/isl.3.6.17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tovar-Méndez A., Miernyk J.A., Randall D.D. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem. 2003;270:1043–1049. doi: 10.1046/j.1432-1033.2003.03469.x. [DOI] [PubMed] [Google Scholar]
- 65.Dumont S., Rivoal J. Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front Plant Sci. 2019;10 doi: 10.3389/fpls.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baud S., Guyon V., Kronenberger J., Wuillème S., Miquel M., Caboche M., et al. Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J. 2003;33:75–86. doi: 10.1046/j.1365-313x.2003.016010.x. [DOI] [PubMed] [Google Scholar]
- 67.Chen C., Li C., Wang Y., Renaud J., Tian G., Kambhampati S., et al. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants. 2017;3:814–824. doi: 10.1038/s41477-017-0023-7. [DOI] [PubMed] [Google Scholar]
- 68.Zhou X., Hua D., Chen Z., Zhou Z., Gong Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009;60:79–90. doi: 10.1111/j.1365-313X.2009.03931.x. [DOI] [PubMed] [Google Scholar]
- 69.AbdElgawad H., Zinta G., Hegab M.M., Pandey R., Asard H., Abuelsoud W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front Plant Sci. 2016;7:276. doi: 10.3389/fpls.2016.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Yan S., Zhao L., Tan J., Zhang Q., Gao F., et al. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014;14:105. doi: 10.1186/1471-2229-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X., Ding A.B., Zhong X. Functions and mechanisms of plant histone deacetylases. Sci China Life Sci. 2020;63:206–216. doi: 10.1007/s11427-019-1587-x. [DOI] [PubMed] [Google Scholar]
- 72.Ding B, Bellizzi MdR, Ning Y, Meyers BC, Wang G-L: HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant cell 24 (2012) 3783–3794. [DOI] [PMC free article] [PubMed]
- 73.Zhao J., Zhang J., Zhang W., Wu K., Zheng F., Tian L., et al. Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci. 2015;5 doi: 10.3389/fpls.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo M., Cheng K., Xu Y., Yang S., Wu K. Plant responses to abiotic stress regulated by histone deacetylases. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ito K., Ito M., Elliott W.M., Cosio B., Caramori G., Kon O.M., et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 76.Taunton J., Hassig C.A., Schreiber S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 77.Rundlett S.E., Carmen A.A., Kobayashi R., Bavykin S., Turner B.M., Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci U S A. 1996;93(14503) doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ageeva-Kieferle A., Rudolf E.E., Lindermayr C. Redox-dependent chromatin remodeling: A new function of nitric oxide as architect of chromatin structure in plants. Front Plant Sci. 2019;10:625. doi: 10.3389/fpls.2019.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi S.-M., Song H.-R., Han S.-K., Han M., Kim C.-Y., Park J., et al. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012;71:135–146. doi: 10.1111/j.1365-313X.2012.04977.x. [DOI] [PubMed] [Google Scholar]
- 80.Liu P., Zhang H., Yu B., Xiong L., Xia Y. Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci Rep. 2015;5:8625. doi: 10.1038/srep08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park J., Lim C.J., Shen M., Park H.J., Cha J.-Y., Iniesto E., et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc Natl Acad Sci U S A. 2018;115(E5400) doi: 10.1073/pnas.1721241115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo M., Wang Y.-Y., Liu X., Yang S., Lu Q., Cui Y., et al. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buszewicz D., Archacki R., Palusiński A., Kotliński M., Fogtman A., Iwanicka-Nowicka R., et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016;39:2108–2122. doi: 10.1111/pce.12756. [DOI] [PubMed] [Google Scholar]
- 84.Mehdi S., Derkacheva M., Ramström M., Kralemann L., Bergquist J., Hennig L. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell. 2016;28:42–54. doi: 10.1105/tpc.15.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim H., Shim D., Moon S., Lee J., Bae W., Choi H., et al. Transcriptional network regulation of the brassinosteroid signaling pathway by the BES1-TPL-HDA19 co-repressor complex. Planta. 2019;250:1371–1377. doi: 10.1007/s00425-019-03233-z. [DOI] [PubMed] [Google Scholar]
- 86.Rajendran R., Garva R., Krstic-Demonacos M., Demonacos C. Sirtuins: molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J Biomed Biotechnol. 2011;2011 doi: 10.1155/2011/368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang J.-Y., Kim J.J., Jang S.Y., Bae Y.-S. The p53–p21Cip1/WAF1 pathway is necessary for cellular senescence induced by the inhibition of protein kinase CKII in human colon cancer cells. Mol Cells. 2009;28:489. doi: 10.1007/s10059-009-0141-9. [DOI] [PubMed] [Google Scholar]
- 88.Alexandrou A.T., Li J.J. Cell cycle regulators guide mitochondrial activity in radiation-induced adaptive response. Antioxid Redox Signal. 2014;20:1463–1480. doi: 10.1089/ars.2013.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau A.W., Liu P., Inuzuka H., Gao D. SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am J Cancer Res. 2014;4:245–255. [PMC free article] [PubMed] [Google Scholar]
- 90.Zee B.M., Levin R.S., Xu B., LeRoy G., Wingreen N.S., Garcia B.A. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kornberg M.D., Sen N., Hara M.R., Juluri K.R., Nguyen J.V., Snowman A.M., et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furukawa A., Tada-Oikawa S., Kawanishi S., Oikawa S. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD+ depletion. Cell Physiol Biochem. 2007;20:45–54. doi: 10.1159/000104152. [DOI] [PubMed] [Google Scholar]
- 93.Kupis W., Pałyga J., Tomal E., Niewiadomska E. The role of sirtuins in cellular homeostasis. J Physiol Biochem. 2016;72:371–380. doi: 10.1007/s13105-016-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Szućko I. Sirtuins: not only animal proteins. Acta Physiol Plant. 2016;38:237. [Google Scholar]
- 95.Soccio M., Laus M.N., Alfarano M., Pastore D. Measuring Activity of Native Plant Sirtuins - The Wheat Mitochondrial Model. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y., Zhang A., Yin H., Meng Q., Yu X., Huang S., et al. Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 2018;217:1582–1597. doi: 10.1111/nph.14933. [DOI] [PubMed] [Google Scholar]
- 97.Ezaki B., Higashi A., Nanba N., Nishiuchi T. An S-adenosyl Methionine Synthetase (SAMS) Gene from Andropogon virginicus L. Confers Aluminum Stress Tolerance and Facilitates Epigenetic Gene Regulation in Arabidopsis thaliana. Front Plant Sci. 2016;7:1627. doi: 10.3389/fpls.2016.01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barozai M.Y.K., Aziz A.N. Recent plant growth and stress management related significant advancements in epigenetics. Ann Agrar Sci. 2018;16:416–421. [Google Scholar]
- 99.Spellmon N., Holcomb J., Trescott L., Sirinupong N., Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He S., Owen D.R., Jelinsky S.A., Lin L.-L. Lysine Methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Sci Rep. 2015;5:14368. doi: 10.1038/srep14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Y., Mukherjee I., Thum K.E., Tanurdzic M., Katari M.S., Obertello M., et al. The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol. 2015;16:79. doi: 10.1186/s13059-015-0640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaurilind E., Xu E., Brosché M. A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genomics. 2015;16:837. doi: 10.1186/s12864-015-1964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gupta K.J., Kolbert Z., Durner J., Lindermayr C., Corpas F.J., Brouquisse R., et al. Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 2020;227:1319–1325. doi: 10.1111/nph.16622. [DOI] [PubMed] [Google Scholar]
- 104.Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Z., Zhang S., Zhang Y., Wang X., Li D., Li Q., et al. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell. 2011;23:396–411. doi: 10.1105/tpc.110.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qian Y., Chen C., Jiang L., Zhang J., Ren Q. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genomics. 2019;20:256. doi: 10.1186/s12864-019-5633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 108.Forneris F., Binda C., Vanoni M.A., Mattevi A., Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 109.Forneris F., Binda C., Vanoni M.A., Battaglioli E., Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 110.Becker D.F., Zhu W., Moxley M.A. Flavin redox switching of protein functions. Antioxid Redox Signal. 2011;14:1079–1091. doi: 10.1089/ars.2010.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Senda T., Senda M., Kimura S., Ishida T. Redox control of protein conformation in flavoproteins. Antioxid Redox Signal. 2009;11:1741–1766. doi: 10.1089/ars.2008.2348. [DOI] [PubMed] [Google Scholar]
- 112.Huang Y., Chen D., Liu C., Shen W., Ruan Y. Evolution and conservation of JmjC domain proteins in the green lineage. Mol Genet Genomics. 2016;291:33–49. doi: 10.1007/s00438-015-1089-4. [DOI] [PubMed] [Google Scholar]
- 113.Luo M., Hung F.-Y., Yang S., Liu X., Wu K. Histone Lysine Demethylases and Their Functions in Plants. Plant Mol Biol Rep. 2014;32:558–565. [Google Scholar]
- 114.Liu P., Zhang S., Zhou B., Luo X., Zhou X.F., Cai B., et al. The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell. 2019;31:430–443. doi: 10.1105/tpc.18.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cimini S., Gualtieri C., Macovei A., Balestrazzi A., De Gara L., Locato V. Redox Balance-DDR-miRNA Triangle: Relevance in Genome Stability and Stress Responses in Plants. Front Plant Sci. 2019;10:989. doi: 10.3389/fpls.2019.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao J., Gulyás Z., Kalapos B., Boldizsár Á., Liu X., Pál M., et al. Identification of a redox-dependent regulatory network of miRNAs and their targets in wheat. J Exp Bot. 2019;70:85–99. doi: 10.1093/jxb/ery339. [DOI] [PubMed] [Google Scholar]
- 117.Jia X.L., Chen Y.K., Xu X.Z., Shen F., Zheng Q.B., Du Z., et al. miR156 switches on vegetative phase change under the regulation of redox signals in apple seedlings. Sci Rep. 2017;7:14223. doi: 10.1038/s41598-017-14671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li T., Li H., Zhang Y.X., Liu J.Y. Identification and analysis of seven H₂O₂-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica) Nucleic Acids Res. 2011;39:2821–2833. doi: 10.1093/nar/gkq1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lv D.W., Zhen S., Zhu G.R., Bian Y.W., Chen G.X., Han C.X., et al. High-Throughput Sequencing Reveals H(2)O(2) Stress-Associated MicroRNAs and a Potential Regulatory Network in Brachypodium distachyon Seedlings. Front Plant Sci. 2016;7:1567. doi: 10.3389/fpls.2016.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ben Chaabane S., Liu R., Chinnusamy V., Kwon Y., Park J.H., Kim S.Y., et al. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013;41:1984–1997. doi: 10.1093/nar/gks1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaucheret H., Vazquez F., Crété P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie Z., Kasschau K.D., Carrington J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 123.Jagadeeswaran G., Li Y.-F., Sunkar R. Redox signaling mediates the expression of a sulfate-deprivation-inducible microRNA395 in Arabidopsis. Plant J. 2014;77:85–96. doi: 10.1111/tpj.12364. [DOI] [PubMed] [Google Scholar]
- 124.Carbonell T., Gomes A.V. MicroRNAs in the regulation of cellular redox status and its implications in myocardial ischemia-reperfusion injury. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Megha S., Basu U., Kav N.N.V. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018;41:1–15. doi: 10.1111/pce.12956. [DOI] [PubMed] [Google Scholar]
- 126.Klemm S.L., Shipony Z., Greenleaf W.J. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 127.Lee J.S., Smith E., Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kang H., Wu D., Fan T., Zhu Y. Activities of Chromatin Remodeling Factors and Histone Chaperones and Their Effects in Root Apical Meristem Development. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roudier F., Ahmed I., Bérard C., Sarazin A., Mary-Huard T., Cortijo S., et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang C., Liu C., Roqueiro D., Grimm D., Schwab R., Becker C., et al. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25:246–256. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lim C.J., Ali A., Park J., Shen M., Park K.S., Baek D., et al. HOS15-PWR chromatin remodeling complex positively regulates cold stress in Arabidopsis. Plant Signal Behav. 2021;16:1893978. doi: 10.1080/15592324.2021.1893978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pien S., Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 133.Kim D.-H., Sung S. Polycomb-mediated gene silencing in Arabidopsis thaliana. Mol Cells. 2014;37:841–850. doi: 10.14348/molcells.2014.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ruta V., Longo C., Boccaccini A., Madia V.N., Saccoliti F., Tudino V., et al. Inhibition of Polycomb Repressive Complex 2 activity reduces trimethylation of H3K27 and affects development in Arabidopsis seedlings. BMC Plant Biol. 2019;19:429. doi: 10.1186/s12870-019-2057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu F., Kuo T., Rosli Y., Liu M.-S., Wu L. Chen L-FO, Fletcher JC, Sung ZR, Pu L: Trithorax Group Proteins Act Together with a Polycomb Group Protein to Maintain Chromatin Integrity for Epigenetic Silencing during Seed Germination in Arabidopsis. Mol Plant. 2018;11:659–677. doi: 10.1016/j.molp.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 136.Flaus A., Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: the means to the end. FEBS J. 2011;278:3579–3595. doi: 10.1111/j.1742-4658.2011.08281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hargreaves D.C., Crabtree G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Himanen S.V., Sistonen L. New insights into transcriptional reprogramming during cellular stress. J Cell Sci. 2019;132(jcs238402) doi: 10.1242/jcs.238402. [DOI] [PubMed] [Google Scholar]
- 139.Menezo Y.J., Silvestris E., Dale B., Elder K. Oxidative stress and alterations in DNA methylation: two sides of the same coin in reproduction. Reprod Biomed Online. 2016;33:668–683. doi: 10.1016/j.rbmo.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 140.Morano K.A., Grant C.M., Moye-Rowley W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ojolo S.P., Cao S., Priyadarshani S.V.G.N., Li W., Yan M., Aslam M., et al. Regulation of Plant Growth and Development: A Review From a Chromatin Remodeling Perspective. Front Plant Sci. 2018;9:1232. doi: 10.3389/fpls.2018.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bäurle I., Trindade I. Chromatin regulation of somatic abiotic stress memory. J Exp Bot. 2020;71:5269–5279. doi: 10.1093/jxb/eraa098. [DOI] [PubMed] [Google Scholar]
- 143.Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Greb T., Mylne J.S., Crevillen P., Geraldo N., An H., Gendall A.R., et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17:73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 145.Sung S., Amasino R.M. Vernalization and epigenetics: how plants remember winter. Curr Opin Plant Biol. 2004;7:4–10. doi: 10.1016/j.pbi.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 146.Sung S., Schmitz R.J., Amasino R.M. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sung S., He Y., Eshoo T.W., Tamada Y., Johnson L., Nakahigashi K., et al. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 148.Zografos B.R., Sung S. Vernalization-mediated chromatin changes. J Exp Bot. 2012;63:4343–4348. doi: 10.1093/jxb/ers157. [DOI] [PubMed] [Google Scholar]
- 149.Schmitz R.J., Sung S., Amasino R.M. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 151.Wood C.C., Robertson M., Tanner G., Peacock W.J., Dennis E.S., Helliwell C.A. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bond D.M., Wilson I.W., Dennis E.S., Pogson B.J., Jean Finnegan E. VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J. 2009;59:576–587. doi: 10.1111/j.1365-313X.2009.03891.x. [DOI] [PubMed] [Google Scholar]
- 153.Gibbs D.J., Tedds H.M., Labandera A.M., Bailey M., White M.D., Hartman S., et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat Commun. 2018;9:5438. doi: 10.1038/s41467-018-07875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hu J., Yang H., Mu J., Lu T., Peng J., Deng X., et al. Nitric Oxide Regulates Protein Methylation during Stress Responses in Plants. Mol Cell. 2017;67:702–710.e704. doi: 10.1016/j.molcel.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 155.Zhang W., Yu R. Molecule mechanism of stem cells in Arabidopsis thaliana. Pharmacogn Rev. 2014;8:105–112. doi: 10.4103/0973-7847.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- 157.Zhou X., Xiang Y., Li C., Yu G. Modulatory Role of Reactive Oxygen Species in Root Development in Model Plant of Arabidopsis thaliana. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.485932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang S., Yu Q., Zhang Y., Jia Y., Wan S., Kong X., et al. ROS: The Fine-Tuner of Plant Stem Cell Fate. Trends Plant Sci. 2018;23:850–853. doi: 10.1016/j.tplants.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 159.Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 160.Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 161.Yang L., Zhang J., He J., Qin Y., Hua D., Duan Y., et al. ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling PLETHORA Expression in Arabidopsis. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shibasaki K., Uemura M., Tsurumi S., Rahman A. Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell. 2009;21:3823–3838. doi: 10.1105/tpc.109.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang L., Mizzen C., Ying C., Candau R., Barlev N., Brownell J., et al. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tanner K.G., Trievel R.C., Kuo M.H., Howard R.M., Berger S.L., Allis C.D., et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 165.Kornet N., Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21:1070–1079. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yao X., Feng H., Yu Y., Dong A., Shen W.-H. SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS ONE. 2013;8:e56537. doi: 10.1371/journal.pone.0056537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aichinger E., Villar C.B., Di Mambro R., Sabatini S., Köhler C. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell. 2011;23:1047–1060. doi: 10.1105/tpc.111.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alemasova E.E., Lavrik O.I. Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019;47:3811–3827. doi: 10.1093/nar/gkz120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ray Chaudhuri A., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen Q., Kassab M.A., Dantzer F., Yu X. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat Commun. 2018;9:3233. doi: 10.1038/s41467-018-05588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Morales J., Li L., Fattah F.J., Dong Y., Bey E.A., Patel M., et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24:15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Murata M.M., Kong X., Moncada E., Chen Y., Imamura H., Wang P., et al. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol Biol Cell. 2019;30:2584–2597. doi: 10.1091/mbc.E18-10-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pellny T.K., Locato V., Vivancos P.D., Markovic J., De Gara L., Pallardó F.V., et al. Pyridine Nucleotide Cycling and Control of Intracellular Redox State in Relation to Poly (ADP-Ribose) Polymerase Activity and Nuclear Localization of Glutathione during Exponential Growth of Arabidopsis Cells in Culture. Mol Plant. 2009;2:442–456. doi: 10.1093/mp/ssp008. [DOI] [PubMed] [Google Scholar]
- 175.Pietrzak J., Spickett C.M., Płoszaj T., Virág L., Robaszkiewicz A. PARP1 promoter links cell cycle progression with adaptation to oxidative environment. Redox Biol. 2018;18:1–5. doi: 10.1016/j.redox.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang D., Hu X., Li J., Liu J., Baks-te Bulte L., Wiersma M., et al. DNA damage-induced PARP1 activation confers cardiomyocyte dysfunction through NAD+ depletion in experimental atrial fibrillation. Nat Commun. 2019;10(1307) doi: 10.1038/s41467-019-09014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Amor Y., Babiychuk E., Inzé D., Levine A. The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 1998;440:1–7. doi: 10.1016/s0014-5793(98)01408-2. [DOI] [PubMed] [Google Scholar]
- 178.Block M.D., Verduyn C., Brouwer D.D., Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41:95–106. doi: 10.1111/j.1365-313X.2004.02277.x. [DOI] [PubMed] [Google Scholar]
- 179.Rissel D., Heym P.P., Thor K., Brandt W., Wessjohann L.A., Peiter E. No Silver Bullet - Canonical Poly(ADP-Ribose) Polymerases (PARPs) Are No Universal Factors of Abiotic and Biotic Stress Resistance of Arabidopsis thaliana. Front Plant Sci. 2017;8:59. doi: 10.3389/fpls.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vanderauwera S., De Block M., Van de Steene N., van de Cotte B., Metzlaff M. Van Breusegem F: Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci U S A. 2007;104:15150–15155. doi: 10.1073/pnas.0706668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kumar S., Mohapatra T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.596236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen H., Tu S., Yuan C., Tian F., Zhang Y., Sun Y., et al. HyperChIP: identification of hypervariable signals across ChIP-seq or ATAC-seq samples. Genome Biol. 2022;23:62. doi: 10.1186/s13059-022-02627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Moshitch-Moshkovitz S., Dominissini D., Rechavi G. The epitranscriptome toolbox. Cell. 2022;185:764–776. doi: 10.1016/j.cell.2022.02.007. [DOI] [PubMed] [Google Scholar]
- 184.Zhu G., Zhu H. Modified Gene Editing Systems: Diverse Bioengineering Tools and Crop Improvement. Front Plant Sci. 2022;13 doi: 10.3389/fpls.2022.847169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang E., Whitcomb L.A., Chicco A.J., Wilson J.W. Transient absorption spectroscopy and imaging of redox in muscle mitochondria. Biomed Opt Express. 2022;13:2103–2116. doi: 10.1364/BOE.452559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.