Abstract
The mytilid mussel Bathymodiolus thermophilus lives in the deep-sea hydrothermal vent regions due to its relationship with chemosynthetic symbiotic bacteria. It is well established that symbionts reside in the gill bacteriocytes of the mussel and can utilize hydrogen sulfide, methane, and hydrogen from the surrounding environment. However, it is observed that some mussel symbionts either possess or lack genes for hydrogen metabolism within the single-ribotype population and host mussel species level. Here, we found a hydrogenase cluster consisting of additional H2-sensing hydrogenase subunits in a complete genome of B. thermophilus symbiont sampled from an individual mussel from the East Pacific Rise (EPR9N). Also, we found methylated regions sparsely distributed throughout the EPR9N genome, mainly in the transposase regions and densely present in the rRNA gene regions. CRISPR diversity analysis confirmed that this genome originated from a single symbiont strain. Furthermore, from the comparative analysis, we observed variation in genome size, gene content, and genome re-arrangements across individual hosts suggesting multiple symbiont strains can associate with B. thermophilus. The ability to acquire locally adaptive various symbiotic strains may serve as an effective mechanism for successfully colonizing different chemosynthetic environments across the global oceans by host mussels.
Subject terms: Evolutionary ecology, Genomics, Comparative genomics, Genome evolution
Introduction
In chemosynthetic habitats such as hydrothermal vents and cold seeps, deep-sea invertebrates rely upon the mutualistic symbiotic partnership with chemoautotrophic bacteria1. Bathymodiolus mussels are among the most prevalent deep-sea animals that contribute to the deep-sea chemosynthetic habitats1,2. Bathymodiolus mussels feed on the nutrients provided by the symbiotic bacteria residing inside specialized cells of the gill tissues3,4. These endosymbiotic bacteria generate organic carbon by fixing CO2 dissolved in the seawater. This reaction is powered by the chemical energy generated from the oxidation of reduced compounds (methane, hydrogen sulfide and so on) which are also sourced from the environment5–7.
Bathymodiolonae mussels have broad geographic distributions and occupy a wide range of chemosynthetic habitats along global mid-ocean ridge system, in back arc basins, in hydrocarbon seeps along continental margins and organic falls such as whale carcasses and wood-falls8–14. Like other deep-sea invertebrates such as siboglinid tubeworms15 and lucinid clams16, Bathymodiolus mussels acquire their chemosynthetic symbionts horizontally from the vicinity15,17 during their early juvenile stage18,19. Indeed, while studies of several Bathymodiolus species such as B. childressi, B. azoricus, B. heckaerae, and B. puteoserpentis have documented the occurrence of gill symbionts during the early life stages of the mussels20,21 these symbionts were not observed in either the gonads or gametes of males and females19,22,23. A large body of work has demonstrated that most Bathymodiolus mussels can host methane-oxidizing and sulfur-oxidizing (SOX) bacteria in their gills, and that multiple SOX symbiont strains can co-exist within a single host24. It was hypothesized that the capacity of host mussels to acquire and exploit locally adapted species and strains of symbiotic bacteria was the reason for such successful distribution of host mussels in diverse deep-sea chemosynthetic habitats globally15,17. However, it is still impossible to directly culture the symbionts and experiment with them. In this context, comparative genomic analyses of Bathymodiolus symbionts can be an alternative to understand diverse symbionts’ metabolic ability with respect to the different environment and locations.
The SOX symbionts in mussels are closely related to the free-living Gammaproteobacteria SUP0525. To date the SOX symbiont genomes of B. azoricus26, B. septemdierum8, B. heckerae10, B. puteoserpentis10, B. brooksi10, B. thermophilus12, and an undescribed Bathymodiolus species from the Mid-Atlantic Ridge (B. sp. 5 South)10 have been reported (Table 1). Genomic analyses suggest that these symbionts use carbon fixation, sulfur oxidation, nitrogen, and hydrogen metabolism to harness energy. However, to adapt in variable geochemical conditions of the surrounding environments, some metabolic genes or pathways (for hydrogen and nitrogen metabolism) are either present or absent in symbionts of the same mussel species, as previously studied in B. septemdierum (from Myojin knoll, Japan)8 and B. azoricus (from Lucky Strike vent field)27. Hydrogen as an potential inorganic energy source other than sulfur or methane was first reported in symbionts of B. puteoserpentis from Logatchev hydrothermal vent field28. In the same study Petersen et al., 2011 found the hydrogenase gene in B. thermophilus from the Axial Dome vent of Pacific-Antarctic Ridge region28. However, previously studied symbiont genomes of B. thermophilus from the EPR9N regions do not possess any hydrogenase genes10,12. Although, it has been studied that hydrogen abundancy in EPR9N regions is similar to the Logatchev hydrothermal vent regions29. In this study, we found that symbiont genome of B. thermophilus studied from the EPR9N region encodes a hydrogenase cluster with additional hydrogenase subunits.
Table 1.
General information about Bathymodiolus thiotrophic symbiont genomes used for phylogenomic analyses.
| Bathymodiolus species | Name of assembly | Location | Latitude | Longitude | References |
|---|---|---|---|---|---|
| B. azoricus | SOX Menez Gwen | Atlantic Ocean | 37°50′40.0"N | 31°31′08.0"W | 10 |
| B. azoricus | BazSymA | Atlantic Ocean | 37°45′34.7"N | 31°38′15.7"W | 26 |
| B. azoricus | BazSymB | Atlantic Ocean | 37°50′40.8"N | 31°31′10.2"W | 26 |
| B. azoricus | SOX ET2 1586I | Atlantic Ocean | 37°17′21.2"N | 32°16′32.2"W | 10 |
| B. brooksi | BBROOK1789B* | Chapopote, GoM | 21°54′00.2"N | 93°26′07.4"W | 10 |
| B. brooksi | BROOK1789C* | Chapopote, GoM | 21°54′00.2"N | 93°26′07.4"W | 10 |
| B. brooksi | BBROOKSOX* | Chapopote, GoM | 21°54′00.2"N | 93°26′07.4"W | 10 |
| B. heckerae | BHECKSOX* | Chapopote, GoM | 21°54′00.2"N | 93°26′07.4"W | 10 |
| B. heckerae | BHECKSOX2* | Chapopote, GoM | 21°53′58.8"N | 93°26′07.2"W | 10 |
| B. puteoserpentis | BPUTEOSOX | Logatchev, MAR | 14°45′18.0"N | 44°59′16.0"W | 10 |
| B. septemdierum | Myojin Knoll | Myojin knoll, Japan:IBA | 32°06′14.8"N | 139°13′09.5"E | 8 |
| B. sp. 5 South | BCLUESOX | 5° South, Clueless, MAR | 4°48′11.8"S | 12°22′18.5"W | 10 |
| B. sp. 5 South | BTURTLESOX | 5° South, Wide Awake, MAR | 4°48′33.5"S | 12°22′27.8"W | 10 |
| B. thermophilus | THERMOS | EPR | 9°50′22.6"N | 104°17′32.0"W | 10 |
| B. thermophilus | THERMOT | EPR | 9°50′22.6"N | 104°17′32.0"W | 12 |
| B. thermophilus | BAT/CrabSpa'14 | EPR | 9°30′00.0"N | 104°10′12.0"W | 10 |
| B. thermophilus | BTHERMOSOX | EPR | 9°50′22.6"N | 104°17′32.0"W | 10 |
| B. thermophilus | EPR9N | EPR | 9°49′12.0"N | 104°18′00.0"W | This study |
GoM Gulf of Mexico, EPR East Pacific Rise, MAR Mid Atlantic Ridge, IBA Izu-Bonin Arc.
*From the cold seep habitat. Remaining assemblies are from the hydrothermal vent habitat.
The SOX symbionts in mussels from different geographical locations may possess distinct genome structures, accessory genes of ecological significance, and foreign genetic material indicative of their exposure to habitat-specific viromes30. Also, symbiont genome possess methylated DNA that may methylate host genome recognition sites31 and can respond to the environmental stresses32. However, the fragmented, incomplete, and contamination-prone nature of most of the published genome draft assemblies complicates comparative investigations of Bathymodiolus SOX symbiont accessory genome, structural rearrangements (such as gene duplications, translocations, or inversions) and hinders the study of mobile genetic elements and horizontal gene transfer. Therefore, in this study, we present a high-quality complete genome of the SOX symbiont associated with a specimen of B. thermophilus from the 9 N hydrothermal vent fields of East Pacific Rise (hereafter the symbiont genome referred to as EPR9N). We sequenced this symbiont genome using PacBio sequencing technology and assembled it with a hierarchical genome-assembly process (HGAP3) pipeline.
Materials and methods
Sample collection and DNA extraction
One Bathymodiolus thermophilus mussel specimen was collected from the Choo Choo site of the East Pacific Rise in 2000 (Dive number: Alvin 3540; latitude 9.82 N and longitude 104.30 W). After reaching onboard, the whole body was immediately preserved at − 80 °C, transported to the laboratory in dry ice, and stored at − 80 °C until further processing. To get high molecular weight DNA, the genomic DNA was extracted and purified from a small piece of the mussel gill tissue using the CHAOS buffer protocol and the phenol–chloroform method33.
Symbiont genome sequencing and assembly
Sequencing was performed using PacBio RS II SMRT sequencing technology (Pacific Biosciences, Menlo Park, CA, USA). An average of 20 kb insert SMRTbell library was constructed and sequenced, yielding 122,925 reads (126 X average genome coverage). De novo assembly was conducted using the hierarchical genome-assembly process (HGAP3) pipeline of the SMRT Analysis v2.3.034 using PacBio CLRs. After gap closing, the newly generated contig served as a reference to which raw PacBio reads were mapped using the resequencing module of the SMRT protocol34. Finally, a consensus assembly was finalized with Trycycler v0.5.335 from the multiple input assemblies generated by HGAP3 pipeline34, Flye v2.936 and Unicycler v3.037 assemblers. Plasmid sequences are screened from the assemblies generated by PlasFlow38 (Table S11).Assembly quality and completeness were assessed with Quast v5.0.039 and CheckM v1.0.1840 based on 280 Gammaproteobacteria specific single copy marker genes.
Assessment of symbiont diversity in the DNA library
The sequencing and assembly of multiple strains of the SOX symbiont would result in a chimeric genome which may not be representative of a true symbiont chromosome. To verify that EPR9N was assembled from a monoclonal symbiont library, we assessed the genome wide SNP frequency distribution and the genetic diversity of the clustered regularly interspaced short palindromic repeats (CRISPR) marker. One CRISPR array was identified in the EPR9N symbiont genome and its spacer sequences extracted with CRISPR-Cas + + webtool41. Following the protocol in42, we characterized the CRISPR haplotypes within our symbiont sequencing library by near matching (identity threshold = 80%) the spacers sequences to the CRISPR-containing PacBio reads using a modified version of the script call_CRISPR_haplotypes.py (available at https://github.com/maepz/CRISPR_distance/tree/master/manuscript_data_and_downstream_analyses_scripts/haplotype_detection_pipelines).
Gene prediction and annotation
We identified protein-coding genes using Prodigal v2.6.143. The predicted CDSs were BLAST-searched against UniProt, Pfam, and COG databases to gain insights into molecular functions and family classification of predicted genes. Signal peptides and transmembrane helices were predicted using SignalP v4.144 and TMHMM v2.045. rRNA, tRNA, and other miscellaneous features were predicted using RNAmmer v1.246, tRNAscan-SE v1.2147, and Rfam48, respectively. KEGG orthology (KO) numbers and functions were retrieved using BlastKOALA online49.
Methylation and mobilome analysis
The DNA base modification analysis was detected with SMRTAnalysis_2.3.0.140936. The mobile genetic elements were identified by webtools of ISfinder (https://isfinder.biotoul.fr/)50, IslandViewer4 (https://pathogenomics.sfu.ca/islandviewer))51, and PHASTER (https://phaster.ca/)52.
Phylogenomic and comparative genome analyses
For phylogenomic analyses, two vesicomyid symbiont genomes of Candidatus Ruthia endofausta and Candidatus Ruthia magnifica53 were used as outgroups. We extracted single-copy orthologous genes (SOGs) from the outgroup and all available Bathymodiolus sp. SOX symbiont genomes (Table 1) with Orthofinder v2.5.454, cleaned the sequences with trimAl v1.4.155, and built a maximum likelihood (ML) tree using the ‘WAG’ general matrix model in IQ-TREE v2.2.0_beta56. The phylogenies of hydrogenase genes (HupUVLS) were analyzed using ML method with the JTT matrix-based model in MEGA-X57. The online iTOL tool58 was used to visualize the resulting ML tree. We used Anvi’o’s Pangenomics, Phylogenomics, and average nucleotide identity (ANI) of Spiroplasma genomes workflow for comparative genome analysis among B. thermophilus symbiont genomes59. We used Mauve v2015022660 and GRIMM web server61 to detect structural variation across B. thermophilus symbiont genomes. The fragmented B. thermophilus symbiont genome assemblies were beforehand aligned to EPR9N and their contigs sorted with D-GENIES webtool62. The 49 CRISPR spacers identified in EPR9N were searched in other B. thermophilus symbiont genome assemblies with blastn by blast + v2.6.063 (e-value cutoff = 1E10−6). Comparisons of hydrogenase gene clusters were visualized by clinker and clustermap.js v1.3264. Hydrogenase operon was detected by Operon-mapper web server65. Hydrogenases were classified using HydDB web server66.
Results
EPR9N symbiont genome assembly and annotations
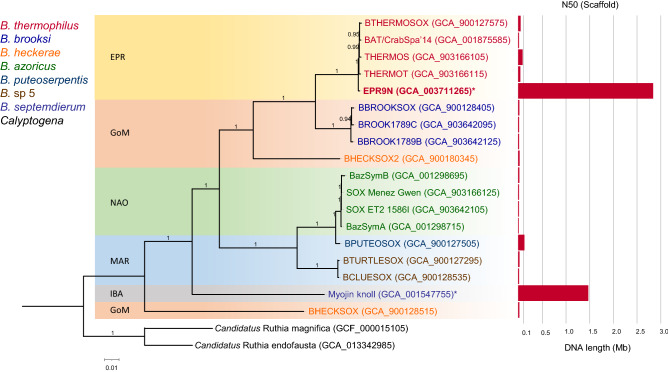
From one single-molecule real-time (SMRT) cell, we obtained 122,925 subreads with an N50 value of 12,539 bp. The longest contig generated by resequencing module of the SMRT protocol was confirmed as circular by Trycycler35. The final EPR9N assembly was a 2,832,685 bp circular contig with a GC content of 38.5%. Its mean coverage was 126 X. Assessment of genome completeness based on 280 proteobacteria-specific single copy marker genes resulted in a completeness score of 97.19% with a low level of contamination (1.99%) (Table 2). EPR9N is the most complete B. thermophilus gill symbiont genome to date (Table 2) and one of two chromosome level assemblies for Bathymodiolus SOX symbionts: the other belongings to B. septemdierum symbionts (Fig. 1). Except for the assembly BPUTEOSOX (B. puteoserpentis symbionts), the other Bathymodiolus mussel symbionts genomes have low contiguity (scaffold N50 < 0.1 Mb) (Fig. 1). Amongst B. thermophilous symbionts, assembly sizes varied from 2.25 Mb (BTHERMOSOX) to 3.09 Mb (BAT/CrabSpa'14). Among all predicted genes, 1,968 genes were functionally annotated by Pfam database, while the remaining 99 genes were unclassified or classified as from the unknown protein domains (Table S4). Predicted proteins were assigned to Clusters of Orthologous Groups numbers and Protein Families domains by querying their sequences against the COG database through the Anvi’o pipeline59.
Table 2.
Genome completeness, contamination, and assembly statistics of B. thermophilus symbionts.
| Name of assembly | Completeness | Contamination | GC | Genome size (bp) | Ambiguous bases | Scaffolds | Longest scaffold | N50 (scaffolds) |
|---|---|---|---|---|---|---|---|---|
| EPR9N | 97.19 | 1.99 | 38.57 | 2,832,685 | 0 | 1 | 2,832,685 | 2,832,685 |
| BTHERMOSOX | 97.85 | 0.66 | 38.40 | 2,249,231 | 0 | 161 | 144,199 | 43,780 |
| THERMOS | 97.19 | 0.66 | 38.19 | 2,414,318 | 95 | 100 | 347,385 | 85,091 |
| THERMOT | 97.85 | 0.99 | 38.42 | 2,333,609 | 100 | 154 | 126,494 | 39,115 |
| CrabSpa'14 | 96.96 | 6.75 | 37.74 | 3,088,407 | 14,424 | 1281 | 61,812 | 10,694 |
Figure 1.
Maximum likelihood phylogeny of single-copy-orthologous genes for deep-sea mussels’ thiotrophic gill endosymbionts. Deep-sea clam symbiont genomes of Candidatus Ruthia magnifica and Candidatus Ruthia endofausta were used as outgroup. Legends represent the host Bathymodiolus species and outgroup Calyptogena. Location of mussel symbionts are presented in background-colored boxes. EPR East Pacific Rise, GoM Gulf of Mexico, NAO North Atlantic Ocean, MAR Mid Atlantic Ridge, IBA Izu-Bonin Arc, Japan. Assembly ID of each symbiont genome is presented in parentheses. The bar chart shows each symbiont genome assembly’s N50 (Scaffold) values. *Represent the complete and circular genomes.
Symbiont genetic homogeneity in EPR9N library
One large CRISPR region (length = 3544 bp) with 49 spacers (average length = 34.6 bp) was identified in EPR9N (Table S1). Of the 263 subreads which mapped to the CRISPR locus, 40 contained the whole CRISPR array (including the leader sequence and 5’ neighboring Fic gene). These 40 sequences were identical in their spacer content and no arrays with putative new spacers or missing spacers were observed amongst all the mapped subreads. This result indicates that the libraries used to assemble EPR9N were monoclonal and thus EPR9N circular contig is biologically sound and representative of the chromosome of a SOX symbiont.
Genome similarity and structural variation
The average nucleotide identity (ANI) of EPR9N was 98% to other B. thermophilus symbiont genomes and 77–84% to other Bathymodiolus SOX symbiont genomes (Table S3). While the genome-wide ANI was high amongst B. thermophilus symbionts, large ANI variations were observed across genes. Indeed, the genome fraction with high identity to EPR9N (nucleotide identity > 75%) was at most 76% with BTHERMOSOX and at least 64% with BAT/CrabSpa'14 (Fig S1) while sequences without any match to EPR9N genome represented 14 to 23% of the other B. thermophilus associated symbiont genomes (Fig. S1).
Furthermore, genome alignment and gene rearrangement analyses showed that the genomes of B. thermophilus symbionts are structurally distinct and are distinguished by translocations and/or inversions events. Indeed, with reference to EPR9N, all but BAT/CrabSpa'14 B. thermophilus symbionts genomes show evidence of reversal events (Fig. S2).
Phylogenomic and comparative genomics
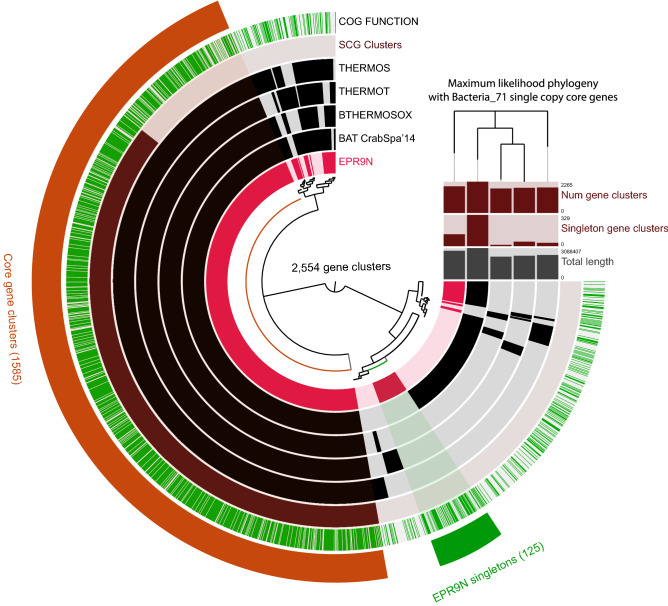
Phylogenetic analyses with 541 single-copy orthologous genes (SOGs) indicated that the B. thermophilus thiotrophic symbionts form a single cluster and are closely related to the cluster of B. brooksi thiotrophic symbionts from Gulf of Mexico (Fig. 1). All B. thermophilus symbiont genomes shared 2554 core gene clusters consisting of 9,365 genes (Fig. 2, Table S5). EPR9N, BAT/CrabSpa'14 and THERMOT contained 125, 131 and 34 unique genes, respectively (Fig. 2, Table S5). The majority of these unique genes did not have clusters of orthologous genes (COG) annotations. The remaining annotated genes include various putative phage and other mobile element remnants (i.e., repeats-containing genes, reverse transcriptases, and transposases), genes for defense mechanism (restriction endonucleases, type I restriction-modification system genes, DNA methylase subunits, RTX toxin-related, and a CRISPR/Cas system (in BAT/CrabSpa'14)). Unique genes in EPR9N also included genes for energy production and conversion such as Ni/Fe-hydrogenases, hydrogenase maturation factors, chaperons like heat shock protein HSP90, rubredoxins (a stress regulating protein) and uncharacterized proteins containing known functional domains (Table S5).
Figure 2.
Pangenomic comparison of B. thermophilus thiotrophic symbiont genomes. The inner dendrogram shows hierarchical relationships among these clusters based on their distribution across genomes. Within layers, dark colors indicate the presence of a gene cluster, while light colors indicate absence. Different gene cluster groups and their abundances are highlighted outside the pangenome graph, including core gene clusters among all B. thermophilus symbiont genomes (red) and EPR9N symbiont strain specific gene clusters (green).
Energy metabolism
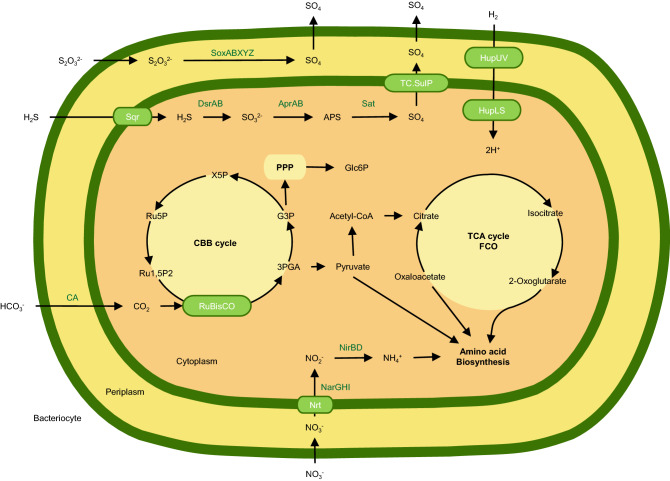
EPR9N encodes genes for the autotrophic CO2-fixation by Calvin-Benson-Bassham (CBB) cycle and the TCA cycle. While key genes for the CBB cycle are found, genes like sedoheptulose-7-phosphatase and fructose-1,6-bisphosphatase are not observed as previously reported12. But an alternative gene to complement the CBB pathway (6-phosphofructokinase) was found in the genome. TCA cycle has genes for first carbon oxidation (FCO), i.e., from oxaloacetate to 2-oxoglutarate (Fig. 3). Several other previously reported genes including malate dehydrogenase and succinate dehydrogenase11 were not found in EPR9N. This genome as well as other B. thermophilus symbiont genomes encodes two complete sets of genes for the SOX system (soxABXYZ), dissimilatory sulfate reduction and oxidation (sat, aprAB, dsrAB genes), assimilatory nitrate reduction, and dissimilatory nitrate reduction (Fig. 3, Tables S4 and S5)12.
Figure 3.
Schematic diagram of key metabolism found in B. thermophilus EPR9N symbiont genome. FCO First carbon oxidation, Glc6P α-D-Glucose-6-phosphate, PPP Pentose Phosphate Pathway, G3P Glyceraldehyde-3-phosphate, 3PGA 3-Phospho-glyceric acid, Ru1, 5P2 Ribulose 1,5-bisphosphate, Ru5P Ribulose 5-phosphate, X5P Xylulose 5-phosphate, Nar Nitrate reductase, Nir nitrite reductase, Hup Ni/Fe hydrogenase. Subcellular localization of bacterial proteins for constructing schematic metabolic pathway were predicted by PSORTb 3.0 webtool93.
Hydrogen metabolism
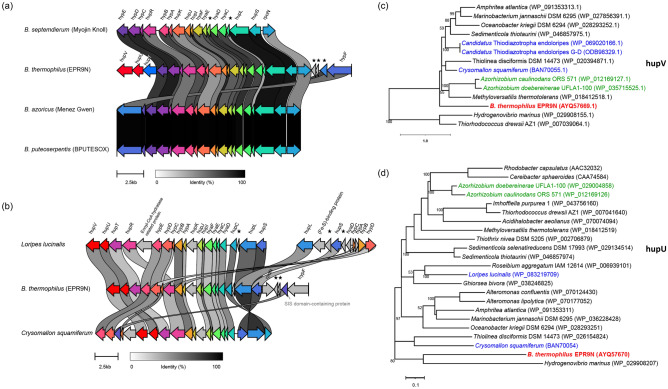
Interestingly, a regulatory hydrogenase cluster, identified as an operon (Table S8) was exclusively found in EPR9N, but not in the other B. thermophilus symbiont strains. (Table S5). However, the hydrogenase cluster was present in a few other Bathymodiolus symbionts with some variation in gene order and content. The arrangement pattern for the hypABCDE, hupIJKLRS, hyaCDEF and rpoN genes was conserved across the symbiont genomes, whereas hypF and another hypothetical gene appeared translocated in the EPR9N genome in comparison to B. azoricus (Menez Gwen) and B. puteoserpentis (BPUTESOX). BLASTP homology inferences showed that EPR9N hupLS and hypDE genes are > 80% identical to these of other Bathymodiolus mussel symbiont strains (Fig. 4a). In contrast, the same homologous genes showed > 97% identity between the B. azoricus (Menez Gwen) and B. puteoserpentis (BPUTESOX) indicating that both symbionts are closely related, which is also evident from the phylogenomic analysis. The phylogenies of hupLS genes reveal the pervasive distribution of these genes across freely living bacteria, legume symbionts and the symbionts of deep-sea invertebrates (i.e., scaly-foot snails, clams, bivalves, and tubeworms) (Fig. S3a,b). Furthermore, two additional subunits of H2-sensing hydrogenase gene (hupUV) along with a histidine kinase (hupT) gene were exclusively found in the EPR9N genome among the Bathymodiolus mussel symbionts (Fig. 4a). The hupUV and hupT genes of EPR9N strain are homologous to those of the symbiont of lucinid bivalve Loripes lucinalis living in low intertidal to about 20 m depth of European and Mediterranean coasts and the symbiont of scaly-foot snail Crysomallon squamiferum living in deep-sea hydrothermal vents of the Indian Ocean with similar gene order (Fig. 4b). BLASTP inference from the hydrogenase cluster gene-rearrangement analysis shows that hupUV genes of EPR9N is > 77% identical to the scaly-foot snail symbiont (Fig. 4b). An extended comparison of the hydrogenase cluster has revealed that the lucinid bivalve has two distinct hydrogenase clusters with a set of hydrogenase subunits (hupLS) in each cluster (Fig. 4b). The hydrogenase cluster in EPR9N genome contains both [NiFe] Group 2b and and 1d hydrogenases (Table S9). Phylogeny analysis confirmed that hupUV genes of EPR9N strain are distantly related to the symbionts of L. lucinalis and C. squamiferum, and to the free-living bacteria Hydrogenovibrio marinus (Fig. 4c,d).
Figure 4.
Schematic visualization of hydrogenase cluster regions in (a) Bathymodiolus mussel thiotrophic symbiont genomes, (b) symbiont genomes of Loripes lucinalis, B. thermophilus EPR9N, and Crysomallon squamiferum. Loripes lucinalis symbiont genome possesses two distinct hydrogen clusters. Star marks are hypothetical genes. Phylogenies of H2-signaling hydrogenases (c) HupV and (d) HupU genes, respectively. The phylogenetic trees were inferred by using the maximum likelihood method with bootstrap values on the tree branches (only over 50 are shown). The taxa in green are of the symbionts of legume host Sesbania, in blue are the symbionts of marine gastropods, and in bold red are the symbiont of B. thermophilus EPR9N. GenBank accession numbers of proteins are presented in parentheses.
Interaction with the host
Our study shows that EPR9N contains a variety of toxin and toxin-antitoxin genes, which are also found in other B. thermophilus symbiont genomes. Toxin genes such as insecticide toxin hcdB, putative AbiEii toxin, RTX toxin, and antitoxin genes such as Antidote-toxin recognition MazE, Antitoxin Phd_YefM, MraZ protein, and ParE toxin of type II toxin-antitoxin system were found. We found various genes coding for Coagulation factor, Cadherin, Phage integrase, Transglycosylase, Transpeptidase, RHS repeat, etratricopeptide repeat, Type II and IV secretion system proteins, Colicin V, hemolysin-D, which could be important for symbiont to attach and invade and colonize the host tissue (Tables S4 and S5).
Interaction with viruses
The CRISPR/Cas immunity was present in all B. thermophilus thiotrophic symbiont genomes and several assemblies appear to contain multiple CRISPR loci (Table S1). Unfortunately, because of the nature of the CRISPR array (short reads do not assemble well over repeats), CRISPR loci were too fragmented to do in-depth characterization. We nonetheless performed BLASTN searches of EPR9N CRISPR spacers against the other B. thermophilus thiotrophic symbiont genome assemblies and found matches for nine of the 49 spacers (Table S2). Interestingly, neighboring spacers different from the ones in the reference array were often found in the CRISPR containing contigs of the B. thermophilus symbiont genomes. Spacer 6 even had multiple matches within the BAT/CrabSpa’14 assembly. This could reflect that the CRISPR immunity in this bacterial species is remarkably active, but further investigation of the CRISPR/Cas evolution and the dynamics of spacer acquisition would be needed to shed further light on the symbiont-virus interactions.
DNA methylation
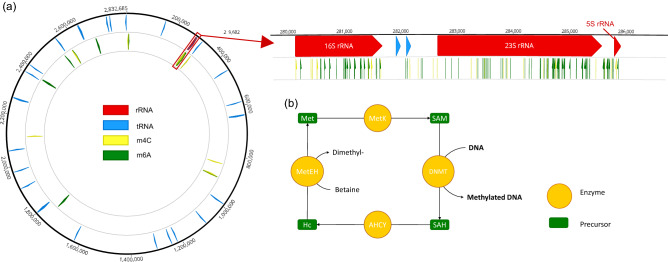
We found the methyl modification in the EPR9N genome corresponding to the m4C and m6A DNA methylation (Fig. 5a). Out of 619 m4C and m6A base modifications, 157 high quality DNA base modification signals (QV ≥ 30) are found in dense patches in rRNA, and in transposase regions. All bacterial methyltransferase genes required for DNA methylation were present in the EPR9N genome (Fig. 5b).
Figure 5.
(a) The methyl modification regions in B. thermophilus EPR9N genome. Zoomed portion shows m4C and m6A base modifications in the rRNA region. (b) Enzymatic pathway involved in DNA methylation process. DNMT DNA methyltransferase, AHCY S-adenosyl-homocysteine hydrolase, Met Methionine, Hc Homocysteine, SAH S-adenosyl-homocysteine, SAM S-adenosyl methionine.
Mobilome
A phage infection region was detected in the EPR9N genome comprising an incomplete ~ 8.5 kb prophage region consisting of 5 phage hit proteins with two bacterial and 2 hypothetical proteins (Fig. S4). Notably, type II toxin-antitoxin system VapC family toxin, tetratricopeptide repeat protein and IS5 family transposase proteins are found in this phage region. Additionally, we have detected genome island regions by IslandViewer4 (Fig. S4 and Table S6). A total of 9 genome island regions were present across the EPR9N genome. Whereas ISfinder detected a total of 58 insertion sequences (IS) of 42 IS classes in the EPR9N genome (Table S7).
Discussion
While a total of 64 symbiont genomes from ten Bathymodiolus mussel species have already been sequenced10,12,26,67, the present single contig with high-quality assembly is only the second complete genome of a Bathymodiolus-associated SOX symbionts and the first for the species B. thermophilus. As such, it represents a valuable resource for further studies on the evolution and biogeography of the deep-sea mussel holobionts.
Symbiont phylogenetic divergence seems correlated to ancient and contemporary geographic locations as the eastern Pacific (the East Pacific Rise and the Gulf of Mexico), Atlantic, and western Pacific symbionts are clustered separately (Fig. 1), as observed in case of phylogenetic relationship of tubeworm symbiont in different locations68. For instance, the relatively close genetic distance between the symbionts of B. brooksi in the Gulf of Mexico and B. thermophilus in the EPR suggests a potential connection between the two locations through ancient pathways in the past. Of course, the possible paths must have disappeared since the geological connection between the north and south Americas. Gene order within the hydrogenase gene cluster further supports this geographical structuring, particularly when the geographic distances between locations are more extended, like the comparison between the Eastern Pacific and the Atlantic symbionts. This pattern is roughly consistent with the phylogeny of Bathymodiolus mussel symbionts based on 16S rRNA gene69 but the host and symbiont phylogenies are not completely concordant. These phylogenetic discrepancies are likely explained by the horizontal symbiont transmission mode in Bathymodiolin mussels70. Indeed, like other symbiont-relying deep-sea invertebrates, Bathymodiolin mussels acquire their chemosynthetic symbiotic partners from their local surroundings19,21,71. Mussel B. heckrae acquires multiple thiotrophic symbiont clades10,72, one is related to the Gulf of Mexico clade and the other one is distinct from the other mussel symbiont clades (Fig. 1), which shows an exception to the broad geographic relationship between mussels and their thiotrophic symbionts. Nevertheless, low sequence similarity across SOX symbionts associated with different Bathymodiolus species occurring from the same locality (B. brooksi and B. heckerae; Table S3) suggests that the partner choice may be to a certain degree influenced by host specificity and/or local niche partition in the free-living symbiont pool.
B. thermophilus symbiont genomes from the EPR showed 98% ANI, suggesting that they belong to the same species according to ANI cutoff values (94–96%)73–75. Still, our comparative genomic analyses revealed high variation in genomic structure and gene composition among the different B. thermophilus symbionts. Mis-assemblies, incompleteness, and contamination in the fragmented genomes can explain some of the observed differences but we suspect that the most of the accessory pangenome reveals actual SOX strain genetic diversity as observed in other deep-sea mussels24,27,76,77, tubeworms78,79, clams80 and snails81,82.
B. thermophilus EPR9N symbiont genome possesses two incomplete gene sets for CO2-fixation, two complete gene sets for sulfur metabolism, and a gene set for nitrogen metabolism, as previously observed11,12. This strain is, however, unique regarding its putative hydrogen oxidation metabolism. Indeed, EPR9N possesses a hydrogenase cluster that is not found in other B. thermophilus symbiont strains. The Ni/Fe-hydrogenases encoded in this cluster are required for utilizing hydrogen as an energy source and are found in other invertebrates SOX symbionts11,28,83. We found that this cluster is also present in some but not in all Bathymodiolus symbionts with variation across symbiont subpopulations for B. azoricus. Different Ni/Fe-hydrogenases content was also observed amongst B. septemdierum symbionts8. Interestingly, in the invertebrate and Bathymodiolus symbiont strains which possess it, the hydrogenase cluster is structurally different from that in EPR9N11,28,83. Hydrogenase subunits in Bathymodiolus symbionts are clustered together with a rubredoxin gene (rpoN), which is required for the biosynthesis of Ni/Fe-hydrogenases84 but is absent in the symbiont hydrogenase cluster of the bivalve Loripes lucinalis and the scaly-foot snail Crysomallon squamiferum. On the other hand, two additional subunits of H2-sensing hydrogenase genes, hupUV, along with a histidine kinase gene hupT are only found in EPR9N, L. lucinalis85, and C. squamiferum symbionts86. These hydrogenase genes are responsible for regulating H2 along with regulatory proteins expressed from the hydrogenase gene cluster87. Histidine kinase, together with hupR, forms a two-component regulatory system required for H2 induction87. The exclusive presence of hupUV genes suggests that the EPR9N symbiont strain has potential to utilize hydrogen sources uniquely among the mussel symbionts. However, further study is needed to assess the presence and activity of hydrogenase clusters in Bathymodiolus symbiont populations in deep-sea environments. The same level of sequence divergence across EPR9N, L. lucinalis and C. squamiferum was observed for the group specific hupUV and hupT genes and the pervasive hupL and hupS genes. This phylogenetic signal suggests hupUV and hupT were not horizontally acquired in EPR9N exclusively but rather lost in the other sequenced Bathymodiolus symbiont strains or missing in their incomplete genome assemblies. The endosymbionts of vesicomyid clams, which are closely related to Bathymodiolus symbionts but vertically transmitted, present similar contrasting patterns of gene conservation across species. These differences are tied to key physiological traits (such as anaerobic respiration and dependency on vitamin B12) and are suspected to be adaptations to divergent host-derived or external environmental contexts88. Additional sampling of Bathymodiolus SOX symbiont strains along with better abiotic characterization of their external and host-associated environment should help us elucidate the evolutive history of the hydrogenase cluster in the mussel symbionts.
DNA methylation in bacteria is involved in diverse defensive biological processes such as DNA mismatch repair, environmental stress responses, or bacteriophages and transposases activity repression89,90. Evidence of DNA methylation together with genes required for methylation were found in EPR9N genome but this epigenetic system do not appear to be ubiquitous amongst deep-sea chemosynthetic symbionts. Indeed, the genome of the tubeworm symbiont Candidatus Endorifita persephone, which like the B. thermophilus symbiont has high host-specificity, does not seem to be methylated and the methyltansferase genes it encodes appear to be inactivated by mobile elements91. This raises questions about the role of epigenetics in host-associated and free-living contexts, and about the evolutive consequences of transitioning towards stricter host-specificity on the symbiont ecology.
Finally, we found that many of the genes unique to each B. thermophilus symbiont strain putatively originated from phages and mobile elements or were associated with viral defense mechanisms. The CRISPR/cas immunity was present across all B. thermophilus symbiont strains and appeared to be functional based on the high diversity and apparent dynamism of spacers. This finding indicates that the symbiont strains in their free-living stage may be under strong selective pressure from the environmental virome and could be more prone to acquire genes through horizontal gene transfer. Presence of genes such as adhesins, cadherins, hemolysin, etc. in EPR9N symbiont genome suggest that the symbiont have capability to attach to host cell, invade and colonize the host tissues as previously studied in the symbiont B. thermophilus92.
The presence of several ISs and genomic islands suggests that B. thermophilus EPR9N symbiont has acquired foreign DNA by horizontal transfer process from the different bacterial populations.
Conclusions
Here we report an improved complete genome assembly of the deep-sea hydrothermal vent mussel B. thermophilus thiotrophic gill symbiont (EPR9N) from the East Pacific Rise, using PacBio sequencing technology with a hierarchical genome-assembly process (HGAP3) pipeline. This symbiont genome is composed of a single circular chromosome which was assembled from a monoclonal symbiont population residing in the gill tissue of a host mussel. The discovery of a hydrogenase cluster with additional hydrogenase subunits in the EPR9N mussel symbiont genome showed that the symbiotic strain may have potential to harness energy efficiently by utilizing H2 resource from the vent fluids. Further study with extensive samples from diverse chemosynthetic environments required to understand the evolutionary history of the hydrogenase cluster in EPR9N symbiont genome. The symbiosis study of the host invertebrates and its symbiotic bacteria is essential to understand the chemosynthetic ecosystem. Even so, genomic studies of symbiotic bacteria at both species and population levels are limited. Therefore, the present mussel symbiont genome will serve as a footstep for further comparative genomic analyses investigating adaptive symbiont genome evolution in deep-sea chemosynthetic environments, particularly with regard to environmental niche partitioning, interactions with viruses, and horizontal transfer of accessory genes.
Supplementary Information
Acknowledgements
We are grateful to Dr. Robert C. Vrijenhoek for providing B. thermophilus sample from EPR9N hydrothermal vent. This research was supported by a grant (No. PE9988B and PEA0083) of Korea Institute of Ocean Science & Technology, KIOST, and was part of a project titled ‘Understanding the deep-sea biosphere on seafloor hydrothermal vents in the Indian Ridge (No. 20170411)’ funded by the Ministry of Oceans and Fisheries, Korea.
Author contributions
A.K.P and M.P. wrote the manuscript and analyzed the comparative genome analysis, S.J.J. prepared DNA for sequencing, and Y.J.W. developed the genome project and revised the manuscript. All authors reviewed the manuscript.
Data availability
The genome data was deposited to BioProject with an accession number PRJNA416436 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). The BioSample accession number for the symbiont genome is SAMN07956337 and assembly accession number GCA_003711265.1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-26669-y.
References
- 1.Sogin EM, Leisch N, Dubilier N. Chemosynthetic symbioses. Curr. Biol. 2020;30:R1137–R1142. doi: 10.1016/j.cub.2020.07.050. [DOI] [PubMed] [Google Scholar]
- 2.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 3.Barry JP, et al. Methane-based symbiosis in a mussel, Bathymodiolus platifrons, from cold seeps in Sagami Bay Japan. Invertebr. Biol. 2002;121:47–54. doi: 10.1111/j.1744-7410.2002.tb00128.x. [DOI] [Google Scholar]
- 4.Le Pennec M, Donval A, Herry A. Nutritional strategies of the hydrothermal ecosystem bivalves. Prog. Oceanogr. 1990;24:71–80. doi: 10.1016/0079-6611(90)90020-3. [DOI] [Google Scholar]
- 5.Rau GH, Hedges JI. Carbon-13 depletion in a hydrothermal vent mussel: Suggestion of a chemosynthetic food source. Science. 1979;203:648–649. doi: 10.1126/science.203.4381.648. [DOI] [PubMed] [Google Scholar]
- 6.Wentrup C, Wendeberg A, Schimak M, Borowski C, Dubilier N. Forever competent: Deep-sea bivalves are colonized by their chemosynthetic symbionts throughout their lifetime. Environ. Microbiol. 2014;16:3699–3713. doi: 10.1111/1462-2920.12597. [DOI] [PubMed] [Google Scholar]
- 7.Dattagupta S, Bergquist D, Szalai E, Macko S, Fisher C. Tissue carbon, nitrogen, and sulfur stable isotope turnover in transplanted Bathymodiolus childressi mussels: Relation to growth and physiological condition. Limnol. Oceanogr. 2004;49:1144–1151. doi: 10.4319/lo.2004.49.4.1144. [DOI] [Google Scholar]
- 8.Ikuta T, et al. Heterogeneous composition of key metabolic gene clusters in a vent mussel symbiont population. ISME J. 2016;10:990–1001. doi: 10.1038/ismej.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takishita K, et al. Genomic evidence that methanotrophic endosymbionts likely provide deep-sea Bathymodiolus mussels with a sterol intermediate in cholesterol biosynthesis. Genome Biol. Evol. 2017;9:1148–1160. doi: 10.1093/gbe/evx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayavedra L, et al. Horizontal acquisition followed by expansion and diversification of toxin-related genes in deep-sea bivalve symbionts. BioRxiv. 2019;110:330. [Google Scholar]
- 11.Ponnudurai R, et al. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 2017;11:463–477. doi: 10.1038/ismej.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponnudurai R, et al. Genome sequence of the sulfur-oxidizing Bathymodiolus thermophilus gill endosymbiont. Stand Genom. Sci. 2017;12:1–9. doi: 10.1186/s40793-017-0266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiel S. The Vent and Seep Biota: Aspects from Microbes to Ecosystems. Springer Science & Business Media; 2010. [Google Scholar]
- 14.Lorion J, et al. Adaptive radiation of chemosymbiotic deep-sea mussels. Proc. R. Soc. B. 2013;280:20131243. doi: 10.1098/rspb.2013.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nussbaumer AD, Fisher CR, Bright M. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature. 2006;441:345–348. doi: 10.1038/nature04793. [DOI] [PubMed] [Google Scholar]
- 16.Gros O, Liberge M, Heddi A, Khatchadourian C, Felbeck H. Detection of the free-living forms of sulfide-oxidizing gill endosymbionts in the lucinid habitat (Thalassia testudinum environment) Appl. Environ. Microbiol. 2003;69:6264–6267. doi: 10.1128/AEM.69.10.6264-6267.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Won Y-J, et al. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl. Environ. Microbiol. 2003;69:6785–6792. doi: 10.1128/AEM.69.11.6785-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laming SR, Gaudron SM, Duperron S. Lifecycle ecology of deep-sea chemosymbiotic mussels: A review. Front. Mar. Sci. 2018;5:282. doi: 10.3389/fmars.2018.00282. [DOI] [Google Scholar]
- 19.Laming SR, Duperron S, Cunha MR, Gaudron SM. Settled, symbiotic, then sexually mature: Adaptive developmental anatomy in the deep-sea, chemosymbiotic mussel Idas modiolaeformis. Mar. Biol. 2014;161:1319–1333. doi: 10.1007/s00227-014-2421-y. [DOI] [Google Scholar]
- 20.Salerno JL, et al. Characterization of symbiont populations in life-history stages of mussels from chemosynthetic environments. Biol. Bull. 2005;208:145–155. doi: 10.2307/3593123. [DOI] [PubMed] [Google Scholar]
- 21.Wentrup C, Wendeberg A, Huang JY, Borowski C, Dubilier N. Shift from widespread symbiont infection of host tissues to specific colonization of gills in juvenile deep-sea mussels. ISME J. 2013;7:1244–1247. doi: 10.1038/ismej.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pennec ML, Beninger PG. Ultrastructural characteristics of spermatogenesis in three species of deep-sea hydrothermal vent mytilids. Can. J. Zool. 1997;75:308–316. doi: 10.1139/z97-039. [DOI] [Google Scholar]
- 23.Eckelbarger K, Young C. Ultrastructure of gametogenesis in a chemosynthetic mytilid bivalve (Bathymodiolus childressi) from a bathyal, methane seep environment (northern Gulf of Mexico) Mar. Biol. 1999;135:635–646. doi: 10.1007/s002270050664. [DOI] [Google Scholar]
- 24.Ansorge R, et al. Diversity matters: Deep-sea mussels harbor multiple symbiont strains. bioRxiv. 2019;99:1039. doi: 10.1038/s41564-019-0572-9. [DOI] [PubMed] [Google Scholar]
- 25.Petersen JM, Wentrup C, Verna C, Knittel K, Dubilier N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Biol. Bull. 2012;223:123–137. doi: 10.1086/BBLv223n1p123. [DOI] [PubMed] [Google Scholar]
- 26.Sayavedra L, et al. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. Elife. 2015;4:e07966. doi: 10.7554/eLife.07966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansorge R, et al. Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nat. Microbiol. 2019;4:2487–2497. doi: 10.1038/s41564-019-0572-9. [DOI] [PubMed] [Google Scholar]
- 28.Petersen JM, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura K, Takai K. Theoretical constraints of physical and chemical properties of hydrothermal fluids on variations in chemolithotrophic microbial communities in seafloor hydrothermal systems. Prog. Earth Planet Sci. 2014;1:1–24. doi: 10.1186/2197-4284-1-5. [DOI] [Google Scholar]
- 30.Perez M, Juniper SK. Insights into symbiont population structure among three vestimentiferan tubeworm host species at eastern Pacific spreading centers. Appl. Environ. Microbiol. 2016;82:5197–5205. doi: 10.1128/AEM.00953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilbanks EG, et al. Metagenomic methylation patterns resolve bacterial genomes of unusual size and structural complexity. ISME J. 2022 doi: 10.1038/s41396-022-01242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Casariego JA, Cunning R, Baker AC, Eirin-Lopez JM. Symbiont shuffling induces differential DNA methylation responses to thermal stress in the coral Montastraea cavernosa. Mol. Ecol. 2022;31:588–602. doi: 10.1111/mec.16246. [DOI] [PubMed] [Google Scholar]
- 33.Triant DA, Whitehead A. Simultaneous extraction of high-quality RNA and DNA from small tissue samples. J. Hered. 2009;100:246–250. doi: 10.1093/jhered/esn083. [DOI] [PubMed] [Google Scholar]
- 34.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 35.Wick RR, et al. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021;22:1–17. doi: 10.1186/s13059-021-02483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 37.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krawczyk PS, Lipinski L, Dziembowski A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 2018;46:e35–e35. doi: 10.1093/nar/gkx1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Couvin D, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez M, Angers B, Young CR, Juniper SK. Shining light on a deep-sea bacterial symbiont population structure with CRISPR. Microbial. Genom. 2021 doi: 10.1099/mgen.0.000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:1–11. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen H. Protein Function Prediction. Springer; 2017. pp. 59–73. [Google Scholar]
- 45.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 46.Lagesen K, et al. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–31C08. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan PP, Lowe TM. Gene Prediction. Springer; 2019. pp. 1–14. [Google Scholar]
- 48.Griffiths-Jones S, et al. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertelli C, et al. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arndt D, et al. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roeselers G, et al. Complete genome sequence of Candidatus Ruthia magnifica. Stand Genomic Sci. 2010;3:163–173. doi: 10.4056/sigs.1103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:1–14. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Letunic I, Bork P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eren AM, et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021;6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling AE, Mau B, Perna NT. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tesler G. GRIMM: Genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- 62.Cabanettes F, Klopp C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018;6:e4958. doi: 10.7717/peerj.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Gilchrist CL, Chooi Y-H. Clinker & clustermap. js: Automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 65.Taboada B, Estrada K, Ciria R, Merino E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics. 2018;34:4118–4120. doi: 10.1093/bioinformatics/bty496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Søndergaard D, Pedersen CN, Greening C. HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.NCBI Genome Browser. https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/. Accessed 12 March 2022.
- 68.Mcmullin ER, Hourdez S, Schaeffer SW, Fisher CR. Review article phylogeny and biogeography of deep sea vestimentiferan tubeworms and their bacterial symbionts. Symbiosis. 2003;34:1–41. [Google Scholar]
- 69.Won Y-J, Jones WJ, Vrijenhoek RC. Absence of cospeciation between deep-sea mytilids and their thiotrophic endosymbionts. J. Shellfish Res. 2008;27:129–138. doi: 10.2983/0730-8000(2008)27[129:AOCBDM]2.0.CO;2. [DOI] [Google Scholar]
- 70.Miyazaki J-I, Martins LdO, Fujita Y, Matsumoto H, Fujiwara Y. Evolutionary process of deep-sea Bathymodiolus mussels. PLoS ONE. 2010;5:e10363. doi: 10.1371/journal.pone.0010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bright M, Bulgheresi S. A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raggi L, Schubotz F, Hinrichs KU, Dubilier N, Petersen JM. Bacterial symbionts of Bathymodiolus mussels and Escarpia tubeworms from Chapopote, an asphalt seep in the southern Gulf of Mexico. Environ. Microbiol. 2013;15:1969–1987. doi: 10.1111/1462-2920.12051. [DOI] [PubMed] [Google Scholar]
- 73.Goris J, et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 74.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:1–14. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ho P-T, et al. Geographical structure of endosymbiotic bacteria hosted by Bathymodiolus mussels at eastern Pacific hydrothermal vents. BMC Evol. Biol. 2017;17:1–16. doi: 10.1186/s12862-017-0966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romero Picazo D, et al. Horizontally transmitted symbiont populations in deep-sea mussels are genetically isolated. ISME J. 2019;13:2954–2968. doi: 10.1038/s41396-019-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez M, Juniper SK. Is the trophosome of Ridgeia piscesae monoclonal? Symbiosis. 2018;74:55–65. doi: 10.1007/s13199-017-0490-7. [DOI] [Google Scholar]
- 79.Polzin J, Arevalo P, Nussbaumer T, Polz MF, Bright M. Polyclonal symbiont populations in hydrothermal vent tubeworms and the environment. Proc. R. Soc. B. 2019;286:20181281. doi: 10.1098/rspb.2018.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Russell SL, Cavanaugh CM. Intrahost genetic diversity of bacterial symbionts exhibits evidence of mixed infections and recombinant haplotypes. Mol. Biol. Evol. 2017;34:2747–2761. doi: 10.1093/molbev/msx188. [DOI] [PubMed] [Google Scholar]
- 81.Breusing C, Genetti M, Russell SL, Corbett-Detig RB, Beinart RA. Horizontal transmission enables flexible associations with locally adapted symbiont strains in deep-sea hydrothermal vent symbioses. Proc. Natl. Acad. Sci. 2022;119:e2115608119. doi: 10.1073/pnas.2115608119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lan Y, et al. Endosymbiont population genomics sheds light on transmission mode, partner specificity, and stability of the scaly-foot snail holobiont. ISME J. 2022 doi: 10.1038/s41396-022-01261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anantharaman K, Breier JA, Sheik CS, Dick GJ. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc. Natl. Acad. Sci. 2013;110:330–335. doi: 10.1073/pnas.1215340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fritsch J, et al. Rubredoxin-related maturation factor guarantees metal cofactor integrity during aerobic biosynthesis of membrane-bound [NiFe] hydrogenase. J. Biol. Chem. 2014;289:7982–7993. doi: 10.1074/jbc.M113.544668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petersen JM, et al. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol. 2016;2:1–11. doi: 10.1038/nmicrobiol.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakagawa S, et al. Allying with armored snails: The complete genome of gammaproteobacterial endosymbiont. ISME J. 2014;8:40–51. doi: 10.1038/ismej.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 2001;25:455–501. doi: 10.1016/S0168-6445(01)00063-8. [DOI] [PubMed] [Google Scholar]
- 88.Perez M, et al. Divergent paths in the evolutionary history of maternally transmitted clam symbionts. Proc. R. Soc. B. 2022;289:20212137. doi: 10.1098/rspb.2021.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S, et al. N 4-cytosine DNA methylation is involved in the maintenance of genomic stability in Deinococcus radiodurans. Front. Microbiol. 2019;10:1905. doi: 10.3389/fmicb.2019.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Oliveira AL, Srivastava A, Espada-Hinojosa S, Bright M. The complete and closed genome of the facultative generalist Candidatus Endoriftia persephone from deep-sea hydrothermal vents. Mol. Ecol. Resour. 2022 doi: 10.1111/1755-0998.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ponnudurai R, et al. Comparative proteomics of related symbiotic mussel species reveals high variability of host–symbiont interactions. ISME J. 2020;14:649–656. doi: 10.1038/s41396-019-0517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome data was deposited to BioProject with an accession number PRJNA416436 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). The BioSample accession number for the symbiont genome is SAMN07956337 and assembly accession number GCA_003711265.1.