Shin and Grueber highlight work of the Jan lab that implicates the GARP complex in sterol transport during dendrite development.
Abstract
Disruptions in membrane trafficking are associated with neurodevelopmental disorders, but underlying pathological mechanisms remain largely unknown. In this issue, O’Brien et al. (2023. J. Cell Biol. https://doi.org/10.1083/jcb.202112108) show how GARP regulates sterol transfer critical for remodeling of dendrites in flies.
Neurons are characterized by their extensive membrane surface area in the form of highly branched dendrites and axons. Neuronal morphology provides a platform for connectivity and function of the nervous system but also creates special challenges for trafficking of proteins, RNAs, and lipids within cells. Thus, many mutations in membrane trafficking pathways are associated with neurodevelopmental disorders and neurodegenerative conditions, including Alzheimer’s disease, Parkinson’s disease, hereditary peripheral neuropathy, and progressive cerebello-cerebral atrophy type 2 (1–3).
In neurons, proteins and sterols are synthesized in the ER and then sent to the Golgi complex, part of the endomembrane system. In anterograde trafficking, incoming vesicles enter the cis side of the Golgi, and outgoing vesicles leave via the TGN. Retrogradely, the Golgi processes cell surface proteins from endolysosomal pathways for recycling and degradation. The Golgi complex also makes contacts with organelles, endosomes, and plasma membrane to counter-exchange lipids, which is a critical process for lipid homeostasis, membrane integrity, and a significant number of cellular processes, such as adhesion and migration (4). These bi-directional exchanges are tightly controlled by multiple tethering proteins in the secretory and endolysosomal pathways, including the Golgi-Associated Retrograde Protein (GARP) and Endosome-Associated Recycling Protein (EARP) complexes. Prior work in non-neuronal cells has provided important insights about the roles of GARP and EARP complexes (5). However, studies of GARP and EARP in neuronal development have been hampered by the lethality of knockout animals. In this issue, O’Brien et al. (6) report complex-specific roles of GARP and EARP in dendrite regrowth during development by using sophisticated genetic approaches in Drosophila, a sterol auxotroph that lacks the ability to synthesize sterols (6).
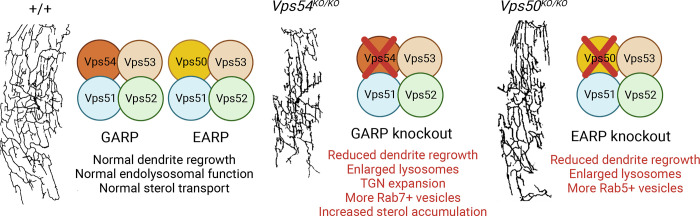
GARP and EARP are conserved membrane complexes each composed of three shared subunits (vacuolar protein sorting 51 [Vps51], Vps52, Vps53) and one unique subunit that determines the localization of GARP to the Golgi complex (Vps54) and EARP to recycling endosomes (Vps50; 7). The authors generated GARP and EARP subunit-specific and common subunit mutant fly strains in which they first evaluated survival. Animals with mutations in complex-specific subunits survived until adult stages, providing an opportunity to study neuronal phenotypes in later life stages in mutant animals. However, mutating the shared subunit Vps53 caused early lethality, thus they used mosaic approaches to circumvent the lethality of Vps53 mutants and study phenotypes in adult stages. In Vps54 and Vps50 mutants, adult fly dendrites became dramatically simplified, with fewer branches and restricted total dendritic arbor length (Fig. 1). Drosophila sensory neurons are born embryonically and progress through embryonic, larval, and adult phases of growth. Embryonic and larval dendritogenesis culminates in complete body wall coverage of the highly branched cIV neurons. These dendrites are pruned in pupae and regrow into adult-specific forms. Surprisingly, larval cIV dendrites developed normally in Vps54 and Vps50 mutants, indicating a selective late-stage dendritic defect, and roles for GARP and EARP in dendritic re-growth after pruning.
Figure 1.
GARP and EARP complexes are required for vesicle trafficking and dendrite remodeling. GARP and EARP complexes share three common units, Vps51, Vps52, and Vps53, and a complex-specific Vps. Mutations in Vps54 of the GARP complex (Vps54KO/KO) induce changes in sensory neurons, including expansion of Rab7-positive vesicles and TGN, and increased sterol accumulation, whereas mutations in Vps50 of the EARP complex (Vps50KO/KO) induce expansion of Rab5-positive vesicles. In the absence of either the GARP or EARP complex–specific component, cIV sensory neurons in adult flies exhibit enlarged lysosomes and reduced regrowth of dendrites. Representative images of dendritic growth in cIV neurons of +/+, Vps54KO/KO, and Vps50KO/KO flies from O’Brien et al. (6). Figure created with BioRender.com.
O’Brien et al. (6) next asked how GARP and EARP regulate dendritogenesis. Because both complexes have distinct roles in lipid metabolism and endolysosomal pathways, the authors focused their cell biological studies on alterations in endolysosomal pathways and sterol localization in the neuronal cell body. They found that, around the time of dendrite regrowth, free sterols accumulated in the TGN of GARP-deficient neurons, indicating increased transfer of sterols to the secretory pathway during this development stage. This observation prompted them to investigate the source of sterol. They first checked whether sterol transfer between the ER and Golgi was affected in the absence of GARP. For that, they deleted the lipid transfer protein Oxysterol-binding protein (Osbp) in GARP-deficient animals. Osbp binds to several different lipid ligands and acts to exchange lipids in the ER for phosphatidylinositol 4-phosphate (PI4P) at ER-Golgi membrane contact sites (8). Consistent with a role for Osbp in dendrite regrowth, loss of a single gene copy of osbp, which would be expected to reduce sterol accumulation at the Golgi, largely rescued dendrite regrowth to normal levels, whereas overexpression of osbp exacerbated the regrowth defect. One key prediction made by the authors is that if GARP regulates Osbp activity, then knockdown of PI4 kinase (known as four wheel drive, fwd, in Drosophila) would reduce levels of PI4P in the Golgi, and decrease Osbp-mediated accumulation of sterols in the Golgi. The authors indeed confirmed with PI4P reporters that RNAi knockdown of fwd reduced levels of PI4P at the TGN, but, contrary to expectations, it aggravated the dendrite morphology defects in Vps54 knockouts. This surprising effect of fwd RNAi on Vps54KO/KO neurons led O’Brien et al. (6) to propose an alternative Osbp-dependent sterol transfer pathway other than the ER-Golgi membrane contact site, such as those between Golgi and endosomes. In fact, Osbp localizes at other membrane contact sites, including ER-endosome and TGN-endosome contacts (8). TGN-endosome contact sites may be mediated by endosomes positive for Rab11, which binds Osbp through RELCH (9) and has been shown to genetically interact with Vps54 in Drosophila (10). Thus, the authors speculated that increased TGN-Rab11+ recycling endosome contacts may underlie sterol accumulation in the absence of GARP, but whether an interaction of Rab11+ endosomes with TGN indeed enables Osbp-mediated sterol transfer remains to be tested. Future studies investigating how secretory and endolysosomal pathways are remodeled in the absence of GARP and EARP complexes, as well as their potential interactions, could shed light on the regulation of dendrite morphogenesis by vesicle trafficking.
The work of O’Brien et al. (6) highlights the importance of tight regulation of sterol transport for dendritogenesis. But how does sterol accumulation in TGN perturb dendritic regrowth? At the first stage of the remodeling of cIV dendrites, dendrites grow rapidly for about 24 h, take a hiatus, and then begin a second stage of the remodeling (6). Therefore, the simplest explanation may be that a disruption in lipid transport impacts delivery to rapidly growing tips of dendrites, which require a considerable supply of lipids while they build plasma membranes. Likewise, perturbation of lipids may have broader consequences, as lipids also serve as structural components of vesicles, form lipid rafts for cell surface proteins, and act as signaling molecules such as hormones (5). This could result in aberrant membrane trafficking of cell surface proteins required for interactions between dendrites and their substrate, which may be inferred from the reduced receptive field coverage by cIV dendrites in GARP and EARP mutants.
Overall, through carefully validating and dissecting cellular mechanisms of GARP and EARP complexes in mediating lipid transport and recycling, this study by O’Brien et al. (6) opens exciting research directions that could contribute to better understand the impact of pathogenic mutations of the GARP and EARP complexes in developmental and degenerative disorders of the nervous system.
Acknowledgments
The authors are supported by National Institutes of Health 5R01NS061908 (W.B. Grueber) and 1R21CA274588 (W.B. Grueber and G.J. Shin). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Feinstein, M., et al. 2014. J. Med. Genet. 10.1136/jmedgenet-2013-101823 [DOI] [Google Scholar]
- 2.Small, S.A., et al. 2005. Ann. Neurol. 10.1002/ana.20667 [DOI] [Google Scholar]
- 3.Zimprich, A., et al. 2011. Am. J. Hum. Genet. 10.1016/j.ajhg.2011.06.008 [DOI] [Google Scholar]
- 4.David, Y., et al. 2021. Contact. 10.1177/25152564211034424 [DOI] [Google Scholar]
- 5.Spang, A. 2016. Front. Cell Dev. Biol. 10.3389/fcell.2016.00035 [DOI] [Google Scholar]
- 6.O’Brien, C.E., et al. 2023. J. Cell Biol. 10.1083/jcb.202112108 [DOI] [Google Scholar]
- 7.Schindler, C., et al. 2015. Nat. Cell Biol. 10.1038/ncb3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsu, F., and Kawasaki A.. 2021. Front. Cell Dev. Biol. 10.3389/fcell.2021.664788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobajima, T., et al. 2018. J. Cell Biol. 10.1083/jcb.201709123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson, E.C., et al. 2021. Front. Genet. 10.3389/fgene.2021.762012 [DOI] [PMC free article] [PubMed] [Google Scholar]