Abstract
Measurable residual disease (MRD) assessment provides a potent indicator of the efficacy of anti-leukemic therapy. It is unknown, however, whether integrating MRD with molecular profiling better identifies patients at risk of relapse. To investigate the clinical relevance of MRD in relation to a molecular-based prognostic schema, we measured MRD by flow cytometry in 189 AML patients enrolled in ECOG-ACRIN E1900 trial (NCT00049517) in morphologic complete remission (CR) (28.8 % of the original cohort) representing 44.4 % of CR patients. MRD positivity was defined as ≥ 0.1 % of leukemic bone marrow cells. Risk classification was based on standard cytogenetics, fluorescence-in-situ-hybridization, somatic gene analysis, and sparse whole genome sequencing for copy number ascertainment. At 84.6 months median follow-up of patients still alive at the time of analysis (range 47.0–120 months), multivariate analysis demonstrated that MRD status at CR (p = 0.001) and integrated molecular risk (p = 0.0004) independently predicted overall survival (OS). Among risk classes, MRD status significantly affected OS only in the favorable risk group (p = 0.002). Expression of CD25 (α-chain of the interleukin-2 receptor) by leukemic myeloblasts at diagnosis negatively affected OS independent of post-treatment MRD levels. These data suggest that integrating MRD with genetic profiling and pre-treatment CD25 expression may improve prognostication in AML.
Keywords: Acute myeloid leukemia, Minimal residual disease, Measurable residual disease, MRD, CD25, Flow cytometry, Genomic profiling
1. Introduction
Recurrent gene mutations in acute myeloid leukemia (AML) patients have biologic, prognostic and therapeutic relevance [1-4]. Mutational profiling can be integrated with cytogenetic analysis and clinical parameters to develop a more robust risk stratification in AML [2,4,5]. However, the risk of relapse and death from recurrent leukemia is not fully predicted by pre-therapy parameters, and there remains a pressing need to develop new biomarkers to guide therapeutic decisions in AML.
Measurable residual disease (MRD) assessment at the time of complete morphologic remission (CR) evaluates the quality of response to induction chemotherapy beyond the level of conventional histopathology. This notion has led to the definition of MRD-negative CR as an important clinical endpoint [5]. Increasingly, studies are suggesting that MRD status serves as a prognostic tool in AML [6-9]. Nevertheless, MRD measurements are not yet routinely used to guide therapeutic decisions in AML, partly due to the lack of uniformity in methodologies used for MRD detection and interpretation [6-8,10-14]. In addition, the independent prognostic relevance of MRD relative to pre-therapeutic prognosticators, such as cytogenetic risk [10,15,16], expression of prognostic antigens, such as CD25 on myeloblasts [17], or molecular profiling [18-25] is not fully understood, particularly in the setting of large clinical trials with uniform treatment.
To investigate the clinical significance of MRD in relation to genetic profiling, we measured MRD by flow cytometry in AML patients age 60 and under enrolled in intergroup phase 3 trial, E1900 (NCT00049517) [2,26]. We previously reported that dose-intensification of daunorubicin during induction therapy improved CR rate and overall survival (OS) [26]. Moreover, expression of CD25, the alpha-chain of the interleukin-2 receptor, had prognostic relevance independent of mutational profiling [17]. We now assess whether MRD has prognostic value irrespective of induction enhancement, mutational profile, cytogenetic status, or CD25 expression.
2. Materials and methods
2.1. Patients
Eligibility criteria for E1900 have been published [26]. Of 657 accrued patients, 629 had baseline specimens submitted to the ECOG-ACRIN Leukemia Translational Research Laboratory (LTRL) for central immunophenotyping by multiparameter flow cytometry. Excluded from central MRD analysis in the LTRL were 83 patients from Rambam Medical Center, Haifa, Israel, for whom only frozen follow-up samples were received. Since submission of follow-up specimens was not mandatory, the LTRL received post-treatment samples for 388/546 remaining patients (71 %) at 1–15 time-points per patient. In 9 patients, only relapse material was submitted and, in 45 patients, baseline specimens, predominantly with monocytic phenotype, did not yield a Leukemia-Associated Immunophenotype (LAIP) suitable for MRD detection. For 189 of the 334 patients with suitable baseline LAIP, MRD data were available at CR, representing 28.8 % of the entire E1900 cohort and 44.4 % of patients who achieved a CR. No statistically significant differences were observed between these 189 patients and patients achieving CR without MRD data with regard to baseline characteristics and outcomes. Of the 189 patients, 87 had MRD samples available from CR as well as various time-points post-consolidation.
2.2. Flow cytometric analysis of baseline immunophenotypes and MRD
Heparinized samples were received and processed by the LRTL within 24 h of collection. MRD was assessed by 4-color flow cytometry in whole, unseparated samples and expressed as percent of nucleated white blood cells (WBC) based on staining with cell-permeant green fluorescent nucleic acid dye (Syto 16) (FACSCanto II flow cytometer, Beckton Dickinson, and FACSDiva analysis software). MRD detection was based on the diagnostic LAIP, using both asynchronous expression of antigens and aberrant antigen intensities in LAIP definition [10,27,28]. Any discrete cell population consistent of at least 10 events which unequivocally expressed LAIP features was considered as MRD. If the quality of bone marrow aspirates precluded us from reliably determining the MRD status, we considered the MRD result as indeterminate. Indeterminate samples were not counted in our MRD analysis. Among Core-Binding-Factor (CBF) leukemias, LAIP features included expression of CD19 and/or CD56, absence of CD11a and CD7 and weak myeloid antigen expression for RUNX1/RUNXT1 AML, and CD2 for CBFβ/MYH11 AML [29]. If more than one leukemic immunophenotypic clone was detected at diagnosis, MRD assessment covered all of the antigen combinations of interest. Antibody combinations for MRD detection were designed individually for each patient based on the diagnostic LAIP. Whenever possible, a minimum of 100,000 events were acquired (median 176,000, range 30,000–400,000). If bone marrow was not submitted, peripheral blood was tested. However, due to decreased confidence in concordant negative MRD results in blood and bone marrow with our assay [30], only positive MRD results from blood specimens were included in the analysis.
2.3. Cytogenetic and mutational analyses
Karyotypes from all cases were centrally reviewed. The definition of favorable, intermediate, indeterminate and unfavorable cytogenetic data followed that published by Slovak et al [31]. Fluorescence-in-situ hybridization (FISH) for t(8;21), inv(16), t(9;22), t(15;17), + 8, – 7/del(7q), – 5/del(5q), rearranged KMT2A and EVI1 genes was performed centrally at the Mayo Cytogenetics Laboratory, Rochester, MN, as described [32]. PCR analysis for RUNX1/RUNX1T1, CBFβ/MYH11, PML/RARα, BCR/ABL transcripts and partial tandem duplication of the KMT2A-gene (KMT2A-PTD) was done centrally at the LTRL [26]. Somatic mutational analysis, including for FLT3 and NPM1 gene aberrations, and integrated mutational/cytogenetic risk stratification were as reported previously [2]. This risk classification was chosen since patients with FLT3-ITD mutations could not be grouped according to the ratio of the mutant to wild-type alleles, as suggested by the 2017 ELN recommendation [5].
2.4. Sparse whole genome sequencing (sWGS) for copy number ascertainment
sWGS was performed using 50 ng of bulk DNA purified from bone marrow or peripheral blood mononuclear cells with a blast count of at least 25 %. TruSeq indexed libraries were generated, sequenced in multiplex fashion targeting a coverage of 2 million sequencing reads per sample, and data subsequently processed for copy number analysis as described [33]. In brief, sequencing reads were mapped to human reference genome hg19 with uniquely mapped reads sorted and indexed. Uniquely mapped reads were then counted in genomic bins while partitioning the genome in 20,000 bins using a described algorithm (Varbin) [34]. Read bin counts were normalized genome wide with subsequent processing using Circular Binary Segmentation to retrieve chromosomal segments that exhibited copy number states deviating from a euploid, 2N state.
2.5. Statistical analysis
Baseline characteristics were compared using Fisher’s exact test if they were categories and Wilcoxon rank sum tests if they were continuous. OS was defined as time from date of CR to death from any cause. Disease-free survival (DFS) was defined as the time from documented CR to relapse or death from any cause. Kaplan-Meier estimates were used to estimate the event-time distributions. Univariate and multivariate Cox models were performed on OS and DFS. Multivariate models were adjusted for induction treatments, age (continuous), gender (binary), cytogenetic risk (categorical), baseline WBC count (continuous), platelet count (continuous), hemoglobin levels (continuous), and allogeneic transplant (time-varying covariate).
To examine whether MRD status after consolidation had prognostic relevance, a time-varying stratum Cox analysis was performed on patients who entered consolidation and had MRD data available at time points after consolidation. As MRD status may change over time, it was included into the Cox models as a time-varying covariate where patients enter the MRD-positive stratum at any time after consolidation when an MRD test is positive and enter back to MRD-negative stratum when an MRD test is negative. All P values were based on 2-sided tests.
3. Results
3.1. Determining the cut-off point for MRD positivity
When determining the clinically most informative cut-off point for defining MRD positivity with our assay sensitivity threshold and patient population, hazard ratios (HR) (MRD negative versus MRD positive) in univariate Cox analysis at CR concerning OS were 0.32 (95 % CI, 0.19–0.56; p < 0.001) when ≥ 0.01 % of blasts defined positive MRD status, 0.35 (95 % CI, 0.21–0.59; p < 0.001) when ≥ 0.05 % of blasts were used, and 0.41 (95 % CI, 0.26–0.65; p < 0.001) when ≥ 0.1 % of blasts defined positive MRD status, indicating a consistently similar worse prognosis with MRD positivity at each of these MRD levels. There was a significant difference in OS between patients with CR MRD levels between undetectable (0 %) and < 0.1 % and those with MRD levels 0.1 % -< 1 % (HR 0.43, 95 % CI, 0.27–0.69, p = 0.002). However, there was no difference in OS between patients with MRD levels of 0.1 %– 1 % and those with MRD levels > =1 % (HR 0.74, 95 % CI, 0.42–1.28, p = 0.28). We, therefore, chose an MRD level of ≥ 0.1 % to define MRD positivity.
3.2. Association of MRD with presenting biologic characteristics
Table 1 summarizes the distribution of MRD-negative and MRD-positive patients by gender, age, presenting WBC count, hemoglobin, platelet count and cytogenetic risk group. For only 6 patients, neither standard cytogenetics nor FISH nor sWGS data were available. There was no significant difference between MRD-positive and -negative patients with respect to clinical or demographic factors. However, MRD-negative patients had a higher likelihood of being in the favorable cytogenetic risk group (32.7 %) and less likely to have unfavorable cytogenetics (7.3 %) than MRD-positive patients (14.9 % and 16.4 %, respectively) (p = 0.03). In agreement with this finding, among all MRD-negative patients, 20 % had favorable RUNX1/RUNX1T1 CBF-leukemia compared with 4 % among MRD-positive patients (p = 0.004). However, no such difference was seen for patients with CBFβ/MYH11 CBF-AML. None of the other genetic abnormalities detected by FISH or sWGS was associated with MRD status. Regarding specific somatic mutations, among 18 genes tested (Supplementary Table S1), DNMT3AR882 mutation was the only mutation significantly associated with the presence of MRD at CR, consistent with what we have previously reported [35].
Table 1.
a Demographics and disease characteristics of the CR patients with MRD status data (continues variables).
| MRD− | MRD+ | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | Min | Max | Median | Min | Max | Median | Min | Max | |
| Age (years) | 45.0 | 19 | 60 | 50.0 | 18 | 60 | 48.0 | 18 | 60 |
| WBC count (/mm3)(x1000) | 9.7 | 1 | 107 | 14.6 | 1 | 176 | 11.2 | 1 | 176 |
| Hemoglobin (g/dl) | 9.2 | 6 | 16 | 9.2 | 5 | 30 | 9.2 | 5 | 30 |
| Platelet count (/mm3)(x1000) | 46.0 | 6 | 246 | 56.0 | 1 | 479 | 54.3 | 1 | 479 |
| b Demographics and Disease Characteristics of the CR patients with MRD status data (categorical variables). | ||||||
|---|---|---|---|---|---|---|
| MRD− | MRD+ | Total | ||||
| N | % | N | % | N | % | |
| Gender | ||||||
| Female | 25 | 45.5 | 66 | 49.3 | 91 | 48.1 |
| Male | 30 | 54.5 | 68 | 50.7 | 98 | 51.9 |
| Cytogenetics | ||||||
| Favorable | 18 | 32.7 | 20 | 14.9 | 38 | 20.1 |
| Indeterminate | 11 | 20.0 | 35 | 26.1 | 46 | 24.3 |
| Intermediate | 22 | 40.0 | 57 | 42.5 | 79 | 41.8 |
| Unfavorable | 4 | 7.3 | 22 | 16.4 | 26 | 13.8 |
3.3. Association of MRD with overall and disease-free survival
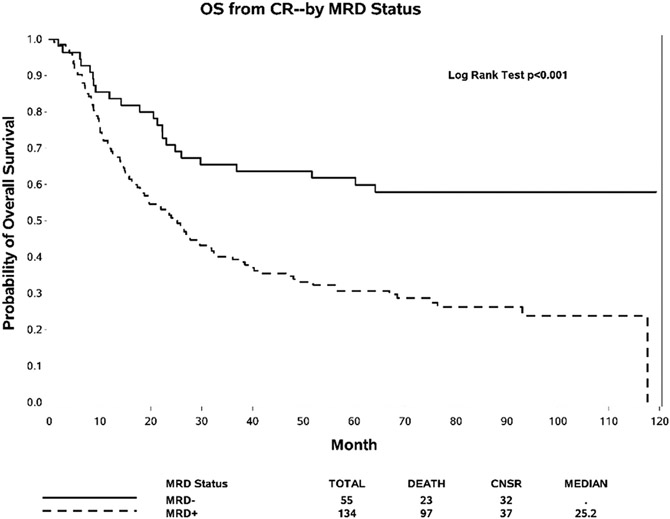
At the time of CR, 55 (29.1 %) patients were MRD-negative and 134 (70.1 %) patients were MRD-positive. At 84.6 months median follow-up of patients still alive at the time of analysis (range 47.0–120 months), the median OS in MRD-negative patients had not yet been reached versus 25.2 months for MRD-positive patients (HR 0.41, 95 % CI: 0.26, 0.65, p < 0.001, Fig. 1). In multivariate analysis, the impact of MRD positivity on OS remained significant (HR 0.45, 95 % CI: 0.28, 0.72, p = 0.0009, Supplementary Table S2). Similarly, the median DFS in MRD-negative patients had not yet been reached versus 12.0 months for MRD-positive patients (univariate analysis HR 0.39, 95 % CI: 0.25, 0.61, p < 0.001, Supplementary Fig. S2; and multivariate analysis HR 0.42, 95 % CI: 0.26, 0.66, p = 0.0002).
Fig. 1.
Overall Survival (OS) according to MRD status at time of complete hematologic remission (CR).
We also assessed the effect of MRD on outcome when measured at any time post-consolidation using Cox models as a time-varying covariate where patients can enter the MRD-positive and MRD-negative strata at any time post-consolidation. MRD data for both CR and post-consolidation therapy were available for 87 patients. Of those, 6 had converted from MRD-negative at CR to positive at the first post-consolidation evaluation and 7 patients who were MRD-positive at CR subsequently converted to MRD-negative status. In both univariate (for OS, HR: 0.24, 95% CI: 0.12, 0.47, p < 0.001; for DFS, HR: 0.21, 95% CI: 0.11, 0.41, p < 0.001) and multivariate analysis (for OS, HR: 0.26, 95% CI: 0.13, 0.52, p < 0.001; for DFS, HR: 0.22, 95% CI: 0.11, 0.44, p < 0.001), data again showed worse outcome for patients who were MRD-positive at any time-point post-consolidation.
3.4. Association of MRD at complete remission with daunorubicin dose
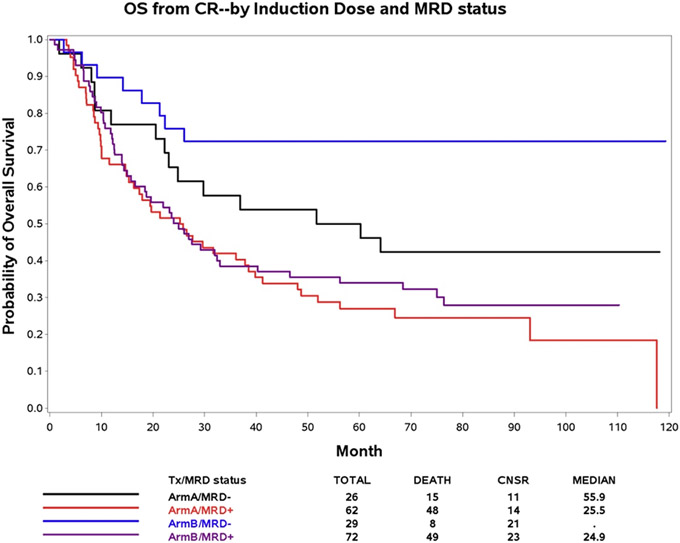
Of patients with MRD data at CR, 88 had been randomized to receive standard-dose daunorubicin (45 mg/m2) and 101 patients to receive high-dose daunorubicin (90 mg/m2) during induction. There was no significant association of MRD status at CR and daunorubicin dose (p = 1.0). However, high-dose, but not standard-dose, daunorubicin was associated with improved OS in MRD-negative (univariate analysis HR: 0.42, 95 % CI: 0.18, 0.99, p = 0.05; multivariate analysis HR: 0.40, 95 % CI: 0.16, 1.06, p = 0.06), but not in MRD-positive patients (univariate analysis HR: 0.87, 95 % CI: 0.58, 1.30; p = 0.49; multivariate analysis HR: 0.93, 95 % CI: 0.61, 1.42, p = 0.75) (Fig. 2), suggesting that dose-intensified induction therapy increased the depth of CR in chemotherapy-sensitive patients. Similar results were observed for DFS (Supplementary Fig. S1).
Fig. 2.
Overall survival (OS) after achievement of complete remission (CR) by MRD status and induction treatment. Arm A− Daunorubicin 45 mg/M2, Arm B− Daunorubicin 90 mg/M2.
3.5. Prognostic effect of MRD in cytogenetic risk groups
Among patients with favorable cytogenetics (n = 38), MRD status had a significant impact on OS (univariate HR: 0.23, 95 % CI: 0.08, 0.71, p = 0.005, Supplementary Fig. S3; multivariate HR 0.20, 95 % CI, 0.05, 076, p = 0.02). The OS for MRD-negative patients with favorable cytogenetics (47 %) has not yet been reached compared with 31.6 months for MRD-positive patients. In patients with intermediate-risk cytogenetics (n = 79), MRD status had a marginally significant impact on OS in univariate (HR 0.51; 95 % CI: 0.25, 1.02, p = 0.051, Supplementary Fig. S4) but not multivariate analysis. Median OS for MRD-negative intermediate-risk patients has not yet been reached compared with 27.5 months in MRD-positive patients. In the unfavorable cytogenetic risk groups (n = 26), OS did not differ by MRD status (HR: 1.05, 95 % CI: 0.31, 3.62, p = 0.94, Supplementary Fig. S5). MRD negativity was achieved in only 4/26 patients with unfavorable cytogenetics.
3.6. MRD status and integrated molecular profiling
Integrated molecular profile combined molecular and cytogenetic risk classes into a single risk stratification schema. The incidence of MRD-positivity at CR was: 56 % in patients with a favorable-risk, 76 % of patients with intermediate-risk and 78 % in patients with unfavorable-risk profile. The frequency of MRD-positivity increased with increasing risk of relapse as stratified by integrated molecular profiling (p = 0.015). Multivariate analysis demonstrated that post-induction MRD status (p = 0.0004) and pre-therapy integrated molecular risk (p < 0.001) independently impacted OS. The effect of MRD status on outcome was significant in all risk classes, though most pronounced in favorable-risk patients (univariate HR: 0.28, 95 % CI: 0.11, 0.69, p = 0.006; multivariate HR: 0.22, 95 % CI: 0.08, 0.61, p = 0.004). Intermediate-risk (univariate HR: 0.34, 95 % CI: 0.12, 0.97, p = 0.04; multivariate HR: 0.30, 95 % CI: 0.09, 1.00, p = 0.05) and integrated adverse-risk MRD-negative patients also had better OS compared to MRD-positive patients (univariate HR: 0.63, 95 % CI: 0.29, 1.37, p = 0.24; multivariate HR: 0.36, 95 % CI: 0.14, 0.92, p = 0.03).
3.7. MRD at CR and CD25 status at baseline
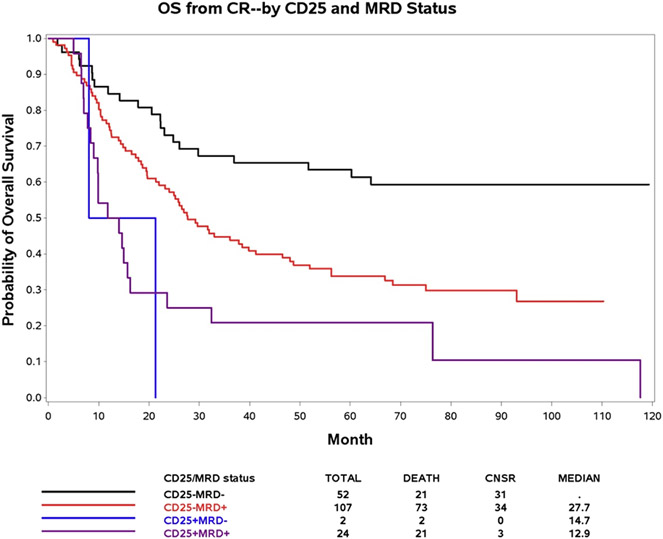
E1900 patients whose myeloblasts at diagnosis expressed CD25, the α-chain of the interleukin 2 receptor, had a significantly poorer outcome than patients without CD25 expression independent of molecular profiling [17]. The presence of a distinct CD25 expressing myeloblast population (range, 19 %–99 %) defined CD25 positivity. CD25 data were available for 185 patients with MRD data at CR. NPM1 (p = 0.014), FLT3-ITD (p < 0.001), and DNMT3AR882 (p = 0.02) were the only mutations tested significantly associated with CD25 positivity (Supplementary Table S3). Of the 185 patients, 26 (14 %) had been CD25-positive at diagnosis, comparable to the rate of CD25-positivity in the entire E1900 cohort (13 %) [17]. Of CD25-positive patients, 92 % were MRD-positive at CR. Among 131 MRD-positive patients at CR, 24 (18.3 %) had been CD25-positive at diagnosis, while only 2 of 54 MRD-negative patients had been CD25-positive (3.7 %) (p = 0.009). Median expression levels of CD25 were 64 % in the two CD25-positive/MRD-negative and 55 % in the 131 CD25-positive/MRD-positive patients. MRD-negative/CD25-negative patients had better OS than MRD-negative/CD25-positive patients (HR: 0.16, 95 % CI: 0.03, 0.72, p = 0.02). MRD-positive/CD25-negative patients also had significantly better OS compared with MRD-positive/CD25-positive patients (HR: 0.55, 95 % CI: 0.34, 0.91, p = 0.02) (Fig. 3). Similar results were observed for DFS (Supplementary Fig. S6). A Cox model confirmed that MRD status at CR (HR: 0.54, 95 % CI: 0.33, 0.87, p = 0.01) and CD25 status at presentation (HR: 0.46, 95 % CI:0.27, 0.77, p = 0.004) were significantly and independently associated with OS (Supplementary Table S4).
Fig. 3.
Overall Survival (OS) according to MRD status and CD25 at time of complete hematologic remission (CR).
4. Discussion
Although molecular, cytogenetic, and clinical features have an important impact on survival in AML, there remains a critical need to identify new predictors of outcome. In adult ALL, MRD status has emerged as a robust measure of therapeutic sensitivity and depth of response with substantive prognostic value [36-38]. In contrast, despite several reports suggesting the prognostic importance of MRD in AML [8, 39,41], it is not considered routine practice in adult AML [40]. To investigate the clinical relevance of MRD in relation to a molecular-based prognostic schema, we measured MRD by flow cytometry in 189 AML patients enrolled in the ECOG-ACRIN E1900 clinical trial (NCT00049517) in morphologic complete remission (CR) (28.8 % of the original cohort) representing 44.4 % of CR patients. We provide evidence that MRD, as assessed by central flow cytometry, has prognostic value in a uniformly treated AML cohort independent of integrated cytogenetic/molecular profiling. This result is particularly noteworthy as our 4-color LAIP-based flow cytometric MRD assay, available during the duration of the E1900 trial (December 2002-November 2008), would nowadays be considered sub-standard [5], confirming the robust nature of MRD as a prognostic parameter. The maturity of the data, on the other hand, provided us with a follow-up of up to 10 years.
To answer the question of optimal timing of MRD measurement, we assessed MRD at the time of hematologic CR and any-time post-consolidation. MRD status at either time-point was strongly predictive of outcome. The other parameter of interest was the cut-off for MRD positivity. Though the standard MRD positivity threshold is 0.1 % [10], we found no difference between several cut-offs (0.1 %, 0.05 % or 0.01 %) with respect to MRD positivity and outcome. These data suggest that it is best to consider MRD as a continuous, quantitative variable, and that whatever cutoff can be enumerated by the flow cytometric method used and retrospectively found to be associated with outcome has prognostic significance in individual AML trials using equivalent treatment strategies. For the majority of patients, the 4-color LAIP-based MRD assessment was not sufficiently sensitive to detect levels below 0.01%. Data with improved methodology suggest that any level of MRD may be prognostically important [41,42].
One important question in the field is how best to incorporate MRD in current risk stratification schema in AML. Previous studies have performed multivariate analyses including both MRD and cytogenetics[43], or MRD and specific mutations, such as FLT3 [44,45] and NPM1 [19,20,44], to show that MRD has independent prognostic value. We show that in the context of comprehensive cytogenetic and molecular risk stratification, MRD has an important prognostic impact. By incorporating sWGS with FISH and standard cytogenetics, we both confirmed recurrent translocations and copy-number alterations which had been identified by cytogenetic analysis and increased the portion of patients with genomic information to 97 %. Because the impact of MRD status on prognosis was significant in all AML patients, whether classified as favorable, intermediate or adverse integrated risk, we conclude that MRD-negativity selects the subset of patients within these risk profiles who have chemo-sensitive disease and thus the most favorable outcome. There is clearly a need of more extensive mutational profiling [46], preferably at the single-cell level [24,47,48], to further underscore this notion.
Surface expression of the CD25 antigen at diagnosis was a powerful adverse prognostic factor in E1900 patients [17]. We now show a significantly higher rate of MRD-positivity at CR in CD25-positive compared to CD25-negative patients. Importantly, CD25 and MRD status were significantly and independently associated with OS, irrespective of age, WBC count and cytogenetic risk. Our data expand on an earlier report by Terwijn et al [49]. which showed a correlation of expression of CD25 on > 10 % myeloblasts at diagnosis with significantly higher MRD frequency after the first cycle of chemotherapy. Based on previous gene expression profiling studies in the E1900 cohort [17] and studies by others on the biology of CD25 in AML [50,51], we hypothesize that CD25 tracks with leukemic stem cell burden/identity, and that MRD-positivity and/or CD25-expression mark AML stem cells which are less sensitive to cytotoxic chemotherapy.
Taken together, we show that the presence of flow cytometric MRD is an independent prognostic factor in newly diagnosed AML patients ≤ 60 years old, both at time of CR and after consolidation. This effect was independent of pre-therapeutic molecular risk and level of CD25 expression, both biomarkers with significant prognostic implications.
Supplementary Material
Acknowledgments
This study was conducted in part by the ECOG-ACRIN Cancer Research Group (Peter J. O’Dwyer, MD and Mitchell D. Schnall, MD, Ph.D., Group Co-Chairs) and supported by the National Cancer Institute of the National Institutes of Health under the following award numbers: U10CA180820, U10CA180794, UG1CA189859, UG1CA233234, UG1CA232760 and UG1CA233290. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government. This work was further supported by National Cancer Institute National Institutes of Health UG1 CA233332 (O.I.A.-W. and R.L.L.), P30 CA008748–55 (S.W.L.), P50 CA254838–01 (S.W.L.), and K08CA169055 (F.G.-B.), the American Society of Hematology (ASHAMFDP-20121) under the ASH-AMFDP partnership with The Robert Wood Johnson Foundation (F.G.-B.), U10 5U10CA180827 (subaward to F.G.-B), William C. and Joyce C. O′Neil Charitable Trust (T.B.), and the Memorial Sloan Kettering Single Cell Sequencing Initiative (T.B.). The authors acknowledge the use of the Integrated Genomics Operation Core, funded by the Memorial Sloan Kettering Cancer Center Support Grant NIH P30 CA008748. The authors also acknowledge the assistance of Xerxes Vevai and other members of the LTRL in the MRD analysis and the technical support of Tak Lee and Caroline Sheridan in the DNA analysis.
Footnotes
Conflict-of-interest disclosure
E.P. does consulting work with Supertechs Inc. and the ECOG-ACRIN Cancer Research Group; R.L.L. is on the supervisory board of Qiagen and is a scientific advisor for Imago, Mission Bio, Zentalis, Ajax, Auron, Prelude, C4 Therapeutics and Isoplexis; he receives research support from and consulted for Celgene and Roche and has consulted for Incyte, Janssen, Astellas, Morphosys and Novartis; he has received honoraria from Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. O.I.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, Prelude Therapeutics and Janssen and is on the scientific advisory board of Envisagenics Inc, Pfizer Boulder and Alchemy Inc; he has received prior research funding from Loxo Oncology and H3B Biomedicine. M.S.T. receives research funding from Abbvie, Cellerant, Orsenix, ADC Therapeutics, Biosight, Glycomimetics, Rafael Pharmaceuticals and Amgen, is on the advisory boards of Abbvie, BioLineRx, Daiichi-Sankyo, Orsenix, KAHR, Rigel, Nohla, Delta Fly Pharma, Tetraphase, Oncolyze, Jazz Pharmaceuticals, Roche, Biosight, Novartis, and receives royalties from UpToDate. H.F.F. serves on the advisory board of Jazz Pharmaceuticals and Incyte. S.W.L. is founder and member of the scientific advisory board of Blueprint Medicines, Mirimus, ORIC Pharmaceuticals and Faeth Therapeutics, and is on the scientific advisory board of Constellation Pharmaceuticals and PMV Pharmaceuticals. H.M.L. is a promotional speaker and consultant for Jazz Pharmaceuticals. The remaining authors declare no competing financial interests.
CRediT authorship contribution statement
Contribution: E.P., R.L.L., Z.S and C.G. prepared the manuscript; E.P., C.G., O.I.A.-W., T.B., F.G.-B., S.W.L., S.M.L and R.L.L. performed experiments and data analyses and helped with data interpretation; Z.S. and M.G. performed statistical analyses; R.K. reviewed institutional cytogenetic data; H.F., H.M.L. and M.S.T. organized the clinical trial; all authors reviewed and approved the manuscript.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.leukres.2022.106971.
References
- [1].N. Cancer Genome Atlas Research, T.J. Ley, C. Miller, et al. , Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia, N. Engl. J. Med 368 (2013) 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Patel JP, Gonen M, Figueroa ME, et al. , Prognostic relevance of integrated genetic profiling in acute myeloid leukemia, N. Engl. J. Med 366 (2012) 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Papaemmanuil E, Gerstung M, Bullinger L, et al. , Genomic classification and prognosis in acute myeloid leukemia, N. Engl. J. Med 374 (2016) 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gerstung M, Papaemmanuil E, Martincorena I, et al. , Precision oncology for acute myeloid leukemia using a knowledge bank approach, Nat. Genet 49 (2017) 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dohner H, Estey E, Grimwade D, et al. , Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel, Blood 129 (2017) 424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ravandi F, Walter RB, Freeman SD, Evaluating measurable residual disease in acute myeloid leukemia, Blood Adv. 2 (2018) 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schwind S, Jentzsch M, Bach E, Stasik S, Thiede C, Platzbecker U, Use of minimal residual disease in acute myeloid leukemia therapy, Curr. Treat. Options Oncol 21 (8) (2020). [DOI] [PubMed] [Google Scholar]
- [8].Short NJ, Zhou S, Fu C, et al. , Association of measurable residual disease with survival outcomes in patients with acute myeloid leukemia: a systematic review and meta-analysis, JAMA Oncol. 6 (2020) 1890–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aitken MJL, Ravandi F, Patel KP, Short NJ, Prognostic and therapeutic implications of measurable residual disease in acute myeloid leukemia, J. Hematol. Oncol 14 (2021) 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schuurhuis GJ, Heuser M, Freeman S, et al. , Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party, Blood 131 (2018) 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ossenkoppele G, Schuurhuis GJ, van de Loosdrecht A, Gloos J, Can we incorporate MRD assessment into clinical practice in AML? Best Pract. Res. Clin. Haematol 32 (2019) 186–191. [DOI] [PubMed] [Google Scholar]
- [12].Buccisano F, Maurillo L, Schuurhuis GJ, et al. , The emerging role of measurable residual disease detection in AML in morphologic remission, Semin. Hematol 56 (2019) 125–130. [DOI] [PubMed] [Google Scholar]
- [13].Maurillo L, Bassan R, Cascavilla N, Ciceri F, Quality of response in acute myeloid leukemia: the role of minimal residual disease, Cancers 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dix C, Lo TH, Clark G, Abadir E, Measurable residual disease in acute myeloid leukemia using flow cytometry: a review of where we are and where we are going, J. Clin. Med 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Buccisano F, Maurillo L, Spagnoli A, et al. , Cytogenetic and molecular diagnostic characterization combined to postconsolidation minimal residual disease assessment by flow cytometry improves risk stratification in adult acute myeloid leukemia, Blood 116 (2010) 2295–2303. [DOI] [PubMed] [Google Scholar]
- [16].Venditti A, Piciocchi A, Candoni A, et al. , GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia, Blood 134 (2019) 935–945. [DOI] [PubMed] [Google Scholar]
- [17].Gonen M, Sun Z, Figueroa ME, et al. , CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900, Blood 120 (2012) 2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Buccisano F, Maurillo L, Del Principe MI, et al. , Prognostic and therapeutic implications of minimal residual disease detection in acute myeloid leukemia, Blood 119 (2012) 332–341. [DOI] [PubMed] [Google Scholar]
- [19].Ivey A, Hills RK, Simpson MA, et al. , Assessment of minimal residual disease in standard-risk AML, N. Engl. J. Med 374 (2016) 422–433. [DOI] [PubMed] [Google Scholar]
- [20].Forghieri F, Comoli P, Marasca R, Potenza L, Luppi M, Minimal/measurable residual disease monitoring in NPM1-mutated acute myeloid leukemia: a clinical viewpoint and perspectives, Int. J. Mol. Sci 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rothenberg-Thurley M, Amler S, Goerlich D, et al. , Persistence of pre-leukemic clones during first remission and risk of relapse in acute myeloid leukemia, Leukemia 32 (2018) 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Press RD, Eickelberg G, Froman A, et al. , Next-generation sequencing-defined minimal residual disease before stem cell transplantation predicts acute myeloid leukemia relapse, Am. J. Hematol 94 (2019) 902–912. [DOI] [PubMed] [Google Scholar]
- [23].Rucker FG, Agrawal M, Corbacioglu A, et al. , Measurable residual disease monitoring in acute myeloid leukemia with t(8;21)(q22;q22.1): results from the AML Study Group, Blood 134 (2019) 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ediriwickrema A, Aleshin A, Reiter JG, et al. , Single-cell mutational profiling enhances the clinical evaluation of AML MRD, Blood Adv. 4 (2020) 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dillon R, Potter N, Freeman S, Russell N, How we use molecular minimal residual disease (MRD) testing in acute myeloid leukaemia (AML), Br. J. Haematol 193 (2021) 231–244. [DOI] [PubMed] [Google Scholar]
- [26].Fernandez HF, Sun Z, Yao X, et al. , Anthracydine dose intensification in acute myeloid leukemia, N. Engl. J. Med 361 (2009) 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zeijlemaker W, Kelder A, Cloos J, Schuurhuis GJ, Immunophenotypic detection of measurable residual (Stem Cell) disease using LAIP approach in acute myeloid leukemia, Curr. Protoc. Cytom 91 (2019), e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rossi G, Giambra V, Minervini MM, et al. , Leukemia-associated immunophenotypes subdivided in "categories of specificity" improve the sensitivity of minimal residual disease in predicting relapse in acute myeloid leukemia, Cytometry B Clin. Cytom 98 (2020) 216–225. [DOI] [PubMed] [Google Scholar]
- [29].Paietta E, Surrogate marker profiles for genetic lesions in acute leukemias, Best Pract. Res. Clin. Haematol 23 (2010) 359–368. [DOI] [PubMed] [Google Scholar]
- [30].Paietta E, Minimal residual disease in acute leukemia: a guide to precision medicine ready for prime time? Treat. Strateg. Haematol 4 (2014) 45–48. [Google Scholar]
- [31].Slovak ML, Kopecky KJ, Cassileth PA, et al. , Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a southwest oncology group/eastern cooperative oncology group study, Blood 96 (2000) 4075–4083. [PubMed] [Google Scholar]
- [32].Vance GH, Kim H, Hicks GA, et al. , Utility of interphase FISH to stratify patients into cytogenetic risk categories at diagnosis of AML in an Eastern Cooperative Oncology Group (ECOG) clinical trial (El900), Leuk. Res 31 (2007) 605–609. [DOI] [PubMed] [Google Scholar]
- [33].Baslan T, Kendall J, Ward B, et al. , Optimizing sparse sequencing of single cells for highly multiplex copy number profiling, Genome Res. 25 (2015) 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Navin N, Kendall J, Troge J, et al. , Tumour evolution inferred by single-cell sequencing, Nature 472 (2011) 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guryanova OA, Shank K, Spitzer B, et al. , DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling, Nat. Med 22 (2016) 1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Berry DA, Zhou S, Higley H, et al. , Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis, JAMA Oncol. 3 (2017), e170580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen X, Wood BL, Monitoring minimal residual disease in acute leukemia: technical challenges and interpretive complexities, Blood Rev. 31 (2017) 63–75. [DOI] [PubMed] [Google Scholar]
- [38].Bassan R, Bruggemann M, Raddiffe HS, Hartfield E, Kreuzbauer G, Wetten S, A systematic literature review and meta-analysis of minimal residual disease as a prognostic indicator in adult B-cell acute lymphoblastic leukemia, Haematologica 104 (2019) 2028–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rubnitz JE, Inaba H, Dahl G, et al. , Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial, Lancet Oncol. 11 (2010) 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bewersdorf JP, Shallis RM, Boddu PC, et al. , The minimal that kills: why defining and targeting measurable residual disease is the "Sine Qua Non" for further progress in management of acute myeloid leukemia, Blood Rev. 43 (2020), 100650. [DOI] [PubMed] [Google Scholar]
- [41].Moffitt AB, Spector MS, Andrews P, et al. , Multiplex accurate sensitive quantitation (MASQ) with application to minimal residual disease in acute myeloid leukemia, Nucleic Acids Res. 48 (2020), e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Godwin CD, Zhou Y, Othus M, et al. , Acute myeloid leukemia measurable residual disease detection by flow cytometry in peripheral blood vs bone marrow, Blood 137 (2021) 569–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nagler A, Labopin M, Canaani J, et al. , Cytogenetic risk score maintains its prognostic significance in AML patients with detectable measurable residual disease undergoing transplantation in remission: on behalf of the acute leukemia working party of the European society for blood and marrow transplantation, Am. J. Hematol 95 (2020) 1135–1141. [DOI] [PubMed] [Google Scholar]
- [44].Thol F, Gabdoulline R, Liebich A, et al. , Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML, Blood 132 (2018) 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Patkar N, Kakirde C, Bhanshe P, et al. , Utility of immunophenotypic measurable residual disease in adult acute myeloid leukemia-real-world context, Front. Oncol 9 (2019) 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hasserjian RP, Steensma DP, Graubert TA, Ebert BL, Clonal hematopoiesis and measurable residual disease assessment in acute myeloid leukemia, Blood 135 (2020) 1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Baslan T, Hicks J, Unravelling biology and shifting paradigms in cancer with single-cell sequencing, Nat. Rev. Cancer 17 (2017) 557–569. [DOI] [PubMed] [Google Scholar]
- [48].Miles LA, Bowman RL, Merlinsky TR, et al. , Single-cell mutation analysis of clonal evolution in myeloid malignancies, Nature 587 (2020) 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Terwijn M, Feller N, van Rhenen A, et al. , Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic blasts is predictive for outcome and level of residual disease in AML, Eur. J. Cancer 45 (2009) 1692–1699. [DOI] [PubMed] [Google Scholar]
- [50].Saito Y, Kitamura H, Hijikata A, et al. , Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells, Sci. Transl. Med 2 (2010) (17ra9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Herrmann H, Sadovnik I, Eisenwort G, et al. , Delineation of target expression profiles in CD34+/CD38− and CD34+/CD38+ stem and progenitor cells in AML and CML, Blood Adv. 4 (2020) 5118–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.