Abstract
Understanding the structure and structure-function relationships of membrane proteins is a fundamental problem in biomedical research. Given the difficulties inherent to performing mechanistic biochemical and biophysical studies of membrane proteins in vitro, we previously developed a facile HeLa cell-based cell-free expression (CFE) system that enables the efficient reconstitution of full-length (FL) functional inner nuclear membrane SUN proteins (i.e. SUN1 and SUN2) in supported lipid bilayers. Here, we provide evidence that suggests that the reconstitution of CFE-synthesized FL membrane proteins in supported lipid bilayers occurs primarily through the fusion of endoplasmic reticulum-derived microsomes present within our CFE reactions with our supported lipid bilayers. In addition, we demonstrate the ease with which our synthetic biology platform can be used to investigate the impact of the chemical environment on the ability of CFE-synthesized FL SUN proteins reconstituted in supported lipid bilayers to interact with the luminal domain of the KASH protein nesprin-2. Moreover, we use our platform to study the molecular requirements for the homo- and hetero-typic interactions between SUN1 and SUN2. Finally, we show that our platform can be used to simultaneously reconstitute three different CFE-synthesized FL membrane proteins in a single supported lipid bilayer. Overall, these results establish our HeLa cell-based CFE and supported lipid bilayer reconstitution platform as a powerful tool for performing mechanistic dissections of the oligomerization and function of FL membrane proteins in vitro. While our platform is not a substitute for cell-based studies, it does provide important mechanistic insights into the biology of difficult-to-study membrane proteins.
Keywords: Membrane protein reconstitution, SUN-KASH interaction, mammalian cell-free expression
Graphical Abstract
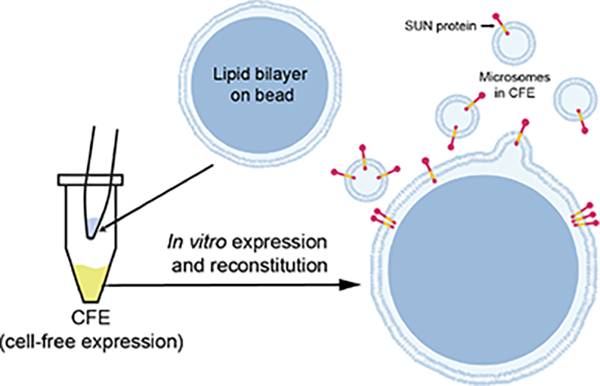
INTRODUCTION
A fundamental aspect of eukaryotic cellular biology is the establishment of membrane-enclosed organellar compartments, such as the endoplasmic reticulum (ER) and the nuclear envelope (NE). These compartments are critically important for the physical segregation of organelle-specific chemical reactions and signal transduction cascades as well as for their movement and positioning within cells. Integral and/or peripheral membrane proteins mediate many of these functions but they are notoriously difficult to study.
Because of their nature, membrane proteins possess hydrophobic surfaces, and they can only be extracted from cell membranes with detergents. Moreover, experimentally isolated membrane proteins are unstable. Consequently, working with full length (FL) membrane proteins is challenging at all levels, including their expression, solubilization, purification, and crystallization(Carpenter et al. 2008). To begin to address these challenges, we previously described the development of a HeLa cell-based cell-free expression (CFE) system that can be used as a simple “bottom-up” synthetic biology platform(Groaz et al. 2021; Sharma et al. 2021) for the rapid reconstitution of FL inner nuclear membrane (INM) Sad1/UNC-84 (SUN) proteins in supported lipid bilayers, which we called artificial nuclear membranes (ANMs)(Majumder et al. 2019).
We originally developed ANMs to be able to reconstitute and mechanistically dissect the assembly of linker of nucleoskeleton and cytoskeleton (LINC) complexes, which are conserved NE-spanning molecular bridges that mediate several fundamental cellular processes, including nuclear integrity, migration, and mechanotransduction(Alam et al. 2014; Chang, Worman, and Gundersen 2015; Tapley and Starr 2013). LINC complexes are formed by the direct transluminal interactions of SUN proteins with the outer nuclear membrane (ONM)-localized Klarsicht/ANC-1/SYNE homology (KASH) proteins(Crisp et al. 2006) (Fig. 1A). In vitro studies performed using the purified C-terminal luminal domains (LDs) of KASH (e.g. the KASH peptide) and SUN proteins revealed that LINC complex assembly depends on the ability of SUN proteins to homo-trimerize, which results in the formation of KASH peptide-binding sites at the interaction interfaces between SUN protein protomers(Sosa et al. 2012; Wang et al. 2012).
Figure 1. ER-derived microsomal structures and the association of FL SUN proteins with membrane fragments in HeLa cell lysates used for CFE.
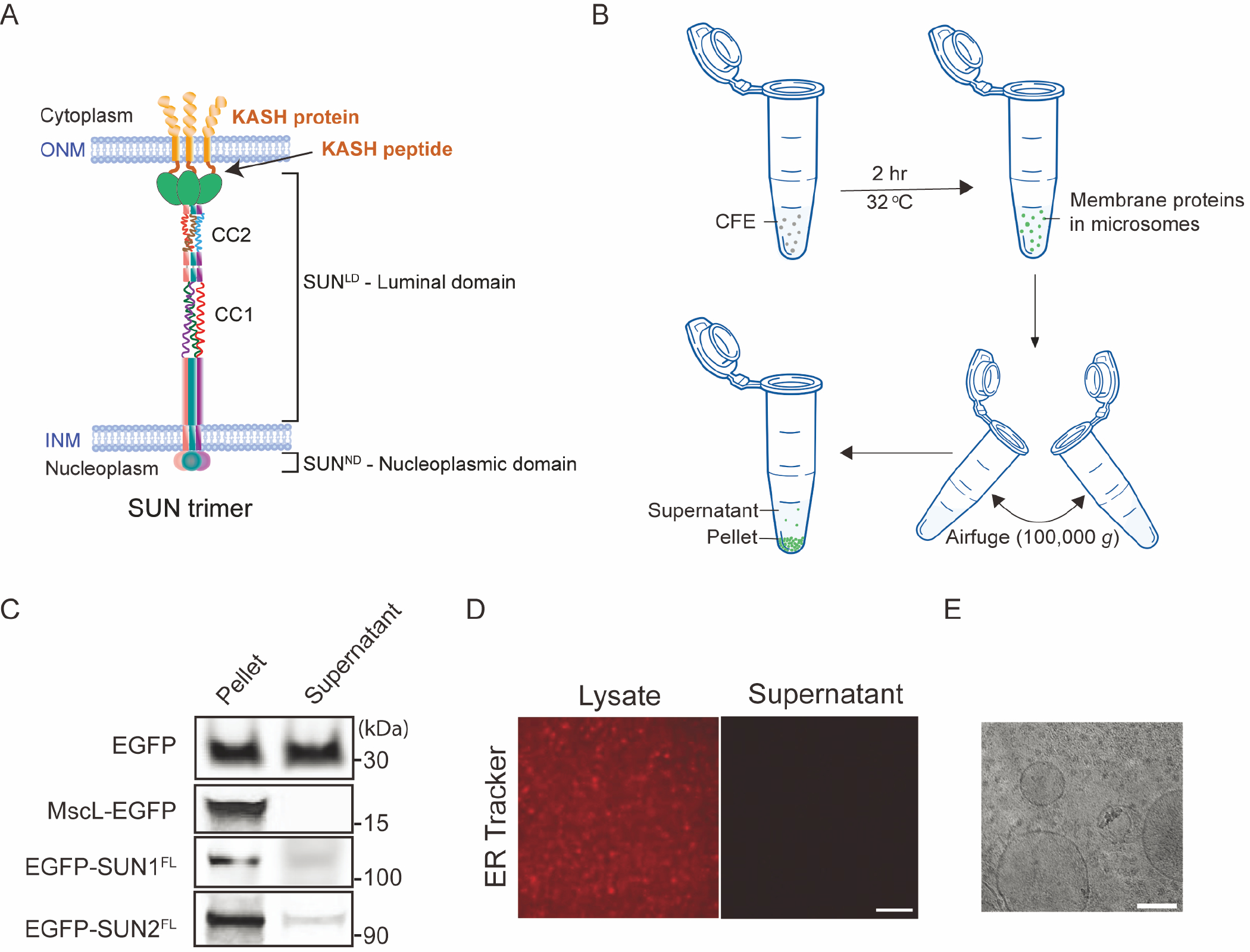
A) Illustration of a SUN2 homo-trimer embedded in the INM that is interacting with KASH proteins embedded in the ONM. B) Schematic illustrating the use of an Airfuge to spin down ER-derived microsomal membranes in bulk CFE reactions. C) Representative in-gel fluorescence images of EGFP and the indicated EGFP-tagged proteins in supernatant and pellet fractions post ultracentrifugation. A self-inserting mechanosensitive membrane channel MscL was also expressed and served as a positive control. D) Representative confocal fluorescence images of ER tracker-labeled HeLa lysate before and after spinning down microsomal membranes. Scale bar: 1 μm. E) Cryo-EM image of HeLa CFE system expressing EGFP-SUN1FL showing vesicular structures. Small structures are visible in the background indicating ribosomes (~ 30 nm). Scale bar: 100 nm.
SUN1 and SUN2 are the most ubiquitously expressed mammalian SUN proteins(Meinke and Schirmer 2015). We previously used ANMs to reconstitute CFE-synthesized N-terminally tagged FL mouse SUN1 and SUN2 (EGFP-SUN1FL and EGFP-SUN2FL) in supported lipid bilayers(Majumder et al. 2019). EGFP-SUN1FL and EGFP-SUN2FL were expressed in the presence of supported lipid bilayers with excess membrane reservoirs (SUPER) templates to mediate their spontaneous reconstitution. SUPER templates are formed on silica beads(Neumann, Pucadyil, and Schmid 2013), which provide a suitable substrate for the incorporation of membrane-spanning proteins with the added advantage of their ease of handling and imaging. By carrying out protease protection assays on ANMs containing reconstituted CFE-synthesized EGFP-SUN1FL or EGFP-SUN2FL, we found evidence for their directional insertion with their LDs facing the solvent accessible side of the SUPER templates. Based on the association of different truncated SUN protein constructs with SUPER templates, we also were able to predict that SUN2 possesses a single transmembrane domain and a hydrophobic membrane-associating domain, while SUN1 possesses three transmembrane domains. Furthermore, we demonstrated our ability to partially reconstitute LINC complex assembly through the interaction of reconstituted EGFP-SUN1FL or EGFP-SUN2FL with synthetic tetramethylrhodamine (TRITC)-labeled wild type (WT) KASH peptides from the mouse KASH protein nesprin-2 (TRITC-KASH2WT). Taken together, these results strongly suggest that our CFE-synthesized FL SUN proteins were directionally reconstituted in the ANMs such that their C-terminal KASH peptide-binding LDs were solvent-exposed and presumably homo-oligomerization competent.
Past studies identified the presence of ER fragments called microsomes in the lysates of Chinese hamster ovary cells used for CFE, which are translationally active and capable of the co-translational insertion of ER-targeted proteins in the microsomal membranes(Dondapati et al. 2019; Thoring and Kubick 2018; Zemella et al. 2019). Given that the orientation of SUN proteins reconstituted in ANMs was not random(Majumder et al. 2019), we hypothesized that the CFE reaction lysates contain ER-derived microsomes that are capable of the co-translational translocation of membrane proteins(Brödel, Wüstenhagen, and Kubick 2014). Consistent with this hypothesis, we previously observed that the lipophilic membrane dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) stained small vesicle-like punctae present within our CFE reactions(Majumder et al. 2019). These punctae exhibited EGFP fluorescence that was presumably produced by the CFE-synthesized EGFP-tagged FL SUN proteins present in these reactions. SUN proteins are known to have multiple regulatory elements at their N-termini, which result in them being targeted to the INM(Katta, Smoyer, and Jaspersen 2014; Laba, Steen, and Veenhoff 2014).. Thus, it is possible that a mammalian CFE system made from HeLa cells could possess ER fragments that can recognize and translocate SUN proteins. Nevertheless, the mechanism underlying the reconstitution of CFE-synthesized FL membrane proteins in SUPER templates requires further investigation.
Our ability to successfully reconstitute CFE-synthesized FL SUN proteins in SUPER templates motivated us to explore the generality of expressing other membrane proteins and the possibility of co-reconstituting multiple membrane proteins in supported lipid bilayers. Having established ANMs as a facile HeLa-based CFE system that enables the efficient reconstitution of FL, KASH-binding competent SUN proteins in supported lipid bilayers in vitro, we sought to use them to experimentally probe three proposed regulators of LINC complex assembly: calcium (Ca2+) ions; redox environment; and the coiled-coil (CC)-containing domains present within the SUN protein LDs. Below, we will briefly discuss what is known about how these three potential regulators influence the assembly of functional LINC complexes.
Ca2+ ions are thought to influence LINC complex assembly in two ways. First, structural studies revealed the presence of a well-defined cation loop in the SUN domain of SUN2 that surrounds and coordinates a bound cation via five backbone carbonyls(Sosa et al. 2012). Importantly, the purified LD of SUN2 lost its ability to interact with KASH peptides when the cation loop was shortened. This result shows that the cation loop, and by extension the cation coordinated by the loop, are important for LINC complex assembly. Second, molecular dynamics modeling predicts that a Ca2+ ion interacts with residue E452 in the first of two CC-containing domains (i.e. CC1) present within the LD of human SUN2(Jahed, Vu, et al. 2018). This residue was proposed to be involved in the monomer-trimer transition of the SUN2LD by mediating the association of the SUN domain with the CC-containing regions of the SUN2LD through Ca2+ ion-binding.
Regarding the potential effect of the redox environment of the NE lumen on LINC complex assembly, two disulfide bonds (DSBs) were detected in the previously reported crystal structure of a truncated LD construct of human SUN2 (amino acids 522-717) in complex with the KASH peptides of nesprin-1 or -2 (KASH1 and KASH2, respectively)(Sosa et al. 2012). While the intermolecular SUN-KASH DSB appeared to be dispensable for the physical interaction of SUN2 and KASH1/2(Sosa et al. 2012), it was shown to be important for maximal mechanotransmission by SUN2 in silico(Jahed, Shams, and Mofrad 2015). While less is known about the role of DSBs during the assembly of SUN1-containing LINC complexes, the conserved intermolecular SUN-KASH disulfide bond-forming cysteine residue found in SUN2 is also present in SUN1(Sosa et al. 2012). In addition, the SUN1 tertiary structure appears to involve interchain SUN1LD-SUN1LD disulfide bonds that may promote the formation of higher-order homo-oligomeric complexes(Lu et al. 2008), such as those predicted in silico(Jahed, Fadavi, et al. 2018) and observed by fluorescence fluctuation spectroscopy within the NE of living cells(Hennen, Angert, et al. 2018; Hennen, Saunders, et al. 2018). Based on these results, DSBs may be potential targets for the regulation of LINC complex assembly and/or function. However, mechanistic experimental support for this hypothesis is currently lacking.
Lastly, the CC-containing domains present within the LD of SUN proteins may regulate LINC complex assembly by controlling their homo- and/or hetero-oligomerization. In vitro data suggests that the CC-containing domains found within the LDs of SUN proteins may act as intrinsic dynamic regulators of their homo-oligomeric state and therefore ability to interact with KASH proteins(Nie et al. 2016). The two CC-containing regions of the mouse SUN2LD were shown to adopt distinct oligomeric states in vitro in solution, with CC1 and CC2 being monomeric and trimeric, respectively. Furthermore, previous cell-based studies demonstrate that SUN1 and SUN2 can undergo hetero-oligomerization via a process that is mediated by both the SUN domain and the adjacent CC-containing regions(Lu et al. 2008; Stewart-Hutchinson et al. 2008; Wang et al. 2006; Xiong et al. 2008).
In this work, we further explore the mechanism underlying the reconstitution of CFE-synthesized FL membrane proteins in SUPER templates and present hypothesis for the observed membrane association of such proteins. This information is crucial for our ability to properly interpret the results of experiments performed using ANMs. We show that Ca2+ ions and oxidizing conditions enhance SUN-KASH interaction. We further find that CC domains are indispensable for homo and heterotypic interaction of SUN proteins. Our work also leverage our initial successes with the ANM platform to mediate the reconstitution of additional mammalian membrane proteins such as voltage-gated ion channels using the same approach with cell-free synthesis and appropriate choice of membrane lipids. The experimental approach described here provides a simple and powerful experimental platform for performing structure/function analyses of any membrane protein, nuclear or otherwise.
METHODS
Materials.
Chemicals.
All lipids were purchased from Avanti Polar Lipids. Other reagents used are previously described8. Dithiothreitol (DTT) was purchased from Sigma-Aldrich. FluoroTect™ Green Lys-tRNA was purchased from Promega. TRITC-KASH peptides were obtained as N-terminally covalently-conjugated peptides from Genscript with greater than 80% purity as previously indicated(Majumder et al. 2019).
Plasmids.
All EGFP tagged and untagged full-length mouse SUN protein plasmids (UniProtKB – Q9D666 and Q8BJS4) were used in our previous study(Majumder et al. 2019). All CC mutants were generated from untagged WT SUN1 and SUN2 constructs by PCR-mediated mutagenesis (NEB site directed mutagenesis kit) (Table S1).
Preparation of SUPER templates.
SUPER template beads were made following a previously published protocol64 and our prior work with SUN proteins(Majumder et al. 2019). ANM composition consists of 45% DOPC, 27% DOPE, 9% DOPS, 2.2% phosphatidic acid, and 16.8% cholesterol. For reconstitution of ion channels, porcine brain polar lipid extract with the composition of 12.6 % PC, 33.1% PE, 4.1% PI, 18.5% PS, 0.8% PA, and 30.9% unknown lipids were used.
CFE of SUN proteins.
A homemade HeLa CFE system was made based on previously published protocols(Lohia, Chatterjee, and Das 1984; Yuan et al. 2005). A typical reaction volume of 10 μl was assembled with 5 nM plasmid in a 1.5 mL microcentrifuge tube and incubated at 30°C for up to 3 hours. Most reactions reached saturation after 2 hours. For the standard reconstitution of SUN proteins, SUPER templates were added to the CFE reaction prior to incubation. When SUPER templates were added to completed CFE reactions, the mixture was incubated at room temperature for ~15 minutes with intermittent shaking prior to subsequent analysis. All reconstituted SUPER templates were washed with 2 ml 1X PBS with or without 6 M urea, as indicated. For fluorescent visualization of non-EGFP-tagged proteins, GreenLys was added to the CFE reaction at a dilution of 1:50 as recommended in the manufacturer’s protocol.
Ultracentrifugation with Airfuge.
An Airfuge (Beckman Coulter) was used to spin down CFE reactions. Completed CFE reactions were collected in a 1.5-mL microcentrifuge tube and then mixed with 30 μl of extraction buffer (20 mM HEPES-KOH, pH 7.5, 45 mM potassium acetate, 45 mM KCl, 1.8 mM magnesium acetate, 1 mM DTT). Next, 40 μl of the mixture was transferred to an ultracentrifuge tube and centrifuged at around 100,000 × g for 15 minutes at room temperature using the Airfuge (Beckman Coulter). After centrifugation, 20 μl of the supernatant were carefully recovered so as not to disturb the pellet and transferred to a 1.5-mL microcentrifuge tube. The remaining 20 μl of pellet fraction was resuspended by pipetting the sample up and down before being transferred to another microcentrifuge tube. To investigate protein incorporation, 2 μl of SUPER templates were added and incubated with the supernatant or pellet fractions for 30 minutes and then centrifuged at 300 × g for 3 minutes. The SUPER templates were visible as a small white pellet, which was gently resuspended by tapping for downstream analysis. For further fractionation as shown in Fig. 5A, after first centrifugation step, 20 μl of supernatant was carefully removed and transferred to a 1.5-mL microcentrifuge tube labelled as S1. Then, 20 μl extraction buffer was added to the remaining 20 μl of pellet fraction and the pellet was resuspended by gently pipetting up and down. Next, the 40 μl mixture was centrifuged again at around 100,000 × g for 15 minutes and 20 μl of supernatant was recovered and transferred to another 1.5-mL microcentrifuge tube labelled as S2. This process was repeated once more and 20 μl fractions of supernatant and pellet were collected as S3 and P, respectively. The protein incorporation was then assessed as described above.
Figure 5. Direct reconstitution of FL membrane proteins mediated by ER-derived microsome fusion is used for the reconstitution of different or multiple membrane proteins.
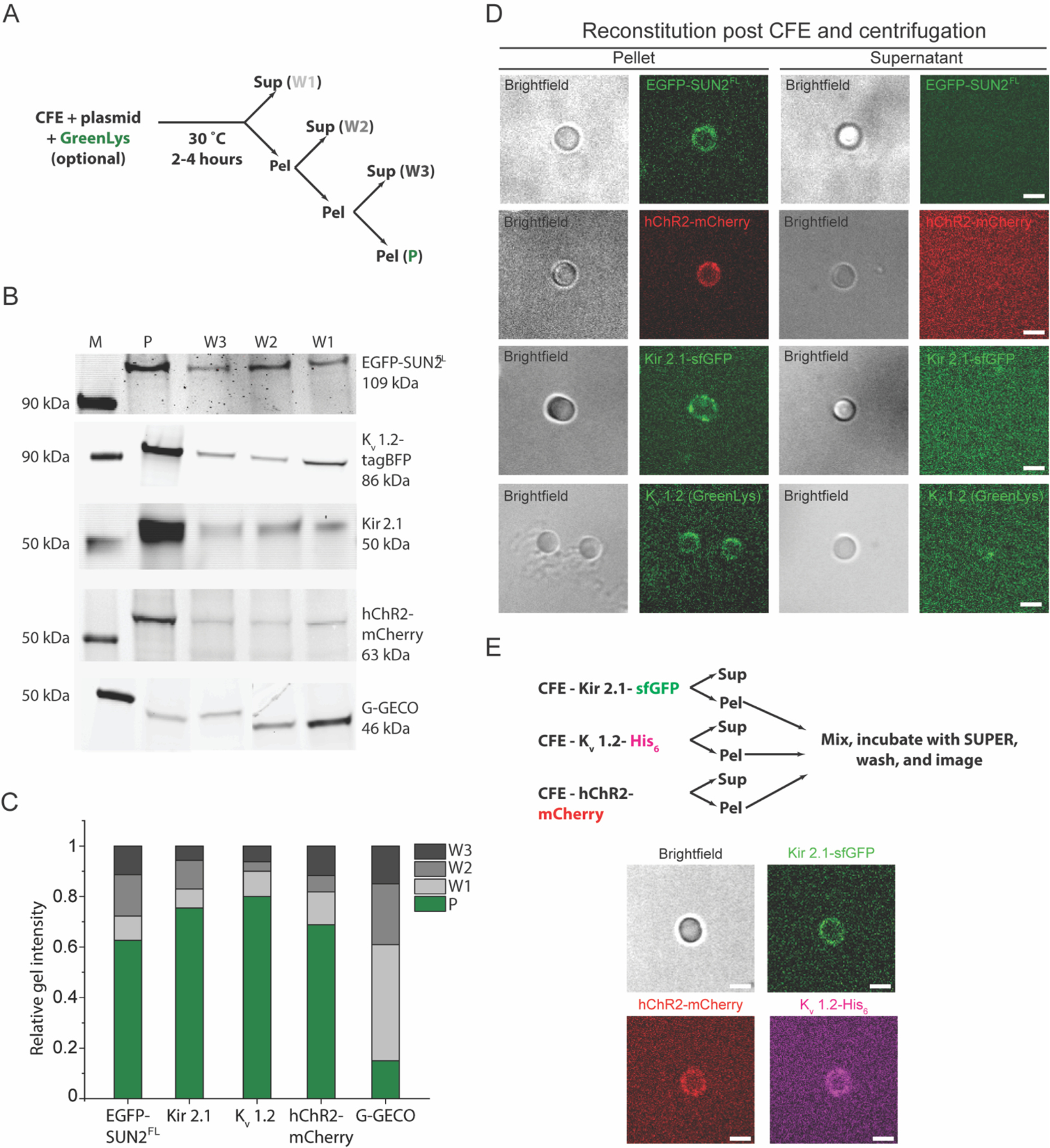
A) Illustration of the experimental workflow used in panels B-E of this figure. B) Representative in-gel fluorescence images of the pellet and supernatant fractions from the microsome enrichment experiment depicted in panel A for the indicated CFE synthesized and GreenLys-labeled protein constructs. Lane M indicates the protein ladder. C) Bar graph of the normalized band intensities quantified from the in-gel fluorescence images presented in panel B. For each protein, band intensities were normalized by the sum of intensities in all lanes for the corresponding protein. D) Representative brightfield and confocal fluorescence images of SUPER templates incubated in and isolated from the pellet (denoted as P in panel A) or the supernatant fraction from the first wash (denoted as W1 in panel A) from microsome enrichment experiments where the indicated membrane protein constructs were synthesized. Scale bars: 5 μm. E) Experimental design for multiple membrane protein reconstitution (top) and representative confocal fluorescence images of SUPER templates incubated with microsome-enriched fractions of completed CFE reactions that synthesized the indicated proteins (bottom). Scale bars: 5 μm.
Protease digestion assay.
Pronase (Roche Cat No.: 10165921001) was dissolved in de-ionized water at a stock concentration of 10 mg/ml. Next, 2 μl of this stock were added to 8 μl of a bulk CFE reaction and incubated at room temperature for 15–20 minutes. Thereafter, 2 μl of 25X Complete mini protease inhibitor cocktail (Millipore-Sigma Cat No.: 11836153001) were added to the digested CFE and incubated for 10 minutes. This mixture was subsequently analyzed by SDS-PAGE or Western blot.
TRITC-KASH2 peptide-binding assay.
Reconstituted SUN proteins on SUPER templates were washed with 1X PBS. The beads were resuspended in 30 μl of 1X PBS and distributed into 9 μl aliquots in 1.5 mL microcentrifuge tubes. To two aliquots, ~300 nM TRITC-KASHWT was added followed by addition of 1 mM (final concentration) calcium chloride (CaCl2) in one and an equal volume of de-ionized water in another. To the third aliquot, ~300 nM of TRITC-KASHΔPPPT were added followed by dilution with CaCl2 to a final concentration of 1 mM. The three tubes were then incubated at room temperature for 10 minutes before subsequent imaging. In order to investigate the effect of DTT on LINC complex assembly, 10 mM DTT (final concentration) was added to the sample after incubation of the TRITC-KASH2 peptide with the SUN protein-reconstituted SUPER templates. All normalization for the bar graphs were carried out with respect to the maximum TRITC intensities observed for the experimental conditions with added Ca2+ ions and 10mM DTT.
In-gel imaging and analysis and Western blots.
In-gel imaging of SUN proteins was carried out on a Sapphire biomolecular imager (Azure Biosystems) for both EGFP-tagged and GreenLys-labeled proteins. Samples were loaded on 4–20% gradient bis-Tris PAGE gels (Bio-Rad) with 4X Laemmeli loading buffer (Bio-Rad). Samples were not heated to retain in-gel EGFP and GreenLys fluorescence. The band intensities in each lane for every protein illustrated in Fig. 5C was measured using ImageJ gel analyzer tool and the intensity values were normalized by the sum of intensities for each protein. A rabbit polyclonal anti-GFP antibody at a dilution of 1:2000 (Abcam ab6556) was used to detect EGFP-tagged SUN mutants by Western blotting. For immunostaining his-tagged proteins in SUPER templates, a 1:2000 dilution of a mouse monoclonal anti-Penta-His antibody conjugated to Alexa Fluor 647 (Qiagen 35370) was used.
Confocal fluorescence microscopy and analysis.
Confocal images of SUPER templates were taken as previously described on a spinning disk confocal microscope and subsequent analysis was carried out using ImageJ as before8. Briefly, 5–10 line scans for each bead for the relevant scenario were taken and the peak intensities recorded. Subsequently, their mean was taken and combined with intensity values from 10–15 beads for background correction and statistical analysis. Pairwise student t-tests and one factor ANOVA for the plots obtained in Fig. 3 were carried out from background subtracted intensity values obtained using ImageJ. The values were then normalized by the maximum intensity of the group before plotting.
Figure 3: The SUN1/2-KASH2 interaction is enhanced by Ca2+ ions and stabilized by oxidizing conditions.
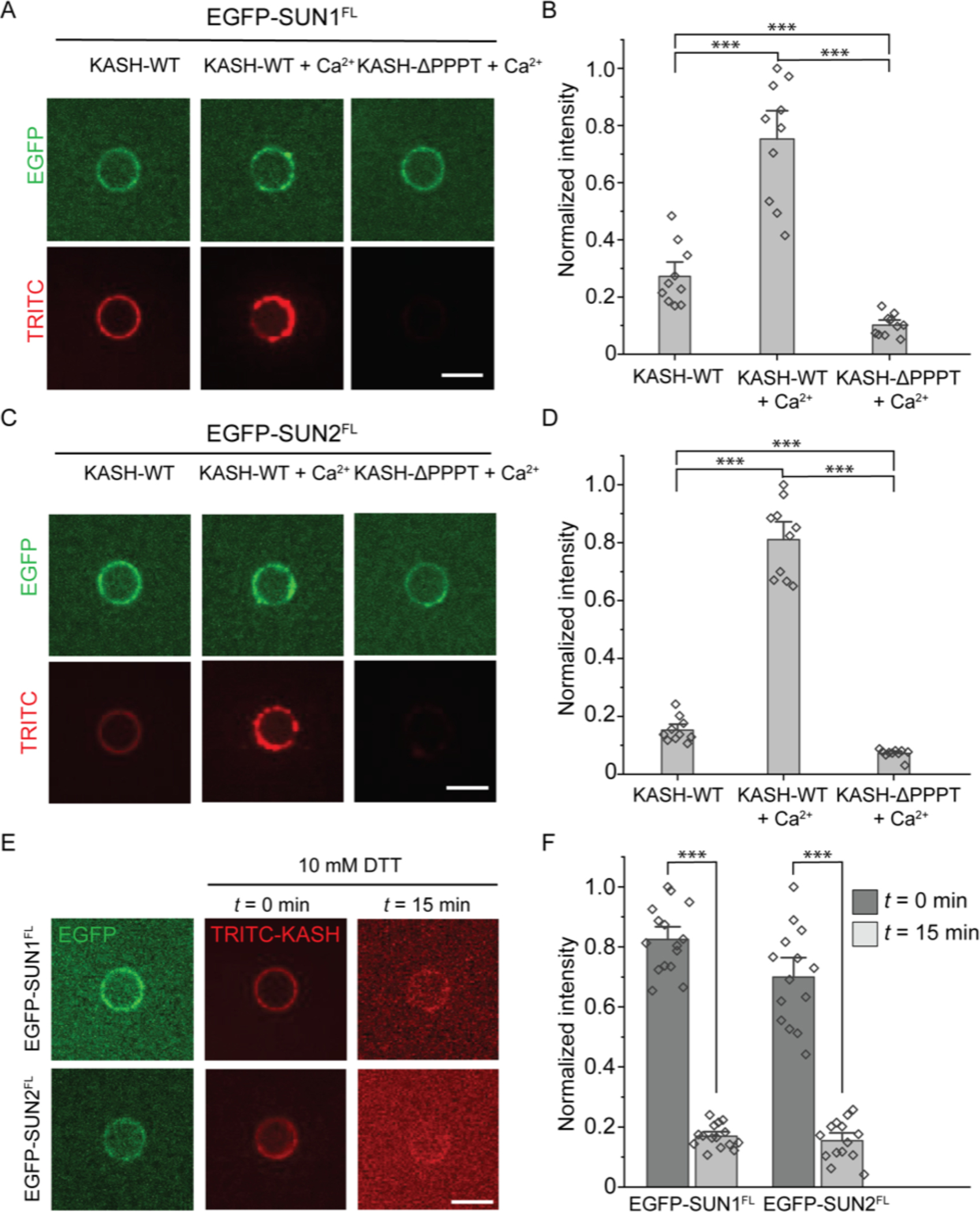
A and C) Representative confocal fluorescence images of SUPER templates containing the indicated reconstituted EGFP-tagged FL SUN protein constructs after incubation with the indicated TRITC-labeled KASH2 peptides in the presence and absence of 1 mM CaCl2. B and D) Bar graphs depicting the normalized intensity of the indicated TRITC-labeled KASH2 peptides recruited to the SUPER templates containing the indicated EGFP-tagged FL SUN protein constructs in the absence or presence of 1 mM CaCl2. E) Representative confocal fluorescence images of SUPER templates containing the indicated reconstituted EGFP-tagged FL SUN protein constructs following incubation with 10 mM DTT. F) Bar graph depicting the normalized intensity of the indicated TRITC-labeled KASH2 peptides recruited to the SUPER templates containing the indicated EGFP-tagged FL SUN protein constructs following incubation with 10 mM DTT. Scale bars: 5 μm. Error bars represent standard error of the mean. 10–15 beads were analyzed for each condition from n = 3 experiments. ***: p < 0.001 by pairwise Student’s t-test.
Cryo-electron microscopy (cryo-EM) grid preparation.
CFE reactions expressing EGFP-SUN1FL were ultracentrifuged as mentioned above and 3.5 μL of the pellet fraction were applied to glow discharged grids (Quantifoil R1.2/1.3 Cu 200 mesh, glow discharged 1 minute, 5 mA) and frozen by plunging into liquid ethane using an FEI Vitrobot Mark IV (100% humidity, temperature: 22°C or 4°C, blot force: 20, blot time: 4 seconds, wait time: 0 seconds). Grids were clipped and stored in liquid nitrogen until analyzed.
Cryo-EM.
Cryo-EM movies were collected using a Thermo Fisher Scientific Glacios cryo-electron microscope operating at 200 kV equipped with a Gatan K2 Summit direct electron detector at the University of Michigan Life Sciences Institute. Movies were collected at 45,000x magnification (pixel size - 0.980 Å) or 22,000x magnification (pixel size - 2.005 Å) using Leginon software package with exposure time of 8 seconds, frame time of 200 ms, and total dose of 63 e−/Å2 (Suloway et al. 2005). Movies were collected at a defocus of either −3.00 μm or −1.80 μm. Representative still images from movies were obtained using the Leginon software package.
RESULTS
FL SUN proteins localize to ER-derived microsomes in HeLa cell-based CFE reactions.
We previously investigated the reconstitution of CFE-synthesized FL SUN proteins in SUPER templates(Majumder et al. 2019). We observed that our EGFP- and 6x histidine (His6)-tagged FL SUN1 (EGFP-SUN1FL-His6; referred to as EGFP-SUN1FL from here out) and SUN2 (EGFP-SUN2FL-His6; referred to as EGFP-SUN2FL from here out) protein constructs were oriented in ANMs with solvent-exposed C-terminal KASH-binding SUN domains. We then demonstrated that both constructs retained the ability to interact with TRITC-KASH2WT peptide in vitro. These results led us to explore the mechanism underlying FL SUN protein reconstitution in ANMs. We did this by using a standard Airfuge-based ultracentrifugation workflow (Fig. 1B), where we spun down CFE reactions with saturated yields (i.e. reactions that reached completion) of EGFP-SUN1FL, EGFP-SUN2FL, or soluble EGFP and tested their presence in equal volumes of supernatant and pellet fractions by in-gel fluorescence imaging (Fig. 1C; Full gel images are shown in Fig. S1). Since the volume of our CFE reactions was small (~10 μl), no significant membrane pellet was visible after ultracentrifugation. However, EGFP fluorescence was clearly visible in the pellet fractions, whereas no signal was detected in the supernatant fractions for any of the samples except for the soluble EGFP construct-containing sample. Furthermore, the expression of the bacterial channel protein mechanosensitive channel of large conductance (MscL) that is capable of self-insertion into lipid membranes(Battle et al. 2009; Jacobs, Boyd, and Kamat 2019; Majumder and Liu 2018) was also enriched in the pellet relative to the supernatant (Fig. 1C).
Staining of the CFE reactions with the ER tracker dye boron-dipyrromethene (BODIPY™)-FL glibenclamide revealed the existence of dense punctate structures in the lysate that were absent in the supernatant following ultracentrifugation (Fig. 1D). The presence of microsomal structures in our CFE lysates is further supported by cryo-EM (Fig. 1E). Lipid bilayer vesicles in the size range of 50–200 nm were clearly visible in our homemade CFE lysates. The results presented above are consistent with the translocation of FL SUN proteins in ER-derived microsomes.
The direct reconstitution of CFE-synthesized FL SUN proteins in SUPER templates is mediated by endogenous ER-derived microsomal structures present in CFE reactions.
Given the presence of endogenous ER-derived microsomes in our HeLa cell-based CFE reactions, we sought to determine if we could enrich these reconstituted FL membrane proteins in ER-derived microsomes by ultracentrifugation. Thus, we developed our Airfuge-based fractionation assay further by washing the pellet fraction after each centrifugation step to remove any proteins that were not associated with microsomes. In this assay, we recovered half of the supernatant after each step and diluted the pellet fraction in a 1:1 volume ratio before centrifuging the mixture again. After enriching for the microsomal fraction, we investigated whether we could directly reconstitute FL membrane proteins by introducing the microsomal fraction to SUPER templates designed to partially mimic the lipid composition of the budding yeast NE with added cholesterol(Majumder et al. 2019; Van Meer, Voelker, and Feigenson 2008).
SUPER templates are made by mixing small unilamellar vesicles (SUVs) containing negatively charged lipids under conditions of high ionic strength with 5 μm diameter silica beads, which results in the fusion of the SUVs around the bead (Fig. 2A). This leads to the formation of supported lipid bilayers with larger areas than the surface of the silica bead such that a thin aqueous layer exists between the bead surface and the lipid bilayer(Pucadyil and Schmid 2010). This aqueous layer provides sufficient spacing between the bead and the lipid bilayer to accommodate membrane proteins with large soluble domains. In addition, fluorescence-based quantification previously demonstrated that the presence of excess membrane on SUPER templates minimizes the effects of substrate interaction(Pucadyil and Schmid 2010). Thus, SUPER templates serve as a superior model membrane system, as compared to tightly substrate-bound supported lipid bilayers, for FL SUN protein reconstitution.
Figure 2. CFE synthesized FL SUN proteins are incorporated into SUPER templates via a potential fusion-based mechanism.
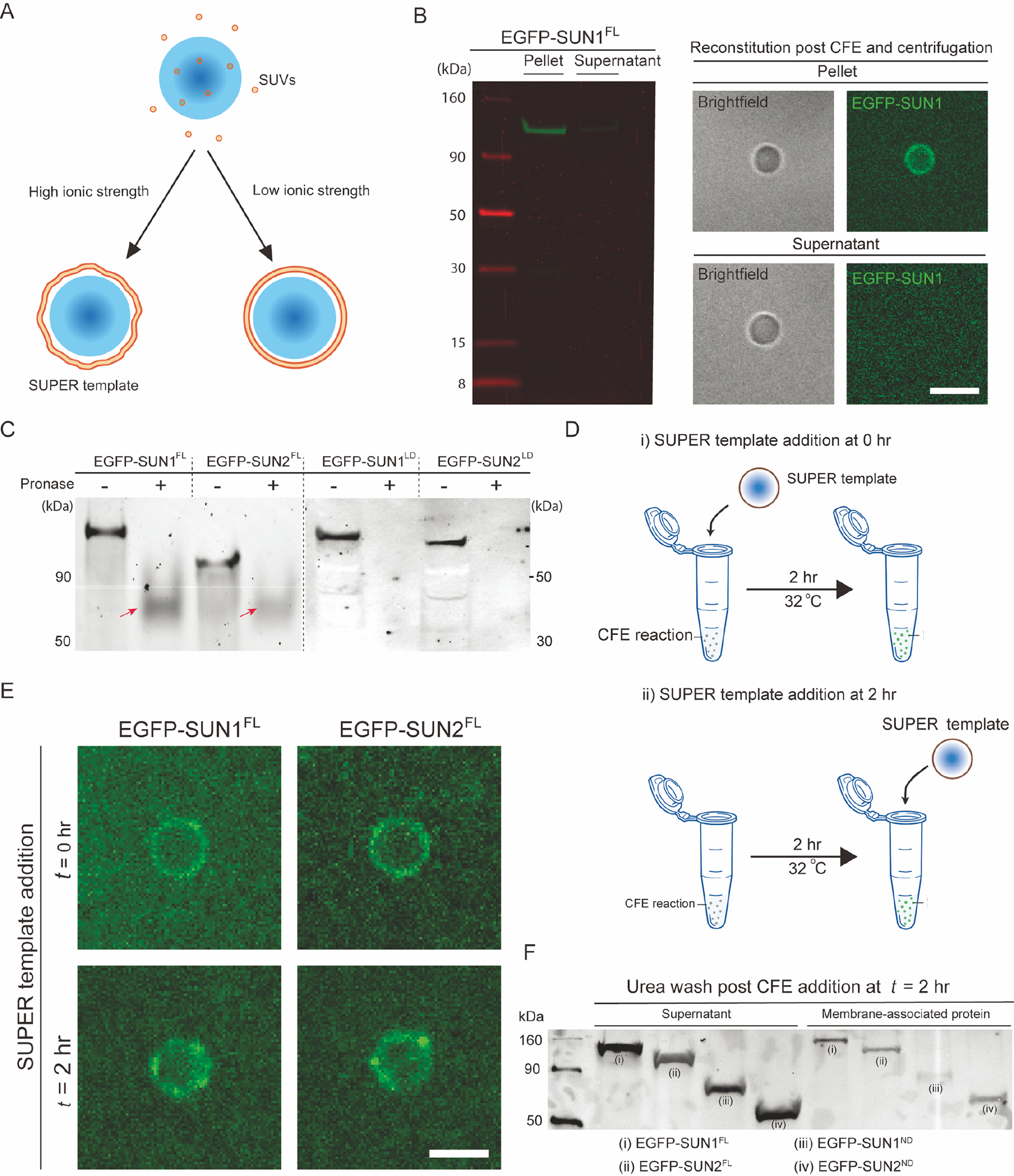
A) Schematic depicting the formation of supported lipid bilayers on silica beads in the presence or absence of low or high ionic strength buffers. As shown, SUPER templates are typically created when the fusion of SUVs results in the formation of loosely bound unilamellar membranes under high salt conditions. B) In-gel fluorescent image of pellet and supernatant fractions of a CFE reaction with SUN1-EGFP. The pellet and supernatant fractions were isolated using the Airfuge fractionation assay and subsequently added to SUPER templates for imaging (right) or directly run on gel (left). Scale bars: 5 μm. C) In-gel fluorescence image of CFE reactions expressing the indicated EGFP-tagged proteins that were or were not exposed to pronase. Arrows: Incompletely pronase-digested proteins. D) Schematic illustrating how the ability of EGFP-tagged FL SUN proteins to be reconstituted in SUPER templates added at the start of the CFE reaction (i) or 2 hours after its initiation (ii). The SUPER templates were washed in 1X PBS with 6M urea before imaging. E) Representative confocal fluorescent images of SUPER templates added to the indicated EGFP-tagged SUN protein CFE reactions at the indicated time points. Scale bars: 5 μm. F) Western blot of the bulk CFE supernatant and membrane-associated protein fractions of CFE reactions to which SUPER templates were added 2 hours after initiating the synthesis of the indicated EGFP-tagged SUN protein constructs, as described in panel D(ii). The supernatant and membrane-associated fractions represent SUN proteins in bulk CFE reactions and SUPER templates, respectively.
To ascertain the enrichment of FL SUN proteins in the CFE pellet fractions, we performed SDS-PAGE analysis and confirmed that the pellet fractions had significantly more EGFP-SUN1FL than the supernatant CFE fractions did after being washed (left panel, Fig. 2B). Consistent with our hypothesis that the direct reconstitution of CFE-synthesized mammalian membrane proteins on SUPER templates is mediated by endogenous ER-derived microsomal structures present in our CFE reactions, we observed EGFP fluorescence from the membranes of the SUPER templates that were incubated with the pellet fraction and not with the supernatant fraction of our centrifuged EGFP-SUN1FL CFE reaction (right panel, Fig. 2B). To control for non-specific interactions between the CFE-synthesized proteins and the SUPER templates, we asked if the soluble green fluorescent genetically encoded Ca2+ indicator for optical imaging (G-GECO)49 behaved similarly to the SUN1 and SUN2 constructs tested above. Indeed, the pellet fraction contained some amount of G-GECO from the residual supernatant buffer CFE fraction (Fig. S2), indicating that this assay simply dilutes non-microsome-associated proteins. These results suggest that FL SUN protein incorporation into the SUPER templates is likely mediated by the fusion of ER-derived microsomes with the supported lipid bilayers.
Fusion between ER-derived microsomes present in CFE reactions and SUPER templates enables the directional reconstitution of CFE-synthesized FL SUN proteins in these model membranes.
To confirm the membrane insertion of EGFP-SUN1FL and EGFP-SUN2FL in ER-derived microsomes following their synthesis in CFE reactions, we performed a protease protection assay followed by an in-gel fluorescence measurement. The correct folding and insertion of FL SUN proteins requires them to span a lipid bilayer with their N- and C-termini on opposite sides of the membrane. Given that our FL SUN1 and SUN2 constructs are N-terminally tagged with EGFP, we were able to probe for EGFP fluorescence post digestion with the protease pronase. If the N-terminal nucleoplasmic domain (ND) of EGFP-SUN1FL or EGFP-SUN2FL were solvent-exposed upon CFE synthesis, one would expect the EGFP to be degraded by the addition of pronase. As controls for pronase activity, we also generated in-gel fluorescence images of N-terminally EGFP-tagged constructs that encode the C-terminal LD of either SUN1 (EGFP-SUN1LD) or SUN2 (EGFP-SUN2LD) following their synthesis by CFE and exposure to protease (Fig. 2C; full gel images are shown in Fig. S3A).
In the absence of pronase (gel lanes labeled “−“; Fig. 2C), we observed fluorescent bands at the sizes predicted for our constructs (e.g. EGFP-SUN1FL: ~130 kD; EGFP-SUN2FL: 110 kD; EGFP-SUN1LD: ~80 kD; and EGFP-SUN2LD: ~80 kD). In contrast, after being digested with pronase (gel lanes labeled “+”; Fig. 2C), we no longer observed fluorescent bands at the same size as in our non-digested samples. Specifically, we detected the presence of much smaller bands (red arrows; Fig. 2C) for EGFP-SUN1FL or EGFP-SUN2FL and the complete absence of fluorescent bands for EGFP-SUN1LD or EGFP-SUN2LD (Fig. 2C). Based on these results, we can conclude that a fraction of our EGFP-tagged FL SUN proteins was successfully translocated into the ER-derived microsomes present in our CFE reactions with their N-terminal NDs being solvent-exposed.
We then asked how the topology of SUN proteins reconstituted in SUPER templates occurs in a unidirectional manner, given their translocation into ER-derived microsomes during their synthesis by CFE. One plausible hypothesis is that SUN proteins are directionally reconstituted in SUPER templates by the spontaneous fusion of ER-derived microsomal membranes with the supported lipid bilayers of the SUPER templates. Previous studies showed that cations and the electrostatic charges of lipids are key determinants of spontaneous vesicle fusion, especially between liposomes with high curvature and planar lipid bilayers(Bentz and Duzgiines 1985; Papahadjopoulos, Nir, and Düzgünes 1990). Since CFE reactions contain mM levels of potassium and magnesium ions and SUPER templates are made with charged lipids (i.e. phosphatidylserine) and lipids that mediate spontaneous fusion at room temperature (i.e. phosphatidylethanolamine)(Allen, Hong, and Papahadjopoulos 1990; Wilschut, Düzgüne, and Papahadjopoulos 1981), the mechanics of membrane fusion can ensure a consistent topology of reconstituted integral membrane proteins as opposed to direct membrane insertion.
To probe this potential mechanism of CFE-synthesized FL SUN protein reconstitution in SUPER templates, we compared the SUPER template localization of EGFP-SUN1FL and EGFP-SUN2FL under two conditions. In the first (Fig. 2Di), the SUPER template was added to the CFE reactions at the beginning of the reaction. In the second (Fig 2Dii), the SUPER template was added to the CFE reactions at the end of the reaction. If the EGFP-tagged FL SUN protein constructs were co-translationally inserted into the SUPER templates, a significant reduction in the levels of SUPER template-associated EGFP fluorescence would be expected for the second condition relative to the first. However, we observed a similar association of both EGFP-SUN1FL and EGFP-SUN2FL with the SUPER templates under both conditions (Fig. 2E). This result indicates that the reconstitution of CFE-synthesized FL SUN proteins in SUPER templates can be carried out by the addition of SUPER templates to saturated CFE reactions. While the direct post-translational insertion of FL SUN proteins in the SUPER templates may explain the lack of an observable difference between the two conditions described above, it is unlikely that SUN proteins with large soluble domains and multi-membrane-associating domains can self-insert into synthetic lipid bilayers while maintaining their specific membrane topologies. Rather, vesicle fusion is the most plausible mechanism of CFE-synthesized FL membrane protein reconstitution in SUPER templates.
To further explore the validity of vesicle fusion as the main mechanism underlying membrane protein reconstitution in SUPER templates, we tested the ability of the chaotropic agent urea to disrupt the SUPER template-association of EGFP-SUN1FL and EGFP-SUN2FL. We also tested the impact of urea on the SUPER template-association of two EGFP-tagged SUN protein ND constructs EGFP-SUN1ND and EGFP-SUN2ND, which we previously demonstrated were peripherally associated with the outer leaflets of SUPER templates(Majumder et al. 2019). Consistent with a urea wash being able to disrupt weak associations of proteins with membranes(Lohia et al. 1984; Yuan et al. 2005), we observed that all four of the constructs we tested exhibited strong interactions with the SUPER templates as demonstrated by their presence in the membrane-associated protein fractions (Fig. 2F). The supernatant fractions described here correspond to the bulk CFE reactions that were added to the SUPER templates at the maximal levels (i.e. completed reactions) of the indicated CFE-synthesized EGFP-tagged FL SUN protein constructs. Altogether, the results presented here suggest that our HeLa cell-based CFE system enables the translocation of FL membrane proteins into ER-derived microsomes that are subsequently directionally reconstituted in SUPER templates most likely through vesicle fusion.
Ca2+ ions and redox state are critical regulators of SUN-KASH interactions in vitro.
Armed with a better understanding of how CFE-synthesized FL SUN proteins are directionally reconstituted in SUPER templates, we next tested the impact of the chemical environment (e.g. Ca2+ ions and redox state) on the ability of FL SUN proteins to interact with KASH peptides. To test the potential role for Ca2+ ions in promoting LINC complex assembly, we took advantage of the negligible levels of Ca2+ ions present in our CFE reactions(Ho et al. 2017; Ho, Murray, and Liu 2015). More specifically, we asked if the addition of 1 mM CaCl2 positively or negatively impacted the ability of SUPER templates containing reconstituted CFE-synthesized EGFP-SUN1FL or EGFP-SUN2FL to recruit TRITC–KASH2WT. Similarly, we asked if either of these CFE-synthesized FL SUN protein-containing SUPER templates were impaired or enhanced in their ability to recruit synthetic TRITC-labeled nesprin-2-derived KASH peptides lacking the four C-terminal amino acids required for the SUN-KASH interaction to normally occur (TRITC-KASH2ΔPPPT)(Padmakumar et al. 2005). We observed that the addition of CaCl2 significantly increased the ability of ANMs containing reconstituted CFE-synthesized EGFP-SUN1FL or EGFP-SUN2FL to recruit TRITC-KASH2WT above standard binding levels measured in the absence of CaCl2 (Figs. 3A–D). In contrast, the recruitment of TRITC-KASH2ΔPPPT to ANMs harboring either CFE-synthesized EGFP-SUN1FL or EGFP-SUN2FL was significantly reduced relative to the recruitment of TRITC-KASH2WT to either type of ANM (Figs. 3A–D). Taken together, these results support the hypothesis that Ca2+ ions promote SUN-KASH interactions.
We next used this experimental platform to investigate the effect of redox state on LINC complex assembly. Specifically, we tested the effect of high concentrations (e.g. 10 mM) of the strong reducing agent dithiothreitol (DTT) on pre-formed SUN-KASH interactions using our in vitro experimental platform. 1–10 mM of DTT is commonly used to disrupt protein disulfide bonds. We found that exposing either CFE-synthesized EGFP-SUN1FL- or EGFP-SUN2FL-containing SUPER templates bound to TRITC-KASH2WT for 15 minutes significantly reduced the amount of bound TRITC-KASH2WT relative to the amount present at the start of the experiment by 4-fold (Figs. 3E–F). In contrast, no significant decrease in SUPER template-associated EGFP fluorescence was detected at the end of these experiments (data not shown). TRITC is covalently attached to KASH2WT, and neither its fluorescence nor stability were affected by DTT. Taken together, these results identify Ca2+ ions and redox state as key regulators of SUN-KASH interactions in vitro. They also demonstrate the ease with which our HeLa cell-based CFE and SUPER template system can be used to investigate mechanistic hypotheses regarding LINC complex assembly and regulation that may be difficult to address in cells.
SUN protein homo- and hetero-oligomerization is mediated by their luminal CC domains.
To further test the hypothesis that the CC domains of SUN proteins act as intrinsic dynamic regulators of SUN protein homo- and hetero-oligomerization, we asked if reconstituted CFE-synthesized FL SUN proteins directionally reconstituted in SUPER templates could form homo- or hetero-oligomers with soluble fragments of their LDs. To do this, we used CFE to synthesize SUN1FL and SUN2FL in the presence of the FluoroTect™ Green Lys in vitro translation labeling system (referred to as GreenLys from here on). GreenLys enables the fluorescent labeling and detection of in vitro synthesized proteins via a lysine-charged tRNA labeled with the fluorophore BODIPY™-FL at its ε position. The CFE-synthesized GreenLys-labeled FL SUN proteins were successfully reconstituted in SUPER templates (Fig. 4A). Next, we designed an assay to investigate the interaction of reconstituted unlabeled FL SUN proteins with their soluble counterparts (i.e. their CFE-synthesized LDs) (Fig. 4B), where the same SUPER templates were exposed to two rounds of CFE reactions expressing different proteins. The SUPER templates were washed with 1X PBS in between each step.
Figure 4: The CC-containing LDs are important for homo- and hetero-typic SUN protein interactions.
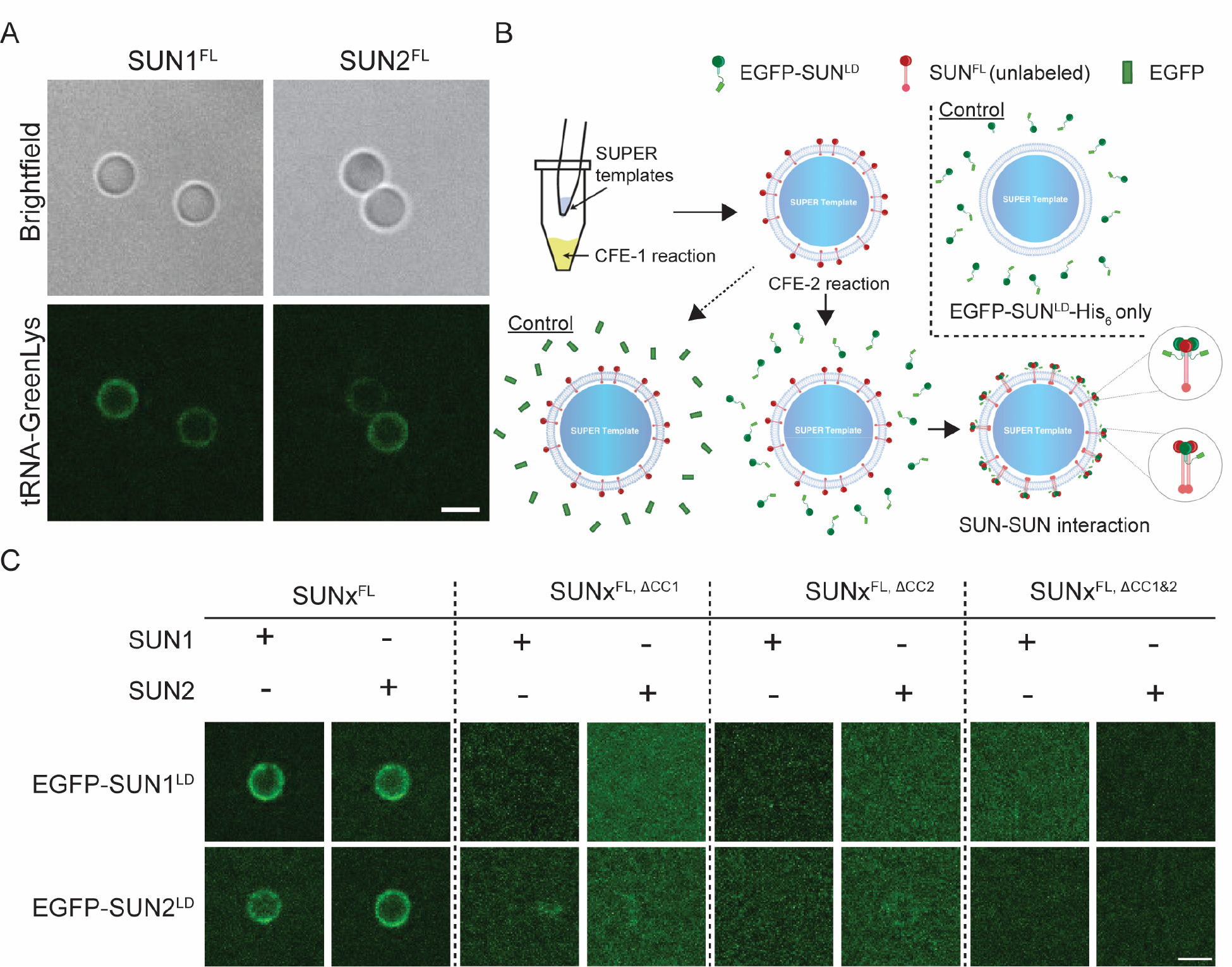
A) Representative confocal fluorescence images of SUPER templates containing the indicated reconstituted CFE synthesized and GreenLys-labeled FL SUN protein constructs. B) Schematic depicting the workflow for testing the ability of CFE synthesized EGFP-tagged SUN protein LD constructs to be recruited to SUPER templates containing reconstituted CFE synthesized unlabeled FL SUN protein constructs. To control for non-specific protein-protein interactions, the recruitment of soluble CFE synthesized EGFP to SUPER templates containing reconstituted CFE synthesized unlabeled FL SUN protein constructs was tested (bottom left). To control for non-specific interactions between the SUPER template and the CFE synthesized EGFP-tagged SUN protein constructs, the ability of these constructs to be recruited to SUPER templates lacking any reconstituted FL SUN protein constructs was tested (top right). C) Representative confocal fluorescence images of SUPER templates containing the indicated reconstituted CFE synthesized unlabeled FL SUN protein constructs and their ability to recruit the indicated CFE synthesized EGFP-tagged LD SUN protein constructs. Experiments were repeated 3 times. Scale bars: 5 μm.
In the first round of CFE and SUPER template reconstitution, CFE-synthesized unlabeled SUN1FL and SUN2FL were reconstituted in SUPER templates, which were then treated with a second round of CFE synthesizing either EGFP or EGFP-SUN1/2LD. Expressing EGFP-SUNLD without expressing SUNFL in the first CFE reaction served as a control (Fig. 4B). As we previously reported8, neither EGFP-SUN1LD nor EGFP-SUN2LD strongly associated with the SUPER templates in the absence of directionally reconstituted CFE-synthesized unlabeled SUN1FL or SUN2FL. Therefore, any EGFP fluorescence detected on the SUPER templates after the second round of CFE with EGFP-SUN1LD or EGFP-SUN2LD indicates the presence of an interaction between the reconstituted FL SUN protein and its corresponding soluble EGFP-tagged LD.
Additionally, given two independent rounds of CFE, it is possible to expose SUN1FL to EGFP-SUN2LD and SUN2FL to EGFP-SUN1LD, in order to study heteromeric SUN protein interactions. Thus, we created CC domain deletion mutants in which either or both of the CC domains were deleted (i.e. SUN1/2FL,ΔCC1, SUN1/2FL,ΔCC2, SUN1/2FL,ΔCC1&2). The membrane association and SUPER template reconstitution of each of the CFE-synthesized SUN protein CC deletion mutant constructs was tested using GreenLys-labeling as described above and all showed successful reconstitution (Fig. S4).
We carried out all combinations of the two CFE reactions with unlabeled FL WT SUN proteins or FL SUN protein mutants that lacked CC1, CC2, or both CC1 and CC2 (Fig. 4C). We found that the EGFP fluorescence detected after the second round of CFE of the EGFP-tagged SUN1LD or SUN2LD constructs was only observed on the SUPER templates when the CFE-synthesized WT SUN1FL or SUN2FL constructs were reconstituted in the first CFE reaction. The presence of EGFP fluorescence in this condition also confirms that EGFP-tagged SUNLD proteins are robustly synthesized by CFE, as expected(Majumder et al. 2019). CFE of soluble EGFP alone in the second round served as a control for the non-specific interaction of EGFP with the SUPER templates (Fig. S5). Hence, for reconstituted CFE-synthesized WT SUN1FL or SUN2FL, we detected robust homo- and hetero-typic interactions between their LDs. In the case of the reconstituted CFE-synthesized CC deletion SUN1 or SUN2 constructs, no significant fluorescence was observed on the SUPER templates for any of the experiments, indicating a lack of homo- or hetero-typic interactions between these SUN protein mutants and the EGFP-tagged LDs of SUN1 or SUN2. Collectively, our findings suggest that the deletion of either CC1 or CC2 from SUN1 or SUN2 is sufficient to disrupt both homo- and hetero-typic SUN protein interactions.
Three different CFE-synthesized FL membrane proteins can be simultaneously reconstituted in a single SUPER template.
Based on the results presented thus far, one might imagine that other mammalian membrane proteins with multiple membrane-spanning domains could be reconstituted using the same strategy undertaken for reconstitution of SUN proteins. Additionally, if the membrane protein reconstitution were mediated by microsome-fusion, multiple membrane proteins could be simultaneously reconstituted on a SUPER template by enriching the microsomal fraction of independent CFE reactions and depositing these fractions together on SUPER templates. To test this possibility, we carried out an experiment following the flowchart depicted in Figure 5A and used CFE to independently synthesize several additional membrane-spanning proteins, including the human codon-optimized channelrhodopsin 2 (hChR2) harboring a C-terminal monomeric Cherry and 6x histidine tag fusion (hChR2-mCherry-His6, referred to as hChR2-mCherry from here on), the inward rectifying potassium channel Kir2.1 harboring a C-terminal superfolder GFP fusion and 6x histidine tag fusion (Kir2.1-sfGFP-His6, referred to as Kir2.1-sfGFP from here on), and the voltage-gated potassium channel Kv1.2 harboring a C-terminal 6x histidine tag (Kv1.2-His6, referred to as Kv1.2 from here on). Note that Kv1.2 has a blue fluorescent protein tag, but proper detection requires a high expression level, so it is labeled with GreenLys unless otherwise noted.
The enrichment of these membrane proteins in the CFE pellet fractions, all synthesized with GreenLys incorporated, was determined by SDS-PAGE, which demonstrated that the pellet fractions had significantly more membrane proteins than the supernatant CFE fractions did after each wash, while the supernatant CFE fractions lost progressively more synthesized membrane protein after each wash (Figs. 5B–C). Importantly, the soluble G-GECO control construct exhibited the opposite behavior from the four membrane proteins tested above. Next, we added the enriched pellet fractions for each CFE reaction using the above described Airfuge fractionation assay to SUPER templates that were made using ANM lipid composition for EGFP-SUN2FL and polar brain extract lipids for the ion channels, as this lipid composition resembles their native lipid environment(Trimmer 2015; Wang, Ou, and Wang 2017). Fluorescent confocal microscopy revealed that each membrane protein including EGFP-SUN2FL was successfully reconstituted in the SUPER templates post CFE and centrifugation (Fig. 5D).
Since all the ion channels used in this experiment had a C-terminal 6x histidine tag, we did not introduce all the pellets at once to the SUPER template to avoid labeling the other CFE-synthesized ion channels during the simultaneous reconstitution of the three ion channels into the same lipid bilayer membrane. After incubating the fluorescent anti-histidine tag antibody with the SUPER templates with Kv1.2, they were washed and then incubated with the mixture of the Kir2.1-sfGFP- and hChR2-mCherry-containing CFE pellet fractions followed by additional washing steps. Using this approach, we were able to successfully reconstitute three different membrane proteins at once in a SUPER template (Fig. 5E). This demonstration further supports the possibility of a fusion-mediated mechanism for reconstitution of CFE-synthesized FL membrane proteins in SUPER templates.
DISCUSSION
In this work, we demonstrate the use of a facile HeLa cell-based CFE system to study FL membrane protein biochemistry in supported lipid bilayers in vitro. We first investigated the mechanism underlying the directional reconstitution of CFE-synthesized FL SUN proteins in SUPER templates that we previously reported8. Next, we investigated the effect of divalent cation (calcium) in the formation of LINC complex in vitro. We further validated past hypothesis of the importance of disulfide bonds in LINC complex formation using DTT to monitor disassociation of SUN-KASH interactions in our model system. Finally, we leveraged the specific topology of CFE-synthesized FL SUN proteins that were directionally reconstituted in SUPER templates to study hetero-typic SUN-KASH interactions as well as homo- and hetero-typic SUN protein interactions.
The use of FL SUN1 and SUN2 labeled with EGFP or incorporated GreenLys and their reconstitution in SUPER templates enabled the direct detection of protein localization and subsequent protein-protein interactions by confocal fluorescence microscopy. With a bead-based platform such as the SUPER templates(Pucadyil and Schmid 2010), it was possible to design simple in vitro biochemical assays to probe protein function owing to the ease of handling the CFE-synthesized and reconstituted FL SUN proteins. The major advantages of a CFE system are its ease and modularity in synthesizing many different proteins(Noireaux and Liu 2020; Perez, Stark, and Jewett 2016). As a result, homo- and hetero-typic SUN protein interactions could be probed through multiple rounds of CFE performed on the same batch of SUPER templates, while expressing different protein constructs. Another powerful advantage of this platform is its ability to simultaneously synthesize and translocate FL SUN proteins into the synthetic membrane of a SUPER template. While the C-terminally exposed topology of FL SUN1 and SUN2 in the SUPER templates was a discovery and not a controlled reconstitution, the evidence of ER-derived microsome-based reconstitution of SUN proteins in SUPER templates, potentially through membrane fusion, provides a way to possibly control the orientation of FL membrane proteins reconstituted in SUPER templates, as previously reported(Cymer and Von Heijne 2013; Cymer, Ismail, and Von Heijne 2014).
Unlike standard supported lipid bilayers, SUPER templates serve as an ideal membrane model in this study because they possess an excess membrane reservoir that is not tightly bound to the bead substrate(Neumann et al. 2013). This is reflected by the ability of reconstituted CFE-synthesized FL WT EGFP-tagged SUN1 or SUN2 to interact with synthetic TRITC-labeled KASH2WT peptides, which should only be possible if the reconstituted SUN proteins present in the SUPER templates were homo-oligomerization competent. Further, the observation of small fluorescent puncta on the SUPER templates could be evidence of SUN protein clustering in these excess membrane reservoirs, suggesting the presence of sufficient protein mobility, which is less readily achievable in conventional supported lipid bilayers(Mashaghi and van Oijen 2014). The short timescale for the entire reconstitution step with sufficient yield of proteins to mediate fluorescence-based detection is a key feature of our HeLa cell-based CFE platform that enabled us to carry out the studies presented here.
Our results only provide a window into the range of experimental possibilities accessible with our HeLa cell-based CFE platform. For instance, one could look at the effect of different concentrations of Ca2+ ions on KASH peptide-binding to reconstituted CFE-synthesized FL SUN proteins. The specific cysteine residues responsible for DSB formation during KASH peptide-binding(Cain et al. 2018) can be mutated and their effect studied without significant changes to the demonstrated approach. Further insights can be gained by the quantitative estimation of the timescale of DTT-based dissociation of the LINC complex. Similar extensions of the current work could be carried out to further investigate the possibility of the formation of hybrid LINC complexes based on heterotypic SUN1-SUN2 interactions. The entire approach can be directly applied to the study of LINC complex formation between SUN1 or SUN2 with non-nesprin-2-derived mammalian genome-encoded KASH peptides, including those from nesprin-1, -3, -4, KASH5, and lymphoid restricted membrane protein (a.k.a. Jaw1)(Meinke and Schirmer 2015). This is especially interesting in light of recent structural studies, which revealed that alternative binding modes exist for the KASH peptides derived from nesprin-3, nesprin-4, and KASH5 with SUN2 homo-trimers relative to the KASH peptides derived from nesprin-1 or nesprin-2(Cruz, Esra Demircioglu, and Schwartz 2020). Moreover, the same study demonstrated that a SUN2 homo-trimer could simultaneously interact with different KASH peptides. While it has been proposed that these differences may have an important role in regulating the SUN-KASH network, the underlying mechanisms remain poorly defined. Finally, our results suggest that CC assembly is highly dynamic as adding SUN LD constructs can trimerize/oligomerize with FL SUN proteins. It would be intriguing to test whether the addition of Ca2+ ions and KASH peptide would stabilize coiled-coil assembly. A more detailed investigation of homo- and hetero-meric assemblies of SUN proteins will be the subject of future studies.
Overall, our findings provide strong support for some of the existing hypotheses about LINC complex assembly, while new insights were gained with respect to the importance of biochemical interactions in SUN-KASH binding. Specifically, we found that Ca2+ ions significantly enhance the ability of reconstituted FL SUN proteins to bind to KASH peptides, but are not necessary for LINC complex assembly. Since the ER is contiguous with the ONM, ER-mediated calcium signaling elicited by mechanical signals(Kim et al. 2015) may strengthen hetero-typic SUN-KASH interactions, thus priming LINC complex-dependent mechanotransduction. On the other hand, our data suggest that DSBs are indispensable for the maintenance of stable SUN1/2-KASH2 interactions. This result is consistent with the recent demonstration that the conserved interchain SUN-KASH DSB is required for actin-dependent nuclear anchorage and movement as well as LINC complex-dependent mechanotransmission(Cain et al. 2018; Jahed et al. 2015). By extension, the redox state of the ER lumen, which is closely linked to ER protein-folding homeostasis(Cao and Kaufman 2014), may be a key regulator of stable LINC complex assembly. Finally, it is likely that homo- and hetero-typic SUN protein interactions require the presence of CC domains within their LDs. Future efforts are needed to identify the exact DSBs that are important for heterotypic SUN-KASH interactions in this system. It will be informative to test the KASH peptide-binding ability of FL SUN protein constructs that harbor mutations designed to inactivate either the intramolecular DSB known to be important for stabilizing the cation loop of SUN2 or the intermolecular SUN-KASH DSB(Sosa et al. 2012).
In summary, we developed a modular platform for the study of in vitro reconstituted CFE-synthesized membrane proteins with an emphasis on their real-time detection through fluorescence microscopy and their potential to reveal novel mechanistic insights into SUN protein biology. Such a platform could be extended to study the biochemical and biophysical properties of other difficult-to-study membrane proteins.
Supplementary Material
Acknowledgement
We thank Dr. Francois St-Pierre of Baylor College of Medicine for providing rKv 1.2, Kir 2.1, and hChR2 plasmids. This research was supported in part by the National Institutes of Health (grants R21GM134167 and R01EB030031 to A.P.L.; R01GM129374 to G.W.G.L.; and R35GM133325 to T.W.G.) and the National Science Foundation (grant 1935265 to A.P.L.).
Footnotes
Competing Interest Statement: The authors declare no competing interests.
REFERENCES
- Alam Samer, Lovett David B., Dickinson Richard B., Roux Kyle J., and Lele Tanmay P.. 2014. “Nuclear Forces and Cell Mechanosensing.” Pp. 205–15 in Progress in Molecular Biology and Translational Science. Vol. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Hong K, and Papahadjopoulos D. 1990. “Membrane Contact, Fusion, and Hexagonal (HII) Transitions in Phosphatidylethanolamine Liposomes.” Biochemistry 29(12):2976–85. doi: 10.1021/bi00464a013. [DOI] [PubMed] [Google Scholar]
- Battle Andrew R, Petrov Evgeny, Pal Prithwish, and Martinac Boris. 2009. “Rapid and Improved Reconstitution of Bacterial Mechanosensitive Ion Channel Proteins MscS and MscL into Liposomes Using a Modified Sucrose Method.” FEBS Letters 583(2):407–12. doi: 10.1016/j.febslet.2008.12.033. [DOI] [PubMed] [Google Scholar]
- Bentz Joe, and Duzgiines Nejat. 1985. “Fusogenic Capacities of Divalent Cations and Effect of Liposome Size.” Biochemistry 24(20):5436–43. doi: 10.1021/bi00341a023. [DOI] [PubMed] [Google Scholar]
- Brödel Andreas K., Wüstenhagen Doreen A., and Kubick Stefan. 2014. “Cell-Free Protein Synthesis Systems Derived from Cultured Mammalian Cells.” Pp. 129–40 in Structural Proteomics: High-Throughput Methods: Second Edition. [DOI] [PubMed] [Google Scholar]
- Cain Natalie E., Jahed Zeinab, Schoenhofen Amy, Valdez Venecia A., Elkin Baila, Hao Hongyan, Harris Nathan J., Herrera Leslie A., Woolums Brian M., Mofrad Mohammad R. K., Gan. Luxton GW, and Starr Daniel A.. 2018. “Conserved SUN-KASH Interfaces Mediate LINC Complex-Dependent Nuclear Movement and Positioning.” Current Biology 28(19):3086–3097.e4. doi: 10.1016/j.cub.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Stewart Siyan, and Kaufman Randal J.. 2014. “Endoplasmic Reticulum Stress and Oxidative Stress in Cell Fate Decision and Human Disease.” Antioxidants and Redox Signaling 21(3):396–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter Elisabeth P., Beis Konstantinos, Cameron Alexander D., and Iwata So. 2008. “Overcoming the Challenges of Membrane Protein Crystallography.” Current Opinion in Structural Biology 18(5):581–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Wakam, Worman Howard J., and Gundersen Gregg G.. 2015. “Accessorizing and Anchoring the LINC Complex for Multifunctionality.” Journal of Cell Biology 208(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp Melissa, Liu Qian, Roux Kyle, Rattner JB, Shanahan Catherine, Burke Brian, Stahl Phillip D., and Hodzic Didier. 2006. “Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex.” Journal of Cell Biology 172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz Victor E., Esra Demircioglu F, and Schwartz Thomas U.. 2020. “Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.” Journal of Molecular Biology 432(23):6028–41. doi: 10.1016/j.jmb.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymer Florian, and Von Heijne Gunnar. 2013. “Cotranslational Folding of Membrane Proteins Probed by Arrest-Peptide- Mediated Force Measurements.” Proceedings of the National Academy of Sciences of the United States of America 110(36):14640–45. doi: 10.1073/pnas.1306787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymer Florian, Ismail Nurzian, and Von Heijne Gunnar. 2014. “Weak Pulling Forces Exerted on Nin-Orientated Transmembrane Segments during Co-Translational Insertion into the Inner Membrane of Escherichia Coli.” FEBS Letters 588(10):1930–34. doi: 10.1016/j.febslet.2014.03.050. [DOI] [PubMed] [Google Scholar]
- Dondapati Srujan Kumar, Lübberding Henning, Zemella Anne, Thoring Lena, Wüstenhagen Doreen A., and Kubick Stefan. 2019. “Functional Reconstitution of Membrane Proteins Derived from Eukaryotic Cell-Free Systems.” Frontiers in Pharmacology 10(JULY). doi: 10.3389/fphar.2019.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groaz Alessandro, Moghimianavval Hossein, Tavella Franco, Giessen Tobias W., Vecchiarelli Anthony G., Yang Qiong, and Liu Allen P.. 2021. “Engineering Spatiotemporal Organization and Dynamics in Synthetic Cells.” Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology 13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen Jared, Angert Isaac, Hur Kwang Ho, Gant Luxton GW, and Mueller Joachim D.. 2018. “Investigating LINC Complex Protein Homo-Oligomerization in the Nuclear Envelopes of Living Cells Using Fluorescence Fluctuation Spectroscopy.” Pp. 121–35 in Methods in Molecular Biology. Vol. 1840. [DOI] [PubMed] [Google Scholar]
- Hennen Jared, Saunders Cosmo A., Mueller Joachim D., and Gan. Luxton GW. 2018. “Fluorescence Fluctuation Spectroscopy Reveals Differential SUN Protein Oligomerization in Living Cells.” Molecular Biology of the Cell 29(9):1003–11. doi: 10.1091/mbc.E17-04-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Kenneth K. Y., Lee Jin Woo, Durand Grégory, Majumder Sagardip, and Liu Allen P.. 2017. “Protein Aggregation with Poly(Vinyl) Alcohol Surfactant Reduces Double Emulsionencapsulated Mammalian Cell-Free Expression.” PLoS ONE 12(3). doi: 10.1371/journal.pone.0174689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Kenneth K. Y., Murray Victoria L., and Liu Allen P.. 2015. “Engineering Artificial Cells by Combining HeLa-Based Cell-Free Expression and Ultrathin Double Emulsion Template.” Methods in Cell Biology 128:303–18. doi: 10.1016/bs.mcb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Miranda L., Boyd Margrethe A., and Kamat Neha P.. 2019. “Diblock Copolymers Enhance Folding of a Mechanosensitive Membrane Protein during Cell-Free Expression.” Proceedings of the National Academy of Sciences of the United States of America 116(10):4031–36. doi: 10.1073/pnas.1814775116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Zeinab, Fadavi Darya, Vu Uyen T., Asgari Ehsaneddin, Gan. Luxton GW, and Mofrad Mohammad R. K.. 2018. “Molecular Insights into the Mechanisms of SUN1 Oligomerization in the Nuclear Envelope.” Biophysical Journal 114(5):1190–1203. doi: 10.1016/j.bpj.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Zeinab, Shams Hengameh, and Mofrad Mohammad R. K.. 2015. “A Disulfide Bond Is Required for the Transmission of Forces through SUN-KASH Complexes.” Biophysical Journal 109(3):501–9. doi: 10.1016/j.bpj.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Zeinab, Vu Uyen T., Fadavi Darya, Ke Huimin, Rathish Akshay, Kim Samuel C. J., Feng Wei, and Mofrad Mohammad R. K.. 2018. “A Molecular Model for LINC Complex Regulation: Activation of SUN2 for KASH Binding.” Molecular Biology of the Cell 29(16):2012–23. doi: 10.1091/mbc.E18-04-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta Santharam S., Smoyer Christine J., and Jaspersen Sue L.. 2014. “Destination: Inner Nuclear Membrane.” Trends in Cell Biology 24(4):221–29. [DOI] [PubMed] [Google Scholar]
- Kim Tae Jin, Joo Chirlmin, Seong Jihye, Vafabakhsh Reza, Botvinick Elliot L., Berns Michael W., Palmer Amy E., Wang Ning, Ha Taekjip, Jakobsson Eric, Sun Jie, and Wang Yingxiao. 2015. “Distinct Mechanisms Regulating Mechanical Force-Induced Ca2+ Signals at the Plasma Membrane and the ER in Human MSCs.” ELife 2015(4). doi: 10.7554/eLife.04876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laba Justyna K., Steen Anton, and Veenhoff Liesbeth M.. 2014. “Traffic to the Inner Membrane of the Nuclear Envelope.” Current Opinion in Cell Biology 28(1):36–45. [DOI] [PubMed] [Google Scholar]
- Lohia A, Chatterjee AN, and Das J. 1984. “Lysis of Vibrio Cholerae Cells: Direct Isolation of the Outer Membrane from Whole Cells by Treatment with Urea.” Journal of General Microbiology 130(8):2027–33. doi: 10.1099/00221287-130-8-2027. [DOI] [PubMed] [Google Scholar]
- Lu Wenshu, Gotzmann Josef, Sironi Lucia, Jaeger Verena-Maren, Schneider Maria, Yvonne Lüke Mathias Uhlén, Cristina Al-Khalili Szigyarto Andreas Brachner, Ellenberg Jan, Foisner Roland, Noegel Angelika A., and Karakesisoglou Iakowos. 2008. “Sun1 Forms Immobile Macromolecular Assemblies at the Nuclear Envelope.” Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1783(12):2415–26. doi: 10.1016/j.bbamcr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Majumder Sagardip, and Liu Allen P.. 2018. “Bottom-up Synthetic Biology: Modular Design for Making Artificial Platelets.” Physical Biology 15(1). doi: 10.1088/1478-3975/aa9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder Sagardip, Willey Patrick T., DeNies Maxwell S., Liu Allen P., and Luxton Gant. 2019. “A Synthetic Biology Platform for the Reconstitution and Mechanistic Dissection of LINC Complex Assembly.” Journal of Cell Science 132(4). doi: 10.1242/jcs.219451. [DOI] [PubMed] [Google Scholar]
- Mashaghi Samaneh, and van Oijen Antoine M.. 2014. “A Versatile Approach to the Generation of Fluid Supported Lipid Bilayers and Its Applications.” Biotechnology and Bioengineering 111(10):2076–81. doi: 10.1002/bit.25273. [DOI] [PubMed] [Google Scholar]
- Van Meer Gerrit, Voelker Dennis R., and Feigenson Gerald W.. 2008. “Membrane Lipids: Where They Are and How They Behave.” Nature Reviews Molecular Cell Biology 9(2):112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke Peter, and Schirmer Eric C.. 2015. “LINC’ing Form and Function at the Nuclear Envelope.” FEBS Letters 589(19):2514–21. [DOI] [PubMed] [Google Scholar]
- Neumann Sylvia, Pucadyil Thomas J., and Schmid Sandra L.. 2013. “Analyzing Membrane Remodeling and Fission Using Supported Bilayers with Excess Membrane Reservoir.” Nature Protocols 8(1):213–22. doi: 10.1038/nprot.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Si, Ke Huimin, Gao Feng, Ren Jinqi, Wang Mingzhu, Huo Lin, Gong Weimin, and Feng Wei. 2016. “Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators.” Structure 24(1):80–91. doi: 10.1016/j.str.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Noireaux Vincent, and Liu Allen P.. 2020. “The New Age of Cell-Free Biology.” Annual Review of Biomedical Engineering 22:51–77. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte Thorsten, Lu Wenshu, Zaim Hafida, Abraham Sabu, Noegel Angelika A., Gotzmann Josef, Foisner Roland, and Karakesisoglou Iakowos. 2005. “The Inner Nuclear Membrane Protein Sun1 Mediates the Anchorage of Nesprin-2 to the Nuclear Envelope.” Journal of Cell Science 118(15):3419–30. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Papahadjopoulos Demetrios, Nir Shlomo, and Düzgünes Nejat. 1990. “Molecular Mechanisms of Calcium-Induced Membrane Fusion.” Journal of Bioenergetics and Biomembranes 22(2):157–79. doi: 10.1007/BF00762944. [DOI] [PubMed] [Google Scholar]
- Perez Jessica G., Stark Jessica C., and Jewett Michael C.. 2016. “Cell-Free Synthetic Biology: Engineering beyond the Cell.” Cold Spring Harbor Perspectives in Biology 8(12). doi: 10.1101/cshperspect.a023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil Thomas J., and Schmid Sandra L.. 2010. “Supported Bilayers with Excess Membrane Reservoir: A Template for Reconstituting Membrane Budding and Fission.” Biophysical Journal 99(2):517–25. doi: 10.1016/j.bpj.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Bineet, Moghimianavval Hossein, Hwang Sung Won, and Liu Allen P.. 2021. “Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions.” Membranes 11(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa Brian A., Rothballer Andrea, Kutay Ulrike, and Schwartz Thomas U.. 2012. “LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins.” Cell 149(5):1035–47. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale Christopher M., Wirtz Denis, and Hodzic Didier. 2008. “Structural Requirements for the Assembly of LINC Complexes and Their Function in Cellular Mechanical Stiffness.” Experimental Cell Research 314(8):1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway Christian, Pulokas James, Fellmann Denis, Cheng Anchi, Guerra Francisco, Quispe Joel, Stagg Scott, Potter Clinton S., and Carragher Bridget. 2005. “Automated Molecular Microscopy: The New Leginon System.” Journal of Structural Biology 151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tapley Erin C., and Starr Daniel A.. 2013. “Connecting the Nucleus to the Cytoskeleton by SUN-KASH Bridges across the Nuclear Envelope.” Current Opinion in Cell Biology 25(1):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoring Lena, and Kubick Stefan. 2018. “Versatile Cell-Free Protein Synthesis Systems Based on Chinese Hamster Ovary Cells.” Pp. 289–308 in Methods in Molecular Biology. Vol. 1850. [DOI] [PubMed] [Google Scholar]
- Trimmer James S. 2015. “Subcellular Localization of K+ Channels in Mammalian Brain Neurons: Remarkable Precision in the Midst of Extraordinary Complexity.” Neuron 85(2):238–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Jun, Ou Shao Wu, and Wang Yun Jie. 2017. “Distribution and Function of Voltage-Gated Sodium Channels in the Nervous System.” Channels 11(6):534–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Qiang, Du Xiulian, Cai Zheng, and Greene Mark I.. 2006. “Characterization of the Structures Involved in Localization of the SUN Proteins to the Nuclear Envelope and the Centrosome.” DNA and Cell Biology 25(10):554–62. doi: 10.1089/dna.2006.25.554. [DOI] [PubMed] [Google Scholar]
- Wang Wenjia, Shi Zhubing, Jiao Shi, Chen Cuicui, Wang Huizhen, Liu Guoguang, Wang Qiang, Zhao Yun, Greene Mark I., and Zhou Zhaocai. 2012. “Structural Insights into SUN-KASH Complexes across the Nuclear Envelope.” Cell Research 22(10):1440–52. doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilschut Jan, Düzgüne Nejat, and Papahadjopoulos Demetrios. 1981. “Calcium/Magnesium Specificity in Membrane Fusion: Kinetics of Aggregation and Fusion of Phosphatidylserine Vesicles and the Role of Bilayer Curvature.” Biochemistry 20(11):3126–33. doi: 10.1021/bi00514a022. [DOI] [PubMed] [Google Scholar]
- Xiong Huajiang, Rivero Francisco, Euteneuer Ursula, Mondal Subhanjan, Mana-Capelli Sebastian, Larochelle Denis, Vogel Annette, Gassen Berthold, and Noegel Angelika A.. 2008. “Dictyostelium Sun-1 Connects the Centrosome to Chromatin and Ensures Genome Stability.” Traffic 9(5):708–24. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- Yuan Xiaoling, Li Jianyong, Shan Yajun, Yang Zhen, Zhao Zhenhu, Chen Bo, Yao Zhenyu, Dong Bo, Wang Shengqi, Chen Jiapei, and Cong Yuwen. 2005. “Subcellular Localization and Membrane Association of SARS-CoV 3a Protein.” Virus Research 109(2):191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemella Anne, Richter Theresa, Thoring Lena, and Kubick Stefan. 2019. “A Combined Cell-Free Protein Synthesis and Fluorescence-Based Approach to Investigate GPCR Binding Properties.” Pp. 57–77 in Methods in Molecular Biology. Vol. 1947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


