Abstract
The hyaluronic acid capsule of Streptococcus uberis has been implicated in conferring resistance to phagocytosis by bovine neutrophils. Construction of a bank of random insertion mutants of S. uberis (strain 0140J) was achieved using the pGh9::ISS1 mutagenesis system (22). Phenotypic screening of approximately 5,000 clones enabled the isolation of 11 acapsular mutants. Southern hybridization indicated that two mutants carried a lesion within a group of genes similar to those involved in the assembly of the hyaluronic acid capsule found in the group A Streptococcus (GAS) has operon. The DNA sequence flanking the points of insertion confirmed the presence of homologues of GAS hasA and hasB in S. uberis. The DNA sequence flanking the ISS1 insertion in another mutant identified a homologue of hasC in S. uberis. The GAS hasABC operon structure was not conserved in S. uberis, and two discrete loci comprising homologues of either hasAB or hasC were identified. Disruption of S. uberis hasA or hasC resulted in the complete cessation of hyaluronic acid capsule production. Correspondingly, these mutants were found to have lost their resistance to phagocytosis by bovine neutrophils. The bactericidal action of bovine neutrophils on S. uberis 0140J was shown unequivocally to depend upon the capsule status of the bacterium.
Streptococcus uberis is the causative agent of a significant proportion of clinical episodes of bovine mastitis worldwide (14a) and, in the United Kingdom, may be responsible for as many as 33% of cases (14). Phagocytosis and killing of bacteria by neutrophils constitute major defense mechanisms of the lactating mammary gland. This system is responsible for controlling Staphylococcus aureus and eliminating Escherichia coli infections within the bovine udder (13, 23). The role of phagocytes is less clear with regard to S. uberis infections. However, strains of S. uberis that resist phagocytosis in vitro can establish infection more effectively than susceptible strains (10, 19). The hyaluronic acid capsule appears to contribute significantly toward the virulence of Streptococcus pyogenes or group A Streptococcus (GAS) (16). Resistance to complement-mediated phagocytic killing was severely decreased in acapsular mutant GAS strains (25). Correspondingly, such mutants displayed greatly reduced virulence when tested in a mouse model (25). In addition, increased capsule production has been reported following reisolation of GAS from systemically infected mice, leading to the suggestion that survival is linked to increased capsule production (15).
A cluster of three genes (hasA, hasB, and hasC) which encode some of the proteins involved in the production of the hyaluronic acid capsule of S. pyogenes has been identified and extensively characterized. Hyaluronan synthase, the gene product of hasA, catalyzes the assembly of hyaluronic acid from N-acetylglucosamine and glucuronic acid (6, 7). hasB, the second gene in the operon, encodes UDP-glucose dehydrogenase (8), while hasC encodes UDP-glucose pyrophosphorylase (3), alternatively known as glucose-1-phosphate uridylyltransferase. The three has genes fall under the control of a single transcriptional promoter located upstream of hasA (4). Further enzymes involved in the synthesis of the hyaluronic acid capsule appear to be drawn from general biochemical pathways. Inactivation of hasC from a heavily encapsulated GAS strain had no effect upon capsule production, suggesting redundancy of the hasC gene product in S. pyogenes (2). Furthermore, the introduction of hasA and hasB into heterologous bacteria, such as Enterococcus faecalis and E. coli, was shown to be sufficient to produce a capsular phenotype, a finding that points toward the availability of alternative sources of UDP-glucose within these species (5).
We report here for the first time the generation of a bank of random mutants of S. uberis and the subsequent identification of genes responsible for capsule formation in a novel genomic arrangement. The effect that the disruption of these loci had upon the ability of S. uberis to resist phagocytosis by bovine neutrophils was determined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A bovine isolate of S. uberis 0140J and a human isolate of S. pyogenes 0358 were routinely grown in Todd-Hewitt broth at 37°C. Highly encapsulated S. uberis 0140J was grown in and harvested from chemically defined medium (CDM) containing casein hydrolysate as described previously (18). E. coli XL1-blue (Stratagene Ltd.) was used as a host strain for cloned DNA and was grown under standard conditions (22).
Generation of a bank of random insertion mutants of S. uberis.
The procedures described previously by Maguin et al. (20) were followed to generate a bank of random mutants. Transformation of S. uberis 0140J with plasmid pGh9::ISS1 was performed using a Gene Pulser apparatus (Bio-Rad) and 0.1-cm-path-length cuvettes. Parameters of 25 μF, 2.4 kV, and 100 Ω generated time constants of the order of 2 ms. Erythromycin-resistant transformants were selected following growth at 28°C. Chromosomal integration of pGh9::ISS1 was achieved by 100-fold dilution of an overnight culture of S. uberis 0140J/pGh9::ISS1 in Todd-Hewitt broth lacking erythromycin and incubation for 3.5 h at 28°C (to an optical density at 550 nm of 0.1). The culture was then transferred to 37.5°C for a further 2.5 h, after which glycerol was added to a final concentration of 15% and aliquots were stored at −70°C. The random nature of the mutant bank was verified using Southern analysis.
Screening of mutants for an acapsular phenotype.
Aliquots from the bank of random insertion mutants were diluted in Todd-Hewitt broth at 37°C and spread on prewarmed sheep blood agar containing esculin (1% [wt/vol]) at a density of approximately 100 CFU per plate. Plates were incubated at 37°C in a moist chamber to enhance the appearance of the capsular phenotype. Colonies exhibiting reduced capsule production were picked and restreaked to check for their acapsular appearance before biochemical characterization.
Capsular material was prepared from selected cultures grown under conditions that promoted capsule production in the wild-type isolate. Briefly, 4 ml of CDM with casein hydrolysate was inoculated with 0.5 ml of an overnight culture and incubated at 37°C for 5 to 6 h. Cells were recovered by centrifugation (5,000 × g for 5 min) and resuspended in 100 μl of phosphate-buffered saline, 2 μl of filter-sterilized bovine testicular hyaluronidase (10,000 U/ml; Sigma) was added, and the mixture was incubated at 37°C for 1 h. Following centrifugation (5,000 × g for 5 min), the supernatant (capsular extract) was recovered; the amount of N-acetylglucosamine was determined using the method of Reissig et al. (21) adapted for a microtiter plate format. Capsular extract (25 μl) was added to 5 μl of reagent A (6.1% [wt/vol] dipotassium tetraborate) and heated to 100°C for 3 min. Samples were cooled rapidly to room temperature, mixed with 150 μl of reagent B [10% (wt/vol) 4-(N,N-dimethylamino)-benzaldehyde dissolved in 1.5% (vol/vol) water; 11% (vol/vol) concentrated HCl; 87.5% (vol/vol) glacial acetic acid], diluted 1:9 with glacial acetic acid prior to use, and incubated at 37°C for 20 min. The absorbance at 595 nm was used to determine the quantity of N-acetylglucosamine present by comparison with a standard curve prepared using purified N-acetylglucosamine (Sigma).
Extraction of genomic DNA.
Streptococcal genomic DNA was prepared using a variation of the method of Hill and Leigh (12). Briefly, 1.5 ml of an overnight culture was centrifuged at 10,000 × g for 2 min, and the cell pellet was washed with 500 μl of 10 mM Tris-Cl–5 mM EDTA (pH 7.8). Bacterial cell walls were disrupted by resuspension in 375 μl of 10 mM Tris-Cl–5 mM EDTA (pH 7.8) containing 30 U of mutanolysin per ml and 10 mg of lysozyme per ml (both from Sigma) and subsequent incubation at 37°C for 30 min. Total cell lysis was achieved by the addition of 20 μl of sodium dodecyl sulfate (SDS) solution (20% [wt/vol] SDS in 50 mM Tris-Cl–20 mM EDTA [pH 7.8]) and proteinase K (Sigma) to a final concentration of 150 μg/ml and further incubation at 37°C for 1 h. Cell wall material was removed by precipitation following the addition of 200 μl of saturated NaCl and subsequent centrifugation at 12,000 × g for 10 min. The supernatant was extracted with phenol-chloroform, and DNA was precipitated by the addition of 2 volumes of absolute ethanol. DNA pellets were washed with 70% aqueous ethanol and air dried prior to resuspension in TE buffer containing 20 μg of RNase A (Sigma) per ml.
DNA amplification and further analysis.
Amplification of DNA was performed using an Omn-E thermal cycler (Hybaid Ltd.) with the conditions and primers listed in Table 1. Digoxigenin (DIG)-labeled probe DNA was generated by thermal cycling using DIG DNA labeling mix (Roche Molecular Biochemicals). Southern analysis of 11 selected mutant clones (designated TRF0-x) was performed by standard procedures (22). Genomic DNA isolated from selected clones was digested using HindIII and hybridized at a reduced stringency (50°C) to a DIG-labeled hasA probe amplified from S. pyogenes 0358. Filters were washed with SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 50°C. DNA sequencing was performed by Cambridge Bioscience, Cambridge, United Kingdom. Sequence analysis was performed using Wisconsin Package software, version 9.1 (Genetics Computer Group, Madison, Wis.).
TABLE 1.
Oligonucleotide primer sequences and their applications
| Designation | Sequence | Application | Template | Annealing temp (°C) |
|---|---|---|---|---|
| ISS1 fora | 5′-GGAGAGAATGGGTTCTGTTGC | ISS1 probe | pGh9::ISS1 | 60 |
| ISS1 reva | 5′-GCTCTAGAGCATTCTCTGGTTC | |||
| P050 | 5′-GTTATCGTTCACCGTTCCC | S. pyogenes hasA probe | S. pyogenes genomic DNA | 47 |
| P052 | 5′-TACGTGTTCCCCATTCCG | |||
| P064a | 5′-AGAACCGAAGAATTCGAACGCTC | Amplification of S. uberis DNA and sequencing primers | Wild-type and mutant S. uberis 0140J genomic DNAs | 50 |
| P067b | 5′-GGTGGAGCGTGCTGCTCA | |||
| P070 | 5′-CAAAACGAATAGATGTATCAATCC | |||
| P082a | 5′-CCAACAGCGACAATAATCACATC | |||
| P084c | 5′-CAGTCCCGACGGTGTAAAACG | |||
| P085c | 5′-GCTAAAGAGGTCCCTAGACTCT |
Primers complementary to ISS1.
A primer complementary to S. pyogenes hasA.
Primers complementary to the pGh9 vector.
Cloning and identification of flanking sequences.
Cloning of the DNA sequence flanking the point of insertion from mutant TRF0-6 was performed as described previously (20). Briefly, genomic DNA from mutant TRF0-6 was cut to completion with restriction endonuclease HindIII to generate a fragment containing the pGh replicon, erythromycin resistance marker, ISS1, and flanking genomic sequence (Fig. 1). The fragment was recircularized by ligation, transformed into E. coli host strain XL1-blue, and selected on Luria-Bertani agar containing erythromycin (100 μg/ml) at 28°C. Plasmid DNA containing the flanking sequence was isolated, and an EcoRI/HindIII fragment consisting of ISS1 and the flanking S. uberis genomic DNA was subcloned into pBluescript II (Stratagene) to facilitate sequence analysis of the cloned DNA.
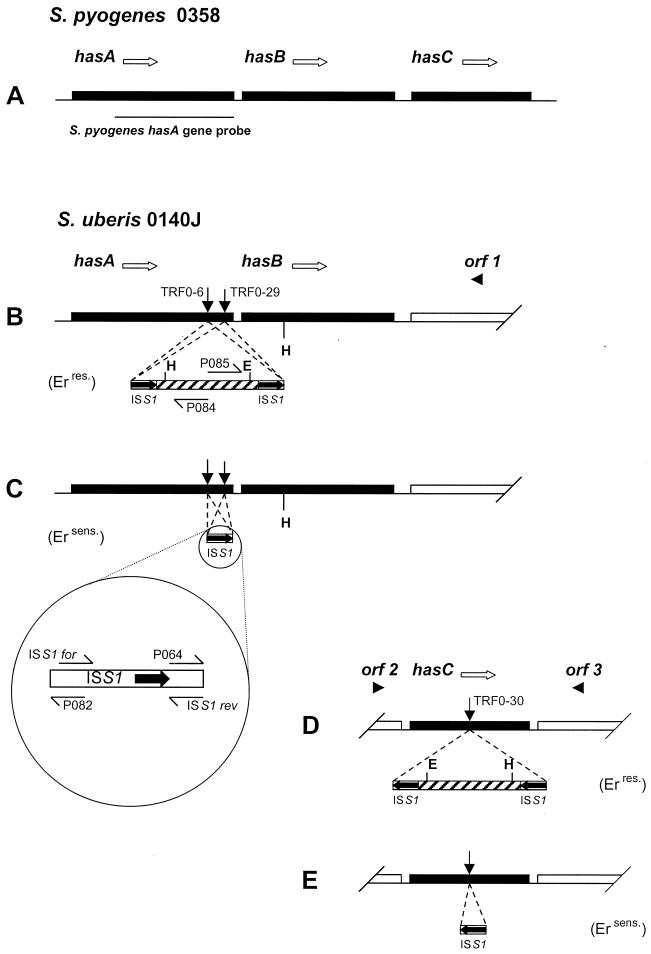
FIG. 1.
Schematic representation of the has locus in S. pyogenes (A), and the homologous regions in S. uberis 0140J (B to E). Open arrows represent the orientations of has genes. Filled arrows indicate the locations and orientations of ISS1-encoded transposase (not shown to scale). Hatched bars represent the pGh9 vector sequence (also not shown to scale). Filled arrowheads indicate the orientation of putative open reading frames (orf). ISS1 insertions were mapped to the points indicated by vertical arrows in strains TRF0-6, TRF0-29, and TRF0-30. (B and D) Arrangements before plasmid excision. (C and E) Arrangements after plasmid excision. Partial and horizontal arrows denote key oligonucleotide primer locations (Table 1). H and E, HindIII and EcoRI restriction sites used in the cloning of flanking sequences, respectively. Erres., erythromycin resistant; Ersens., erythromycin sensitive.
Identification of flanking sequences from other mutants was achieved by one of two methods. Prior to vector excision (see below), a template generated by self-ligation of HindIII- or EcoRI-digested chromosomal DNA was subjected to inverse PCR using primers complementary to the pGh9 vector sequence. For HindIII-generated templates, primer pair P064-P084 (Table 1) was used, while EcoRI-generated templates were amplified using primers P082 and P085 (Table 1).
Alternatively, following vector excision (see below), Vectorette (Sigma-Genosys) libraries were constructed using genomic DNA. These were subjected to PCR using Vectorette, ISS1 (P064 or P082), or additional chromosomal walking primers as described in the text (Table 1). Sequencing reactions were performed directly with DNA amplified by inverse PCR and with DNA obtained from Vectorette libraries following purification using DNA purification kit II spin columns (Hybaid).
Stabilization of insertion mutants.
The plasmid vector sequence was excised from the chromosome of selected mutant strains by permitting plasmid replication during overnight growth in broth cultures at 28°C without antibiotic selection. Broth cultures were diluted, and plate counts were determined with selective and nonselective solid media at the nonpermissive temperature. Excision of the vector sequence from the bacterial chromosome was indicated by loss of the erythromycin resistance marker. Replica plating enabled the isolation of stabilized mutant strains.
Bactericidal assay.
The resistance of S. uberis 0140J to the bactericidal action of bovine neutrophils was determined as described previously (18). Briefly, 0.1 ml of bacterial suspension (6 × 108 CFU/ml) was mixed with 0.2 ml of neutrophils (107 cells/ml) and 0.3 ml of normal pooled bovine serum (20% [vol/vol] in Hanks balanced salt solution). The mixture was rolled at 120 rpm and 37°C for 90 min. Numbers of viable bacteria remaining were determined by plating dilutions of the mixture on esculin-blood agar plates and incubating the plates overnight at 37°C. Percent survival was determined by expressing the number of bacteria remaining after 90 min as a percentage of those present at the start. Data were collected in triplicate from two independent broth cultures of each strain used (n = 6).
Nucleotide sequence accession numbers.
The nucleotide sequences of the regions containing the S. uberis hasAB and hasC homologues have been submitted to the EMBL nucleotide database under accession numbers AJ242946 and AJ400707, respectively.
RESULTS
Generation of a bank of random insertion mutants of S. uberis.
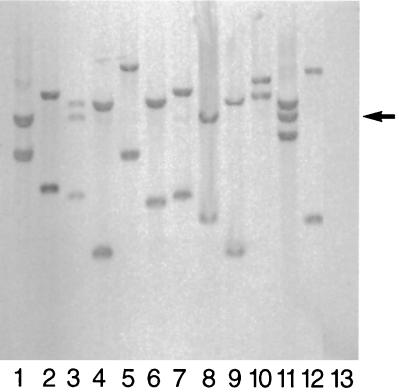
The temperature-sensitive plasmid pGh9::ISS1, consisting of a temperature-sensitive pG+host replicon, a lactococcal insertion sequence, ISS1, and an erythromycin resistance marker (20), was introduced into S. uberis 0140J by electroporation. Chromosomal integration of pGh9::ISS1 was achieved by subsequent growth at the nonpermissive temperature. The level of integration was determined to be 1.1% by comparison of plate counts on solid media at 37°C both with erythromycin (to select for integrants) and without erythromycin (to determine total viability). The random nature of integration was ascertained by Southern analysis of HindIII-digested genomic DNA prepared from 12 randomly selected integrants (Fig. 2). ISS1 was shown to be present in all 12 DNA preparations and absent from the parental strain 0140J. In 10 of the integrants, single copies of the integrated plasmid flanked on either side by ISS1 occupied different chromosomal locations. The remaining two preparations contained tandem copies of the vector and ISS1 (i.e., a minimum of two copies of pGh9, each flanked by ISS1, inserted in a single locus), again situated at different chromosomal locations.
FIG. 2.
Southern analysis of integrants picked randomly from the S. uberis 0140J ISS1 mutant bank. Genomic DNAs from 12 mutant clones (lanes 1 to 12) and the parent strain 0140J (lane 13) were digested using HindIII and probed with DIG-labeled ISS1. Two clones (lanes 3 and 11) showed three bands, one of which corresponded to a 4.6-kb linearized pGh9::ISS1 fragment (arrow) characteristic of tandem pGh9::ISS1 integration.
Isolation of acapsular 0140J mutants.
A primary screening of random 0140J mutants was carried out on the basis of altered colony morphology following growth on solid media in a moist environment. A total of 190 colonies exhibiting a nonmucoid phenotype were selected from a total number of approximately 5,000. The quantity of N-acetylglucosamine released from the selected mutants following hyaluronidase treatment was compared with that released from the parent strain when the bacteria were grown under conditions that promoted capsule production. Eleven of the mutants were found to have significantly reduced amounts (≤6%) of hyaluronidase-released N-acetylglucosamine compared with the parent strain (data not shown). These mutants were used in subsequent experiments.
Mapping of insertions to a homologue of S. pyogenes hasA.
Genomic DNA was isolated from the 11 acapsular mutants, and Southern hybridization was carried out at reduced stringency using a hasA probe amplified from S. pyogenes strain 0358 (Fig. 1A). Comparison of the banding pattern produced by parent strain 0140J with those produced by the mutant strains showed a common band present in all but two of the samples (TRF0-6 and TRF0-29) (data not shown). Cloning of genomic DNA located immediately 3′ of the point of insertion in mutant strain TRF0-6 was facilitated by restriction digestion with HindIII and rescue of a plasmid consisting of the pGh vector, ISS1, and flanking genomic DNA up to the point where the same restriction site occurred in the chromosome (Fig. 1B). The sequence adjacent to the ISS1 element displayed significant homology (68% identity) to the 3′ end of S. pyogenes hasA. A further open reading frame that displayed 63% identity to S. pyogenes hasB was also identified within the same fragment.
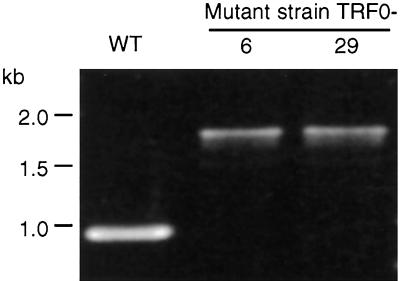
Further analysis of DNA flanking the ISS1 sequence was carried out using chromosomal DNA prepared from derivatives of the TRF0-6 and TRF0-29 clones from which the pGh plasmid had been excised (see below). Genomic DNA located 5′ of ISS1 in mutant TRF0-6 was amplified using oligonucleotide primers complementary to S. pyogenes hasA (P067) and the 5′ end of ISS1 (P082) and was subjected to sequence analysis. Further amplification reactions using templates from both mutants (TRF0-6 and TRF0-29) and primers complimentary to S. pyogenes hasA (P067) and the S. uberis-derived hasA homologue (P070) generated products that spanned the ISS1 elements and thereby enabled the insertion in TRF0-29 to be mapped to the same region (Fig. 3). Sequence analysis showed that the insertion points in mutants TRF0-6 and TRF0-29 were spaced 169 and 83 bp, respectively, from the 3′ end of the predicted S. uberis hasA coding sequence. The orientation of ISS1 (encoding transposase) was the same as that of the hasA genes in both mutant TRF0-6 and mutant TRF0-29 (Fig. 1B and C). Two 8-bp transposition target sequences, 5′-ATTATTTT from mutant TRF0-6 and 5′-GATACATC from mutant TRF0-29, were duplicated accurately and appeared to be flanking either end of the ISS1 elements, as described previously (20).
FIG. 3.
PCR amplification products from the 3′ end of the S. uberis homologue of hasA demonstrating band shifts due to the insertion of ISS1 in mutants TRF0-6 and TRF0-29, compared to the wild-type (WT) strain 0140J.
The full coding sequence of S. uberis hasA and hasB was determined using chromosomal walking techniques (1). Comparison of DNA amplified from this region with that amplified from the parent strain also confirmed that single copies of the hasA and hasB homologues were present in S. uberis (Fig. 3). The predicted hasA homologue-ISS1 chimeric translation products of mutants TRF0-6 and TRF0-29 included 7 and 12 ISS1-encoded amino acids, respectively, before termination was achieved (Table 2). The genomic DNA sequence located further 3′ of S. uberis hasB showed no homology to hasC. A putative open reading frame (orf 1) to which no function in capsule biosynthesis could be attributed was identified in this region (Fig. 1B).
TABLE 2.
Putative translation products for mutant S. uberis hasA and hasC homologues
| Strain | Gene | Predicted amino acid sequencea |
|---|---|---|
| TRF0-6 | hasA | … LVAFLVIILV LLQSFLISLF * |
| TRF0-29 | hasA | … LVAFLVIIFI VALCRNVHYM VKHPFAFLLS PFYGLIHRFC CKVF** |
| TRF0-30 | hasC | … DDLMDITNTV LLQSLKIKYK VYNPSCS* |
ISS1-encoded amino acids are denoted by underlining. An asterisk denotes a stop codon.
Identification of a discrete locus encoding a homologue of S. pyogenes hasC.
Inverse PCR and chromosomal walking techniques were used to determine DNA sequences flanking the points of ISS1 insertion in the remaining nine acapsular mutant strains. One such sequence (TRF0-30) displayed high homology (77%) to GAS hasC. In this strain, the ISS1 insertion point was mapped to a central location, 417 bp from the 5′ end of the predicted S. uberis hasC coding sequence. The orientation of the insertion cassette was opposite that of the hasC homologue in mutant TRF0-30 (Fig. 1D and E). The predicted hasC homologue-ISS1 chimeric translation product of mutant TRF0-30 included 18 ISS1-encoded amino acids before termination was achieved (Table 2). However, the flanking sequence was not consistent with that of a GAS hasABC operon. Indeed, a search of the EMBL nucleotide sequence database suggested a high degree of homology not only to the coding sequence but also to a downstream flanking sequence of a putative open reading frame (orf 3; Fig. 1D) encoding Streptococcus mutans glucose-1-phosphate uridylyltransferase. Additionally, a further putative open reading frame (orf 2; Fig. 1D) identified upstream of the S. uberis hasC gene displayed homology to the glycerol-3-phosphate dehydrogenase genes from Streptococcus pneumoniae and Bacillus subtilis. The remaining mutants had ISS1 insertions located in either novel sequences or regions for which no direct link to capsule biosynthesis could be made.
Stabilization of ISS1-generated mutant strains by excision of the vector DNA sequence.
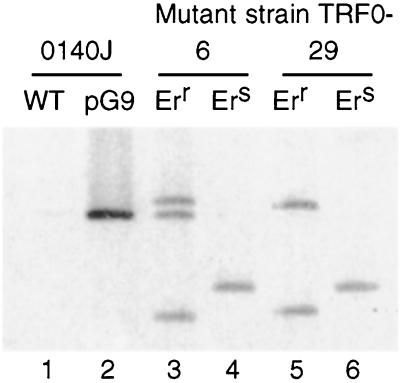
Vector excision through recombination between insertion sequence elements located on either side of the chromosomally integrated plasmid sequence was promoted by growth of selected mutants in broth cultures at the permissive temperature without antibiotic selection. The efficiency of the excision process, as indicated by the loss of an antibiotic resistance marker, varied widely across the 11 mutant strains tested. Six of these became sensitive to erythromycin after a maximum of two cycles of growth at 28°C and another strain became sensitive after four cycles. However, two mutants required 13 cycles of growth before losing the marker, while another two strains remained resistant to the antibiotic after 16 cycles. Southern hybridization of genomic DNA using a probe amplified from ISS1 was used to confirm the loss of the vector sequence and the retention of a single copy of ISS1 within the chromosomal DNAs of mutants TRF0-6 and TRF0-29 (Fig. 4) and mutant TRF0-30 (data not shown). Throughout the study, comparisons of growth, capsule production, and resistance to phagocytosis were made between mutant strains carrying the fully integrated plasmid sequence and their respective derivative strains, i.e., those from which the vector DNA had been excised. The observed phenotypes of these strains showed no differences (Table 3), further supporting the utility of the pGh9::ISS1 mutagenesis system.
FIG. 4.
Southern analysis of S. uberis 0140J mutants TRF0-6 and TRF0-29. Genomic DNA was digested with HindIII and hybridized with a DIG-labeled ISS1 probe at 65°C. Lane 1, wild type (WT); lane 2, wild-type 0140J transformed with pGh9::ISS1 and grown at 28°C (pG9); lanes 3 and 4, mutant TRF0-6 before (Err) and after (Ers) excision of the plasmid vector sequence, respectively; lanes 5 and 6, mutant TRF0-29 before (Err) and after (Ers) excision of the plasmid vector sequence, respectively. Lane 2 shows the linearized preintergrated free-plasmid form of pGh9::ISS1. Lanes 3 and 5 show that mutant TRF0-6 contained a tandem insertion, whereas mutant TRF0-29 had a single copy of pGh9::ISS1 inserted in the chromosome.
TABLE 3.
Resistance to phagocytosis and capsular status of mutants TRF0-6, TRF0-29, and TRF0-30 compared to wild-type S. uberis 0140J grown in CDM containing casein hydrolysate
| Straina | Mean (SD)b
|
|
|---|---|---|
| % Survivalc | μg of capsuled | |
| Wild-type 0140J | 103.02 (15.21) | 145.775 (27.89) |
| TRF0-6 Err | 0.118 (0.06) | 0.325 (0.26) |
| TRF0-6 Ers | 0.440 (0.12) | 0.625 (0.46) |
| TRF0-29 Err | 0.250 (0.15) | 0.750 (0.31) |
| TRF0-29 Ers | 0.135 (0.05) | 0.700 (0.37) |
| Wild-type 0140J | 71.83 (33.67) | 122.35 (18.02)e |
| TRF0-30 Err | 0.16 (0.06) | 1.29 (2.00)e |
| TRF0-30 Ers | 0.22 (0.02) | 1.63 (1.54)e |
Mutant strains were tested before (Err) and after (Ers) pGh plasmid excision.
Unless otherwise indicated, data from triplicate samples from two independently grown broth cultures were pooled (n = 6).
Resistance to phagocytosis is shown as the percent survival of cells relative to cells at time zero.
Capsule status is shown as the amount of hyaluronidase-released N-acetylglucosamine per 1010 bacteria.
Data from quadruplicate samples from two independently grown cultures were pooled (n = 8).
Sequence and operon structure of has genes are only partly conserved between S. pyogenes WF50 and S. uberis 0140J.
The predicted protein sequence encoded by the S. uberis hasA homologue displays 71% identity with the S. pyogenes hasA gene product, 75% identity with the Streptococcus equisimilis hasA gene product, but no significant homology to the hyaluronan synthase of Pasteurella multocida. Sequence analysis confirmed the presence of a gene homologous to S. pyogenes hasB (8) immediately 3′ of the putative S. uberis hasA coding sequence. The S. uberis hasB homologue gene product displays high levels of homology with numerous UDP-glucose dehydrogenases listed in the database, specifically, 65% identity with the S. pyogenes hasB gene product and 57% identity with S. pneumoniae UDP-glucose dehydrogenase. No homologue of hasC was found immediately 3′ of S. uberis hasB. Similarly, it was not possible to attribute a putative function to the open reading frame (orf 1; Fig. 1A) identified in this region. The predicted protein sequence of the S. uberis hasC homologue displays 89% identity with the S. pyogenes hasC gene product, 87% identity with the S. pneumoniae galU gene product (UDP-glucose pyrophosphorylase), and 85% identity with S. mutans glucose-1-phosphate uridylyltransferase (encoded by gluA).
Resistance to phagocytosis of selected acapsular 0140J mutants.
The ability of acapsular mutants displaying reduced amounts of hyaluronidase-released N-acetylglucosamine to resist the bactericidal action of bovine neutrophils was determined and compared with that of the parent strain. All strains were grown under conditions that promoted capsule production as described previously (18a). Cultures of the parent strain were highly encapsulated and displayed a high degree of phagocytic resistance compared with those of mutants TRF0-6, TRF0-29, and TRF0-30 grown under similar conditions (Table 3).
DISCUSSION
Attempts to define the pathogenic mechanisms of S. uberis at the genetic level have been hampered by the lack of reliable molecular tools to enable the comparison of mutants against an isogenic background. Poor transformation frequencies (typically ≤104 CFU/μg) of S. uberis and a lack of suitable plasmid vectors have contributed to this situation. A highly efficient (>0.5% insertional frequency) transposition mutagenesis system based on the delivery of ISS1 by the thermosensitive plasmid pG+host, developed for lactococci (20), was applied to S. uberis. This system was found to be efficient, and it provided mechanisms for the further analysis of mutants at the genetic level.
The bank of random mutants was generated readily in S. uberis 0140J with a transposition frequency of 1.1%. This frequency compares favorably with reported transposition frequencies of >0.5% in Lactococcus lactis (20) and 0.5 to 1.8% in Streptococcus agalactiae (24). An assessment of the overall content of the mutant bank with respect to the number and type of insertions was made using Southern analysis in combination with an ISS1 probe. Both this screening and subsequent independent screenings of the mutant bank (data not shown) suggested that tandem insertional events at a particular locus occurred at a frequency of about 10%, while no instances of individual clones containing insertions at multiple loci were found. If the latter situation arose, Southern analysis would enable its detection. This method represents a significant advantage over some of the more traditional random mutagenesis procedures. Attempts to generate similar mutant banks in strains of Streptococcus suis and Streptococcus dysgalactiae using pGh9::ISS1 were not successful due to transposition occurring during growth at the permissive temperature for plasmid replication (28°C). This situation resulted in apparently elevated chromosomal integration frequencies ranging from 25 to 90%. Southern analysis of individual mutants picked randomly from the S. suis and S. dysgalactiae preparations with the ISS1 probe showed common banding patterns suggestive of a nonrandom distribution (P. N. Ward et al., unpublished data).
The utility of this versatile tool was supported further by the finding that S. uberis mutants containing the fully integrated pGh9::ISS1 insertion showed no phenotypic differences from their vector-lacking derivatives, apart from resistance to erythromycin. With the exception of lethal mutations, the mutant bank appeared to represent a comprehensive coverage of the S. uberis genome. The following argument supports this claim. Given a probable genome size of ∼1.8 Mb (based upon the sizes of the S. pneumoniae and S. pyogenes genomes) and a target of approximately 3 kb of has-related coding sequence, an unbiased distribution would generate 1 has mutant in every 600. Thus, the phenotypic identification of three different acapsular has mutants from a primary screening of 5,000 colonies is consistent with the theoretical expected value of 8.3 after the possibilities of genetic redundancy and silent mutations are taken into consideration.
Screening of approximately 5,000 mutants yielded 11 phenotypically acapsular mutants, mutations of which 1 mapped to a homologue of GAS hasC, while mutations of a further 2 mapped to within 1 kb of each other in a different locus containing the genetic homologue of GAS hasA. The finding that a further open reading frame with a putative capsule-related role lay adjacent to the S. uberis hasA homologue was consistent with the arrangement found in S. pyogenes (4, 8). It was apparent from this study that the level of sequence divergence between the predicted gene products of hasA and hasB from S. uberis and their counterparts in S. pyogenes might well be utilized to identify common regions critical to the function of these enzymes. Furthermore, it is of interest that the S. uberis hasAB locus does not contain a homologue of hasC, particularly since the equivalent gene has been shown to be redundant with respect to capsulation in GAS (2). The finding that S. uberis hasC and a significant portion of its flanking sequence were homologous to a locus encoding glucose-1-phosphate uridylyltransferase in S. mutans prompted a search of the University of Oklahoma streptococcal genome sequence database for strain M1 GAS (http://dna1.chem.ou.edu/strep.html) using the WF51, T18 GAS hasC sequence (accession number U33452). An open reading frame displaying 99% nucleotide identity and 98% amino acid identity with hasC in M1 GAS was identified lying adjacent to hasA and hasB on the minus strand, as expected. Interestingly, another open reading frame, displaying 78% nucleotide identity (90% amino acid identity), was identified outside this region on the plus strand. It is therefore conceivable that the apparent redundancy of hasC suggested by inactivating targeted mutagenesis of GAS hasC lying within the has operon could be compensated for by the presence of this closely related gene lying elsewhere in the genome. Correspondingly, in this study, interruption of a hasC-like gene in S. uberis resulted in an acapsular phenotype, suggesting that only one hasC-like gene occurs in S. uberis and consequently that no such compensatory mechanism is available to this bacterium. We can speculate that some of the remaining eight acapsular mutants might have mutations that map proximal to the hasAB or hasC regions or that lie within regulatory elements controlling the expression of these capsule biosynthesis genes. Alternatively, some mutations may lie within other genes that contribute to the production of capsule precursors, such as UDP-N-acetylglucosamine. Flanking sequences from these mutants are currently under investigation.
An S. uberis hasAB gene fragment was used to investigate the distribution of these genes across a panel of 10 S. uberis strains. Seven of the strains tested were found to possess the hasAB gene fragment (data not shown); six of these exhibited a capsular phenotype. The acapsular strain found positive with the hasAB gene probe might be explained by the possibility of minor changes to the coding sequence, regulatory mutations governing the expression of hasAB, or a dysfunctional hasC gene. Correspondingly, the three strains found to lack hasAB-homologous DNA exhibited an acapsular phenotype. The resistance of S. uberis strains to phagocytosis by bovine neutrophils can be induced in vitro when the organism is grown in the presence of casein-derived peptides, an observation that is consistent with those made in vivo following experimental infection of the lactating mammary gland (11). Capsular strains of S. uberis appear to be resistant to phagocytosis, whereas acapsular strains are more readily phagocytosed and killed by bovine neutrophils (17). Interestingly, the amount of immunoglobulin bound to S. uberis was not affected by the presence of a capsular layer, contrary to the situation observed for E. coli and S. aureus (18). The question of whether an antiphagocytic component associated with the S. uberis capsule is responsible for the resistance phenotype observed has also been raised (9, 18a); however, gross morphology of neutrophils appears unchanged following exposure to capsular (phagocytosis-resistant) S. uberis (18). The induction of a highly encapsulated phenotype via nutritional factors also raises the possibility of the concomitant expression of additional cell surface-associated proteins that could influence the interaction of the bacterium with phagocytes.
The aim of this study was to determine the contribution that the capsule plays in conferring resistance to phagocytosis through the disruption of genes directly involved in the assembly of the hyaluronic acid capsule. The acapsular phenotype of S. uberis 0140J mutants TRF0-6, TRF0-29, and TRF0-30, which all displayed markedly reduced resistance to phagocytosis, is therefore considered likely to be due to the aberrant expression of gene products homologous to those of S. pyogenes hasA and hasC. This finding links the correlation of the production of the hyaluronic acid capsule with resistance to phagocytosis by neutrophils in this bacterium. However, the precise mechanism by which the capsule layer confers resistance to phagocytosis remains unclear.
ACKNOWLEDGMENT
We acknowledge funding from the Ministry of Agriculture, Fisheries and Food.
REFERENCES
- 1.Arnold C, Hodgson I J. Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl. 1991;1:39–42. doi: 10.1101/gr.1.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh C D, Alberti S, Wessels M R. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J Bacteriol. 1998;180:4955–4959. doi: 10.1128/jb.180.18.4955-4959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crater D L, Dougherty B A, van de Rijn I. Molecular characterization of hasC from an operon required for hyaluronic acid synthesis in group A streptococci. Demonstration of UDP-glucose pyrophosphorylase activity. J Biol Chem. 1995;270:28676–28680. doi: 10.1074/jbc.270.48.28676. [DOI] [PubMed] [Google Scholar]
- 4.Crater D L, van de Rijn I. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J Biol Chem. 1995;270:18452–18458. doi: 10.1074/jbc.270.31.18452. [DOI] [PubMed] [Google Scholar]
- 5.DeAngelis P L, Papaconstantinou J, Weigel P H. Isolation of a Streptococcus pyogenes gene locus that directs hyaluronan biosynthesis in acapsular mutants and in heterologous bacteria. J Biol Chem. 1993;268:14568–14571. [PubMed] [Google Scholar]
- 6.DeAngelis P L, Papaconstantinou J, Weigel P H. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993;268:19181–19184. [PubMed] [Google Scholar]
- 7.Dougherty B A, van de Rijn I. Molecular characterization of hasA from an operon required for hyaluronic acid synthesis in group A streptococci. J Biol Chem. 1994;269:169–175. [PubMed] [Google Scholar]
- 8.Dougherty B A, van de Rijn I. Molecular characterization of hasB from an operon required for hyaluronic acid synthesis in group A streptococci. Demonstration of UDP-glucose dehydrogenase activity. J Biol Chem. 1993;268:7118–7124. [PubMed] [Google Scholar]
- 9.Field T R, Norton P M, Bland A P, Leigh J A. Changes in bovine neutrophils induced by the capsule of Streptococcus uberis. Adv Exp Med Biol. 1997;418:957–960. doi: 10.1007/978-1-4899-1825-3_225. [DOI] [PubMed] [Google Scholar]
- 10.Hill A W. Pathogenicity of two strains of Streptococcus uberis infused into lactating and non-lactating bovine mammary glands. Res Vet Sci. 1988;45:400–404. [PubMed] [Google Scholar]
- 11.Hill A W, Finch J M, Field T R, Leigh J A. Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow. FEMS Immunol Med Microbiol. 1994;8:109–117. doi: 10.1111/j.1574-695X.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 12.Hill A W, Leigh J A. DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol Infect. 1989;103:165–171. doi: 10.1017/s0950268800030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill A W, Shears A L, Hibbitt K G. The elimination of serum-resistant Escherichia coli from experimentally infected single mammary glands of healthy cows. Res Vet Sci. 1978;25:89–93. [PubMed] [Google Scholar]
- 14.Hillerton J E, Shearn M F, Teverson R M, Langridge S, Booth J M. Effect of pre-milking teat dipping on clinical mastitis on dairy farms in England. J Dairy Res. 1993;60:31–41. doi: 10.1017/s0022029900027321. [DOI] [PubMed] [Google Scholar]
- 14a.Hogan J S, Smith K L. Proceedings of the Symposium on Udder Health Management for Environmental Streptococci. Arlington, Va: National Mastitis Council Inc; 1997. Occurrence of clinical and sub-clinical environmental streptococcal mastitis; pp. 36–41. [Google Scholar]
- 15.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson D R, Stevens D L, Kaplan E L. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J Infect Dis. 1992;166:374–382. doi: 10.1093/infdis/166.2.374. [DOI] [PubMed] [Google Scholar]
- 17.Leigh J A, Field T R. Killing of Streptococcus uberis by bovine neutrophils following growth in chemically defined media. Vet Res Commun. 1991;15:1–6. doi: 10.1007/BF00497784. [DOI] [PubMed] [Google Scholar]
- 18.Leigh J A, Field T R. Streptococcus uberis resists the bactericidal action of bovine neutrophils despite the presence of bound immunoglobulin. Infect Immun. 1994;62:1854–1859. doi: 10.1128/iai.62.5.1854-1859.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Leigh J A, Field T R. Pathogenic streptococci: present and future. Proceedings of the 12th Lancefield Symposium on Streptococci and Streptococcal Diseases. St. Petersburg, Russia: Lancer Publications; 1994. Streptococcus uberis resists phagocytosis despite the presence of bound immunoglobulin; pp. 196–197. [Google Scholar]
- 19.Leigh J A, Field T R, Williams M R. Two strains of Streptococcus uberis, of differing ability to cause clinical mastitis, differ in their ability to resist some host defence factors. Res Vet Sci. 1990;49:85–87. [PubMed] [Google Scholar]
- 20.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reissig J L, Strominger J L, Leloir L F. A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem. 1955;217:959–960. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schalm O W, Lasmanis J, Jain N C. Conversion of chronic staphylococcal mastitis to acute gangrenous mastitis after neutropenia in blood and bone marrow produced by an equine anti-bovine leukocyte serum. Am J Vet Res. 1976;37:885–890. [PubMed] [Google Scholar]
- 24.Spellerberg B, Pohl B, Haase G, Martin S, Weber-Heynemann J, Lutticken R. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J Bacteriol. 1999;181:3212–3219. doi: 10.1128/jb.181.10.3212-3219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessels M R, Goldberg J B, Moses A E, DiCesare T J. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect Immun. 1994;62:433–441. doi: 10.1128/iai.62.2.433-441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]