Abstract
Diseases are manifestations of complex changes within protein-protein interaction (PPI) networks whereby stressors, genetic, environmental, and combinations thereof, alter molecular interactions and perturb the individual from the level of cells to tissues to the entire organism. Targeting stressor induced dysfunctions in PPI networks has therefore become a promising yet technically challenging frontier in therapeutics discovery. This Opinion provides a new framework, based upon disrupting epichaperomes, pathological entities that enable dysfunctional rewiring of PPI networks, as a mechanism to revert context-specific PPI network dysfunctions to normative states. It speculates on the implications of recent research in this area for a precision medicine approach to detecting and treating complex diseases, including cancer and neurodegenerative disorders.
Keywords: protein-protein interaction networks, protein network dysfunction, epichaperome, neurodegenerative disorders, cancer, network medicine, Alzheimer’s disease
From targeting a protein to targeting a network
A classic, albeit one-dimensional premise of drug discovery approaches is that a disease state can be effectively treated by developing small molecule candidates that can inhibit, activate, degrade, and/or stabilize specific protein targets. Alternatively, a drug candidate can change the interaction fate of a protein with another protein (i.e., a protein-protein interaction, see Glossary). In this context, an identified and rigorously tested small molecule effectively eliminates the aberrant action of the offending protein and prevents disease. Although well intentioned, this basic hypothesis of ‘single drug targeting a single protein’, perhaps emerging from an inadvertent bias to oversimplify to aid in general understanding of a complex problem, is too narrow to be the sole driver of experimental therapeutics [1,2].
Proteins, akin to individuals, do not function in isolation. They interact with, impact and are impacted, by other proteins. They often behave in systems, termed protein-protein interaction (PPI) networks, with redundancies that have inherent adaptability capabilities [3]. This translates all too often into a drug failing to produce a desired effect, especially in disease states (e.g., cancer) where treatments are effectively evaded through cellular plasticity [4].
Importantly, the overwhelming majority of human diseases have a complex etiology with both internal and external perturbations negatively impacting, directly or indirectly, proteins in the context of cells, tissues, organs, and ultimately organisms. In this Opinion we refer to these pathologic perturbers as stressors. Stressors can be acute or chronic, and responses to stressors are complex in biological systems. For example, stressor adaptation, or conversely stressor maladaptation, in the context of stressor to phenotype, is modulated by the duration, intensity, controllability, and ability to predict a specific stressor or stressors, as well as the genetic makeup, environment, and life history [5,6]. Therefore, individuals, and their internal structures (e.g., cells, tissues, and organs) experience stressors and respond to them in different ways.
Thus, a paradigm shift in thinking about how to realistically develop drugs and therapies to target complex, biologically relevant PPI networks is top of mind [3,7-9]. In this context, the disease PPI network, a map of how individual or a combination of stressors alter cellular PPIs and perturb the system, from organ to organism, as a whole, becomes the target of therapeutic intervention. From a network perspective, proteins are not considered like individuals. Their behavior cannot be predicted based on a reductionist understanding of component parts. Rather, the number and architecture of connections (i.e. edges) within elements in the network (i.e. nodes) defines significance [10,11] (see Box 1).
Box 1. The basics of biological networks.
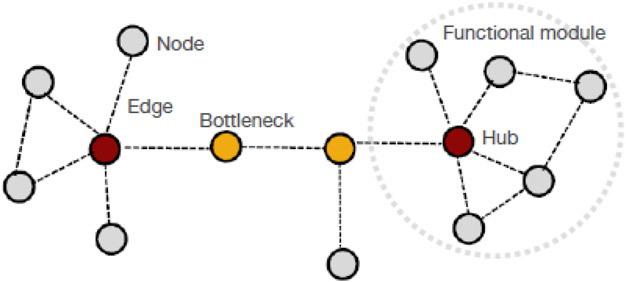
To address the complexity of biological systems, the science of “network biology” has taken root, where the architecture of intricate interactions among cellular component molecules, internal milieu, and external environment defines a phenotype [10]. Network biology uses concepts from the mathematical field of graph theory, systems biology, and statistical analyses [10]. An interaction network is a graphical representation of the interactome, where each component (e.g., a given protein, metabolite, or other molecule) is a node, and the interaction between components is an edge (Figure I). Thus, networks include multiple nodes that are analyzed assessing their concurrent interactions within the global structure of the graph. The topological structure of the network provides basic and direct information regarding the network and its functions [10].
Biological networks, including PPI networks, exhibit a near scale-free organization where a few proteins exhibit high connectivity while most are weakly connected to the network [11]. This architecture enables short across network distances, an advantageous property for malleable biological systems as it enables efficient response to external stimuli (i.e., facilitation of biochemical diversity at minimal energy cost) while preserving homeostasis upon perturbation (i.e., robustness, the ability to accommodate stressor perturbations with minimal functional consequences).
Topological parameters are fundamental measures of nodes in network theory [10]. Topological analysis identifies the significance of proteins in a network, and in turn the vulnerability of a network to elimination of proteins, for example hub proteins or bottleneck proteins (Figure I). These nodes play key roles in mediating communication within a given network.
Network analyses also inform on network functionality [10,11]. The topology of a network is a direct reflection of its function, and functional mapping can be used as a representation of the PPI network topology (i.e., the topological organization of proteins in a network has a specific functional outcome as reflected in the protein pathways impacted by such topological organization) [32,33]. In a network, proteins that form clusters are communities of nodes with an increased degree of interconnection relative to the whole network and may represent functional modules or protein complexes (Figure I).
Network models and quantitative tools querying underlying network biology have led to important information i) how cellular elements are organized and ii) how specific perturbations impact complex PPI networks [38]. Network analyses have also addressed controllability of PPI networks, which may serve from a pharmacotherapeutic perspective to direct disease relevant biological systems from a drug-resistant state to a state of vulnerability [70-72].
Figure I. Elements in a hypothetical protein-protein interaction network.
Protein-protein interaction (PPI) networks are graphical representations of contacts between proteins in a specific cellular context. From this perspective, proteins are not viewed as individual elements, but rather constituent components of a network of proteins. Thus, it is the number and architecture of connections of a protein with other proteins in the networks that defines significance and function of a given protein [10,11]. Nodes, proteins; Edges, protein-protein interactions; Hub proteins, proteins with many interaction partners in the PPI network map; Bottlenecks, proteins that enable the flow of information within the PPI network.
Understanding and exploiting individual biological networks as they are shaped by their combinatorial genetic and environmental stressors may be a daunting yet effective strategy to consider implementing from a pharmacotherapeutic perspective. This requires movement from the unitary protein approach to a goal of targeting PPI networks, specifically context-specific dysfunctions in such networks, as a means of disease control. Previously erroneously labeled as not druggable, PPI networks can in fact be modulated, albeit not as readily as solitary protein-ligand or protein-protein interactions. Herein, we share our view how disease-causing dysfunctions within PPI networks can be effectively detected, targeted, and manipulated for therapeutic efficacy. We start by providing an overview of mechanisms by which stressors rewire PPI networks in disease (Section 1). We then present our opinion on an emerging concept whereby PPI network dysfunctions may be corrected by disrupting epichaperomes, scaffolding platforms that act as architects of PPI reorganization in disease (Section 2). We conclude by presenting our view on how targeting PPI network dysfunctions via epichaperomes may lend itself to precision medicine (Section 3).
1. Features of PPI networks under stressor conditions
PPI networks are key components of biological networks as they encode the flux of information that links stressors to phenotype [12,13]. Not surprisingly, cells exploit alterations to PPI networks as a mechanism for adaptation under stressor conditions. However, when stressors are persistent or excessive, remodeling of PPI networks by stressors can be detrimental (i.e., maladaptive), leading to cellular dysfunction, and ultimately disease states [14-20] (Figure 1).
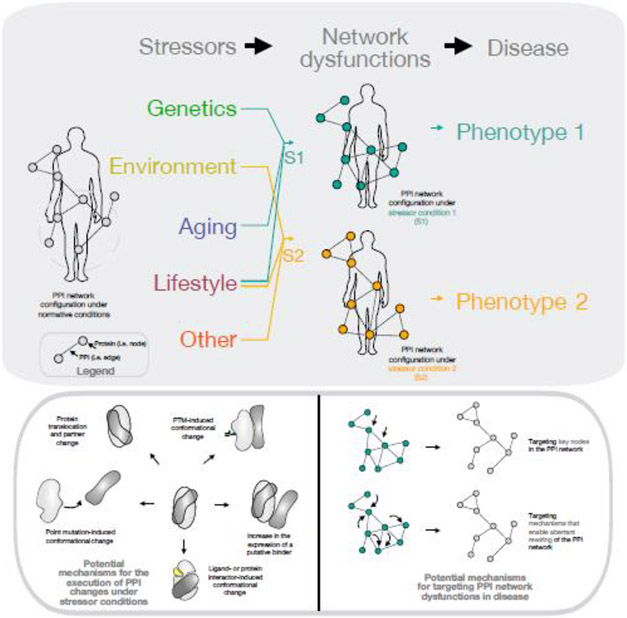
Figure 1. Disease as the manifestation of perturbation severity to the complex network of molecular interactions within an individual organism.
The overwhelming majority of human diseases have a complex etiology with both internal (e.g., genetic mutations, epigenetic alterations, proteotoxic stress) and external (e.g., chemical/environmental exposure, lifestyle, socioeconomic status, psychosocial factors, healthcare access disparities, gut-microbiome diversity, among others) perturbations negatively impacting, directly or indirectly, specific cells, tissues, organs, and ultimately organisms. Disease states are the outcome of how a combination of such perturbers (which we call stressors) alter cellular interactomes (i.e., the complex network of molecular interactions) and perturb from organ to organism, the individual. In this context, it is the complex network of molecular interactions that becomes the target of therapeutic intervention. PPI networks are an important component of cellular interactomes as they encode and execute the flux of information that link, at the cellular level, stressors to phenotype. As cells do not function in isolation, the effect of PPI networks reverberates and extends beyond individual cells, and into tissue and whole organism levels. A large number of PPIs rewire in disease states, with PPI network changes being context-dependent and context-specific. Disease associated modules in PPI networks map to dynamic regions in the PPI maps (exemplified by the gray, dashed circle). PPI network rewiring may be executed by a variety of protein-modifying mechanisms, such as posttranslational modifications (PTMs), protein conformational selection or protein mislocalization, and other mechanisms, as exemplified in the lower solid gray oval. PPI networks may be targeted through proteins centrally placed in the network or by disabling mechanisms that enable aberrant PPI network rewiring.
Several elegant studies based both on biological investigation and computationally derived predictions have framed the principles of complex changes occurring within PPI networks [3,15,21-23] to conclude that broad-scale rewiring of PPI networks drives the cell to execute different biological processes imposed by stressors [14,15,21]. It is now estimated that <10% of human PPIs remain unchanged upon stressor perturbations, and that rewiring of a large set of PPIs is necessary for evolution for lineage-specific adaptation, but also as a mechanism of network maladaptation in disease [13,21,22,24,25]. PPI changes in disease can be driven by changes in protein expression, but most often tend to be modulated by alterations in the strength of interactions and in cellular mislocalization, both which in turn can be influenced by alterations in post-translational modifications (PTMs), stabilization of disease-enriched protein conformations, and other protein-modifying mechanisms [21,26-28].
For example, yeast (S. cerevisiae) is a model widely used to understand intricacies of PPI changes. The yeast protein interactome (i.e., PPI network) is estimated to contain 37,600-75,500 detectable interactions under standard growth conditions [29]. Liu et al., 2020 analyzed changes induced in the yeast interactome under nine stressor conditions (e.g., salt, hydroxyurea, and hydrogen peroxide induced stress, among others) [21]. They quantified the relative PPI abundance of 1.6 million protein pairs for a total of 44 million measurements to identify 13,764 pairwise PPIs to find i) PPI networks contain highly dynamic regions that reorganize to drive or respond to cellular changes, ii) mutable PPIs (i.e., PPIs that change across stressor conditions) far outnumbered immutable PPIs, with PPIs identified in only one stressor outnumbering all other PPIs combined, and iii) changes in the dynamic PPI module were driven by post-translational events such as condition-specific protein localization, binding, or other mechanisms, rather than by frank changes in protein abundance [21].
Confirming predictions from co-expression data, Liu et al., 2020 found proteins in mutable PPIs were more likely to contain intrinsically disordered regions (i.e., regions that by themselves do not fold into unique three-dimensional structures), suggesting they may adopt different conformations in a context-dependent manner to promote a specific PPI [21]. French-Pacheco et al., 2022 found transcription factors and signaling proteins involved in stress response pathways had a higher content of structural disorder compared to the rest of the proteins, and that intrinsically disordered transcription factors participated in a higher number of PPIs than those with low structural disorder [30]. Thus, proteins in mutable PPIs are prone and able to adapt to context-dependent conformations to promote a specific PPI.
PPI alterations across stressor conditions may reflect both loss and gain in interactions compared to the inherent interactions found under normal physiological conditions [11,15,28,31-34]. For example, Qiu et al., 2021 found among important hub proteins across the cancer interactome, 757 gained new connections, 532 lost connections whereas 4 both gained and lost connections [35]. Proteins gaining new PPIs in cancer-related networks were enriched in oncogenic signaling pathways including MAPK, PI3K-Akt, and Wnt, whereas proteins losing PPIs were enriched in pathways for general functions including ribosome biogenesis and the spliceosome [35]. Examining postmortem human brains of sporadic Alzheimer’s disease (AD) patients and age matched counterparts with no cognitive impairment (NCI), Inda et al., 2020 identified 942 proteins that lost interactions with at least one partner while 1,191 proteins formed new interactions in the progression from NCI to AD [32]. These PPI changes negatively impacted protein pathways with key functions in synaptic plasticity, such as signaling by second messenger, Galpha(i) signaling, signaling by Rho GTPases, response to elevated cytosolic Ca2+, MAPK signaling, Wnt signaling, adhesion-regulatory networks including extracellular matrix organization and cell–cell communication, and protein translation-related networks [32]. Thus, both short-term memories that rely on post-translational modification of pre-existing synaptic proteins via the aforementioned signaling networks, and long-term memories, requiring production of new synaptic proteins, are vulnerable to, and negatively affected by, pervasive PPI network changes in AD [14].
In sum, a large number of PPIs rewire under stressor conditions to support disease-specific phenotypic changes. PPI network changes are context-dependent and context-specific, underscoring the need to understand and target proteins and their networks within specific disease-associated contexts. Disease-associated modules in PPI networks map to dynamic regions within PPI maps, and PPI network rewiring is executed by protein-modifying mechanisms rather than frank changes in protein expression. In aggregate, these features propose points of vulnerability in disease-associated PPI networks are provided by i) protein location within the network, and ii) mechanisms that execute and enable PPI rewiring (Figure 1).
For the former, the idea is that modulating key proteins in a network will impact the interaction profiles of multiple proteins synchronously, which in turn may impact disease [39-41]. For example, hubs and bottlenecks are believed to be crucial (i.e., central) candidate proteins (reviewed in [23,36-38]). The guiding principle behind centrality is that if a few major hubs or bottlenecks from the network are lost, the network is turned into a set of isolated graphs, preventing the proper flow of information (i.e., cellular function) to manifest effectively [41,42]. Supporting the network centrality concept as a means of target discovery, Viacava Follis, 2021 found viable targets are likely to be gateway proteins to self-standing networks [43]. Targets of select FDA approved small molecule drugs exhibit high node centrality within protein networks relative to a broader set of investigational targets, as well as higher centrality than other proteins within their respective functional classes [43]. Therefore, modulating the function of these key proteins may be less susceptible to network robustness, a mechanism which could enable redundant or overlapping signaling mechanisms, resulting in compensation of on-target drug effects.
2. Targeting epichaperomes: entities that enable pathologic rewiring of PPI networks
It is critically important to recognize that PPI networks are context dependent. A single protein may carry out different functions with different partners in different biological contexts under various stressors [44,45]. Therefore, PPIs discovered under a single condition or cell type represent a fraction of the full protein interactome [11,12,21,46,47], and nonspecific, broad-based surveys are likely to generate uninformed, largely inaccurate results when attempting to derive context-dependent PPI network information [34,45]. It is also important to remember changes in PPIs are seldom driven by protein expression changes, and thus using datasets from ‘omics that inventory proteins may provide limited information on context-specific PPI networks, and thus on proteins central to key PPI networks. Technology that enables direct identification, analysis, and modulation of PPI network dysfunctions in native (e.g., unengineered) cells and tissues has yet to be fully developed and implemented. Methods that target PPI network dysfunctions, irrespective of context, are thus needed to move the field forward.
In this Opinion we propose a new framework for targeting and controlling PPI networks in disease. It takes advantage of entities that enable dysfunctional rewiring of PPI networks in the first place, the epichaperomes, thus we propose a mechanism of PPI network rewiring, rather than specific network proteins, as a means of therapeutic intervention. Therefore, this approach is not contingent on knowing the identity and connectivity of all proteins in the specific context. It only depends on the knowledge that the execution of aberrant PPIs is conducted though epichaperomes (Figure 2, Key Figure), which can be targeted pharmacotherapeutically.
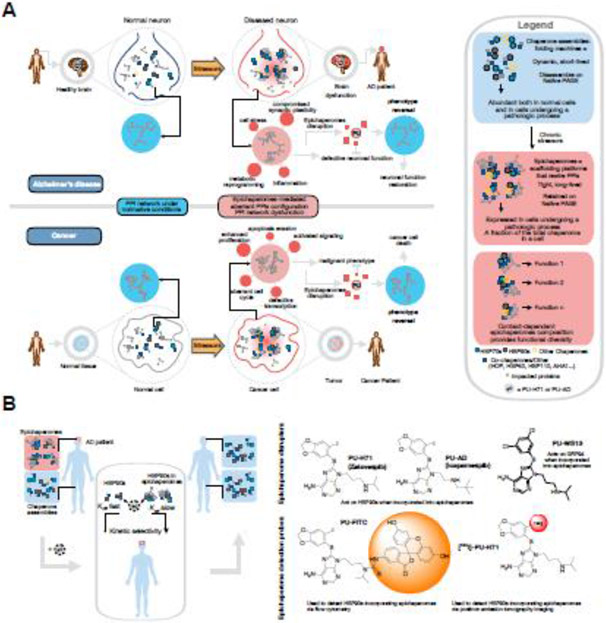
Figure 2. Key Figure. Treating patients by disrupting epichaperomes, the scaffolding structures that enable pathologic restructuring of PPI networks under disease-causing stressor conditions.
A The topological reorganization of proteins impacted by epichaperome formation in cancer and Alzheimer’s disease (AD) is shown schematically. Epichaperome disruptors revert such organization to pre-stressor states, highlighting a causative relationship between epichaperomes and PPI network dysfunctions. In neurodegenerative disorders, epichaperomes result in defects within intrinsic PPI networks involved in brain cell function (e.g., synaptic plasticity, innate immune responses, metabolic programming), and also intercellularly, where they disrupt intrinsic network connectivity within cells and brain circuits. Pharmacologically disrupting epichaperomes may thus be a therapeutic approach to restore PPI networks, and revert brain cells, and in turn brain connectomes, to pre-stressor normative states. In cancer, epichaperomes impact the connectivity of proteins critical to maintaining context-dependent malignant phenotypes. Epichaperome disassembly in cancer is efficacious in the context of PPI network hyperconnectivity, and also as a means to create therapeutic vulnerability to current therapies by controlling the connectivity of PPI networks (see Box 2).
B Designing molecules that selectively target epichaperomes over chaperones is feasible by regulating the kinetics of target engagement {rates of association (kon) and dissociation (koff)}, as exemplified here for the presented PU-type ligands (e.g., epichaperome-disruptors and epichaperome detection probes). Binding kinetics are important for drug efficacy and safety as they may affect pharmacodynamics (i.e., efficacy at the site of action), selectivity (i.e., impact on similar targets) and therapeutic index (i.e., safety profile during administration).
Epichaperomes are oligomeric structures composed of tightly bound chaperones, co-chaperones, and other factors [48]. Not to be confused with chaperones, which are abundantly expressed in all cells and across normal and disease conditions, epichaperomes are principally localized to diseased cells and tissues [27,32,48,49]. Functionally, epichaperomes are also distinct from chaperones. They act as scaffolds remodeling PPIs at the proteome-wide level, rather than serving as folders of proteins in protein synthesis and degradation pathways [14,22,27,32,49]. The more epichaperomes are present in cells and tissues, the higher the number of proteins negatively impacted upon, and in turn, the higher the severity of perturbation to the complex network of molecular interactions [48,49].
Specific mechanisms for epichaperome formation and remodeling of PPIs by epichaperomes are not fully understood. PTMs may play an important role to stabilize specific chaperone conformations that facilitate both epichaperome incorporation and alteration of binding with individual proteins or protein complexes [27]. These conformations may become stabilized by PTMs within constituent chaperones [27], interacting proteins, or both. For example, the heat-shock protein 90 (HSP90) paralog, glucose-regulated protein 94 (GRP94), is a chaperone confined to the endoplasmic reticulum where it folds client proteins through transient interactions [27]. Yan et al., 2020 reported that N-glycosylation on N62 promotes a conformational state in GRP94 that favors epichaperome formation and allows for plasma membrane localization and stable interactions with plasma membrane proteins [27]. Through this complexation stabilization, the functions of these plasma membrane proteins are enhanced, and downstream cellular protein pathways are aberrantly remodeled [27].
Epichaperome composition represents a stressor-specific fingerprint [49], meaning that a complement of epichaperome structures, each with distinct composition, forms and affects specific PPIs, and in turn cellular functions. For example, Kishinevsky et al., 2018 discovered that within midbrain dopaminergic neurons HSP90 and HSP60 recruitment into epichaperomes was enhanced by toxic stressors (e.g., rotenone), and that these epichaperome structures aberrantly rewired the connectivity of proteins involved in dopamine synthesis pathways [49]. Genetic stressors, on the other hand, led to significant increase in the recruitment of HSP90, heat-shock cognate 70 (HSC70), HSP70-HSP90 organizing protein (HOP), HSP40, and several other co-chaperones to epichaperomes, to essentially rewire the interaction of numerous protein involved in inflammatory signaling pathways [49]. How specific epichaperome composition impacts individual proteins and protein assemblies remains to be elucidated. The nature and identity of epichaperome component chaperones and co-chaperones and their organization could possibly dictate function (i.e., confers an epichaperome structure with specificity for individual protein interactors), but this remains to be demonstrated.
Systems level investigations of proteins impacted by epichaperomes indicate epichaperome formation alters the connectivity of individual proteins critical in maintaining context-dependent disease phenotypes ([6,22,26,32,49] and see detailed below). Functional reversal of phenotypes to pre-stressor states by epichaperome disruptors support epichaperomes as key orchestrators of context-dependent PPI network dysfunctions ([6,22,32,33,49-51] and see below).
Indirectly supporting epichaperomes as key components of PPI networks that form under pathologic stressor conditions are computational methods that analyzed the centrality of chaperones within PPI networks [8,52,53] or examined PPI networks in the context of dementia, diabetes, macular degeneration, and other diseases, searching for points of vulnerability [54-57]. For example, in a landmark paper, the Csermely laboratory examined yeast and human interactomes and found chaperones are inter-modular integrators of PPI networks, which often bridge hubs [22]. Moreover, they found chaperones became more central in the organization of PPI modules in yeast PPI networks under stressor conditions [22]. Even though these investigations were not experimentally equipped to differentiate chaperones from epichaperomes, they highlight a role for these entities as a mechanism of maladaptation under stressors.
Neurodegenerative disorders
In neurodegenerative disorders, epichaperome formation impacts the connectivity of proteins important for neuronal function, such as proteins involved in synaptic plasticity, cell-to-cell communication, protein translation, cell cycle re-entry, axon guidance, metabolic processes and inflammation, leading to network-wide dysfunction and cognitive decline ([14,32,34,51] and Figure 2A). Epichaperomes form in neurodegenerative disorders in cells and tissues exposed to maladaptive stressor conditions that are associated with disease. For example, epichaperomes were detected in postmortem human AD brain but not age-matched NCI subjects [32], in APP duplication-expressing iPSC-derived neurons (i.e., where phenotype is caused by a genetic stressor) but not in wildtype iPSC-derived neurons, PS19 transgenic mouse brains (i.e., where phenotype is caused by mutant tau overexpression, a proteotoxic stressor) but not wildtype littermates [32], mutant PARKIN expressing iPSC derived dopaminergic neurons (i.e., where phenotype is caused by a genetic stressor) but not wildtype iPSC-derived dopaminergic neurons [49], and iPSC derived neuron-astrocyte spheroids exposed to toxic human tau oligomers (i.e., where phenotype is caused by a proteotoxic stressor) [51].
Pharmacologically disrupting epichaperomes in neurodegenerative models resulted in reversal of phenotype to pre-stressor levels [32,49,51]. For example, Inda et al., 2020 found introduction of human tau into N2a cells led to epichaperomes that rewired the connectivity of proteins within synaptic protein networks [32]. Importantly, PU-AD, a small molecule epichaperomes disruptor (see Figure 2B and [32,50]), reverted these pathologic effects on synaptic protein pathways to the pre-tau stressor state [32].
A similar effect of PU-H71, a PU-AD analog, was noted by Rickner et al., 2022 in a 3D neuron-astrocyte coculture model exposed to toxic oligomeric tau as the stressor [51]. Application of oligomeric tau induced a PPI network profile that closely recapitulated late-stage changes in adult neurodegeneration, supported by the transcriptomic profile and measures of tau pathology, neurodegeneration, and reactive astrogliosis [51]. PU-H71 treatment revealed a striking reduction in tau pathology and corresponding reduction of neurodegeneration (e.g., reduction in tau phosphorylation, misfolding, oligomerization and tangle formation) and decreases in neuronal cell death and reactive astrogliosis [51]. Importantly, transcriptomic analyses supported the putative benefit observed for PU-H71, with the expression profile shifting to one observed early in the disease process, prior to the point where the system was overwhelmed by oligomeric tau [51]. For example, the astrocyte population showed a reduction in upregulation of the A1 toxic astrocytic signature and an increase in the upregulation of the A2 protective astrocytic signature [51]. Neurons no longer demonstrated upregulation of AD-associated neuronal signatures [51]. Additionally, PU-H71 reversed the upregulation of neuroinflammatory signatures within glutamatergic excitatory neurons, supporting the proposed mechanism of protection [51].
Analogous restorative effects were found in transgenic mice [32]. In PS19 mice, mutant tau overexpression is one of the major stressors that imbalances synaptic protein networks. Treatment of PS19 mice with the epichaperome disruptor PU-AD resulted in a significant rebalance in the activity of synaptic protein networks to levels observed in wildtype mice [32]. PU-AD treatment restored postsynaptic responsiveness and repaired compromised synaptic plasticity, as interrogated via slice electrophysiology [32]. Functionally, epichaperomes disruption resulted in the reversal of cognitive decline, as measured by the Barnes maze [32]. Studies conducted in both preventive and interventional treatment settings in PS19 mice demonstrated cognitive improvement to baseline levels seen in wildtype mice [32].
Aggregating these findings, pharmacologically dismantling epichaperomes may be a viable and potentially transformative therapeutic approach to restore PPI networks, and revert brain cells, and in turn brain connectomes, to pre-stressor normative states [14].
Cancer
In cancer, epichaperomes formation impacts the connectivity of proteins critical in maintaining context-dependent malignant phenotypes, such as proteins involved in signaling and metabolic pathways, among others ([22,27,58-62] and Figure 2A). Approximately 50%–60% of tumors express variable levels of HSP90-incorporating epichaperomes, independent of tissue origin, tumor subtype, or genetic background, with ~10–15% being high epichaperome expressors [48]. The higher the epichaperome levels, the higher the number of proteins being negatively impacted, translating to a higher number of PPIs being rewired [22,33,48]. Putting this in the context of network centrality discussed above, disassembling epichaperomes is most detrimental to cancer cells with high epichaperome expression, as a critical sector of the PPI network map is affected upon epichaperome disruption. This cellular state of vulnerability to therapy was coined a state of ‘interactome hyperconnectivity’ or ‘PPI network hyperconnectivity’ [33]. Joshi et al, 2021 demonstrated no rebound pathways can be activated in the PPI network hyperconnectivity state, enabling maximal vulnerability to targeting [33]. This cellular state is identifiable and measurable both preclinically and in the clinic via epichaperome positron emission tomography (PET) imaging or flow cytometry ([59,63,64] and see Figure 2B for epichaperome detection agents).
Preclinical and clinical studies support the notion that epichaperome disassembly in cancer is efficacious (i.e., lethal) in the context of PPI network hyperconnectivity (e.g., whereby no bypass pathways are available and therefore none can be deployed) [33,48,59,60,63-65]. As observed in preclinical studies using multiple mouse models of epichaperome-high tumors, irrespective of tumor type, epichaperomes disruption resulted in rebalancing of PPI network activity (e.g., reduction of overactive signaling to physiologic levels), which led to tumor regression and an increase in mouse survival [33,48,60,62,64,65]. In such tumors, Pillarsetty et al., 2019 showed tumor regression with no tumor regrowth upon PU-H71 treatment, even after treatment cessation in mice [64]. In a human clinical trial conducted in metastatic breast cancer patients, majorly triple negative breast cancer, Jhaveri et al., 2020 found a positive significant correlation between baseline epichaperomes levels (determined by epichaperome PET imaging) and time-to-progression on epichaperome therapy [63]. Tumor regression was observed in epichaperome high tumors upon treatment [63]. In refractory acute myeloid leukemia transformed from myeloproliferative neoplasms, a poor prognosis disorder, Sugita et al., 2021 reported complete remission, which was retained beyond 4 years in a patient with high baseline epichaperomes levels, detected in both blasts and granulocytes [59]. Following 16 doses in the span of 3 months, the patient reached normalization of peripheral blood counts, resolved splenomegaly and constitutional symptoms [59]. The epichaperome agent PU-H71 was administered initially twice per week for 2 weeks at 300 mg/m2, a dose reported to maximally engage the target in epichaperome-positive tumors [64]. An immediate decrease in the number of blasts present in peripheral blood and a decrease in total white blood cell count to normal levels was observed, paralleled by decreased epichaperome levels and a rebalance in the activity of PPI networks, as evidenced by decreased activity of signaling pathways [59]. This effect seen in blasts was also observed in acute myeloid leukemia progenitor and stem cell populations [59]. While effective at keeping the disease in check, no negative effects were observed for PU-H71 delivery on the immune system in this study. Notably, Sugita et al., 2021 monitored immune populations during treatment, and suggest a possible role for PU-H71 in decreasing an immune-suppressive microenvironment, thus allowing donor-derived T-lymphocytes to exert immune surveillance [59].
Aggregating these results in cancer, pharmacologic epichaperomes modulation could be used i) as monotherapy in hyperconnected tumors (i.e., tumors with intrinsically high epichaperome levels) and ii) in combination, as means to create therapeutic vulnerability to currently approved therapies by controlling the connectivity of PPI networks (see Box 2). Effects in clinic seen in poor prognosis malignancies provide proof-of-principle that epichaperome inhibition may lead to durable responses. Moreover, this novel approach is safe when applied using precision oncology principles for patient selection and tumor pharmacometrics measurements, the hallmarks of modern precision medicine.
Box 2. Potential network control through epichaperomes.
Since epichaperomes function as scaffolds to rewire PPI networks, it may be possible to develop therapeutic strategies redirecting this intrinsic mechanism towards novel applications controlling PPI networks (i.e. shifting the PPI network from a drug-insensitive to a drug-sensitive state) (Figure II). This question was recently addressed in the context of pancreatic cancer where the dynamic and redundant nature of PPI networks represents a challenge for monotherapy. Demonstrating proof-of-concept, Joshi et al., 2021 showed tumors do not possess an infinite number of redundant pathways for signal rerouting, and a biological limit exists where no further PPls can be physically and functionally actionable within a cell [33]. This implies even cancer cells have a finite number of possible epichaperome-mediated functional PPI permutations in a specific cellular context for any given stressor to phenotype situation. In this regard, Joshi et al., 2021 demonstrated tumors can be pharmacologically forced into a PPI hyperconnectivity cellular state by manipulating epichaperomes via PU-H71 [33].
Joshi et al., 2021 also defined the mechanisms of PPI hyperconnectivity, in that i) is performed by increasing the number of interactions HSP90 establishes with the proteome through recruitment of HSP70 chaperones and co-chaperones, and ii) retains intrinsic protein pathway activity and cellular phenotype by reactivating protein pathways already in existence, and constitutively active, at baseline [33]. Whereas new connections were established at the PPI hyperconnectivity state, the goal of these PPIs was to retain overall signaling through constitutively active pathways.
Applying these mechanistic findings, Joshi et al., 2021 demonstrated pancreatic tumors engineered into the hyperconnectivity state became highly vulnerable to conventional FDA-approved inhibitors of intrinsic protein pathways, such as of the RAF/MEK/ERK pathway (e.g., trametinib) [33]. In contrast, standard RAF/MEK/ERK pathway inhibitors are not feasible for treatment of these tumors because of compensatory increases in alternate signaling activity, including the AKT/mTOR pathway [73].
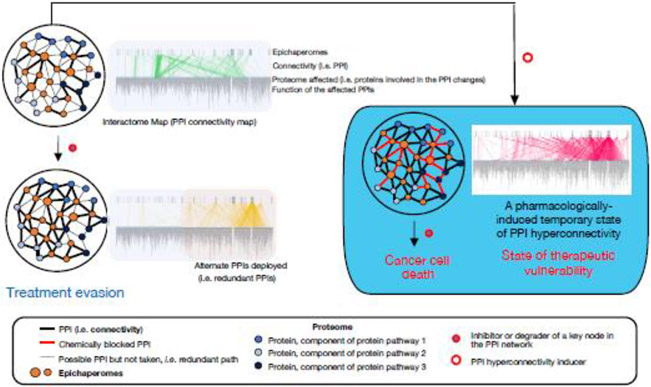
Figure II. Controlling protein-protein interaction networks in cancer via epichaperomes.
PPI network plasticity in cancer, which arises from highly redundant signaling pathways, poses a challenge to therapy and accounts for treatment resistance. Pharmacologically rewiring PPI networks into a hyperconnected state can be a modality for inducing therapeutic vulnerability. Existing treatments may be more effective than previously observed if PPI network hyperconnectivity, whereby cancerous cells are forced into a state devoid of redundancy, is created prior to drug treatment. Several inhibitors and degraders of PPI network nodes are already in clinical use or in development, making this approach both timely and potentially transformative. This treatment paradigm is not a combination method per se because the hyperconnectivity inducer is used once to prime the tumor, followed by current therapy. This approach also differs from synthetic lethality where simultaneous perturbation of two or more genes is required for cell death. Figure adapted from ref. [33].
3. Epichaperome targeting drugs and diagnostics
The chaperone HSP90 is a central epichaperome component in several disorders, including AD, PD, and cancer [48,49]. Therefore, HSP90 provides a common point of vulnerability for the disruption of epichaperomes, and in turn, for reversal of aberrant PPI networks which may be exploited for therapeutic use in several diseases (Figure 2B). Epichaperome therapeutics that bind a chaperone but kinetically select for epichaperome-incorporated chaperone over other chaperone pools, such as PU-H71 (zelavespib) (binds HSP90-incorporating epichaperomes) [59], PU-AD (icapamespib) (binds HSP90-incorporating epichaperomes) [32,50], and PU-WS13 (binds GRP94-incorporating epichaperomes) [27], were tested and demonstrated therapeutic benefit in several models of cancer, neurodegenerative disorders, infection, and inflammation [22,32,33,50,64-67]. Notably, zelavespib and icapamespib have translated to clinic in cancer and AD [50,59,63]. These epichaperome disruptors stabilize epichaperomes upon binding, and this ‘trapped’ drug-epichaperome intermediate is then followed by disassembly of the epichaperome, and in turn, de-coupling of aberrant PPI network connections [27,32,33,50,59,64]. We provide several examples throughout the text (see Section 2) to evidence PPI network restoration to a pre-stressor state by epichaperome disruptors.
Labeled derivatives of PU-H71 and PU-AD can be used to detect and quantify epichaperomes in vivo [64,68,69] (Figure 2B). Furthermore, these molecules have potential as diagnostic tools, as epichaperome levels positively correlate with cancer sensitivity to epichaperome-disrupting compounds [48,50,63]. For example, a fluorescein isothiocyanate labeled PU-H71, PU-FITC, was used in combination with flow cytometry for single cell measurements of epichaperomes in the context of hematologic malignancies [59]. Pillarsetty et al., 2019 used a radiolabeled PU-H71, [124I]-PU-H71, combined with PET imaging to demonstrate detection of epichaperome-positive tumors is feasible in clinical settings [64]. This agent was used to monitor target engagement by PU-H71 in patient tumors at single-lesion resolution in real time, and demonstrated quantitative evaluation at the level of individual tumors can be used to optimize dose and schedule selection for epichaperome-based therapy [64].
In sum, exploiting epichaperome vulnerability provides a precision medicine approach for targeting stressor-induced dysfunctions within PPI networks with a biomarker for use in patient selection (i.e., intrinsic epichaperome levels) and diagnostics assays (i.e., PU-PET and PU-FITC) for both patient selection and precise measurements of target engagement that are readily available for use in clinic. Patient selection based on biomarker identification, namely epichaperome presence detection and expression level quantitation [48], is a primary consideration for epichaperome therapy. Treating a patient at a dose and schedule regimen based on target engagement, rather that maximal tolerated dose, is yet another consideration to factor into individualized precision medicine treatment [59,63,64].
Concluding remarks and future perspectives
In this Opinion, we highlight a causal link between epichaperomes formation and context-specific stressor-induced PPI network dysfunctions in a variety of treatment refractory and/or currently incurable diseases, including cancer, AD, and PD. Epichaperomes pathologically rewire PPIs through scaffolding functions in these diverse diseases. This maladaptation to stressors causes thousands of proteins to aberrantly organize inside cells, negatively affecting cellular phenotypes. We posit epichaperomes serve as cornerstones of malfunctioning PPI networks in disease. Accordingly, we propose the development of drug candidates to disrupt epichaperomes may have an important role in the treatment of a broad spectrum of diseases. The target is the aberrant complement of PPIs within a specific disease context, i.e., a cell-specific interactome dysfunction, which may be corrected through epichaperomes disruption. Thus, epichaperome disruptors represent a unique approach to ‘PPI network-based therapy’ acting specifically on pathologic epichaperome scaffolds.
To date, only networks modulated by HSP90-containing epichaperomes are targetable, highlighting the need for discovery of alternate modalities (e.g., other key components of epichaperomes) for target engagement. Expanding mechanistic insights into context-dependent epichaperomes composition, structure, and function will be necessary to move this field forward in order to develop and broaden the toolkit and applicability. Future investigations into potential epichaperomes presence, abundance, composition and function in other diseases beyond cancer, AD, and PD as well as in chronic disorders (e.g., pain, addiction, and inflammation, among others) is warranted (see Outstanding Questions).
Outstanding Questions Box.
Epichaperomes composition is context dependent. Therefore, is HSP90 an ubiquitous component of epichaperomes? If not, what other chaperones are at the core of epichaperomes formation? What defines the composition of individual epichaperomes?
Specific epichaperome composition impacts distinct proteins. Is the nature and identity of component proteins and their organization dictating epichaperome function (i.e., does it confer epichaperome structure with specificity for individual protein interactors)? Are epichaperomes component proteins enriched in mediators of PPIs acting as hubs in PPI networks?
Current epichaperomes disruptors act via binding to HSP90. Are epichaperome components other than HSP90 useful targets for pharmacologic modulation of PPI network dysfunctions?
PTMs play a role in epichaperome formation. What is the complement of PTMs associated with the switch of chaperones into epichaperomes? What mechanisms other than PTMs play a regulatory role in the switch of chaperones into epichaperomes?
Epichaperomes formation impacts the connectivity of proteins critical in maintaining context-dependent pathologic phenotypes. Are there any structural or conformational commonalities among epichaperome interactors?
Biological networks have evolutionarily inbuilt redundancies. What are the redundancies, if any, associated with targeting PPI network dysfunctions via epichaperomes?
To date, epichaperomes have been studied mainly in the context of cancers and neurodegenerative disorders. Is epichaperome formation a mechanism of PPI network dysfunction that characterizes other disease states and disorders? If yes, is epichaperome disruption a viable therapeutic approach in such settings? Can targeting epichaperomes in these states be employed for precision medicine?
Targeting networks is a new frontier. Application of epichaperome therapy in clinic thus should be combined with principles of precision medicine, as developed preclinically, but this remains to be rigorously implemented. Dialogue between principals in systems biology, discovery biomedical science, and clinical practice needs to be streamlined in order to translate discoveries in modulating aberrant PPI networks to the clinic. This pharmacologic approach targeting stressor-induced PPI network dysfunctions to rebalance cellular networks may be a novel and effective therapeutic strategy, and may complement current drugs, whether monotherapy, polypharmacology, or multitarget, in a variety of complex diseases with limited treatment options and/or efficacy.
Highlights.
Most diseases have a complex etiology with internal and external stressors negatively impacting, directly or indirectly, specific cells, tissues, organs, and ultimately organisms.
The disease network, a map of how individual or a combination of stressors alter cellular PPIs and in turn, perturb the system, is a target for therapeutic intervention.
A paradigm shift in developing therapies is needed that requires movement from the unitary protein approach to a goal of targeting context-specific PPI network dysfunctions for disease control.
Disassembling epichaperomes, scaffolding platforms that enable pathologic rewiring of PPI networks, may restore PPI networks to a pre-stressor state, and has emerged as an approach to target context-specific disease PPI networks in neurodegenerative disorders and cancers.
Targeting stressor-induced PPI network dysfunctions via epichaperomes is amenable to a precision medicine approach.
Acknowledgements:
This work was supported by the NIH (R01 CA172546, P01 CA186866, R56 AG061869, R01 AG067598, P01 AG014449, P01 AG017617, R01 AG074004, R56 AG072599, RF1 AG071805, and P30 CA08748), The Breast Cancer Fund, Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Experimental Therapeutics Center of MSKCC. S.S. acknowledges support from the Bright Focus Foundation. The authors acknowledge Dr. Tai Wang for his contribution to figure design.
Glossary
- Adaptive and maladaptive response to stressors
adaptive responses of biological systems to stressors enhance resilience and facilitate coping mechanisms against future stressors. In contrast, maladaptive responses represent a failure to cope with stressors and/or return to a normative state leading to vulnerability to stressor-associated pathology. In this context, successful coping with stressors implies a stress response is activated quickly when needed but also efficiently terminated when the stressor condition is removed. However, under conditions where stress occurs repeatedly, persists chronically, or is of such magnitude that a biological system fails to re-establish homeostasis, an overload state results that often leads to maladaptive responses and pathological consequences.
- Bottlenecks
nodes with high betweenness centrality (i.e., network nodes that have many "shortest paths" going through them, analogous to major bridges on a highway map). In a PPI network, bottlenecks are proteins that enable the flow of information within the PPI network.
- Chaperone
protein that through one-on-one dynamic interactions assists in the conformational folding or unfolding as well as the assembly or disassembly of other macromolecular structures so they can work correctly.
- Connectome
an interaction network, from intercellular to regional to circuit level, that drives specific tissue, organ and/or organism functions.
- Epichaperomes
malfunctioning long-lived assemblies of chaperones and other factors that act as aberrant scaffolding structures leading to misorganized proteins including loss of normal protein-protein interactions and gain of abnormal protein-protein interactions within a vulnerable cell, exacerbating disease onset and progression.
- Epichaperome disruptors
pharmacologic agents (i.e., small molecules or other entities) that interfere with epichaperome function by any of the possible mechanisms: disassembly and/or trapping of epichaperome scaffolding platforms, alteration in composition, and/or other putative mechanisms.
- Hubs
the most connected nodes within the network that are requisite to sustain network connectivity. In a PPI network, hubs are proteins with many interaction partners.
- Interactome
the complement of interactions between molecules in a specific biological context (e.g., cell, organism). For protein-protein interactions, the term refers to physical interactions. In the context of full protein interactomes, it refers to all possible PPIs across a species or over all species.
- Precision medicine
a medical model designed to optimize efficiency or therapeutic benefit by selecting particular groups of patients based on molecular profiling (as opposed to, for example, tumor type) and by proposing the customization of treatment based on target engagement principles (as opposed to, for example, maximal tolerated dose).
- Protein-protein interaction
the interaction established between two proteins.
- Protein-protein interaction network
the complement of protein-protein interactions in the cell.
- Protein-protein interaction network dysfunction
the complement of aberrant protein-protein interactions in the cell that manifest in a pathologic phenotype.
- Stressor
internal (e.g., genetic mutations, epigenetic alterations, proteotoxic stress) and external (e.g., chemical/environmental exposure, lifestyle, socioeconomic status, psychosocial factors, healthcare access disparities, gut-microbiome diversity, among others) perturbations that cause responses by the organism.
Footnotes
Conflict of interests: Memorial Sloan Kettering Cancer Center holds the intellectual rights to the epichaperome portfolio. G.C. and L.N. share partial ownership and are members of the board of directors at Samus Therapeutics Inc, which has licensed this portfolio. G.C. and S.S. are inventors on the licensed intellectual property. Other disclosers for L.N. are listed at https://www.mskcc.org/cancer-care/doctors/larry-norton. S.D.G. declares no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greene JA and Loscalzo J (2017) Putting the Patient Back Together - Social Medicine, Network Medicine, and the Limits of Reductionism. N. Engl. J. Med 377, 2493–2499. [DOI] [PubMed] [Google Scholar]
- 2.Picard M (2022) Why Do We Care More About Disease than Health? Phenomics 2, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barabasi AL et al. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genet 12, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavi O (2015) Redundancy: a critical obstacle to improving cancer therapy. Cancer Res. 75, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novais A et al. (2017) How age, sex and genotype shape the stress response. Neurobiol Stress 6, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsberg SD et al. (2021) The penalty of stress - Epichaperomes negatively reshaping the brain in neurodegenerative disorders. J. Neurochem 159, 958–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conte F et al. (2020) A paradigm shift in medicine: A comprehensive review of network-based approaches. Biochim Biophys Acta Gene Regul Mech 1863, 194416. [DOI] [PubMed] [Google Scholar]
- 8.Csermely P et al. (2013) Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol Ther 138, 333–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny JC and Collins FS (2021) Precision medicine in 2030-seven ways to transform healthcare. Cell 184, 1415–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutrouli M et al. (2020) A Guide to Conquer the Biological Network Era Using Graph Theory. Front Bioeng Biotechnol 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidal M et al. (2011) Interactome networks and human disease. Cell 144, 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luck K et al. (2020) A reference map of the human binary protein interactome. Nature 580, 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nussinov R et al. (2019) Protein ensembles link genotype to phenotype. PLoS Comput. Biol 15, e1006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsberg SD et al. (2021) The penalty of stress - Epichaperomes negatively reshaping the brain in neurodegenerative disorders. J. Neurochem 159, 958–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahni N et al. (2015) Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai B et al. (2020) Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron 105, 975–991 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson ECB et al. (2022) Large-scale deep multi-layer analysis of Alzheimer's disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat. Neurosci 25, 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomkins JE and Manzoni C (2021) Advances in protein-protein interaction network analysis for Parkinson's disease. Neurobiol. Dis 155, 105395. [DOI] [PubMed] [Google Scholar]
- 19.Zuo Y et al. (2021) Unveiling the Pathogenesis of Psychiatric Disorders Using Network Models. Genes (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee LY et al. (2021) Network medicine in Cardiovascular Research. Cardiovasc. Res 117, 2186–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z et al. (2020) A large accessory protein interactome is rewired across environments. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi S et al. (2018) Adapting to stress - chaperome networks in cancer. Nat. Rev. Cancer 18, 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouhaddou M et al. (2019) Mapping the protein-protein and genetic interactions of cancer to guide precision medicine. Curr Opin Genet Dev 54, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper JW and Bennett EJ (2016) Proteome complexity and the forces that drive proteome imbalance. Nature 537, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghadie MA et al. (2018) Interactome evolution: insights from genome-wide analyses of protein-protein interactions. Curr. Opin. Struct. Biol 50, 42–48. [DOI] [PubMed] [Google Scholar]
- 26.Ginsberg SD et al. (2022) Disease-specific interactome alterations via epichaperomics: the case for Alzheimer's disease. FEBS J. 289, 2047–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan P et al. (2020) Molecular Stressors Engender Protein Connectivity Dysfunction through Aberrant N-Glycosylation of a Chaperone. Cell Rep. 31, 107840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennington KL et al. (2018) The dynamic and stress-adaptive signaling hub of 14-3-3: emerging mechanisms of regulation and context-dependent protein-protein interactions. Oncogene 37, 5587–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambourg L and Thierry-Mieg N (2010) New insights into protein-protein interaction data lead to increased estimates of the S. cerevisiae interactome size. BMC Bioinformatics 11, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French-Pacheco L et al. (2022) Intrinsically disordered signaling proteins: Essential hub players in the control of stress responses in Saccharomyces cerevisiae. PLoS One 17, e0265422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z et al. (2017) The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat Commun 8, 14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inda MC et al. (2020) The epichaperome is a mediator of toxic hippocampal stress and leads to protein connectivity-based dysfunction. Nat. Commun 11, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi S et al. (2021) Pharmacologically controlling protein-protein interactions through epichaperomes for therapeutic vulnerability in cancer. Commun. Biol 4, 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginsberg SD et al. (2021) Disease-specific interactome alterations via epichaperomics: the case for Alzheimer’s disease. FEBS J. 289, 2047–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu J et al. (2021) Network-based protein-protein interaction prediction method maps perturbations of cancer interactome. PLoS Genet. 17, e1009869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeger-Lotem E and Sharan R (2015) Human protein interaction networks across tissues and diseases. Front Genet 6, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalili M et al. (2018) Unveiling network-based functional features through integration of gene expression into protein networks. Biochim Biophys Acta Mol Basis Dis 1864, 2349–2359. [DOI] [PubMed] [Google Scholar]
- 38.Karimi MR et al. (2022) Prospects and challenges of cancer systems medicine: from genes to disease networks. Brief. Bioinform 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manzoni C et al. (2020) Network Analysis for Complex Neurodegenerative Diseases. Current Genetic Medicine Reports 8, 17–25. [Google Scholar]
- 40.Popescu VB et al. (2022) Network controllability solutions for computational drug repurposing using genetic algorithms. Sci. Rep 12, 1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X et al. (2020) Computational methods for identifying the critical nodes in biological networks. Brief. Bioinform 21, 486–497. [DOI] [PubMed] [Google Scholar]
- 42.Jalili M et al. (2015) CentiServer: A Comprehensive Resource, Web-Based Application and R Package for Centrality Analysis. PLoS One 10, e0143111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viacava Follis A (2021) Centrality of drug targets in protein networks. BMC Bioinformatics 22, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haigis KM et al. (2019) Tissue-specificity in cancer: The rule, not the exception. Science 363, 1150–1151. [DOI] [PubMed] [Google Scholar]
- 45.Hekselman I and Yeger-Lotem E (2020) Mechanisms of tissue and cell-type specificity in heritable traits and diseases. Nat. Rev. Genet 21, 137–150. [DOI] [PubMed] [Google Scholar]
- 46.Huttlin EL et al. (2021) Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040 e3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahni N et al. (2013) Edgotype: a fundamental link between genotype and phenotype. Curr Opin Genet Dev 23, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodina A et al. (2016) The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 538, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishinevsky S et al. (2018) HSP90-incorporating chaperome networks as biosensor for disease-related pathways in patient-specific midbrain dopamine neurons. Nat. Commun 9, 4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolaender A et al. (2021) Chemical tools for epichaperome-mediated interactome dysfunctions of the central nervous system. Nat. Commun 12, 4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rickner HD et al. (2022) Single cell transcriptomic profiling of tauopathy in a novel 3D neuron-astrocyte coculture model. bioRxiv, 2022.2005.2003.490513. [Google Scholar]
- 52.Echeverria PC et al. (2011) An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One 6, e26044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizzolo K and Houry WA (2019) Multiple functionalities of molecular chaperones revealed through systematic mapping of their interaction networks. J. Biol. Chem 294, 2142–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hadizadeh Esfahani A et al. (2018) A systematic atlas of chaperome deregulation topologies across the human cancer landscape. PLoS Comput. Biol 14, e1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lalwani AK et al. (2022) Network Theoretical Approach to Explore Factors Affecting Signal Propagation and Stability in Dementia's Protein-Protein Interaction Network. Biomolecules 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soofi A et al. (2020) Centrality Analysis of Protein-Protein Interaction Networks and Molecular Docking Prioritize Potential Drug-Targets in Type 1 Diabetes. Iran J Pharm Res 19, 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xing C et al. (2021) Hsp90-associated DNA replication checkpoint protein and proteasome-subunit components are involved in the age-related macular degeneration. Chin. Med. J. (Engl.) 134, 2322–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moulick K et al. (2011) Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat. Chem. Biol 7, 818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugita M et al. (2021) Targeting the Epichaperome as an Effective Precision Medicine Approach in a Novel PML-SYK Fusion Acute Myeloid Leukemia. . NPJ Precis Oncol 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zong H et al. (2015) A Hyperactive Signalosome in Acute Myeloid Leukemia Drives Addiction to a Tumor-Specific Hsp90 Species. Cell Rep. 13, 2159–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calvo-Vidal MN et al. (2021) Oncogenic HSP90 Facilitates Metabolic Alterations in Aggressive B-cell Lymphomas. Cancer Res. 81, 5202–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nayar U et al. (2013) Targeting the Hsp90-associated viral oncoproteome in gammaherpesvirus-associated malignancies. Blood 122, 2837–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jhaveri KL et al. (2020) Measuring Tumor Epichaperome Expression Using [(124)I] PU-H71 Positron Emission Tomography as a Biomarker of Response for PU-H71 Plus Nab-Paclitaxel in HER2-Negative Metastatic Breast Cancer. JCO Precis Oncol 4, 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pillarsetty N et al. (2019) Paradigms for Precision Medicine in Epichaperome Cancer Therapy. Cancer Cell 36, 559–573.e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kourtis N et al. (2018) Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat. Med 24, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouchard A et al. (2021) The GRP94 Inhibitor PU-WS13 Decreases M2-like Macrophages in Murine TNBC Tumors: A Pharmaco-Imaging Study with (99m)Tc-Tilmanocept SPECT. Cells 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sumitomo T et al. (2021) GP96 Drives Exacerbation of Secondary Bacterial Pneumonia following Influenza A Virus Infection. mBio 12, e0326920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merugu S et al. (2020) Chemical probes and methods for single-cell detection and quantification of epichaperomes in hematologic malignancies. Methods Enzymol. 639, 289–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma S et al. (2022) Synthesis of (124)I-labeled epichaperome probes and assessment in visualizing pathologic protein-protein interaction networks in tumor bearing mice. STAR Protoc 3, 101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y-Y et al. (2011) Controllability of complex networks. Nature 473, 167–173. [DOI] [PubMed] [Google Scholar]
- 71.Wuchty S et al. (2017) Links between critical proteins drive the controllability of protein interaction networks. Proteomics 17, e1700056. [DOI] [PubMed] [Google Scholar]
- 72.Kanhaiya K et al. (2017) Controlling Directed Protein Interaction Networks in Cancer. Sci. Rep 7, 10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crawford HC et al. (2019) Signaling Networks That Control Cellular Plasticity in Pancreatic Tumorigenesis, Progression, and Metastasis. Gastroenterology 156, 2073–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]