Abstract
Medically important yeasts of the genus Candida secrete aspartic proteinases (Saps), which are of particular interest as virulence factors. Like Candida albicans, Candida tropicalis secretes in vitro one dominant Sap (Sapt1p) in a medium containing bovine serum albumin (BSA) as the sole source of nitrogen. Using the gene SAPT1 as a probe and under low-stringency hybridization conditions, three new closely related gene sequences, SAPT2 to SAPT4, encoding secreted proteinases were cloned from a C. tropicalis λEMBL3 genomic library. All bands identified by Southern blotting of EcoRI-digested C. tropicalis genomic DNA with SAPT1 could be assigned to a specific SAP gene. Therefore, the SAPT gene family of C. tropicalis is likely to contain only four members. Interestingly, the SAPT2 and SAPT3 gene products, Sapt2p and Sapt3p, which have not yet been detected in C. tropicalis cultures in vitro, were produced as active recombinant enzymes with the methylotrophic yeast Pichia pastoris as an expression system. As expected, reverse transcriptase PCR experiments revealed a strong SAPT1 signal with RNA extracted from cells grown in BSA medium. However, a weak signal was obtained with all other SAPT genes under several conditions tested, showing that these SAPT genes could be expressed at a basic level. Together, these experiments suggest that the gene products Sapt2p, Sapt3p, and Sapt4p could be produced under conditions yet to be described in vitro or during infection.
During the past two decades, Candida infections have increased in number and severity (33). In immunocompromised patients, Candida species have the potential to invade all host organs and cause severe systemic infections. Although Candida albicans is the organism most often associated with serious fungal infections, other Candida species have emerged as clinically important pathogens of these opportunistic infections (19, 31). The asexual diploid yeast C. tropicalis is the second most pathogenic of the Candida species (18, 19, 31). Unlike C. albicans, which is a normal commensal on human mucous membranes, the detection of C. tropicalis is more often associated with the development of deep fungal infections (10, 13).
To assist invasion of host tissues, many pathogenic microbes posses constitutive and inducible hydrolytic enzymes that destroy, alter, or damage membrane integrity, leading to dysfunction or disruption of host structures. Pathogenic species of Candida produce a large variety of secreted hydrolases, and among various potential virulence factors proposed, the secreted aspartic proteinases (Sap) have been intensively investigated. It is now well established that the ability of C. albicans to adhere to mucosae in the oral and vaginal tracts, to invade in deep organs, and to resist phagocytic cells apparently requires the use of several different proteinases suitable to each particular condition during the infection (3, 4, 6, 24, 25). Eight genes (SAP1 to SAP8) encoding true Saps and two others (SAP9 and SAP10) encoding putative GPI-anchored proteinases have been cloned from C. albicans to date (12, 15; A. Felk, W. Schaefer, and B. Hube, SAP10 GenBank accession no. AF146440).
Like C. albicans, Candida tropicalis secretes in vitro Sap activity in a medium containing bovine serum albumin (BSA) as the sole source of nitrogen. One Sap, called Sapt1p, was purified from culture supernatant, biochemically characterized, and crystallized (26, 28). However, previous data suggested the existence of a SAPT gene family in the genome of C. tropicalis (16). The presence of aspartic proteinases secreted by C. tropicalis has also been demonstrated on the surface of fungal elements penetrating tissues during disseminated infection and evading macrophages after phagocytosis of yeast cells (1, 2, 22). It has become apparent that the use of different members of a gene family by microorganisms is linked to the process of pathogenesis (3, 4, 11, 16). In the present work, we have further characterized the C. tropicalis gene family in order to elucidate the molecular basis of the virulence of this pathogenic yeast.
(This work was done by C. Zaugg in partial fulfillment of the requirements for a Ph.D. degree from the University of Lausanne, Lausanne, Switzerland.)
MATERIALS AND METHODS
Strains and plasmids.
C. tropicalis ATCC 750, DSM 4959, and four clinical isolates (CHUV 740.00, GO 25896, GO 26110, and GO 28861) from deep-seated candidiasis of different patients at the University Hospital in Lausanne (Switzerland) and Göttingen (Germany) were used. All strains were identified by the use of Chromagar plates, on which they developed a dark blue color, and by sugar fermentation with the API system galleries, where the identification was certified at 99%.
E. coli LE392 was used for the propagation of the bacteriophage λEMBL3 (Promega). All plasmid subcloning experiments were performed in Escherichia coli DH5α with plasmid pMTL21 (5). Pichia pastoris GS115 and the expression vector pKJ113 (3) were used to express recombinant Sapt enzymes.
Growth media.
All C. tropicalis strains were maintained on YPD (1% yeast extract, 2% peptone, 2% dextrose, 1.5% agar) agar plates. Solid and liquid BSA media were used to promote Sapt1p expression (28). Yeast was also grown in modified Lee's medium containing 5% (vol/vol) fetal calf serum (17).
C. tropicalis genomic library and gene cloning.
A genomic DNA library was prepared with DNA of C. tropicalis ATCC 750. The isolated DNA was partially digested with Sau3A, and DNA fragments ranging from 12 to 20 kb were isolated from low-melting-point agarose (Bio-Rad); these fragments were inserted into bacteriophage λEMBL3 cloning system (Promega).
Recombinant plaques (2 × 104) of the genomic library were immobilized on GeneScreen nylon membranes (Dupont). The filters were hybridized with a 32P-labelled C. tropicalis SAPT1 probe under low-stringency conditions (16). The probe was obtained by PCR amplification of SAPT1 by using oligonucleotides 1 and 2 (Table 1) and plasmid pMTL21-E4 (28) as the targeted DNA. All positive plaques were purified and the associated bacteriophage DNAs were isolated as described by Grossberger (9). Hybridizing fragments from λEMBL3 bacteriophages were subcloned into pMTL21 by standard procedures (23). Microsynth (Balgach, Switzerland) performed the sequencing of the SAPT genes.
TABLE 1.
Materials used for the expression of the different Saptp proteins in P. pastoris and for RT-PCR
| Genes | Oligonucleotide primer (sequence) | Orientation | Encoded amino acid sequencea | Template | PCR product (with cloning sites) |
|---|---|---|---|---|---|
| SAPT1 | 1 (GGA A/GA TCT GAT GTG CCA ACT ACA TTG A) | Senseb | pMTL21-E4 or cDNA | SAPT1 (181–1,186 bp); BglII----NotI | |
| 2 (CGT GC/GGC CGC T CTA CAA AGC CGA GAT GTC T) | Antisenseb | ||||
| 3 (GTA C/TC GAG AAA AGA TTG TTT GCT CAA GGT CTT ACT) | Sensec | (L)(E)(K)(R)LFAQGLT | pMTL21-E4 | SAPT1 (55–1,185 bp); XhoI----NotI | |
| 4 (CAT GC/GC CGC CTA CAA AGC CGA GAT GTC TGA AGA) | Antisensec | SSDISALSTOP | |||
| SAPT2 | 5 (GTA C/T CGA GTT TCC AAG CTC AAC GAT CGT) | Sensec | (R)(V)SKLNDR | pCT2 | SAPT2 (55–511 bp); XhoI----Asp718I |
| 6 (C TTT GAC CCA G/GT ACC ACT AGA AGA A) | Antisensec | SSSGTWVK | |||
| 7 (T TCT TCT AGT G/GT ACC TGG GTC AAA G) | Sensebc | SSSGTWVK | pCT2 or cDNA | SAPT2 (486–1,248 bp); Asp718I----BglII | |
| 8 (CAT A/GA TCT CTA AAC AAT AGT GAC ATT AGA) | Antisensebc | SNVTIVSTOP | |||
| SAPT3 | 9 (GTA C/T CGA GTG GTT TTA AAT GCT GGT TCA) | Sensebc | (R)(V)VLNAGS | pCT3 or cDNA | SAPT3 (52–1,170 bp); XhoI----BglII |
| 10 (CAT A/GA TCT CTA CAT AAT TGC TCT AAT TTT) | Antisensebc | KIRAIMSTOP | |||
| SAPT4 | 11 (GTA C/T CGA GCT CCT ACA ACT TCA CCT CCT) | Sensebc | (R)(A)PTTSPP | pCT4 or cDNA | SAPT4 (55–1,185 bp); XhoI---- BamHI |
| 12 (CAT G/GA TCC CTA TGT AAG TGG AAG TAT GTT) | Antisensebc | NILPLTSTOP |
In parentheses are shown amino acids encoded by the XhoI restriction site sequence and added to the N-terminal extremity of the Saptp prosequences.
Primers used for RT-PCR.
Primers used for expression in P. pastoris.
Expression of recombinant aspartic proteinases.
Expression plasmids were constructed by cloning a PCR product of the C. tropicalis SAPT genes in the multiple cloning site of the Escherichia coli-P. pastoris shuttle vector pKJ113. PCR was performed according to standard conditions with homologous primers derived from DNA sequences of the different SAPT genes (Table 1).
For cloning into pKJ113, the PCR products were purified by using a PCR purification kit (Roche Diagnostic) and digested by restriction enzymes for which a site was previously designed at the 5′ extremity of the primers (Table 1). P. pastoris GS115 was transformed by electroporation with 10 μg of plasmid DNA linearized by SmaI. Transformants selected on histidine-deficient medium were screened for insertion of the construct at the AOX1 site on minimal methanol plates. The transformants unable to grow on media containing only methanol as a carbon source were assumed to contain the construct at the correct yeast genomic location by integration events in the AOX1 locus displacing the AOX1 coding region. They were grown to near saturation (optical density of 20 at 600 nm) at 30°C in 10 ml of glycerol-based yeast medium. Cells were harvested and resuspended in 2 ml of the same medium with 0.5% (vol/vol) methanol instead of glycerol and incubated for 2 days. Subsequently, the supernatant was harvested and tested for protein production on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Recombinant Saptp enzymes were produced in large quantities from 400 ml of cell culture supernatant. The volume was reduced to 20 ml by ultrafiltration with the Amicon 8050 system (Millipore). Salts and low-molecular-weight solutes were removed by passing through Sephadex G-75 column (Pharmacia) with 10 mM sodium citrate buffer (pH 6.5). The recombinant proteinases recovered from active fractions were finally concentrated by ultrafiltration with the Centricon 30 system (Amicon).
Protein extract analysis.
Protein concentrations were measured by the method of Bradford (4a). Protein extracts were analyzed by SDS-PAGE with a separation gel of 12% polyacrylamide. Gels were stained with Coomassie brilliant blue R-250 (Bio-Rad). N-Glycosidase F digestion and N-terminal sequencing analyses were performed as previously described (8).
Proteolytic assays.
The proteolytic activity of Sap isoenzymes was measured with 0.02% resorufin-labeled casein as a substrate at different pH values (2.0 to 7.0) in 50 mM sodium citrate buffer in a total volume of 0.5 ml. After incubation at 37°C, the undigested substrate was precipitated by trichloroacetic acid (4% final concentration) and separated from the supernatant by centrifugation. The absorbance of the supernatant was measured in the alkaline range at 574 nm after addition of 30 μl of 4 N NaOH. For practical purposes, 1 U of Sap activity was defined as that producing an absorbance of 0.001 per min in a proteolytic assay at an optimum pH of activity.
Standard PCR.
PCRs were performed by using a Perkin-Elmer DNA Thermal Cycler. Two hundred nanograms of genomic DNA or 10 ng of plasmidic DNA in 5 μl of 10 mM Tris-HCl (pH 8.0), 10 μl of each of the sense and antisense oligonucleotides at a concentration of 42 mM, and 8 μl of deoxynucleotide mix (containing 10 mM of each deoxynucleoside triphosphate [dNTP]) were dissolved in 100 μl of PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). To each reaction mixture, 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer) was added. The reaction mixtures were incubated for 5 min at 94°C; subjected to 25 cycles of 0.5 min at 94°C, 0.5 min at 55°C, and 0.5 min at 72°C; and finally incubated for 10 min at 72°C.
RT-PCR.
Total RNA was isolated and purified from C. tropicalis by using the yeast I protocol of a Qiagen RNeasy Mini kit. Yeast cells (5 × 107) were lysed with 250 U of lyticase for 20 min in Y1 buffer (1 M sorbitol, 0.1 M EDTA [pH 7.4]) and then by vortexing with 0.6 g of calcinated glass beads for 1 min. Reverse transcriptase PCR (RT-PCR) was performed with a Qiagen OneStep RT-PCR kit. Briefly, 1 μg of total RNA, 10 μl of supplied 5× OneStep RT-PCR buffer (12.5 mM MgCl2 [pH 8.7]), 2 μl of deoxynucleotide mix (containing 10 mM each dNTP), 5 μl of sense and antisense primers at a concentration of 6 μM, and 2 μl of OneStep RT-PCR enzyme mix were mixed on ice and subsequently incubated at 50°C for 30 min and 95°C for 15 min. The reaction mixtures were subjected to 35 cycles of 0.5 min at 94°C, 0.5 min at 55°C, and 1 min at 72°C and finally were incubated for 10 min at 72°C. The PCR products were visualized on a 0.8% agarose gel.
RESULTS
C. tropicalis-secreted proteolytic activity.
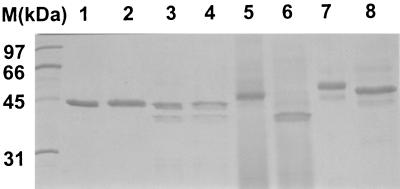
The proteolytic activities of six C. tropicalis strains were tested on BSA agar plates and in BSA liquid medium after 72 h of growth at 30°C. Except for the DSM 4959 strain, all strains examined exhibited similar proteolytic activities. SDS-PAGE of total protein extract from culture supernatant of all proteolytically active strains revealed only one protein with a molecular mass of about 44 kDa (Fig. 1, lane 1). This protein corresponded to Sapt1p, which has been previously characterized.
FIG. 1.
Protein profile of native Sapt1p obtained from a culture of C. tropicalis ATCC 750 in BSA medium (lanes 1 and 2) and recombinant Sapt1p (lanes 3 and 4), Sapt2p (lanes 5 and 6), and Sapt3p (lanes 7 and 8) produced in P. pastoris. The gel was stained with Coomassie brilliant blue R-250. Lanes 2, 4, 6, and 8 show proteins deglycosylated after N-glycosidase F treatment. Molecular mass markers (M) are shown in the leftmost lane.
The DSM 4959 strain did not grow on solid or in liquid BSA medium. No proteolytic activity was detected, and SDS-PAGE analysis revealed that the BSA in liquid medium remained intact (data not shown). However, a SAPT1 gene of C. tropicalis DSM 4959 could be amplified by PCR with SAPT1-specific primers (Table 1) and DSM 4959 DNA as a template. Sequencing of the PCR product revealed 100% identity with the previously published sequence of SAPT1 cloned from the type strain of C. tropicalis ATCC 750 (28). These results suggest that the lack of proteolytic activity in the DSM 4959 strain cannot be attributed to mutations in the SAPT1 coding region.
Cloning of three new members of the C. tropicalis SAPT gene family.
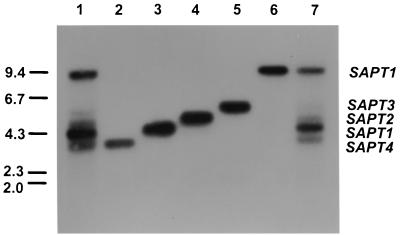
A screening of a C. tropicalis λEMBL3 genomic library was performed with the whole SAPT1 gene as a probe and under low-stringency conditions of hybridization. Among 2 × 104 individual recombinant bacteriophage plaques, corresponding to 20 yeast genome equivalents, all hybridizing clones (100 in total) were purified. The DNA obtained from these clones was restricted with EcoRI, gel electrophoresed, blotted onto membranes, and hybridized with the selecting SAPT1 probe. Five groups of clones containing 8.8-, 5.0-, 4.4-, 4.0-, and 3.8-kb EcoRI fragments (Fig. 2) were retained and further analyzed. The 4.0-kb EcoRI fragment corresponded to the SAPT1 fragment previously isolated and partially sequenced (28). The signal intensity of the 8.8-kb fragment was as strong as that of the 4.0-kb fragment (Fig. 2). PCR of bacteriophage DNA with SAPT1 primers amplified a fragment the sequence of which was 100% identical to SAPT1. C. tropicalis DNA was subsequently digested by other enzymes and analyzed by Southern blotting with the SAPT1 probe under high-stringency conditions. Only one band of 4.2 kb and one band of 9.0 kb were revealed with DNA digested with BglII and HindIII, respectively (data not shown). These results suggested that the two SAPT1 alleles were on two different EcoRI fragments. The three other EcoRI fragments were subcloned into pMTL21, generating the plasmids pCT2, pCT3, and pCT4, for which the map of the inserts is shown in Fig. 3.
FIG. 2.
Southern blots of EcoRI digestions of C. tropicalis ATCC 750 DNA (lanes 1 and 7), pMTL21-E4 (lane 3), pCT2 (lane 4), pCT3 (lane 5), pCT4 (lane 2), and λEMBL3 phage DNA containing the second allele of SAPT1 (lane 6) were hybridized to the SAPT1 gene used as a probe. The molecular size markers (kilobases) are indicated to the left.
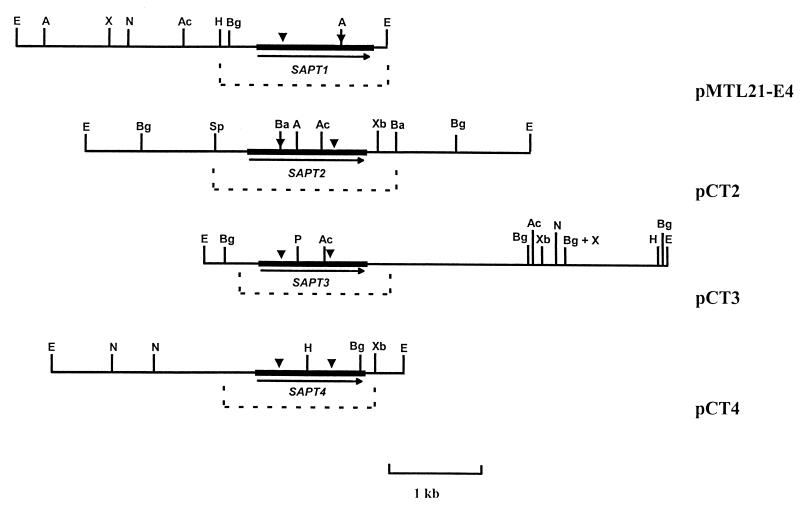
FIG. 3.
Restriction maps of inserts of the plasmids containing SAPT1, SAPT2, SAPT3, and SAPT4. The SAPT ORFs are represented by solid arrows. The domains of the catalytic sites of the enzymes are indicated by solid triangles. The GenBank accession numbers of the sequenced regions of pMTL21-E4 (29), pCT2, pCT3, and pCT4 (represented by a dashed line) are X61438, AF115320, AF115321, and AF115322, respectively. A, Asp718; Ac, AccI; Ba, BamHI; Bg, BglII; E, EcoRI; H, HindIII; N, NcoI; P, PstI; Sp, SphI; X, XhoI; Xb, XbaI.
The nucleotide sequence of areas hybridizing with the SAPT1 probe and flanking regions in plasmids pCT2, pCT3, and pCT4 revealed three long open reading frames (ORFs) of 1,245, 1,167, and 1,182 bp, respectively. The amino acid sequence deduced from these genes showed significant similarities to that of the other aspartic proteinases, in particular within the regions that contained the two reactive aspartic acid residues. Four cysteine residues were also conserved (Fig. 4). The cysteine residues in homologous positions form disulfide bridges in all aspartic proteinases (27). The deduced amino acid sequences suggested the existence, like in Sapt1p and in the C. albicans Saps, of both a signal peptide with putative signal peptidase cleavage sites and a prosequence with one or two KR sequences (16). These tandems of basic amino acids are known to be proteolytic processing sites by the proconvertase Kex2p (32). The genes corresponding to the 5.0-, 4.4-, and 3.8-kb EcoRI fragments were called SAPT2, SAPT3, and SAPT4, respectively. The Sapt2p, Sapt3p, and Sap4p polypeptide chains generated by cleavage at the Kex2p-like cleavage site have calculated molecular masses of 36, 37, and 37 kDa, respectively (Table 2).
FIG. 4.
Comparison of deduced amino acid sequences of the secreted aspartic proteinases Sapt1p to Sapt4p from C. tropicalis. ▾, aspartic acid residues corresponding to those found in the active site of the pepsin family; ✻, conserved cysteine residues involved in disulfide bridge formation in the three-dimensional structure of aspartic proteinases. The putative signal peptidase cleavage site is indicated by a vertical arrowhead. An arrow delimits the N-terminal positions of the mature secreted proteinases just after a KR sequence known to be a proteolytic processing site. The alignment was performed with the PileUp algorithm implemented in the GCG package of the Genetics Computer Group, University of Wisconsin, Madison, and reformatted with Boxshade 3.2.
TABLE 2.
Characteristics of C. tropicalis-secreted aspartic proteinases
| Characteristic | Result for aspartic proteinase
|
|||
|---|---|---|---|---|
| Sapt1p | Sapt2p | Sapt3p | Sapt4p | |
| Preproprotein length (amino acids) | 394 | 415 | 389 | 394 |
| Mature domain of the protein (amino acids) | 334 | 341 | 338 | 342 |
| Putative KR processing site (amino acid positions) | 32–33, 59–60 | 38–39, 73–74 | 50–51 | 51–52 |
| Theoretical molecular mass of the polypeptide domain of the mature domain (kDa)a | 35.8 | 36.5 | 37.1 | 37.4 |
| Apparent molecular mass determined by SDS-PAGE (kDa) | 44 | 49 | 48/55b | NDc |
| Apparent molecular mass after N-glycosidase treatment by SDS-PAGE (kDa) | 44 | 40 | 44/50b | ND |
| No. of putative glycosylation sites | 0 | 4 | 2 | 5 |
| Calculated pIa | 4.23 | 4.09 | 4.95 | 5.69 |
| Optimum pH of activity experimentally determined | 3.5 | 5.0 | 5.0 | ND |
The theoretical molecular mass of the mature domain and the pI were calculated with the program Compute pI/Mw tool (http://www.expasy.ch/ch2d/pi_tool.html).
Mature form and proprotein.
ND, not done. (No recombinant SAPT4 translation product was obtained from P. pastoris transformants.)
Southern blotting of EcoRI-digested DNA hybridized at low stringency with the SAPT1 probe revealed five bands for all C. tropicalis strains tested, including DSM 4959. These bands corresponded to the DNA fragments harboring SAPT2, SAPT3, and SAPT4 and the two alleles of SAPT1 (Fig. 2). The same band pattern was observed with six C. tropicalis strains isolated from different patients (data not shown). Therefore, four members are likely to constitute the C. tropicalis SAPT gene family.
Properties of recombinant Sapt2p and Sapt3p (Table 2).
Sapt2p and Sapt3p, which have never been detected in C. tropicalis culture supernatants, were produced by P. pastoris as active proteinases. The yields of recombinant Sapt2p and Sapt3p were 3.7 and 3 U ml−1, corresponding to about 2.5 and 5 μg of protein ml−1, respectively. No recombinant SAPT4 translation product was obtained from P. pastoris transformants. As a control, Sapt1p was recovered in P. pastoris culture supernatant as an active proteinase with a yield of 15 U ml−1 corresponding to 50 μg of protein ml−1. However, two bands were detected by SDS-PAGE (Fig. 1, lane 3). The recombinant Sapt1p product with the lowest electrophoretic mobility comigrated with Sapt1p produced from C. tropicalis (Fig. 1, lane 1). The N-terminal sequences of both proteins were identical and determined to be SDVPTTLKN. Western blotting analysis showed that the Sapt1p product with higher electrophoretic mobility cross-reacted with anti-Sapt1p antibodies and was likely a degradation product.
Sapt2p showed a single protein band in an SDS-PAGE gel with an estimated molecular mass of 49 kDa (Fig. 1, lane 5). The Sapt2p N-terminal sequence was determined to be DDYKSVELY, which corresponded to amino acids 61 to 70 of the translation product, just after a Kex2p processing site, KR. Two bands were observed on SDS-PAGE for recombinant Sapt3p (Fig. 1, lane 7). The N-terminal sequences of the 55- and 48-kDa products were determined to be RVVLNAG and GFYRTQDLI, respectively. These sequences corresponded to amino acids 19 to 25 and 75 to 83 of the SAPT3 translation product, after a signal peptidase processing site and after the Kex2p processing site KR, respectively. Our results suggest that Sapt3p was secreted by P. pastoris as a mix of proprotein and mature protein. Both recombinant Sapt2p and Sapt3p were glycosylated as demonstrated by reduction of the molecular mass after N-glycosidase F treatment (Fig. 1, lanes 5 to 8).
Sapt2p and Sapt3p, like Sapt1p, were shown to be inhibited by the classical aspartic proteinase inhibitor pepstatin A. The pH-dependent enzymatic activities of the recombinant Saps were determined in citrate buffer between pH 2.0 and 7.0. Sapt2p and Sapt3p had a pH optimum at 5.0, whereas both native and recombinant Sapt1p were optimally active at pH 3.5.
SAPT gene expression in C. tropicalis.
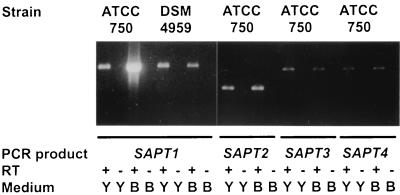
Total RNA was isolated and purified from C. tropicalis at different times of culture at 30°C in YPD medium, BSA medium, and in modified Lee's medium containing 5% fetal calf serum. A strong SAPT1 signal was obtained from culture in BSA medium with 35 cycles of PCR, while only faint SAPT2, SAPT3, and SAPT4 signals comparable to that of SAPT1 under noninduced conditions (YPD medium) were detected (Fig. 5). Interestingly, a faint SAPT1 was also detected with RNA isolated from C. tropicalis DSM 4959 grown under inducing and noninducing conditions. Only faint signals of each SAPT were also obtained with RNA extracted from cells grown in modified Lee's medium containing 5% fetal calf serum at different times of culture (data not shown). As a control in all experiments, RT-PCR performed in the absence of RT still showed no SAPT signals after 35 cycles of PCR. These negative results ascertained the absence of genomic DNA in the PCR.
FIG. 5.
RT-PCR performed with RNA extracted from cells grown for 24 h in YPD (Y) and BSA (B) media. For each reaction, a control without the reverse transcription step (RT −) was done. The sizes of the SAPT1, SAPT2, SAPT3, and SAPT4 PCR products are 1,005, 762, 1,118, and 1,130 bp, respectively. The primers used are described in Table 1.
DISCUSSION
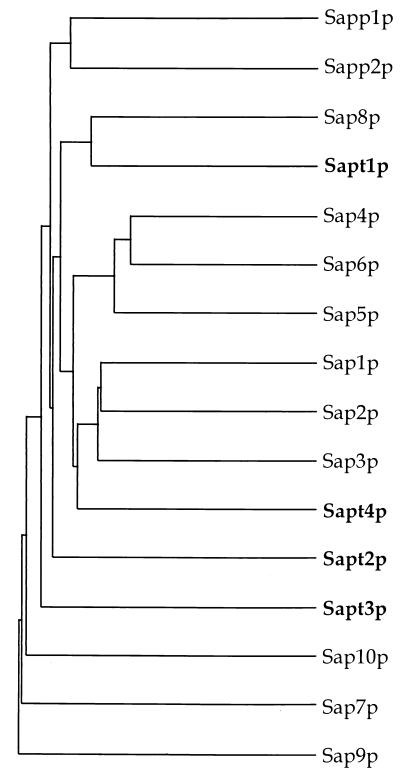
A total of four SAPT genes have now been cloned from C. tropicalis. All bands identified by Southern blotting of EcoRI-digested genomic DNA and low-stringency hybridization with SAPT1 could be assigned to a specific SAPT gene. Therefore, the likelihood of finding additional SAPT genes in the C. tropicalis genome is rather low. A dendrogram deduced from the alignments of Sapt1p to Sapt4p together with C. albicans Sap1p to Sap10p is shown in Fig. 6. Sapt1p to Sapt4p form distinct branches and are not clustered in subgroups like Sap1p to Sap3p and Sap4p to Sap6p from C. albicans (15, 16). The percentage of similarity between two members of the Saptp protein family does not exceed 63% (Table 3). However, Sapt1p and Sapt4p appeared to be clustered with C. albicans Sap8p and Sap1p to Sap3p, respectively. SAPT1 and SAP8 in one side and SAPT4 and SAP1-3 in another side could be the descendants of two different SAP genes in a species from which C. albicans and C. tropicalis split before further specialization.
FIG. 6.
Dendrogram of the SAP gene family in Candida species. The branch lengths are proportional to the similarity between amino acid sequences. The dendrogram was created by the PileUp program implemented in the GCG package of the Genetics Computer Group, University of Wisconsin, Madison.
TABLE 3.
Pairwise comparisons of putative Sap isoenzymes from C. tropicalis to which were added C. albicans Sap2p, Sap6p, and Sap8p
| Isoenzyme | % Similarity or identitya
|
||||||
|---|---|---|---|---|---|---|---|
| Sapt1p | Sapt2p | Sapt3p | Sapt4p | Sap2p | Sap6p | Sap8p | |
| Sapt1p | 61 | 50 | 63 | 67 | 56 | 73 | |
| Sapt2p | 49 | 42 | 60 | 58 | 58 | 61 | |
| Sapt3p | 38 | 31 | 48 | 55 | 52 | 56 | |
| Sapt4p | 55 | 48 | 35 | 67 | 63 | 60 | |
| Sap2p | 63 | 53 | 46 | 60 | 67 | 66 | |
| Sap6p | 50 | 50 | 46 | 53 | 60 | 58 | |
| Sap8p | 69 | 54 | 48 | 55 | 61 | 52 | |
The percent similarity (top right-hand corner) and percent identity (bottom left-hand corner) values were obtained with the program Gap implemented in the GCG package of the Genetics Computer Group, University of Wisconsin, Madison.
So far, only Sapt1p produced in BSA medium has been shown to be produced by C. tropicalis. One strain, DSM 4959, did not secrete detectable acid proteolytic activity. However, no difference was found between the regions coding for Sapt1p from strains ATCC 750 and DSM 4959. RT-PCR revealed that SAPT1 from strain DSM 4959 is expressed at a basic level in rich medium as well as in BSA medium in which SAPT1 of other strains is induced. Therefore, the lack of secreted proteolytic activity of DSM 4959 strain appears to be due to a defect in induction of SAPT1 and not to any mutation or deletion in this gene.
Several pieces of evidence suggest that the other SAPT genes are expressed under conditions that remain to be discovered in vitro or during infection, as described below.
(i) SAPT2 and SAPT3 gene products could be obtained from P. pastoris. They are highly active secreted aspartic proteases completely inhibited by pepstatin A.
(ii) It can be assumed that the weak signals obtained by RT-PCR reflect a residual level of expression. A control for the absence of genomic DNA involved an additional PCR in the absence of RT with selected RNA samples. No amplification product was detected in these control reactions. Likewise, no signal was detected when 100 ng of wild-type C. tropicalis DNA was added to RNA of mutants before DNase treatment.
(iii) Immunoblot analysis performed with sera of mice infected with C. tropicalis SAPT1 disruptants still revealed antibodies reacting with Sapt1p. This observation suggested that other isoenzymes were produced during infection with C. tropicalis (29).
(iv) It is likely that the SAPT genes expressed by C. tropicalis inside and outside the macrophages are different from SAPT1. Indeed, after ingestion of yeast cells by phagocytic cells, Sap antigens have been shown to be expressed by C. albicans and C. tropicalis, but not by C. parapsilosis (1, 20). However, under in vitro conditions, when serum albumin is the major nitrogen source in the growth medium, C. parapsilosis also produces large amounts of one dominant Sap (e.g., Sapp1p, like C. albicans and C. tropicalis, which produce Sap2p and Sapt1p, respectively (7, 14, 16, 20, 21, 28). Furthermore, the genes encoding these three isoenzymes are coordinately regulated (30). We have demonstrated in C. albicans that genes other than SAP2 (i.e., SAP4, SAP5, and SAP6) were induced after engulfment of yeast cells by macrophages (3). Therefore, it is likely that another gene different from SAPT1 is expressed in macrophages after phagocytosis of the yeast cells. Experiments are under way to identify what Sap or Saps among Sapt1p to Sapt4p are secreted under similar conditions by C. tropicalis.
The relevance of putative virulence attributes of pathogenic Candida species can be based on comparisons with nonpathogenic yeasts, like Saccharomyces cerevisiae and other less pathogenic species. For instance, many genes of C. albicans shown to be involved in virulence, especially those organized in large gene families (11, 16), have no homologous counterpart in the closely related yeast S. cerevisiae. Expansions of genes to form a gene family could reflect selection during evolution to allow organisms a better adaptation to different conditions of their environment. The use of several specific proteinases suitable to each particular condition during the infection could allow C. tropicalis, like C. albicans, to adhere to mucosae, resist phagocytic cells, and invade deep organs.
ACKNOWLEDGMENTS
We thank H. Pelloux, B. Lechenne, and S. Jaccoud for technical assistance and M. Holdom for critical review of the manuscript and assistance with English.
REFERENCES
- 1.Borg M, Ruchel R. Demonstration of fungal proteinase during phagocytosis of Candida albicans and Candida tropicalis. J Med Vet Mycol. 1990;28:3–14. [PubMed] [Google Scholar]
- 2.Borg M, Rüchel R. Expression of extracellular acid proteinase by proteolytic Candida spp. during experimental infection of oral mucosa. Infect Immun. 1988;56:626–631. doi: 10.1128/iai.56.3.626-631.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol. 1998;28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- 4.Borg-von Zepelin M B, Meyer I, Thomssen R, Wurzner R, Sanglard D, Telenti A, Monod M. HIV-protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J Investig Dermatol. 1999;113:747–751. doi: 10.1046/j.1523-1747.1999.00747.x. [DOI] [PubMed] [Google Scholar]
- 4a.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chambers S P, Prior S E, Barstow D A, Minton N P. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 6.De Bernardis F, Arancia S, Morelli L, Hube B, Sanglard D, Schafer W, Cassone A. Evidence that members of the secretory aspartyl proteinase gene family, in particular SAP2, are virulence factors for Candida vaginitis. J Infect Dis. 1999;179:201–208. doi: 10.1086/314546. [DOI] [PubMed] [Google Scholar]
- 7.de Viragh P A, Sanglard D, Togni G, Falchetto R, Monod M. Cloning and sequencing of two Candida parapsilosis genes encoding acid proteases. J Gen Microbiol. 1993;139:335–342. doi: 10.1099/00221287-139-2-335. [DOI] [PubMed] [Google Scholar]
- 8.Doumas A, van den Broek P, Affolter M, Monod M. Characterization of the prolyl dipeptidyl peptidase gene (dpplV) from the koji mold Aspergillus oryzae. Appl Environ Microbiol. 1998;64:4809–4815. doi: 10.1128/aem.64.12.4809-4815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossberger D. Minipreps of DNA from bacteriophage lambda. Nucleic Acids Res. 1987;15:6737. doi: 10.1093/nar/15.16.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn R, Wong B, Kiehn T E, Armstrong D. Fungemia in a cancer hospital: changing frequency, earlier onset, and results of therapy. Rev Infect Dis. 1985;7:646–655. doi: 10.1093/clinids/7.5.646. [DOI] [PubMed] [Google Scholar]
- 11.Hoyer L L, Hecht J E. The ALS6 and ALS7 genes of Candida albicans. Yeast. 2000;16:847–855. doi: 10.1002/1097-0061(20000630)16:9<847::AID-YEA562>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Hube B. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 1996;7:55–69. [PubMed] [Google Scholar]
- 13.Komshian S V, Uwaydah A K, Sobel J D, Crane L R. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;11:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald F, Odds F C. Inducible proteinase of Candida albicans in diagnostic serology and in the pathogenesis of systemic candidosis. J Med Microbiol. 1980;13:423–435. doi: 10.1099/00222615-13-3-423. [DOI] [PubMed] [Google Scholar]
- 15.Monod M, Hube B, Hess D, Sanglard D. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology. 1998;144:2731–2737. doi: 10.1099/00221287-144-10-2731. [DOI] [PubMed] [Google Scholar]
- 16.Monod M, Togni G, Hube B, Sanglard D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994;13:357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 17.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odds F C. Candida and candidosis. 2nd ed. London, United Kingdom: Bailliére Tindall; 1988. [Google Scholar]
- 19.Rangel-Frausto M S, Wiblin T, Blumberg H M, Saiman L, Patterson J, Rinaldi M, Pfaller M, Edwards J E, Jr, Jarvis W, Dawson J, Wenzel R P. National epidemiology of mycoses survey (NEMIS): variations in rates of bloodstream infections due to Candida species in seven surgical intensive care units and six neonatal intensive care units. Clin Infect Dis. 1999;29:253–258. doi: 10.1086/520194. [DOI] [PubMed] [Google Scholar]
- 20.Rüchel R, Böning B, Borg M. Characterization of a secretory proteinase of Candida parapsilosis and evidence for the absence of the enzyme during infection in vitro. Infect Immun. 1986;53:411–419. doi: 10.1128/iai.53.2.411-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruchel R, Uhlemann K, Boning B. Secretion of acid proteinases by different species of the genus Candida. Zentbl Bakteriol Mikrobiol Hyg A. 1983;255:537–548. [PubMed] [Google Scholar]
- 22.Ruchel R, Zimmermann F, Boning-Stutzer B, Helmchen U. Candidiasis visualised by proteinase-directed immunofluorescence. Virchows Arch A Pathol Anat Histopathol. 1991;419:199–202. doi: 10.1007/BF01626348. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Schaller M, Korting H C, Schafer W, Bastert J, Chen W, Hube B. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol. 1999;34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 25.Schaller M, Schafer W, Korting H C, Hube B. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol Microbiol. 1998;29:605–615. doi: 10.1046/j.1365-2958.1998.00957.x. [DOI] [PubMed] [Google Scholar]
- 26.Symersky J, Monod M, Foundling S I. High-resolution structure of the extracellular aspartic proteinase from Candida tropicalis yeast. Biochemistry. 1997;36:12700–12710. doi: 10.1021/bi970613x. [DOI] [PubMed] [Google Scholar]
- 27.Tang J, Wong R N. Evolution in the structure and function of aspartic proteases. J Cell Biochem. 1987;33:53–63. doi: 10.1002/jcb.240330106. [DOI] [PubMed] [Google Scholar]
- 28.Togni G, Sanglard D, Falchetto R, Monod M. Isolation and nucleotide sequence of the extracellular acid protease gene (ACP) from the yeast Candida tropicalis. FEBS Lett. 1991;286:181–185. doi: 10.1016/0014-5793(91)80969-a. [DOI] [PubMed] [Google Scholar]
- 29.Togni G, Sanglard D, Monod M. Acid proteinase secreted by Candida tropicalis: virulence in mice of a proteinase negative mutant. J Med Vet Mycol. 1994;32:257–265. doi: 10.1080/02681219480000331. [DOI] [PubMed] [Google Scholar]
- 30.Togni G, Sanglard D, Quadroni M, Foundling S I, Monod M. Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiology. 1996;142:493–503. doi: 10.1099/13500872-142-3-493. [DOI] [PubMed] [Google Scholar]
- 31.van't Wout J W. Fluconazole treatment of candidal infections caused by non-albicans Candida species. Eur J Clin Microbiol Infect Dis. 1996;15:238–242. doi: 10.1007/BF01591361. [DOI] [PubMed] [Google Scholar]
- 32.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberger M, Sacks T, Sulkes J, Shapiro M, Polacheck I. Increasing fungal isolation from clinical specimens: experience in a university hospital over a decade. J Hosp Infect. 1997;35:185–195. doi: 10.1016/s0195-6701(97)90206-1. [DOI] [PubMed] [Google Scholar]