Abstract
Previous reports indicated a correlation between loss of plasmids and decreased infectivity of Borrelia burgdorferi strain B31, suggesting that plasmids may encode proteins that are required for pathogenesis. In this study, we expand on this correlation. Using the B. burgdorferi genomic sequence, we designed primers specific for each plasmid, and by using PCR we catalogued 11 linear and 2 circular plasmids from 49 clonal isolates of a mid-passage B. burgdorferi strain B31, initially derived from infected mouse skin, and 20 clones obtained from mouse skin infected with a low-passage isolate of B. burgdorferi strain B31. Among the 69 clones analyzed, nine distinct genotypes were identified relative to wild-type B. burgdorferi strain B31. Among the nine clonal genotypes obtained, only the 9-kb circular plasmid (cp9), the 25-kb linear plasmid (lp25), and either the 28-kb linear plasmid 1 or 4 (lp28-1 and lp28-4, respectively) were missing, in different combinations. We compared the infectivity of the wild-type strain, containing all known B. burgdorferi plasmids, with those of single mutants lacking either lp28-1, lp28-4, or lp25 and a double mutant missing both cp9 and lp28-1. The infectivity data indicated that B. burgdorferi strain B31 cells lacking lp28-4 were modestly attenuated in all tissues analyzed, whereas samples missing lp25 were completely attenuated in all tissues, even at the highest inoculum tested. Isolates without lp28-1 infected the joint tissue yet were not able to infect other tissues as effectively. In addition, we have observed a selection in vivo in the skin, bladder, and joint for cells containing lp25 and in the skin and bladder for cells containing lp28-1, indicating that lp25 and lp28-1 encode proteins required for colonization and short-term maintenance in these mammalian tissues. In contrast, there was no selection in the joint for cells containing lp28-1, suggesting that genes on lp28-1 are not required for colonization of B. burgdorferi within the joint. These observations imply that the dynamic nature of the B. burgdorferi genome may provide the genetic heterogeneity necessary for survival in the diverse milieus that this pathogen occupies in nature and may contribute to tropism in certain mammalian host tissues.
Lyme borreliosis, or Lyme disease, is a multisystemic inflammatory disorder transmitted by the bites of ticks infected with the spirochetal bacterium Borrelia burgdorferi sensu lato (13, 25). Over the past 15 years, Lyme disease has become the most frequent arthropod-borne disease in the United States and has been identified in a latitudinal strip in the Northern hemisphere including Europe, Eurasia, and Asia (2, 4, 5, 29). Determination of the genomic sequence of B. burgdorferi strain B31 by The Institute of Genomic Research (TIGR), along with several other investigators, in 1997 revealed that this spirochete contained a unique genomic organization consisting of a linear chromosome of 910 kb in size and several plasmids (reference 8 and the TIGR website [http://www.tigr.org/tdb/mdb/bbdb/bbdb.html]). Complete annotation of the genome sequence indicated that B. burgdorferi contained 21 plasmids, i.e., 12 linear plasmids ranging in size from 5 to 56 kb (designated by lp followed by their size in kilobase pairs) and 9 circular plasmids with sizes between 9 and 32 kb (designated by cp followed by the size in kilobase pairs) (5; TIGR website). Previous studies with other pathogens, including Shigella flexneri, Yersinia spp., and Salmonella enterica serovar Typhimurium (7, 15), indicated that virulence determinants were often plasmid encoded and that, in some instances, loss of the virulence-associated plasmid resulted in either a decrease or an abrogation of pathogenicity. The hypothesis that B. burgdorferi plasmids carry infection-associated genes is supported by the fact that prolonged serial in vitro cultivation of infectious nonclonal isolates changes the plasmid and protein profiles of the organism and results in the loss of infectivity (9, 21, 22, 30). Reports published before the genome project was completed indicated a reduction in infectivity when several plasmids in the 24- to 28-kb range, as well as a 9-kb circular plasmid designated cp9, were lost (30). Since then, a 28-kb linear plasmid, designated lp28-1, has been linked to a reduced-infectivity phenotype (32).
The purpose of this study was to identify plasmids, including lp28-1, that correlate with the infectious phenotype and to determine whether tissue tropism is related to plasmid content in B. burgdorferi strain B31. With the advent of the B. burgdorferi strain B31 genome sequence information, we designed PCR primers specific to 11 linear and 2 circular plasmids and, in conjunction with PCR, determined the plasmid content of 69 separate clones of B. burgdorferi strain B31. Of the 69 clones tested, 20 were from mouse skin infected with a low-passage isolate and 49 were from a mid-passage isolate initially obtained from an infected mouse. We then assessed the requirement for specific plasmids in the pathogenesis of B. burgdorferi by testing appropriate clones in the mouse model of Lyme borreliosis and quantitatively assessing the infectivity deficits of these mutants relative to wild-type B. burgdorferi strain B31. Furthermore, we determined the plasmid content of clones obtained from our infectivity studies to determine whether genotypic changes occurred in vivo. The results presented in this report challenge the paradigm of clonality as it applies to B. burgdorferi, as cells containing different plasmid combinations were obtained from an individual colony, perhaps due to deficits in plasmid partitioning upon replication. Finally, the observation that B. burgdorferi clones are genetically variable when isolated from different infected animal tissues suggests that genetic mosaics of B. burgdorferi are tolerated in vivo and result in variants with differential infectivity phenotypes and tissue-tropic properties.
MATERIALS AND METHODS
Bacterial strains.
B. burgdorferi sensu stricto strain B31 (ATCC 35210) was used exclusively in this study. This isolate of strain B31 was originally isolated from ticks on Shelter Island, New York, and appears, based on Southern blotting, PCR, and nucleotide sequencing (reference 23 and data not shown), to be identical to the TIGR isolate previously sequenced (TIGR website). The derivatives of strain B31 obtained in this study and their relevant genotypes are indicated in Table 1. All B. burgdorferi cultures were grown in 1% CO2 at 32°C either in BSK-II liquid medium lacking gelatin (3) or on BSK-II agarose plates as described previously (20), both supplemented with 6% normal rabbit serum (Pel-Freez Biologicals, Rogers, Ark.). A polyclonal B. burgdorferi strain B31 passage 2 isolate was used to infect mice intradermally at an inoculum of 103 cells, and skin samples were cultured at 4 weeks postinfection in BSK-II medium. B. burgdorferi cells were enumerated from these cultures using dark-field microscopy, serially diluted in BSK-II medium, and plated onto BSK-II agarose in overlays as previously described (14). Individual colonies were obtained and cultured in BSK-II medium to mid- to late-log phase (approximately 108 B. burgdorferi cells per ml), and their corresponding plasmid profiles were determined using PCR as outlined below. In addition, a B. burgdorferi strain B31 isolate that had been passaged 48 times in vitro (designated P48) was plated on BSK-II agarose plates, individual colonies were picked, and template DNA prepared and subjected to PCR as outlined below. Clones obtained from the P48 infectivity analysis (see “ID50 determinations” below) were isolated from infected mouse tissues by plating samples diluted from BSK-II liquid cultures in BSK-II agarose overlays as previously described (3, 14, 20).
TABLE 1.
B. burgdorferi strain B31 genotypic variants used in this study
| B31 variant(s) | Description | Source or reference |
|---|---|---|
| MSK5 | Clone obtained from the skin of a mouse infected with low passage B. burgdorferi B31 and determined to have all of the known plasmids | This study |
| MSK7 | Clone obtained from the skin of a mouse infected with low-passage B. burgdorferi B31 and determined to be lp28-4− | This study |
| MSK10 | Clone obtained from the skin of a mouse infected with low-passage B. burgdorferi B31 and determined to be lp28-1− | This study |
| P48 | B. burgdorferi B31 isolate originally obtained from a mouse infected with low-passage B. burgdorferi B31 and serially passaged in vitro 48 times | 6 |
| ML28 | Clone isolated from nonclonal P48 sample and determined to be cp9− and lp28-1− | This study |
| ML23 | Clone isolated from nonclonal P48 sample and determined to be lp25− | This study |
| MJ1 to -20 | Clones from tibiotarsal joint tissue of mice infected with clone ML28 passage 2 | This study |
| OMB1 to -20 | Clones isolated from bladder tissue of mice infected with nonclonal P48 sample | This study |
| OMS1 to -20 | Clones isolated from skin tissue of mice infected with nonclonal P48 sample | This study |
| OMJ1 to -99 | Clones isolated from tibiotarsal joint tissue of mice infected with nonclonal P48 sample | This study |
PCR analysis.
The complete sequences of all of the open reading frames in each plasmid were retrieved from the B. burgdorferi strain B31 genome database (TIGR website) and saved as MacVector version 6.5 files (Oxford Molecular Group, Inc., Campbell, Calif.). Primer sets, specific for an entire gene coding sequence, were designed for all of the plasmids with the exception of the 32-kb circular plasmids (cp32), due to their inherent similarities, and linear plasmid 21 (lp21), because of its similarity to linear plasmid 5 (lp5) (5, 28). The primers used in this study are listed in Table 2. Primers specific for lp36 (BBK19F and BBK19R) were predicted to yield a 610-bp fragment (Table 2). However, we found that the BBK19F oligonucleotide primer alone was capable of amplifying a 316-bp PCR product. BBK19F hybridizes to both strands in the proper orientation, yielding this PCR amplimer, even though the annealing site in the reverse orientation contains five mismatches. In all samples tested, the 316-bp band was observed (see Fig. 1 and 2). No other oligonucleotide primer sets yielded anomalous PCR products.
TABLE 2.
Oligonucleotides used in this study
| Plasmid | Genome designation | Sequence (5′ to 3′) | Predicted fragment size (bp) |
|---|---|---|---|
| cp9 | BBC10F | GAACTATTTATAATAAAAAGGAGAGC | 459 |
| BBC10R | ATCTTCTTCAAGATATTTTATTATAC | ||
| BBC07F | GATGAAAATCTTTCACAAAAG | 291 | |
| BBC07R | AACAATAATAGAGATAAAGGG | ||
| cp26 | BBB19F | AATAATTCAGGGAAAGATGGG | 573 |
| BBB19R | AGGTTTTTTTGGACTTTCTGCC | ||
| lp5 | BBTF | CTTGCTTTAAGCCCTATTTCAC | 645 |
| BBTR | GCACACTACCCATTTTTGAATC | ||
| lp17 | BBD10F | CAAACTTATCAAATAGCTTATC | 519 |
| BBD10R | ACTGCCACCAAGTAATTTAAC | ||
| lp25 | BBE16F | ATGGGTAAAATATTATTTTTTGGG | 618 |
| BBE16R | AAGATTGTATTTTGGCAAAAAATTTTC | ||
| lp28-1 | BBF20F | ATGAACAAAAAATTTTCTATTTC | 291 |
| BBF20R | GTTGCTTTTGCAATATGAATAGG | ||
| BBFF | CGCTGGACAGTTTATATTGGGG | 1,723 | |
| BBFR | GTTGATAATGCTAATGCTGGGGAC | ||
| lp28-2 | BBG02F | TCCCTAGTTCTAGTATCTACTAGACCG | 810 |
| BBG02R | TTTTTTTTGTATGCCAATTGTATAATG | ||
| lp28-3 | BBH06F | GATGTTAGTAGATTAAATCAG | 651 |
| BBH06R | TAATAAAGTTTGCTTAATAGC | ||
| lp28-4 | BBI16F | CAGGCCGGATTTTAATATCGA | 1,300 |
| BBI16R | GTTTATATTTTGACACTATAAG | ||
| BBI16(II)F | GCAGGCCGGATTTTAATATCGATC | 732 | |
| BBI16(II)R | GCTCATTAGATAGCGTATTTTTTAG | ||
| lp36 | BBK19F | AAGTTTATGTTTATTATTGC | 316 (610)a |
| BBK19R | ATTGTTAGGTTTTTCTTTTCC | ||
| lp38 | BBJ34F | AAATTCTATGGAAGTGATG | 1,010 |
| BBJ34R | TTTCTATTTATTTTTAGGC | ||
| lp54 | BBA16F | GCACAAAAAGGTGCTGAG | 840 |
| BBA16R | TTTTAAAGCGTTTTTAAGC | ||
| lp56 | BBQ56F | AAGATTGATGCAACTGGTAAAG | 975 |
| BBQ56R | CTGACTGTAACTGATGTATCC |
BBK19F anneals to both strands, yielding a 316-bp amplimer; see text for details.
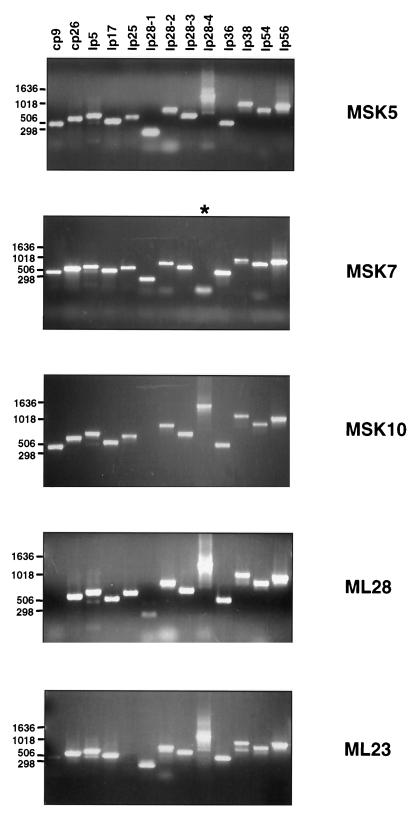
FIG. 1.
Plasmid profiles of B. burgdorferi strain B31 clones MSK5, ML28, ML23, ML28, and MSK10. PCR analysis was performed as detailed in Materials and Methods with oligonucleotide primers listed in Table 2. PCR products were resolved by electrophoresis on a 1.5% agarose gel. Molecular size markers (in base pairs) are indicated on the left. The asterisk in the MSK7 panel indicates the location of a putative primer dimer observed with the lp28-4 oligonucleotide primers.
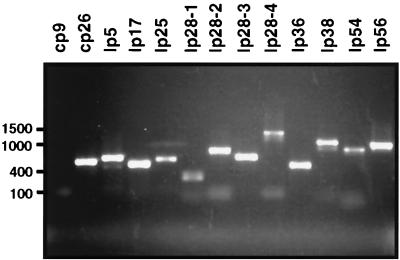
FIG. 2.
Plasmid profile of the polyclonal B. burgdorferi strain B31 passage 48 (P48) sample. PCR analysis was performed as detailed in Materials and Methods using the primers shown in Table 2. PCR products were resolved by electrophoresis in a 1.5% agarose gel. Molecular size markers (in base pairs) are indicated on the left.
Total DNA was extracted from a liquid culture of each isolate once it had reached mid-log phase. Template was obtained by pelleting B. burgdorferi at 4,000 × g for 15 min, washing the bacteria in an equal volume of phosphate-buffered saline (PBS) (pH 7.4), repelleting the cells as before, and resuspending the resulting pellet in water such that the final concentration was 5 × 106 B. burgdorferi equivalents per μl. The samples were then boiled for 5 min, the insoluble material was pelleted, and the supernatant containing the DNA was removed. One microliter of the resulting supernatant was used as a DNA template (i.e., 5 × 106 B. burgdorferi equivalents), and each primer was used at a concentration of 0.2 μM in a 20-μl PCR mixture containing PCR Supermix (GIBCO-BRL, Gaithersburg, Md.) as the source of Mg2+, deoxynucleotides, and Taq polymerase. The PCR parameters used were as follows: a denaturation step at 94°C for 1 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 2 min; and a final extension step at 72°C for 6 min. PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Southern blots.
Southern blotting was conducted essentially as described previously (23) with the following modifications. The template for the random priming reaction was either a PCR product or purified plasmid DNA. Preparation of probes using plasmid DNA was done as previously outlined (23). When a PCR amplimer was used, roughly 500 ng of the PCR product was resolved on an agarose gel and the band of the appropriate size was excised. The DNA was removed and incubated with Tris-HCl buffer-saturated phenol. The sample was flash frozen and separated into phases by centrifugation. The upper aqueous phase was extracted with 2:1 phenol-chloroform and separated by centrifugation, and the resulting aqueous phase was extracted again with chloroform. The DNA was then precipitated, and the final washed pellet was resuspended in 20 μl of sterile water. A 19-μl portion of the final sample was used as a template with either the Gene Images system (Amersham Pharmacia Biotech, Piscataway, N.J.) or the Renaissance system (NEN Life Science Products Inc., Boston, Mass.), which resulted in the incorporation of fluoresceinated dUTP into a randomly primed probe. The DNA samples to be tested were isolated from approximately 2 × 1010 B. burgdorferi cells using Qiagen (Chatsworth, Calif.) tip 100 or tip 500 columns. Subsequently, the DNA was digested and either transferred or dot blotted onto a nylon membrane (Hybond-N; Boehringer Mannheim Inc., Indianapolis, Ind.) and subjected to Southern blotting as previously described (23). Probes that hybridized to immobilized DNA were detected by incubating the blots with antifluorescein antibodies conjugated to horseradish peroxidase (HRP) and were exposed to Kodak XAR-5 X-ray film following incubation with the appropriate chemiluminescent substrate (enhanced chemiluminescence [ECL] developing reagents [Amersham Pharmacia Biotech]).
ID50 determinations.
Three 8-week-old C3H/HeN female mice (Charles River Laboratories, Wilmington, Mass.) were separately infected by intradermal inoculation with 10-fold serial dilutions of the different B. burgdorferi B31 mutant isolates to be tested, at levels of 10, 102, 103, 104, and 105 B. burgdorferi strain B31 variants. Two weeks later, mice were sacrificed and the spleen, tibiotarsal joint, lymph node, heart, bladder, and abdominal skin were removed aseptically. Tissues were processed using sterile pestles and placed in liquid BSK-II medium. Cultures were passaged blindly 5 days later into fresh BSK-II medium to minimize toxicity associated with host tissue. Tubes were examined for borrelial growth every week thereafter using dark-field microscopy, and the 50% infective dose (ID50) was calculated using the method of Reed and Muench (16) as outlined by Norris et al. (14).
PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was conducted as described previously (24). After SDS-PAGE, the resulting gel was stained with Coomassie brilliant blue or transferred to an Immobilon-P polyvinyl difluoride membrane and immunoblotted as described below.
Immunoblot analysis.
Immunoblotting was conducted as described previously (23) using the following antisera as primary antibodies: normal mouse serum, pooled sera from mice infected for 6 weeks with 103 B. burgdorferi strain B31 derivative MSK5 cells (contains all known borrelial plasmids; see above and Results for details), serum from a patient with late Lyme disease manifestations (kindly provided by Allen Steere, Tufts University, Boston, Mass.), adsorbed serum from infection-immune rabbits (6), or antiserum against VlsE (generously provided by Steven Norris, University of Texas Health Science Center, Houston). Following transfer, the polyvinylidene difluoride membrane was blocked in 10% powdered milk in PBS (pH 7.4)–0.2% Tween 20 (PBST) for 1 h at room temperature. The blot was then incubated separately with each primary antibody (indicated above) diluted 1:1,000 (or, for the anti-VlsE serum, diluted 1:5,000) in 10% powdered milk in PBST for 1 h and subsequently washed in PBST. After the wash step, each blot was incubated with a 1:1,000 dilution of protein A conjugated to HRP (Amersham Corp., Arlington Heights, Ill.). The blots were then washed with PBST to eliminate unbound protein A-HRP, developed using the ECL system (Amersham Corp.), and exposed to Kodak XAR-5 film for times ranging from 1 to 30 min.
Statistical analysis.
Infectivity data were analyzed using a nonparametric chi-square test. Significant differences were accepted when the P value was <0.05.
RESULTS
Design of PCR primers.
Several investigators have previously demonstrated that B. burgdorferi loses plasmid DNA in response to in vitro passage and that the loss of plasmids correlates with a decrease in infectivity, implying that genetic loci within these plasmids encode virulence determinants (14, 21, 22, 30; J. E. Purser and S. J. Norris, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. D-214, p. 275, 2000). The large number of plasmid species (12 linear and 9 circular) have made it difficult to definitively determine which of the plasmids were missing from B. burgdorferi isolates, due to their low copy number (10) and because several of these species are similar in size (for example, four linear plasmids of approximately 28 kb designated lp28-1, lp28-2, lp28-3, and lp28-4 [8]). Also, the possibility that circular plasmids comigrate with linear plasmids contributed to the inability to accurately resolve these different species. To circumvent these issues, we used the available genomic sequence of Borrelia burgdorferi strain B31 (TIGR website) to design specific PCR primers for 11 linear and 2 circular plasmids. We excluded the seven 32-kb circular plasmids (cp32-1, -3, -4, -6, -7, -8, and -9 [5; TIGR website]) from consideration since they are nearly identical at the nucleotide level, with the notable exceptions of the ospEF/elp (also called erp) loci, the mlp genes, and the bdr paralogues (17, 18, 28, 31, 33). More importantly, no correlation in infectivity was seen in strains containing various combinations of the cp32 plasmids, both in our laboratory (data not shown) and by others (J. V. McDowell, S. Y. Sung, and R. T. Marconi, unpublished data). The 21-kb linear plasmid (lp21) was also eliminated, since it is nearly identical to the 5-kb linear plasmid (lp5) with the exception of a nonencoding repetitive domain that contributes approximately 12.8 kb of the lp21 sequence (the TIGR sequence indicated that lp21 was 18,753 nucleotides total [5; TIGR website]). For the remaining plasmids, a single gene was chosen and analyzed using the MacVector 6.5 software package to predict PCR primers that would generate amplicons diagnostic for each plasmid. Due to the frequency of paralogous genes within the B. burgdorferi genome (5, 8; TIGR website), we identified genes that were unique to each plasmid to prevent the coamplification of multiple PCR products unlinked to the gene or plasmid of interest. Primers were chosen for a given plasmid-encoded gene and tested against a composite file of the B. burgdorferi plasmid DNA to ensure that the primer pair was specific (data not shown). Oligonucleotide primers that had significant homology with other paralogous sequences (i.e., contained three or four mismatches) were eliminated from consideration. Oligonucleotide primer pair sequences, the genes they amplify, and the expected sizes of the DNA fragments they amplify are indicated in Table 2.
Stability of clonal isolates.
To begin analyzing the genotypic differences and their potential linkage to infectivity in B. burgdorferi strain B31, we infected C3H/HeN mice intradermally and isolated 20 independent colonies from skin samples cultivated on BSK-II agarose plates. DNA from these 20 clones was subjected to PCR analysis, and two mutants were obtained, one missing lp28-1 (designated MSK10) and one lacking lp28-4 (designated MSK7) (Fig. 1). The remaining 18 isolates contained all plasmids tested, and one of these samples, termed MSK5, has been designated our wild-type strain (Fig. 1). We then subjected MSK5, MSK7, and MSK10 to numerous in vitro passages in an attempt to isolate mutant strains lacking different plasmids, since this approach has been used by several investigators to generate genetically distinct populations of B. burgdorferi (14, 21, 22). Using PCR to evaluate these potential changes, we did not observe the loss of any additional plasmids from MSK5, MSK7, or MSK10 after as many as 27 in vitro passages (data not shown), indicating that B. burgdorferi is genetically stable when cultivated for numerous generations within our laboratory.
Isolation of clones from mid-passage B. burgdorferi strain B31.
Our inability to isolate genetically diverse populations of B. burgdorferi strain B31 isolate MSK5, MSK7, or MSK10 after extensive in vitro passages prompted us to utilize a different methodology to obtain genetically diverse B. burgdorferi clones. We plated and isolated colonies from B. burgdorferi strain B31 P48, which had been passaged 48 times in vitro; this strain was less virulent in the rabbit model of Lyme borreliosis and was incapable of completely protecting against challenge with a low-passage, infectious isolate (6). This phenotype implied that P48 was attenuated due to genetic differences relative to wild-type, low-passage B. burgdorferi strain B31. In support of this contention, PCR analysis of the polyclonal P48 sample indicated that the cp9 plasmid was not present (Fig. 2). Given that cp9 has been associated with infectious isolates, we suspected that this genetic difference might, in part, explain the phenotype associated with P48. We subsequently plated P48 into agarose overlays and incubated the resulting samples until colonies were apparent (approximately 10 to 14 days). After this time, 49 individually isolated colonies were picked and cultured in BSK-II liquid medium, and the DNA from each sample was prepared for PCR as described in Materials and Methods. PCR-amplified DNA from the 11 linear plasmids and 2 circular plasmids from all 49 samples was resolved on agarose gels, and eight genotypic groups of B. burgdorferi strain B31 P48 were identified (Table 3). Table 3 shows that of the 13 plasmids analyzed, only cp9, lp25, lp28-1, or lp28-4 were missing from the clones characterized, in various combinations and at different frequencies. Furthermore, cp9 was detected by PCR in 4.1% of the clones, indicating that the initial plasmid profile of P48 yielded a false negative for cp9 compared with the overall genetic complexity of the polyclonal population. All other plasmids tested (i.e., cp26, lp5, lp17, lp28-2, lp28-3, lp36, lp38, lp54, and lp56) were present in every sample tested (data not shown). The most common isolate observed lacked cp9, lp25, and lp28-4, followed by clones that were missing all four plasmids (Table 3). Our data indicate that of the four plasmid species whose presence was variable, lp28-1 was most commonly observed in the different clonal isolates (69.4%), followed distantly by lp25 (10.2%), lp28-4 (10.2%), and cp9 (4.1%). In some instances cp9, lp25, lp28-1, and lp28-4 signals appeared as faint bands in various clones following PCR relative to wild-type MSK5, to which the bands were normalized following agarose gel electrophoresis (as indicated in Table 3). This result suggested that loss of plasmids might be occurring over time, thereby converting a genetically identical clone into a mixed collection of genetically variable cells. From the clones characterized, we selected four mutants missing either cp9, lp25, lp28-1, or lp28-4 to characterize further.
TABLE 3.
Genotypic compositions of ML clones derived from a nonclonal P48 isolate
| Plasmid(s) absent | No. of clones (%) (n = 49) |
|---|---|
| cp9, lp25, lp28-1, lp28-4 | 13a (26.5) |
| cp9, lp25, lp28-4 | 26b (53.1) |
| cp9, lp28-1, lp28-4 | 1 (2.0) |
| cp9, lp25 | 4c (8.2) |
| cp9, lp28-4 | 2 (4.1) |
| cp9, lp28-1 | 1d (2.0) |
| lp25, lp28-4 | 1 (2.0) |
| lp28-4 | 1 (2.0) |
Two clones had a low PCR signal for lp25, five clones had a low PCR signal for lp28-1, and one clone had a low PCR signal for lp28-4.
Three clones had a low PCR signal for lp25, and two clones had a low PCR signal for cp9.
Two clones had a low PCR signal for cp9.
The clone had a low PCR signal for lp28-1.
Genetic characterization of mutant clones.
Several of the B. burgdorferi strain B31 clones that we identified were lacking a single plasmid based on PCR analysis, including a putative cp9− mutant designated ML28 and an lp25− mutant designated ML23 (Fig. 1). Since we already had mutants lacking either lp28-1 or lp28-4 alone (MSK10 and MSK7, respectively [Fig. 1]), we next wanted to determine the infectivity deficit associated with each of these clones relative to the wild-type strain, MSK5. C3H/HeN mice were infected with 10-fold serial dilutions of all five strains, i.e., MSK5 (wild-type strain), ML28 (cp9−), ML23 (lp25−), MSK10 (lp28-1−), and MSK7 (lp28-4−), and various tissues from each infected mouse were removed aseptically at 2 weeks postinfection. The tissues were cultured in BSK-II medium and scored for growth at each dilution. The results are summarized in Table 4 and indicate that the infectious dose calculated for the wild-type strain, MSK5, was identical to that reported previously for B. burgdorferi strain Sh-2-82 by Norris et al. (14). Furthermore, we determined that the strain lacking lp28-4 alone, MSK7, showed a slight but detectable infectivity deficit relative to wild-type MSK5, as seen by a 1-log-unit increase in the ID50 for all tissues tested except the heart. In contrast, MSK10, which lacks lp28-1, was significantly attenuated in its ability to colonize the spleen and lymph node and showed low-level infectivity in the heart, bladder, and skin, yet it demonstrated significant infectivity in joint tissue, albeit less than wild-type MSK5 (ID50 of 3.2 × 103 in the joint for MSK10-infected mice versus an ID50 of 1.8 × 102 for the joints of mice infected with MSK5 [Table 4]). Similarly, strain ML28 was attenuated in C3H/HeN mice in all tissues except the tibiotarsal joint. However, the most dramatic effect was seen in strain ML23, lacking lp25, which was completely noninfectious for all challenge inocula tested, suggesting that lp25 encodes virulence determinants required for normal colonization and/or dissemination in C3H/HeN mice.
TABLE 4.
ID50 determination for B. burgdorferi strain B31 clones
| Isolate and inoculum | No. of cultures positive/total no.
|
No. of mice positive/total no. of mice | Calculated ID50
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen | Joint | Heart | Bladder | Skin | Lymph node | All sites | Spleen | Joint | Heart | Bladder | Skin | Lymph node | ||
| MSK5a | 1.8 × 102 | 1.8 × 102 | 1.8 × 102 | 1.8 × 102 | 1.8 × 102 | 1.8 × 102 | ||||||||
| 105 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | ||||||
| 104 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | ||||||
| 103 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | ||||||
| 102 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 6/18 | 1/3 | ||||||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| MSK7b | 1.9 × 103 | 3.2 × 103 | 5.6 × 102 | 1.9 × 103 | 1.9 × 103 | 1.9 × 103 | ||||||||
| 105 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | ||||||
| 104 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | ||||||
| 103 | 1/3 | 1/3 | 2/3 | 1/3 | 1/3 | 1/3 | 7/18 | 2/3 | ||||||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| MSK10b | >105 | 104 | 3.4 × 104 | 105 | 105 | >105 | ||||||||
| 105 | 0/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 1/18 | 1/3 | ||||||
| 104 | 0/3 | 3/3 | 0/3 | 1/3 | 1/3 | 0/3 | 5/18 | 3/3 | ||||||
| 103 | 0/3 | 1/3 | 2/3 | 0/3 | 0/3 | 0/3 | 3/18 | 2/3 | ||||||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| ML28c | >105 | 3.2 × 103 | >105 | >105 | >105 | >105 | ||||||||
| 105 | 0/3 | 3/3 | 0/3 | 0/3 | 0/3 | 0/3 | 3/18 | 3/3 | ||||||
| 104 | 0/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 1/18 | 1/3 | ||||||
| 103 | 0/3 | 2/3 | 0/3 | 0/3 | 0/3 | 0/3 | 2/18 | 2/3 | ||||||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| ML23c | >105 | >105 | >105 | >105 | >105 | >105 | ||||||||
| 105 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 104 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 103 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||||||
Wild type (refers to a clone not derived from nonclonal P48 that has been determined to have all of the known plasmids).
Both of these clones were isolated from the skin from a mouse infected with low-passage B. burgdorferi strain B31.
Both of these clones were derived from nonclonal P48.
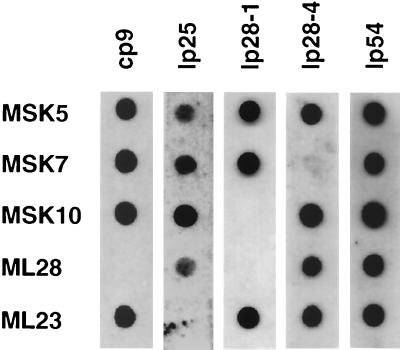
The similar infectivity deficits and tissue-tropic defects observed for MSK10 and ML28 prompted us to reevaluate the genotypic profiles of the mutant strains we had isolated. Because we had used PCR initially to assess the plasmid content of our clones, it is conceivable that we had identified false positives due to the inherent sensitivity of this method. To assess this, we used DNA purified from each strain and subjected these samples to either conventional Southern blot or dot blot analyses using PCR-amplified probes specific for cp9, lp25, lp28-1, lp28-4, and, as a positive control for all strains tested, lp54 (see Table 2 for plasmid-specific probes used). Using this approach, we determined that ML28, a clone missing cp9, was also lp28-1− (Fig. 3). All other clones contained the plasmid DNA determined previously (Fig. 1). Our results indicate that the threshold for detection in our dot blot analyses is between 104 and 105 B. burgdorferi cells (data not shown) and is independent of the probe used. In contrast, our PCR sensitivity data demonstrate that a positive PCR signal requires DNA derived from only 50 to 50,000 B. burgdorferi cells, depending on the primer set used (data not shown), and in some instances resulted in the erroneous characterization of the plasmid content of the clones tested (see, e.g., lp28-1 lane for ML28 in Fig. 1).
FIG. 3.
Dot blot analysis of B. burgdorferi strain B31 clones. Fifty nanograms of total plasmid DNA from each clone was blotted onto a nylon membrane and hybridized with probes specific to the plasmids indicated above the lanes. The clones tested are listed vertically on the left (see Table 1 for details). The plasmid tested and the probe from each plasmid are specified as follows (TIGR designations are used): cp9, region including bbc10 (rev paralogue); lp25, bbe16; lp28-1, a 1.7-kb region including bbf30, bbf31, and part of the bbf32 cassette region described previously (32); lp28-4, bbi16 (vraA); and lp54, bba16 (ospB).
ML28 and MSK10 have similar infectivity phenotypes, implying that the correlation between cp9 and any marked effect related to infectivity is not substantial. Instead, our results were only able to indicate that lp25 and lp28-1 are important for infectivity in the C3H/HeN mouse. The other plasmids that were missing from our clones, cp9 and lp28-4, have only subtle effects on the establishment and dissemination of B. burgdorferi strain B31. Our results do not exclude the possibility that other plasmids are important for pathogenesis, but, in the absence of such additional mutants in this study, it is impossible to evaluate these phenotypes (see Discussion). Taken together, our results indicate that lp25, either directly or indirectly, encodes determinants that drastically alter the infectivity phenotype of B. burgdorferi strain B31, presumably due to a colonization defect. Also, strains lacking lp28-1 show a dramatic infectivity deficit as well, being capable of effectively colonizing only the joint, although other tissues are also infected at low levels (see MSK10 data in Table 4). This phenotype is accentuated in ML28, which lacks both lp28-1 and cp9, suggesting that the tropism for additional tissue compared to what is observed in MSK10, albeit subtle, is due to cp9-derived sequences.
Characterization of protein and antigenic content.
We next wanted to determine whether differences at the genetic level would yield notable differences in the protein or antigen produced by these clones. The loss of a given plasmid could result in the loss of proteins encoded by that particular plasmid or could affect the global expression of genes at distal sites. To determine whether this occurred, we subjected each isolate to SDS-PAGE and ECL immunoblotting using serum from either infected mice, rabbits, or humans. Coomassie blue-stained gels showed no overt differences (data not shown), and no change was seen in the antigenic profiles, with the notable and expected exception of the antigenic variant VlsE (32), which was missing from the lp28-1− strains MSK10 and ML28 (data not shown). In addition, a single 24-kDa antigen was missing from MSK7 in blots probed with sera from infected mice, implying that the expression of this antigen is directly or indirectly related to the presence of lp28-4 (data not shown).
Genotype of B. burgdorferi strain B31 variant ML28.
Initially our PCR analysis defined ML28 as a cp9 mutant. However, after further characterization (Table 4 and Fig. 3), ML28 was missing additional plasmids. Furthermore, its infectivity pattern was similar to that observed for MSK10 (lp28-1−). If a given plasmid is missing from the majority of cells in a culture derived from a clonal isolate yet is present at levels sufficient to yield a positive PCR signal, then one would expect that upon infection and isolation from the infected host, the plasmid profile would most likely reflect the genotypic profile of the challenge inoculum. To address this possibility, we took joint tissue from mice infected with B. burgdorferi strain B31 isolate ML28, plated the sample to isolate individual colonies, and conducted PCR on 20 separate clones designated MJ1 through MJ20 (Table 1). Our results indicated that 12 of these clones were missing cp9, lp28-1, and lp28-4, while 5 lacked cp9 and lp28-1 (data not shown). The remaining three clones were either lp28-1− (two total) or a double mutant lacking both lp28-1 and lp28-4 (data not shown). Seven of the 20 ML28 clones from infected joints were subjected to PCR with all of the primer sets (Table 2), and, as before, no additional plasmids were missing from these isolates (data not shown); therefore, none of the remaining 13 clones were subjected to additional PCR with the remaining primer sets. These results confirmed the data obtained from our dot blot analyses (i.e., that ML28 did not have lp28-1 or cp9 [Fig. 3]) and are consistent with those reported by Purser and Norris (Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) for clones of B. burgdorferi strain B31 that do not have lp28-1 in terms of their infectivity deficit. In light of the minimal correlation observed for cp9 (compare MSK10 and ML28 [Table 4] and the modest effect seen for lp28-4 (MSK7 [Table 4]), the infectivity defect and retention of tropism for joint tissue observed for ML28 appears to be mediated either directly or indirectly by lp28-1.
In vivo selection of polyclonal B. burgdorferi strain B31 P48.
We established that our polyclonal P48 isolate was composed of numerous genotypic variants (Table 3) and determined the infectivity and tissue-tropic defect associated with some of these clones as indicated above. We next wanted to elucidate whether the mouse could be used to select specifically for variants that were of the high-infectivity phenotype. This type of positive selection would both validate our previous results and corroborate which of the plasmids were required for colonization in C3H/HeN mice. C3H/HeN mice were infected with serial dilutions of the polyclonal B. burgdorferi strain B31 P48 isolate, and an ID50 was determined. The results indicated that the P48 isolate had the same infectivity phenotype as the low-passage, wild-type MSK5 strain for all tissues tested except the skin, suggesting that a selection for the infectious, highly virulent B. burgdorferi cells had occurred within these mice (Table 5).
TABLE 5.
ID50 determination for nonclonal P48 B. burgdorferi strain B31
| Inoculum | No. of cultures positive/total no.
|
No. positive mice/total no. of mice | Calculated ID50
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Spleen | Joint | Heart | Bladder | Skin | All sites | Skin | Other sites | ||
| 105 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 14/15 | 3/3 | 6.8 × 102 | 1.6 × 102 |
| 104 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 14/15 | 3/3 | ||
| 103 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 3/3 | ||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0/3 | ||
| 101 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0/3 | ||
To prove that the infectious forms of strain B31 P48 were present within these tissue samples, we plated out tissue from skin, bladder, and joint and screened individual colonies for their plasmid content via PCR amplification. One would predict that if the mouse (i.e., in vivo) positively selects for infectious isolates of B. burgdorferi from a mixed polyclonal pool, then the clones obtained from infected tissues should contain plasmids that are required for maintenance of the spirochetes within these tissues. The results are summarized in Table 6 and indicate that all 139 clones contained lp25, even though only 10% of the polyclonal population of P48 contained this plasmid (Tables 3 and 6). The selection observed for lp25 in the mouse relative to in vitro cultivation was highly significant, with a probability (P) value of less than 0.001 (Table 6). In contrast to lp25, the data for cp9 and lp28-4 show that there was no dramatic selection for cells carrying these plasmids, particularly for cp9 in that only 1 of the 139 clones tested contained this plasmid (see Table 6 for statistical significance). Interestingly, lp28-1 appeared to be present in vivo overall at a frequency commensurate with the value calculated for in vitro-cultivated cells (80.6% in vivo relative to 69.4% in vitro [Table 6]). However, when the tissues are compared against one another, a statistically significant selection for lp28-1 is evident within the bladder tissue (P < 0.001) and to a lesser extent in the skin (P = 0.001). In contrast, there is no statistically significant difference when the joint tissue is compared to the in vitro-grown clones (75.7% lp28-1+ in the joint relative to 69.4% in vitro). The statistical significance of these data is shown in Table 6. The overall value calculated for the in vivo selection of lp28-1 is thus skewed due to the reduced selection within the joint, with an additional minor effect mediated by the skin samples. These results demonstrate that B. burgdorferi variants lacking lp28-1 are able to infect and disseminate to joint tissue and presumably other organs but are capable of persisting only within the joint.
TABLE 6.
Comparison of genotypic compositions of B. burgdorferi strain B31 P48 clones following either in vitro cultivation or in vivo selection in C3H/HeN mice
| Plasmidd | Presencea in P48
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| In vitrob (n = 49)c
|
In vivo
|
|||||||||
| + | − | Skin (OMS)e (n = 20)
|
Bladder (OMB)e(n = 20)
|
Joint (OMJ)e(n = 99)
|
Total (n = 139)
|
|||||
| + | − | + | − | + | − | + | − | |||
| cp9 | 4.1 (2) | 95.9 (47) | 0 (0)f | 100 (20) | 0 (0)f | 100 (20) | 1 (1)i | 99 (98) | 0.7 (1) | 99.3 (138) |
| lp25 | 10.2 (5) | 89.8 (44) | 100 (20)g | 0 (0) | 100 (20)g | 0 (0) | 100 (99)g | 0 (0) | 100 (139) | 0 (0) |
| lp28-1 | 69.4 (34) | 30.6 (15) | 85 (17)g | 15 (3) | 100 (20)g | 0 (0) | 75.7 (75)i | 24.3 (24) | 80.6 (112) | 19.4 (27) |
| lp28-4 | 10.2 (5) | 89.8 (44) | 0 (0)h | 100 (20) | 10 (2)h | 90 (18) | 4 (3)f | 96 (96) | 3.6 (5) | 96.4 (134) |
The values indicate the percentage of samples that contained the given plasmid (+) versus the percentage that lacked the given plasmid (−). Actual numbers of samples are indicated parenthetically.
Reflects the eight different genotypic variants shown in Table 3.
Number of individual colonies (clones) tested.
Plasmids lost either individually or as a group in the genotypic variants shown in Table 3.
See Table 1.
χ2 analysis indicates a selection (P < 0.05) against this plasmid when this tissue is compared to the in vitro sample.
χ2 analysis indicates a significant selection (P < 0.001) for this plasmid when this tissue is compared to the in vitro sample.
χ2 analysis indicates a significant selection (P < 0.001) against this plasmid when this tissue is compared to the in vitro sample.
χ2 analysis indicates no significant selection for or against this plasmid when this tissue is compared to the in vitro sample.
DISCUSSION
The hypothesis that B. burgdorferi plasmids carry infection-associated genes is supported by the fact that prolonged serial in vitro cultivation of infectious uncloned isolates changes the plasmid profiles of this organism and results in the loss of infectivity (21, 22, 30). Our observation that various forms of B. burgdorferi can be isolated from infected mouse tissues indicates that such heterogeneity is also permissive in vivo (Table 6). These results raise the question as to how B. burgdorferi become genetically variable at the plasmid level. One possible explanation involves the asymmetric distribution of plasmid DNA following replication and partitioning to daughter cells or, alternatively, it may reflect an in vitro adaptation resulting in the selection against cells carrying all or most of the plasmids. One might predict that partitioning of so many plasmid DNAs could be a stochastic process; however, we see a distinct bias toward the loss of the cp9, lp25, lp28-1, or lp28-4 plasmid. The observation that B. burgdorferi strains show variability in their plasmid contents in vitro and in vivo indicates that a significant degree of genomic plasticity for plasmid DNA is tolerated in this bacterium. However, despite the differences observed at the DNA level, very few changes in the total protein profile have been observed previously (21, 22, 30), and such changes were not observed in our analyses here (data not shown).
There is a possibility that the phenotype we observe for our clonal mutants is due to subtle changes within the genomic DNA, i.e., point mutations or rearrangements. One argument against this comes from concurrent studies of Purser and Norris (Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) that indicate a similar phenotype for the mutants we have reported in this study. A recent report by Anguita et al. (1) demonstrated that B. burgdorferi strain N40 passaged 75 times in vitro was incapable of causing carditis and arthritis and gave rise to altered gene expression but was still infectious in all tissues tested. None of the plasmids tested were missing from the passage 75 isolate, suggesting that other subtle genomic changes were responsible for the altered gene expression and the resulting phenotypes observed; however, the authors evaluated only 6 of the 21 plasmid species (1). Therefore, it is possible that the decrease in pathology and gene expression observed was linked to the absence of one of the 15 plasmids not accounted for in their analysis. The different phenotypes observed with our mutant clones may be explained by differential gene expression due to the loss of a regulatory locus that either directly or indirectly affects an operon or regulon. Studies to address this hypothesis are in progress.
In contrast to earlier studies with other animal models (30), our study also evaluated joint tissue from infected animals. This is particularly important inasmuch as B. burgdorferi infection can result in arthritic manifestations, particularly late in infection (25–27). Norris et al. reported that a low-infectivity clonal isolate of strain Sh-2-82 was capable of colonizing only the joints and hearts of infected C3H/HeN mice (14); however, the molecular basis of this defect was not reported at that time. In light of our results and those of Purser and Norris (Abstr. 100th Gen. Meet. Am. Soc. Microbiol.), this low infectivity clone was almost certainly missing the lp28-1 linear plasmid, as this phenotype is evident for both variants MSK10 and ML28 (Table 4). We have also followed up on the ID50 analyses by determining the genotypic contents of clones obtained from the infected tissues (Table 6). This assessment was crucial in order to exclude or confirm the genotypes of the clones we had identified from our polyclonal P48 isolate. Our results indicated that in instances when a specific plasmid yielded a weakly reactive product, that plasmid was most likely missing from clones obtained from the infected tissue(s). This was due to the inherent sensitivity of PCR (data not shown), which was capable of detecting plasmid species even if they were present in only a minority of the cells. For example, the B. burgdorferi strain B31 variant ML28 was purported to lack only cp9 following PCR, with a weakly reactive lp28-1 signal, implying that some of the cells were missing lp28-1 even though this isolate was obtained from an individual colony. Upon infection of this isolate and subsequent plating of the joint tissue, the genotypic analysis of the clones obtained indicated that all of the samples tested (20 clones total) did not contain lp28-1, suggesting either that the great majority of the cells used in the infection (at least greater than 1 in 20) were lp28-1− or that, due to the lack of selective pressure within the joint, lp28-1 was lost from these B. burgdorferi cells. Surprisingly, lp28-4 was also missing from the bulk (65%) of the isolates, implying a similar type of selection against cells carrying this plasmid. These results indicate that a clonal isolate of B. burgdorferi may be genetically diverse, and therefore, one must be cautious when attempting to link the phenotype observed to the genotype of the sample obtained.
Clearly the lack of selection observed in vitro does not mimic what is seen in vivo, especially for lp25 and lp28-1. For these plasmids, there is a selection within the mouse relative to cells grown in vitro in BSK-II medium. This was particularly evident in the mouse positive-selection screen, which indicated that lp25 was present in all 139 clones tested, including samples taken from skin, bladder, and joint tissues (Table 6). In contrast, there was only partial selection evident for either cp9 or lp28-4, depending on the tissue tested (Table 6). This is in agreement with a recent study showing that the cp9 plasmid is not selected for in the Peromyscus mouse yet is in the Ixodes arthropod vector (19), suggesting that genes on cp9 are more important for survival and/or transmission in the tick vector than for colonization and maintenance in the reservoir host. The data on lp28-1 were somewhat more complicated due to a reduced selection in the joint (∼76% lp28-1 positive) relative to the skin (85% lp28-1 positive) and bladder (100% lp28-1 positive). The frequency of B. burgdorferi containing lp28-1 in joint tissue was approximately the same as that observed in vitro (Table 6). Statistical analysis indicated that no significant difference occurred in vitro versus within the joint, consistent with the phenotype observed for MSK10 and ML28. That is, since lp28-1− spirochetes were able to colonize the joint more efficiently than other tissues, it is not surprising that there is an apparent lack of selection for B. burgdorferi containing this plasmid. Taken together, one possible interpretation for these results would be that all of the different plasmid combinations for the P48 sample (as shown in Table 3) infect and disseminate throughout the mouse but only those that contain lp25, and to a lesser extent lp28-1, are able to colonize and persist for the period tested. The noted exception is in the joint, where B. burgdorferi, whether it contains lp28-1 or not, is able to colonize this tissue.
The fact that lp28-1− cells are capable of persisting in joint tissue suggests that some protein or proteins on lp28-1 are required for maximal survival at other sites within the host, e.g., skin, bladder, or lymph nodes. Presumably the antigenic variant VlsE (32), encoded by lp28-1, is required for maintenance within tissues other than the joint (except when lp25 is also missing [Table 4]). This result implies that the ability to evade the host immune response via VlsE-specific antigenic variation is apparently not required within the joint. One possibility is that lp28-1 encodes an adhesin essential for colonization in all tissues excluding the joint. B. burgdorferi is believed to be an extracellular pathogen, and since only a small number of spirochetes are presumed to be present within most infected tissues, their ability to evade host immunity requires that they migrate to immunoprotected niches. This is particularly important since the infected host mounts a potent humoral response that does not clear B. burgdorferi in vivo yet elicits killing of B. burgdorferi in vitro. Some protein or proteins encoded on lp28-1, other than VlsE, may provide B. burgdorferi with the proper molecular components to survive in all of the microenvironments in which the spirochete resides except the joint. Another possibility could be the access or exposure to interstitial fluid, not found in the joint, which contains complement proteins. It is possible that lp28-1 encodes some protein or proteins that are required for serum resistance in B. burgdorferi and that survival within the joint could be tolerated without lp28-1 in the absence of complement proteins. We are currently in the process of testing whether the lp28-1− strains MSK10 and ML28 are serum sensitive relative to wild-type B. burgdorferi. Alternatively, it may be that survival of lp28-1− cells in the joint is related to the time frame (2 weeks) of our study. Studies of later time points may indicate that lp28-1− B. burgdorferi cells are cleared from joint tissue, indicating that cells lacking lp28-1 are protected from the host immune response in joints for longer times than in other tissues.
The positive selection observed for lp25 and lp28-1 implies that genes present on these plasmids and their requisite protein products are required for the establishment of infectious foci in different tissues throughout the mouse. That lp25 and lp28-1 are required for maximal infectivity is somewhat surprising, as these two plasmids have been categorized as “poorly behaved,” along with eight other B. burgdorferi plasmids (5). This is because lp25 and lp28-1 each have only 10 open reading frames greater than 300 bp in length and have 28 or 39 genes that are either smaller than 300 bp or appear to be pseudogenes, respectively. This lack of genetic information was interpreted to imply that these plasmids might be undergoing an evolutionary process that could ultimately result in their elimination (5). Our results, however, argue that lp25 and lp28-1 are important for B. burgdorferi survival within the mouse. As such, some of these genes, whether large or small, appear to be important for wild-type levels of infectivity.
Although lp25 and lp28-1 are the only plasmids missing that result in a reduced-infectivity phenotype in this study, McDowell et al. (unpublished) demonstrated that other clonal mutants result in noninfectious variants of B. burgdorferi strain B31. The differences in these studies are perhaps related to the time frame of the infection in C3H/HeJ mice, i.e., only 2 weeks for the study reported here and greater than 3 months in the related analysis. The results of the long-term study indicate that some clones obtained from infected mouse skin lacked lp28-3, along with other plasmids (lp5 and cp9 in most samples tested), but contained lp25 and lp28-1, yet were noninfectious upon subsequent intradermal challenge in C3H/HeJ mice (McDowell et al., unpublished). These results imply that several plasmids in the correct combinations may be required to mediate mammalian infection. For example, lp25 and some other plasmid, perhaps lp28-3, may be required together for wild-type infectivity in B. burgdorferi strain B31. If either lp25 or lp28-3 is missing, the B. burgdorferi cells are no longer infectious. It is important to note that clones obtained at 3 months postinfection were capable of persisting within the mouse but are unable to recolonize and/or disseminate upon reinfection. It seems unlikely that a selection against plasmids would occur, inasmuch as the clonal mutants lacking lp28-3 are noninfectious and would have a selective disadvantage for downstream propagation. Such a negative selection in vivo would ultimately result in a population of B. burgdorferi that would be dead-end pathogens, that is, unable to spread to new hosts, and would thereby be eliminated from the infectious cycle that moves between arthropods and small rodent species. Therefore, it seems more likely that the loss of plasmids is a stochastic event that permits persistent infection and results in a significant degree of genomic plasticity in vivo over time. One might envision that some mutant clones might have a short-term selective advantage in vivo if the plasmid they are missing encodes a target for humoral immune clearance yet might be attenuated for downstream infectivity due to the absence of plasmids required for virulence. Evidence provided by several investigators indicates that B. burgdorferi has laterally transferred plasmids, specifically cp26 and lp38, which encode OspC and OspD, respectively (11, 12). These results suggest that B. burgdorferi can transfer plasmids; however, the mechanism that B. burgdorferi uses to exchange DNA is not known at this time. It is thus tempting to speculate that B. burgdorferi may be capable of reacquiring previously lost plasmids required for maximal infectivity. This mechanism, if operative in vivo, would represent a novel modality for B. burgdorferi to modulate its genome and regain virulence.
ACKNOWLEDGMENTS
We thank Vernon Tesh, James Samuel, J. Seshu, and Kristen L. Swingle for critical evaluation of the manuscript; Andreas Bäumler and Renée Tsolis for helpful comments and suggestions; Tom Champney for help with the statistical analysis; and Deanna Moore for technical assistance. We also thank Michael Kaiser for assistance with some of the PCR analyses reported in this paper.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01-AI42345) and the American Heart Association (both to J.T.S.).
ADDENDUM IN PROOF
As a follow-up to the abstract cited in this study, Purser and Norris have demonstrated a correlation between reduction in infectivity in C3H/HeN mice and loss of either lp25 or lp28-1 from Borrelia burgdorferi clonal isolates (J. E. Purser and S. J. Norris, Proc. Natl. Acad. Sci. USA, in press).
REFERENCES
- 1.Anguita J, Samanta S, Revilla B, Suk K, Das S, Barthold S W, Fikrig E. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect Immun. 2000;68:1222–1230. doi: 10.1128/iai.68.3.1222-1230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 3.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens S, Delange M, Ley III H L, Rosa P, Huang W M. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J Bacteriol. 1995;177:2769–2780. doi: 10.1128/jb.177.10.2769-2780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C M. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 6.Foley D M, Gayek R J, Skare J T, Wagar E A, Champion C I, Blanco D R, Lovett M A, Miller J N. Rabbit model of Lyme borreliosis: erythema migrans, infection-derived immunity, and identification of Borrelia burgdorferi proteins associated with virulence and protective immunity. J Clin Invest. 1995;96:965–975. doi: 10.1172/JCI118144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formal S B, Hale T L, Sansonetti P J. Invasive enteric pathogens. Rev Infect Dis. 1983;5(Suppl. 4):S702–S707. doi: 10.1093/clinids/5.supplement_4.s702. [DOI] [PubMed] [Google Scholar]
- 8.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 9.Golde W T, Dolan M C. Variation in antigenicity and infectivity of derivatives of Borrelia burgdorferi, strain B31, maintained in the natural, zoonotic cycle compared with maintenance in culture. Infect Immun. 1995;63:4795–4801. doi: 10.1128/iai.63.12.4795-4801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch J, Barbour A G. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J Bacteriol. 1992;174:5251–5257. doi: 10.1128/jb.174.16.5251-5257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jauris-Heipke S, Liegl G, Preac-Mursic V, Rossler D, Schwab E, Soutschek E, Will G, Wilske B. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J Clin Microbiol. 1995;33:1860–1866. doi: 10.1128/jcm.33.7.1860-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176(15):4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadelman R B, Wormser G P. Lyme borreliosis. Lancet. 1998;352:557–565. doi: 10.1016/S0140-6736(98)01146-5. [DOI] [PubMed] [Google Scholar]
- 14.Norris S J, Howell J K, Garza S A, Ferdows M S, Barbour A G. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun. 1995;63:2206–2212. doi: 10.1128/iai.63.6.2206-2212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Portnoy D A, Martinez R J. Role of a plasmid in the pathogenicity of Yersinia species. Curr Top Microbiol Immunol. 1985;118:29–51. doi: 10.1007/978-3-642-70586-1_3. [DOI] [PubMed] [Google Scholar]
- 16.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 17.Roberts D M, Carlyon J A, Theisen M, Marconi R T. The bdr gene families of the Lyme disease and relapsing fever spirochetes: potential influence on biology, pathogenesis, and evolution. Emerg Infect Dis. 2000;6:110–122. doi: 10.3201/eid0602.000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts D M, Theisen M, Marconi R T. Analysis of the cellular localization of Bdr paralogs in Borrelia burgdorferi, a causative agent of Lyme disease: evidence for functional diversity. J Bacteriol. 2000;182:4222–4226. doi: 10.1128/jb.182.15.4222-4226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan J R, Levine J F, Apperson C S, Lubke L, Wirtz R A, Spears P A, Orndorff P E. An experimental chain of infection reveals that distinct Borrelia burgdorferi populations are selected in arthropod and mammalian hosts. Mol Microbiol. 1998;30:365–379. doi: 10.1046/j.1365-2958.1998.01071.x. [DOI] [PubMed] [Google Scholar]
- 20.Samuels D S, Marconi R T, Huang W M, Garon C F. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J Bacteriol. 1994;176:3072–3075. doi: 10.1128/jb.176.10.3072-3075.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson W J, Garon C F, Schwan T G. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 23.Skare J T, Foley D M, Hernandez S R, Moore D C, Blanco D R, Miller J N, Lovett M A. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect Immun. 1999;67:4407–4417. doi: 10.1128/iai.67.9.4407-4417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skare J T, Mirzabekov T A, Shang E S, Blanco D R, Erdjument-Bromage H, Bunikis J, Bergstrom S, Tempst P, Kagan B L, Miller J N, Lovett M A. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect Immun. 1997;65:3654–3661. doi: 10.1128/iai.65.9.3654-3661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 26.Steere A C, Gibofsky A, Patarroyo M E, Winchester R J, Hardin J A, Malawista S E. Chronic Lyme arthritis. Clinical and immunogenetic differentiation from rheumatoid arthritis. Ann Intern Med. 1979;90:896–901. doi: 10.7326/0003-4819-90-6-896. [DOI] [PubMed] [Google Scholar]
- 27.Steere A C, Schoen R T, Taylor E. The clinical evolution of Lyme arthritis. Ann Intern Med. 1987;107:725–731. doi: 10.7326/0003-4819-107-5-725. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, van Dam A P, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12:633–653. doi: 10.1128/cmr.12.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Kodner C, Coleman L, Johnson R C. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 33.Zückert W R, Meyer J, Barbour A G. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect Immun. 1999;67:3257–3266. doi: 10.1128/iai.67.7.3257-3266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]