Abstract
As the second-largest neurodegenerative disease in the world, Parkinson’s disease (PD) has brought a severe economic and medical burden to our society. Growing evidence in recent years suggests that the gut microbiome may influence PD, but the exact pathogenesis of PD remains unclear. In addition, the current diagnosis of PD could be inaccurate and expensive. In this study, the largest meta-analysis currently of the gut microbiome in PD was analyzed, including 2269 samples by 16S rRNA gene and 236 samples by shotgun metagenomics, aiming to reveal the connection between PD and gut microbiome and establish a model to predict PD. The results showed that the relative abundances of potential pro-inflammatory bacteria, genes and pathways were significantly increased in PD, while potential anti-inflammatory bacteria, genes and pathways were significantly decreased. These changes may lead to a decrease in potential anti-inflammatory substances (short-chain fatty acids) and an increase in potential pro-inflammatory substances (lipopolysaccharides, hydrogen sulfide and glutamate). Notably, the results of 16S rRNA gene and shotgun metagenomic analysis have consistently identified five decreased genera (Roseburia, Faecalibacterium, Blautia, Lachnospira, and Prevotella) and five increased genera (Streptococcus, Bifidobacterium, Lactobacillus, Akkermansia, and Desulfovibrio) in PD. Furthermore, random forest models performed well for PD prediction based on 11 genera (accuracy > 80%) or 6 genes (accuracy > 90%) related to inflammation. Finally, a possible mechanism was presented to explain the pathogenesis of inflammation leading to PD. Our results provided further insights into the prediction and treatment of PD based on inflammation.
Subject terms: Microbiota, Clinical microbiology
Introduction
Parkinson’s disease (PD) is an incurable, progressive, and chronic neurodegenerative disease characterized by the formation of Lewy bodies (mainly formed by misfolded α-synuclein) and the loss of dopaminergic neurons in the substantia nigra1. More than 6 million individuals worldwide were diagnosed with PD. PD alters dopaminergic, noradrenergic and serotonergic neurons in the brain, causing a drop in dopamine levels and premotor and non-motor symptoms, including akinesia, rigidity, balance difficulties, tremor, as well as neuropsychiatric, cognitive, autonomic and sensory disturbances. These non-motor symptoms can appear years or even decades before motor symptoms appear, but are often unrecognized2, resulting in PD patients not receiving timely treatment. Although PD has brought great medical and social burdens, its specific pathogenesis is still unclear. Numerous measurements have been used to diagnose PD and mainly include positron emission tomography, cerebrospinal fluid tests, and clinical symptoms3. However, positron emission tomography is quite costly and the reproducibility and reliability of the cerebrospinal fluid tests have been suspected. Therefore, it is imperative to further explore the pathogenesis of PD and find reliable and cheap biomarkers.
In recent years, it has been proposed that the human gastrointestinal microbiota is one of the most important pathogenic mechanisms of many neurodegenerative diseases4,5. Gut microbiota encodes millions of genes and produces thousands of metabolites, affecting the metabolism of the host4,6,7. Substantial evidence suggests a bidirectional interaction between the gastrointestinal microbiota and the central nervous system, known as the “gut–microbiota–brain axis”8. Multiple “gut–microbiota–brain axis” pathways exist, including molecules with neuroendocrine activity produced by microbes (such as gamma-aminobutyric acid and serotonin) and the gut microbial community influenced by the central nervous system8. These connections form a feedback loop between human physiology and the state of the microbial community. In recent years, the gut microbiota has been proven to play a vital role in the progression of PD in animal models through the gut–microbiota–brain axis9,10.
Multiple studies have described prodromal symptoms (gastrointestinal motility disorders) affecting the quality of life of patients with PD, including delayed gastric emptying and chronic constipation11. Intestinal symptoms (e.g., constipation) often precede motor symptoms, indicating a possible pathogen in the gut of PD patients. Based on neuropathology, Braak et al.12 suggested that PD may be caused by an enteric pathogen that can cross the intestinal mucosal barrier and enteric neurons, and ultimately enter the central nervous system via the vagus nerve. The hallmarks of PD, Lewy bodies and misfolded α-synuclein proteins, were found in both the central and enteric nervous systems13. The transport of α-synuclein in the gut–microbiota–brain axis and the newly discovered vagal pathway may induce or accelerate the progression of PD11. Removing the vagus nerve appears to reduce the risk of PD14,15. In a landmark study, Sampson et al. demonstrated that the microbiome itself can trigger or delay the motor symptoms of PD in mice9. Microbiota may facilitate α-synuclein diffusion, since the gut microbiota can secrete extracellular amyloid, and proteins such as PrPSc, Tau, and α-synuclein can spread in the body like prions16. Aggregation-prone proteins such as α-synuclein and Tau spread throughout the body during microbial colonization, biofilm formation and infection17. In PD patients, several indicators of symptom severity were positively correlated with microbial alpha and beta diversity indices18. The above evidence suggests that dysbiosis of gastrointestinal microbiota may provide an interesting clue to explore the pathogenesis of PD and become a new diagnostic and therapeutic target.
We hypothesized that PD patients in different countries and regions share common microbial and metabolic characteristics. In this study, the bacterial communities and metabolic pathways of PD were characterized by collecting massive open-access 16S rRNA gene and shotgun metagenomic data from extensive studies. Our goal is to understand the underlying microbial community and metabolic patterns in the gut of PD patients, then reveal the pathogenesis of PD and construct a model to predict PD.
Results
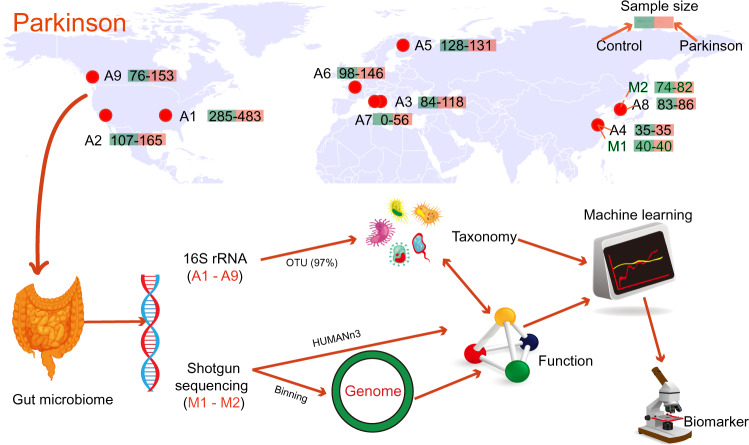
Changes in bacterial communities
After quality control, nine datasets (A1–9) including 2269 16S rRNA gene amplicon samples (1373 PD and 896 healthy controls) and two datasets (M1-2) including 236 shotgun sequencing metagenomic samples (122 PD and 114 healthy controls) were collected by searching the keywords “Parkinson” and “microbes” in the National Center for Biotechnology Information (NCBI) SRA database and Google Scholar (Fig. 1 and Supplementary Table 1). The resulting merged operational taxonomic unit (OTU) table contained 3847 taxa from 2269 16S rRNA gene samples (Supplementary Table 2). To explore whether the composition of the gut bacteria differed between the PD patients and healthy control, firstly, three α-diversity indexes (Shannon, Simpson, Pielou) were calculated, and Principal coordinates analysis (PCoA) was performed. The results (Supplementary Fig. 1) showed that there was no significant difference in α-diversity (0.89 > p > 0.62, Wilcoxon rank-sum test) and β-diversity (p = 0.72, analysis of similarities) between PD and Healthy control.
Fig. 1. Study design.
Nine datasets (A1–9) including 2269 16S rRNA gene samples and two datasets (M1-M2) including 236 shotgun metagenomic samples were collected from 11 studies. Taxonomy was inferred from 16s rRNA gene based on the operational taxonomic unit (OTU), the function was inferred from shotgun sequencing, and the genome was obtained by binning (metagenome-assembled genomes). Finally, the biomarker was found using machine learning.
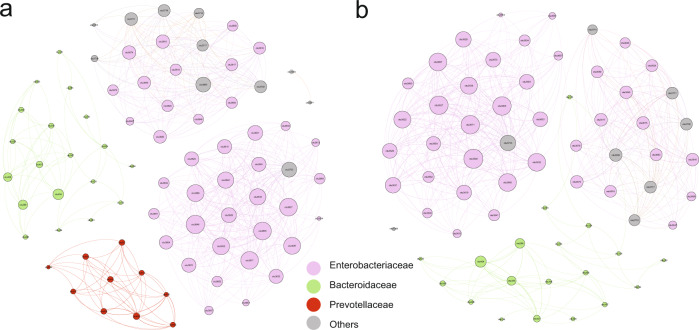
To explore the potential taxon co-occurrence pattern in PD, Spearman’s correlations between the microbial taxa (OTU) were calculated and visualized based on the combined dataset (A1–9). There was an obvious difference in the network structure between PD and healthy control (Fig. 2 and Supplementary Fig. 2). The results revealed a less number of nodes and links in the PD network (Supplementary Table 3). After removing nodes with few connections (<5), the network of healthy control contained four main modules while PD had only three (Fig. 2). The nodes in the network were dominated by Enterobacteriaceae, Bacteroidaceae and Prevotellaceae. It is worth noting that Prevotellaceae (Prevotella) did not appear in the PD network (Fig. 2).
Fig. 2. Co-occurrence networks.
Co-occurrence networks were calculated based on Spearman correlations at the operational taxonomic unit (OTU) level. The more connections, the larger the node (degree). Only strong (r > 0.7) and significant (p < 0.05) correlations are shown in the figure and nodes with few connections (<5) were removed. a Healthy controls. b Patients with Parkinson’s disease (PD). Nodes were colored by family.
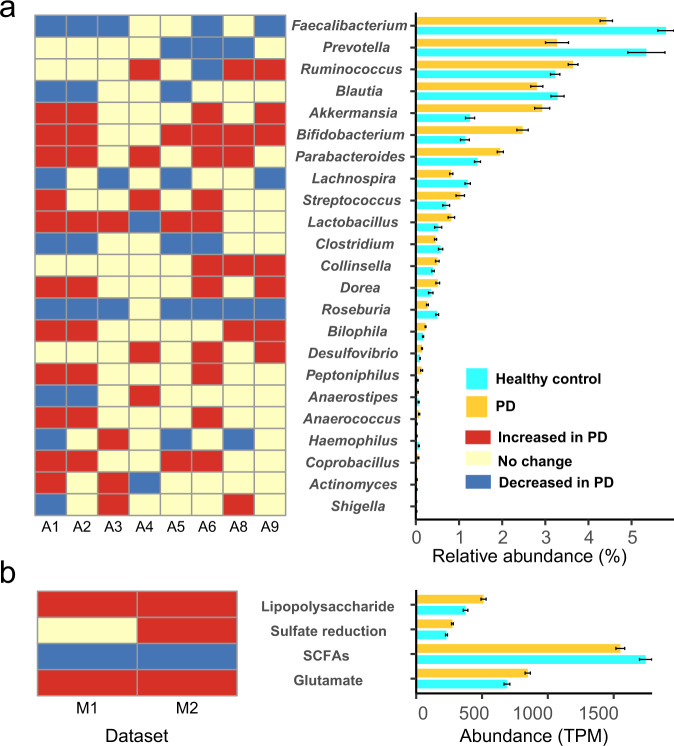
At the genus level, the relative abundances of 23 genera (Fig. 3a) were significantly different between healthy control and PD in at least three datasets (p < 0.05, Wilcoxon rank-sum test). Amplicon dataset 7 (A7) can not be analyzed separately for difference statistics because it only contains PD samples. These 23 genera also share the same variance in the combined dataset (A1–9) except Collinsella, Ruminococcus, Dorea, Shigella, and Anaerostipes (Supplementary Fig. 3). Five genera (Roseburia, Faecalibacterium, Blautia, Lachnospira, and Prevotella) are well-known producers of short-chain fatty acids (SCFAs) in the gut19,20, and their abundances were significantly reduced in PD. These five genera may be associated with anti-inflammation in PD. Streptococcus21 is an opportunistic pathogen, and its relative abundance was significantly increased in PD. Three genera (Bifidobacterium22, Lactobacillus23, and Akkermansia24) are probiotics, but their abundances were significantly increased in PD patients. Desulfovibrio25 predominates among intestinal sulfate-reducing bacteria with the ability to produce hydrogen sulfide (H2S), and its abundance was significantly increased in PD. These five genera may be associated with pro-inflammation in PD. It may seem ironic that these probiotics were elevated in PD patients, but they may also act as opportunistic pathogens and even cause damage in immunocompromised individuals under certain conditions22–24. SCFAs are anti-inflammatory under certain conditions, while H2S promotes intestinal inflammation. Except for these ten genera, there seemingly are no reports related to PD in other genera. The results above suggested that inflammation may play a key role in the pathogenesis of PD. Previous studies have similarly shown a strong link between PD and inflammation-associated bacteria26,27. Therefore, metabolic pathways related to inflammatory metabolism and the ten potential inflammation-related genera mentioned above were further analyzed.
Fig. 3. Differences in microorganisms and metabolic pathways between healthy controls and patients with Parkinson’s disease (PD).
a Genera with significant differences in relative abundance. Amplicon datasets (A1–9). b Metabolic pathways (related to inflammation) with significant differences in transcripts per million (TPM) abundance. Metagenomic datasets (M1–2). SCFAs: short-chain fatty acids. The error bar represents the standard error of the mean abundance in each bar plot.
Significant changes in inflammatory metabolic pathways
Functional annotation was performed using HUMANn3 based on the metagenomic combined dataset (M1–2), and then potential inflammatory metabolism-related pathways and genes were selected for further analysis. By reviewing the literature on Google Scholar with “Parkinson” and “inflammation” as keywords, four commonly reported metabolic pathways (SCFAs, sulfate reduction, lipopolysaccharide, and glutamate) related to intestinal inflammation were identified18,19. SCFAs resist intestinal inflammation, while sulfate reduction, lipopolysaccharide, and glutamate promote intestinal inflammation. The genes of these four metabolic pathways were determined through the MetaCyc database28. Finally, 7958 genes (UniRef90) for these four metabolic pathways were extracted from the results of HUMANn3. The transcripts per million (TPM) abundances29 of genes belonging to the same metabolic pathway were summed as the abundance of that metabolic pathway.
The TPM abundances of 63 genes were significantly changed (p < 0.01, Wilcoxon rank-sum test, Supplementary Fig. 3b) in the combined dataset (M1–2). Among them, 19 SCFAs genes were significantly decreased, and all sulfate reduction (4), lipopolysaccharide (4) metabolism and glutamate metabolism (18) genes were increased in PD. Moreover, the SCFAs pathway was significantly decreased, while the sulfate reduction, lipopolysaccharide and glutamate metabolism pathways were increased in PD (p < 0.01, Wilcoxon rank-sum test, Fig. 3b).
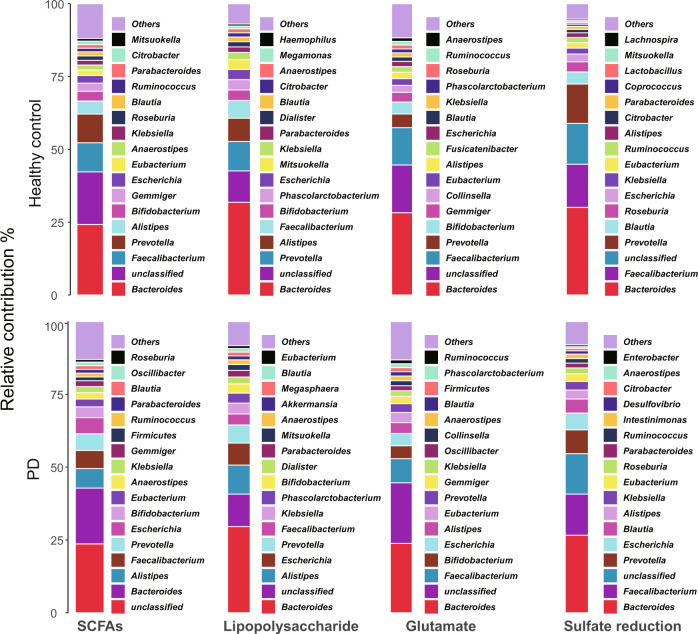
HUMANn3 provided the correspondence between genes and microorganisms, which allows us to analyze the source of genes29. The source of the genes of the four metabolic pathways was shown in Fig. 4 and Supplementary Table 4 at the genus level in the metagenomic combined dataset (M1-2). Bacteroides, Faecalibacterium, Prevotella, and Alistipes had the greatest contribution to the four metabolic pathway genes. 128 genera can provide SCFAs metabolism genes, including Roseburia, Faecalibacterium, Blautia, Lachnospira, and Prevotella. 114 genera can provide glutamate metabolism genes, 89 genera can provide lipopolysaccharide metabolism genes, and 73 genera can provide sulfate reduction genes, including Streptococcus, Bifidobacterium, Lactobacillus, Akkermansia, and Desulfovibrio. It is worth noting that Roseburia not only provided a large number of SCFAs genes, but its contribution to SCFAs genes decreased (p < 0.05, Wilcoxon rank-sum test) in PD, from 1.5% in healthy control to 0.9% in PD, and the contribution of Desulfovibrio to sulfate reduction genes was significantly increased (p < 0.05, Wilcoxon rank-sum test), from 0.04% in healthy control to 1% in PD (Fig. 4 and Supplementary Table 4).
Fig. 4. The contribution rate of genera to the four inflammation-related metabolic genes.
The value is the mean contribution rate in two Metagenomic datasets (M1–2). Parkinson’s disease (PD). SCFAs short-chain fatty acids.
Genome reconstruction
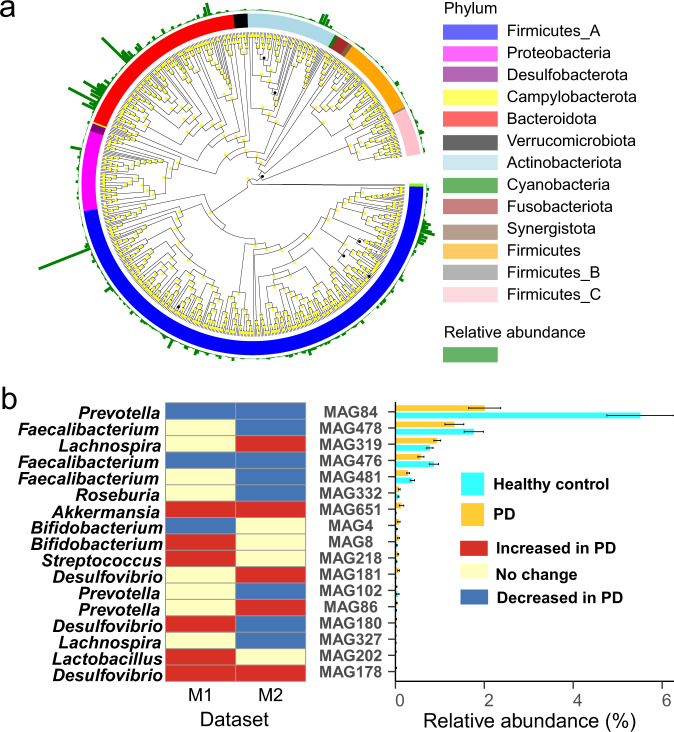
To further explore the correspondence between genes and microorganisms, binning analysis was performed. Binning yielded 654 metagenome-assembled genomes (MAGs) with high-quality (completeness > 80%, contaminate < 5%, 652 bacterial MAGs and 2 archaeal MAGs) from the combined dataset (M1-2), including 13 phyla (Fig. 5a and Supplementary Table 5). Most MAGs are Firmicutes_A (318) and Bacteroidota (116), and most Bacteroidota MAGs had high relative abundance. For the combined dataset (M1-2), the relative abundances of 242 MAGs were significantly different between PD and healthy groups (p < 0.05, Wilcoxon rank-sum test), and the number of pro-inflammatory and anti-inflammatory genes contained in each MAG is shown in Supplementary Fig. 4 and Supplementary Table 5.
Fig. 5. Metagenome-assembled genomes analysis.
a The maximum-likelihood phylogenetic tree of the 654 metagenome-assembled genomes (MAGs). The tree was based on the 120 bacterial (122 archaeal) concatenated ribosomal proteins, colored by phyla. Bootstrap values were calculated based on 1000 replicates, and the value higher than 80% were marked in yellow. b The 17 MAGs with significant differences between Parkinson’s disease (PD) and healthy groups in at least one Metagenomic dataset (M1–2). The error bar represents the standard error of the mean relative abundance in each bar plot.
As shown in Fig. 5b, 17 MAGs related to inflammation were significantly different between PD and healthy groups (p < 0.05, Wilcoxon rank-sum test). The results of MAGs analysis (Fig. 5b) were generally consistent with amplicon analysis (Fig. 3a) that the relative abundances of potential anti-inflammatory MAGs were significantly decreased in PD in at least one dataset (M1 and M2) and potential pro-inflammatory MAGs were significantly increased.
Predicting PD with inflammatory microbes and genes
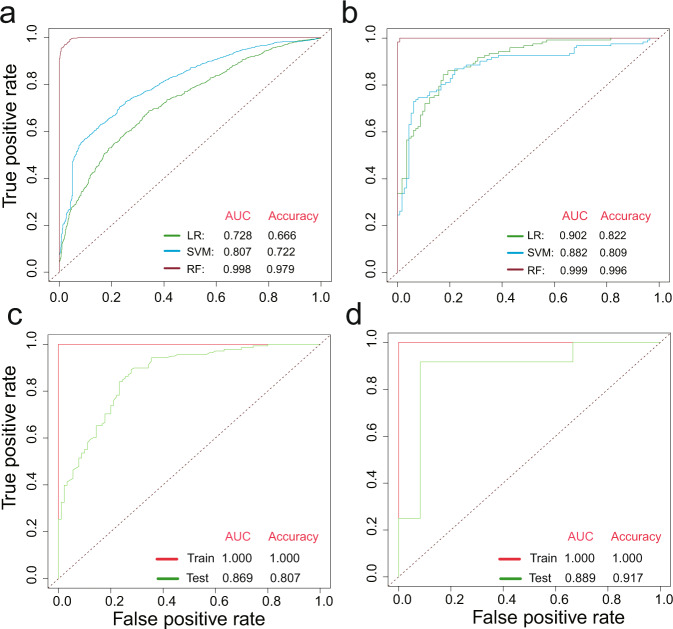
The above results have demonstrated that the relative abundances of potential inflammation-related microorganisms and genes in PD changed significantly. These differential microorganisms and genes were then used to build classification models through three machine learning methods (logistic regression (LR), support vector machines (SVM), and random forests (RF)), and the receiver operating characteristic (ROC) curve and the area under the curve (AUC) were used to evaluate the model performance. Based on the 32 genera (Supplementary Fig. 3a) which significantly changed in PD, the classification model with high accuracy can be obtained (Fig. 6a), and the performance of RF (AUC = 0.99, Accuracy = 97%) was better than that of SVM (AUC = 0.80, Accuracy = 72%) and LR (AUC = 0.72, Accuracy = 66%). Based on the 63 genes (Supplementary Fig. 3b) which significantly changed in PD, the classification model with high accuracy also can be obtained (Fig. 6b), and the performance of RF (AUC = 0.99, Accuracy = 99%) was better than that of SVM (AUC = 0.88, Accuracy = 80%) and LR (AUC = 0.90, Accuracy = 82%). Therefore, RF was chosen for further analysis.
Fig. 6. The prediction models constructed by machine learning.
The receiver operating characteristic (ROC) curve of models was calculated based on 32 genera (a) and 63 genes (b) with three different methods (i.e., logistic regression (LR), support vector machines (SVM), and random forests (RF)). The performance of the optimal model using 11 genera (c) or 6 genes (d) on training and test sets.
Considering that it is necessary to minimize the measured indicators to reduce the cost in the actual diagnosis process, the model was further optimized. Firstly, the MeanDecreaseGini index of the 32 genera and the 63 genes was calculated. The larger the MeanDecreaseGini, the more important it is to the model. Then, the genes were sorted according to MeanDecreaseGini from large to small (Supplementary Fig. 5), and different numbers of genera or genes from the front were selected for modeling. Ultimately, the optimal model was obtained based on 11 genera or 6 genes (Supplementary Fig. 6). The optimized models (Fig. 6c, d) had good performance on both the training set (AUC = 1, Accuracy = 100%, based on 11 genera or 6 genes) and the test set (AUC = 0.869, Accuracy = 80.7%, based on 11 genera; AUC = 0.889, Accuracy = 91.7%, based on 6 genes). The importance of each variable in the optimal model is shown in Supplementary Fig. 7.
Discussion
This study is the largest meta-analysis of the gut microbiome in PD to date, which provided for the first time an integrative analysis of 16S rRNA gene and shotgun metagenomic data on PD and a detailed exploration of how alterations in gut bacterial composition and function affect PD. Firstly, there was no significant difference in bacterial alpha and beta diversity between PD patients and healthy controls (Supplementary Fig. 1). Since the samples in this study came from various countries and regions, the difference may be masked by some confounding factors such as dietary habits, region, gender, sampling method, etc23. However, the co-occurrence networks (Fig. 2 and Supplementary Fig. 2) showed that the co-occurrence network of PD was obviously changed. Of note, Prevotella was only present in the network of healthy control after removing the nodes with few connections (<5). In addition, Fig. 3a and Supplementary Fig. 3a showed that the relative abundances of potential anti-inflammatory bacteria were decreased and potential pro-inflammatory bacteria were increased in PD.
Therefore, we hypothesized that inflammation is a factor contributing to PD. Then, the genes of metabolic pathways associated with inflammation were further analyzed. Here, SCFAs metabolism was a potential anti-inflammatory metabolic pathway, and potential pro-inflammatory metabolic pathways include lipopolysaccharide, H2S and Glutamate metabolism. Our results showed that (Supplementary Fig. 3b and Fig. 3b) more than half of the potential anti-inflammatory genes and pathways were significantly decreased in PD, while that of all the potential pro-inflammatory genes and pathways were significantly increased. Furthermore, the gene source and binning analysis (Figs. 4 and 5) clarified which microorganisms provided these genes at the genus and genome level. These results further demonstrated that the significantly altered gut microbes mentioned above indeed have potential anti- or pro-inflammatory functions. Finally, the optimal RF models were obtained with high accuracy (>80%) to distinguish PD from healthy control based on the 11 genera or the 6 genes related to inflammation. Interestingly, the model based on the 6 genes outperformed the model based on the 11 genera (Fig. 6). However, the diagnosis of PD remains a problem since many clinical characteristics of PD overlap with other neurodegenerative diseases30.
SCFAs31,32 are the most common gut microbial metabolites, of which over 95% are composed of butyrate, propionate and acetate. SCFAs have numerous physiological functions such as manipulating the maturation of microglia (immune effector cells) in the central nervous system, strengthening intestinal epithelial cells, and reducing inflammation risk19. SCFAs also can bind to G protein-coupled receptors such as GPR41, GPR109A and GPR43, and exert anti-inflammatory effects by activating regulatory T cells33. Previous studies also demonstrated that the SCFAs-producing bacteria were reduced in PD32. A study in Germany showed that PD patients had reduced SCFAs in their feces34. Reduced SCFAs (i.e., butyrate, acetate, and propionate) in feces were also found using both a targeted gas chromatography platform and an untargeted nuclear magnetic resonance metabolomics platform35. In this study, five potential SCFAs producers (Roseburia, Faecalibacterium, Blautia, Lachnospira, and Prevotella) were decreased in PD, which was consistent with previous studies19,20,32. However, the change of Prevotella in PD was controversial in previous literature. For example, the previous meta-analysis19,32 reported that most studies found a decreased Prevotella in PD, but opposite results were also obtained in some studies. Wallen et al.23 claimed that this contradiction may be due to the use of different taxonomic classifiers in different studies (Supplementary Table 1). To avoid this contradiction, the same taxonomic classifier was used for the nine amplicon datasets in this study.
Lipopolysaccharide is a component of the outer wall of Gram-negative bacteria. Bacterial lipopolysaccharide was proven to alter miRNA expression in macrophages, resulting in a cascade of inflammatory responses22. This process will lead to mitochondrial dysfunction, iron accumulation, dopamine depletion, and neuroinflammation that may further drive the development of PD36. Lipopolysaccharide can also lead to toll-like receptor (TLR) activation, causing gut and brain inflammation and barrier deficiency in PD. PD mouse models indicated that lipopolysaccharide can result in a loss (34%) of dopamine neurons and a heavy pro-inflammatory response through glial activation and increased TNF-α, IL-10, IL-6, and IL-120. Pietrucci et al.37 also reported a high level of lipopolysaccharide synthesis in PD, which is consistent with the findings of this study. However, it should be noted that not all bacteria that produce lipopolysaccharides will promote inflammation. Therefore, in this study, these bacteria which increased in PD and contain lipopolysaccharides synthesis genes were thought to have the potential to promote inflammation in PD. However, it is worth noting that these bacteria can not be considered as “classical intestinal pro-inflammatory bacteria”, because there was no direct evidence to prove that they have a pro-inflammatory effect. Similarly, the other changed bacteria mentioned in this paper can not simply be considered as “classic inflammation-associated bacteria”, such as Lactobacillus and Bifidobacteria.
As a gas neurotransmitter, H2S is produced by sulfate reduction of certain gut microbes (such as Desulfovibrio), which can affect neuronal signaling at low concentrations and be severely toxic at high concentrations25. High concentrations of H2S can help to release mitochondrial cytochrome c into the cytoplasm, where the cytochrome can then form α-synuclein free radicals, ultimately triggering the aggregation of α-synuclein38. In addition, H2S can increase the level of iron in the cytoplasm, which will further lead to α-synuclein aggregation. High concentrations of H2S also can inhibit intestinal motility and cause constipation, serious central nervous system dysfunction and even death39. H2S can reduce disulfide bonds in the mucosal layer of the enteric epithelium, thereby disrupting the intestinal barrier40,41. Desulfovibrio is the dominant sulfate reduction bacteria in the human gut25, also producing lipopolysaccharide and Fe3O4. Desulfovibrio has the capacity to reduce ferric iron to ferrous iron by the periplasmic [FeFe]-hydrogenase, which is present in almost all Desulfovibrio, and thus can produce Fe3O442. Exposed Fe3O4 nanoparticles have been proven to stimulate α-synuclein aggregation43. Of note, multiple lines of evidence in this study (Figs. 3, 4 and 5) have repeatedly confirmed that Desulfovibrio can provide sulfate reduction genes and its relative abundance is significantly increased in PD which was consistent with the previous study25.
Glutamate acts as an excitatory neurotransmitter, causing excitatory responses44. Glutamate is the richest excitatory neurotransmitter in the human brain, which is 1000 times higher than other important excitatory neurotransmitters such as serotonin, dopamine, and norepinephrine45. Excessive glutamate induces overstimulation of glutamate receptors and increases intracellular Na+ and Ca2+ concentrations, which can directly lead to neuronal damage and cell death. Inflammation is known to induce glutamate excitotoxicity, and a high level of glutamate will cause elevated harmful amino acid metabolite (phenylacetylglutamine) that further exacerbate inflammation46. For these reasons, glutamate synthesis was defined as a potential pro-inflammatory pathway in the present study. However, this does not imply that glutamate is a formal inflammatory factor.
The combination of a mucus layer composed of mucins with the gut microbiota is considered as a gut biofilm. The gut biofilm can prevent intestinal damage, thereby preventing intestinal permeability. Decreased Blautia, Roseburia, and Faecalibacterium in this study (Fig. 3 and Supplementary Fig. 3) are commensal bacteria involved in gut biofilm24. Increased Akkermansia (Figs. 3 and 5) which has been reported47 may lead to intestinal permeability, as this genus requires mucus for energy, leading to biofilm disruption. Defects in the gut barrier increase the risk of systemic exposure to inflammatory microbial products such as lipopolysaccharide18. In addition, increased lipopolysaccharide and decreased lipopolysaccharide-binding protein were detected in the blood, supporting the existence of a defect in the intestinal barrier48.
Growing research supported inflammation as a hallmark of PD49,50. Raised numerous inflammatory molecules in the brain and blood were founded in PD patients1,49. Excess inflammatory microbial products (such as lipopolysaccharides) may cause damage to the intestinal barrier, further leading to systemic inflammation51. Compared with healthy controls, PD patients had higher levels of zonulin and alpha-1-antitrypsin, markers of intestinal permeability. Researchers found that the longer the course of PD, the less anti-inflammatory bacteria and more pathogenic bacteria20. High-level intestinal inflammation can activate glial cells and enteric neurons, and lead to α-synuclein misfolding and aggregation31.
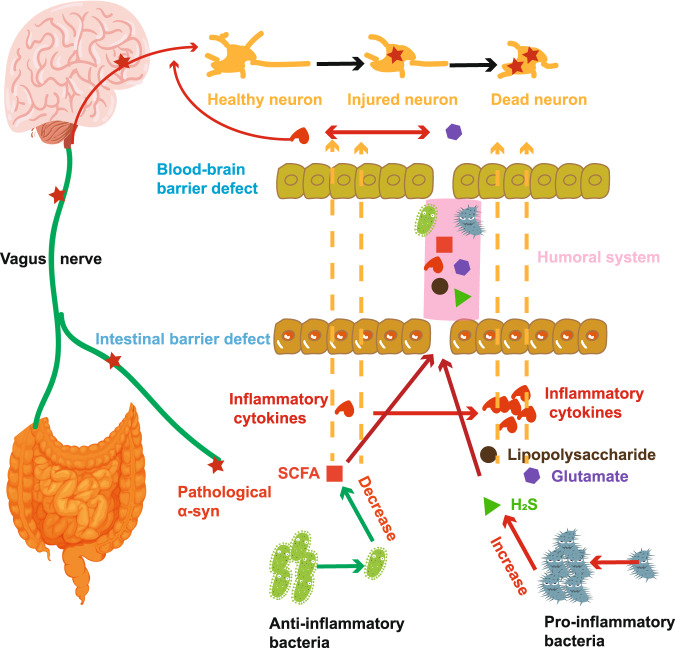
Based on the above analysis, we proposed a potential model to elucidate how inflammation contributes to PD (Fig. 7). In the gut of PD patients, the changed bacteria may lead to a decrease of anti-inflammatory factors (such as SCFAs) and an increase of pro-inflammatory factors (such as lipopolysaccharide, H2S and glutamate), causing intestinal inflammation and intestinal barrier damage. Intestinal barrier defect induces leakage of the microbiota and its metabolites (such as lipopolysaccharide, H2S and glutamate) into the body, prompting the production of inflammatory cytokines and pathological α-synuclein, further causing blood-brain barrier deficiency. These microorganisms and their metabolites can cross the blood-brain barrier through the humoral system, resulting in microglia and astrocytes activation and brain neuroinflammation9. Pathologic α-synuclein may be transmitted to the brain through the vagus nerve or other pathways14,15,47. These inflammatory factors, pathological α-synuclein, and microbial metabolites lead to the dysfunction and even death of dopaminergic neurons, eventually causing PD.
Fig. 7. The potential pathogenesis of Parkinson’s disease based on inflammation.
Changes in bacterial abundance may lead to decreased anti-inflammatory substances, such as short-chain fatty acids (SCFAs), and increased pro-inflammatory substances (Lipopolysaccharide, Glutamate, H2S), resulting in the accumulation of pathological α-synuclein (α-syn), increased inflammatory cytokines, intestinal inflammation and intestinal barrier defects. These substances may reach the brain through the vagus nerve or humoral system, and may eventually cause Parkinson’s disease (PD).
In conclusion, we presented the largest-to-date meta-analysis of the microbial community in the gut of PD, including 16S rRNA gene and shotgun metagenomic data simultaneously. The results showed that potential pro-inflammatory bacteria and genes in PD were significantly increased, while potential anti-inflammatory bacteria and genes were significantly reduced. These changes may result in decreased levels of SFCAs, which may have anti-inflammatory effects, and increased levels of lipopolysaccharides, H2S and glutamate, which may have pro-inflammatory effects. Furthermore, RF models can predict PD with high accuracy based on 11 genera (>80%) or 6 genes (>90%) associated with inflammation. Finally, we proposed a potential mechanism to clarify how inflammation contributes to PD. We believe that inflammation may be a future therapeutic target for PD.
Methods
Data collection
After quality control (for details, see below), metadata related to PD from 7 countries was collected from 11 studies with 2269 16S rRNA gene amplicon samples (1373 PD and 896 healthy controls) and 236 shotgun sequencing metagenomic samples (122 PD and 114 healthy controls) by searching the keywords “Parkinson” and “microbes” in the National Center for Biotechnology Information (NCBI) SRA database and Google Scholar (Fig. 1). Raw data in this study were obtained from two open-access databases: The European Nucleotide Archive and NCBI SRA database. Details of metadata are provided in Supplementary Table 1, such as BioProject number, country, database, primers, sequencing platform, etc. For the nine 16S rRNA gene studies, four different primer pairs (515F:806R; 314F:806 R; 520F:907R; and 341F:785R) were identified from the metadata, using the V3-V4, V4, and V4-V5 regions to produce amplicons. However, only four of the nine 16S rRNA gene studies we collected provided some confounding factors. Therefore, this study did not control for confounders in our subsequent analysis.
16S rRNA gene data processing
According to previous research52,53, adapter, barcodes, and low-quality reads (quality score below 20) were screened using Cutadpt v3.454 and paired-end reads were joined using VSEARCH v2.755. To avoid the interference caused by different sequencing regions, the V4 region of all 16S rRNA gene data was extracted using Cutadpt v3.4 with the primer set 520F-785R. Reads <150 bp or samples with fewer than 10,000 reads were removed before OTU clustering. After quality control, all reads were mapped to Greengenes database 13.8 with 97% identity using VSEARCH v2.7 to create the OTU table and assign taxonomy to reference sequences based on the taxonomic information in the Greengenes database52. The Greengenes database is comprised of full-length sequences which can further reduce the biased result from different 16 S rRNA gene regions. The OTUs that only appeared in less than one-tenth of all samples were deleted to address PCR biases.
Shotgun data processing
The quality control process of shotgun data was the same as above. Besides, human reads were removed using KneadData software (https://huttenhower.sph.harvard.edu/kneaddata) with the default parameters. Functional profiling was performed with HUMANn329 using clean reads with default settings based on the UniRef90 database. The associations between genes and microorganisms were obtained from the result of HUMANn3.
MEGAHIT v1.2.9 was used to assemble the clean reads into contigs with the parameters (--min-contig-len 500, --presets meta-sensitive)56. Contigs larger than 1500 bp were automated binned by MetaWRAP v1.3.2 (Binning module) with the parameters (--metabat2 --maxbin2 --concoct) to MAGs57. dRep v1.4.358 was used to evaluate the completeness and contamination of MAGs. MAGs were first dereplicated using dRep v1.4.3 with the parameters (-comp 80 -con 5), and only high-quality MAGs (completeness > 80% and contamination < 5%) were selected for further analysis. Open reading frames were predicted from MAGs using Prodigal v2.6.3 with default parameters59, and genes were assigned to the UniRef90 database for functional annotation using Diamond v2.0.660 with an e-value cutoff of 10−5. The taxonomic classifications of MAGs were inferred using GTDB-Tk v1.561 with default parameters. The Maximum-likelihood phylogenetic tree was generated using the IQ-TREE v1.6.1262 based on the 120 bacterial (122 archaeal) concatenated ribosomal proteins extracted by GTDB-Tk v1.5 and visualized using Evolview363. Bootstrap values were calculated based on 1000 replicates.
Statistical analysis
Group differences in taxonomy and gene profile were analyzed using the Wilcoxon rank-sum test. In this study, the results of all multiple comparisons with p < 0.05 were considered statistically significant, using the Benjamini-Hochberg (BH) method for p-value correction. The correlations among OTUs were calculated using R based on Spearman’s rank correlation (r > 0.7 and p < 0.05). The OTUs that only appeared in less than one-tenth of all samples were removed before the calculation of correlations. Co-occurrence networks were established using Gephi v0.9.264. PCoA was performed using the R package “vegan” based on the Bray-Curtis distance and analysis of similarities (ANOSIM) was used to determine whether the difference is significant after PCoA.
Model building based on machine-learning
To better distinguish PD patients from healthy controls, three well-established machine-learning algorithms (i.e., LR, SVM, and RF52) were performed using tenfold cross-validation by R to construct models using the abundances of genus or genes. In the process of model construction, the combined amplicon (A1–9) or metagenomic (M1–2) data were first divided into ten parts. Then nine parts (training set) were randomly selected and used for model construction, and then the remaining independent part (test set) was used for model validation. After that, the ROC curve and AUC were used to evaluate the model performance. The importance of each feature in the model was assessed by the R package “randomForest”. Then, the features were sorted by importance, and different numbers of features were selected for modeling to determine the most concise model. The detailed code and documentation for model building are available on GitHub (https://github.com/Yuange-lab/Shiqing-Nie).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (42177274, 31961133025), and State Key Laboratory of Urban and Regional Ecology (SKLURE2022-1-3).
Author contributions
S.N. and Y.G. designed the research and wrote the manuscript. S.N. analyzed the data. J.W., Y.D., and Z.Y. provided advice on data analysis and manuscript writing. Y.G. supervised the project. All authors revised the manuscript and approved the final manuscript.
Data availability
654 MAGs have been deposited into the China National GeneBank DataBase (CNGBdb) with accession number CNP0002780.
Code availability
The detailed code and documentation for model building are available on GitHub (https://github.com/Yuange-lab/Shiqing-Nie).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-022-00367-z.
References
- 1.Armstrong MJ, Okun MS. Diagnosis and treatment of Parkinson disease: a review. Jama-J. Am. Med Assoc. 2020;323:548–560. doi: 10.1001/jama.2019.22360. [DOI] [PubMed] [Google Scholar]
- 2.Lubomski M, et al. Parkinson’s disease and the gastrointestinal microbiome. J. Neurol. 2020;267:2507–2523. doi: 10.1007/s00415-019-09320-1. [DOI] [PubMed] [Google Scholar]
- 3.Adams-Carr KL, et al. Constipation preceding Parkinson’s disease: a systematic review and meta-analysis. J. Neurol. Neurosur Ps. 2016;87:710–716. doi: 10.1136/jnnp-2015-311680. [DOI] [PubMed] [Google Scholar]
- 4.Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: A review. Ann. Neurol. 2017;81:369–382. doi: 10.1002/ana.24901. [DOI] [PubMed] [Google Scholar]
- 5.Yan, Y. P. et al. Gut microbiota and metabolites of alpha-synuclein transgenic monkey models with early stage of Parkinson’s disease. Npj Biofilms Microbiol. 7, 69 (2021). [DOI] [PMC free article] [PubMed]
- 6.Lubomski, M. et al. Parkinson’s disease and the gastrointestinal microbiome: clinicopathological correlations and controversies. J. Neurol. Neurosur Ps. 90, e7 (2019).
- 7.Stanislawski M. A., Dabelea D., Lange L. A., Wagner B. D. & Lozupone C. A. Gut microbiota phenotypes of obesity. Npj Biofilms Microbiol. 5, 18 (2019). [DOI] [PMC free article] [PubMed]
- 8.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 9.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X. D., Qian Y. W., Xu S. Q., Song Y. Y. & Xiao Q. Longitudinal analysis of fecal microbiome and pathologic processes in a rotenone induced mice model of Parkinson’s disease. Front. Aging Neurosci. 9, 441 (2018). [DOI] [PMC free article] [PubMed]
- 11.Travagli RA, Browning KN, Camilleri M. Parkinson disease and the gut: new insights into pathogenesis and clinical relevance. Nat. Rev. Gastro Hepat. 2020;17:673–685. doi: 10.1038/s41575-020-0339-z. [DOI] [PubMed] [Google Scholar]
- 12.Braak H, Rub U, Gai WP, Del Tredici K. Idiopathic Parkinson’s disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural Transm. 2003;110:517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- 13.Barrenschee, M. et al. Distinct pattern of enteric phospho-alpha-synuclein aggregates and gene expression profiles in patients with Parkinson’s disease. Acta Neuropathol. Com. 5, 1 (2017). [DOI] [PMC free article] [PubMed]
- 14.Zheng WX, et al. Regulation of immune-driven pathogenesis in Parkinson’s disease by gut microbiota. Brain Behav. Immun. 2020;87:890–897. doi: 10.1016/j.bbi.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Svensson E, et al. Vagotomy and subsequent risk of Parkinson’s disease. Ann. Neurol. 2015;78:522–529. doi: 10.1002/ana.24448. [DOI] [PubMed] [Google Scholar]
- 16.Luk KC, et al. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hijaz BA, Volpicelli-Daley LA. Initiation and propagation of alpha-synuclein aggregation in the nervous system. Mol. Neurodegener. 2020;15:19. doi: 10.1186/s13024-020-00368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aho, V. T. E. et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 16, 6 (2021). [DOI] [PMC free article] [PubMed]
- 19.Romano, S. et al. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. Npj Parkinsons Dis. 7, 27 (2021). [DOI] [PMC free article] [PubMed]
- 20.Chen, Z. J. et al. Association of Parkinson’s disease with microbes and microbiological therapy. Front. Cell Infect. Microbiol. 11, 619354 (2021). [DOI] [PMC free article] [PubMed]
- 21.Li Z, et al. Oral, nasal, and gut microbiota in Parkinson’s disease. Neuroscience. 2022;480:65–78. doi: 10.1016/j.neuroscience.2021.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Haikal, C., Chen, Q. Q. & Li, J. Y. Microbiome changes: an indicator of Parkinson’s disease? Transl. Neurodegener. 8, 38 (2019). [DOI] [PMC free article] [PubMed]
- 23.Wallen, Z. D. et al. Characterizing dysbiosis of gut microbiome in pd: evidence for overabundance of opportunistic pathogens. Npj Parkinsons Dis. 6, 11 (2020). [DOI] [PMC free article] [PubMed]
- 24.Lorente-Picon, M. & Laguna, A. New avenues for Parkinson’s disease therapeutics: disease-modifying strategies based on the gut microbiota. Biomolecules11, 433 (2021). [DOI] [PMC free article] [PubMed]
- 25.Murros, K. E., Huynh, V. A., Takala, T. M. & Saris, P. E. J. Desulfovibrio bacteria are associated with Parkinson’s disease. Front. Cell Infect. Microbiol. 11, 652617 (2021). [DOI] [PMC free article] [PubMed]
- 26.Heintz-Buschart A, et al. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2018;33:88–98. doi: 10.1002/mds.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tansey, M. G. et al. Inflammation and immune dysfunction in Parkinson disease. Nat Rev Immunol. 22, 657–673 (2022). [DOI] [PMC free article] [PubMed]
- 28.Caspi R, et al. The metacyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48:D445–D453. doi: 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with biobakery 3. Elife10, e65088 (2021). [DOI] [PMC free article] [PubMed]
- 30.Tolosa E, Garrido A, Scholz SW, Poewe W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021;20:385–397. doi: 10.1016/S1474-4422(21)00030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, S. Y. et al. Potential roles of gut microbiota and microbial metabolites in Parkinson’s disease. Ageing Res. Rev. 69, 101347 (2021). [DOI] [PubMed]
- 32.Nishiwaki H, et al. Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord. 2020;35:1626–1635. doi: 10.1002/mds.28119. [DOI] [PubMed] [Google Scholar]
- 33.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 34.Unger MM, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. D. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Toh TS, et al. Gut microbiome in Parkinson’s disease: new insights from meta-analysis. Parkinsonism Relat. D. 2022;94:1–9. doi: 10.1016/j.parkreldis.2021.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Chu CQ, Yu LL, Chen W, Tian FW, Zhai QX. Dietary patterns affect Parkinson’s disease via the microbiota-gut-brain axis. Trends Food Sci. Tech. 2021;116:90–101. doi: 10.1016/j.tifs.2021.07.004. [DOI] [Google Scholar]
- 37.Pietrucci D, et al. Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Parkinsonism Relat. D. 2019;65:124–130. doi: 10.1016/j.parkreldis.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Kumar A, Ganini D, Mason RP. Role of cytochrome c in alpha-synuclein radical formation: Implications of alpha-synuclein in neuronal death in maneb- and paraquat-induced model of Parkinson’s disease. Mol. Neurodegener. 2016;11:70. doi: 10.1186/s13024-016-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haouzi, P., Sonobe, T. & Judenherc-Haouzi, A. Hydrogen sulfide intoxication induced brain injury and methylene blue. Neurobiol. Dis. 133, 104474 (2020). [DOI] [PubMed]
- 40.Mao, L. W. et al. Cross-sectional study on the gut microbiome of Parkinson’s disease patients in central China. Front. Microbiol. 12, 728479 (2021). [DOI] [PMC free article] [PubMed]
- 41.Ijssennagger N, van der Meer R, van Mil SWC. Sulfide as a mucus barrier-breaker in inflammatory bowel disease? Trends Mol. Med. 2016;22:190–199. doi: 10.1016/j.molmed.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Pereira, I. A. C. et al. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front. Microbiol. 2, 69 (2011). [DOI] [PMC free article] [PubMed]
- 43.Murros, K. et al. Magnetic nanoparticles in human cervical skin. Front. Med-Lausanne. 6, 123 (2019). [DOI] [PMC free article] [PubMed]
- 44.Reiner A, Levitz J. Glutamatergic signaling in the central nervous system: Ionotropic and metabotropic receptors in concert. Neuron. 2018;98:1080–1098. doi: 10.1016/j.neuron.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J., Wang, F. S., Mai, D. M. & Qu, S. G. Molecular mechanisms of glutamate toxicity in Parkinson’s disease. Front. Neurosci-Switz. 14, 585584 (2020). [DOI] [PMC free article] [PubMed]
- 46.Cirstea MS, et al. Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Mov. Disord. 2020;35:1208–1217. doi: 10.1002/mds.28052. [DOI] [PubMed] [Google Scholar]
- 47.Yemula N, Dietrich C, Dostal V, Hornberger M. Parkinson’s disease and the gut: Symptoms, nutrition, and microbiota. J. Parkinson Dis. 2021;11:1491–1505. doi: 10.3233/JPD-212707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Waal, G. M. et al. Correlative light-electron microscopy detects lipopolysaccharide and its association with fibrin fibres in Parkinson’s disease, alzheimer’s disease and type 2 diabetes mellitus. Sci Rep-Uk. 8, 16798 (2018). [DOI] [PMC free article] [PubMed]
- 49.Lin, C. H. et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 16, 129 (2019). [DOI] [PMC free article] [PubMed]
- 50.Houser, M. C. & Tansey, M. G. The gut-brain axis: Is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? Npj Parkinsons Dis. 3, 3 (2017). [DOI] [PMC free article] [PubMed]
- 51.Brown, G. C. The endotoxin hypothesis of neurodegeneration. J. Neuroinflamm. 16, 180 (2019). [DOI] [PMC free article] [PubMed]
- 52.Yuan J, et al. Predicting disease occurrence with high accuracy based on soil macroecological patterns of fusarium wilt. Isme J. 2020;14:2936–2950. doi: 10.1038/s41396-020-0720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, T. Y., Wu, P. H., Lin, Y. T. & Hung, S. C. Gut dysbiosis and mortality in hemodialysis patients. Npj Biofilms Microbiol. 7, 20 (2021). [DOI] [PMC free article] [PubMed]
- 54.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:3. [Google Scholar]
- 55.Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahe, F. Vsearch: a versatile open source tool for metagenomics. Peerj. 4, e2584 (2016). [DOI] [PMC free article] [PubMed]
- 56.Li DH, Liu CM, Luo RB, Sadakane K, Lam TW. Megahit: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 57.Uritskiy GV, DiRuggiero J, Taylor J. Metawrap-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olm MR, Brown CT, Brooks B, Banfield JF. Drep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. Isme J. 2017;11:2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics11, 119 (2010). [DOI] [PMC free article] [PubMed]
- 60.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 61.Parks DH, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 62.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian B, Gao SH, Lercher MJ, Hu SN, Chen WH. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bastian, M., Heymann, S. & Jacomy, M. Gephi: An open source software for exploring and manipulating networks. ICWSM. 361–362 (2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
654 MAGs have been deposited into the China National GeneBank DataBase (CNGBdb) with accession number CNP0002780.
The detailed code and documentation for model building are available on GitHub (https://github.com/Yuange-lab/Shiqing-Nie).