Abstract
Recently we described the isolation of spontaneous bacteriophage K139-resistant Vibrio cholerae O1 El Tor mutants. In this study, we identified phage-resistant isolates with intact O antigen but altered core oligosaccharide which were also affected in galactose catabolism; this strains have mutations in the galU gene. We inactivated another gal gene, galE, and the mutant was also found to be defective in the catabolism of exogenous galactose but synthesized an apparently normal lipopolysaccharide (LPS). Both gal mutants as well as a rough LPS (R-LPS) mutant were investigated for the ability to colonize the mouse small intestine. The galU and R-LPS mutants, but not the galE mutant, were defective in colonization, a phenotype also associated with O-antigen-negative mutants. By investigating several parameters in vitro, we could show that galU and R-LPS mutants were more sensitive to short-chain organic acids, cationic antimicrobial peptides, the complement system, and bile salts as well as other hydrophobic agents, indicating that their outer membrane no longer provides an effective barrier function. O-antigen-negative strains were found to be sensitive to complement and cationic peptides, but they displayed significant resistance to bile salts and short-chain organic acids. Furthermore, we found that galU and galE are essential for the formation of a biofilm in a spontaneous phage-resistant rugose variant, suggesting that the synthesis of UDP-galactose via UDP-glucose is necessary for biosynthesis of the exopolysaccharide. In addition, we provide evidence that the production of exopolysaccharide limits the access of phage K139 to its receptor, the O antigen. In conclusion, our results indicate involvement of galU in V. cholerae virulence, correlated with the observed change in LPS structure, and a role for galU and galE in environmental survival of V. cholerae.
The causative agent of the intestinal disease cholera is Vibrio cholerae, a gram-negative motile bacterium. Of the more than 150 known serogroups, only the noncapsulated O1 and the encapsulated O139 serogroup have been found to be associated with epidemic cholera. Epidemic O139 strains are related to and were derived from O1 El Tor strains after genetic alterations of the O-antigen biosynthesis gene cluster (16). The ongoing seventh pandemic, which began in 1961, is caused by O1 El Tor strains (3). V. cholerae is a natural inhabitant of aquatic ecosystems and is known to attach to environmental surfaces such as plants, filamentous green algae, zooplankton, crustaceans, or insects (8). Recently, V. cholerae O1 El Tor was found to form a three-dimensional biofilm on abiotic surfaces (70). Biofilm formation may be important in the life cycle of pathogenic V. cholerae strains, because they reside within natural aquatic habitats during interepidemic periods. O1 El Tor strains are also able to switch to a rugose colony phenotype. This morphology correlates with the constitutively production of an exopolysaccharide allowing biofilm formation on abiotic surfaces (65, 72). Such rugose variants are chlorine resistant and fully virulent in humans (43).
Important steps of the noninvasive disease process include ingestion of V. cholerae along with contaminated food or water, passage through the gastric acid barrier of the stomach, adherence to and penetration through the intestinal mucus lining, adherence to intestinal epithelial cells, multiplication, and cholera toxin (CT) production (25). In the course of the transition from the external environment to the human body, the bacteria are exposed to a series of changes, such as in temperature, acidity, and osmolarity, and must survive in the intestine, an environment which is enriched in growth-inhibitory substances like bile salts and organic acids. They are also exposed to antimicrobial components of the innate immune system, like complement factors secreted by intestinal epithelial cells (2) or defensins produced by Paneth cells (37). Several V. cholerae gene products have been shown to be important for colonization of the small intestine. These include the toxin-coregulated pili (TCP) (62), accessory colonization factors (49), regulatory proteins (e.g., ToxR/ToxS, TcpP/TcpH, and ToxT) and outer membrane proteins (21, 55), metabolic factors, biotin, purine biosynthetic genes, and the O antigen of the lipopolysaccharide (LPS) (6). Some factors are suggested to be involved in adhesion to the small intestine; however, no specific V. cholerae adhesins or specific mucosal receptors have been identified.
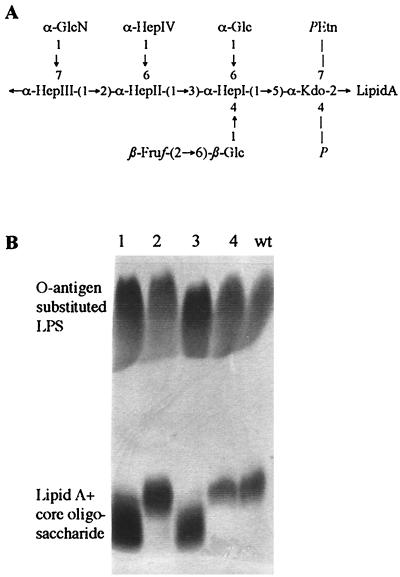
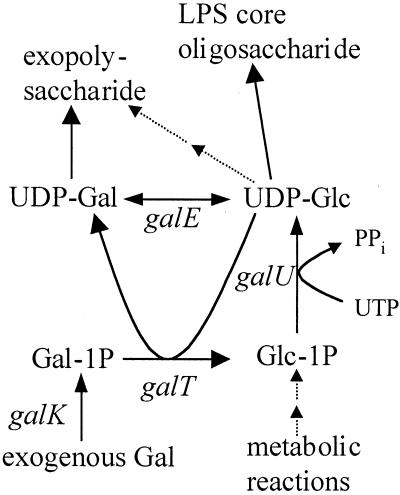
In gram-negative bacteria, LPS is the major integral component of the outer membrane with three general features: O antigen, core oligosaccharide, and lipid A. The O antigen of V. cholerae O1 consists of a homopolymer of approximately 18 perosamine residues which are substituted with tetronate (30). Recently, the structure of the LPS core oligosaccharide was reported for O1 and O139 mutants (33, 64); the results from these investigators are summarized in Fig. 1A. The site of attachment for the O1 antigen is unknown, whereas the O139 antigen is linked to the heptosyl III (HepIII) residue (33). Furthermore, the linkage of the carbohydrate quinovosamine, also detected in the LPS of O1 strains, is not known (59). In contrast to the well-investigated O1-antigen biosynthesis gene cluster (rfb) (59), little is known about the genes and corresponding enzymes involved in the biosynthesis of the core oligosaccharide. In gram-negative bacteria, the major core oligosaccharide biosynthesis genes are clustered on the chromosome in the waa (former rfa) locus, encoding specific transferases and enzymes involved in the synthesis of activated carbohydrates (23). Some activated moieties like UDP-glucose and UDP-galactose are often involved in the synthesis of different surface structures, and hence enzymes controlling their biosynthesis are not necessarily genetically linked with LPS biosynthesis gene clusters. Enzymes for the biosynthesis of UDP-glucose and UDP-galactose are UDP-glucose-pyrophosphorylase, encoded by galU, and UDP-glucose-4-epimerase, encoded by galE (36). In some bacteria, GalE and GalU are also important in the catabolism of exogenous galactose together with enzymes encoded by galT and galK (36). In many gram-negative pathogens, mutations in galE or galU lead to attenuated virulence, mainly if changes in LPS or capsular structures were observed (18). However, nothing is known about the role of galU and galE in V. cholerae O1 virulence.
FIG. 1.
LPS structure of V. cholerae. (A) LPS core oligosaccharide backbone proposed for V. cholerae O1 and O139. The representation is based on the structural analysis of Knirel et al. (33) and Vinogradov et al. (64), indicating a conserved core oligosaccharide for both serogroups. The site of the attachment of the O1 antigen has not been determined. The O139 antigen was found to be attached to the HepIII residue. Hep, l-glycero-d-manno-heptose; Kdo, 3-deoxy-d-manno-octulosonic acid; PEtn, 2-aminoethyl phosphate. (B) Analysis of LPS by SDS-PAGE and Silver staining. Lanes: 1, P27459res29/pACYC177; 2, P27459res29/pACYCgalU; 3, P27459galU; 4, P27459ΔgalE::cm; wt, P27459.
Recently, temperate phage K139 (51) was found to use the O antigen of V. cholerae O1 as its receptor (46). A collection of spontaneous phage-resistant O1 El Tor P27459 strains were investigated according to their LPS pattern on silver-stained polyacrylamide (PAA) gels, and different groups of altered LPS structures were described. These included O-antigen-negative strains, where one mutant was characterized for the loss of O antigen due to transposition of IS1004 into wbeW, encoding a putative glycosyltransferase. Another class identified were rough LPS (R-LPS) mutants comprising an altered core oligosaccharide with no O antigen attached; however, the mutation(s) responsible remains unidentified. In this study, we investigated the genetic nature of a phage-resistant mutant with intact O antigen but altered core oligosaccharide composition and found that this strain contained a mutation in galU. We also compared galU with galE and LPS-defective (R-LPS and O-antigen-negative) mutants with regard to LPS synthesis, galactose catabolism, the ability to colonize the small intestine of mice, and the ability to survive in the presence of bactericidal substances. Among our spontaneous phage-resistant mutants, we have also found rugose colony variants (unpublished results). In this work, we provide evidence that galU and galE are involved in rugose polysaccharide production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
V. cholerae strains used in this study are listed in Table 1. Escherichia coli K-12 strain LE392 (54) was used for all genetic constructions, unless the vector being used was a derivative of pCVD442 (13); in this case E. coli SM10λpir (42) was used. All strains were grown in Luria broth (LB) at 37°C except as noted otherwise. Antibiotics were used at the following concentrations: kanamycin, 50 μg/ml; and chloramphenicol, 2.5 μg/ml (V. cholerae) or 30 μg/ml (E. coli); ampicillin, 50 or 100 μg/ml; and streptomycin, 100 μg/ml. Plasmids used in this study are listed in Table 1.
TABLE 1.
V. cholerae strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| CO970 | O1 El Tor Ogawa, India 1994, Smr Cmr | J. J. Mekalanos |
| CO970galU | CO970 galU::pCVD442, Apr | This study |
| O395pCTX-Km | Kmr | 66 |
| P27459-S | Wild type (O1 El Tor, Inaba), spontaneous Smr | 48 |
| P27459ΔgalE::cm | ΔgalE::cm in P27459-S, Cmr | This study |
| P27459galU | galU::pCVD442 in P27459-S, Apr | This study |
| P27459lacZ | lacZ::pMD13 in P27459-S, Apr | 46 |
| P27459res29 | Altered core oligosaccharide, intact O antigen; mutant of P27459-S | 46 |
| P27459res46 | R-LPS mutant of P27459-S | This study |
| P27459res105 | Rugose colony variant of P27459-S | This study |
| P27459res105ΔgalE::cm | ΔgalE::cm in P27459res105, Cmr | This study |
| P27459res105galU | galU::pCVD442 in P27459res105, Apr | This study |
| P27459res118 | R-LPS mutant of P27459-S | 46 |
| P27459ΔwbeW | ΔwbeW in P27459-S, O-antigen negative | This study |
| Plasmids | ||
| pACYC177 | Apr Kmr | 52 |
| pACYCgalE | galE allele in bla of pACYC177; Kmr | This study |
| pACYCgalU | galU allele in bla of pACYC177; Kmr | This study |
| pBR322 | Apr Tcr | 71 |
| pCVD442 | oriR6K, mobRP4, sacB, Apr | 13 |
| pCVDgalU | ′galU′ in pCVD442; Apr | This study |
| pJNwbeW | wbeW allele in bla of pACYC177; Kmr | 46 |
| pKEK229 | pCVD442 with alternative MCS; Apr | 9 |
| pKEKΔgalE::cm | orf′ ′galE-cm-′galE orf′ in pKEK229; Apr Cmr | This study |
| pKEKΔwbeW | orf35′ ′wbeW-′wbeW orf′ in pKEK229; Apr | This study |
| pTrckan | Kmr | 45 |
Oligonucleotides, PCR, and Southern hybridization.
All oligonucleotides used for PCR are listed in Table 2. PCRs were performed as described by Mullis and Faloona (44). Southern blotting was performed according to Southern (56). Briefly, chromosomal DNA was prepared according to Grimberg et al. (19), digested with appropriate restriction enzymes, fractionated on a 0.7% agarose gel, and transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech, Freiburg, Germany). Hybridization with horseradish peroxidase-labeled probe and detection of hybridizing bands were carried out according to the procedure provided by the manufacturer of the ECL (enhanced chemiluminescence) system (Amersham Pharmacia Biotech, Freiburg, Germany).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| cm1 | GCAAGTCGACGGGCCCGCACCTCAAAAACACCATCATACA |
| cm2 | AGTCGTCGACAGGCGTTTAAGGGCACCAATAACTG |
| galEPstI | AAAACTGCAGAAAAGCCGTTACAATCGCCG |
| galEFspI | GAAAATGCGCAGGGCGTTTGTATAGCAACAG |
| 5′galESmaI | TCCCCCGGGCTATCGTACGGCTCATAACA |
| 5′galESalI | TACGCGTCGACATACCCGCTTGGATCATCTG |
| 3′galESalI | TACGCGTCGACCGTAATCTACACACAATGGC |
| 3′galEApaI | GTAAGGGCCCAATCGGCCACGGTGGAAATT |
| galUPstI | AACTGCAGCTAGCACACCATAGAGCAA |
| galUFspI | CCCTGCGCATGCATTGTTTAGATAGCGAAT |
| galU1 | AAGAGCTCAGCTATTGGGTGATATCCGTA |
| galU2 | GATCTAGACTTCGATATAGCCTTCAACGC |
| 5′wbeWSmaI | TCCCCCGGGAATTCCCTTACCTCTTGGTGC |
| 5′wbeWSalI | ACGGCTCGACCAGAAAATAGGGGAGCCAGT |
| 3′wbeWSalI | ACGCGTCGACGACACCAGTAAAGCTTGCAG |
| 3′wbeWApaI | GAGGGCCCCGATGTGCGCATGAGTTAAG |
Underlined nucleotides are not exact matches to the sequence and were introduced to add restriction enzyme sites. Nucleotide sequences were derived from the publicly available sequences from the V. cholerae genome project (http://www.tigr.org) (22) and accession no. X59954 and Y07788.
Computer analysis.
The publicly available sequences from the V. cholerae genome project (http://www.tigr.org) (22) were examined for the presence of gal genes. The amino acid sequences of E. coli GalE and GalU, obtained from the database, were used as query sequences. Homology analysis was performed with Basic BLAST Search 2.0 (1).
Construction of plasmids.
To construct complementing plasmids, gene-specific oligonucleotides (Table 2) were designed to introduce PstI and FspI sites at the 5′ and 3′ ends, respectively, of the genes galU and galE. Following PCR amplification, the products were digested and ligated into the PstI- and FspI-digested plasmid pACYC177. The resulting plasmids pACYCgalU and pACYCgalE express the corresponding genes from the bla promoter. To construct suicide vector pCVDgalU, an internal fragment of galU, obtained after PCR with primers galU1 and -2, was ligated into the SacI/XbaI-digested plasmid pCVD442 (13); this plasmid was used to construct an insertion mutation in the galU gene. Suicide plasmids to introduce chromosomal in-frame deletions of the genes wbeW and galE were constructed in the same manner. DNA fragments of about 500 bp encompassing the regions upstream of the corresponding gene were PCR amplified using primer pairs 5xSmaI and 5xSalI, x is the gene of interest (Table 2). The downstream DNA region of the corresponding gene was PCR amplified using primer pairs 3xSalI and 3xApaI (Table 2). The PCR products were digested with SalI and ligated overnight (O/N). The ligation mix was digested with SmaI and ApaI, purified, and ligated into pKEK229 (9) that had been digested with SmaI and ApaI to give plasmids pKEKΔgalE and pKEKΔrfbW. In addition a PCR-derived cat fragment (with oligonucleotides cm1 and -2 [Table 2]) was digested with SalI and ligated together with the SalI-digested plasmid pKEKΔgalE to give the chloramphenicol-resistant (Cmr) plasmid pKEKΔgalE::cm.
Construction of bacterial strains.
To construct strains containing a mutation in galU, plasmid pCVDgalU was mated by conjugation from E. coli SM10λpir into V. cholerae P27459-S, P27459res105, or CO970, selecting for streptomycin and ampicillin resistance (Smr and Apr) (42). The resulting strains obtained a chromosomal insertion caused by the integration of the plasmid through homologous recombination via the internal galU fragment. Strains harboring in-frame deletion mutations were constructed by allelic exchange (13). Plasmid pKEKΔwbeW was mated into P27459-S from E. coli SM10λpir by selecting for Smr and Apr. Single colonies were grown for successive generations in LB with no antibiotic selection and then plated on LB without NaCl but containing 10% sucrose at 30°C. Several sucrose-resistant, Aps colonies were isolated and screened for loss of wbeW in cross-streaking experiments using lytic phage K139.cm9 (45). Strain P27459ΔwbeW was furthermore shown to synthesize no O antigen in Western blot analysis. O-antigen biosynthesis was restored in P27459ΔwbeW containing a plasmid carrying wbeW (46). The sucrose counterselection method failed to introduce galE in-frame deletions. In this case, plasmid pKEKΔgalE::cm was mated from E. coli SM10λpir into V. cholerae P27459-S and P27459res105, selecting for Smr and Cmr. Single colonies were tested for Aps, indicating that the plasmid was absent and a double crossover had occurred. The correct chromosomal insertion for all mutants was confirmed by Southern blot analysis.
LPS analysis.
The isolation, separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and silver staining of LPS were performed as described previously (46).
Serum resistance assay.
Cells were grown to mid-logarithmic phase in LB, washed, and mixed to a final concentration of 20% with normal human serum (obtained and pooled from four donors) or 20% heat-inactivated normal human serum in phosphate-buffered saline with 0.1% peptone. Approximately 108 CFU was used in each assay. After incubation at 37°C for 1 h, the cells were harvested, washed, and resuspended in phosphate-buffered saline with 0.1% peptone. Viable cell counts were determined by plating serial dilutions onto LB agar.
Mouse colonization assays.
The infant mouse colonization assay has been described previously (32). Briefly, mutant strains (lac+) were mixed with the strain P27459lacZ (lac) and given in a peroral inoculum ratio of approximately 107 CFU mutant to 107 CFU of wild type to 5- to 6-day-old CD-1 suckling mice. After a 24-h period of colonization, intestinal homogenates were collected and the ratio of mutant to wild type was determined by plating appropriate dilutions on LB agar containing streptomycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
CTX-KmΦ transduction and GM1-ELISA.
For optimal CT and TCP expression, the strains were incubated under inducing conditions. O/N cultures grown in AKI medium (27) were diluted 1:100 in AKI without NaHCO3, grown for 4 h at 37°C under static growth conditions, and then transferred to vigorous shaking for 4 h at 37°C (40). For CT measurement, 1 ml of the culture was spun down, and the culture supernatant was filtered (0.2-μm-pore-size filter; Schleicher & Schuell, Dassel, Germany), and frozen at −20°C. The level of CT present in 200 μl of the supernatant was measured by GM1-ganglioside enzyme-linked immunosorbent assay (GM1-ELISA) (61) using purified CT (Sigma, Deisenhofen, Germany) to generate a standard curve.
For phage transduction, 100 μl of the same culture was mixed with 100 μl of phage CTX-KmΦ (66). After standing for 30 min at room temperature, the mixture was diluted and plated onto LB agar containing kanamycin. The transduction frequency was calculated from the CFU of kanamycin-resistant (Kmr) transductants correlated with the optical density at 600 nm (OD600) of the recipient strain. An additional CTX-KmΦ transduction experiment was performed to enumerate the numbers of both transductants and potential recipients. Strain P27459ΔtoxR was used as a negative control for both assays. The ΔtoxR1 mutation was introduced into the chromosome of P27459-S by plasmid pMD60, as previously described (31), to form strain P27459ΔtoxR. Phage CTX-KmΦ was prepared from a 100-ml O/N culture (LB, 30°C) of strain O395pCTX-Km by filtering the culture supernatant through a 0.2-μm-pore-size filter.
Determination of MICs antimicrobial cationic peptides.
Standard MIC testing of susceptibility to polymyxin B (50 μg/ml to 780 ng/ml; Sigma) and protamine (675 to 10.5 μg/ml; Sigma) was done as described by Steinberg et al. (57). The MIC was defined as the lowest concentration of drug which prevented visible turbidity after incubation for 18 h at 37°C without shaking.
Sensitivity to hydrophobic agents.
Sensitivity to SDS and novobiocin was assessed by determination of the MIC. O/N cultures of strains to be tested were diluted in LB to give 1 × 105 to 5 × 105 CFU/ml. After 100-μl aliquots of the diluted cultures were dispensed into each well of a 96-well microtiter plate (Greiner, Frickenhausen, Germany), 11 μl of test compound was added. SDS (Merck, Hamburg, Germany) and novobiocin (Sigma) were dissolved in water at 10-fold the desired test concentration and then 2-fold serially diluted in water. The final concentrations tested ranged from 600 to 9.3 μg/ml for SDS and 1.5 μg/ml to 23 ng/ml for novobiocin. The sensitivity to bile was tested on thiosulfate citrate bile sucrose (TCBS) agar plates, prepared as instructed for the commercially available TCBS (Difco, Heidelberg), containing various amounts of bile (0 to 0.8%).
Biofilm formation.
Quantitative biofilm measurement was performed in a microtiter assay as described previously (50), with minor modifications. Briefly, bacteria were grown O/N in LB and diluted 1:100 in fresh LB containing appropriate antibiotics; then 100-μl aliquots were placed in a 96-well microtiter plate (polystyrene; Greiner) and incubated for 24 h at room temperature. Bacterial cultures were poured out, washed three times with H2O, fixed with 2.5% glutaraldehyde, washed twice, and stained with 200 μl of a 0.4% crystal violet solution. After solubilization of the crystal violet with 300 μl of ethanol-acetone (80:20), the OD was determined at 570 nm in an ELISA reader (Bio-Rad, Munich, Germany). Qualitative biofilm formation was assessed by incubating 1:100-diluted O/N cultures (LB, with appropriate antibiotics) in borosilicate tubes for 48 h without shaking at room temperature. Strains were noted as biofilm positive when floating pellicles with closely packed bacteria were formed; strains growing as a suspension were designated as biofilm negative. Cultures were then poured out, and adherent bacteria were stained with crystal violet.
Acid tolerance response assay.
The ability of cells to survive exposure to organic acids (cocktail of 2.5 mM butyric acid, 8.7 mM acetic acid, and 3.7 mM propionic acid) in LB (pH 4.4) was determined as described by Merrell and Camilli (41) except that no organic acids were added to the adaptation medium.
Plaque inhibition assay.
The phage neutralizing capacity of purified LPS was determined as described previously (46).
β-Lactamase detection by immunoblotting analysis.
Cells were harvested at OD600 1.5 to 2.5 and diluted to give 2 ml of OD600 = 1. After centrifugation of 1 ml of cell suspension, the cell pellets were resuspended in 100 μl of sample buffer. The supernatants were filtered, and proteins were precipitated with 10% (final concentration) trichloracetic acid. The protein pellet was solubilized in 100 μl of sample buffer. Samples (20 μl) were separated by SDS-PAGE (34), transferred to a nitrocellulose membrane, and reacted with an antibody directed against BlaM (5 Prime→3 Prime Inc., Boulder, Colo.).
RESULTS
Identification of a spontaneous galU mutant expressing an altered LPS core oligosaccharide.
The recently described spontaneous phage K139.cm9-resistant mutant P27459res29 showed an altered LPS core oligosaccharide with an intact O antigen (Fig. 1B, lane 1) (46). The observation that this strain could not grow on M9 medium containing galactose as a carbon source and the fact that glucose is a component of the LPS core oligosaccharide (Fig. 1A) implicated a possible galU mutation in this strain. To test this hypothesis, we constructed a plasmid encoding galU, based on nucleotide sequence information obtained from the V. cholerae genome project (http://www.tigr.org) (22). Plasmid pACYCgalU complemented all observed phenotypes in strain P27459res29, showing a normal LPS pattern on a silver-stained PAA gel (Fig. 1B, lane 2) as well as restoring sensitivity against phage K139.cm9 and growth on M9 galactose plates (data not shown).
Construction and characterization of gal mutants.
To determine whether a possible virulence effect of the galU mutant might be caused by a general defect in galactose utilization, we constructed a V. cholerae strain with a deletion/insertion in another galactose utilization gene, galE. Additionally, we generated a galU strain by plasmid insertion (Fig. 2; Materials and Methods) and compared the phenotypes of the two strains. Neither mutant could grow on M9 galactose plates, confirming that galE is also involved in galactose utilization as predicted. Growth could be restored in trans by a plasmid encoding galE in strain P27459ΔgalE::cm and by a plasmid encoding galU in strain P27459galU (data not shown). In E. coli and Salmonella enterica serotype Typhimurium, the genetic loss of galE and galU causes bacteriolysis when growing cells are exposed to high concentrations of d-galactose (36). This property was tested in V. cholerae galE and galU mutants on LB plates containing 2% galactose. The galE mutant could not grow on such plates, whereas both galU mutants (spontaneous and constructed) grew initially but eventually lysed. This phenotype could be complemented by introduction of galU or galE in trans. The LPS patterns of both galE and galU mutants were then analyzed by SDS-PAGE. In a silver-stained PAA gel, the galU mutant showed an altered core with an apparent intact O antigen (Fig. 1B, lane 3). In contrast, the LPS of the galE mutant migrated similarly to the LPS of the wild-type (wt) strain (Fig. 1B, lane 4), revealing that GalE does not affect LPS biosynthesis. Another evidence for the presence of intact LPS, was the finding that the galE mutant is as sensitive as the wt strain in cross-streak analysis versus phage K139.cm9 (data not shown).
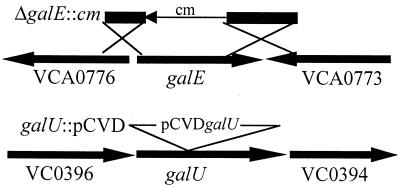
FIG. 2.
Chromosomal organization of investigated gal genes and surrounding ORFs. While this report was under review, the complete genomic sequence of V. cholerae was published (22). The representation is therefore based on data from the V. cholerae genomic database (http://www.tigr.org). galE (VCA0774) is located on chromosome 2, whereas galU (VC0395) is located on chromosome 1. Based on homology analysis, the surrounding ORFs seem not to be involved in galactose-catabolism or LPS biosynthesis. Knockout constructions introduced into strain P27459-S are illustrated schematically (Materials and Methods).
Colonization of gal and LPS mutants in infant mice.
The ability of gal mutants to colonize the small intestine was assessed in a competition assay using perorally infected CD-1 suckling mice. To compare the colonization behaviors of the gal mutants and other LPS mutants, we also investigated the spontaneous R-LPS mutant P27459res46 and a defined O-antigen-negative strain (ΔwbeW; Materials and Methods). As shown in Table 3, strain P27459ΔwbeW was significantly reduced in its ability to colonize the mouse small intestine, a result also found for other O-antigen-deficient V. cholerae strains (gmd [67] and manB [6]). No mutant bacteria were recovered from mice inoculated with the R-LPS mutant P27459res46, harboring an altered LPS core oligosaccharide without attached O antigen. The core oligosaccharide-defective galU mutants (P27459 res29, P27459 galU) were approximately 50- to 100-fold attenuated in the ability to colonize, although they carry the O antigen. The colonization defect of the spontaneous mutant P27459res29 was fully complemented by the presence of a plasmid expressing galU, revealing that the colonization defect of this strain is due to a mutation in galU. Importantly, the galE mutant with intact LPS colonized as well as the wt strain, indicating that the colonization defect of galU-deficient strains is due to neither their galactose sensitivity nor their defect in utilizing exogenous galactose. These results suggest that the core oligosaccharide defect in galU mutants attenuates V. cholerae intestinal colonization.
TABLE 3.
Intestinal colonization, CTX-KmΦ phage transduction, and CT release
| Strain | Mouse CIa (no. of mice) | CTX-KmΦ transductionb | CT productionc | HAP secretiond |
|---|---|---|---|---|
| P27459-S (lac+) | 1.6 (3) | + | + | + |
| P27459galU | 0.03 (5) | + | + | + |
| P27459res29 | 0.01 (6) | + | + | + |
| P27459res29/pACYC177 | 0.01 (6) | NDe | ND | ND |
| P27459res29/pACYCgalU | 1.94 (5) | ND | ND | ND |
| P27459ΔgalE::cm | 0.96 (6) | + | + | + |
| P27459ΔwbeW | 0.024 (5) | + | + | + |
| P27459ΔwbeW/pJNwbeW | 0.8 (4) | ND | ND | ND |
| P27459res46, P27459res118f | <10−4 (5) | + | + | + |
The competitive index (CI) for colonization is defined as the output ratio of mutant to wt bacteria divided by the input ratio of mutant (lac+) to wt bacteria (lac). Strains P27459galU, P27459res29, P27459res29/pACYC177, P27459ΔwbeW, and P27459res46 had significant differences in colonization compared to the wt strain (P27459) as determined by Student's two-tailed t test (P < 0.01); all other strains showed less significant differences (P > 0.01). In vitro competition assays were also done for galU mutants: P27459res29, CI 0.7; P27459res29/pACYC177, CI 4.7; P27459galU, CI 1. The in vitro CIs demonstrate that galU mutants had no handicap or growth defect versus the wt under in vitro conditions.
Determined in triplicate (Materials and Methods). The frequency of transduction ranged from 10−4 to 10−5, with no significant differences between the strains tested.
Measured in duplicate (Materials and Methods). The levels of CT present found in the culture supernatants were between 0.5 and 2 μg ml−1/OD600, with no significant differences between the strains tested.
Detected as a zone of clearing on 1% nonfat milk agar plates.
ND, not determined.
Mouse colonization assays were initially performed with strain P27459res46, an R-LPS mutant which lacks O antigen and has an altered core, like P27459res118. All other experiments were done with strain P27459res118. To preserve mice, we did not repeat the colonization assays with strain P27459res118.
Outer membrane integrity of LPS mutants.
Misassembly of outer membrane protein multimers in LPS mutants, and the significance of LPS to transporters, has been described for some gram-negative bacteria (35, 68). In V. cholerae, the presence of the type IV pilus TCP during infection is important for colonization (62). We measured TCP expression by a transduction assay using CTX-KmΦ, a Kmr version of phage CTXΦ, which is known to use TCP as its receptor (66). As indicated in Table 3, there were no significant differences in the frequency of phage transduction in galU, wbeW, and R-LPS strains. These data indicated that there is functional TCP on the surface of all three LPS-deficient mutants, allowing at least an efficient phage CTXΦ transduction. In addition, we investigated the presence of the main virulence factor CT and hemagglutinin-protease (HAP) in the extracellular medium, since both enzymes are known to be secreted by the type II secretion apparatus eps (extracellular protein secretion) (53). CT concentration in the supernatant was measured by GM1-ELISA, and HAP activity was determined on agar plates containing 1% nonfat milk. As shown in Table 3, no significant differences were found between the LPS mutants and the wt strain.
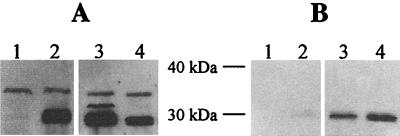
Outer membrane integrity was further studied by investigating the localization of the periplasmic enzyme β-lactamase, expressed from plasmid pBR322, in Western blot analysis. Very little β-lactamase was detected in culture supernatants of the wt strain, (Fig. 3B, lane 2), the galU mutant exhibited slightly more (lane 3), and the spontaneous R-LPS mutant produced enhanced amounts (lane 4). The latter observation correlates with less β-lactamase in corresponding whole-cell extracts (Fig. 3A, lane 4). However, no significant difference of β-lactamase production was observed between wt and the galU mutant in whole-cell extracts (Fig. 3A, lane 2 and 3). We have found no evidence that the increased level of β-lactamase in the supernatant is due to enhanced cell lysis (e.g., by observing normal growth abilities in liquid cultures and the inability to detect activity of the cytoplasmic enzyme LacZ in the supernatants [data not shown]). The results suggest that the core-deficient mutants are leaky for periplasmic proteins, enhanced for the R-LPS and less leaky for the galU mutant.
FIG. 3.
Effect of LPS alteration on the localization of β-lactamase. BlaM was immunodetected by ECL as indicated in Materials and Methods. (A) Whole-cell extracts; (B) supernatant extraction. Lanes: 1, wt P27459 without plasmid; 2, wt P27459/pBR322; 3, spontaneous galU mutant SP27459res29/pBR322; 4, R-LPS mutant SP27459res118/pBR322. The experiment was performed in duplicate with independently growing cells. Samples were matched by equivalent OD600 units.
Survival in the presence of organic acids.
V. cholerae may be exposed to high concentrations of weak organic short-chain acids present in the gastrointestinal tracts of humans (12). It is well known that weak organic acids can permeate the cytoplasmic membrane in the protonated form (depending on environmental pH), deprotonate, and accumulate intracellularly in the harmful anionic form (17). We investigated the survival of the three different types of LPS mutants after exposure to short-chain acids. The O-antigen mutant (ΔwbeW) survived after exposure for 1 h to the shock medium (LB containing 2.5 mM butyric acid, 8.7 mM acetic acid, and 3.7 mM propionic acid [pH 4.4]) as well as the wt strain (Table 4). In contrast, mutant strains with an incomplete LPS core oligosaccharide are apparently more susceptible to organic acids. The galU mutant (res29) was about 75-fold attenuated, while the R-LPS mutant (res118) showed the most severe defect. Merrell and Camilli (41) reported that V. cholerae is capable of mounting an acid tolerance response which protects against such substances. Therefore, we tested the survival of the LPS core oligosaccharide mutants after adaptation for 1 h in LB (pH 5.7) before placement in the shock medium. All mutants survived as well as the wt strain after a 1-h exposure, but after longer exposure (>2 h), the R-LPS mutant was approximately 50-fold attenuated (data not shown). These results suggest that an intact LPS core region, in addition to the inducible acid tolerance response, provides protection against protonated weak acids.
TABLE 4.
Resistance against organic acids, hydrophobic agents, antimicrobial peptides, and the complement
| Strain | Resistance to organic acids (% survival)a | MIC (μg/ml)b
|
Serum resistance (% survival)c | |||
|---|---|---|---|---|---|---|
| SDS | Novobiocin | Polymyxin B | Protamine | |||
| P27459-S | 5.7 × 10−3 ± 2.9 × 10−3 | 300 | 0.75 | 12.5 | 337 | 85 ± 13 (5) |
| P27459galU | NDd | 150 | 0.187 | 1.56 | 84 | ND |
| P27459galU/pTrcAkan | ND | 150 | 0.187 | ND | ND | 0.21 (1) |
| P27459galU/pACYCgalU | ND | 300 | 0.75 | 12.5 | 337 | 100 (3) |
| P27459res29 | 7.6 × 10−5 ± 8.9 × 10−5 | 150 | 0.187 | 1.56 | 84 | 0.6 ± 1 (4) |
| P27459res29/pACYC177 | ND | 150 | 0.187 | ND | ND | 1.1 ± 1.6 (2) |
| P27459res29/pACYCgalU | ND | 300 | 0.75 | 12.5 | 337 | 100 (3) |
| P27459ΔgalE::cm | ND | 300 | 0.75 | 12.5 | 337 | 81 ± 26 (2) |
| P27459ΔwbeW | 5.8 × 10−3 ± 1.7 × 10−3 | 300 | 0.75 | 6.25 | 168 | 0.006 ± 0.0056 (2) |
| P27459ΔwbeW/pACYC177 | ND | ND | ND | 6.25 | 168 | 0.001 ± 0.0019 (2) |
| P27459ΔwbeW/pJNwbeW | ND | ND | ND | 12.5 | 337 | 100 (2) |
| P27459res118 | 0 | 150 | 0.047 | 0.78 | 21 | 0.0003 ± 0.0004 (2) |
Survival of wt and mutant strains in response to short-chain organic acid challenge. Cells were grown in LB (pH 7) and subsequently shocked in LB (pH 4.4) containing 8.7 mM acetic acid, 2.5 mM butyric acid, and 3.7 mM propionic acid. Approximately 109 CFU/ml was used in each experiment. Percent survival at 60 min was calculated by comparison with the initial number of cells measured at time zero. Data shown are the average from triplicate experiments ± standard deviation.
All experiments were performed in triplicate.
Calculated as 100 × (number of CFU after cells were incubated for 1 h in 20% normal human serum/number of CFU after cells were incubated for 1 h in 20% heat-inactivated normal human serum). Indicated is the mean ± standard deviation. The numbers of independent experiments are given in parentheses.
ND, not determined.
Sensitivity to bile and other hydrophobic agents.
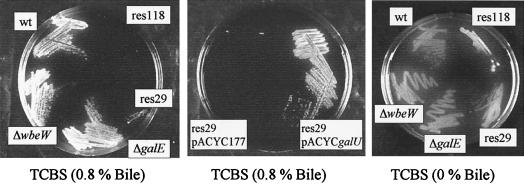
The LPS core oligosaccaride region of gram-negative bacteria appears to act as an effective outer membrane barrier against hydrophobic agents with charged groups (47). To determine whether this is also true for V. cholerae, we tested growth on TCBS plates containing 0.8% bile. In the human intestine, the concentration of bile salts is estimated to be ∼1% (depending on nutrition status) (24). As shown in Fig. 4, galU (res29) and R-LPS (res118) mutants did not grow on these plates. However, the R-LPS mutant also grew poorly on the same plates lacking bile, while the galU mutant grew similar to the wt strain. O-antigen-negative (wbeW) mutants, galE mutants, and galU mutants complemented with a galU expression plasmid were able to grow on TCBS containing bile. A growth defect of galU and R-LPS mutants was also observed in LB containing >0.1% of the bile component deoxycholate (data not shown). In addition, the sensitivity of the galU and R-LPS mutant strains to anionic hydrophobic agents was confirmed by determining the MICs of SDS and novobiocin. As indicated in Table 4, the R-LPS mutant (res118) and both galU mutants (res29, galU) were more sensitive to SDS and novobiocin than wt, O-antigen-negative, or galE strains. These results demonstrate a role for the V. cholerae core oligosaccharide in resistance to bile and other hydrophobic agents.
FIG. 4.
Growth phenotypes of V. cholerae O1 El Tor and mutant strains on TCBS containing bile at the indicated concentrations.
Resistance to bactericidal substances of the innate immune system.
Antimicrobial cationic peptides are widespread in nature and are produced by many organisms. For example, in the human small intestine, at least two such peptides (defensins) are expressed from Paneth cells (37). Cationic peptides have been shown to permeabilize the outer membrane of gram-negative bacteria followed by a depolarization of the cytoplasmic membrane (20). Since cationic peptides possess similar features and a similar mode of action, the role of V. cholerae LPS in resistance to two such peptides was investigated by determining the MICs of polymyxin B and protamine. As shown in Table 4, sensitivity to these peptides decreased in the following order: R-LPS mutant (res118) > galU mutants (res29, galU) > O-antigen-negative strain (ΔwbeW) > wt = galE mutant = galU mutants containing plasmid pACYCgalU. These results indicate that the LPS core oligosaccharide is more important than the O-antigen for resistance against cationic peptides.
Recently, it was demonstrated that V. cholerae O-antigen-negative strains are sensitive to complement activity (5, 67), as determined by serum resistance assays. To analyze whether the LPS core oligosaccharide also contributes to serum resistance, the galU mutants (res29, galU) were investigated in a serum bactericidal assay (Materials and Methods). galU mutants exhibited markedly greater serum sensitivity than wt or galE strains (Table 4). Survival was restored in galU mutants harboring complementation plasmid pACYCgalU. Strains lacking the O antigen (ΔwbeW, R-LPS) were about 100-fold more sensitive to human serum than galU mutants, indicating that while an intact core oligosaccharide plays a critical role, the O antigen is the most important factor in complement resistance.
Role of galE and galU in biofilm formation.
Among our spontaneous phage K139.cm9-resistant P27459 isolates, we found strains with a rugose colony morphology. Such variants have been described previously (72) to be constitutively synthesizing an exopolysaccharide (VPS) allowing the bacteria to form a biofilm on abiotic surfaces. The composition of the VPS from two different V. cholerae O1 El Tor rugose isolates has recently been published. Wai et al. found a VPS consisting of N-acetyl-d-glucosamine, d-mannose, 6-deoxy-d-galactose, and d-galactose (65), whereas Yildiz and Schoolnik described the composition of the VPS as glucose, galactose, N-acetylglucosamine, mannose, and xylose (72). These findings prompted us to investigate a possible role of galE and galU in production of the VPS.
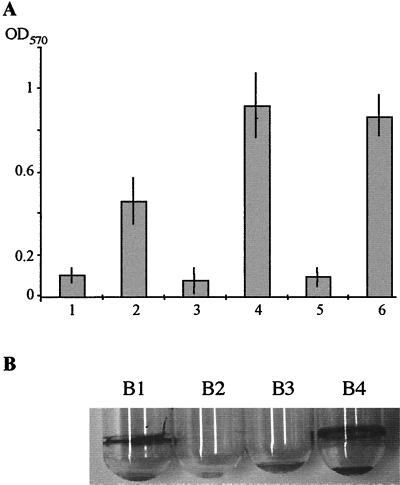
The parental wt strain P27459 was unable to form a biofilm on polystyrene or borosilicate glass, in contrast to the rugose variant P27459res105. To study the influence of galU and galE mutations on biofilm formation, we introduced these mutations into the rugose variant P27459res105. Both mutations yielded smooth colony forms, suggesting that galU and galE mutants are unable to synthesize the VPS. Introducing plasmids carrying galU or galE restored the rugose colony morphology. The phenotypes were confirmed in a quantitative biofilm measurement assay (Fig. 5A). To confirm the galU phenotype in a nonrugose wt V. cholerae O1 El Tor strain, we screened several clinical isolates for the ability to produce a biofilm in polystyrene microtiter dishes. Strain CO970, which formed a biofilm, was chosen for further investigation. As shown in Fig. 5B, a galU mutant of strain CO970 was no longer able to form a biofilm on borosilicate glass tubes. Our results indicate that UDP-glucose or UDP-galactose is necessary for biosynthesis of the VPS.
FIG. 5.
Biofilm formation ability of V. cholerae and its mutants. (A) OD570 quantifies the amount of crystal violet associated with the biofilm on polystyrene microtiter plates after staining. Columns: 1, P27459 (wt); 2, rugose strain P27459res105; 3, P27459res105galU; 4, P27459res105galU/pACYCgalU; 5, P27459res105ΔgalE::cm; 6, P27459res105ΔgalE::cm/pACYCgalE. Shown is the average of four independent experiments (each experiment performed in triplicate). Error bars represent standard deviations. Strains containing control plasmids (columns 3 and 5) were measured separately, but the data are summarized together with those for strains without control plasmids. (B) Biofilms on borosilicate glass tubes stained with crystal violet. Tubes: B1, CO970 (wt); B2, CO970galU; B3, CO970galU/pTrcAkan; B4, CO970galU/pACYCgalU.
Rugose phenotype and phage K139 accessibility.
We also wished to determine why spontaneous rugose variants of strain P27459 are phage resistant. Since the O antigen is the receptor of phage K139, we first analyzed the LPS structure of rugose strain P27459res105 by SDS-PAGE analysis (silver staining and Western blotting). Furthermore, the ability of the phage to bind to purified LPS from P27459res105 was determined in a plaque inhibition assay (46). Neither type of analysis revealed differences from the wt LPS, indicating that the phage could potentially bind to strain P27459res105 LPS. Second, we investigated the sensitivity to phage K139.cm9 of the nonrugose mutant P27459res105ΔgalE::cm in cross-streaking and plaque formation experiments. The nonrugose mutant is phage sensitive, in contrast to its parent rugose variant (data not shown). All results together provide evidence that production of the VPS limits the access of phage K139 to its receptor, the O antigen, hence leading to attenuated phage susceptibility.
DISCUSSION
In this work, we identified a mutation in galU that caused the spontaneous bacteriophage K139.cm9-resistant phenotype of strain P27459res29. The O antigen of galU mutants is able to be adsorbed by phage K139; nevertheless, these strains are phage resistant (46). The mechanism responsible for this phenotype is unresolved. The structure of the core oligosaccharide in galU-deficient strains is not known; however, preliminary evidence suggests that it contains no glucose. We predict that galU-deficient strains lack the α-Glc and β-Fru-β-Glc branches on the HepI residue (Fig. 1A), but proof of this awaits structural analysis.
We demonstrated that galU mutants were significantly reduced in the ability to colonize the mouse small intestine. This phenotype could be caused by a pleiotropic effect, because galU mutants are predicted to be unable to synthesize UDP-glucose. Besides its function as a substrate for glucosyltransferases resulting in glucosylated surface structures, UDP-glucose plays a well-established biochemical role as a glycosyl donor in the enzymatic biosynthesis of carbohydrates. Some examples in gram-negative bacteria are synthesis of the osmoprotectants trehalose (under conditions of high osmolarity) and membrane-derived oligosaccharide (under conditions of low osmolarity) in E. coli (11); synthesis of UDP-arabinose, which is incorporated into lipid A of S. enterica serovar Typhimurium to protect against antimicrobial cationic peptides, especially polymyxin B (14); and synthesis of UDP-galactose, which serves as a donor for several surface structures, including glycosylated pili in Neisseria meningitidis (58). It is less clear whether UDP-glucose additionally acts as an intracellular signal molecule. For one study, it was reported that UDP-glucose controls the expression of ςs in E. coli (4).
Investigating both galU and galE mutants, we ruled out the possibility that the observed colonization defect of V. cholerae O1 galU mutants is due to a conferred galactose sensitivity or to the inability to catabolize galactose, both of which are phenotypes of galE and galU mutants (Fig. 6). Unlike the galU strain, the galE mutant was found to colonize the mouse small intestine as well as the wt strain. The most apparent difference between the galE and galU mutants lies in their LPS structures. galE-deficient strains show an apparently normal LPS, while galU-deficient strains have altered core oligosaccharide. Two observations do not support the possibility that UDP-galactose, synthesized from nontoxic concentrations of exogenous galactose in galE mutants, could be incorporated into the LPS, as known for other gram-negative bacteria (38). First, compositional and structural analyses of O1 LPS have failed to detect galactose (29, 30, 64). Second, a V. cholerae O1 galEK double mutant, insensitive to galactose and unable to synthesize UDP-galactose from exogenous galactose (Fig. 6), apparently produced wt LPS as determined by SDS-PAGE analysis (silver staining and Western blotting) and was able to colonize the small intestines of mice similar to a wt strain (unpublished results).
FIG. 6.
Synthesis and possible functions of UDP-glucose (UDP-Glc) and UDP-galactose (UDP-Gal) in V. cholerae. Strains lacking galE are predicted to accumulate the toxic products UDP-Gal and galactose-1-phosphate (Gal-1P) in the presence of high exogenous galactose. Strains grown in the absence of exogenous galactose must synthesize UDP-Gal via GalE.
According to the V. cholerae genome sequences (22), open reading frame (ORF) VC0262, located within the rfb gene cluster, is annotated as a second galE gene. However, in contrast to the galE gene investigated in this study (VCA0774; 65.4% identity to galE of E. coli in 338 amino acids), ORF VC0262 shows only low homology to the galE gene of E. coli (26.6% identity in 169 amino acids). In addition, Fallarino et al. (15) suggested that ORF VC0262 is unlikely a galE gene, showing that it could not complement an S. enterica serovar Typhimurium galE mutant. Furthermore, they found that V. cholerae mutants within ORF VC0262 synthesize a normal LPS. Taking the data together, we conclude that our constructed galEK double mutant could not synthesize UDP-galactose at all, which suggests that no galactose-containing surface structures in V. cholerae O1 El Tor are involved in colonization of the small intestine in mice.
One explanation for the colonization defect of galU mutants is that alteration of the LPS core region reduced their ability to survive within the intestine. To address the role of an intact LPS core oligosaccharide in V. cholerae intestinal survival, we tested the effects of several substances known to be present in the human intestine. Three different types of LPS mutants were compared: one lacking the O antigen (wbeW), one with an altered core (galU), and one lacking an O antigen and expressing an altered core (R-LPS). These comparative studies demonstrated several functions of the V. cholerae LPS substructures. An intact core oligosaccharide, without attached O antigen, is sufficient for survival in the presence of anionic detergents (e.g., bile salts) and short organic acids. However, effective protection against these substances is no longer provided by galU and R-LPS mutants. These results suggest that an altered core oligosaccharide increases the susceptibility to short protonated organic acids and anionic detergents. The observed growth inhibition seen in anionic detergent MIC assays is probably due to the disruption of the inner membrane integrity. Enteric bacteria like E. coli are known to need additional efflux pumps for sufficient protection against hydrophobic substances such as bile salts (73), and recently one such system has been identified in V. cholerae (7).
Both V. cholerae LPS substructures, core and O antigen, were found to have roles in resistance to bactericidal substances of the innate immune system. The core oligosaccharide region was found to be more important than the O antigen in resisting the outer membrane-disturbing action of cationic peptides. Resistance against cationic peptides can depend on several factors, such as the charge of the LPS molecules or LPS concentration in the membrane. In addition, it is known that some gram-negative bacteria are capable of activating an inducible resistance mechanism, such as the modification of lipid A and core oligosaccharides with aminoarabinose and ethanolamine, which reduces the negative charge of inner anionic regions and thereby reduces binding of the cationic peptides (14). It is not known if V. cholerae induces modifications of lipid A; hence, the sensitivity of galU mutants to cationic peptides such as polymyxin B could also be due to an inability to synthesize aminoarabinose from UDP-glucose (see above). Interestingly, the most important LPS structure for resistance against the complement system is the O antigen (67) rather than the core LPS. However, our experiments demonstrated that for effective protection against complement components, an intact core oligosaccharide is also necessary.
The results of the in vitro assays suggest that in galU mutants the integrity of the outer membrane is compromised and it no longer represents an effective barrier. The instability of the outer membrane in galU mutants could be due to weakened LPS core-LPS core interactions. Disruption or alterations of the outer membrane integrity could eventually cause enhanced penetration of proteins and diffusion of periplasmic material, resulting in periplasmic leakage (47, 63). Evidence for the loss of periplasmic proteins of a galU mutant strain was supported by the observation that β-lactamase, but not the cytoplasmic enzyme β-galactosidase (data not shown), was found in slightly increased amounts in the supernatant. This may suggest that active HAP and CT, normally secreted via the eps system, could also escape from the periplasm due to penetration or diffusion across the outer membrane. However, other than slightly increased β-lactamase in supernatants and sensitivity against hydrophobic substances, we found no evidence for outer membrane damage, since outer membrane preparations of galU strains indicated no visible protein alterations compared to the wt strain (data not shown). Additionally, we found TCP, which could serve as phage receptor for CTXΦ phage transduction. Since TCP is also required for colonization of the small intestine, we suggest that the colonization defect of the galU mutants is not due to alterations of TCP. However, it could also be possible that the quantity or function of TCP is altered in the mutant strain within the harsh environment of the small intestine. The secretion of virulence factors and the assembly of TCP in galU mutants remain to be addressed.
The spontaneous R-LPS mutants showed the most severe defect in colonization and survival in in vitro assays. The mutation in strain P27459res118 remains unidentified, but we speculate that it is affected in the HepII-transferase (Fig. 1A), since it has been hypothesized that V. cholerae O1 heptoseless mutants are nonviable (60). The alteration in the core oligosaccharide in this R-LPS mutant (res118) was found to have a more drastic effect on outer membrane integrity. It was expected that this type of mutant could not colonize the small intestine because it lacks the O antigen. The exact role of the LPS O antigen in colonization is unknown. An early report hypothesized that a lack of the pilus TCP leads to attenuated colonization (26); however, recently it was reported that O-antigen-negative strains express TCP (5). We can also demonstrate that TCP function as the CTXΦ phage receptor is maintained in the O-antigen-negative strains, which is consistent with the finding of Chiang and Mekalanos (5). One known phenotype of O-antigen mutants is a remarkable sensitivity against the complement system (67). Our data in this report showed that O-antigen-negative strains are in addition slightly more sensitive to cationic peptides. It was recently reported that the Helicobacter pylori O antigen is involved in resistance to inorganic pH stress, a condition found in the stomach (39). However, we found no evidence that V. cholerae O-antigen-negative strains (or the core mutants) are more sensitive to inorganic acids (data not shown).
We reported also that galU and galE are involved in the production of the VPS necessary for biofilm formation on abiotic surfaces. Biofilm formation is likely to be important in the life cycle of pathogenic V. cholerae strains, since these strains survive and persist in aquatic ecosystems (8, 16). Commonly, bacteria in aquatic environments are rarely found in the planktonic or free-swimming phase but instead are organized in a three-dimensional biofilm on aquatic surfaces. This may provide an advantage for the bacteria since they are better protected against bactericidal agents, and nutrients may be available (10). Recently, it was reported that V. cholerae O1 El Tor strains develop a three-dimensional biofilm on abiotic surfaces. Furthermore, it was shown that for accelerated attachment to abiotic surfaces, V. cholerae needs a functional flagellum as well as the type IV mannose-sensitive hemagglutinin pili, and that the development of the three-dimensional biofilm requires production of the VPS (69). The regulation of this complex process is not fully understood.
Constitutive expression of VPS by V. cholerae O1 El Tor leads to a rugose colony phenotype, and these cells are able to form a biofilm on abiotic surfaces (72), probably without the need of the regulatory cascade necessary in wt bacteria. We could enrich for rugose strains by exposure of the motile, biofilm-negative O1 El Tor strain P27459 to the lytic phage K139.cm9. One nonmotile rugose isolate was further characterized, and our results suggested that production of the VPS is responsible for enhanced phage resistance, possibly because the phage cannot penetrate the VPS well enough to bind its receptor, the O antigen. The mutation(s) responsible for constitutive VPS production and the underlying regulatory mechanisms are only poorly understood. However, nonmotility of strain P27459res105 may be the cause of the rugose phenotype, because recent results suggest that the absence of a flagellum leads to constitutive VPS expression, at least in O139 strains (P. Watnick, C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter, submitted for publication). We discounted an alternate possibility, namely, that the loss of HapR, the activator of HAP, caused the rugose phenotype (28), since we could observe HAP activity in this strain (data not shown).
Our data suggest that UDP-glucose is a precursor in the synthesis of the VPS, since the introduction of a galU mutation into a previously rugose strain led to a nonrugose phenotype, and this strain failed to form a biofilm. Also, the O1 El Tor strain CO970 with a galU mutation was shown to be biofilm negative. The introduction of a mutation in galE into rugose strains also renders them nonrugose, and they are unable to form biofilms on abiotic surfaces, suggesting that VPS synthesis requires UDP-galactose to be synthesized via UDP-glucose in the absence of exogenous galactose (Fig. 6). Thus, we identified in this work two additional enzymes necessary for biosynthesis of the VPS and the formation of V. cholerae biofilms. They are encoded by galU and galE and are required in addition to those encoded by the vps (Vibrio polysaccharide synthesis) region (72).
ACKNOWLEDGMENTS
We thank J. Schmidt-Brauns for critical reading of the manuscript. For the clinical V. cholerae strain used in this study, we thank J. J. Mekalanos.
This work was funded by BMBF grant 01KI8906 and NIH grant AI43486 to K.E.K.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang Z J, Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andoh A, Fujiyama Y, Sakumoto H, Uchihara H, Kimura T, Koyama S, Bamba T. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol. 1998;111:477–483. doi: 10.1046/j.1365-2249.1998.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake P A. Historical perspectives on pandemic cholera. In: Wachsmuth I K, Blake P A, Olsvik O R, editors. Vibrio cholerae and cholera. Washington, D.C.: American Society for Microbiology; 1994. pp. 293–295. [Google Scholar]
- 4.Böhringer J, Fischer D, Mosler G, Hengge-Aronis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of ςs-dependent genes in Escherichia coli. J Bacteriol. 1995;177:413–422. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang S L, Mekalanos J J. rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization. Infect Immun. 1999;67:976–980. doi: 10.1128/iai.67.2.976-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang S L, Mekalanos J J. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 7.Colmer J A, Fralick J A, Hamood A N. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol Microbiol. 1998;27:63–72. doi: 10.1046/j.1365-2958.1998.00657.x. [DOI] [PubMed] [Google Scholar]
- 8.Colwell R R. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 9.Correa N E, Lauriano C M, McGee R, Klose K E. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol. 2000;35:743–755. doi: 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 10.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: Cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1210–1223. [Google Scholar]
- 12.Cummings J H, Pomare E W, Branch W J, Naylor C P E, Macfarlane G T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst R K, Guine T, Miller S I. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J Infect Dis. 1999;179(Suppl. 2):S326–S330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 15.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster J W. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 18.Fry B N, Feng S, Chen Y Y, Newell D G, Coloe P J, Korolik V. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect Imm. 2000;68:2594–2601. doi: 10.1128/iai.68.5.2594-2601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimberg J, Maguire S, Belluscio L. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 1989;17:8893. doi: 10.1093/nar/17.21.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman E A. How bacteria resist killing by host-defense peptides. Trends Microbiol. 1994;2:444–449. doi: 10.1016/0966-842x(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 21.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann A F. Bile secretion and the enterohepatic circulation of bile acids. In: Feldman M, Scharschmidt B F, Sleisenger M H, editors. Gastrointestinal and liver disease. W. B. Philadelphia, Pa: Saunders Co.; 1998. pp. 937–948. [Google Scholar]
- 25.Holmgren J, Svennerholm A M. Mechanisms of disease and immunity in cholera: a review. J Infect Dis. 1977;136(Suppl.):S105–S112. doi: 10.1093/infdis/136.supplement.s105. [DOI] [PubMed] [Google Scholar]
- 26.Iredell J R, Manning P A. Outer membrane translocation arrest of the TcpA pilin subunit in rfb mutants of Vibrio cholerae O1 strain 569B. J Bacteriol. 1997;179:2038–2046. doi: 10.1128/jb.179.6.2038-2046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 28.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 29.Kabir S. Characterization of the lipopolysaccharide from Vibrio cholerae O395 (Ogawa) Infect Immun. 1982;38:1263–1272. doi: 10.1128/iai.38.3.1263-1272.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenne L, Lindberg B, Unger P, Gustafsson B, Holme T. Structural studies of the Vibrio cholerae O-antigen. Carbohydr Res. 1982;100:341–349. doi: 10.1016/s0008-6215(00)81047-2. [DOI] [PubMed] [Google Scholar]
- 31.Klose K E, Mekalanos J J. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klose K E, Mekalanos J J. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 33.Knirel Y A, Widmalm G, Senchenkova S N, Jansson P-E, Weintraub A. Structural studies on the short-chain lipopolysaccharide of Vibrio cholerae O139 Bengal. Eur J Biochem. 1997;247:402–410. doi: 10.1111/j.1432-1033.1997.00402.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Larid M W, Kloser A W, Misra R. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J Bacteriol. 1994;176:2259–2264. doi: 10.1128/jb.176.8.2259-2264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 307–342. [Google Scholar]
- 37.Mallow E B, Harris A, Salzman N, Russell J P, DeBerardinis R J, Ruchelli E, Bevins C L. Human enteric defensins. J Biol Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- 38.Maskell D J, Szabo M J, Deadman M E, Moxon E R. The gal locus from Haemophilus influenzae: cloning, sequencing and the use of gal mutants to study lipopolysaccharide. Mol Microbiol. 1992;6:3051–3063. doi: 10.1111/j.1365-2958.1992.tb01763.x. [DOI] [PubMed] [Google Scholar]
- 39.McGowan C C, Necheva A, Thompson S A, Cover T L, Blaser M J. Acid-induced expression of an LPS associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 40.Medrano A I, DiRita V J, Castillo G, Sanchez J. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator ToxT in response to culture conditions. Infect Immun. 1999;67:2178–2183. doi: 10.1128/iai.67.5.2178-2183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrell S D, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 42.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris J G, Sztein M B, Rice E W, Nataro J P, Losonsky G A, Panigrahi P, Tacket C O, Johnson J A. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J Infect Dis. 1996;174:1364–1368. doi: 10.1093/infdis/174.6.1364. [DOI] [PubMed] [Google Scholar]
- 44.Mullis K B, Faloona F. Specific synthesis of DNA in vitro via a polymerase chain reaction. Methods Enzymol. 1987;155:335–340. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 45.Nesper J, Blaβ J, Fountouloakis M, Reidl J. Characterization of the major control region of Vibrio cholerae bacteriophage K139: immunity, exclusion, and integration. J Bacteriol. 1999;181:2902–2913. doi: 10.1128/jb.181.9.2902-2913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nesper J, Kapfhammer D, Klose K E, Merkert H, Reidl J. Characterization of Vibrio cholerae O1 antigen as bacteriophage K139 receptor, and identification of IS1004 insertions aborting O1 antigen biosynthesis. J Bacteriol. 2000;182:5097–5104. doi: 10.1128/jb.182.18.5097-5104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikaido H. Outer membrane. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magsanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 29–47. [Google Scholar]
- 48.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 51.Reidl J, Mekalanos J J. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini transposon to identify a phage-encoded virulence factor. Mol Microbiol. 1995;18:685–701. doi: 10.1111/j.1365-2958.1995.mmi_18040685.x. [DOI] [PubMed] [Google Scholar]
- 52.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandkvist M, Michel O, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 55.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 56.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;51:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Steinberg D A, Hurst M A, Fujii C A, Kung A H, Ho J F, Cheng F-C, Loury D J, Fiddes J C. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stimson E, Virji M, Makepeace K, Dell A, Morris H R, Payne G, Saunders J R, Jennings M P, Barker S, Panico M, et al. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamid-2,4,6-trideoxyhexose. Mol Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 59.Stroeher U H, Jedani K E, Manning P A. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene. 1998;223:269–282. doi: 10.1016/s0378-1119(98)00407-7. [DOI] [PubMed] [Google Scholar]
- 60.Stroeher U H, Karageorgos L E, Morona R, Manning P A. In Vibrio cholerae serogroup O1, rfaD is closely linked to the rfb operon. Gene. 1995;155:67–72. doi: 10.1016/0378-1119(94)00923-g. [DOI] [PubMed] [Google Scholar]
- 61.Svennerholm A M, Holmgren J. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbant assay (GM1-ELISA) procedure. Curr Microbiol. 1978;1:19–23. [Google Scholar]
- 62.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinogradov E V, Bock K, Holst O, Brade H. The structure of the lipid A-core region of the lipopolysaccharides from Vibrio cholerae O1 smooth strain 569B (Inaba) and rough mutant strain 95R (Ogawa) Eur J Biochem. 1995;233:152–158. doi: 10.1111/j.1432-1033.1995.152_1.x. [DOI] [PubMed] [Google Scholar]
- 65.Wai S N, Mizunoe Y, Takade A, Kawabata S-I, Yoshida S-I. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waldor K W, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 67.Waldor M K, Colwell R, Mekalanos J J. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc Natl Acad Sci USA. 1994;91:11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wandersman C, Letoffe S. Involvement of lipopolysaccharide in the secretion of Escherichia coli α-haemolysin and Erwinia chrysanthemi proteases. Mol Microbiol. 1993;7:141–150. doi: 10.1111/j.1365-2958.1993.tb01105.x. [DOI] [PubMed] [Google Scholar]
- 69.Watnick P, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- 72.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zgurskaya H I, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]