Abstract
We have used signature-tagged mutagenesis to identify mutants of the host-specific Salmonella enterica serotype Dublin which were avirulent in calves and/or BALB/c mice. A mutant with a transposon insertion in the sseD gene of Salmonella pathogenicity island 2 (SPI-2), which encodes a putative secreted effector protein, was identified. This mutant was recovered from the bovine host but not from the murine host following infection with a pool of serotype Dublin mutants. However, a pure inoculum of the sseD mutant was subsequently shown to be attenuated in calves following infection either by the intravenous route or by the oral route. The sseD mutant was fully invasive for bovine intestinal mucosa but was subsequently unable to proliferate to the same numbers as the parental strain in vivo. Both the sseD mutant and a second SPI-2 mutant, with a transposon insertion in the ssaT gene, induced significantly weaker secretory and inflammatory responses in bovine ligated ileal loops than did the parental strain. These results demonstrate that SPI-2 is required by serotype Dublin for the induction of both systemic and enteric salmonellosis in calves.
Within the species Salmonella enterica there are more than 2,000 different serotypes, which include bacteria of tremendous medical and veterinary importance. The pathology of Salmonella infections can vary from mild enteritis to severe systemic salmonellosis and is largely dependent on the particular combination of serotype and host species. The ubiquitous Salmonella enterica serotype Typhimurium induces a systemic, typhoid-like disease in mice (34). However, serotype Typhimurium is considered a broad-host-range serotype, as it is capable of infecting many diverse host species, usually causing self-limiting diarrhea, although more severe and even life-threatening infections can occur in young or immunocompromised animals (reviewed in reference 2). Other serotypes have a more restricted host range and are predominantly associated with severe systemic disease in a single host species. For example, the host-specific serotype Salmonella enterica serotype Dublin is primarily associated with infections of cattle, inducing both systemic and enteric symptoms of salmonellosis (24, 29, 40, 46). Adult cattle which survive serotype Dublin infection often continue to be carriers for significant periods of time, leading to sporadic, repeated outbreaks of disease among herds (24). Occasional outbreaks of serotype Dublin infection in other species, including humans, are considered most likely to have originated from infected cattle (5). Genetically susceptible Itys mice infected experimentally with serotype Dublin develop a severe systemic infection similar to that produced by serotype Typhimurium (19, 28).
Some of the virulence factors that influence Salmonella-induced enteric and systemic disease have been characterized. It is widely accepted that in relatively recent evolutionary history Salmonella has acquired large pieces of DNA by horizontal gene transfer that confer virulence-associated functions upon the host bacteria (26, 32). These genetic loci have been termed pathogenicity islands. Furthermore, it is becoming apparent that these pathogenicity islands are able to influence different stages of pathogenesis. For example, Salmonella pathogenicity island 1 (SPI-1), which encodes the Inv-Spa type III secretion system (TTSS-1), is involved in both invasion of intestinal epithelial cells (reviewed in reference 15) and the induction of fluid secretion and inflammatory responses in bovine ligated ileal loops (17, 50). SPI-5 encodes several proteins, including the TTSS-1-dependent secreted effector protein SopB, which also influence the induction of intestinal inflammation and fluid secretion (17, 53). Disruption of the expression of SPI-1 or SPI-5 genes attenuates enteropathogenesis but not systemic pathogenesis (1, 3, 16, 23, 36, 53). However, other genetic loci have been shown to primarily influence systemic disease. The slyA gene influences the systemic pathogenesis of Salmonella in mice but has no significant effect upon the induction of enteropathogenic responses in bovine ileal loops (27, 52). The precise role of slyA remains unclear, but this gene has been implicated in gene regulation (35). Similarly, SPI-2, which encodes a second type III secretion system (TTSS-2), has been shown to influence systemic virulence in mice but appears not to be involved in serotype Typhimurium-induced enteritis in calves (43) or in the induction of enteropathogenic responses to serotype Typhimurium in rabbit ileal loops (13). Again, the precise mechanism of action of the SPI-2 effectors is unclear, and it has been proposed that TTSS-2 influences both net intracellular growth (8, 22) and survival within macrophages (33, 45).
The recent advent of large-scale in vivo screening techniques such as signature-tagged mutagenesis (STM) has enabled identification of many bacterial genes which are expressed in vivo and potentially involved in pathogenesis (reviewed in references 7, 37, and 38). STM involves the generation of large numbers of individually “tagged” transposon insertion mutants which are then screened in pools for loss of virulence using an appropriate animal model. Attenuated mutants are identified by the loss of their DNA tag from the pool of DNA isolated from bacteria recovered postinfection. Both classical virulence genes and essential housekeeping genes can be identified by STM. Thus, STM combines the potential of random insertion mutagenesis to identify novel candidate genes with the benefits of negative selection in vivo while dramatically reducing the number of experimental animals required for screening mutants. Furthermore, it is possible to modulate the screening procedure to identify mutants with specific defects by manipulating parameters such as the choice of host species or the route of inoculation (7). We are screening an STM mutant bank of serotype Dublin in two different host species. We have used calves as the natural host species for serotype Dublin and genetically susceptible Itys mice as an alternative host. This approach enables identification and comparison of genes essential for serotype Dublin virulence in two different host species. In this report, we describe the identification of serotype Dublin mutants with altered virulence phenotypes and further characterization of the mutants by their ability to cause enteric and systemic forms of salmonellosis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Serotype Dublin strain SD3246 was originally isolated from a case of Salmonella-induced abortion in cattle and is highly virulent for calves, causing severe systemic and enteric disease with high mortality (18, 49). A spontaneously occurring nalidixic acid-resistant mutant, SD3246 Nalr, was used in this work. Escherichia coli strain DH5α was commercially obtained (GIBCO/BRL). The E. coli strains S17.1 λpir and CC118 and the mini-Tn5 Km2 signature-tagged transposons were kindly provided by David Holden (Imperial College, London, United Kingdom). The plasmid pBluescript KS(+) was commercially obtained (Stratagene). Bacterial strains were stored in Luria-Bertani (LB) medium containing 14% glycerol at −70°C and grown routinely at 37°C in LB broth, or on LB agar, containing the appropriate antibiotics.
Experimental animals.
All animal experiments were conducted according to the requirements of the Animal Scientific Procedures Act (1986). BALB/c female mice, aged 8 to 10 weeks, which had been bred in the animal facilities of IAH (Compton, United Kingdom) were used. Groups of mice were housed in separate cages with free access to dried food pellets and water. Following inoculation, mice were monitored for signs of disease at least twice daily. These included a hunched posture, staring coat, and unwillingness to open eyes or move around. Mice which displayed predefined symptoms approaching moderate severity were judged to have reached their end point and were humanely killed.
Twenty-eight-day-old Friesian calves were housed in single cubicles in a disease-secure animal unit and fed on a diet of powdered milk supplemented with unmedicated calf weaner pellets. Calves were not fed for 18 h before oral inoculation or surgical procedures, but all calves had free access to water. None of the calves excreted Salmonella prior to infection, as assessed by enrichment of fecal samples in Rappaport broth (at 37°C for 18 h) and Selenite brilliant green broth (at 42°C for 18 h) followed by incubation of the enrichment cultures on modified brilliant green agar (Oxoid) containing 120 mg of sulfadiazine/liter. Following inoculation, calves were monitored for signs of disease at least twice daily. These included an increase in rectal temperature, dullness, dry mucous membranes, anorexia, and diarrhea. Calves which were anorexic and dehydrated, with persistent diarrhea leading to a cumulative scour score of 20 or greater (49), or which were reluctant to stand unaided were judged to have reached their end point and were humanely killed.
Generation and screening of the transposon mutant bank.
A bank of approximately 5,000 signature-tagged mutants of serotype Dublin SD3246 Nalr was generated and maintained essentially as described previously for serotype Typhimurium (20). “Input pool” DNA was prepared from pools of 96 mutants as described previously (20). For preparation of inocula, 96 individual mutants (stored in LB medium containing 14% glycerol, in a 96-well plate, at −70°C) were subcultured into a second 96-well plate containing 150 μl of LB broth plus antibiotics (20 μg of nalidixic acid/ml and 50 μg of kanamycin/ml) and grown overnight, with gentle shaking, at 37°C. The 96 mutants were then pooled, and the concentration was adjusted with sterile 0.9% NaCl. Mice were infected by the intravenous route (tail vein) with approximately 105 CFU in a total volume of 100 μl. Calves were infected by the intravenous route (jugular vein) with approximately 5 × 106 CFU in a total volume of 1 ml. After 3 days the animals were humanely killed, bacteria were recovered from the spleens, and “output pool” DNA was prepared and analyzed as described previously (20).
Mapping of transposon insertion sites and sequence analysis.
Procedures were essentially those described by Sambrook et al. (39). DNA flanking the site of transposon insertion was cloned by ligation of EcoRI- or EagI-restricted genomic DNA into either EcoRI- or EagI-restricted cloning vector pBluescript KS(+), transformation into E. coli strain DH5α, and selection for the kanamycin resistance marker of mini-Tn5 Km2. Plasmid DNA was isolated using a QIAFilter plasmid isolation kit (Qiagen), and the sequence of genomic DNA was obtained using one of two mini-Tn5 Km2-specific primers, P6 (5′-CCTAGGCGGCCAGATCTGAT-3′) or P7 (5′-GCACTTGTGTATAAGAGTCAG-3′). Sequences were obtained commercially from Cambridge BioScience Ltd. (Cambridge, United Kingdom). Sequence homologies were determined using the BLAST 2.0 search algorithm at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST).
Mouse infection studies.
To prepare the inoculum, LB broth containing 20 μg of nalidixic acid/ml was inoculated with several bacterial colonies from a fresh agar plate and incubated at 37°C, with shaking, for approximately 15 h. Mice were infected by the intravenous route with approximately 40 CFU of each bacterial strain suspended in 100 μl of sterile 0.9% NaCl. Mice were monitored as described above. Mice infected with the parental strain, SD3246 Nalr, reached their end point after 4 days, at which point all mice were humanely killed. Bacteria were recovered from spleens and livers and enumerated on LB agar containing appropriate antibiotics as described previously (53).
Calf infection studies.
The inoculum was prepared as described for mouse infection studies. For oral infection, approximately 5 × 108 CFU of each bacterial strain was added to 20 ml of sterile deionized water containing 5% (wt/vol) Mg(SiO3)3, 5% (wt/vol) NaHCO3, and 5% (wt/vol) MgCO3 immediately before administration. This mixture was fed to each calf by using a syringe before the morning feeding. For intravenous infection, approximately 2.5 × 105 CFU of each bacterial strain was suspended in 1 ml of sterile 0.9% NaCl, which was then injected into the jugular vein. Calves were monitored as described above. Calves infected with the parental strain, SD3246 Nalr, reached their end point between 5 and 7 days postinfection, at which point all calves were humanely killed. A postmortem was conducted as described previously (50). Bacteria were recovered from both intestinal and systemic sites and were enumerated on LB agar containing appropriate antibiotics.
In vitro gentamicin protection assays for invasion and persistence.
Int407 epithelial cells were seeded at 5 × 105 cells per well in 24-well cell culture plates in Eagle minimal essential medium (Gibco BRL) supplemented with 10% fetal calf serum and incubated overnight at 37°C in an atmosphere of 5% CO2 and 90% relative humidity. RPMI medium (without phenol red and l-glutamine; Gibco BRL) containing 5% fetal calf serum, 2 mM l-glutamine, and 18 mM HEPES buffer was prepared and prewarmed to 37°C. Immediately prior to infection, the cells were washed once with RPMI medium and 900 μl of medium was added to each well. The density of mid-log-phase bacterial cultures was adjusted to 5 × 106 CFU/ml by addition of RPMI medium, and 100 μl of diluted culture was added to each well (infection ratio of 1:1). Cells were incubated for 1 h (37°C, 5% CO2, 90% relative humidity) and then washed once with RPMI medium before addition of a further 1 ml of RPMI medium containing 100 μg of gentamicin/ml. Cells were incubated under the same conditions for a further 1 h to kill extracellular bacteria, after which time the medium was replaced with RPMI medium containing 10 μg of gentamicin/ml. Intracellular bacteria were recovered 2 h postinfection to assess invasion and either 8, 12, or 24 h postinfection to assess persistence. Cells were lysed and intracellular bacteria were recovered, after two washings with RPMI medium, by repeated pipetting with 1 ml of phosphate-buffered saline containing 0.1% sodium deoxycholate. Bacteria were enumerated on LB agar containing appropriate antibiotics. At each time point the monolayers were examined microscopically for deterioration and cell lysis was quantified by measurement of lactate dehydrogenase release using the CytoTox 96 nonradioactive assay kit (Promega).
Bovine ligated ileal loop assay for enteropathogenesis.
This assay has been described in detail elsewhere (49). Briefly, calves were anesthetized with pentobarbital, and intestinal loops, 6 cm in length and spaced 1 to 2 cm apart, were constructed in the ileum using braided surgical silk. To prepare the inoculum, bacterial strains were grown overnight, with shaking, at 25°C. The cultures were diluted approximately 1:3 in fresh LB medium and incubated at 37°C, with shaking, for 90 min. The optical density at 600 nm was adjusted by addition of LB broth to give a concentration of approximately 8.5 log10 CFU/ml. A total of 5 ml of this suspension was injected into each loop. The same volume of sterile LB broth was used as a negative control. All bacterial strains and controls were tested in three loops per animal. Polymorphonuclear cells (PMNs) were isolated from 50 ml of blood removed from the calf, labeled with 111In, and reinjected into the jugular vein. Twelve hours after inoculation the anesthetized animal was humanely killed and all loops were exteriorized. Fluid secretion was measured as a ratio of volume of accumulated fluid to loop length. PMN influx was measured as a ratio of 111In activity in test loops to that in control loops.
Bovine ligated ileal loop assay for invasion and persistence.
The assay for invasion has been described in detail elsewhere (51). Briefly, calves were anesthetized with pentobarbital and intestinal loops, 9 cm in length and spaced 1 to 2 cm apart, were constructed in the distal ileum using braided surgical silk. Bacterial strains were prepared as described above except that the optical density was adjusted to approximately 6.5 log10 CFU/ml. A total of 5 ml of this suspension was injected into each loop. The same volume of sterile LB broth was used as a negative control. All bacterial strains and controls were tested in three loops per animal. After infection, loops were left either for 2 h, to assess invasion of bacteria, or for 8 h, to assess net growth within the ileal mucosa. Loops which were used to assess persistence were infected at 0 h. One hour postinfection these loops were exteriorized, 5 ml of GC/Tcm10 solution containing 300 μg of gentamicin/ml was injected, and the loops were returned to the abdominal cavity. At 6 h the remaining loops, which were used to assess invasion, were infected. Again, 1 h postinfection these loops were exteriorized, 5 ml of GC/Tcm10 solution containing 300 μg of gentamicin/ml was injected, and the loops were returned to the abdominal cavity. At 8 h the anesthetized animal was humanely killed and all loops were exteriorized. Biopsy samples were removed and processed as described previously (51).
RESULTS
Identification of serotype Dublin SPI-2 mutants with altered virulence using STM.
A mutant bank consisting of 5,280 signature-tagged serotype Dublin 3246 Nalr mini-Tn5 Km2 transposon mutants was constructed and assembled in 96-well microtiter plates. Pools of 96 mutants were used to infect individual mice and calves via the intravenous route. After 3 days, bacteria were recovered from the spleens of these animals. The presence of an individual signature-tagged transposon within each mutant enabled a comparison of the input pool of mutants with the output pools recovered from mice and calves. Mutants that were absent from the output pools of both mice and calves were detected at a rate of 1 to 2 per plate. This is similar to the rate of detection reported previously for a Salmonella mutant bank screened in mice (20). Mutants that were absent from the output pool of only one host species were identified less frequently.
The site of transposon insertion in serotype Dublin mutants with altered virulence was characterized by DNA sequencing. Two mutants with insertions in SPI-2, which encodes TTSS-2 (33, 42), were identified. One mutant carried the transposon in an open reading frame (ORF) showing 93% nucleotide sequence identity to the sseD gene of serotype Typhimurium. After 3 days, this mutant was recovered from the bovine host but not from the murine host. The sseD gene is predicted to encode a secreted effector protein which shows weak homology to the enteropathogenic E. coli (EPEC) secreted protein EspB (12, 22). Although STM has successfully identified a large number of virulence associated genes in a variety of bacterial species, there have been very few reports of the identification of putative secreted virulence factors using this technique (6, 9). The insertion in sseD may have affected expression of other genes in the operon. However, as nonpolar mutations in sseE, sseF, and sseG have little or no detectable effect upon virulence in mice (22), the attenuation of this mutant is almost certainly caused by disruption of sseD.
A second mutant carried the transposon in the 3′ end of an ORF showing 92% nucleotide sequence identity to the ssaT gene of serotype Typhimurium. This SPI-2 gene is predicted to encode a structural component of TTSS-2 (21). The ssaT mutant was not recovered from either host species. The insertion may have also disrupted expression of ssaU, the final gene in the ssaK-U operon, as these genes are thought to be transcriptionally coupled (21).
Serotype Dublin SPI-2 mutants are attenuated in mice following intravenous infection.
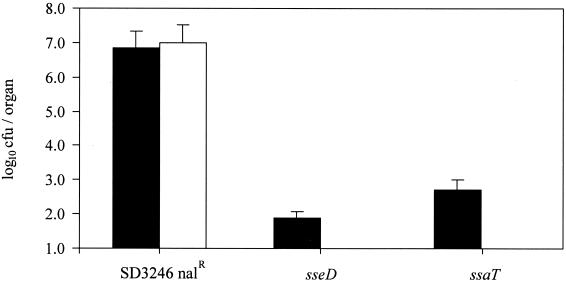
The SPI-2 type III secretion system was originally identified by the isolation of serotype Typhimurium mutants which failed to proliferate and cause systemic infections in mice (33, 42). Accordingly, the virulence of both serotype Dublin SPI-2 mutants in mice was assessed following intravenous inoculation. Animals were infected with approximately 4 × 101 CFU of the sseD mutant, the ssaT mutant, or the parental strain. After 4 days the mice were killed, livers and spleens were recovered, and the bacteria within these organs were enumerated (Fig. 1). Mice infected with the parental strain rapidly developed visible symptoms of systemic salmonellosis and, after 4 days, high numbers of bacteria were recovered from livers and spleens. In contrast, mice infected with either SPI-2 mutant showed no signs of disease. Low numbers of bacteria were detected in the spleens of these animals, and none were isolated from the livers. This result agrees with previous reports showing that serotype Typhimurium SPI-2 mutants are attenuated in mice and also confirms the “mouse-negative” phenotype of both mutants from the original screening procedure.
FIG. 1.
Recovery of bacteria from the spleens (▪) and livers (□) of BALB/c mice 4 days after intravenous infection with SD3246 Nalr or the SPI-2 (sseD and ssaT) mutant strains. Each bar represents the mean recovery of bacteria from three mice and is presented with the standard error of the mean.
A serotype Dublin SPI-2 mutant is attenuated in calves following intravenous infection.
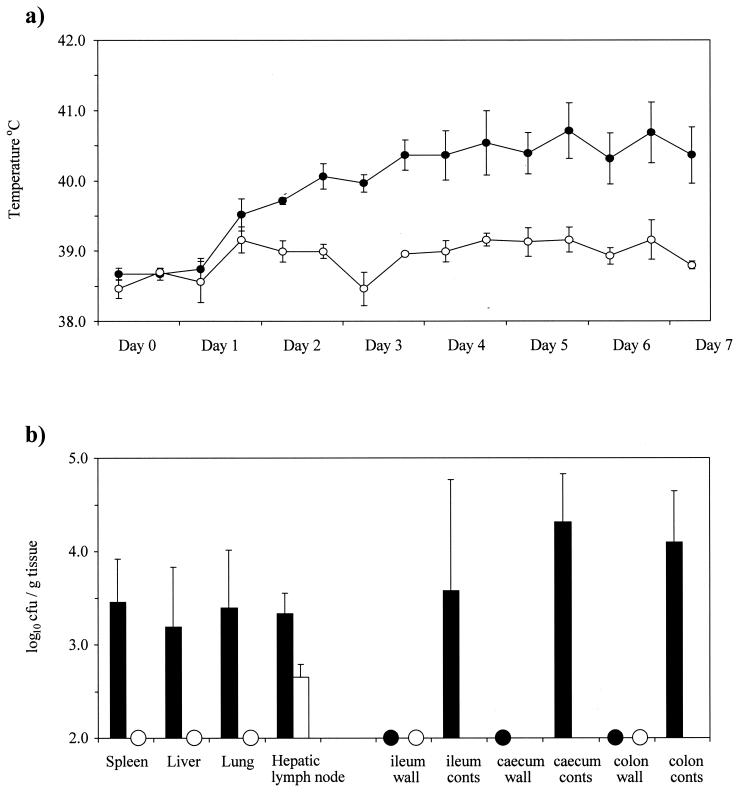
The initial STM screen, which used an inoculum comprised of 96 different insertion mutants, did not identify sseD as a gene essential for systemic virulence of serotype Dublin in calves. This result indicated a potential host-specific difference, as the sseD mutant strain is clearly avirulent in mice following intravenous infection. Consequently, the effect of the insertion in sseD was investigated in calves by using an inoculum consisting solely of the sseD mutant strain. Calves were infected intravenously with 2.5 × 105 CFU of either the parental strain, SD3246 Nalr (four animals), or the sseD mutant (three animals). Calves infected with the parental strain experienced a rise in temperature accompanied by a dull appearance and loss of appetite. These symptoms were sustained throughout the course of the infection (Fig. 2a). Two of the four animals developed diarrhea between days 5 and 7 of the experiment. In contrast, the calves infected with the sseD mutant strain experienced little change in temperature and appeared healthy in all other respects. After 7 days the calves were killed, and the bacteria within intestinal and systemic sites were enumerated (Fig. 2b). The parental strain was recovered at a level of >3.0 log10 CFU/g from the livers, spleens, and lungs of all four animals, whereas the sseD mutant strain was either not recovered or recovered only after enrichment culture of these tissues. Both strains were isolated in more comparable numbers from the hepatic lymph nodes. The parental strain was also present in the intestinal mucosae and contents, while the mutant strain was either not recovered or recovered only after enrichment culture of these enteric sites. These results demonstrate that disruption of the sseD gene of SPI-2 does in fact reduce the virulence of systemic serotype Dublin infections in calves.
FIG. 2.
Intravenous infection of calves with SD3246 Nalr (four calves) or the sseD mutant strain (three calves). (a) Mean rectal temperature responses of calves following infection with either the parental strain (●) or the sseD mutant strain (○) for 7 days. Each datum point is presented with the standard error of the mean. (b) Recovery of bacteria from systemic sites and intestinal sites of calves 7 days after infection with the parental (▪) or the sseD mutant strain (□). Each bar represents the mean recovery of bacteria and is presented with the standard error of the mean. Samples which were positive only after enrichment culture are indicated with circles.
Disruption of SPI-2 genes influences the enteropathogenesis of serotype Dublin in cattle.
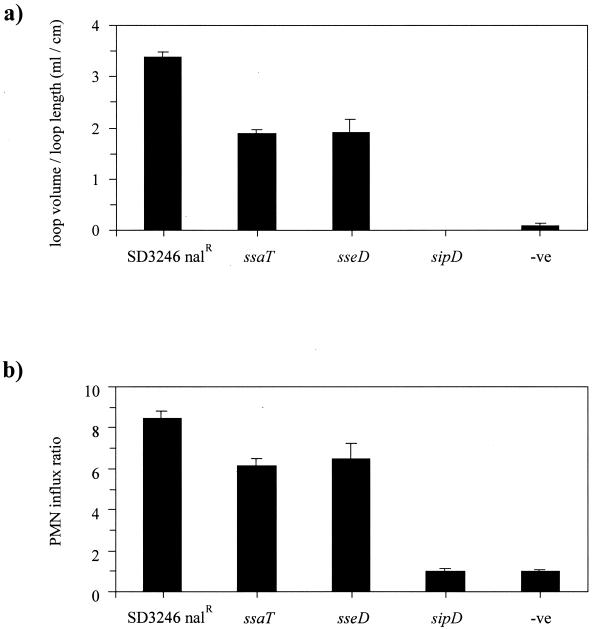
SPI-2 mutant strains of serotype Typhimurium are unable to induce systemic infection in mice (33, 41, 42) but have been shown to induce wild-type enteropathogenic responses in rabbits (13) and to cause diarrhea in calves (43). Taken together, these results suggest that disruption of SPI-2 affects systemic salmonellosis but has no effect on serotype Typhimurium-induced enteritis. Little is known about the interaction between systemic and intestinal forms of disease during serotype Dublin infection of cattle. Accordingly, the effect of a mutation in SPI-2 on the enteropathogenic response induced by SD3246 Nalr was assessed in both bovine ligated ileal loops and orally inoculated calves. The fluid secretion and PMN influx into ligated ileal loops infected with either an SPI-2 mutant or the parental strain were quantified. An SPI-1 mutant carrying a transposon insertion in the sipD gene was also included in the experiment as a control. This mutant had previously been shown to be poorly invasive for Int407 cells (data not shown), thus confirming the SPI-1 mutant phenotype (15, 25). After 12 h, the parental strain had induced potent secretory and inflammatory responses whereas the SPI-1 mutant had produced responses similar to those seen in uninfected loops (Fig. 3). Perhaps surprisingly, the responses stimulated by the sseD and ssaT mutants, which were similar in magnitude, were significantly lower than those induced by the parental strain (P < 0.05). This result was reproduced in two additional calves.
FIG. 3.
Bovine secretory (a) and inflammatory (b) responses in ileal loops 12 h after infection with SD3246 Nalr or the SPI-1 (sipD) and SPI-2 (sseD and ssaT) mutant strains. Negative control loops (-ve) were injected with LB medium. The secretory response is expressed as the volume of fluid within a loop divided by the length of the loop. The PMN influx ratio is expressed as the PMN influx within a test loop divided by the PMN influx in the control loops. Each bar is derived from the mean secretory or inflammatory response from three loops and is presented with the standard error of the mean. The SPI-1 mutant strain was tested in two calves, and all other strains were tested in three calves. The results from one representative calf are presented.
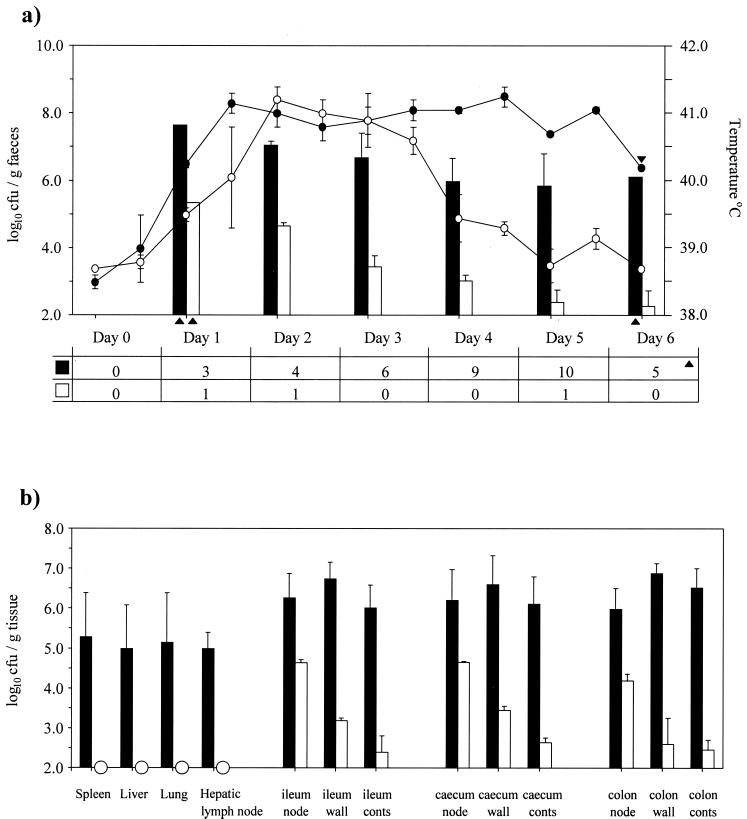
To further investigate the effect of the transposon insertion in sseD upon enteropathogenesis, the virulence of the serotype Dublin sseD mutant following oral inoculation of calves was investigated. Calves were infected with either the parental strain (SD3246 Nalr; two animals) or the sseD mutant strain (two animals). Calves infected with the parental strain rapidly developed severe diarrhea together with a pyrexic response, both of which persisted until the animals reached a predefined clinical end point after 5 or 6 days. The cumulative scour score for these animals was 37 out of a potential maximum of 40. Daily quantification of bacteria in the feces showed that these animals maintained high numbers of bacteria in the intestines despite the rapid transit and dilution of the intestinal contents. In contrast, calves infected with the SPI-2 mutant strain suffered a transient form of infection, as rapid in onset as that induced by the parental strain but not sustained. The cumulative scour score for these animals was only 3. The SPI-2 mutant bacteria, although initially present in the feces in high numbers, were almost cleared from the gut over the course of infection (Fig. 4a). Bacteria were recovered from systemic sites of calves infected with the parental strain in high numbers (>5.0 log10 CFU/g of tissue), whereas the sseD mutant bacteria were detected only after enrichment culture of systemic tissues (Fig. 4b). With the exception of the hepatic lymph node, this reflected the pattern of recovery from calves infected intravenously with these strains (Fig. 2b). The parental strain was also recovered from intestinal sites in consistently higher numbers than the mutant strain. The sseD mutant strain was recovered in the highest numbers from the intestinal lymph nodes.
FIG. 4.
Oral infection of calves with SD3246 Nalr (two calves) or the sseD mutant strain (two calves). (a) Mean rectal temperature responses, mean bacterial recovery per gram of feces, and daily scour scores of calves following infection with either the parental strain (● and ▪) or the sseD mutant strain (○ and □) for 6 days. Each bar or datum point is presented with the standard error of the mean. Each scour score represents the daily sum of scores from two animals. Points where data are taken from one animal only are indicated with triangles. (b) Recovery of bacteria from systemic sites and intestinal sites of calves 5 or 6 days after infection with the parental strain (▪) or the sseD mutant strain (□). Each bar represents the mean recovery of bacteria and is presented with the standard error of the mean. Samples that were positive only after enrichment culture are indicated with circles.
Serotype Dublin SPI-2 mutants are defective in net growth within Int407 epithelial cells in vitro and within intestinal mucosa in vivo.
Most of the SPI-2 genes investigated to date have been shown to influence the net growth of Salmonella within epithelial cells and macrophage-like cell lines (8, 22, 33). The possibility that the observed reduction in enteropathogenicity could be attributed to this previously characterized effect of SPI-2 on net intracellular growth was considered. The invasion and persistence of the parental strain, SD3246 Nalr, and the SPI-2 mutant strains were initially examined in vitro using Int407 epithelial cells. The poorly invasive sipD mutant strain was also included in the experiment as a control. After 2 h, comparable numbers of SD3246 Nalr bacteria and SPI-2 mutant bacteria had invaded Int407 cells, whereas the sipD mutant strain was recovered in much lower numbers (Fig. 5). The net intracellular growth of the parental and SPI-2 mutant strains was similar between the 2- and 8-h time points, and comparable numbers of all three strains were recovered after 8 h. However, between the 8- and 24-h time points the net intracellular growth of SD3246 Nalr was greater than that of the two SPI-2 mutant strains, and after 24 h the numbers of SPI-2 mutant bacteria recovered were significantly lower than the numbers of the parental strain (P < 0.05). The differences in bacterial recovery were not caused by differential damage to the epithelial cell monolayer, as confirmed both by microscopic examination and by measurement of lactate dehydrogenase release (data not shown). Thus, the sseD and ssaT mutants of serotype Dublin had reduced net intracellular growth rates within epithelial cells compared with the parental strain.
FIG. 5.
Recovery of gentamicin-protected bacteria from Int407 epithelial cells over 24 h following infection with SD3246 Nalr (▪), the sipD mutant (□), the sseD mutant (●), or the ssaT mutant (▵). Each datum point represents the mean recovery of bacteria from three wells and is presented with the standard error of the mean.
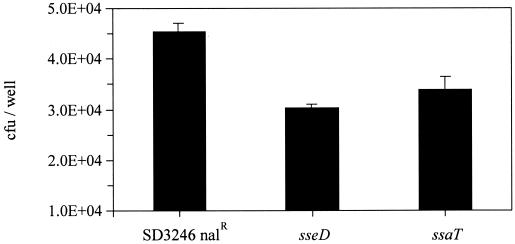
The differences in induction of enteropathogenesis by SD3246 Nalr and the SPI-2 mutant strains were assessed after 12 h. To enable a corresponding evaluation of the effect of the SPI-2 mutations on net intracellular growth and enteropathogenicity, the Int407 epithelial cells were infected with both the mutant and parental strains for 12 h. The results are presented on a linear scale to allow a direct comparison with the enteropathogenicity data to be made. After 2 h equal numbers of all three strains had invaded the cells. However, the difference in numbers of parental strain and mutant bacteria recovered after 12 h (Fig. 6) was directly comparable to the differences in magnitude of the secretory response in the ligated ileal loops (Fig. 3a).
FIG. 6.
Recovery of gentamicin-protected bacteria from Int407 epithelial cells 12 h after infection with SD3246 Nalr or the SPI-2 (sseD and ssaT) mutant strains. Each bar represents the mean recovery of bacteria from six wells and is presented on a linear scale with the standard error of the mean.
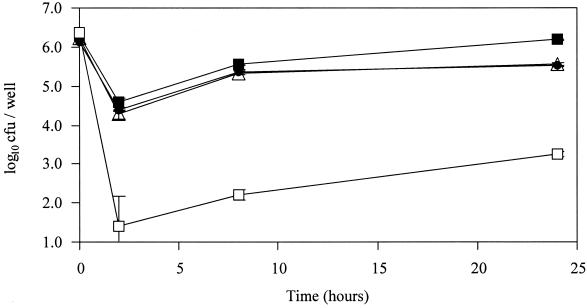
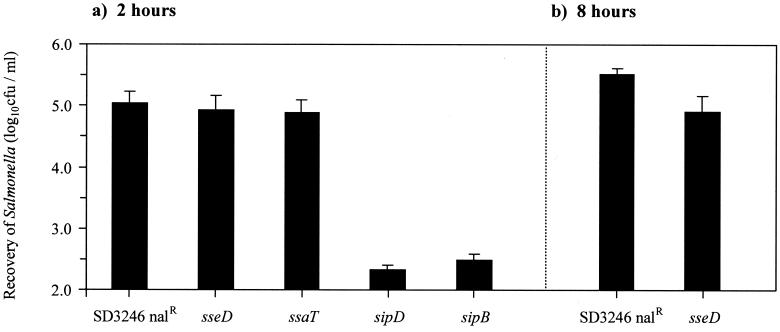
The ability of the SPI-2 mutants to invade and proliferate within bovine ileal mucosa in vivo was assessed. Ligated loops were infected with approximately 107 CFU of either the parental strain, an SPI-2 mutant strain, or the sipD mutant strain for up to 8 h. Gentamicin was added 1 h after infection to kill extracellular Salmonella in the lumen of the gut. Histological examination confirmed that under these conditions the overall integrity of the gut epithelium was maintained. Equal numbers of the parental and SPI-2 mutant strains were recovered from the ileal mucosa after 2 h, demonstrating that SPI-2 has no effect on intestinal invasion in vivo (Fig. 7a). As expected, the invasion-deficient SPI-1 mutant was recovered in much lower numbers than the parental strain. After 8 h, a significant reduction (P < 0.05) in the recovery of gentamicin-protected sseD mutant bacteria was detected compared with recovery of the parental strain (Fig. 7b).
FIG. 7.
Recovery of gentamicin-protected bacteria from bovine ileal loops 2 h (a) and 8 h (b) after infection with SD3246 Nalr or the SPI-1 (sipD and sipB) and SPI-2 (sseD and ssaT) mutant strains. Each bar is derived from the mean secretory or inflammatory response from six loops (two calves) with the exception of the sipD bar, which is derived from the mean of three loops (one calf). Each bar is presented with the standard error of the mean.
DISCUSSION
We have applied the technique of STM to investigate the virulence of serotype Dublin in calves and mice and identified a mutant which was recovered from the bovine host but not from the murine host in the initial screening procedure. Sequence analysis demonstrated that the transposon had inserted itself within the ORF of the sseD gene of SPI-2. SPI-2 encodes a type III secretion system that is considered essential for survival and proliferation of Salmonella within the intracellular environment (8, 22, 33). SPI-2 genes that have been previously identified by STM contained transposon insertions in either structural (ssa) or regulatory (ssr) components of SPI-2 or, in one case, in the gene encoding a putative chaperone, sscA (22, 42). To date, no STM mutants have been reported with insertions in any of the seven sse genes predicted to code for the secreted effector proteins of SPI-2, although at least four of these genes are essential for a fully virulent phenotype (22, 30). Here we demonstrate that STM is also able to detect attenuated strains carrying transposon insertions in the sse genes.
Our initial screen suggested that the mutant with an insertion in sseD was attenuated in mice but not in calves. However, a pure inoculum of the sseD mutant was subsequently shown to be almost avirulent when given intravenously to calves, which appeared to contradict the result from the calf screening procedure. It is possible that the original result in calves was an experimental artifact or a false-positive result. Cross-hybridization of the randomly generated tags within a pool of mutants can occur during the screening procedure, and this has the potential to conceal a negative result. However, if cross-hybridization generated a false result during screening of the pool of mutants recovered from calves, it inexplicably did not occur during screening of the same pool of mutants recovered from mice. The predicted amino acid sequence of the sseD gene product shares some homology with that of the EPEC secreted protein EspB (12, 22). Another product of the Salmonella sse operon, SseB, shares homology with the EPEC secreted protein EspA and has been shown to be secreted under certain conditions by an SPI-2-dependent mechanism (4). Consequently, it has been suggested that SseD might also be a secreted protein. Within the growing catalogue of genes identified by STM in a variety of bacterial species, there have been relatively few reports of genes encoding secreted proteins, toxins, or other extracellular virulence factors (6, 9). This has led to some debate on the ability of these mutants to be rescued in trans by the presence of virulent strains (7, 31, 37, 38). Potentially, an attenuated mutant which lacks an essential secreted virulence factor may not be identified by STM if the defect can be complemented by the presence of other bacteria secreting this factor. Thus, an alternative explanation for the apparent virulence of the sseD mutant in calves during the STM screening procedure may be trans complementation by other mutants secreting wild-type levels of SseD. It is evident that trans complementation of the serotype Dublin sseD mutant did not occur in mice, and further investigation of this phenomenon in vivo may contribute to our understanding of the responses of different species to Salmonella infection.
The second SPI-2 mutant used in this study carried an insertion in the ssaT gene, which encodes a structural component of TTSS-2. Such a mutation would disrupt the entire secretion apparatus at a fundamental level, and it is therefore less likely that the effects of this mutation could be trans complemented in a mixed infection. This mutant was not recovered from either host species during the STM screening procedure. These results clearly demonstrate that the ability of any candidate mutant to cause disease in an animal can be accurately assessed only by quantitative infection studies using a pure inoculum of the strain of interest. The serotype Dublin sseD mutant was identified by virtue of differential virulence within a mixed pool of mutants in two distinct host species. This indicates that the selective screening power of STM has the potential to be a useful molecular tool for investigating serotype Dublin serotype host specificity. This feature of STM has been noted previously during an investigation of putative host range factors of the broad-host-range serotype, serotype Typhimurium (44).
The characterization of SPI-2 function in vivo has largely depended on the murine model of salmonellosis (22, 30, 33, 41, 42). However, as mice do not develop quantifiable symptoms of diarrhea, this model cannot be used to investigate Salmonella-induced enteritis, which is so often a feature of nontyphoid Salmonella infection in humans and domestic animals. Having confirmed the systemic attenuation of the SPI-2 mutants in both mice and calves, we then used the bovine ligated ileal loop model to compare intestinal inflammatory and secretory responses triggered by the serotype Dublin parental strain and the SPI-2 mutants in vivo. Surprisingly, disruption of SPI-2 by a transposon insertion into genes encoding TTSS-2 structural or secreted effector proteins caused a significant reduction in both fluid secretion and PMN influx after 12 h. This result is in contrast to recent work which suggested that disruption of SPI-2 by mutation of the regulatory gene ssrA did not affect serotype Typhimurium-induced fluid secretion in the rabbit ileum (13). One possible explanation for this apparent discrepancy lies with the preparation of the loop inocula. Previous studies in the rabbit model have demonstrated that log-phase bacteria, such as those used in this bovine loop experiment, elicit a more potent secretory response than stationary-phase organisms (48). As the serotype Typhimurium study specified inoculation of loops with overnight bacterial cultures, it is unlikely that the secretory response elicited by either the wild-type or the SPI-2 mutant strain was optimal, and so potential differences between the enteropathogenic phenotypes of these strains may have been missed. However, this laboratory has previously demonstrated that serotype Typhimurium elicits comparatively greater enteropathogenic responses than serotype Dublin in bovine ligated ileal loops (50). Thus, it is also possible that undefined serotype-specific effectors could have masked the enteropathogenic phenotype of the SPI-2 mutant in the serotype Typhimurium background.
The virulence of the sseD mutant was further assessed in calves following oral inoculation. The mutant was attenuated, confirming that SPI-2 influences Salmonella-induced enteritis in cattle. This observation appears to contradict recent work which concluded that calves infected with an SPI-2 mutant of serotype Typhimurium developed a fatal, acute enteric Salmonella infection (43). Again, it is possible that these are serotype-specific differences. However, the animals in the latter study were infected orally with approximately 1010 CFU of serotype Typhimurium and died between 1 and 3 days postinfection. Two calves which survived infection with 109 CFU of the serotype Typhimurium SPI-2 mutant nevertheless developed acute diarrhea comparable to that observed in calves infected with the wild-type strain. It is possible that because of the large inocula the infection rapidly overwhelmed the animals and prevented the detection of the attenuated TTSS-2 mutant phenotype. Here, we have shown that calves infected with an SPI-2 mutant strain of serotype Dublin suffered only a mild, transient infection. The early stages of serotype Dublin pathogenesis do not appear to be severely affected by disruption of SPI-2, as calves infected either with the sseD mutant strain or with the parental strain initially exhibited similar pyrexic responses and all animals developed diarrhea. However, between days 3 and 6 postinfection, the calves infected with the parental strain maintained these responses and deteriorated while the calves infected with the sseD mutant strain recovered and showed no further symptoms of disease. Previous work with isogenic mutant strains has demonstrated a reliable correlation between the ileal loop assay and experimentally induced enteritis following infection of calves by the oral route (50, 51, 52). These results, obtained with the SPI-2 sseD mutant, confirm this assay as an effective means of assessing the role of putative virulence genes in the initial stages of Salmonella-induced enteritis.
Following colonization of intestinal mucosa, Salmonella can be seen to invade intestinal epithelial cells, interact with macrophages, and evoke an inflammatory cell influx (14). TTSS-1-dependent secreted effector proteins that are translocated into target cells play key roles in all three of these stages of pathogenesis (47). Disruption of TTSS-1 generates a Salmonella phenotype which is less invasive and less enteropathogenic in the bovine ileal loop model (1, 17, 51). There is some evidence that mutations in SPI-2 can affect the expression of certain SPI-1-encoded TTSS-1 proteins. Previously, an ssaT mutant strain of serotype Typhimurium was shown to be defective in expression of the SPI-1 gene, sipC (11). However, this mutant was originally identified by a reduction in the ability to invade cultured cell lines (21). In this work we clearly show that both serotype Dublin SPI-2 mutants invaded cultured cell lines in vitro and intestinal mucosal tissues in vivo at wild-type levels, indicating that TTSS-1 secreted invasins are not affected by these mutations in SPI-2.
It is possible that effector proteins secreted via TTSS-2 influence the enteropathogenic response by directly modulating host cell function. Indeed, recent work has proposed a role for SPI-2 in subverting the bactericidal mechanisms of macrophages (45). However, SPI-2 is also known to influence the net growth of Salmonella within epithelial cells in vitro (8, 33). Thus, an alternative explanation is that the reduction in enteropathogenic responses elicited by the SPI-2 mutant strains is a consequence of reduced net intracellular growth within the intestinal mucosa, which may indirectly affect the delivery of virulence factors to target cells. We therefore assessed the effect of the sseD and ssaT mutations on invasion and net growth within epithelial cells. The results presented in this work obtained with Int407 cells confirm previous studies showing that SPI-2 mutants have no defect in epithelial cell uptake but are unable to proliferate to the same level as the parental strain over 24 h. We also adapted the bovine ileal loop model to examine invasion and intracellular growth of Salmonella in vivo. The results obtained were consistent with those from the epithelial cell assay in vitro. Initially, both the parental strain and the SPI-2 mutant strains invaded the ileal mucosa in equal numbers but after 8 h there was a significant reduction in the numbers of gentamicin-protected SPI-2 mutant bacteria recovered compared with the numbers in the parental strain, demonstrating that net growth within the mucosa in vivo was attenuated.
The differences in bovine intestinal secretory and inflammatory responses induced by the SPI-2 mutant and parental strains were observed in ligated ileal loops after 12 h. The Int407 cell assay was correspondingly modified to establish the difference in net growth between the parental strain and the SPI-2 mutant strains after 12 h. The relative differences in intracellular growth of the mutant and parental strains in vitro mirrored the relative differences in the enteropathogenic responses induced by these strains in vivo. These observations are consistent with SPI-2 influencing enteropathogenesis through an effect on growth within epithelial cells, although they do not exclude a potential effect of SPI-2 on the growth and survival of Salmonella within other cells in the intestinal mucosa.
The intestinal inflammatory response, exfoliation of epithelial cells, and associated secretion of fluids into the lumen of the gut which characterize the response of cattle to Salmonella infection (14) may be considered a host defense mechanism, rapidly clearing both unattached and invading bacteria from the gut. In this study, calves infected orally with SD3246 Nalr had severe diarrhea, resulting in the fecal shedding of high numbers of bacteria, throughout the course of infection. In the natural situation this extensive fecal shedding would expose more potential hosts to serotype Dublin and thus further disseminate the pathogen in the external environment. Consequently, the induction of enteritis can also be perceived as being advantageous to a pathogen such as Salmonella. Calves infected with a fully invasive serotype Dublin SPI-2 mutant strain suffered only transient diarrhea, and the numbers of Salmonella bacteria in the feces dropped rapidly. There is no evidence to suggest that SPI-2 mutants are less able to survive and proliferate within the lumen of the gut. Indeed, because the environmental conditions necessary to trigger expression of SPI-2 are thought to correspond to conditions within intracellular vacuoles (4, 10), it is possible that SPI-2 genes are not ordinarily expressed in the intestinal lumen. Taken together, these results suggest that serotype Dublin maximizes fecal shedding by intracellular proliferation of bacteria within infected mucosa, which continually reseeds the intestinal lumen, rather than by the proliferation of bacteria within the lumen itself. Hence, it is possible that intracellular proliferation of serotype Dublin is the key to persistent and lethal diarrheal disease in calves, which may have evolved as a strategy to perpetuate fecal shedding of Salmonella into the environment and thus maximize host-to-host transmission. This work provides the first demonstration of the importance of SPI-2 in Salmonella-induced enteritis and systemic salmonellosis in cattle.
ACKNOWLEDGMENTS
This work was supported by MAFF, BBSRC, and EU Framework4 Fair3 grant CT96–1743. B. N. Tripathi was sponsored by DFID as part of the UK-India TOMBIT project.
We are grateful to Sue Paulin and Annette Benmore (IAH) for performing the surgical procedures.
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 2.Bäumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutation in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuzon C R, Banks G, Deiwick J, Hensel M, Holden D W. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 5.Bulgin M S. Salmonella dublin: what veterinarians should know. J Am Vet Med Assoc. 1983;182:116–118. [PubMed] [Google Scholar]
- 6.Carmacho L R, Ensergueix D, Perez E, Giquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 7.Chiang S L, Mekalanos J J, Holden D W. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 9.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 10.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 11.Deiwick J, Nikolaus T, Shea J E, Gleeson C, Holden D W, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI-2) genes affecting transcription of SPI-1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–4780. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliot S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 13.Everest P, Ketley J, Hardy S, Douce G, Khan S, Shea J E, Holden D W, Maskell D, Dougan G. Evaluation of Salmonella typhimurium mutants in a model of experimental gastroenteritis. Infect Immun. 1999;67:2815–2821. doi: 10.1128/iai.67.6.2815-2821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost A J, Bland A P, Wallis T S. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet Pathol. 1997;34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- 15.Galán J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 16.Galán J E, Curtiss R. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 18.Hall G A, Jones P W. An experimental study of Salmonella dublin abortion in cattle. Br Vet J. 1976;132:60–65. doi: 10.1016/s0007-1935(17)34788-7. [DOI] [PubMed] [Google Scholar]
- 19.Heffernan E J, Fierer J, Chikami G, Guiney D. The natural history of oral Salmonella dublin infection in BALB/c mice: effect of an 80-kilobase pair plasmid on virulence. J Infect Dis. 1987;155:1254–1259. doi: 10.1093/infdis/155.6.1254. [DOI] [PubMed] [Google Scholar]
- 20.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 21.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 22.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Beuzon C R, Gleeson C, Fang F C, Holden D W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 23.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P W. Salmonellosis. In: Andrews A H, Blowley R W, Boyd H, Eddy R G, editors. Bovine medicine: diseases and husbandry of cattle. Oxford, United Kingdom: Blackwell Scientific Publications; 1992. pp. 181–193. [Google Scholar]
- 25.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning E J, Baird G D, Jones P W. The role of plasmid genes in the pathogenicity of Salmonella dublin. J Med Microbiol. 1986;21:239–243. doi: 10.1099/00222615-21-3-239. [DOI] [PubMed] [Google Scholar]
- 29.McDonough P L, Fogelman D, Shin S J, Brunner M A, Lein D H. Salmonella enterica serotype Dublin infection: an emerging infectious disease for the northeastern United States. J Clin Microbiol. 1999;37:2418–2427. doi: 10.1128/jcm.37.8.2418-2427.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina E, Paglia P, Nikolaus T, Müller A, Hensel M, Guzman C A. Pathogenicity island 2 mutants of Salmonella typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect Immun. 1999;67:1093–1099. doi: 10.1128/iai.67.3.1093-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller V L. Signature-tagged mutagenesis and the hunt for virulence factors: response. Trends Microbiol. 1999;7:388. doi: 10.1016/s0966-842x(99)01583-8. [DOI] [PubMed] [Google Scholar]
- 32.Ochman H, Groisman E A. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ørskov J, Moltke O. Studien über den Infektions-mechanismus bei verschiedenen Paratyphus-infektionen aus weissen Mäusen. Z Immunitaetsforsch. 1929;59:357–405. [Google Scholar]
- 35.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 36.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 37.Perry R D. Signature-tagged mutagenesis and the hunt for virulence factors. Trends Microbiol. 1999;7:385–388. doi: 10.1016/s0966-842x(99)01582-6. [DOI] [PubMed] [Google Scholar]
- 38.Perry R D. Signature-tagged mutagenesis and the hunt for virulence factors: response. Trends Microbiol. 1999;7:388. doi: 10.1016/s0966-842x(99)01593-0. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Scottish Agricultural College [SAC Veterinary Science Division] Salmonella dublin continues to cause problems in Scottish cattle. Vet Rec. 2000;146:5–6. [PubMed] [Google Scholar]
- 41.Shea J E, Beuzon C R, Gleeson C, Mundy R, Holden D. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsolis R M, Adams L G, Ficht T A, Bäumler A J. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsolis R M, Townsend S M, Miao E A, Miller S I, Ficht T A, Adams L G, Bäumler A J. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden D W, Lucia S M, Dinauer M C, Mastroeni P, Fang F C. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 46.Veterinary Laboratories Agency [VLA] Salmonella dublin causes a variety of problems for cattle in September. Vet Rec. 1999;145:537–538. [PubMed] [Google Scholar]
- 47.Wallis T S, Galyov E E. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 48.Wallis T S, Hawker R J H, Candy D C A, Qi G-M, Clarke G J, Worton K J, Osbourne M P, Stephen J. Quantification of the leukocyte influx into rabbit ileal loops induced by strains of Salmonella typhimurium of different virulence. J Med Microbiol. 1989;30:149–156. doi: 10.1099/00222615-30-2-149. [DOI] [PubMed] [Google Scholar]
- 49.Wallis T S, Paulin S M, Plested J S, Watson P R, Jones P W. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun. 1995;63:2755–2761. doi: 10.1128/iai.63.7.2755-2761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson P R, Paulin S M, Bland P, Jones P W, Wallis T S. Characterization of intestinal invasion by Salmonella dublin and effect of a mutation in the invH gene. Infect Immun. 1995;63:2743–2754. doi: 10.1128/iai.63.7.2743-2754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson P R, Paulin S M, Bland A P, Libby S J, Jones P W, Wallis T S. Differential regulation of enteric and systemic salmonellosis by slyA. Infect Immun. 1999;67:4950–4954. doi: 10.1128/iai.67.9.4950-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood M W, Jones M A, Watson P R, Hedges S, Wallis T S, Galyov E E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]