Abstract
Objective(s):
This research was designed to study the prevalence of OqxAB efflux pump genes and also to investigate the relationship between efflux pump and resistance to antibiotics, especially to fluoroquinolones, evaluate the expression levels of OqxAB genes, and molecular typing of Klebsiella pneumoniae isolated from hospitalized patients in Hamadan hospitals, west of Iran.
Materials and Methods:
In a cross-sectional study, 100 clinical strains of K. pneumoniae were isolated from hospitalized patients in three major teaching hospitals from January to June 2021. The antibiotic susceptibility of isolates was evaluated by the disk-diffusion agar method. The frequency of genes encoding oqxA and oqxB of efflux pump genes was investigated by PCR, and the expression of the oqxA efflux pump gene was investigated by the Real-time PCR method. The genetic relationship of K. pneumoniae isolates was analyzed by the Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR technique.
Results:
According to our results, the multi-drug resistance phenotype (MDR) in 65% and high prevalence resistance to ciprofloxacin in 89% of K. pneumoniae isolates was detected. The higher prevalence of oqxA (95%) and oqxB (98%) was also detected. There was a significant relationship between ciprofloxacin resistance and the oqxB gene as well as between ceftriaxone and chloramphenicol resistance and the oqxA gene. The expression of the oqxA gene was higher in ciprofloxacin-resistant isolates.
Conclusion:
The results of this study suggest a potential reservoir for the spread of OqxAB genes among hospital-acquired bacteria. Infection control strategies should be used prudently to reduce the spread of resistant strains of K. pneumoniae in hospitals.
Key Words: Antimicrobial, Ciprofloxacin, Efflux, K. pneumoniae, Pump, Resistance
Introduction
Klebsiella pneumoniae is one of the most important nosocomial pathogens that cause various infections in the urinary tract, bloodstream, and respiratory tract. This bacterium has the ability to develop resistance to cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, and even polymyxins (1, 2). K. pneumoniae may also produce a thick layer of extracellular biofilm, which helps them adhere to various surfaces. Infections caused by biofilm-forming strains of K. pneumoniae are difficult to treat (3).
The increasing rate of multi-drug resistance (MDR) K. pneumoniae is a major challenge in hospitals and has been reported in several studies in Iran (4-7). Treatment of MDR K. pneumoniae infections is often challenging due to the lack of available therapeutic choices, increasing length of hospitalization morbidity, mortality, and healthcare-associated costs (8).
Infections caused by K. pneumoniae are mainly treated with beta-lactams and fluoroquinolones (9, 10). Ciprofloxacin belongs to fluoroquinolones used to treat different bacterial infections (10). Resistance to fluoroquinolones is mainly caused by mutations in genes encoding qyrA and topoisomerase IV. However, low-level resistance due to plasmid (Plasmid-mediated quinolone resistant=PMQR) is caused by qnr genes, QepA, and OqxAB efflux pumps (11-14).
Efflux pumps are found in almost all species of bacteria and play a role in the intrinsic and acquired resistance to many antibiotics. Most of the efflux pump genes are located on the chromosome, but some of them are located on the plasmid. In 2004, efflux pump OqxAB was detected in plasmid-encoded multi-drug resistance in Escherichia coli in Denmark. In recent years, there have been reports of increased prevalence of efflux pump OqxAB in the Enterobacteriaceae family (15). A high prevalence of OqxAB efflux pump has been reported in K. pneumoniae. The main genetic origin of this efflux pump has been detected in the K. pneumoniae chromosome. In several studies, this efflux pump has only been reported in clinical isolates of K. pneumoniae, and it has been hypothesized that this efflux pump is widely present among ESBL (extended-spectrum beta-lactamase) producing and carbapenemase-producing isolates (15-18).
A repetitive element palindromic (REP) PCR method such as ERIC-PCR is a quick, reliable, and cost-effective technique for molecular typing of the Enterobacteriaceae family, therefore we used ERIC-PCR for the analysis of genetic linkage among K. pneumoniae isolates (19).
The main purpose of this study was to investigate the frequency and expression of OqxAB efflux pump genes as well as antibiotic resistance patterns especially ciprofloxacin and molecular typing of K. pneumoniae isolates from clinical samples of patients in Hamadan hospitals, west of Iran.
Materials and Methods
Bacterial isolation and identification
In a cross-sectional study, a total of 100 K. pneumoniae isolates were isolated from different clinical samples of patients in different wards of three major hospitals in Hamadan city, from January to June 2021. K. pneumoniae isolates were confirmed by conventional and molecular methods (20).
Antimicrobial susceptibility testing
Antimicrobial susceptibility to 12 different antibiotics including amikacin (30 µg), ampicillin-sulbactam (10/10 µg), ceftriaxone (30 µg), cefotaxime (30 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), imipenem (10 µg) meropenem (10 µg), gentamicin (10 µg), nitrofurantoin (300 µg), piperacillin-tazobactam (100/10 µg), tobramycin (10 µg), and chloramphenicol (30 µg) was detected by the disk diffusion method according to CLSI 2021 criteria (21). The antibiotic disks were supplied by Condalab Company from Spain. Escherichia coli ATCC 25922 was used as the quality control strain. MICs of ciprofloxacin in K. pneumoniae isolates were determined by the micro broth dilution method over a range of dilutions from 0.5 to 256 μg/ml using ciprofloxacin (Sigma–Aldrich). According to CLSI guidelines, breakpoints were used for the interpretation of ciprofloxacin MIC results (susceptible: ≤1 μg/ml; resistant: >1 μg/ml).
DNA extraction and PCR
Genomic DNAs of K. pneumoniae isolates were extracted by boiling after alkaline (NaOH) treatment of cells (22). Molecular detection of oqxA and oqxB genes encoding the OqxAB efflux pump was performed by PCR and using specific primers as described previously (23). Amplification reactions were performed in a final volume of 25 μl including 12 μl of 2X master mix (Ampliqon, Demark), 0.5 μM each primer, 2 μl DNA template, and 13 μl of double-distilled water (ddH2O). The PCR conditions were as follows: oqxA gene: initial denaturation (2 min at 94 °C), followed by 25 cycles of denaturation (15 sec at 94 °C), annealing (30 sec at 56 °C), and extension (1 min at 72 °C), followed by the final extension at 72 °C for 7 min. oqxB gene: initial denaturation (2 min at 94 °C), followed by 32 cycles of denaturation (30 sec at 94 °C), annealing (30 sec at 55 °C), and extension (1 min at 72 °C), followed by the final extension at 72 °C for 10 min. PCR products were subjected to electrophoresis in 1.2% agarose gel (Invitrogen, US) containing DNA Safe Stain (CinnaGen Co, Iran) at 70 V for 1 hr, and the band patterns were visualized in a Gel Doc.
RNA Extraction and cDNA synthesis for RT-PCR
Real-time PCR (RT-PCR) was used to investigate the expression level of the oqxA gene. A total of 16 K. pneumoniae strains were selected based on ciprofloxacin MICs, MDR phenotype, susceptibility to ciprofloxacin, type of clinical sample, and also the presence of the oqxA and oqxB genes. Total RNA was extracted using an RNA extraction kit (SinaClone, Iran), and converted into cDNA using the cDNA synthesis kit (AddBio, Korea) according to the manufacturer’s instruction. The quality and purity of the RNA obtained was evaluated using a spectrophotometer.
Real-Time PCR reaction
Real-time quantification of cDNA was carried out in a detection system (Roche, Germany) using the SYBR green PCR master mix. The optimized reaction consisted of a master mix (10X), 1 µl of each primer (10 pmol), 2 µl of cDNA (100 μg/ml), and 6 µl of DEPC water in a total volume of 20 µl. ureD gene primer was used as the internal control. Expression values (R) were determined using the ΔΔCt method. Expressions of all genes were calculated using the 2−ΔΔCt method (fold). The real-time PCR procedure was programmed as follows: initial denaturation at 95 °C (15 min) followed by 40 cycles of 94 °C (10 sec), 55 °C (60 sec), 72 °C (30 sec), and melt curve at 60 °C (15 sec) and 94 °C for 15 sec.
The analysis of the results was done using the SPSS software, version 22.0 (IBM Co., Armonk, NY, USA). Categorical variables were compared by the Chi-square or Fisher’s exact test and continuous variables were compared by the Mann-Whitney test. The difference in the expression level of oqxA was analyzed using a t-test for two independent means.
ERIC-PCR
The ERIC-PCR technique was used for the analysis of the genetic link among 100 K. pneumoniae isolates. This technique was carried out in a thermal cycler machine (Bio-Rad, Inc. USA) using the primer ERIC (F):5ʹ-ATG TAA GCT CCT GGG GAT TCAC-3ʹ and ERIC (R): 5ʹ-AAG TAA GTG ACT GGG GTG AGC G3ʹ (Metabion Co, Germany) according to the protocol described previously (24). The electrophoresis of PCR products was done on a 2% agarose gel (Sigma-Aldrich) at 70 V for 1 hr, and the patterns of ERIC bands were visualized on gel documentation. The patterns of ERIC bands were analyzed by an online data analysis service (inslico.ehu.es) which compared the ERIC profiles using the Dice method and clustered by the PGMA program.
Results
Out of 100 K. pneumoniae clinical isolates, 33% were isolated from the urine culture, 31% from the trachea, 13% from the blood culture, 12% from the wound, and 1% from Bronchoalveolar lavage (BAL), Cerebrospinal fluid (CSF), Pleural fluid, and gastric lavage. Out of 100 K. pneumoniae, 52% of K. pneumoniae strains were isolated from female and 48% from male patients. In this study, the range age of patients was 20-92 years. The prevalence of K. pneumoniae was higher in the age range of 71-80 years (24%). Most (74%) K. pneumoniae-positive samples were isolated from patients in the ICUs of the hospitals.
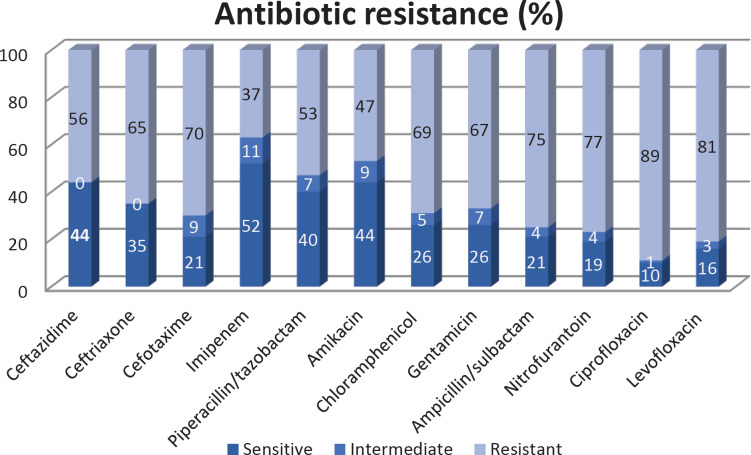
According to the results of antimicrobial susceptibility testing by disk diffusion (Figure 1), the highest antibiotic resistance was to ciprofloxacin (89%) and levofloxacin (81%). The MDR phenotype was detected in 65% of isolates. Ciprofloxacin MIC ranged from 0.5 to 256 µg/ml. The MIC ranges of ciprofloxacin were from 0.5-256 µg/ml. MICs equal to 128 µg/ml (18%), 256 µg/lm (26%), 64 µg/lm (23%), 32 µg/ml (18%), 16 µg/ml 16(%), 4 µg/ml (2%), 1 µg/ml (4%), and 0.5 µg/ml (1%) were detected.
Figure 1.
Antibiotic resistance (%) of Klebsiella pneumoniae isolated from Hamadan hospitals
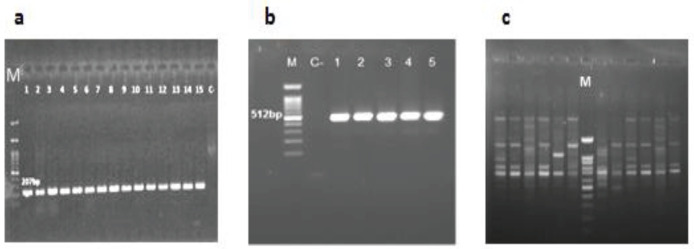
According to PCR results the prevalence of oqxA and oqxB was 95% and 98%, respectively (Figure 2). Among the 10 isolates susceptible to ciprofloxacin, 3 (30%) and 1 (10%) of isolates were positive for oqxA and oqxB, respectively. There was a significant relationship between resistance to ciprofloxacin and oqxB gene (P-value=0.04) and also between resistance to ceftriaxone (P-value=0.01), chloramphenicol (P-value= 0.02), and oqxA gene.
Figure 2.
Gel electrophoresis of PCR products of OqxAB encoding genes and ERIC-PCR typing in Klebsiella pneumoniae isolates from hospitalized patients in Hamadan hospitals
a: oqxA (207bp), b: oqxB (512bp), c: ERIC-PCR, Lane M: 100 bp DNA size marker
The results of Real-time PCR showed that the oqxA gene was expressed in both ciprofloxacin-susceptible and ciprofloxacin-resistant strains (Figure 3). However, oqxA expression was higher in ciprofloxacin-resistant strains than in ciprofloxacin-sensitive strains (Table 1). Statical analysis showed that there was a significant relationship between resistance to ciprofloxacin and oqxA gene expression (P-value=0.02).
Figure 3.
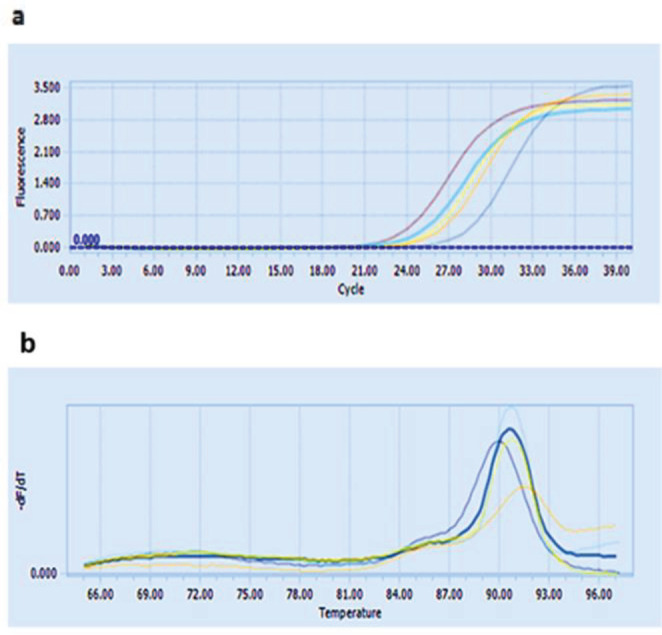
a: Specific amplification of the oqxA gene in Klebsiella pneumoniae isolates during Real-time PCR, b: melting curve of primers
Table 1.
Comparison of gene expression in the studied strains of Klebsiella pneumoniae
| ΔCT | MIC g/mlµ |
MDR | CIP resistance | Sample | Number |
|---|---|---|---|---|---|
| 4.08 | 32 | + | + | blood | 37 |
| 3.54 | 64 | + | + | Urine | 107 |
| 7.2 | 256 | - | + | blood | 7 |
| 4.58 | 32 | + | + | tracheal | 60 |
| 4.84 | 1 | + | - | urine | 137 |
| 2.96 | 256 | - | + | urine | 28 |
| 5.23 | 1 | + | - | urine | 103 |
| 7.11 | 1 | - | - | urine | N4 |
| 8.62 | 128 | + | + | urine | 20 |
| 5.57 | 1 | - | - | urine | 5 |
| 0.26 | 256 | + | + | urine | N5 |
| 5.66 | 64 | + | + | sputum | 65 |
| 1.9 | 128 | + | + | tracheal | N8 |
| 4.4 | 32 | + | + | blood | 128 |
| 7.67 | 1 | + | - | urine | 134 |
| 0.76 | 256 | + | + | tracheal | 4 |
Analysis of genetic linkage among isolates by ERIC-PCR showed ≤50-100% similarity among K. pneumoniae isolates (Figure 4). Genetic diversity was established among K. pneumoniae isolates by detecting 46 different ERIC types with a similarity cutoff ≥95%. 46 different ERIC profiles, including 21 common types (including more than one strain) and 25 unique types (including one strain), were identified. The largest common type includes 13 different strains. These strains were isolated from three different hospitals and different wards which indicates the circulation of clones that are genetically related. There was no significant difference between ERIC groups and antibiotic resistance, hospital, and wards (P-value≥0.005). However, most strains in ERIC types were positive for oqxA and oqxB. The negative oqxA and oqxB strains were placed in single types and they were placed in no common ERIC type.
Figure 4.
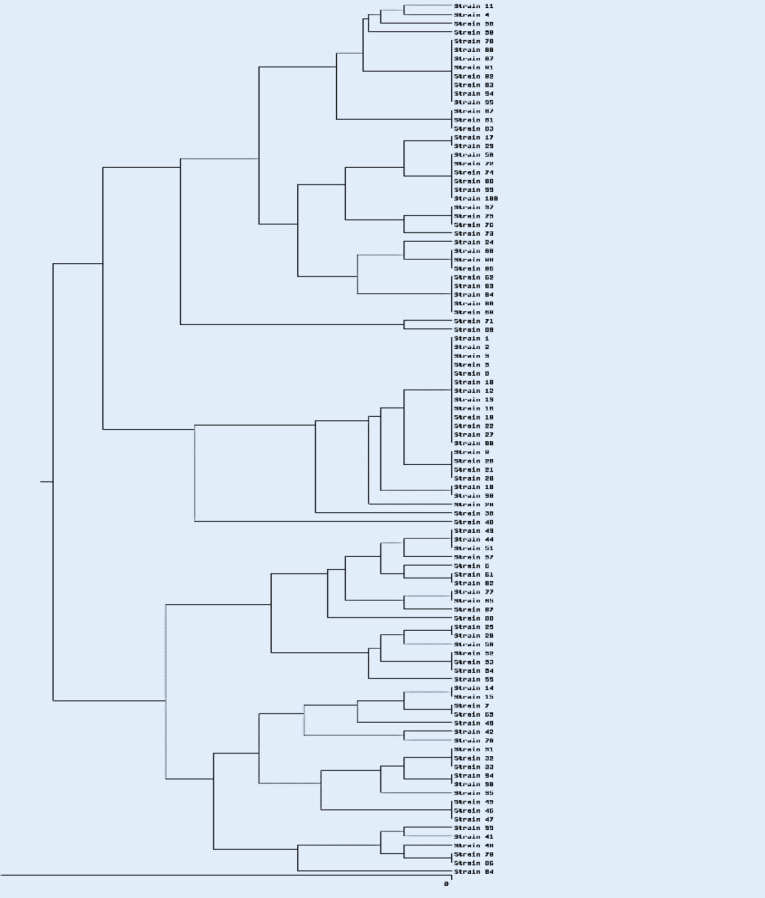
Dendrogram of ERIC-PCR patterns of 100 Klebsiella pneumoniae isolates from Hamadan hospitals
Discussion
The results of this study have shown the high prevalence resistance to fluoroquinolones as well as other classes of antibiotics in K. pneumoniae isolated from patients in Hamadan hospitals. More than 60% of isolates showed MDR phenotypes. The rate of resistance to ciprofloxacin and levofloxacin in our study is higher than the results of other studies conducted in different regions of Iran (4, 7, 25). A review article reported that the average resistance to ciprofloxacin in Iran was 34.8% from 2002 to 2014 (4). While, according to the results of recent studies, resistance to ciprofloxacin has increased in recent years (6). Resistance to ciprofloxacin is higher than the resistance reported from other countries (26-28). The reason for this difference in the results can be due to differences in the sampling area, population under study, sample size, and the type of strains circulating in different hospitals. The high antibiotic resistance in this study could be due to the fact that most K. pneumoniae strains have been isolated from ICUs. The prevalence of resistance to ciprofloxacin and levofloxacin was not significantly different from the earlier study in Hamadan hospitals. In the previous study in Hamadan hospitals, the prevalence of resistance to ciprofloxacin and levofloxacin in K. pneumoniae isolates was 80% and 85%, respectively (6). These results demonstrated that appropriate strategies have not been adopted to control or reduce resistant strains in hospitals. The results of our study are significant because our sampling was done during the COVID-19 pandemic; therefore, the presence of highly resistant K. pneumoniae strains, especially in ICUs provided a potential threat to patients.
These results also indicate the lack of appropriate efficacy of these antibiotics against K. pneumoniae, which is a worrying result, and physicians should pay more attention to the prescription of these antibiotics for the treatment of patients, especially hospitalized patients, and prescribe antibiotics based on the results of microbiology laboratories.
In our study, approximately 33% of isolates were related to urine samples, and about 31% of isolates were obtained from tracheal samples. Based on these results, the frequency of urine and respiratory samples is nearly equal or contrary to the results reported by Hamadan hospitals and other studies (24, 29). In the previous study from Hamadan hospitals, about 60% of isolates were related to the respiratory system, and about 20% of isolates were obtained from urine samples (24). In a study from Iran, 70% of K. pneumoniae isolates were related to urine samples and 14% were extracted from tracheal samples (29). Nirwati et al. isolated 51.5% of K. pneumoniae stains from respiratory specimens (30). According to the results of various studies, urinary tract and respiratory tract infections are two major infections in patients that are caused by K. pneumoniae.
The detection of oqxA and oqxB genes in most K. pneumoniae isolates was another major finding in this study. Most studies relating to oqxAB have concentrated on the contribution of the OqxAB efflux pump to quinolone resistance. The OqxAB efflux pump is known as one of the mechanisms of resistance to quinolones and fluoroquinolones (31). The OqxAB gene has been commonly detected in quinolone-resistant bacteria such as E. coli and K. pneumoniae (31). As demonstrated by Martinez et al. the expression levels of OqxAB in K. pneumoniae with reduced susceptibility to quinolones were 4-fold higher than in the susceptible strains (11). In our study, resistance to ciprofloxacin was significantly associated with expression levels of the oqxA gene. These results indicate a significant relationship between the presence of OqxAB efflux pump genes and resistance to ciprofloxacin. The prevalence of oqxA and oqxB genes in our study was higher compared with other Iranian studies. (32-34). High resistance to ciprofloxacin may be associated with a high level of efflux pump genes in this study.
In a report from China, it was found that the OqxAB gene is located in clinical isolates of Enterobacteriaceae and K. pneumoniae on a chromosome or plasmid. It plays a role in low and moderate resistance to quinoxalines, fluoroquinolones, tigecycline, nitrofurantoin, and many detergents (31). We also identified OqxAB genes in ciprofloxacin-susceptible strains. In this study, there was a significant relationship between resistance to ciprofloxacin and the oqxB gene and also between resistance to ceftriaxone and chloramphenicol with the oqxA gene. Based on the results of a study nitrofurantoin resistance in K. pneumoniae was relatively high; by studying the resistance mechanisms, it was found that the OqxAB efflux pump can play a role in nitrofurantoin resistance (35). Resistance to nitrofurantoin was also high in our study, with 77% of isolates exhibiting resistance to nitrofurantoin. In a study from China, 12 (2%) of 546 human clinical Salmonella Typhimurium were co-resistant to both ciprofloxacin and ceftriaxone, and four of the 12 resistant isolates carried the OqxAB gene (36). Based on the results of our study and other studies, the OqxAB efflux pump is not only associated with resistance to fluoroquinolones but can also be associated with resistance to other antibiotics. The high prevalence of K. pneumoniae strains producing efflux pump genes containing OqxAB genes has been considered an important issue in hospitals which causes serious problems for infection control and antibiotic treatment. High expression of OqxAB pump seems to contribute OqxAB to ciprofloxacin resistance in K. pneumoniae. The presence of OqxAB efflux genes and resistance to antibiotics were also significantly correlated. Considering the possibility of efflux pump genes on plasmids and transferring them to other bacteria, controlling resistant strains and inhibiting the efflux pumps of bacteria is very important. According to these results, the use of natural efflux pump inhibitors in clinical cases and chemical inhibitor efflux pumps in environmental cases in bacteria as an alternative or antibiotic supplement can be beneficial and effective in reducing the incidence of antibiotic resistance, reducing hospitalization time, and also reducing mortality.
The results indicated that 46 different types of ERIC were detected by ERIC-PCR, demonstrating the genetic diversity of K. pneumoniae isolates in Hamadan hospitals. Our findings indicated that there was no significant relationship between the ERIC groups and antibiotic resistance patterns, and the OqxAB efflux pump genes. Studies in Iran and other countries have demonstrated the genetic diversity of K. pneumoniae isolates according to the PCR-ERIC analysis (20, 23, 28). Our results indicate various K. pneumoniae colons with different antibiotic resistance profiles that may cause problems controlling and treating K. pneumoniae infections in hospitals.
Conclusion
To conclude, the prevalence of OqxAB efflux pump is high in K. pneumoniae in Hamadan hospitals and constitutes a potential reservoir for the spread of OqxAB among nosocomial bacteria. The high expression of this pump appears to reduce the sensitivity of clinical K. pneumoniae isolates to fluoroquinolones. Further studies on the various mechanisms of antibiotic resistance as well as efflux systems are suggested.
Authors’ Contributions
LSH and MRA Conceived and designed the experiments. FA Performed the experiments and analyzed the data. LSH and FA Prepared figures and tables and wrote the manuscript. MRA and LSH critically reviewed the manuscript. All authors have read and approved the final manuscript.
Ethical Approval
The present study was ethically approved by the Institutional Review Board of Hamadan University of Medical Sciences (IR.UMSHA.REC.1399.980).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgment
The results presented in this paper were part of a student thesis. This research was funded by a grant from Hamadan University of Medical Sciences, Hamadan, Iran (Grant Number: 140003252507). We would like to thank all members of the microbiology laboratory of Hamadan hospitals
References
- 1.Onori R, Gaiarsa S, Comandatore F, Pongolini S, Brisse S, Colombo A, et al. Tracking nosocomial Klebsiella pneumoniae infections and outbreaks by whole-genome analysis: Small-scale Italian scenario within a single hospital. J Clin Microbiol. 2015;9:2861–2868. doi: 10.1128/JCM.00545-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shadkam S, Goli HR, Mirzaei B, Gholami M, Ahanjan M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann Clin Microbiol Antimicrob. 2021;20:1–7. doi: 10.1186/s12941-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidary M, Nasiri MJ, Dabiri H, Tarashi S. Prevalence of drug-resistant Klebsiella pneumoniae in Iran: A review article. Iran J Public Health. 2018;47:317. [PMC free article] [PubMed] [Google Scholar]
- 5.Mirzaei B, Babaei R, Bazgir ZN, Goli HR, Keshavarzi S, Amiri E. Prevalence of Enterobacteriaceae sp and its multidrug-resistant rates in clinical isolates: A two-center cross-sectional study. Mol Biol Rep. 2021;48:665–675. doi: 10.1007/s11033-020-06114-x. [DOI] [PubMed] [Google Scholar]
- 6.Karimi K, Zarei O, Sedighi P, Taheri M, Doosti-Irani A, Shokoohizadeh L. Investigation of antibiotic resistance and biofilm formation in clinical isolates of Klebsiella pneumoniae. Int J Microbiol. 2021;14: 2021. doi: 10.1155/2021/5573388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhadi M, Ahanjan M, Goli HR, Haghshenas MR, Gholami M. High frequency of multidrug-resistant (MDR) Klebsiella pneumoniae harboring several β-lactamase and integron genes collected from several hospitals in the north of Iran. Ann Clin Microbiol Antimicrob. 2021;20:1–9. doi: 10.1186/s12941-021-00476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galani I, Karaiskos I, Giamarellou H. Multidrug-resistant Klebsiella pneumoniae: mechanisms of resistance including updated data for novel β-lactam-β-lactamase inhibitor combinations. Expert Rev Anti Infect Ther. 2021;19:1457–1468. doi: 10.1080/14787210.2021.1924674. [DOI] [PubMed] [Google Scholar]
- 9.Endimiani A, Luzzaro F, Perilli M, Lombardi G, Colì A, Tamborini A, et al. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum β-lactamase: Treatment outcome of patients receiving imipenem or ciprofloxacin. Clin Infect Dis. 2004;38:243–251. doi: 10.1086/380645. [DOI] [PubMed] [Google Scholar]
- 10.Andersson MI, MacGowan AP. Development of the quinolones. J Antimicrob Chemother. 2003;51:1–11. doi: 10.1093/jac/dkg212. [DOI] [PubMed] [Google Scholar]
- 11.Martı´nez JL, Alonso A, Go´ mez-Go´ mez JM. Quinolone resistance by mutations in chromosomalgyrase genes just the tip of the iceberg? J Antimicrob Chemother. 1998;42:683–688. doi: 10.1093/jac/42.6.683. [DOI] [PubMed] [Google Scholar]
- 12.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–25. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordmann P, Poirel L. Emergence of plasmidmediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 14.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. 2019;8:44. doi: 10.1186/s13756-019-0489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernando-Amado S, Blanco P, Alcalde-Rico M, Corona F, Reales-Calderon JA, Sanchez MB, et al. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist Updat. 2016;28:13–27. doi: 10.1016/j.drup.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Perez F, Rudin SD, Marshall SH, Coakley P, Chen L, Kreiswirth BN, et al. OqxAB, aquinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob Agents Chemother. 2013;57:4602–4603. doi: 10.1128/AAC.00725-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bharatham N, Bhowmik P, Aoki M, Okada U, Sharma S, Yamashita E, et al. Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae. Nature Commun. 2021;12:1–2. doi: 10.1038/s41467-021-25679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Belkum A TP, Dijkshoorn L, Haeggman S, Cookson B, Fry N, Fussing V, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 20.Mahon C, Lehman D, Manuselis G. Textbook of Diagnostic Microbiology. 5th Edidtion. Missouri: Elsevier; 2014. [Google Scholar]
- 21.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100; 2021. [Google Scholar]
- 22.Oliveira CF, Paim TG, Reiter KC, Rieger A, D’azevedo PA. Evaluation of four different DNA extraction methods in coagulase-negative Staphylococci clinical isolates. Rev Inst Med Trop Sao Paulo. 2014;56:29–33. doi: 10.1590/S0036-46652014000100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Martínez JM, Díaz de Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2013;68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 24.Sedighi P, Zarei O, Karimi K, Taheri M, Karami P, Shokoohizadeh L. Molecular typing of Klebsiella pneumoniae clinical isolates by Enterobacterial repetitive intergenic consensus polymerase chain reaction. Int J Microbiol. 2020;2020:1–5. [Google Scholar]
- 25.Moosavian M, Emam N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect Drug Resist. 2019;12:1001–1010. doi: 10.2147/IDR.S192597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madahiah BM, Noor US, Abdul S, Ali Abbas Q. Klebsiella pneumoniae urinary tract infections associated with long-term catheterization and spinal cord injuries. J Med Sci. 2002;2:227–229. [Google Scholar]
- 27.Cepas V, López Y, Muñoz E, Rolo D, Ardanuy C, Martí S, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb Drug Resist. 2019;25:72–79. doi: 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- 28.Aditi Priyadarshini B, Mahalakshmi K, Naveen Kumar V. Mutant prevention concentration of ciprofloxacin against Klebsiella pneumoniae clinical isolates: An ideal prognosticator in treating multidrug-resistant strains. Int J Microbiol. 2019;2019:6850108. doi: 10.1155/2019/6850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsaie Mehr V, Shokoohizadeh L, Mirzaee M, Savari M. Molecular typing of Klebsiella pneumoniae isolates by enterobacterial repetitive intergenic consensus (ERIC)–PCR. Infect Epidemiol Microbiol. 2017;3:112–116. [Google Scholar]
- 30.Nirwati H, Sinanjung K, Fahrunissa F, Wijaya F, Napitupulu S, Hati VP, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13:1–8. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z, Hao H. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. 2019;8:1–3. doi: 10.1186/s13756-019-0489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadpour Bishak F, Ashrafi F, Moradi Bidhendi S, Mirzaie A, Noorbazargan H. The impact of Grammosciadium platycarpum Boiss & Hausskn extract on oqxA efflux pump gene expression in antibiotic resistant clinical isolates of Klebsiella pneumoniae using real time PCR. J Med Plants. 2020;19:291–304. [Google Scholar]
- 33.Taherpour A, Hashemi A. Detection of OqxAB efflux pumps, OmpK35 and OmpK36 porins in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates from Iran. Hippokratia. 2013;17:355. [PMC free article] [PubMed] [Google Scholar]
- 34.Jamshidi MR, Zandi H, Eftekhar F. Correlation of quinolone-resistance, qnr genes and integron carriage in multidrug-resistant community isolates of Klebsiella spp. Iran J Basic Med Sci. 2019;22:1387–1391. doi: 10.22038/IJBMS.2019.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Q, Jiang J, Zhu Z, Xu T, Sheng ZK, Ye M, et al. Efflux pumps AcrAB and OqxAB contribute to nitrofurantoin resistance in an uropathogenic Klebsiella pneumoniae isolate. Int J Antimicrob Agents. 2019;54:223–227. doi: 10.1016/j.ijantimicag.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother. 2014;58:3752–3756. doi: 10.1128/AAC.02770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]