Abstract
The abundance and types of yeasts in the wet and dry sand of three recreational beaches in South Florida were determined. Samples were collected on 17 occasions between August 2001 and July 2002. After analyzing 102 sand samples, a total of 21 yeast species were identified by molecular methods. These isolates comprised four Basidiomycetes and 17 Ascomycetes and included eight species that had previously been reported from humans. The most frequently encountered yeasts were Candida tropicalis and Rhodotorula mucilaginosa. A greater diversity of species (16 species) was found in the dry sand above the high tide mark compared with the wet sand in the intertidal zone (11 species). Densities were also highest in the dry sand relative to wet sand (20-fold higher at Hobie beach, 6-fold higher at Fort Lauderdale Beach and 1.3-fold higher at Hollywood beach). There were no clear temporal patterns in the data and overall densities were greatest at the busiest bathing beach (Hobie Beach) where total yeasts averaged 37,720 cfu 100 g -1 dry sand and 1852 cfu 100 g-1 in the wet sand. This concentration of yeast was significantly higher than populations at the less populated beaches. Fort Lauderdale beach had a mean count of 4130 cfu 100 g-1 dry sand and 705 cfu 100 g-1 in the wet sand while the least populated beach, Hollywood Beach averaged 1945 cfu 100 g-1 dry sand and 1483 cfu 100 g-1 wet sand. While definitive statements cannot be made, high levels of yeasts may have a deleterious bearing on human health and the presence of such a diverse aggregation of species suggests that yeasts could have a role as indicators of beach health.
Keywords: Fungi, Non-traditional indicators, Recreational waters, Beach quality
1. Introduction
The occurrence and activity of yeasts in near-shore marine waters is well established (Meyers and Ahearn, 1974; Kohlmeyer and Kohlmeyer, 1979) due to their role in the decomposition of organic substrates, nutrient recycling, biodegradation of hydrocarbons, and as prey for a variety of marine organisms (Siepmann and Hoehnk, 1962; Meyers and Ahearn, 1974; Lachance et al., 1976). More recently, there has been an interest in yeast populations inhabiting recreational beaches, particularly those concerned with human health. Papadakis et al. (1997) found a positive correlation between the numbers of swimmers and the presence of yeasts of human origin. Increasingly, it is being realized that traditional fecal indicator organisms, such as enterococci, can be poor measures of bathing water quality and the inclusion of non-traditional indicators, such as yeasts, may be desirable when testing recreational waters (Papadakis et al., 1997).
Open-ocean waters can contain between 10 and 100 yeast cells per liter, while near-shore waters support much larger populations, i.e. up to several thousand cells per liter (Fell, 1976), because these waters receive significant input from terrestrial yeasts (van Uden and Fell, 1968) and sewage-derived yeasts (Cooke et al., 1960; Fell, 1976. Fell et al. (1960) contend that in estuarine regions the majority of yeast species are of non-marine origin. In Biscayne Bay, Florida, Fell and van Uden (1963) demonstrated that population densities of intestinal yeasts often fluctuated according to the degree of pollution.
Beach sand receives constant microbiological inputs from the water, through the filtering action during tidal cycles, and from washout from land during rain events. Beach users also contribute directly to the microbial quality of beach sand. Consequently, recreational beaches may harbor significant yeast populations, particularly, if sand particles provide a micro-habitat for the enhanced survival of yeasts as suggested by Ghinsberg et al. (1994). Sand might be an important reservoir of yeasts and afford a health risk for beach users (Papadakis et al., 1997), since some 150 yeast species are pathogenic to humans and animals (Kwon-Chung and Bennett, 1992). Candida spp. infections, in particular, have increased dramatically in recent years due to the increase in immunocompromised individuals in the population (Cooke et al., 2002).
A few prior studies have demonstrated the presence of yeasts in sand. For example, Kishimoto and Baker (1969) found dermatophytes to be common in beach sands in Hawaii and (Papadakis et al. (1997) isolated yeasts from water and sand samples collected from a bathing beach in Greece. At least one study isolated human pathogenic fungi from beach sand (Anderson, 1979), while another reported pathogenic species from coastal areas of California (Dabrowa et al., 1964). However, the present study is the first to document the nature and abundance of yeast populations in subtropical beach sands.
It was therefore, the purpose of this study, to identify and enumerate the yeast species, which inhabit the sediments at three bathing beaches to determine if yeast species may have value as indicator organisms and to gain insight in the ecology of yeasts in subtropical intertidal environments.
2. Materials and methods
2.1. Sampling
Sand samples were collected from three recreational beaches in South Florida between August 2001 and July 2002. Hobie Beach in Miami (25°44′22.5″N, 80° 10′18.7″W) represented a ‘heavy use’ beach because of the high density of users per unit area. Fort Lauderdale (26°07′17.35″N, 80°06′14.24″W) represented a ‘moderate use’ beach and Hollywood Beach (26°02′02.56″N, 80°06′50.36″W) was a ‘light use’ beach. At each beach, two transects (100 m apart) running perpendicular to the water line were identified (transects 1 and 2). These beaches were sampled on alternate outings over the study period. Wet sand samples were collected midway between high and low tide level (along the transect), and dry sand samples were collected 5 m above the high tide level. Each sand sample was a composite of the top 12 cm of sand. At each location, three replicate samples (spaced 0.5 m apart) were collected. Sand was placed in sterile plastic bags in a cooler for transport to the laboratory and processed within 4 h of collection.
The 17 sampling dates were as follows: 8/28/2001; 9/11/2001; 9/25/2001; 10/9/2001; 10/23/2001; 11/6/2001; 12/4/2001; 1/14/2002; 1/28/2002; 2/11/2002; 2/25/2002; 3/11/2002; 3/25/2002; 4/23/2002; 5/29/2002; 6/25/2002; 7/23/2002. On each outing, temperature and salinity of the interstitial water was measured.
2.2. Enumeration of yeasts
Samples were processed for yeasts using the membrane filtration method of Sherry and Qureschi (1981). Twenty-five grams of sand were added to 200 ml of sterile phosphate-buffered saline (PBS) and shaken vigorously for 1 min to dislodge attached yeasts. Aliquots of supernatant (1–50 ml) were filtered through 0.45μm sterile Millipore filters and placed on Sabouraud dextrose agar (SDA) containing 150 mg/l streptomycin sulfate to suppress bacterial growth. After incubation at room temperature for 3–4 d, the numbers of yeasts (CFU) were counted. Colonies differing in morphology were subcultured and maintained on SDA at 4 °C until they could be identified by molecular methods.
2.3. Identification of yeasts
Axenic cultures for identification were grown in 2 ml micro-centrifuge tubes for 24–48 h in liquid YPD medium (10 g yeast extract, 10 g peptone, 10 g glucose in 1 l distilled water) with agitation at 30 °C. DNA was extracted using a QIAmp tissue kit (Qiagen, Valencia, CA) and amplified by PCR according to the methods outlined in Fell et al. (2000) and Scorzetti et al (2002). Depending on the strains, the following forward and reverse primer pairs were used: LR6/NS7 (CGC CAG TTC TGC TTA CC/GAC GCA ATA ACA GGT CTG TGA TGC) was initially used while the primer pair LR6/ITS5 (CGC CAG TTC TGC TTA CC/GGA AGT AAA AGT CGT AAC AAG G) or LR6/F63 (CGC CAG TTC TGC TTA CC/GCA TAT CAA TAA GCG GAG GAA AAG) was utilized when the initial primer pair failed to produce an amplicon. These pairs targeted the ITS region and the D1/D2 regions (part of the large subunit rDNA) of the genome. Amplicons of the positive PCR products were sequenced by the University of Florida DNA sequencing laboratory (Gainesville, FL). The primer for unidirectional sequencing was MLF (TAA GCG GAG GAA AAG). Acquired sequences were compared to yeast sequences in GenBank (using a BLAST search).
3. Results
Hobie Beach sand in Miami had the highest counts of yeasts throughout the year (averaging 19,786 CFU 100 g-1 sand), regardless of sand type [wet or dry]) followed by Fort Lauderdale beach (2418 CFU 100 g-1 sand) and Hollywood beach (1715 CFU 100 g-1 sand). Hobie Beach, the most populated of the beaches studied, had significantly more yeasts (p<0.001) than the two other less populated beaches. Comparing the numbers of yeasts in the dry and wet sand for all three beaches, dry sand harbored significantly more yeasts (14,599 CFU 100 g-1 sand) than wet sand (1347 CFU 100 g-1 sand). Much of this difference was attributable to the markedly more abundant yeast populations in Hobie beach dry sand relative to the other beaches (Table 1). Significance could not be demonstrated for seasonal differences. Statistical analysis was performed based on least-squares means (LSM) and Tukey’s test to determine whether or not the differences in yeast counts between beaches and between wet and dry sand were statistically significant. An R2 value of 0.55 for the overall data set indicates that 55% of the differences between yeast counts can be attributed to differences in the beaches and sand types, and that 45% of the differences are due to other factors, such as temperature, rainfall and nutrient availability.
Table 1.
Mean yeast counts (100 g-1 sand) in dry sand and wet sand at Hobie beach, Fort Lauderdale beach and Hollywood beach. Samples collected between August 2001 and July 2002
| Beach | CFU 100 g-1 sand |
|
|---|---|---|
| Dry sand | Wet sand | |
| Hobie | 37,720.14 | 1852.55 |
| Fort Lauderdale | 4130.35 | 705.10 |
| Hollywood | 1945.73 | 1483.94 |
There was no clear evidence of temporal trends in the data (Fig.1) despite the fact that temperature of the sand was lowest in winter (November–January, ca. 20–25°C) relative to mid summer (July–August, ca. 30–35°C). Trends may have been masked by two factors. Firstly, salinity varied markedly in the sand, which may have influenced the survival (and abundance) of yeasts in the sand. Generally, the salinity in dry sand fluctuated from 0 to 80 ppt and wet sand between 10 and 65 ppt. Secondly, as evident from the error bars in Fig. 1, yeast populations varied between replicate samples indicating that distributions were heterogeneous. This patchiness suggests that yeasts may have been growing in the sand.
Fig. 1.
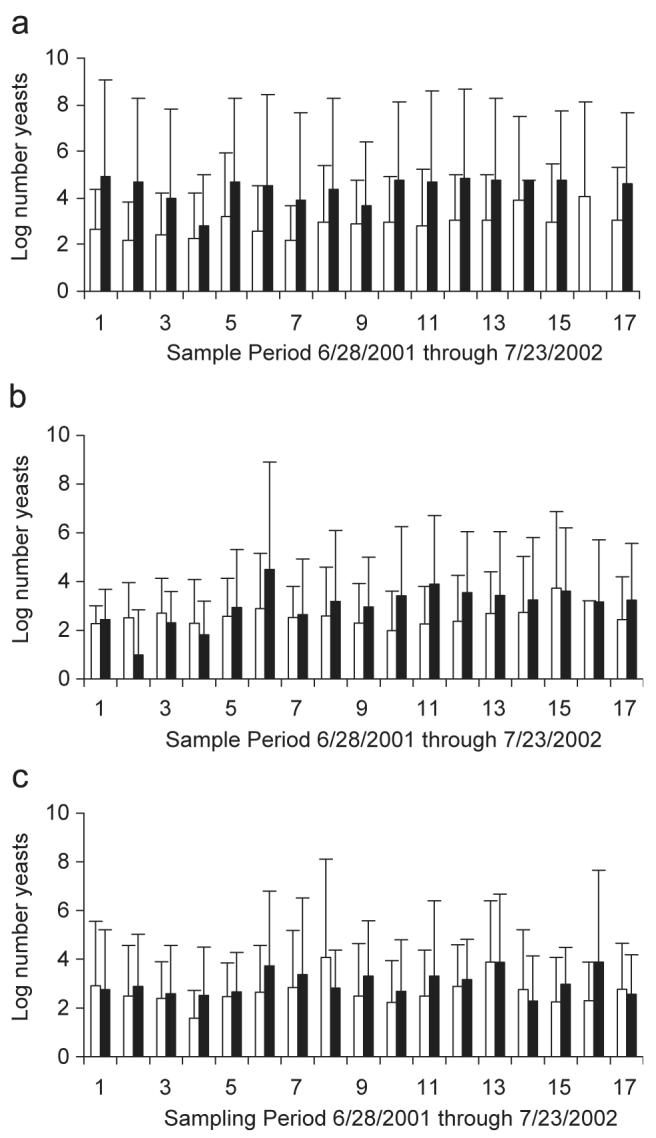
Number of yeasts (log10) per 100 g of sand (CFU) at Hobie Beach (top), Fort Lauderdale Beach (middle) and Hollywood Beach (bottom). Data as means (n = 3) with S.E. Open bars are wet sand samples; black bars are dry sand samples. Samples were collected between August 2001 and July 2002 every 2–3 weeks (specific sampling dates [1–17] given in Section 2).
A total of 21 different yeast species (four Basidiomycetes, 17 Ascomycetes) were identified by molecular methods from the sand samples (Table 2), along with three filamentous fungi (Aspergillus, Penicillium, Fusarium). These filamentous fungi were present in every collection and at all of the sampling sites. The most frequently isolated yeasts were Candida tropicalis and Rhodotorula mucilaginosa. All other yeasts were isolated on only a few occasions from the sand. In total, 11 yeast species were isolated from wet sand and 16 species from dry sand (Table 2).
Table 2.
Identity of yeasts in wet sand and dry sand from three beaches (Hobie Beach, Fort Lauderdale Beach and Hollywood Beach)
| Wet sand | Dry sand |
|---|---|
| Candida catenulate a | Candida albicans a |
| Candida tropicalis a | Candida tropicalis a |
| Candida ishiwadae | Candida catenulata a |
| Candida sp. | Candida naeodendra |
| Metschnikowia bicuspidata | Metschnikowia bicuspidata |
| Yarrowia lipolytica a | Yarrowia lipolytica a |
| Rhodotorula mucilaginosa a | Rhodotorula mucilaginosa a |
| Trichosporon asahii a | Trichosporon asahii a |
| Trichosporon coremiiforme a | Trichosporon coremiiforme a |
| Pichia anomala | Pichia anomala a |
| Pichia onychis a | Pichia ohmeri (Kodamaea ohmeri) |
| Clavispora lusitaniae a | Galactomyces sp. |
| Cryptococcus sp. | |
| Rhodosporidium paludigenum | |
| Torulaspora delbruckii | |
| Issatchenkia orientalis |
Species that have been isolated from human sources.
4. Discussion
This study is the first to provide quantitative data showing high levels of yeast populations surviving in sub-tropical beach sands. Kishimoto and Baker (1969) and Papadakis et al. (1997), reported on the occurrence of yeasts in beach sand but these studies provided no information on the abundance of the populations. In a study of yeasts in the waters off Hobie Beach, Miami, (Ahearn et al. (1968) reported yeast populations ranging from ca. 500 to 1000 cells per 100 ml. Although these abundances in water samples (as volume) cannot be compared directly with sand samples (as weight) they do suggest significant accumulations of yeasts in beach sand particularly at the most populated beach. At this beach (Hobie), the mean count over the sampling period was 19,787 CFU per 100 g sand. If the counts are normalized to account for the water content of the sand (i.e. ml of interstitial water in which the yeasts reside), the counts are dramatically increased. For example, taking the mean count of yeasts at Hobie beach over the sampling period, yeasts averaged 1852 and 37,720 yeasts per 100 g sand for wet and dry sand, respectively. In addition to a high level of human activity at Hobie Beach a number of users bring their dogs, which may also be a contributing factor. From a health perspective, these high concentrations might pose a health hazard although we have no conclusive evidence for this possibility.
The accumulation of yeasts in sand is not surprising since sand can filter microbes from the water column during tidal cycles and trap cells in surface water runoff. Moreover, the nature of sand particles with their protected microhabitats (i.e. cracks and crevices) rich in nutrients probably provide sites for the enhanced survival, and perhaps growth, of sequestered yeasts. In a similar way, in situ growth of fecal indicator bacteria on the beaches of South Florida has been demonstrated to result in significant accumulations of indicator bacteria in both wet and dry sand relative to the water column (A. Hartz and T. Bonilla, pers. comm.). These observations are confirmed by Shibata et al. (2004), who demonstrated highest concentrations of several microbial indicators (enterococci, Escherichia coli, fecal coliform, total coliform and Clostridium perfringens) in the swash zone along the high tide mark at Hobie Beach and progressively decreasing concentrations of indicators upon sampling in offshore waters. Solo-Gabrielle et al. (2000) in a study of sources of E. coli in a nearby coastal area also found high levels of E. coli at high tide and suggested this was the result of the emergence of newly grown cells washed up from the intertidal sediment. Although in situ growth of yeasts was not addressed in this study, the high numbers of readily culturable yeasts and their heterogeneous distribution on the beach suggests that they may indeed grow in the sand. Moreover, since yeasts of human origin are frequently found on the skin, many of these “shed” yeasts are probably tolerant of elevated salinities and could survive the elevated salinities found in beach sand (salinity ranged from 0 to 80 ppt over the sampling period).
Papadakis et al. (1997) noted that the survival of pathogenic yeasts in wet sand might have health implications for beach users. They also suggested that since yeasts of human origin correlated with the numbers of swimmers at popular beaches, the use of non-fecal indicators should also be considered when assessing beach quality. Indeed, Wade et al. (2003) indicated the need for additional microbiological indicators for assessing recreational waters.
The genus Candida is composed of more than 250 species (M.A. Lachance pers. comm.) of which eight species were recovered in the course of this study. Of these, only C. tropicalis had a common occurrence possibly because this species is prevalent in soil of warm locales (Kwon-Chung and Bennett, 1992). C. tropicalis can be pathogenic. The species has been found in infected nails, blood and feces, as a common intestinal inhabitant in gulls and terns (Buck, 1983), fish (Roth et al. 1962), and from sewage-polluted waters (Cooke and Matsuura, 1963), sand (Papadakis et al., 1997) and coastal waters off Miami (Fell et al. 1960; van Uden and Fell, 1968; Combs et al., 1971). R. mucilaginosa was commonly isolated from beach sand in this study reflecting a cosmopolitan distribution in terrestrial, freshwater, marine habitats as well as sewage (Cooke et al., 1960; Cooke and Matsuura, 1963). It has been isolated in waters around Miami and in beach sands from elsewhere (Ahearn et al., 1968; Papadakis et al., 1997). R. mucilaginosa is often isolated from humans and is currently the only species within the genus to cause human cutaneous and systemic infections (Kwon-Chung and Bennett, 1992).
The remaining 19 yeast species isolated in the course of this study were found only occasionally and never from all three beaches. The source of these yeasts is unclear since many are common in soil although some can also come from human sources. The higher diversity of yeasts in the dry sand at the top of the beach is consistent with inputs from soil and beach users. For example, C. albicans, C. ishiwadae and C. naeodendra have been isolated from soil and humans, soil, and insect frass, respectively (van Uden and Fell, 1968). C. catenulata is a human pathogen which has also been reported from estuarine waters, soil and beach sand (van Uden and Fell, 1968; Papadakis et al., 1997). Metschnikowia is frequently isolated from ephemeral flowers and their associated insects as well as from marine invertebrates, fish, and seagrasses (Lachance et al., 1998; Lachance and Starmer, 1998). Yarrowia lipolytica has been found in soil, meat products, petroleum products and the human cornea (Kurtzman, 1998) and Trichosporon spp. are prevalent in soils, estuarine waters, sewage and sandy beaches (Cooke and Matsuura, 1963; van Uden and Fell, 1968; Kishimoto and Baker, 1969; Papadakis et al. 1997) and some species cause severe human diseases (Gueho et al., 1998, Kwon-Chung and Bennett 1992, Diaz and Fell 2004). Pichia spp. have been isolated from sand (Papadakis et al. 1997), plants, infected human nails and dust (Kurtzman, 1998). Clavispora lusitaniae has been found in clinical specimens and cacti (Lachance and Phaff, 1998), Rhodosporidium paludigenum from mangroves in South Florida and Cryptococcus spp. from numerous sources including soil, polluted water and humans (Fell and Statzell-Tallman, 1998a, b). Torulaspora spp. are common in soil (Kurtzman, 1998) and Issatchenkia has been reported from seawater, feces, and food products (Kurtzman, 1998).
As indicated above, the most frequently isolated yeast species from beach sand were C. tropicalis and R. mucilaginosa. (Sherry and Qureschi (1981) state that while Rhodotorula species are ubiquitous, Candida species predominate in areas high in organically rich waters contaminated with industrial and domestic wastes. They proposed that the presence of pathogenic species of the genus Candida might serve as an indicator of fecal contamination in recreational areas. The presence of species of Candida in both wet and dry sand of all three beaches sampled supports the view that species of pathogenic Candida may be appropriate indicators of pollution in near-shore aquatic systems and beaches. In fact, C. albicans has been shown to be a primary etiologic agent in fungal infections in immunocompromised hosts, diabetics, neonates as well as postoperative patients (Bendel et al., 1993). In a study of fungal infections in these respective populations, Bendel et al. (1993) found C albicans represented 60% of the isolates and C. tropicalis 12–20% of the isolates whereas less pathogenic Candida species including C. parapsilosis, C. glabrata and C. krusei were cumulatively identified less than 20% of the time. Future studies may demonstrate that both C. albicans and C. tropicalis are reliable and accurate indicators of fecal contamination. Based on the rates of occurrence on the human body, one would anticipate that C. albicans would be in high incidence in the sand, along with C. tropicalis. Potentially C. albicans may have a high rate of die off and, therefore, could be an indicator of immediate contamination. In contrast, C. tropicalis may survive longer in the environment and possibly re-grow in the sand. Moreover, Desmarais et al. (2002) suggest it may not be appropriate to utilize traditional fecal indicator organisms such as Escherichia coli in subtropical recreational beaches. Further studies are required to determine the parameters necessary to correlate yeast populations with measurable levels of pollution and/or health risk.
5. Conclusions
More populated recreational beaches harbor greater numbers and species of potentially pathogenic yeast organisms.
High concentrations of yeast species may pose a health risk to human populations.
Selected yeast species such as C. albicans and C. tropicalis, may be useful, measurable indicators of the health risk associated with pollution and/or health risk of recreational beaches.
Acknowledgments
This work was partially funded by the United States Environmental Protection Agency (Grant R828830) to AR. The research results have not been subjected, however, to the EPA’s required peer review and therefore do not necessarily reflect the view of the Agency. No official endorsement should be inferred. Participation by RSMAS (AT & JF) was funded, in part, by the NSF-NIEHS Oceans and Human Health Centers Program (NSF 0432368 and NIEHS P50ES12736).
REFERENCES
- Ahearn DG, Roth FJ, Meyers SP. Ecology and characterization of yeasts from aquatic regions of South Florida. Mar. Biol. 1968;1:291–308. [Google Scholar]
- Anderson JH. vitro survival of human pathogenic fungi in seawater. Sabouraudia. 1979;7:1–12. [PubMed] [Google Scholar]
- Bendel CM, Hostetter MK, McClellan M. Distinct mechanisms of epithelial adhesion for Candida albicans and Candida tropicalis: identification of the participating ligands and development of inhibitory peptides. J. Clin. Invest. 1993;92(4):1840–1849. doi: 10.1172/JCI116775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck JD. Occurrence of Candida albicans in fresh gull feces in temperate and subtropical areas. Microb. Ecol. 1983;9:171–176. doi: 10.1007/BF02015129. [DOI] [PubMed] [Google Scholar]
- Combs JL, Murchelano RA, Jurgen F. Yeasts isolated from Long Island Sound. Mycologia. 1971;63:178–181. [PubMed] [Google Scholar]
- Cooke WB, Matsuura GS. A study of yeast populations in a waste stabilization pond system. Protoplasma. 1963;57:163–187. [Google Scholar]
- Cooke WB, Phaff HJ, Miller MW, Shifrine M, Knapp EP. Yeasts in polluted water and sewage. Mycologia. 1960;52:210–230. [Google Scholar]
- Cooke WB, Miles RJ, Price RG, Midgley G, Khamry W, Richardson AC. New chromogenic agar medium for the identification of Candida spp. Appl. Environ. Microbiol. 2002;68:3622–3627. doi: 10.1128/AEM.68.7.3622-3627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowa N, Landau JW, Newcomer VD, Plunkett OA. A survey of tide-washed coastal areas of southern California for fungi potentially pathogenic to man. Mycopathol. Appl. Mycol. 1964;24:137–150. doi: 10.1007/BF02075556. [DOI] [PubMed] [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 2002;68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MR, Fell JW. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J. Clin. Microbiol. 2004;42:3696–3706. doi: 10.1128/JCM.42.8.3696-3706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JW, Statzell-Tallman A. Rhodsporidium Banno. In: Kurtzman CP, Fell JW, editors. The Yeasts, A Taxonomic Study. Elsevier Science; Amsterdam: 1998. pp. 678–692. [Google Scholar]
- Fell JW, Statzell-Tallman A. Cryptococcus Vuillemin. In: Kurtzman CP, Fell JW, editors. The Yeasts, A Taxonomic Study. Elsevier Science; Amsterdam: 1998a. pp. 742–762. [Google Scholar]
- Fell JW, van Uden N. Yeasts in marine environments. In: Oppenheimer CH, editor. Symposium on Marine Microbiology. C.C. Thomas; Springfield, Illinois: 1963. pp. 329–340. [Google Scholar]
- Fell JW, Ahearn DG, Meyers SP, Roth FJ., Jr. Isolation of yeasts from Biscayne Bay, Florida and adjacent benthic areas. Limnol. Oceanogr. 1960;5:366–371. [Google Scholar]
- Fell JW. Yeasts in oceanic regions. In: Jones EBG, editor. Recent Advances in Aquatic Mycology. Elek Science; London: 1976. pp. 93–124. [Google Scholar]
- Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large subunit rD1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Ghinsberg RC, BarDov L, Rotol M, Sheinberg Y, Nitzan Y. Monitoring of selected bacteria and fungi in sand and sea water along the Tel Aviv coast. Microbios. 1994;77:29–40. [PubMed] [Google Scholar]
- Gueho E, Smith MT, deHoog GS. Trichosporon Behrend. In: Kurtzman CP, Fell JW, editors. The Yeasts, A Taxonomic Study. Elsevier Science; Amsterdam: 1998. pp. 854–872. [Google Scholar]
- Kishimoto RA, Baker GE. Pathogenic and potentially pathogenic fungi isolated from beach sands and selected soils of Oahu, Hawaii. Mycologia. 1969;61:537–548. [PubMed] [Google Scholar]
- Kohlmeyer J, Kohlmeyer E. Marine Mycology: The Higher Fungi. Academic Press; New York: 1979. p. 690pp. [Google Scholar]
- Kurtzman CP. Yarrowia van der Walt and von Arx. In: Kurtzman CP, Fell JW, editors. The Yeasts, A Taxonomic Study. Elsevier Science; Amsterdam: 1998. pp. 420–432. [Google Scholar]
- Kwon-Chung KJ, Bennett JE. Medical Mycology. Lea and Febiger; Philadelphia: 1992. p. 866pp. [Google Scholar]
- Lachance MA, Phaff HJ. Clavispora Rodrigues de Miranda. In: Kurtzman CP, Fell JW, editors. The Yeasts, A Taxonomic Study. Elsevier Science; Amsterdam: 1998. pp. 148–152. [Google Scholar]
- Lachance MA, Starmer WT. Ecology and yeasts. The Yeasts, A Taxonomic Study. In: Kurtzman CP, Fell JW, editors. Elsevier Science; Amsterdam: 1998. pp. 21–30. [Google Scholar]
- Lachance MA, Miranda M, Miller MW, Phaff HJ. Dehiscence and active spore release in pathogenic strains of the yeast Metschnikowia bicuspidata var. australis : possible predatory implications. Can. J. Bot. 1976;22:1756–1761. doi: 10.1139/m76-259. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Rosa CA, Starmer WT, Schlag-Edler B, Barker JSF, Bowles JM. Metschnikowia continentalis var. borealis, Metschnikowia continentalis var. continentalis, and Metschnikowia hibisci, new heterothallic haploid yeasts from ephemeral flowers and associated insects. Can. J. Microbiol. 1998;44:279–288. [Google Scholar]
- Meyers SP, Ahearn DG. Implications of yeasts and yeast-like fungi in marine processes. Veroeff. Inst. Meeresforsch. Bremerh. 1974;(Suppl. 5):321–338. [Google Scholar]
- Papadakis JA, Mavridou A, Richardson SC, Lampiri M, Marcelou U. Bather-related microbial and yeast populations in sand and seawater. Water Res. 1997;31:799–804. [Google Scholar]
- Roth FJ, Jr., Ahearn DG, Fell JW, Meyers SP, Meyers SA. Ecology and taxonomy of yeasts isolated from various marine substrates. Limnol. Oceanogr. 1962;7:178–185. [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. Systematics of basidiomycetous yeasts: a comparison of large sub-unit D1D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Sherry JP, Qureschi AA. Isolation and enumeration of fungi using membrane filtration. In: Dutka BJ, editor. Membrane Filtration—Applications, Techniques, and Problems. Marcel Dekker, Inc.; New York: 1981. pp. 189–212. [Google Scholar]
- Shibata T, Solo-Gabriele HM, Fleming LE, Elmir S. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepmann R, Hoehnk W. Ueber hefen und einige pilze (Fungi imp., Hyphales) aus dem Nordatlandtik. Veroff. Inst. Meeresforschung Bremerhaven. 1962;8:79–97. [Google Scholar]
- Solo-Gabrielle HM, Wolfert MA, Desmarais TR, Palmer CJ. Sources of E. coli in a coastal subtropical environment. Appl. Environ. Microbiol. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uden N.van., Fell JW. Marine yeasts. In: Droop MR, Ferguson Wood EF, editors. Advances in Microbiology of the Sea. Academic Press; London: 1968. pp. 167–201. [Google Scholar]
- Wade TJ, Pai N, Eisenberg JNS, Colford JM., Jr. Do US environmental protection agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 2003;111:1102–1109. doi: 10.1289/ehp.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]


