Abstract
Immune checkpoint inhibitor (ICI) therapy has revolutionised the treatment of various cancer types. ICIs reinstate T-cell function to elicit an anti-cancer immune response. The resulting immune response can however have off-target effects which manifest as autoimmune type serious immune-related adverse events (irAE) in ~10–55% of patients treated. It is currently challenging to predict both who will experience irAEs and to what severity. Identification of patients at high risk of serious irAE would revolutionise patient care. While the pathogenesis driving irAE development is still unclear, host genetic factors are proposed to be key determinants of these events. This review presents current evidence supporting the role of the host genome in determining risk of irAE. We summarise the spectrum and timing of irAEs following treatment with ICIs and describe currently reported germline genetic variation associated with expression of immuno-modulatory factors within the cancer immunity cycle, development of autoimmune disease and irAE occurrence. We propose that germline genetic determinants of host immune function and autoimmune diseases could also explain risk of irAE development. We also endorse genome-wide association studies of patients being treated with ICIs to identify genetic variants that can be used in polygenic risk scores to predict risk of irAE.
Subject terms: Predictive markers, Cancer genomics, Cancer immunotherapy
Introduction
Immune checkpoint inhibitors (ICI) are monoclonal antibodies that release the brakes off immune checkpoints such as cytotoxic T lymphocyte antigen (CTLA-4), programmed cell death 1 (PD-1), programmed cell death ligand 1 (PD-L1) and lymphocyte-activation gene 3 (Lag-3). They prevent the immune escape of cancer cells and ultimately cause cancer cell death1. They have revolutionised the management of several cancer types including melanoma, lung and urological cancers, which have historically had dismal prognoses in the metastatic setting. Commonly used ICIs include anti-PD-1 (nivolumab, pembrolizumab), anti-PD-L1 (atezolizumab, durvalumab, avelumab) and anti-CTLA-4 (ipilimumab) agents. Despite their notable successes in improving patients’ survival, ICI-induced toxicities, also known as immune-related adverse events (irAEs), can be life-changing and in some cases fatal. As the indication for using ICIs expands and moves earlier in the treatment pathway in neoadjuvant and adjuvant settings, identifying patients at risk of developing irAEs is important to minimise the risk of serious toxicities while maximising the treatment benefit gained by patients.
There are currently no established predictive tools or biomarkers that can help detect patients at risk of irAEs. Clinicians mainly rely on patients’ past clinical history including presence of autoimmune disease (AD) to provide a risk estimation. However, as patients with ADs have been excluded from most clinical trials of ICIs, it is hard to determine the effectiveness of this strategy. Given the estimated overall prevalence of autoimmune diseases of 7.6–9.4%, a high proportion of patients would miss out on potentially lifesaving cancer treatment were this to be used as a screening tool2.
While the mechanism of irAEs is still unclear, host genetic factors are hypothesized to be key determinants. Associations between germline genetic polymorphisms and toxicity have already been established for several chemotherapeutic agents3. For example, variants in the dihydropyrimidine dehydrogenase (DPYD) gene have been shown to be associated with severe toxicities and are now screened for prior to administration of fluorouracil-based chemotherapy3.
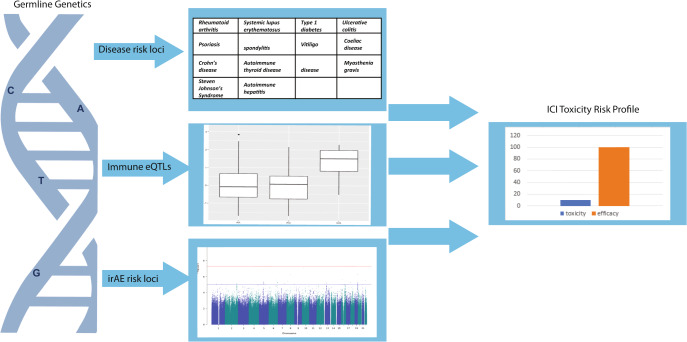
This review aims to summarise the clinical presentation of ICI-induced irAEs and the current evidence regarding germline factors and predisposition towards irAEs. As shown in Fig. 1, germline genetics may explain a significant proportion of the variation in patients’ risk of irAEs. We hypothesise that risk is conferred via associated or intermediate phenotypes such as autoimmune diseases and levels of proteins involved in the cancer immunity cycle. Novel genetic determinants will only be uncovered by large-scale studies of common and rare variants in patients treated with ICIs.
Fig. 1. Germline genetic profiling may assist in generating patient ICI induced toxicity profiles.
An ICI toxicity risk profile incorporating germline genetic factors linked with autoimmune disease risk loci, immuno-modulatory eQTLs and irAE associated loci would aid in toxicity management and decision-making to maximise treatment benefit while reducing the risk of serious irAE.
The spectrum and challenges of immune-related adverse events
Given their similar presentation to autoimmune type events, irAEs are thought to be caused by an over-active immune response and disruption to immune homeostasis1. IrAEs can affect multiple organ systems including skin (manifesting as rash, pruritus, vitiligo), endocrine (thyroid, pituitary, adrenal disorders), gastrointestinal (colitis), lung (pneumonitis), musculoskeletal (arthralgia, myalgia) and liver (hepatitis). Toxicities are graded based on the Common Terminology Criteria (CTCAE) for adverse events and are usually managed using oral, intravenous high-dose steroids, or immuno-modulatory agents. Early recognition and treatment of irAE is essential for symptom resolution. Some irAEs present similarly to chemotherapy or targeted therapy induced adverse events but their underlying cause and treatment approach may differ. For example, diarrhoea induced by ICIs is likely due to immune-mediated colitis requiring treatment with steroids, whereas chemotherapy causes direct damage to the intestinal mucosa itself. Skin toxicities are known adverse events of chemotherapies, targeted agents (BRAF inhibitors and EGFR inhibitors) and ICIs. Some skin toxicity presentations are more treatment-type specific, such as hand-foot syndrome with chemotherapy and vitiligo with ICIs. Rash and pruritis are common to both chemotherapy and ICIs, but the risk of both toxicities are higher with ICI4. Pneumonitis, more common following treatment with anti-PD-1 than other ICI, is an example of an irAE that is likely immune related in patients on concurrent ICI and chemotherapy.
Toxicity profile of different ICI therapies
The incidence and severity of irAEs varies according to the type of ICI therapy being used (e.g. single agent PD-1/PDL-1 inhibitors or CTLA-4 inhibitors) and whether single-agent or combination regimens are administered. Table 1 shows the frequencies of organ-specific irAEs in the context of single and combination agent ICI treatment5–7. Anti-PD-L1 treatment generally leads to fewer any grade or ≥grade 3 irAEs compared to anti-PD-1, possibly due to lack of inhibition of PD-L2 ligand which is involved in T cell regulation8. Colitis, hypophysitis and rash are more common with anti-CTLA-4 therapy, while pneumonitis, hypothyroidism, musculoskeletal toxicities and vitiligo occurred more often with anti-PD-1 therapy5. Higher incidences of serious ≥ grade 3 irAEs have been observed in patients treated with combination ICI regimens compared to monotherapy ICI regimens. In a phase 3 melanoma trial of 945 patients, 55% of patients receiving a doublet ICI regime of anti-CTLA-4 and anti-PD-1 experienced ≥grade 3 irAEs9. The incidence of irAEs following single-agent ICI treatment is lower, particularly with anti-PD-1 or anti-PD-L1 agents (~14% ≥ grade 3 events)5,6,10. A meta-analysis reported fatality rates related to irAEs ranged from 0.36–1.23% in patients receiving monotherapy and combination ICI treatment respectively11. Colitis and myocarditis were frequent causes of deaths from combination ICI therapy, whereas pneumonitis, hepatitis and neuro-toxicities most commonly contributed to anti-PD-1 or PD-L1 related fatalities11.
Table 1.
Frequency of different side effects by ICI treatment type.
| ICI agents | Anti-PD-1 | Anti-PD-L1 | Anti-CTLA-4 | Anti-CTLA-4 and anti-PD-1 | ||||
|---|---|---|---|---|---|---|---|---|
| irAE | Max frequency of any grade events (%) | Max frequency of ≥Grade 3 events (%) | Max frequency of any grade events (%) | Max frequency of ≥Grade 3 events (%) | Max frequency of any grade events (%) | Max frequency of ≥grade 3 events (%) | Max frequency of any grade events (%) | Max frequency of ≥Grade 3 events (%) |
| Pneumonitis | 2.4–3.6% | 0.70% | 0.00% | 0.00% | 0.40% | 0.1% | 5.3–7.8% | 0.2–1.1% |
| Cutaneous | 4–28.5% | 0–1.3% | <0.1–16% | 0–0.6% | 1.1–25.9% | 0–1.3% | 37.7–65% | 0.3–7.6% |
| Diarrhoea | 12.1–13% | 1–1.5% | 7.3–16.3% | 0–0.6% | 27.00% | 5–7.4% | 22.3–46.7% | 0–15.8% |
| Colitis | 0.7–2% | 0.4–2% | 0.90% | 0.30% | 5.7–8% | 4.1–5% | 46.00% | 16.00% |
| Hepatitis | 0.1–7% | 0–1.5% | 0.4.% | 0–0.4% | 0–2.6% | 0–0.9% | 13.4–29.5% | 1.1–15.4% |
| Thyroiditis | 1.6–9.7% | 0.10% | 0–2.2% | 0.00% | 0.5–2% | 0.00% | 24–27.2% | 0.3–1.3% |
| Pituitary | 0.20% | 0.10% | 0.00% | 0.00% | 1.2–4% | 0.80% | 1.4–4% | 0.4–2.4% |
| Adrenal | 0.00% | 0.00% | 0.70% | 0.4%% | 0.40% | 0.20% | 0.3–3.5% | 0.2–1.4% |
| Diabetes | 0.10% | 0.10% | 0.00% | 0.00% | 0.00% | 0.00% | 0.5% | 0.5% |
| Neurological | 6.00% | 2.00% | <0.1% | <0.1% | 3.80–4.5% | 1.9% | 12.00% | – |
| Renal | 2.7% | <0.1–0.4% | 0.3% | <0.1% | – | – | 5.1–8.6% | 0.5–1.7% |
| Rheumatological | 0.1–43% | 0–0.2% | 0–6.2% | 0.00% | 0–2.1%% | 0–0.2% | 5–14% | <1–1% |
| Ocular | All ICI < 1% | – | – | – | – | – | – | – |
Patient and tumour characteristics that may impact risk of irAEs
Chennamadhavuni et al have reviewed the patient and tumour characteristics that are associated with risk of irAEs12. It is difficult to make general statements about individual risk factors as their influence is context-dependent, varying by type of ICI used and organs at risk. There is a general agreement across studies that endocrine toxicities and pneumonitis are more common in younger patients while skin toxicities are more frequent in older patients13,14. There are reports that sex influences risk, with thyroid irAEs being more common in women while neurological, dermal and vascular events occurred more in men15. Higher body mass index (BMI) and performance status have also been associated with increased risk of irAE16,17. Specific co-morbidities can increase the risk of certain irAEs, for example, chronic obstructive pulmonary disease is associated with an increased risk of pneumonitis18.
One study reported different frequencies of anti-PD-1 related irAEs in patients with different cancer types, where more gastrointestinal and skin irAEs were found in melanoma patients while pneumonitis was more common in lung and renal cancer patients5. When considering incidence of any grade or ≥grade 3 irAEs, Wang et al found no significant difference in incidence by cancer type, although melanoma patients did experience the highest incidence of any grade irAEs10.
Time to onset of irAE
The timing of irAEs vary according to the type of toxicity and ICI treatment. In an analysis of 8436 patients treated with ICI therapies, the pooled median time to onset of any toxicity of any grade ranged from 2.2 to 14.8 weeks19. Infusion reaction, skin and gastrointestinal events had the shortest median onset time, whereas renal toxicities occurred the latest following all ICI therapies19. When considering all ≥grade 3 events, the median time to onset for a serious event was 7.9 weeks for patients treated with a doublet ICI regime, 7 weeks for anti-CTLA-4 and 27.5 weeks for anti-PD-1/PDL-1 therapies19. An ongoing multi-centre study conducted in the UK also found that patients on combination ICI therapy tend to experience irAEs earlier than those who are on monotherapy (Fig. 2).
Fig. 2. Time to onset of irAE based on the ICI GENETICS Study dataset (IRAS 237779, CPMS 39433).
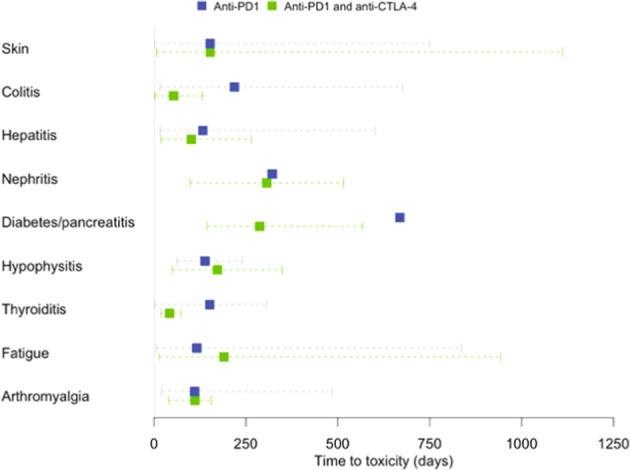
Time to toxicity for different irAE according to whether patients in the ICI Genetics study were treated with single anti-PD-1 or combined anti-CTLA-4 and anti-PD-1 therapy. The square box and dotted lines refer to the mean and range of the time taken from the treatment start date to development of toxicity in patients treated with anti-PD1 therapy (blue) (N = 92) and anti-PD1 and anti-CTLA-4 therapy (green) (N = 21).
Some irAEs can persist and have long-term effects. The median time to resolution of irAEs can range from 0.1 to 54.3 weeks, with the longest to resolve being endocrine events19. In a real world analysis of 437 patients, 35.2% of irAEs reported lasted ≥6 months20. The lasting impact of ICIs was also demonstrated objectively in a study of patients with anti-CTLA-4 induced enterocolitis, where colitis was still evident on endoscopy for a median of 4 months after symptoms onset21. Other irAEs that can continue beyond treatment completion include ICI-induced type 1 diabetes and inflammatory arthritis22,23. These toxicities cause considerable morbidity, reliance on lifelong medication and expensive costs to healthcare systems.
ICI-induced toxicities have also been reported up to almost one year after cessation of ICI therapy6. Nigro et al assessed 436 patients treated with anti-PD-1/PD-L1 therapies and found that late irAEs occurring >12 months after cessation of treatment were experienced in a third of patients, with 4.8% of cases being serious events24. A review analysing registration trials that lead to ICI approval by the U.S Food and Drug Administration (FDA) and real-world data from melanoma patients identified that 6.9% of irAEs occur more than one year after treatment initiation20. Late-onset toxicities that have been reported include Raynaud’s phenomenon, which was observed >20 months after starting combination immunotherapy25.
Steroids, the main treatment used to counteract irAEs can cause debilitating effects on bone health, adrenal insufficiency and diabetes. Various immuno-modulatory therapies may be required to treat serious recurrent irAEs which can cause profound immunosuppression. The significant impact of irAEs on patients’ long-term health and quality of life need to be considered alongside the potentially longer survival benefit gained from ICI therapy. Predictive tests able to estimate both a patient’s likelihood of experiencing toxicity and a patient’s likelihood of a survival benefit from ICIs would be very helpful in treatment decision-making.
ICI therapy in patients with autoimmune disease
Having concurrent AD is not uncommon in cancer patients. 13.5% of lung cancer patients in the US were found to be diagnosed with AD26. Studies have begun to investigate whether single-agent ICIs can be tolerated by these patient groups. Menzies et al found 38% of patients with AD treated with anti-PD-1 had an exacerbation of their AD requiring immunosuppression, but these were mainly mild events27. In this study, 29% of patients with AD developed an irAE and 10% experienced a grade 3 event, which is comparable to toxicity rates observed in clinical trials27. However, one study found 33% of patients with AD developed grade 3 irAEs on ipilimumab, indicating that patients with AD receiving anti-CTLA-4 may require more stringent monitoring28.
Correlation between any grade irAE and response to ICI treatment response
There have been several reports evidencing the link between any grade ICI-induced toxicity and treatment efficacy29,30. A meta-analysis of 4324 patients treated with ICI found that development of all-grade irAE was correlated with a reduced risk of death (HR 0.49, 95% CI 0.38–0.62, p < 0.001), less risk of disease progression (HR 0.51, 95% CI 0.42–0.64, p < 0.001) and an odds of treatment response of 4.56 (95% CI 3.72–5.59, p < 0.001) compared to patients with no irAE30. Cancer type and drug type did not influence the results in this analysis30. However, a subgroup analysis found no significant correlation between grade 3/4 events or any grade events of pneumonitis with overall survival30. It is important to predict these more severe irAEs that are not associated with treatment benefit but instead concomitant with significant morbidity to help deliver better cancer care.
Potential germline determinants of serious immune-related adverse events
Germline genetic factors are strong determinants of immune homeostasis and our immunological status31. Inherited genetic variation may influence ICI treatment outcomes, including irAE development, and explain the apparent disparity in immune responses seen between different patients receiving similar ICI agents. We highlight below genetic loci that have been found to be associated with immune traits and autoimmune diseases which warrant association testing in relation to irAEs.
Host genetics influence levels of immuno-modulatory cells and molecules within the cancer immunity cycle
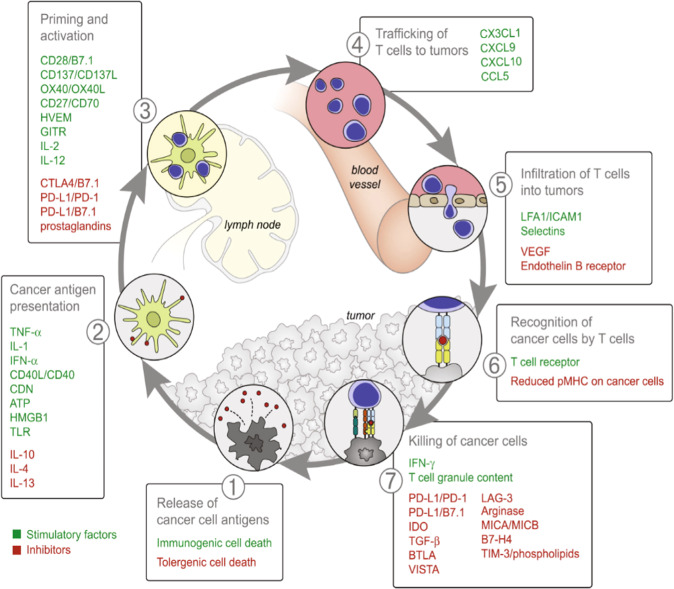
The cancer immunity cycle consists of seven step-wise events delineating the anti-cancer immune response (Fig. 3)32. Stimulatory and inhibitory immune factors expressed by dendritic cells, T cells and B cells regulate immune feedback mechanisms to activate the immune response while also preventing autoimmunity32. Host genetics explain variation in the expression and activity of various immune cell markers31. Many immune traits have been shown to be heritable. Those with the highest heritability include CD39 on CD4 T cells functioning as regulatory T cells and CD32 expression on dendritic cells33. Dendritic cells have the largest proportion of highly heritable traits followed by CD4 T cells and CD8 cells34. Hundreds of single nucleotide polymorphisms (SNPs) have been identified that are associated with the abundance of blood cells, immune cell marker expression and levels of molecules such as growth factors, cytokines and MHC-associated proteins35–37. Table 2 shows the number of genetic loci that reached GWAS significance for expression of immuno-modulatory traits from the GWAS catalogue from March to April 202137.
Fig. 3. Cancer immunity cycle (Reprinted from Immunity, Vol. 39 Issue 1, Daniel S. Chen, Ira Mellman, Oncology meets immunology: the cancer-immunity cycle, page 10, Copyright (2013)32, with permission from Elsevier (Supplementary Material)).
The cycle consists of step-wise events characterising the immune response against cancer cells. Various stimulatory and inhibitory immune factors play a role in each event by activating or suppressing the anti-cancer immune response. Immune checkpoints such as CTLA-4, PD-1, PD-L1 and LAG-3 act as inhibitory factors to suppress T cell activation and prevent the killing of cancer cells.
Table 2.
Genetic variants associated with immune-modulatory traits in the cancer immunity cycle.
| Immune traits | No. of SNPs reported in the GWAS catalogue reaching GWAS significance (P ≤ 5 × 10–8) and in linkage equilibrium | Average effect sizes (beta) of alleles | Largest discovery cohort’s sample size (ethnic population) | Largest replication cohort sample size (ethnic population) | Reference |
|---|---|---|---|---|---|
| TNFα | 1 | 2.13 | 13,577 (Finnish) | NA | 89 |
| IL-1α | 1 | 0.74 | 790 (Hispanic, European, Asian, South Asian or African American) | NA | 90 |
| IL-1β | 1 | 0.33 | 997 (European) | 338 (Arab, South Asian and Filipino) | 91 |
| IFNα | 1 | 1.80 | 410 (European) | 1165 (European, African American) | 92 |
| CD40 | 2 | 0.89 | 982 (Scottish) | NA | 93,94 |
| CD40L | 1 | 0.96 | 653 (European) | 317 | 95 |
| IL-10 | 8 | 0.08 | 764 (Hispanic, European, Asian, South Asian or African American) | 425 (West African) | 90,96 |
| IL-4 | 1 | 0.40 | 764 (Hispanic, European, Asian, South Asian or African American) | NA | 90 |
| IL-13 | 1 | 0.42 | 3557 (Finnish) | NA | 97 |
| aCD27/CD70 | 2 | 0.64 | 997 (European) | 338 (Arab, South Asian and Filipino) | 91 |
| IL-2 | 1 | 0.27 | 764 (Hispanic, European, Asian, South Asian or African American) | NA | 90 |
| aIL-12 | 5 | 0.21 | 9263 (Finnish) | NA | 94,97 |
| CCL5 | 1 | 0.33 | 3421 (Finnish) | NA | 97 |
| aLFA1/ICAM1 | 6 | 3.47 | 22,435 (European) | 9813 | 89,91,98,99 |
| aVEGF | 18 | 0.17 | 13,577 (Finnish) | 2800 (European, Sorbian) | 36,89,97,100,101 |
| IFNγ | 2 | 0.31 | 7701 (Finnish) | NA | 97 |
| aMHC class I polypeptide-related sequence A (MICA) | 27 | 0.77 | 997 (European) | 338 (Arab, South Asian and Filipino) | 91,93 |
| MHC class I polypeptide-related sequence B (MICB) | 8 | 0.63 | 997 (European) | 338 (Arab, South Asian and Filipino) | 91 |
Variants that reached GWAS significance (P ≤ 5 × 10–8) were identified from the GWAS catalogue from March to April 202137. The sample sizes of the largest discovery cohort used to identify the SNP at GWAS significance are provided. Where an independent validation cohort had also been used details of the largest replication cohort are also provided. For a SNP to be included in this table, the following summary statistics had to be available: effect allele, non-effect allele, beta and P-value (minimal statistics required for including a variant in a polygenic risk score). Linkage disequilibrium (LD) between the lead SNP in a region and any other SNPs in the region had to be <0.8. The average absolute beta value of all SNPS is reported for traits with >1 correlated SNP. SNPs that had effect sizes reported in units measuring change of gene/protein expression were excluded.
GWAS genome-wide association study, NA not available, SNPs single nucleotide polymorphism.
aSome SNPs for this trait have beta effect sizes that are inverse transformed and/or scaled to standard deviation units.
Immune checkpoints (CTLA-4, PD-1, PD-L1) targeted by ICI therapies elicit an inhibitory effect in the priming (step 3) and cancer-cell killing (step 7) phases of the cycle32. CTLA-4 gene polymorphisms have been shown to contribute to susceptibility to Graves’ disease, autoimmune hypothyroidism and type 1 diabetes38. The most likely causal polymorphisms, in a non-coding 3’UTR region, affect CTLA-4 expression and are presumed to increase T-cell reactivity38. A missense variant in CTLA-4, Y60C was also found to be associated with early-onset Crohn’s disease39. Variants in the IL-23 and Th1 helper pathways, which are involved in immune regulation of exogeneous antigens have also been implicated in immune-mediated and autoimmune conditions40. These include variants at the TYK2 locus, IL-12 and IL-12R pathways, STAT3 and STAT4 and NFKB1 family40.
Two SNPs, rs6673928 and rs6695772, which are expression quantitative trait loci (eQTLs) influencing IL19 (an immuno-modulatory cytokine) and BATF3 (essential for CD8 + dendritic cell development) expression respectively, were shown to be significantly associated with overall survival in melanoma patients41. Such SNPs could potentially be used to predict prognosis for an immunogenic malignancy like melanoma.
Genetics factors associated with autoimmune diseases
The close phenotypic similarity between patients with irAE and patients with autoimmune disease (AD) justifies further investigation into whether the resemblance is explained by shared genetic factors. Genome-wide association studies (GWAS) have identified hundreds of risk loci for over sixty autoimmune conditions31. Each locus is marked by one or more polymorphism each with small to moderate effect sizes. The loci tend to map to non-coding regions enriched in distant regulatory elements42. Table 3 shows the number of genetic loci in linkage equilibrium that reached GWAS significance for common autoimmune diseases traits from the GWAS catalogue from March to April 202137.
Table 3.
Genetic variants associated with common autoimmune disease traits.
| Autoimmune disease trait | No. of SNPs reported in the GWAS catalogue reaching GWAS significance (P ≤ 5 × 10–8) and in linkage equilibrium | Average effect sizes (OR) of alleles | Largest discovery cohort’s sample size (ethnic population) | Largest replication cohort sample size (ethnic population) |
|---|---|---|---|---|
| Rheumatoid arthritis | 186 | 1.16 | 22,628 cases, 288,664 cases (European, East Asian) | 24,107 cases, 79,295 controls (East Asian, European) |
| Psoriasis | 127 | 1.75 | 11,988 cases, 275,335 controls (European) | 11,301 cases, 19,879 controls (European) |
| Crohn’s disease | 180 | 1.23 | 19,085 cases, 34,213 controls (European) | 16,619 cases, 31,766 controls (European, Iranian, Indian, East Asian) |
| Ulcerative colitis | 80 | 1.32 | 12,924 cases, 21,442 controls (European) | 25,683 cases, 17,015 controls (European) |
| Systemic lupus erythematosus | 184 | 1.41 | 11,590 cases, 15,984 controls (European, African American, Hispanic) | 1387 cases, 28,564 controls (Japanese) |
| Ankylosing spondylitis | 55 | 3.74 | 8726 cases, 34,213 controls (European) | 2111 cases, 4483 controls (European) |
| Autoimmune thyroid disease | 141 | 1.48 | 30,234 cases, 724,172 controls (European) | 7891 cases, 8351 controls (Han Chinese) |
| Coeliac disease | 59 | 1.29 | 11,489 cases, 22,308 controls (European) | 12,041 cases, 12,228 controls (European, Indian) |
| Type 1 diabetes | 66 | 1.30 | 9934 cases, 16,956 controls (European) | 4329 cases, 9543 controls (European) |
| Vitiligo | 70 | 1.37 | 2853 cases, 37,405 controls (European) | 6623 cases, 10,740 controls (East Asian) |
| Addison’s disease | 10 | 2.23 | 1223 cases, 4097 controls (European) | NA |
| Myasthenia gravis | 8 | 1.68 | 972 cases, 1977 controls (European) | 423 cases, 467 controls (European) |
| Steven Johnson’s Syndrome | 5 | 2.53 | 424 cases, 1881 controls (European) | 102 cases, 469 controls (East Asian, Indian, Brazilian) |
| Autoimmune hepatitis | 1 | 2.90 | 649 cases, 13,436 controls (European) | 451 cases, 4103 controls (European) |
Variants that reached GWAS significance (P ≤ 5 × 10–8) were identified from the GWAS catalogue from March–April 202137. The sample sizes of the largest discovery cohort used to identify the SNP at GWAS significance is provided. Where an independent validation cohort had also been used, details of the largest replication cohort are also provided. For a SNP to be included in this table, the following summary statistics had to be available: effect allele, non-effect allele, OR, and P-value (minimal statistics required for including a variant in a polygenic risk score). LD between the lead SNP in a region and any other SNPs in the region had to be <0.8. The average absolute OR of all SNPS associated with the relevant trait is reported.
GWAS genome-wide association study, OR odds ratio, NA not available, SNPs single nucleotide polymorphism.
The genetics of AD is complicated by genetic pleiotropy, which describes a single gene or variant affecting multiple traits31. The A allele of rs2476601, mapping to PTPN22 has been identified as a susceptibility allele in genome-wide studies of multiple ADs43,44. Other variants have been associated with multiple ADs at genome significance but with opposite effects45.
HLA genes
The human leucocyte antigen (HLA) region, a 3.6 Mb region on chromosome 6 has been linked to multiple ADs42. Disease-linked variants at this locus often have the strongest effect sizes of all variants detected by GWAS. Variants in the genes encoding class I HLA proteins (HLA-A, HLA-B and HLA-C, expressed on all nucleated cells) have been shown to be strongly associated with ankylosing spondylitis (e.g. HLA-B27), Graves’ Disease (e.g. HLA-C*07), Type I diabetes (e.g. B*39) and multiple sclerosis (e.g. C*05)46. Associations between variants in class II proteins (HLA-DP, HLA-DM, HLA-DOA, HLA-DOB, HLA-DQ and HLA-DR expressed by B cells, antigen-presenting cells and activated T cells) have been identified for Graves’ disease (HLA-DR3), systemic lupus erythematosus (HLA-DR3), Hashimoto’s thyroiditis, myasthenia Gravis (HLA-DR3), Addison’s disease (HLA-DR3), rheumatoid arthritis (e.g. HLA-DRB1), coeliac disease (e.g. HLA-DR3-DQ2), multiple sclerosis (HLA-DR3) and type 1 diabetes (DR3)40,46. HLA variation is also known to be important in explaining a patient’s response to specific drugs; HLA-B*57:01 is screened for because of its association with severe hypersensitivity reaction to Abacavir47.
Pharmacoethnicity
A large proportion of GWAS of ADs have studied European populations and while many of the identified susceptibility alleles have been replicated in Asian and African populations, some polymorphisms related to ADs such as rheumatoid arthritis have disease susceptibility effects that are variable across different ethnic groups48. Pharmacoethnicity needs to be taken into consideration when designing studies to identify variants associated with response to treatments like ICIs. Strong associations of HLA-B*1502 with carbamazepine-induced Steven Johnson’s Syndrome have been observed in the Han Chinese population but not in European populations49,50. Other pharmacogenetically relevant variants such as the DPYD variants recommended for testing by the clinical pharmacogenomics implementation consortium (CPIC) and Dutch Pharmacogenetics working group have lower frequencies in Asian and African populations compared to European populations51,52. These differences limit their utility in multi-ethnic populations and demonstrate the need for identification of relevant variants in diverse populations.
Are genetic variants associated with response to ICIs also determinants of risk of irAEs?
Given the association between toxicity and response to ICIs discussed above, it would be interesting to determine whether SNPs associated with response are also associated with irAEs. Chat et al investigated 25 SNPs associated with three or more ADs in 436 melanoma patients and found that rs17388568 was significantly associated with favourable response to anti-PD-1 treatment and rs1893217 was significantly associated with worse outcome in patients treated with anti-CTLA-453. rs17388568 maps to a locus containing IL-2, IL-21 and ADAD1 that have been associated with allergy, colitis and type 1 diabetes and rs1893217 maps to PTPN2 which has a negative regulatory role in cytokine signalling and has been linked with rheumatoid arthritis42,54. rs2282055 and rs4143815, mapping to PD-L1 have also been shown to be associated with better treatment responses in non-small cell lung cancer (NSCLC) patients treated with nivolumab55. These SNPs have been associated with increased PD-L1 expression and several AD but further functional evaluation is required55. In a study of 166 SNPs mapping to immune-related genes in 94 NSCLC patients, Refae et al found distinct SNPs associated with response and toxicity56. They found that treatment response was significantly correlated with SNPs related to the tumour microenvironment, whereas SNPs associated with toxicity were predominantly target cell-related56. Large, well-powered studies will be required to determine whether germline variants impact on both toxicity and outcome.
Current efforts to identify genetic factors influencing irAE development
Table 4 summarises the existing studies that have tested genetic variants for associations with irAEs and highlights variants that have been found to be significantly associated with irAE development.
Table 4.
Genetic variants associated with irAE development.
| Author | Sample size | Cancer type | Type of ICI studied | Genetic analysis approach | Toxicity type | Chr | SNPs | Effect size (95% CI) | P value | Mapped gene/nearest genes |
|---|---|---|---|---|---|---|---|---|---|---|
| Abdel-Wahab102 | 89 (44 cases, 45 controls) | Melanoma | Anti-CTLA-4, anti-PD-1/PDL1, combined ICI | All tagging SNPs | ≥Grade 2 irAE | 5 | rs11743438 | 4.30 (2.3–8.0) | 5.56 × 10–6 | GABRP |
| 5 | rs11743735 | 4.50 (2.3–8.8) | 8.34 × 10–6 | GABRP | ||||||
| 20 | JHU_20.57183980 | 6.90 (2.7–17.6) | 8.85 × 10–6 | DSC2 | ||||||
| 2 | rs56328422 | 4.20 (2.1–8.4) | 4.14 × 10–5 | BAZ2B | ||||||
| 3 | rs35807769 | 4.90 (2.2–11.1) | 6.07 × 10–5 | Near genes ZPLD1 and MIR548AB | ||||||
| 5 | rs3026321 | 19.80 (2.6–152.7) | 6.31 × 10–5 | SEMA5A | ||||||
| 5 | rs1276216 | 6.40 (2.3–17.6) | 8.18 × 10–5 | Near genes LOC105374704 and CDH6 | ||||||
| 2 | rs919682 | 4.60 (2.1–10.1) | 8.29 × 10–5 | OSBPL6 | ||||||
| 2 | JHU_2.179128562 | 4.60 (2.1–10.1) | 8.29 × 10–5 | AGPS | ||||||
| 21 | rs239731 | 11.30 (2.5–50.4) | 8.56 × 10–5 | LOC102724355 | ||||||
| 5 | JHU_5.9269447 | 3.50 (1.9–6.5) | 8.61 × 10–5 | SEMA5A | ||||||
| 2 | rs359975 | 8.50 (2.4–29.9) | 9.41 × 10–5 | Near genes CFAP65 and LOC100129175 | ||||||
| 3 | rs11711517 | 0.16 (0.07–0.37) | 2.39 × 10–6 | LOC105377125 | ||||||
| 15 | JHU_15.93602126 | 0.24 (0.13–0.46) | 8.89 × 10–6 | RGMA | ||||||
| 15 | JHU_15.93604000 | 0.24 (0.13–0.46) | 8.89 × 10–6 | RGMA | ||||||
| 18 | rs470753 | 0.16 (0.07–0.40) | 1.65 × 10–5 | Near genes LINC01924 and CDH7 | ||||||
| 15 | rs4778080 | 0.24 (0.12–0.47) | 1.92 × 10–5 | RGMA | ||||||
| 12 | rs2117997 | 0.22 (0.10–0.45) | 2.44 × 10–5 | ANKRD42 | ||||||
| 6 | rs55733913 | 0.16 (0.06–0.42) | 3.98 × 10–5 | PACRG | ||||||
| 12 | rs7954686 | 0.16 (0.06–0.42) | 3.98 × 10–5 | FAR2 | ||||||
| 3 | rs6440251 | 0.13 (0.04–0.40) | 5.15 × 10–5 | LOC105374140 | ||||||
| 14 | JHU_14.101799748 | 0.11 (0.03–0.38) | 5.20 × 10–5 | Near genes LINC02285 and LINC00524 | ||||||
| 18 | rs4800887 | 0.25 (0.13–0.50) | 5.74 × 10–5 | Near genes CDH2 and MIR302F | ||||||
| 3 | rs162263 | 0.24 (0.12–0.49) | 5.88 × 10–5 | ROBO1 | ||||||
| 9 | rs10814859 | 0.24 (0.12–0.49) | 5.88 × 10–5 | GLIS3 | ||||||
| 3 | kgp3960064 | 0.28 (0.15–0.53) | 6.76 × 10–5 | ROBO1 | ||||||
| 3 | rs2062059 | 0.28 (0.15–0.53) | 7.37 × 10–5 | Near genes DCBLD2 and MIR548G | ||||||
| 8 | rs12548560 | 0.28 (0.14–0.53) | 8.95 × 10–5 | PVT1 | ||||||
| 6 | rs66502444 | 0.16 (0.06–0.43) | 9.11 × 10–5 | PACRG | ||||||
| 8 | rs6993547 | 0.16 (0.06–0.43) | 9.11 × 10–5 | PREX2 | ||||||
| Bins57 | 322 (96 cases, 226 controls) | NSCLC | Anti-PD-1 | Small scale candidate study | Elevated liver transaminases | 12 | rs2301756* | 2.31 (1.02–5.21)a | 0.041 | PTPN11 (SHP2) |
| Rheumatological toxicity | 12 | rs2069705* | 6.04 (1.53–23.86)a | 0.019 | IFNG | |||||
| Treatment-related adverse events | 2 | rs2227981* | 0.45 (0.21–0.98)a | 0.041 | PDCD1 804C>T (PD-1) | |||||
| Queirolo103 | 173 (10 cases, 163 control) | Melanoma | Anti-CTLA-4 | Small scale candidate study | Endocrinopathies | 2 | rs4553808 (A/G or G/G genotypes) | 0.09 (0.02–0.43)a,b | 0.003 | CTLA-4–1661A>G |
| Cappelli59 | 753 (27 cases, 726 controls) | Mixed (largest cohort Melanoma) | Anti-PD-1/CTLA-4, anti-PD-1/PD-L1+/− anti-CD73 or anti-LAG3 | HLA specific approach | Inflammatory arthritis | 6 | HLA DRB1*04: 05 | 8.60 (1.70–43.40) | 0.040 | HLA |
| Magis60 | 163 (5 cases, 158 controls) | Melanoma | Anti-PD-1 | HLA specific approach | Acute type 1 diabetes | 6 | HLA DRB1*04:01, DRB1*04:05, DRB1*03:01 | NA | NA | HLA |
| Refae56 | 94 (15 cases, 79 controls) | Advanced cancer (largest cohort NSCLC) | Anti-PD-1/PDL1 | Medium scale candidate study | Serious ≥grade 3 irAE | 12 | rs246079 | 0.09 (0.02–0.36)c | <0.001 | UNG |
| 9 | rs10964859 | 6.08 (1.52–24.33)b | 0.014 | IFNW1 | ||||||
| 9 | rs4143815 | 8.11 (2.09–31.40)b | 0.003 | PD-L1 | ||||||
| 19 | rs12979860 | 3.72 (1.09–12.68)b | 0.036 | IFNL4 | ||||||
| 2 | rs3087243 | 3.19 (1.01–10.27)b | 0.048 | CTLA4 | ||||||
| 2 | rs11571302 | 4.06 (1.23–13.38)b | 0.018 | CTLA4 | ||||||
| 2 | rs7565213 | 3.73 (1.14–12.19)b | 0.026 | CTLA4 | ||||||
| Iafolla62 | 101 (23 cases, 78 controls) | Mixed (largest cohort triple negative breast cancer) | Anti-PD-1 | HLA specific approach | ≥Grade 2 irAE | 6 | HLA-A | 1.03 (0.23–4.62)d | 0.970 | HLA |
| 6 | HLA-B | 1.32 (0.24–7.26)d | 0.750 | HLA | ||||||
| 6 | HLA-C | 0.81 (0.24–2.78)d | 0.740 | HLA | ||||||
| 6 | HLA-A, -B and -C | 0.58 (0.21–1.62)d | 0.300 | HLA | ||||||
| Hasan Ali58 | 102 (59 cases, 43 controls) | Advanced cancer | Anti-PD-1 | HLA specific approach | Pruritus | 6 | HLA-DRB1*11:01 | 4.53 (X21,95 = 9.45) | 0.002 | HLA |
| Colitis | 6 | HLA-DQB1*03:01 | 3.94 (X21,95 = 5.67) | 0.017 | HLA | |||||
| Luo69 | 729 (95 cases, 634 controls) | NSCLC | Anti-PD-1 or combined anti-PD-1 and CTLA-4 | All tagging SNPs | Thyroid disorders | 6 | rs9268543 | NA | 7.5 × 10–7 | HLA |
| Kirchhoff104 | 69 (30 cases, 39 controls | Melanoma | Anti-CTLA-4 | Whole exome sequencing and detection of ~1000 autoimmune variants | Serious ≥grade 3 irAE | 19 | rs504963 | 2.57 | 0.006 | 3’UTR of FUT2 |
| 6 | rs1738074 | 2.21 | 0.025 | TAGAP | ||||||
| Montaudié70 | 57 (33 cases, 24 controls) | Melanoma | Anti-PD-1, Anti-CTLA-4, combined anti-PD-1 and anti-CTLA-4 | All tagging SNPs | Any grade irAE | 2 | rs782637 | NA | 4.10 × 10–8 | PNPT1 |
| 2 | rs78259 | NA | 3.40 × 10–7 | PNPT1 | ||||||
| 2 | rs7882572 | NA | 3.80 × 10–7 | PNPT1 | ||||||
| 2 | rs3762513 | NA | 6.60 × 10–7 | CFAP36 | ||||||
| 11 | rs7481951 | NA | 3.10 × 10–5 | ANO5 | ||||||
| 2 | rs4988956 | NA | 3.80 × 10–4 | IL1RL1 | ||||||
| Yano61 | 11 cases | Melanoma, NSCLC, gastric | Anti-PD-1, anti-CTLA-4 | HLA specific approach | Pituitary disorders | 6 | HLA-DR15 | NA | 1.40 × 10–3 | HLA |
| 6 | HLA-DR1502 | NA | 0.002 | HLA | ||||||
| 6 | HLA-Cw12 | NA | 0.001 | HLA | ||||||
| 6 | HLA-B52 | NA | 0.003 | HLA | ||||||
| Marschner105 | 167 (39 cases. 128 controls) | Melanoma, lung | Anti-PD-1/PDL1 | Small scale candidate study | Serious ≥grade 3 irAE | 5 | rs2910164 (C/C genotype) | 6.78 (1.87–24.60) | 0.004 | MIR146A |
| Groha65 | 1751 (339 cases, 1412 controls) | Mixed (largest cohort NSCLC) | Anti-PD-1/PD-L1, combined ICI, anti-CTLA-4 | All tagging SNPs | All grade irAE | 8 | rs16906115 | HR = 2.0 (1.6–2.5) | 3.8 × 10–9 | IL-7 |
| 1 | rs75824728* | HR = 1.9 (1.5–2.4) | 8.4 × 10–9 | IL22RA1 | ||||||
| 4 | rs113861051* | HR = 2.0 (1.6–2.6) | 1.1 × 10–8 | Near genes LINC02484 and ARAP2 | ||||||
| Weidhaas68 | 161 (45 cases, 116 controls) | Mixed (largest cohort melanoma) | Anti-PD-1/PD-L1 | Candidate study (50 common variants mapping to regions expected to disrupt miRNAs) | ≥Grade 2 irAE | 7 | rs9374 | AUC 0.82 (in a panel) | <0.001 | RAC1 |
| Udagawa66 | 622 (520 cases, 102 controls) | Mixed (largest cohort lung) | Anti-PD-1 | All tagging SNPs | All ≥grade 1 irAE | 21 | rs469490 | 5.15 (2.39–13.22) | 2.97 × 10–7 | Near genes APP and CYYR1-AS1 |
| ≥Grade 1 Thyroid irAE | 15 | rs8023690 | 4.09 (2.21–8.15) | 2.59 × 10–7 | Near genes RGMA and LINC02207 |
Studies investigating on genetic determinants of irAE were reviewed up till October 2022. Effect sizes for all studies are reported in odds ratios and 95% CI unless indicated otherwise.
Chr chromosome, CI confidence interval, HR hazard ratio, ICI immune checkpoint inhibitor, irAE immune-related adverse events, NA not available, NSCLC non-small cell lung cancer, SNPs single nucleotide polymorphism.
aUnivariate analysis.
bUnder recessive model.
cUnder dominant model.
dSummary statistics based on HLA-1 loci heterozygosity relative to homozygosity.
*Did not validate in independent sample sets.
Majority of the existing studies involved sample sizes of <200 patients and analysed variants in candidate genes or regions associated with immune response autoimmunity and response to systemic stress. The small sample sizes studied to date limit the chance of identifying true associations and indeed one of the larger of the existing studies, involving 322 NSCLC patients on nivolumab was unable to validate associations between lower odds of any grade toxicity in patients homozygous for PDCD1 804C>T (rs2227981) and higher odds of rheumatological toxicity in patients with one or more copy of IFNG −1616T>C in a validation cohort57.
As shown in Table 4, the association between HLA alleles and irAE occurrence has been explored. HLA variation tends to be associated with organ-specific autoimmune toxicities as opposed to a combined measure of irAE occurrence. HLA alleles have been shown to be associated with ICI-induced pruritus (HLA-DRB1*11:01, OR 4.53, p = 0.002) colitis (HLA-DQB1*03:01, OR 3.94, p = 0.017)58, arthritis (HLA-DRB1*04:05, OR 8.6, p = 0.04)59, type 1 diabetes (HLA-DRB1*03 and HLA-DRB1*04 haplotypes)60 and pituitary irAE (81.8% HLA-DR15 vs 33.5% in healthy controls, p = 0.0014)61. In contrast, analysis of cases of severe irAEs such as fulminant type 1 diabetes have failed to identify any association between HLA alleles and irAEs62–64. Validating the hypothesis that germline variants associated with AD or immunomodulation may be important in explaining risk of irAEs; three of the variants identified as associated with irAE (Table 4) were associated with ADs (Table 3); rs1738074 (coeliac disease risk), HLA-DQB1*03:01 (systemic lupus erythematosus risk) and rs3087243 (rheumatoid arthritis risk).
The first GWAS of irAE was posted on MedRXiv in April 202265. This study of 1751 patients on ICIs identified a genome-wide significant association between a SNP mapping to IL7, rs16906115, and any grade irAE toxicities65. rs16906115 replicates in two independent cohorts and is the first genetic variant associated with irAEs to have been identified using large sample sizes and validation cohorts65. The vast majority of patients included in this study were on PD1/PD-L1 inhibitors as single agents65. It remains to be determined if rs16906115 is also associated with CTLA-4 driven irAEs. A second GWAS of any grade irAE toxicity has also recently been published66. The study identified 27 SNPs associated with any grade nivolumab induced irAE with a P < 1 × 10–4 including rs469490, which lies upstream to APP which has been linked to Crohn’s disease67. A subgroup analysis identified rs8023690 as a potential predictive marker for hypothyroidism (OR 4.09, 95% CI 2.21–8.15, P = 2.58 × 10–7)66. Majority of patients included in the study experienced an irAE; only 86 and 16 controls were included in the discovery and replication phases, respectively66. The study was only powered (at 80%) to detect large effect sizes of common SNPs (minor allele frequency >0.1 and OR > 2.2).
Weidhaas et al tested a panel of germline variants predicted to disrupt miRNA binding in 62 melanoma patients and 99 patients with other cancer types including prostate cancer treated with anti-PD-1/PD-L1. The 50 variant panel, applied using four different classifiers achieved an AUC of ~0.80 for the prediction of ≥grade 2 irAEs in their training and validation cohorts68. One of the markers included in the classifier rs9374 was associated with a nine-fold increased risk of ≥grade 2 irAEs. It would be interesting to examine this marker in larger datasets. A polygenic risk score for hypothyroidism developed using UK Biobank data consisting of 1502 SNPs was found to predict thyroid irAE in NSCLC patients treated with ICIs69. This finding suggests the potential utility of applying risk scores to generate irAE risk profiles. Exome sequencing studies of patients treated with ICIs have also only been performed in small cohorts of patients. Montaudié et al performed exome sequencing of 57 melanoma patients with 57 patients also available for validation70. The authors concluded that germline variants had limited impact on irAE occurrence, however much larger cohorts are required to test this70.
Future directions
55% of patients treated with combination ICIs and 14–33% of patients treated with single-agent ICIs experience one or more ≥grade 3 irAEs across their treatment course. We have highlighted above the rationale for utilising germline testing to predict those at risk of developing these serious toxicities. As mentioned, any grade toxicity has also been associated with improved outcome and so the risk of toxicity will need to be balanced with chance of efficacy to optimise cancer care. Thus far the majority of studies testing individual genetic factors for associations with irAEs have been limited by their sample size and lack of independent validation of their findings. Large cohort studies with selection of an appropriate methodology are vital to produce meaningful associations that can be translated to clinical practice.
Methods of identifying genetic variants associated with irAE
One established hypothesis-free method of determining potential genetic determinants of human traits is GWAS71. GWAS have contributed towards the understanding of disease susceptibility, biomarker discovery and personalised therapeutic options72. Most genetic studies of AD have implemented GWAS to identify common predisposing variants of specific AD. This approach can similarly be applied to identify common variants predisposing to irAE risk using either SNP array technology or low pass (0.5–1x) whole genome sequencing73. Standard depth (~30x) whole genome sequencing (WGS) is a more comprehensive genotyping modality and allows rare variants to be accurately assessed. It is however a considerably higher cost approach compared to SNP array genotyping or low-pass sequencing both in terms of experimental cost and analytical complexity74. While whole exome sequencing is more cost-effective than WGS, it does not allow assessment of non-coding regulatory regions that could be functionally important74.
GWAS do not normally identify causal variants or target genes, these are identified by fine mapping variants in linkage disequilibrium with the SNPs associated with the phenotype of interest to identify the variants that should be studied further in functional analyses72. GWAS have also identified novel pathways and mechanisms important in conferring phenotypes and disease risk. While GWAS do require large sample sizes, we note that pharmacogenetics studies have reported larger per allele effect sizes than have been detected in studies of complex diseases75. Studies of >500 patients with irAEs will therefore be powered to pick up large effect sizes in excess of an odds ratio of 2 (assuming a grade 3 toxicity incidence of ~10%). Larger studies will be required to detect the full spectrum of variants explaining an individual’s risk of developing irAEs. Encouraging results from the first GWAS with a sample size of over 1000 patients suggest that there are likely to be further germline genetic variants influencing the occurrence of irAEs65. As the incidence of irAE may be influenced by various treatment, disease and patient-related factors, covariates including type of ICI regime, cancer type, age, gender, BMI and performance status may need to be considered in these studies12.
Close consideration of the definition of cases in GWAS of irAE is also important. Identifying variants predictive of serious ≥grade 3 irAE would likely have more beneficial translational value in practice compared to predicting any grade events, although large sample sizes would be required to avoid being underpowered.
Polygenic risk score analysis
A useful adjunct to GWAS is polygenic risk profiling using polygenic risk scores (PRS), which measure an individual’s genetic susceptibility to a disease determined by the sum of the risk alleles (weighted by the effect size of each variant) they carry76. As mentioned above a PRS for hypothyroidism was able to significantly predict thyroid irAE in 729 NSCLC patients (HR per SD 1.34, 95% CI 1.08–1.66, P = 9.73 × 10–3, AUROC = 0.6)69. In addition, Khan et al identified an association between the development of skin irAE with PRS for psoriasis in bladder cancer patients receiving anti-PD-L177. Existing risk scores for relevant autoimmune diseases may have power in predicting patients at high risk of specific toxicities. Risk scores however are only going to be powerful if they explain a large proportion of the genetic risk of developing a toxicity. If only a few genetic causes of a trait are known about, the PRS has limited value. PRS would likely need to be used alongside non-genetic risk factors that are also associated with the risk of developing irAEs.
Developing a predictive model of irAE for use in clinical practice
The challenge of identifying patients at serious risk of irAE and the potentially harmful short and long-term adverse effects warrant further investigation onto the development of a predictive model of irAE. The potential treatment benefit also needs to be considered in order to not deprive patients of an effective cancer treatment. Balance between treatment efficacy and toxicity will need to be achieved by developing predictive models to estimate both treatment outcomes to help with risk stratification.
Research findings on host genetics should be complemented with those from other ‘omics’ such as the microbiome, immunome, metabolome and the tumour microenvironment. Numerous studies have detected increased levels of cytokines and chemokines including IL-1a, IL-2, IFNα2, IL-6, IL-17 and post-treatment CXCL9 and CXCL10 as probable predictors of irAE occurrence78–81. Early diversification of T cell repertoire with decline in T cell clonality, increase in Th17 cells or early B cell changes have all been found to be associated with early development of irAE82–85. Deficiency in certain regulatory B cell phenotypes was found in patients who developed serious irAE86. Chaput et al showed that the composition of gut microbiota with Faecalibacterium and other Firmicutes has been associated with ipilimumab-induced colitis, whereas, the presence of Bacteroidetes was protective against colitis87. Tahir et al also found a correlation with autoantibodies anti-GNAL and anti-ITM2B with hypophysitis and anti-CD-74 with pneumonitis induced by ICI88.
An effective predictive model of toxicity can guide clinical decisions on personalised treatment options and safety monitoring of toxicities based on the patient’s risk of irAE. Earlier detection of patients at high risk of a particular toxicity with allow for stricter monitoring and earlier intervention to reduce the risk.
Conclusion
A large number of genetic variants have been identified which explain variation in immune modulation and risk of AD development. These variants may also be genetic determinants of irAEs. Large genome-wide studies of thousands of patients on ICIs are needed to detect common and rare variants with clinically relevant effect sizes that can contribute towards the development of a candidate gene panel in a predictive risk model of toxicity.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
I.S.C. acknowledges grant funding from the Birmingham Cancer Research UK Centre. C.P. acknowledges funding from Bowel Cancer UK.
Author contributions
I.S.C. and C.P. contributed to writing the manuscript. C.P., I.S.C. and A.K. contributed to collecting the data and reviewing the literature required for this manuscript. C.P. and G.M. conceived the idea for the manuscript. All authors contributed to editing and critical review of the manuscript.
Data availability
The datasets analysed during the current study are available in the NHGRI-EBI GWAS Catalog repository [https://www.ebi.ac.uk/gwas/].
Competing interests
S.P. declares honoraria from BMS, MSD, Roche, Amgen, Remedy Bio, Zelluna, GSK. A.O.-B. received grant support from Roche, Bristol-Myers Squibb, Eli Lily, Novartis, and UCB Pharma, and personal fees from Roche and Bristol-Myers Squibb, all outside the submitted work. G.M. declares honoraria from AZ and Pfizer. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-022-00345-6.
References
- 1.Martins F, et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 2019;16:563–580. doi: 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
- 2.Cooper GS, Bynum MLK, Somers EC. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hertz DL, Rae J. Pharmacogenetics of cancer drugs. Annu. Rev. Med. 2015;66:65–81. doi: 10.1146/annurev-med-053013-053944. [DOI] [PubMed] [Google Scholar]
- 4.Yang, W., Li, S. & Yang, Q. Risk of dermatologic and mucosal adverse events associated with PD-1/PD-L1 inhibitors in cancer patients: a meta-analysis of randomized controlled trials. Medicine98, e15731 (2019). [DOI] [PMC free article] [PubMed]
- 5.Khoja L, Day D, Wei-Wu Chen T, Siu LL, Hansen AR. Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: a systematic review. Ann. Oncol. 2017;28:2377–2385. doi: 10.1093/annonc/mdx286. [DOI] [PubMed] [Google Scholar]
- 6.Haanen JBAG, et al. Management of toxicities from immunotherapy: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017;28:iv119–iv142. doi: 10.1093/annonc/mdx225. [DOI] [PubMed] [Google Scholar]
- 7.Electronic Medicines Compendium (emc). https://www.medicines.org.uk/emc/. (2021).
- 8.Solinas C, et al. Programmed cell death-ligand 2: a neglected but important target in the immune response to cancer? Transl. Oncol. 2020;13:100811. doi: 10.1016/j.tranon.2020.100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: a systematic review and meta-analysis. JAMA Oncol. 2019;5:1008–1019. doi: 10.1001/jamaoncol.2019.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang DY, et al. Fatal toxic effects associated with immune checkpoint inhibitors: a systematic review and meta-analysis. JAMA Oncol. 2018;4:1721–1728. doi: 10.1001/jamaoncol.2018.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chennamadhavuni, A., Abushahin, L., Jin, N., Presley, C. J. & Manne, A. Risk factors and biomarkers for immune-related adverse events: a practical guide to identifying high-risk patients and rechallenging immune checkpoint inhibitors. Front. Immunol. 13, 1–14 (2022). [DOI] [PMC free article] [PubMed]
- 13.Samani A, et al. Impact of age on the toxicity of immune checkpoint inhibition. J. Immunother. Cancer. 2020;8:e000871. doi: 10.1136/jitc-2020-000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asada M, et al. The risk factors associated with immune checkpoint inhibitor-related pneumonitis. Oncology. 2021;99:256–259. doi: 10.1159/000512633. [DOI] [PubMed] [Google Scholar]
- 15.Triggianese P, et al. Immune checkpoint inhibitors-induced autoimmunity: the impact of gender. Autoimmun. Rev. 2020;19:102590. doi: 10.1016/j.autrev.2020.102590. [DOI] [PubMed] [Google Scholar]
- 16.Guzman-Prado Y, Ben Shimol J, Samson O. Body mass index and immune-related adverse events in patients on immune checkpoint inhibitor therapies: a systematic review and meta-analysis. Cancer Immunol. Immunother. 2021;70:89–100. doi: 10.1007/s00262-020-02663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada N, et al. Risk factors of immune checkpoint inhibitor-related interstitial lung disease in patients with lung cancer: a single-institution retrospective study. Sci. Rep. 2020;10:13773. doi: 10.1038/s41598-020-70743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atchley WT, et al. Immune checkpoint inhibitor-related pneumonitis in lung cancer: real-world incidence, risk factors, and management practices across six health care centers in North Carolina. Chest. 2021;160:731–742. doi: 10.1016/j.chest.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S-Q, et al. The pattern of time to onset and resolution of immune-related adverse events caused by immune checkpoint inhibitors in cancer: a pooled analysis of 23 clinical trials and 8,436 patients. Cancer Res. Treat. 2021;53:339–354. doi: 10.4143/crt.2020.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghisoni E, et al. Late-onset and long-lasting immune-related adverse events from immune checkpoint-inhibitors: an overlooked aspect in immunotherapy. Eur. J. Cancer. 2021;149:153–164. doi: 10.1016/j.ejca.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Marthey L, et al. Cancer immunotherapy with anti-CTLA-4 monoclonal antibodies induces an inflammatory Bowel disease. J. Crohns. Colitis. 2016;10:395–401. doi: 10.1093/ecco-jcc/jjv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clotman K, Janssens K, Specenier P, Weets I, De Block CEM. Programmed cell death-1 inhibitor-induced type 1 diabetes mellitus. J. Clin. Endocrinol. Metab. 2018;103:3144–3154. doi: 10.1210/jc.2018-00728. [DOI] [PubMed] [Google Scholar]
- 23.Calabrese LH, Calabrese C, Cappelli LC. Rheumatic immune-related adverse events from cancer immunotherapy. Nat. Rev. Rheumatol. 2018;14:569–579. doi: 10.1038/s41584-018-0074-9. [DOI] [PubMed] [Google Scholar]
- 24.Nigro O, et al. Late immune-related adverse events in long-term responders to PD-1/PD-L1 checkpoint inhibitors: a multicentre study. Eur. J. Cancer. 2020;134:19–28. doi: 10.1016/j.ejca.2020.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Khan S, et al. Late‐onset immunotherapy toxicity and delayed autoantibody changes: checkpoint inhibitor-induced Raynaud’s‐like phenomenon. Oncologist. 2020;25:e753–e757. doi: 10.1634/theoncologist.2019-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan SA, Pruitt SL, Xuan L, Gerber DE. Prevalence of autoimmune disease among patients with lung cancer: implications for immunotherapy treatment options. JAMA Oncol. 2016;2:1507–1508. doi: 10.1001/jamaoncol.2016.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menzies AM, et al. Anti-PD-1 therapy in patients with advanced melanoma and preexisting autoimmune disorders or major toxicity with ipilimumab. Ann. Oncol. 2016;28:368–376. doi: 10.1093/annonc/mdw443. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DB, et al. Ipilimumab therapy in patients with advanced melanoma and preexisting autoimmune disorders. JAMA Oncol. 2016;2:234–240. doi: 10.1001/jamaoncol.2015.4368. [DOI] [PubMed] [Google Scholar]
- 29.Ricciuti B, et al. Impact of immune-related adverse events on survival in patients with advanced non-small cell lung cancer treated with nivolumab: long-term outcomes from a multi-institutional analysis. J. Cancer Res. Clin. Oncol. 2019;145:479–485. doi: 10.1007/s00432-018-2805-3. [DOI] [PubMed] [Google Scholar]
- 30.Petrelli F, et al. Immune-related adverse events and survival in solid tumors treated with immune checkpoint inhibitors: a systematic review and meta-analysis. J. Immunother. 2019;43:1. doi: 10.1097/CJI.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 31.Kirchhoff, T. & Ferguson, R. in Biomarkers for Immunotherapy of Cancer: Methods and Protocols (eds. Thurin, M., Cesano, A. & Marincola, F. M.) 93–117 (Springer New York, 2020).
- 32.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Roederer M, et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangino M, Roederer M, Beddall MH, Nestle FO, Spector TD. Innate and adaptive immune traits are differentially affected by genetic and environmental factors. Nat. Commun. 2017;8:13850. doi: 10.1038/ncomms13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nath AP, et al. Multivariate genome-wide association analysis of a cytokine network reveals variants with widespread immune, haematological, and cardiometabolic pleiotropy. Am. J. Hum. Genet. 2019;105:1076–1090. doi: 10.1016/j.ajhg.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res.47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed]
- 38.Ueda H, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 39.Zeissig S, et al. Early-onset Crohn’s disease and autoimmunity associated with a variant in CTLA-4. Gut. 2015;64:1889–1897. doi: 10.1136/gutjnl-2014-308541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 41.Vogelsang M, et al. The expression quantitative trait loci in immune pathways and their effect on cutaneous melanoma prognosis. Clin. Cancer Res. 2016;22:3268–3280. doi: 10.1158/1078-0432.CCR-15-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat. Rev. Genet. 2016;17:160–174. doi: 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper JD, et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLOS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gough SCL, Simmonds MJ. The HLA region and autoimmune disease: associations and mechanisms of action. Curr. Genomics. 2007;8:453–465. doi: 10.2174/138920207783591690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin MA, Kroetz DL. Abacavir pharmacogenetics—from initial reports to standard of care. Pharmacotherapy. 2013;33:765–775. doi: 10.1002/phar.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada R, Yamamoto K. Mechanisms of disease: genetics of rheumatoid arthritis—ethnic differences in disease-associated genes. Nat. Clin. Pract. Rheumatol. 2007;3:644–650. doi: 10.1038/ncprheum0592. [DOI] [PubMed] [Google Scholar]
- 49.Chung W-H, et al. A marker for Stevens–Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 50.Lonjou C, et al. A marker for Stevens-Johnson syndrome …: ethnicity matters. Pharmacogenomics J. 2006;6:265–268. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 51.Amstutz U, et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther. 2018;103:210–216. doi: 10.1002/cpt.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White C, et al. Ethnic diversity of DPD activity and the DPYD gene: review of the literature. Pharmgenomics. Pers. Med. 2021;14:1603–1617. doi: 10.2147/PGPM.S337147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chat V, et al. Autoimmune genetic risk variants as germline biomarkers of response to melanoma immune-checkpoint inhibition. Cancer Immunol. Immunother. 2019;68:897–905. doi: 10.1007/s00262-019-02318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of Janus family kinases 1 and 3. Curr. Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 55.Nomizo T, et al. Clinical impact of single nucleotide polymorphism in PD-L1 on response to nivolumab for advanced non-small-cell lung cancer patients. Sci. Rep. 2017;7:45124. doi: 10.1038/srep45124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Refae S, et al. Germinal Immunogenetics predict treatment outcome for PD-1/PD-L1 checkpoint inhibitors. Invest. N. Drugs. 2019 doi: 10.1007/s10637-019-00845-w. [DOI] [PubMed] [Google Scholar]
- 57.Bins S, et al. Association between single-nucleotide polymorphisms and adverse events in nivolumab-treated non-small cell lung cancer patients. Br. J. Cancer. 2018;118:1296–1301. doi: 10.1038/s41416-018-0074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasan Ali O, et al. Human leukocyte antigen variation is associated with adverse events of checkpoint inhibitors. Eur. J. Cancer. 2019;107:8–14. doi: 10.1016/j.ejca.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 59.Cappelli LC, Dorak MT, Bettinotti MP, Bingham CO, Shah AA. Association of HLA-DRB1 shared epitope alleles and immune checkpoint inhibitor-induced inflammatory arthritis. Rheumatology. 2019;58:476–480. doi: 10.1093/rheumatology/key358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magis, Q. et al. Diabetes and blood glucose disorders under anti-PD1. J. Immunother. 41, 232–240 (2018). [DOI] [PubMed]
- 61.Yano S, et al. Human leucocyte antigen DR15, a possible predictive marker for immune checkpoint inhibitor-induced secondary adrenal insufficiency. Eur. J. Cancer. 2020;130:198–203. doi: 10.1016/j.ejca.2020.02.049. [DOI] [PubMed] [Google Scholar]
- 62.Iafolla MAJ, et al. Predicting toxicity and response to pembrolizumab through germline genomic HLA class 1 analysis. JNCI Cancer Spectr. 2020;5:pkaa115–pkaa115. doi: 10.1093/jncics/pkaa115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lowe JR, et al. Genetic risk analysis of a patient with fulminant autoimmune type 1 diabetes mellitus secondary to combination ipilimumab and nivolumab immunotherapy. J. Immunother. Cancer. 2016;4:89. doi: 10.1186/s40425-016-0196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolchok JD, et al. Ipilimumab efficacy and safety in patients with advanced melanoma: a retrospective analysis of HLA subtype from four trials. Cancer Immun. 2010;10:9. [PMC free article] [PubMed] [Google Scholar]
- 65.Groha, S. et al. Germline variants associated with toxicity to immune checkpoint blockade. Nat Med. 10.1038/s41591-022-02094-6 (2022). [DOI] [PMC free article] [PubMed]
- 66.Udagawa C, et al. Association between genetic variants and the risk of nivolumab-induced immune-related adverse events. Pharmacogenomics. 2022 doi: 10.2217/pgs-2022-0113. [DOI] [PubMed] [Google Scholar]
- 67.Li H, et al. Integrated bioinformatics analysis identifies ELAVL1 and APP as candidate crucial genes for Crohn’s disease. J. Immunol. Res. 2020;2020:3067273. doi: 10.1155/2020/3067273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weidhaas J, et al. Germline biomarkers predict toxicity to anti-PD1/PDL1 checkpoint therapy. J. Immunother. Cancer. 2022;10:e003625. doi: 10.1136/jitc-2021-003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo J, et al. Immunotherapy-mediated thyroid dysfunction: genetic risk and impact on outcomes with PD-1 blockade in non-small cell lung cancer. Clin. Cancer Res. 2021 doi: 10.1158/1078-0432.CCR-21-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montaudié H, et al. Germline variants in exonic regions have limited impact on immune checkpoint blockade clinical outcomes in advanced melanoma. Pigment Cell Melanoma Res. 2021;34:978–983. doi: 10.1111/pcmr.12958. [DOI] [PubMed] [Google Scholar]
- 71.What are genome-wide association studies (GWAS)? EMBL-EBI Train Online (2020). https://www.ebi.ac.uk/training-beta/online/courses/gwas-catalogue-exploring-snp-trait-associations/what-is-gwas-catalog/what-are-genome-wide-association-studies-gwas/ (2020).
- 72.Tam V, et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 73.Li JH, Mazur CA, Berisa T, Pickrell JK. Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays. Genome Res. 2021;31:529–537. doi: 10.1101/gr.266486.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chat V, Ferguson R, Kirchhoff T. Germline genetic host factors as predictive biomarkers in immuno-oncology. Immuno-Oncol. Technol. 2019;2:14–21. doi: 10.1016/j.iotech.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maranville JC, Cox NJ. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J. 2016;16:388–392. doi: 10.1038/tpj.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi SW, Mak TS-H, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan Z, et al. Polygenic risk for skin autoimmunity impacts immune checkpoint blockade in bladder cancer. Proc. Natl Acad. Sci. USA. 2020;117:12288–12294. doi: 10.1073/pnas.1922867117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim SY, et al. Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clin. Cancer Res. 2019;25:1557 LP–1551563. doi: 10.1158/1078-0432.CCR-18-2795. [DOI] [PubMed] [Google Scholar]
- 79.Ozawa Y, et al. Impact of early inflammatory cytokine elevation after commencement of PD-1 inhibitors to predict efficacy in patients with non-small cell lung cancer. Med. Oncol. 2019;36:33. doi: 10.1007/s12032-019-1255-3. [DOI] [PubMed] [Google Scholar]
- 80.Tarhini AA, et al. Baseline circulating IL-17 predicts toxicity while TGF-β1 and IL-10 are prognostic of relapse in ipilimumab neoadjuvant therapy of melanoma. J. Immunother. Cancer. 2015;3:39. doi: 10.1186/s40425-015-0081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan S, et al. Immune dysregulation in cancer patients developing immune-related adverse events. Br. J. Cancer. 2019;120:63–68. doi: 10.1038/s41416-018-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh DY, et al. Immune toxicities elicted by CTLA-4 blockade in cancer patients are associated with early diversification of the T-cell repertoire. Cancer Res. 2017;77:1322–1330. doi: 10.1158/0008-5472.CAN-16-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koenen HJPM, et al. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17–producing cells. Blood. 2008;112:2340–2352. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 84.von Euw E, et al. CTLA4 blockade increases Th17 cells in patients with metastatic melanoma. J. Transl. Med. 2009;7:35. doi: 10.1186/1479-5876-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Das R, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Invest. 2018;128:715–720. doi: 10.1172/JCI96798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patel AJ, et al. Regulatory B cell repertoire defects predispose lung cancer patients to immune-related toxicity following checkpoint blockade. Nat. Commun. 2022;13:3148. doi: 10.1038/s41467-022-30863-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chaput N, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 88.Tahir SA, et al. Autoimmune antibodies correlate with immune checkpoint therapy-induced toxicities. Proc. Natl Acad. Sci. USA. 2019;116:22246 LP–22222251. doi: 10.1073/pnas.1908079116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sliz E, et al. Genome-wide association study identifies seven novel loci associating with circulating cytokines and cell adhesion molecules in Finns. J. Med. Genet. 2019;56:607–616. doi: 10.1136/jmedgenet-2018-105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Traglia M, et al. Cross-genetic determination of maternal and neonatal immune mediators during pregnancy. Genome Med. 2018;10:67. doi: 10.1186/s13073-018-0576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kariuki SN, et al. Genetic analysis of the pathogenic molecular sub-phenotype interferon-alpha identifies multiple novel loci involved in systemic lupus erythematosus. Genes Immun. 2015;16:15–23. doi: 10.1038/gene.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahsan M, et al. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS Genet. 2017;13:e1007005. doi: 10.1371/journal.pgen.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hillary RF, et al. Multi-method genome- and epigenome-wide studies of inflammatory protein levels in healthy older adults. Genome Med. 2020;12:60. doi: 10.1186/s13073-020-00754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Enroth S, Johansson A, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat. Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tekola Ayele F, et al. Genome-wide associated loci influencing interleukin (IL)-10, IL-1Ra, and IL-6 levels in African Americans. Immunogenetics. 2012;64:351–359. doi: 10.1007/s00251-011-0596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahola-Olli AV, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Paré G, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barbalic M, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum. Mol. Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi SH, et al. Six novel loci associated with circulating VEGF levels identified by a meta-analysis of genome-wide association studies. PLOS Genet. 2016;12:e1005874. doi: 10.1371/journal.pgen.1005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Debette S, et al. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ. Res. 2011;109:554–563. doi: 10.1161/CIRCRESAHA.111.243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abdel-Wahab N, et al. Genetic determinants of immune-related adverse events in patients with melanoma receiving immune checkpoint inhibitors. Cancer Immunol. Immunother. 2021;70:1939–1949. doi: 10.1007/s00262-020-02797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Queirolo P, et al. CTLA-4 gene variant -1661A>G may predict the onset of endocrine adverse events in metastatic melanoma patients treated with ipilimumab. Eur. J. Cancer. 2018;97:59–61. doi: 10.1016/j.ejca.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 104.Kirchhoff T, et al. Germline determinants of immune related adverse events (irAEs) in melanoma immunotherapy response. Ann. Oncol. 2017;28:v407–v408. [Google Scholar]
- 105.Marschner D, et al. MicroRNA-146a regulates immune-related adverse events caused by immune checkpoint inhibitors. JCI Insight. 2020;5:e132334. doi: 10.1172/jci.insight.132334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study are available in the NHGRI-EBI GWAS Catalog repository [https://www.ebi.ac.uk/gwas/].