Abstract
Regeneration, the ability to replace lost body parts, is a widespread phenomenon in the animal kingdom often connected to asexual reproduction or fission, since the only difference between the two appears to be the stimulus that triggers them. Both developmental processes have largely been characterized; however, the molecular toolkit and genetic mechanisms underlying these events remain poorly unexplored. Annelids, in particular the oligochaete Pristina leidyi, provide a good model system to investigate these processes as they show diverse ways to regenerate, and can reproduce asexually through fission under laboratory conditions. Here, we used a comparative transcriptomics approach based on RNA‐sequencing and differential gene expression analyses to understand the molecular mechanisms involved in anterior regeneration and asexual reproduction. We found 291 genes upregulated during anterior regeneration, including several regeneration‐related genes previously reported in other annelids such as frizzled, paics, and vdra. On the other hand, during asexual reproduction, 130 genes were found upregulated, and unexpectedly, many of them were related to germline development during sexual reproduction. We also found important differences between anterior regeneration and asexual reproduction, with the latter showing a gene expression profile more similar to that of control individuals. Nevertheless, we identified 35 genes that were upregulated in both conditions, many of them related to cell pluripotency, stem cells, and cell proliferation. Overall, our results shed light on the molecular mechanisms that control anterior regeneration and asexual reproduction in annelids and reveal similarities with other animals, suggesting that the genetic machinery controlling these processes is conserved across metazoans.
Keywords: Annelida, differential gene expression, paratomic fission, Pristina leidyi, regeneration, reproduction
Summary of the workflow developed in this study.

Research Highlights
Regeneration and asexual reproduction are molecularly distinct events in the annelid Pristina leidyi, even though they share a limited number of genes, some of them pluripotency‐related genes that play a relevant role during both processes.
1. INTRODUCTION
Regeneration and asexual reproduction are two postembryonic developmental processes that have attracted the interest of biologists for centuries (Bely & Wray, 2001; Sanchez Alvarado, 2000). Regeneration, the ability to replace lost body parts, is a widespread phenomenon in the animal kingdom, present in several metazoan lineages (Bely & Nyberg, 2010) including annelids or segmented worms (Annelida). The ability to regenerate lost body parts is widely distributed across annelids and typically occurs through epimorphosis, which involves cell proliferation in a newly formed specialized structure called blastema that differentiates to restore the missing parts (e.g., Bely & Nyberg, 2010; de Jong & Seaver, 2018; Kostyuchenko & Kozin, 2021; Sanchez Alvarado, 2000). On the other hand, asexual reproduction or fission implies offspring production without the involvement of germ cells or gametes, and it is also a process that has evolved independently in numerous lineages throughout the tree of life (Campagna et al., 2016; Dolmatov et al., 2018; Zattara & Bely, 2016). Within the phylum Annelida, asexual reproduction can be achieved through two main ways: (1) architomy or fragmentation, when the worm first splits apart and then each fragment regenerates the missing structures; or (2) paratomy or paratomic fission, when a new head and tail is developed in a specific segment with high proliferative activity (i.e., fission zone) and subsequently the worm splits apart (Bely & Wray, 2001).
Both abilities (regeneration and asexual reproduction) have often been connected in many organisms, since under a morphological perspective, both processes show great similarities, and thus the main difference between the two events appears to be the stimulus triggering them. While regeneration occurs following a deleterious incident such as an injury, an internal stimulus triggered by environmental factors such as food availability or temperature is necessary for asexual reproduction to happen (Sanchez Alvarado, 2000). In fact, it has been proposed that fission derives from regeneration, since agametic reproduction has evolved mainly in groups with extensive regeneration abilities, and all animals capable to reproduce asexually are also able to regenerate (Bely & Wray, 2001). Specifically in annelids, it has been suggested that asexual reproduction is a novel developmental process evolved through the cooption of the anterior regeneration process (Bely & Wray, 2001; Zattara & Bely, 2011, 2016), and several studies have tried to shed light into the molecular framework underlying these two developmental processes. However, most previous studies focused on the characterization of a few candidate genes (Kostyuchenko & Kozin, 2021 and the references herein), or samples from different stages of regeneration and fission pooled together (Nyberg et al., 2012), hindering the possibility of differentiating the specific mechanisms that regulate each event. In our study, we have analyzed the two processes independently to be able to describe the genetic toolkit underlying each of these developmental mechanisms. However, due to limited resources, we have only been able to include one stage of differentiation, which might offer limited but highly valuable information since this is the first study of its kind. In future studies, we will describe more stages to understand the changes in gene expression throughout the whole duration of these developmental processes.
Additionally, the source of cell production for the proliferative activity entailing the generation of new tissue in annelids is still unclear. It has been suggested that, similarly to other invertebrates with huge regenerative abilities such as planarian worms (e.g., Brockes & Kumar, 2008), pluripotent cells replace the missing structures during annelid regeneration. However, in contrast to the typical self‐renewal capacities of planarian stem cells (i.e., neoblasts), it seems that in annelids the pluripotent cells migrate from pre‐existing segments to the wound site, proliferate in the blastema, and redifferentiate in the missing tissues (Bilello & Potswald, 1974; de Jong & Seaver, 2018; Sugio et al., 2012; Zattara & Turlington, & Bely, 2016). In the case of asexual reproduction, it has been suggested that cells of already differentiated tissues dedifferentiate and migrate from neighboring areas to the fission zone where active cell proliferation occurs (Kostyuchenko et al., 2016; Özpolat & Bely, 2015).
The freshwater oligochaete Pristina leidyi (Clitellata, Naididae) represents an outstanding annelid model to study regeneration and asexual reproduction, due to its remarkable anterior and posterior regenerating abilities, asexual reproduction by paratomic fission, and continuous growth under favorable laboratory conditions (Bely & Wray, 2001; Özpolat & Bely, 2015; Smith, 1896; Zattara & Bely, 2011). Previous studies performed by Zattara and Bely (2011) have characterized morphologically the regeneration (Figure 1a) and fission (Figure 1b) processes of P. leidyi recognizing different stages easily identified using a stereomicroscope. Therefore, the phenotypic features and morphological changes that take place during regeneration and asexual reproduction in P. leidyi are well characterized. However, our knowledge about the underlying molecular mechanisms in this species in particular, and in annelids in general, is very limited.
Figure 1.
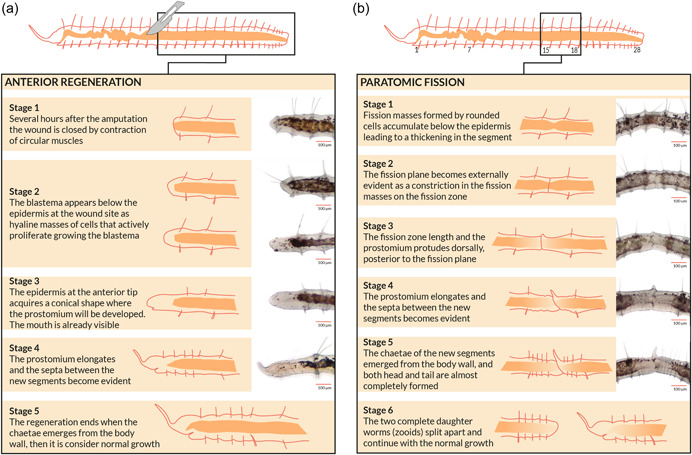
Anterior regeneration and asexual reproduction in Pristina leidyi. Brief description of major events, schematic representation and bright‐field microscopy images of the different stages defined for (a) anterior regeneration and (b) asexual reproduction through paratomic fission in the annelid P. leidyi. Scale bars = 100 μm (based on Zattara & Bely, 2011).
Thus, the present study aims to further investigate the molecular machinery involved in regeneration and asexual reproduction in the species P. leidyi, using a transcriptomics approach to study the genes potentially involved in each process, with special attention to pluripotency and stem cells gene markers. To unravel the mechanisms underlying these developmental processes and to evaluate the hypothesized link between the two, we have characterized gene expression patterns during anterior regeneration and paratomic fission, identifying candidate genes that may regulate these processes. Moreover, we have compared both events and identified genes involved in both anterior regeneration and asexual reproduction, to assess whether these two processes are evolutionarily linked. Finally, we have identified candidate genes related to cell pluripotency and/or multipotency that might be involved in these two developmental processes. Moreover, the abundant transcriptome data generated here can provide new insights into annelid development, setting the base for future comparative studies that will enable us to understand how these events, and the cellular types involved in them, have evolved within Annelida and across metazoans.
2. RESULTS AND DISCUSSION
2.1. General characterization of de novo assembled transcriptome of P. leidyi
Three libraries were prepared for each of the three experimental conditions (i.e., anteriorly regenerating, asexually reproducing, and control individuals). Nonetheless, one anterior regeneration library failed the quality controls, and therefore only eight of the nine libraries were sequenced (see Materials and Methods section). Therefore, sequencing of eight complementary DNA (cDNA) libraries (anterior regeneration, n = 2; asexual reproduction, n = 3; control, n = 3) resulted in approximately 223.6 million raw reads (31.6 GB), of which a total of 218,666,879 (~98%) were retained after trimming low‐quality portions and reads shorter than 30 bp (Supporting Information: Table S1). The filtered reads were assembled de novo using Trinity software generating a reference transcriptome for P. leidyi with 360,928 transcripts, an average GC content of 44.45% and N50 = 3545 bp (Supporting Information: Table S2). Of the total assembled transcripts, 73,959 (20.49%) had a BLAST hit in the UniProt Knowledgebase (UniProtKB) database and were functionally annotated. More detailed information on raw sequencing data, assembly statistics, and functional annotation of the reference transcriptome can be found in Supporting Information: Tables S1 and S2.
Our assembled transcriptome was highly complete, with a BUSCO score of 97.8%. The analysis recovered 943 of the total 954 metazoan single‐copy orthologs. These corresponded to 933 complete BUSCOs (160 single‐copy BUSCOs, and 773 duplicated BUSCOs) and 10 fragmented BUSCOs (1% of the recovered BUSCOs). Only 1.2% (11 BUSCOs) of the total metazoan BUSCOs were missing in our transcriptome. These high levels of completeness are similar to those found in other annelid species (Álvarez‐Campos et al., 2019). A summary of the transcriptome completeness statistics can be found in Supporting Information: Table S2.
2.2. Gene expression patterns during anterior regeneration and asexual reproduction are largely different
Over the last few decades, several authors have pointed out developmental similarities between regeneration and asexual reproduction (e.g., Bely & Wray, 2001; Zattara & Bely, 2011). In fact, a recent study that examined the presence of regeneration and fission capabilities across Annelida proposed that fission is a process derived from regeneration and that anterior regeneration specifically must be present for fission to evolve (Zattara & Bely, 2016). This would suggest that gene expression patterns of the two processes at a comparable time point should be somewhat similar. Indeed, expression of some genes such as orthodenticle (otx) and engrailed (en), as well as a lumbriculid Lan 3‐2 glycoepitope, has been reported in both regeneration and asexual reproduction in several annelid species (Bely & Wray, 2001; Martinez et al., 2005).
We detected a total of 1496 differentially expressed genes (up‐ and downregulated) in the pairwise comparisons performed (Supporting Information: Figure S1). Of those, 1285 genes were differentially expressed in the comparison of anteriorly regenerating and control individuals (291 upregulated and 994 downregulated in anterior regeneration), while 211 genes were found differentially expressed in the comparison of asexually reproducing and control individuals (130 upregulated and 81 downregulated in asexual reproduction) (Supporting Information: Table S3). Hierarchical clustering of samples, according to gene expression patterns, showed that asexually reproducing individuals have a genetic profile more similar to control samples (nonregenerating, nonreproducing individuals), compared to anteriorly regenerating individuals (Figure 2a). This better correlation between asexual reproducing and control worms is to be expected since both conditions show complete and entire worms. Conversely, anteriorly regenerating worms lack most of the anterior structures and therefore the genes herein expressed may explain the high number of downregulated genes in this regeneration condition, and thus it would be reasonable to find several anterior identities and head markers downregulated in this condition. However, we have found no differential expression of several anterior neural markers previously reported in adult and larval annelids and other invertebrates as aristaless‐related homeobox, dachshund, empty spiracles homeobox, forebrain zinc‐finger, orthopedia, paired box 3/6, retinal homeobox, six 3/6 homeobox, or synaptotagmin 1 (Gąsiorowski et al., 2021; Kerbl et al., 2016; Santagata et al., 2012). Even though these genes have been reported in our transcriptome, none of them shows a significatively distinctive expression with the defined thresholds, which could suggest that brain structures are developing in the 72 hpa anterior blastema. In any case, the anterior head gene expression within the downregulated genes in the anterior regenerating comparison needs further research. Moreover, it is worth noting that individuals of P. leidyi maintained under optimal conditions are continuously undergoing fission, and therefore it is possible that the molecular toolkit necessary for asexual reproduction is triggered even before the associated morphological changes can be observed. This could contribute to even larger similarities between the two conditions. In fact, recruitment of the developmental programs involved in asexual reproduction has already been observed in the earthworm Lumbriculus sp. 1 week before the process starts (Martinez et al., 2005).
Figure 2.
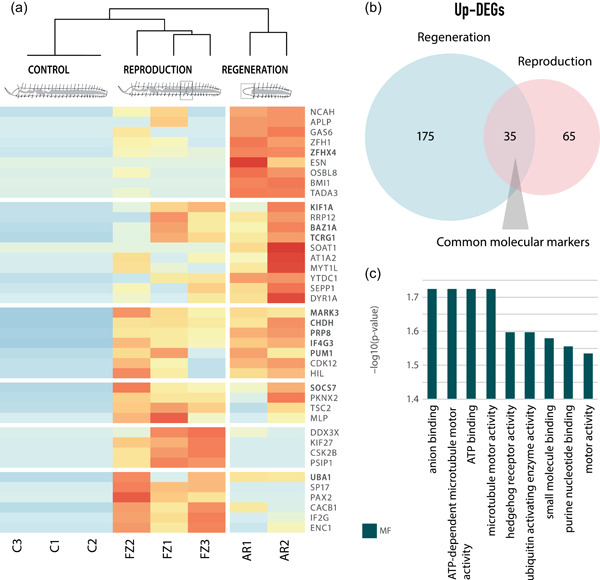
Gene expression patterns during anterior regeneration and asexual reproduction. (a) Hierarchically clustered heatmap of the 40 most upregulated, annotated, and nonredundant genes from pairwise comparisons of anteriorly regenerating, asexually reproducing and control individuals. Upregulated genes in both anteriorly regenerating and fissioning comparisons are shown in bold. (b) Scaled Venn diagram showing the overlapped annotated genes upregulated in both regeneration and asexual reproduction conditions. (c) Enriched Gene Ontology terms are associated with the 35 upregulated genes shared between anterior regeneration and asexual reproduction (molecular function category).
It should be noted that this great difference in the number of differentially expressed genes during regeneration and asexual reproduction when compared to control individuals (1285 genes were differentially expressed during anterior regeneration, whereas only 211 genes were differentially expressed during asexual reproduction; Supporting Information: Table S3) could be suggesting blastema formation during anterior regeneration, which implies the reestablishment of anterior identity and de novo renewal of anterior‐specific structures, and may demand a more complex molecular toolkit than the development of a new individual from preexisting tissues during asexual reproduction. Nonetheless, further research of the downregulated genes in this anteriorly regenerating comparison, and especially those anterior identity markers, would be needed to explain the great differences found here. Overall, these results indicate that although regeneration and asexual reproduction are substantially similar morphologically, the molecular toolkits controlling them seem to be rather different. These results agree with recent studies suggesting that even though these two processes might share a common origin, there are marked differences that might be due to their divergent evolution (Zattara & Bely, 2011).
On the other hand, several common features between the two processes have been noted. By comparing both upregulated subsets, that is, anterior regeneration upregulated genes and asexual reproduction upregulated genes, we have found a statistically significant overlap of 35 genes that are upregulated during anterior regeneration and asexual reproduction processes when compared to control individuals (Figure 2a,b and Supporting Information: Tables S4, S5, and S6A). Among these 35 genes, some of them have been reported in other invertebrates and mammals to show a relevant role during regeneration, such as vitamin D3 receptor A (vdra; Chen et al., 2021), and germline development, including ubiquitin‐like modifier‐activating enzyme 1 (uba1; Yi et al., 2010). Nevertheless, the importance of these overlapped genes relies on those stem‐cell‐related markers that seem to play a role during anterior regeneration and asexual reproduction, including connector enhancer of kinase suppressor of Ras 2 (cnkr2), cyclin‐Y‐like (ccnyl1), HECT, UBA, and WWE domain‐containing E3 ubiquitin protein ligase 1 (huwe1), and pumilio homolog 1 (pum1) orthologs (Bose et al., 2017; Henderson et al., 2015; Juliano et al., 2010; Vizziano‐Cantonnet et al., 2018; Zeng et al., 2016). The upregulation of these stem cell genes may suggest the presence of pluripotent‐like cell populations mediating both processes.
Enrichment analyses showed that all shared upregulated genes were associated with Gene Ontology (GO) terms in the Molecular Function (MF) category, including nucleoside phosphate binding, nucleotide binding, adenyl nucleotide binding, or purine ribonucleotide binding (Figure 2c and Supporting Information: Table S6B). Interestingly, these metabolic pathways related to the biosynthesis of purine nucleotides and DNA/RNA are also enriched in rapidly proliferating cancer cells (Ngoka, 2008; Weber, 1983). Thus, the high demand for nucleic acids during regeneration and asexual reproduction in P. leidyi might indicate an elevated proliferative activity during the development of the blastema and the fission zone, respectively. This is consistent with previous findings showing epidermal cell proliferation and blastema/fission zone growth as the major shared events occurring during these stages (Zattara & Bely, 2011). The GO term motor activity was also found enriched in both processes, illustrating the importance of cellular reorganization. Other GO terms enriched during these stages of anterior regeneration and asexual reproduction included ubiquitin‐activating enzyme activity, due to the upregulation of uba1 ortholog, which is involved in maintaining genomic integrity during cell proliferation, as well as in different neuronal processes such as neuron differentiation, growth, and development (Lambert‐Smith et al., 2020). This suggests that nervous system development is already an important process during these stages of postembryonic development. Remarkably, we also found a B9 domain‐containing protein 1 (b9d1) ortholog upregulated during both regeneration and asexual reproduction. This gene is responsible for cilia biogenesis and is involved in the hedgehog signaling pathway (hedgehog receptor activity, MF category), showing an important role during regeneration, segmentation, and fission in several organisms (e.g., Dolmatov et al., 2018; Dray et al., 2010; Schnapp et al., 2005).
2.3. Gene expression patterns and functional enrichment during anterior regeneration
We detected a total of 1285 genes differentially expressed during the second stage of anterior regeneration, of which 291 (~23%) were upregulated and 994 (~77%) downregulated (Figure 3a,b and Supporting Information: Table S3). Of these, 210 (~72%) of the upregulated genes and 529 (~53%) of the downregulated genes had a blast hit in the UniProtKB database, and thus were functionally annotated (Supporting Information: Table S4). Among the 210 upregulated genes, 42 have been previously cited in different regeneration processes in other species (Supporting Information: Table S7) and 7 have been specifically reported during annelid regeneration. For instance, we found frizzled class receptor 1 (fzd1) upregulated, one of the major receptors in the wnt signaling pathway, which is involved in several developmental processes including regeneration. This gene has already been reported during fission and regeneration in P. leidyi and in several other organisms such as sea cucumbers and newts (e.g., Girich et al., 2017; Nyberg et al., 2012; Singh et al., 2018). Our results also showed upregulation of brain tumor (brat), glutamate dehydrogenase 1 (GLUD1), multifunctional protein ADE2 (PUR6, also known as paics), ABC transporter G family member 21 (AB21G), vdra, and some genes from the Sox family (i.e., Sox‐14 and Sox‐21), which have been previously found upregulated during regeneration in other metazoans, including several annelid species such as Enchytraeus japonensis, Eisenia fetida, Syllis gracilis, Sphaerosyllis hystrix, and Lumbriculus variegatus (Bhambri et al., 2018; Gómez et al., 2018; Myohara et al., 2006; Ribeiro et al., 2019; J. Tao et al., 2019; Tellez‐Garcia et al., 2021).
Figure 3.
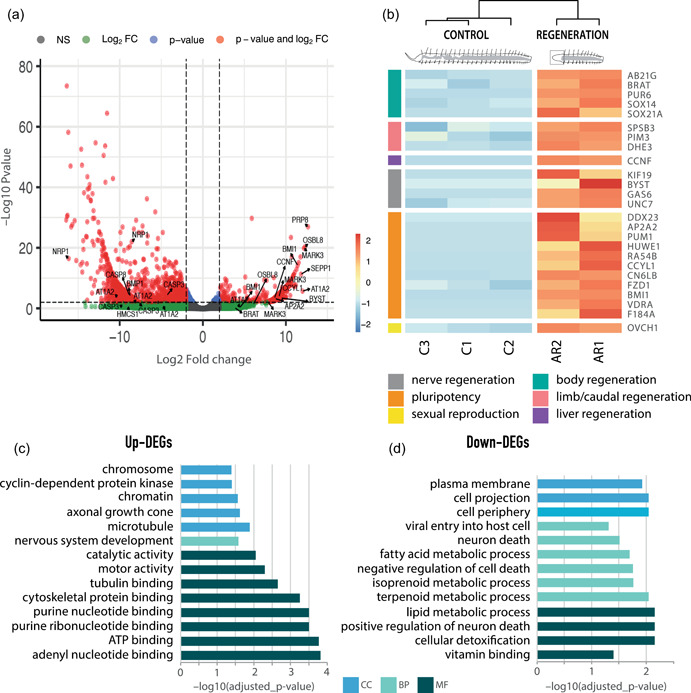
Gene expression patterns during anterior regeneration. (a) Volcano plot displaying the –log10 p value (false discovery rate [FDR]) as a function of fold change in the regenerating and control individuals. Labeled genes are discussed in the text. (b) Hierarchically clustered heatmap of the most important upregulated annotated and nonredundant genes in this comparison, and their categories according to the main function discussed. (c) Gene Ontology enrichment analysis of the upregulated annotated genes. (d) Gene ontology enrichment analysis of the downregulated annotated genes.
Several regeneration‐related genes previously reported in nonannelid species, including fish, amphibians, mammals, or planarians, were also found upregulated during anterior regeneration in P. leidyi. Some of these genes include pim3, splA/ryanodine receptor domain‐containing SOCS box 3 (spsb3), or vdra, which have been reported in limb, fin, or caudal regeneration of amphibians and zebrafish (Baddar et al., 2021; Chen et al., 2021; Ivanova et al., 2018). We also found upregulation of cyclin‐F (ccnf), ccnyl1, and growth arrest‐specific 6 (gas6), which have been also reported during mammals' liver regeneration (Couchie et al., 2005; L. Huang et al., 2015; Pibiri, 2018). In addition, we also found upregulated genes involved in different aspects of muscle and nerve development in planarians, zebrafish, amphibians, and mammals, including bystin, innexin Unc‐7 (unc‐7), kif19, GLUD1, and gas6 (Gibbs et al., 2011; Güiza et al., 2018; T. C. Huang et al., 2020; Sheng et al., 2004; Stratton et al., 2018). The upregulation of these genes highlights the importance of nervous system development during this stage of regeneration, as suggested in previous studies (Zattara & Bely, 2011). Furthermore, we also found upregulation of glutamine/glutamate metabolism genes (glutamine synthetase, GLUD, glutaminase), which recent transcriptomic studies in other annelid species have revealed to be important in the early stages of regeneration (e.g., Ribeiro et al., 2019; J. Tao et al., 2019; Tellez‐Garcia et al., 2021).
Interestingly, we have also found several upregulated genes that have been previously linked to neoblast and stem cell regulation and development. For instance, rad54B, huwe1, and DEAD‐box helicase 23 (ddx23) are neoblast‐specific genes required for different aspects of neoblast cell cycle progression and regulation in planarias (Galloni, 2012; Henderson et al., 2015; Lei et al., 2016). In addition, our results showed upregulation of bmi1, an epigenetic regulator of cell cycle and self‐renewal capacity of mammalian stem cells, required for neoblast function in planarians (Supporting Information: Table S7) (e.g., Önal et al., 2012; Robson et al., 2011), tissue regeneration in several mammals (e.g., Fukuda et al., 2012), and recently reported during early regeneration of the annelid L. variegatus (Tellez‐Garcia et al., 2021). In addition, we also found upregulation of the translational repressor pum1, whose expression in planarians seems to be restricted to neoblasts and is related to cell self‐renewal in mammals (Salvetti et al., 2005; Spassov & Jurecic, 2003). Additional markers of pluripotency from other cell types have also been found upregulated in regenerating P. leidyi individuals, including adaptor‐related protein complex 2 (ap2a2),alpha 2 subunit, CCR4‐NOT transcription complex subunit 6‐like‐B (CN6LB), and fam184A (Elmén et al., 2020; Kokkaliaris et al., 2012; Tauran et al., 2019). The presence of genes related to stem cell maintenance and pluripotency, and more specifically those markers exclusive to neoblasts, suggests the presence of stem cells and their relevance during P. leidyi regeneration. These results provide additional evidence supporting previous studies that indicate the presence of pluripotent cell populations able to migrate and participate in regeneration processes in annelids (Supporting Information: Table S7) (e.g., Bilello & Potswald, 1974; de Jong & Seaver, 2018; Sugio et al., 2012). However, the presence of these genes does not imply that stem cells are the only source of cellular production during tissue renewal, and it might also be possible that dedifferentiated tissues make a significant contribution during annelid regeneration as it has been previously suggested (Bely, 2014).
Surprisingly, we also found an ovochymase‐1 (ovch1) ortholog upregulated in anteriorly regenerating P. leidyi individuals, a gene typically involved in oogenesis in other invertebrates including the annelid Syllis magdalena, where it may assist maturation of oocytes and prevent self‐fertilization (Álvarez‐Campos et al., 2019). In addition, bmi1 and pum1, required for germ stem cell maintenance and germline development and maturation in several species (Juliano et al., 2010; Komai et al.,2014), were also found upregulated. Germ cell precursors have been identified during anterior regeneration in other annelids (Tadokoro et al., 2006), and thus the upregulation of ovch1, bmi1, and pum1 may indicate the restoration of lost gonadal tissue that takes place during anterior regeneration in P. leidyi.
GO analysis of the upregulated genes once again shows a significant enrichment of purine metabolism (Figure 3b and Supporting Information: Table S8), which is required for nucleotide biosynthesis (Ngoka, 2008; Weber, 1983) and is also necessary for the high proliferative activity that takes place in the blastema during annelid regeneration (Bely, 2014). In addition, other enriched GO terms in the Cellular Component (CC) category, such as cyclin/CDK‐positive transcription elongation factor complex, chromatin, and chromosome, confirm the importance of this proliferative activity through the regulation of the cell cycle (Malumbres & Barbacid, 2009). Other upregulated GO terms in the MF category, including catalytic activity and kinase activity and cell junction in the CC category, have also been previously reported during the regeneration of several annelid species (Paul et al., 2021; Ribeiro et al., 2019). The enrichment of GO terms such as nervous system development in the Biological Process (BP) category, as well as other related GO terms in the MF category (e.g., axonal growth cone) point out the importance of neural development during this stage of P. leidyi regeneration. In fact, 72 h after amputation, the horizontal nerves become evident and the central nervous system starts to develop in the new head (Zattara & Bely, 2011).
As anteriorly regenerating worms lack most of the anterior tissues and structures, as well as the anterior brain, the downregulation of different anterior identity markers is to be expected. However, none of the genes previously reported (Gąsiorowski et al., 2021; Kerbl et al., 2016; Santagata et al., 2012) has been found to be significantly differentially expressed in this condition. However, among the 529 genes downregulated during anterior regeneration (Supporting Information: Table S5), 14 of them were identified as regeneration‐related genes previously reported in other species (reviewed by Zhao et al., 2016). For instance, Willebrand factor D and EGF domains (vwde), important for blastema formation in some vertebrates (Leigh et al., 2020), was also found downregulated during early regeneration in the annelid L. variegatus (Tellez‐Garcia et al., 2021), although its role in this process is still unknown. The downregulation of other genes, such as casp3, casp8, or epidermal grow factor receptor (egfr), which modulate apoptosis in annelids and regulate blastema growth in planarias (Campagna et al., 2016; Fraguas et al., 2011; Shao et al., 2020), may indicate the important role of apoptotic processes in the control of cell proliferation (Kostyuchenko & Kozin, 2021) during P. leidyi regeneration. Actually, the downregulation of these pro‐ and antiapoptotic genes and other regeneration‐related genes, such as neuropilin and receptor‐interacting serine‐threonine kinase 3, led to the enrichment of several GO terms (BP category) related to the regulation of cell death, which have been shown to be important during anterior regeneration in cnidarians (Chera et al., 2011) (Figure 3c and Supporting Information: Table S8). In addition, we found downregulation of bone morphogenetic protein (bmp) and neuronal nicotinic acetylcholine receptor (chrna), two genes involved in signaling pathways controlling differentiation and development of the nervous system. Contrary to our results, both genes have been previously reported upregulated in other regenerating metazoan species, including annelids (Bhambri et al., 2018; Cho et al., 2009; Nyberg et al., 2012; Gómez et al., 2018), the former in late blastema developing stages during patterning (Bandyopadhyay et al., 2006), and the latter especially upregulated within the first 24 h after amputation (Cho et al., 2009). This suggests that these genes may show a dynamic expression that decreases during the second stage of P. leidyi regeneration, that is, when the blastema is completely formed and still elongating.
Other downregulated GO terms in the BP category such as cellular hormone metabolic processes were also found enriched. It is well known that the annelid brain secretes hormones that promote and regulate posterior regeneration (e.g., de Jong & Seaver, 2018). However, during anterior regeneration the brain is absent and thus is incapable of secreting hormones, which might explain the downregulation of these hormone metabolic pathways. The GO term fatty acid metabolic process (BP category) was also enriched due to the downregulation of fatty acid desaturase 2 (fads2), among others, which maintains the structure and function of cell membranes (Los & Murata, 1998) (Figure 3c and Supporting Information: Table S8). This suggests the importance of cell reorganization during P. leidyi regeneration, probably due to the high proliferative activity.
2.4. Gene expression patterns and functional enrichment during asexual reproduction
We detected 211 differentially expressed genes during the second stage of asexual reproduction, of which 130 (~62%) were upregulated, whereas 81 (~38%) were downregulated (Figure 4a and Supporting Information: Table S3). Among these, 100 (~77%) of the upregulated genes and 59 (~73%) of the downregulated genes had a blast hit in the UniProtKB database and thus were functionally annotated (Supporting Information: Table S5).
Figure 4.
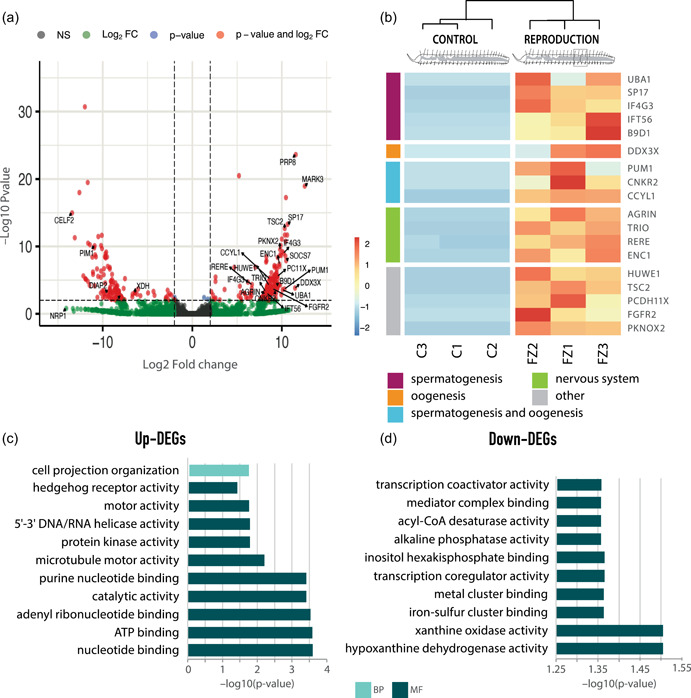
Gene expression patterns during asexual reproduction by paratomic fission. (a) Volcano plot displaying the –log10 p value (false discovery rate [FDR]) as a function of fold change in fissioning and control individuals. Labeled genes are discussed in the text. (b) Hierarchically clustered heatmap of the most important upregulated annotated and nonredundant genes in this comparison, and their categories according to the main function discussed. (c) Gene Ontology enrichment analysis of upregulated annotated genes. (d) Gene Ontology enrichment analysis of downregulated annotated genes.
Little is known about asexual reproduction in P. leidyi in particular, and annelids in general, with only a few molecular studies carried out in the phylum (Bely & Wray, 2001; Martinez et al., 2005; Nyberg et al., 2012; Özpolat & Bely, 2015). In this study, we provide a first overview of the molecular toolkit involved in annelid asexual reproduction and greatly expand the knowledge regarding the genetic machinery underlying this process, as only four of the upregulated genes we identified have been previously reported during asexual reproduction in other metazoans and only one of them in annelids. For instance, fibroblast growth factor receptor 2 (fgfr2) was the only gene previously found highly expressed during P. leidyi asexual reproduction (Nyberg et al., 2012). However, its expression cannot be exclusively attributed to asexual reproduction given that it was also found in regenerative worms. In fact, fgfr2 seems to be essential in the initial formation and outgrowth of the vertebrate's limb and fin regeneration blastema (Brockes & Kumar, 2008). A member of the cadherin family (protocadherin‐11 X‐linked, pcdh11x) and tuberin (TSC complex subunit 2, tsc2), a tumor suppressor gene related to stem cell behavior, were found upregulated in this condition and have also been reported during the asexual cycle of the chordate Botryllus schlosseri (Campagna et al., 2016; Ricci et al., 2016). Finally, Pbx/knotted 1 homeobox 2 (pknox2) was also found upregulated, a gene whose upregulation has also been reported in fissioning individuals of the sea cucumber Cladolabes schmeltzii (Dolmatov et al., 2018). Although the expression of these genes has been reported during asexual reproduction, their specific role in the process is still unknown. Agrin, a structural component of the extracellular matrix, which also shows a relevant role in the nervous system (Daniels, 2012), was found upregulated during P. leidyi asexual reproduction. Conversely, this gene has been reported to be upregulated in C. schmeltzii not‐fissioning individuals (Dolmatov et al., 2018), which might suggest that in this species, unlike in P. leidyi, nervous system development continues after fissioning has finished. Other genes found upregulated during asexual reproduction were also involved in neurogenesis, nervous system development, neural differentiation, and muscle formation. These genes include ectoderm‐neural cortex 1 (enc1), arginine‐glutamic acid dipeptide repeats (rere), and triple functional domain (trio), among others (B. J. Kim & Scott, 2014; S. G. Kim et al., 2009; T. Tao et al., 2020). Once again, the expression of these genes may indicate that nervous system development occurs early during asexual reproduction, as suggested in previous studies (Zattara & Bely, 2011).
Additionally, we also found several upregulated genes in asexually reproducing individuals that play a relevant role not only during germline maintenance but also during germ cell development and differentiation in sexual reproduction. For instance, DEAD‐box helicase 3 X‐linked (ddx3x) is expressed during both oogenesis and spermatogenesis processes in mice and fish (Matsumura et al., 2019; L. Sun et al., 2020). Genes preferentially expressed during spermatogenesis in different species such as mammals, fish, and invertebrates include some previously reported upregulated, anteriorly regenerating P. leidyi individuals including pum1, eukaryotic translation initiation factor 4 gamma 3 (eIF4G3), uba1, and bromodomain adjacent to zinc finger domain protein 1A (baz1a) (Dowdle et al., 2013; F. Sun et al., 2010; Yi et al., 2010; Zheng et al., 2005). Besides, other genes exclusively upregulated during this condition such as sperm surface protein Sp17 and intraflagellar transport protein 56 (Dias et al., 2020; Teves et al., 2020) are also required for spermatogenesis. In addition, genes such as pum1 and socs7 have been reported during female germ cell development (oogenesis) (Assou et al., 2006; Juliano et al., 2010; Virant‐Klun et al., 2013). Finally, other stem‐cell‐related genes (e.g., cnkr2, huwe1, ccnyl1) involved in the regulation, differentiation, and/or maintenance of germline precursors (Bose et al., 2017; Vizziano‐Cantonnet et al., 2018; Zeng et al., 2016) were also upregulated during regeneration. Although P. leidyi seems to reproduce only asexually by paratomic fission under laboratory conditions, gonads are established during postembryonic development (Juliano et al., 2010; Smith, 1896) and therefore the germline must be transmitted over asexual generations (Özpolat & Bely, 2015). Little is known about postembryonic germline development, yet some studies have highlighted the importance of germline precursors in the maintenance of stem germ cells (Marescalchi et al., 2020; Özpolat & Bely, 2015), including genes such as piwi, vasa, and nanos (Juliano et al., 2010). The expression of these genes has been observed using in situ hybridization in germinal cells of several annelid species (Marescalchi et al., 2020; Tadokoro et al., 2006) and during fission zone development in P. leidyi individuals, which might indicate that asexually reproducing individuals of this species routinely form gonads and germline precursors that are transferred to the new zooid through the fission zone (Özpolat & Bely, 2015). Our results suggest that precursors of stem germ cells are present during asexual reproduction, and also that gonads are fully developing. It should be noted that some of these genes (e.g., ddx23, huwe1 and pum1) have also been reported in planarian neoblasts (Galloni, 2012; Henderson et al., 2015; Salvetti et al., 2005) and were also found upregulated during anteriorly regenerating worms, which may indicate the presence of “stem‐cell‐like” populations mediating the asexual reproduction and regeneration processes, being the cellular source for the proliferative activity occurring in the fission zone and blastema.
Enrichment analyses of the upregulated genes (Figure 4b and Supplementary Information: Table S9) show the importance of hedgehog signaling (MF category) during this process (Vladislavovich, 2021), as well as glutamine metabolism, relevant under high proliferation conditions (Tellez‐Garcia et al., 2021). GO terms ATP binding and microtubule motor activity (MF category) were also found enriched, as previously reported during the asexual cycle of B. schlosseri (Campagna et al., 2016). Other enriched GO terms including purine biosynthesis and binding, helicase and transferase activity of phosphorus‐containing groups, and kinase activity are evidence of the intense proliferation activity during this stage of development of the fission zone, similar to what happens during the second stage of anterior regeneration. The enrichment of ATPase and catalytic and hydrolytic activity highlights the elevated energetic demand of this process. GO terms related to cell projection and plasma membrane‐bounded cell projection organization (CC category), which reflect cellular reorganization, were also found to be significantly enriched at this stage.
Among the 58 downregulated genes (Figure 4a and Supporting Information: Table S5), we have found several genes related to apoptosis, including death‐associated inhibitor of apoptosis 2 (diap2). Previous studies have reported the downregulation of another inhibitor of apoptosis during the asexual cycle of the chordate B. schlosseri (Campagna et al., 2016), which may indicate a positive regulation of cell death during fission zone development. Other key regulator of apoptosis also found downregulated in our analysis is pim‐1 proto‐oncogene, serine/threonine kinase (pim1), which has an important role in the regulation of cell proliferation during ovarian cancer (Ngoka, 2008). The importance of the control of cell proliferation during this stage of reproduction is also reflected in the downregulation of several transcription factors such as sem‐2 and 7‐like (Yin et al., 2020).
The enrichment analysis of the downregulated genes in this condition (Figure 4c and Supporting Information: Table S9) revealed several enriched GO terms in the MF category including metal cluster binding, transcription coregulator, and coactivator activity (Figure 4c). Among them, the most enriched metabolic process is related to xanthine/hypoxanthine oxidase and dehydrogenase activity due to the downregulation of the xanthine dehydrogenase (xdh). This is a catabolic enzyme involved in the degradative pathway of purine metabolism, whose expression is sharply decreased in liver cancer (Ngoka, 2008; Weber, 1983). Once again this suggests the importance of cell proliferation during this process, which may be occurring not only in the fission zone where the new zooid is developing but also in the posterior growth zone, as normal growth is also taking place and individuals are still adding new segments.
3. MATERIALS AND METHODS
3.1. Animal culturing
Pristina leidyi specimens were provided by Dr. Alexandra Bely who originally obtained them from Carolina Biological Supply (Bely & Wray, 2001). Animals were cultured at room temperature (RT) in fish tanks with 50 L of 1% filtered artificial seawater that was changed every 2 weeks. Worms were fed with 0.03 g/L of dried spirulina powder and were repeated every 2 weeks. Under these conditions, worms reproduce continuously by paratomic fission (Bely & Wray, 2001).
3.2. Sampling and establishment of experimental groups
Individuals with a length between 21 and 25 segments were selected and kept under starvation for 1 week before starting the experiments. Following this starvation period, three experimental conditions were established (Figure 5): (1) anteriorly regenerating individuals, which were amputated at the 7th segment and maintained for 72 h at RT until they reached the second stage of anterior regeneration (Figure 1a); (2) asexually reproducing individuals, which were specifically selected in the second stage of paratomic fission (Figure 1b); and (3) nonregenerating and nonreproducing individuals, which were used as control. For each experimental condition, three biological replicates were generated to account for interindividual variability in gene expression. Due to the reduced size of P. leidyi individuals, and to have enough genetic material to carry out sequencing, each replicate consisted of a pool of 20 clonal individuals in the same experimental stage. Synchronizing regeneration and specifically fission stages after a feeding pulse could be a limitation in our experimental design, as the timing of this process is highly variable. However, the morphological stage of each pooled individual was carefully evaluated under the stereomicroscope, to select only those individuals in the same fissioning timepoint. All samples were fixed in RNAlater solution (Sigma), stored at 4°C overnight and then at –80°C until RNA extraction was performed. Although three replicates are standard and commonly used for differential expression analyses with nonmodel organisms, a higher number would provide more statistical power; however, due to limited resources, we had to limit our replicates to three. Nevertheless, it is worth mentioning that this study represents the only one so far that uses replicates to compare different and nonpooled stages of asexual reproduction and regeneration in an annelid species.
Figure 5.
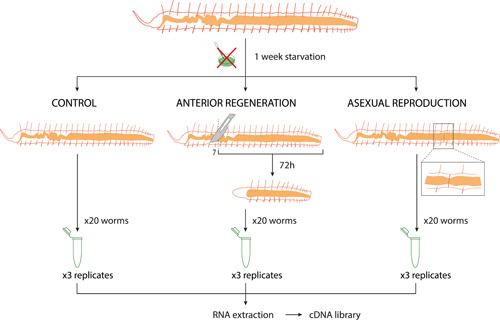
Experimental design and sampling for RNA‐sequencing. After a 1‐week starvation period, worms were selected for the three experimental groups: (1) control group, with nonregenerative and nonreproducing individuals, (2) anterior regenerating group, with worms amputated at segment 7 and fixed 72 h post amputation, and (3) asexually reproducing group, with individuals showing a fission zone at the second stage. Three replicates per condition were prepared, consisting of a pool of 20 individuals each, at the same experimental stage. Following RNA extraction and complementary DNA library construction, the samples were sequenced using Illumina technology.
3.3. RNA extraction and sequencing
Total RNA was extracted from each replicate using TRIzol™ Reagent (Invitrogen™) following the manufacturer's instructions. Quantity and integrity of total RNA were assessed with an Agilent 2100 BioAnalyzer pico RNA assay (Agilent Technologies). All nine replicates except one, corresponding to the anteriorly regenerating condition, passed the required quality thresholds established by the sequencing facility Novogene (Cambridge). Therefore, a total of eight cDNA libraries (anterior regeneration, n = 2; asexual reproduction, n = 3; control, n = 3) were prepared and sequenced on the Illumina HiSeq. 2500 v4 sequencing platform, with an output of 150 bp paired‐end reads.
3.4. Sequence processing and transcriptome assembly
The quality of the raw reads corresponding to each of the eight replicates was assessed and visualized using FASTQC v 0.11.5 (Andrews, 2010). Adapter sequences and low‐quality bases (phred scores < 30) were trimmed off with Trimmomatic v.0.39 (Bolger et al., 2014) using the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. Subsequently, de novo transcriptome assembly was performed with Trinity v.2.4 (Haas et al., 2013) using the resulting clean reads from all replicates, including controls, regenerating and fissioning individuals. Raw reads have been deposited in the BioProject PRJNA830434. Completeness of the assembled transcriptome was assessed in the webserver gVolante (https://gvolante.riken.jp/; Nishimura et al., 2019), with default settings using the ortholog search pipeline BUSCO v.5.1.2 and the metazoan database (metazoa_odb10, last accessed October 17, 2021).
3.5. Functional annotation and GO
The resulting reference transcriptome was annotated using BLAST v.2.6.0 (Basic Local Alignment Search Tool; Altschul et al., 1990) against the UniProtKB (The UniProt Consortium, 2021), with an expected value (E value) cutoff of 1e − 5 in Blast homology searches. To explore the biological functions associated to the differentially expressed genes and to better understand the molecular processes that take place during anterior regeneration and asexual reproduction, a GO enrichment analysis on the differentially expressed genes was performed using g:Profiler (version e104_eg51_p15_3922dba) GOSt function, with Benjamini–Hochberg false discovery rate (FDR) method applying a significance threshold of 0.05 (Raudvere et al., 2019).
3.6. Differential gene expression analyses
Gene expression levels for each condition were obtained by mapping the reads from each individual library to the reference transcriptome using BOWTIE2 v.2.4.4 (Langmead & Salzberg, 2012) and estimating transcript abundance with RSEM v.1.2.12 (Li & Dewey, 2011), as implemented in the Trinity module (Grabherr et al., 2011). The package edgeR (Robinson et al., 2010) was then used to perform pairwise comparisons between the experimental conditions (Regeneration vs. Control; Reproduction vs. Control) and to extract the differentially expressed genes. The package edgeR was run with the following parameters: false discovery rate (FDR) p < 0.001 and min abs(log 2(a/b)) change of 2 (a minimal fourfold change), indicating that the expression value is four times higher than the reference value. Visualization of the differentially expressed genes (upregulated and downregulated) between the conditions was graphically represented with a hierarchically clustered heatmap generated with the pheatmap package in R (https://www.r-project.org/). Volcano plots were generated using the EnhancedVolcano package in R (Blighe et al., 2021).
4. CONCLUSIONS
In this study, we used the RNA‐seq technology to analyze the gene expression profiles during the mid‐early stages of anterior regeneration and asexual reproduction by paratomic fission in the annelid P. leidyi. The results obtained show large differences between anterior regenerating and control individuals, with both fissioning and nonfissioning worms showing more similar expression. This is probably due to the absence of the anterior part in those regenerating worms; however, further research is needed to clarify these differences. Nonetheless, we have identified 35 common genes involved in both processes, some of them previously reported as neoblast markers in planarias, related to different aspects of stem cells and pluripotency, as well as germline development (e.g., ddx23, huwe1, and pum1). This highlights the relevant role of pluripotency‐like cell populations in the development and cell proliferation of the blastema and fission zone. Our work provides a new insight into annelid development programs and brings new research lines for future comparative studies. Nevertheless, further work is needed to better understand the complexity of the molecular programs underlying these processes.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jez.b.23143
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We are grateful to Dr. Alexandra Bely for providing us with the Pristina leidyi cultures. In addition, we would like to thank Dr. Zattara and an anonymous reviewer for their invaluable time in reviewing the manuscript and providing suggestions for improvement. Irene del Olmo was supported by the program “Ayudas para el Fomento de la Investigación en Estudios de Máster‐UAM 2020 en la Facultad de Ciencias” financed by the Universidad Autónoma de Madrid (3931‐326F‐726DP6A38‐4F77). Aida Verdes was funded by the European Union's Horizon 2020 research and innovation program through a Marie Skłodowska‐Curie individual fellowship (grant 841576). This study was funded through the Madrid Government (Comunidad de Madrid‐Spain) under the Multiannual Agreement with Universidad Autónoma de Madrid in the line of action encouraging youth research doctors, in the context of the V PRICIT (Regional Program of Research and Technological Innovation) (SI1/PJI/2019‐00532), and the EMBO Long Term Fellowship (ALTF‐217‐2018) to Patricia Álvarez‐Campos.
del Olmo, I. , Verdes, A. , & Álvarez‐Campos, P. (2022). Distinct patterns of gene expression during regeneration and asexual reproduction in the annelid Pristina leidyi . Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 338, 405–420. 10.1002/jez.b.23143
DATA AVAILABILITY STATEMENT
Raw sequence files are available at NCBI SRA under BioProject PRJNA830434.
REFERENCES
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Álvarez‐Campos, P. , Kenny, N. J. , Verdes, A. , Fernández, R. , Novo, M. , Giribet, G. , & Riesgo, A. (2019). Delegating sex: Differential gene expression in stolonizing syllids uncovers the hormonal control of reproduction. Genome Biology and Evolution, 11, 295–318. 10.1093/gbe/evy265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S . (2010). FastQC: Una herramienta de control de calidad para datos de secuencia de alto rendimiento. Accessed February 12, 2021. https://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Assou, S. , Anahory, T. , Pantesco, V. , Le Carrour, T. , Pellestor, F. , Klein, B. , Reyftmann, L. , De Vos, J. , & Hamamah, S. (2006). The human cumulus–oocyte complex gene‐expression profile. Human Reproduction, 21, 1705–1719. 10.1093/humrep/del065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddar, N. W. A. H. , Dwaraka, V. B. , Ponomareva, L. V. , Thorson, J. S. , & Voss, S. R. (2021). Chemical genetics of regeneration: Contrasting temporal effects of CoCl2 on axolotl tail regeneration. Developmental Dynamics, 250, 852–865. 10.1002/dvdy.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, A. , Tsuji, K. , Cox, K. , Harfe, B. D. , Rosen, V. , & Tabin, C. J. (2006). Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genetics, 2, e216. 10.1371/journal.pgen.0020216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely, A. E. (2014). Early events in annelid regeneration: A cellular perspective. American Zoologist, 54, 688–699. 10.1093/icb/icu109 [DOI] [PubMed] [Google Scholar]
- Bely, A. E. , & Nyberg, K. G. (2010). Evolution of animal regeneration: Re‐emergence of a field. Trends in Ecology & Evolution, 25, 161–170. 10.1016/j.tree.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Bely, A. E. , & Wray, G. A. (2001). Evolution of regeneration and fission in annelids: Insights from engrailed‐ and orthodenticle‐class gene expression. Development, 128, 2781–2791. [DOI] [PubMed] [Google Scholar]
- Bhambri, A. , Dhaunta, N. , Patel, S. S. , Hardikar, M. , Bhatt, A. , Srikakulam, N. , Shridhar, S. , Vellarikkal, S. , Pandey, R. , Jayarajan, R. , Verma, A. , Kumar, V. , Gautam, P. , Khanna, Y. , Khan, J. A. , Fromm, B. , Peterson, K. J. , Scaria, V. , Sivasubbu, S. , & Pillai, B. (2018). Large scale changes in the transcriptome of Eisenia fetida during regeneration. PLoS One, 13, e0204234. 10.1371/journal.pone.0204234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello, A. A. , & Potswald, H. E. (1974). A cytological and quantitative study of neoblasts in the naid Ophidonais serpentina (Oligochaeta). Wilhelm Roux'Archiv für Entwicklungsmechanik der Organismen, 174, 234–249. 10.1007/BF00573227 [DOI] [PubMed] [Google Scholar]
- Blighe, K. , Rana, S. , & Lewis, M. (2021). EnhancedVolcano: Publication‐ready volcano plots with enhanced colouring and labeling. R package version 1.12.0. https://github.com/kevinblighe/EnhancedVolcano
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, R. , Sheng, K. , Moawad, A. R. , Manku, G. , O'Flaherty, C. , Taketo, T. , Culty, M. , Fok, K. L. , & Wing, S. S. (2017). Ubiquitin ligase Huwe1 modulates spermatogenesis by regulating spermatogonial differentiation and entry into meiosis. Scientific Reports, 7, 1–13. 10.1038/s41598-017-17902-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes, J. P. , & Kumar, A. (2008). Comparative aspects of animal regeneration. Annual Review of Cell and Developmental Biology, 24, 525–549. 10.1146/annurev.cellbio.24.110707.175336 [DOI] [PubMed] [Google Scholar]
- Campagna, D. , Gasparini, F. , Franchi, N. , Vitulo, N. , Ballin, F. , Manni, L. , Valle, G. , & Ballarin, L. (2016). Transcriptome dynamics in the asexual cycle of the chordate Botryllus schlosseri . BMC Genomics, 17, 1–17. 10.1186/s12864-016-2598-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A. , Han, Y. , & Poss, K. D. (2021). Regulation of zebrafish fin regeneration by vitamin D signaling. Developmental Dynamics, 250, 1330–1339. 10.1002/dvdy.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera, S. , Ghila, L. , Wenger, Y. , & Galliot, B. (2011). Injury‐induced activation of the MAPK/CREB pathway triggers apoptosis‐induced compensatory proliferation in hydra head regeneration. Development, Growth & Differentiation, 53, 186–201. 10.1111/j.1440-169X.2011.01250.x [DOI] [PubMed] [Google Scholar]
- Cho, S. J. , Lee, M. S. , Tak, E. S. , Lee, E. , Koh, K. S. , Ahn, C. H. , & Park, S. C. (2009). Gene expression profile in the anterior regeneration of the earthworm using expressed sequence tags. Bioscience, Biotechnology, and Biochemistry, 73, 29–34. 10.1271/bbb.80391 [DOI] [PubMed] [Google Scholar]
- Couchie, D. , Lafdil, F. , Martin–Garcia, N. , Laperche, Y. , Zafrani, E. S. , & Mavier, P. (2005). Expression and role of Gas6 protein and of its receptor Axl in hepatic regeneration from oval cells in the rat. Gastroenterology, 129, 1633–1642. 10.1053/j.gastro.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Daniels, M. P. (2012). The role of agrin in synaptic development, plasticity and signaling in the central nervous system. Neurochemistry International, 61, 848–853. 10.1016/j.neuint.2012.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, T. R. , Agarwal, A. , Pushparaj, P. N. , Ahmad, G. , & Sharma, R. (2020). Reduced semen quality in patients with testicular cancer seminoma is associated with alterations in the expression of sperm proteins. Asian Journal of Andrology, 22, 88–93. 10.4103/aja.aja_17_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmatov, I. Y. , Afanasyev, S. V. , & Boyko, A. V. (2018). Molecular mechanisms of fission in echinoderms: Transcriptome analysis. PLoS One, 13(13), e0195836. 10.1371/journal.pone.0195836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle, J. A. , Mehta, M. , Kass, E. M. , Vuong, B. Q. , Inagaki, A. , Egli, D. , Jasin, M. , & Keeney, S. (2013). Mouse BAZ1A (ACF1) is dispensable for double‐strand break repair but is essential for averting improper gene expression during spermatogenesis. PLoS Genetics, 9, e1003945. 10.1371/journal.pgen.1003945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray, N. , Tessmar‐Raible, K. , Le Gouar, M. , Vibert, L. , Christodoulou, F. , Schipany, K. , Guillou, A. , Snyman, H. , Béhague, J. , Vervoort, M. , Arendt, D. , & Balavoine, G. (2010). Hedgehog signaling regulates segment formation in the annelid Platynereis . Science, 329, 339–342. 10.1126/science.1188913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén, L. , Volpato, C. B. , Kervadec, A. , Pineda, S. , Kalvakuri, S. , Alayari, N. N. , Foco, L. , Ocorr, K. , Rossini, A. , Cammarato, A. , Colas, A. R. , Hicks, A. A. , & Bodmer, R. (2020). Silencing of CCR4–NOT complex subunits affects heart structure and function. Disease Models & Mechanisms, 13, dmm044727. 10.1242/dmm.044727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraguas, S. , Barberán, S. , & Cebrià, F. (2011). EGFR signaling regulates cell proliferation, differentiation and morphogenesis during planarian regeneration and homeostasis. Developmental Biology, 354(1), 87–101. 10.1016/j.ydbio.2011.03.023 [DOI] [PubMed] [Google Scholar]
- Fukuda, A. , Morris, J. P., IV , & Hebrok, M. (2012). Bmi1 is required for regeneration of the exocrine pancreas in mice. Gastroenterology, 143(821‐831), e2–831. 10.1053/j.gastro.2012.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni, M. (2012). Global irradiation effects, stem cell genes and rare transcripts in the planarian transcriptome. International Journal of Developmental Biology, 56, 103–116. 10.1387/ijdb.113455mg [DOI] [PubMed] [Google Scholar]
- Gibbs, K. M. , Chittur, S. V. , & Szaro, B. G. (2011). Metamorphosis and the regenerative capacity of spinal cord axons in Xenopus laevis . European Journal of Neuroscience, 33, 9–25. 10.1111/j.1460-9568.2010.07477.x [DOI] [PubMed] [Google Scholar]
- Girich, A. S. , Isaeva, M. P. , & Dolmatov, I. Y. (2017). Wnt and frizzled expression during regeneration of internal organs in the holothurian Eupentacta fraudatrix . Wound Repair and Regeneration, 25, 828–835. 10.1111/wrr.12591 [DOI] [PubMed] [Google Scholar]
- Gómez, C. M. A. , Woodcock, R. M. , Smith, J. J. , Voss, R. S. , & Delgado, J. P. (2018). Using transcriptomics to enable a plethodontid salamander (Bolitoglossa ramosi) for limb regeneration research. BMC Genomics, 19, 1–12. 10.1186/s12864-018-5076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M. G. , Haas, B. J. , Yassour, M. , Levin, J. Z. , Thompson, D. A. , Amit, I. , Adiconis, X. , Fan, L. , Raychowdhury, R. , Zeng, Q. , Chen, Z. , Mauceli, E. , Hacohen, N. , Gnirke, A. , Rhind, N. , di Palma, F. , Birren, B. W. , Nusbaum, C. , Lindblad‐Toh, K. , … Regev, A. (2011). Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology, 29, 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güiza, J. , Barría, I. , Sáez, J. C. , & Vega, J. L. (2018). Innexins: Expression, regulation, and functions. Frontiers in Physiology, 9, 1414. 10.3389/fphys.2018.01414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gąsiorowski, L. , Børve, A. , Cherneva, I. A. , Orús‐Alcalde, A. , & Hejnol, A. (2021). Molecular and morphological analysis of the developing nemertean brain indicates convergent evolution of complex brains in Spiralia. BMC Biology, 19, 175. 10.1186/s12915-021-01113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J. , Papanicolaou, A. , Yassour, M. , Grabherr, M. , Blood, P. D. , Bowden, J. , Couger, M. B. , Eccles, D. , Li, B. , Lieber, M. , MacManes, M. D. , Ott, M. , Orvis, J. , Pochet, N. , Strozzi, F. , Weeks, N. , Westerman, R. , William, T. , Dewey, C. N. , … Regev, A. (2013). De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8, 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, J. M. , Nisperos, S. V. , Weeks, J. , Ghulam, M. , Marín, I. , & Zayas, R. M. (2015). Identification of HECT E3 ubiquitin ligase family genes involved in stem cell regulation and regeneration in planarians. Developmental Biology, 404, 21–34. 10.1016/j.ydbio.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Huang, L. , Damle, S. S. , Booten, S. , Singh, P. , Sabripour, M. , Hsu, J. , Jo, M. , Katz, M. , Watt, A. , Hart, C. E. , Freier, S. M. , Monia, B. P. , & Guo, S. (2015). Partial hepatectomy induced long noncoding RNA inhibits hepatocyte proliferation during liver regeneration. PLoS One, 10, e0132798. 10.1371/journal.pone.0132798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T. C. , Wu, H. L. , Chen, S. H. , Wang, Y. T. , & Wu, C. C. (2020). Thrombomodulin facilitates peripheral nerve regeneration through regulating M1/M2 switching. Journal of Neuroinflammation, 17, 1–14. 10.1186/s12974-020-01897-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova, A. S. , Korotkova, D. D. , Ermakova, G. V. , Martynova, N. Y. , Zaraisky, A. G. , & Tereshina, M. B. (2018). Ras‐dva small GTPases lost during evolution of amniotes regulate regeneration in anamniotes. Scientific Reports, 8, 1–16. 10.1038/s41598-018-30811-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, D. M. , & Seaver, E. C. (2018). Investigation into the cellular origins of posterior regeneration in the annelid Capitella teleta . Regeneration, 5, 61–77. 10.1002/reg2.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano, C. E. , Swartz, S. Z. , & Wessel, G. M. (2010). A conserved germline multipotency program. Development, 137, 4113–4126. 10.1242/dev.047969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbl, A. , Martín‐Durán, J. M. , Worsaae, K. , & Hejnol, A. (2016). Molecular regionalization in the compact brain of the meiofaunal annelid Dinophilus gyrociliatus (Dinophilidae). EvoDevo, 7, 20. 10.1186/s13227-016-0058-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B. J. , & Scott, D. A. (2014). Mouse model reveals the role of RERE in cerebellar foliation and the migration and maturation of Purkinje cells. PLoS One, 9, e87518. 10.1371/journal.pone.0087518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. G. , Jang, S. J. , Soh, J. , Lee, K. , Park, J. K. , Chang, W. K. , Park, E. W. , & Chun, S. Y. (2009). Expression of ectodermal neural cortex 1 and its association with actin during the ovulatory process in the rat. Endocrinology, 150, 3800–3806. 10.1210/en.2008-1587 [DOI] [PubMed] [Google Scholar]
- Kokkaliaris, K. D. , Loeffler, D. , & Schroeder, T. (2012). Advances in tracking hematopoiesis at the single‐cell level. Current Opinion in Hematology, 19, 243–249. 10.1097/MOH.0b013e32835421de [DOI] [PubMed] [Google Scholar]
- Komai, Y. , Tanaka, T. , Tokuyama, Y. , Yanai, H. , Ohe, S. , Omachi, T. , Atsumi, N ., Yoshida, N. , Kumano, K. , Hisha, H. , Matsuda Ueno, H . (2014). Bmi1 expression in long‐term germ stem cells. Scientific Reports, 4(1), 1‐12. 10.1038/srep06175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko, R. P. , & Kozin, V. V. (2021). Comparative aspects of annelid regeneration: Towards understanding the mechanisms of regeneration. Genes, 12, 1148. 10.3390/genes12081148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko, R. P. , Kozin, V. V. , & Kupriashova, E. E. (2016). Regeneration and asexual reproduction in annelids: Cells, genes, and evolution. Biology Bulletin, 43, 185–194. 10.1134/S1062359016030067 [DOI] [Google Scholar]
- Lambert‐Smith, I. A. , Saunders, D. N. , & Yerbury, J. J. (2020). The pivotal role of ubiquitin‐activating enzyme E1 (UBA1) in neuronal health and neurodegeneration. The International Journal of Biochemistry & Cell Biology, 123, 105746. 10.1016/j.biocel.2020.105746 [DOI] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, K. , Vu, H. T. K. , Mohan, R. D. , McKinney, S. A. , Seidel, C. W. , Alexander, R. , Gotting, K. , Workman, J. L. , & Alvarado, A. S. (2016). Egf signaling directs neoblast repopulation by regulating asymmetric cell division in planarians. Developmental Cell, 38, 413–429. 10.1016/j.devcel.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, N. D. , Sessa, S. , Dragalzew, A. C. , Payzin‐Dogru, D. , Sousa, J. F. , Aggouras, A. N. , Johnson, K. , Dunlap, G. S. , Haas, B. J. , Levin, M. , Schneider, I. , & Whited, J. L. (2020). von Willebrand factor D and EGF domains is an evolutionarily conserved and required feature of blastemas capable of multitissue appendage regeneration. Evolution & Development, 22, 297–311. 10.1111/ede.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , & Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics, 12, 1–16. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los, D. A. , & Murata, N. (1998). Structure and expression of fatty acid desaturases. Biochimica et Biophysica Acta (BBA)—Lipids and Lipid Metabolism, 1394, 3–15. 10.1016/S0005-2760(98)00091-5 [DOI] [PubMed] [Google Scholar]
- Malumbres, M. , & Barbacid, M. (2009). Cell cycle, CDKs and cancer: A changing paradigm. Nature Reviews Cancer, 9, 153–166. 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- Marescalchi, O. , Gargiulo, G. , & Falconi, R. (2020). Evidence of germline precursors in asexually reproducing Aeolosoma hemprichi and Aeolosoma viride (Annelida, Aeolosomatidae). Invertebrate Reproduction & Development, 64, 75–81. 10.1080/07924259.2019.1699610 [DOI] [Google Scholar]
- Martinez, V. G. , Menger, G. J., III , & Zoran, M. J. (2005). Regeneration and asexual reproduction share common molecular changes: Upregulation of a neural glycoepitope during morphallaxis in Lumbriculus . Mechanisms of Development, 122, 721–732. 10.1016/j.mod.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Matsumura, T. , Endo, T. , Isotani, A. , Ogawa, M. , & Ikawa, M. (2019). An azoospermic factor gene, Ddx3y and its paralog, Ddx3x are dispensable in germ cells for male fertility. Journal of Reproduction and Development, 65, 121–128. 10.1262/jrd.2018-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myohara, M. , Niva, C. C. , & Lee, J. M. (2006). Molecular approach to annelid regeneration: cDNA subtraction cloning reveals various novel genes that are upregulated during the large‐scale regeneration of the oligochaete, Enchytraeus japonensis . Developmental Dynamics, 235, 2051–2070. 10.1002/dvdy.20849 [DOI] [PubMed] [Google Scholar]
- Ngoka, L. C. (2008). Dramatic down‐regulation of oxidoreductases in human hepatocellular carcinoma hepG2 cells: Proteomics and gene ontology unveiling new frontiers in cancer enzymology. Proteome Science, 6, 1–21. 10.1186/1477-5956-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, O. , Hara, Y. , & Kuraku, S. (2019). Evaluating genome assemblies and gene models using gVolante. Methods in Molecular Biology, 1962, 247–256. 10.1007/978-1-4939-9173-0_15 [DOI] [PubMed] [Google Scholar]
- Nyberg, K. G. , Conte, M. A. , Kostyun, J. L. , Forde, A. , & Bely, A. E. (2012). Transcriptome characterization via 454 pyrosequencing of the annelid Pristina leidyi, an emerging model for studying the evolution of regeneration. BMC Genomics, 13, 1–14. 10.1186/1471-2164-13-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önal, P. , Grün, D. , Adamidi, C. , Rybak, A. , Solana, J. , Mastrobuoni, G. , Wang, Y. , Rahn, H. P. , Chen, W. , Kempa, S. , Ziebold, U. , & Rajewsky, N. (2012). Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. The EMBO Journal, 31, 2755–2769. 10.1038/emboj.2012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özpolat, B. D. , & Bely, A. E. (2015). Gonad establishment during asexual reproduction in the annelid Pristina leidyi . Developmental Biology, 405, 123–136. 10.1016/j.ydbio.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Paul, S. , Balakrishnan, S. , Arumugaperumal, A. , Lathakumari, S. , Syamala, S. S. , Arumugaswami, V. , & Sivasubramaniam, S. (2021). The transcriptome of anterior regeneration in earthworm Eudrilus eugeniae . Molecular Biology Reports, 48, 259–283. 10.1007/s11033-020-06044-8 [DOI] [PubMed] [Google Scholar]
- Pibiri, M. (2018). Liver regeneration in aged mice: New insights. Aging, 10, 1801–1824. 10.18632/aging.101524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere, U. , Kolberg, L. , Kuzmin, I. , Arak, T ., Adler, P. , Peterson, H. , & Vilo, J. (2019). g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Research, 47(W1), W191‐W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, R. P. , Ponz‐Segrelles, G. , Bleidorn, C. , & Aguado, M. T. (2019). Comparative transcriptomics in Syllidae (Annelida) indicates that posterior regeneration and regular growth are comparable, while anterior regeneration is a distinct process. BMC Genomics, 20, 1–13. 10.1186/s12864-019-6223-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci, L. , Chaurasia, A. , Lapébie, P. , Dru, P. , Helm, R. R. , Copley, R. R. , & Tiozzo, S. (2016). Identification of differentially expressed genes from multipotent epithelia at the onset of an asexual development. Scientific Reports, 6, 1–10. 10.1038/srep27357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D. , McCarthy, D. J. , & Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, L. G. , Di Foggia, V. , Radunovic, A. , Bird, K. , Zhang, X. , & Marino, S. (2011). Bmi1 is expressed in postnatal myogenic satellite cells, controls their maintenance and plays an essential role in repeated muscle regeneration. PLoS One, 6, e27116. 10.1371/journal.pone.0027116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti, A. , Rossi, L. , Lena, A. , Batistoni, R. , Deri, P. , Rainaldi, G. , Locci, M. T. , Evangelista, M. , & Gremigni, V. (2005). DjPum, a homologue of Drosophila pumilio, is essential to planarian stem cell maintenance. Development, 132, 1863–1874. 10.1242/dev.01785 [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado, A. (2000). Regeneration in the metazoans: Why does it happen? BioEssays, 22, 578–590. [DOI] [PubMed] [Google Scholar]
- Santagata, S. , Resh, C. , Hejnol, A. , Martindale, M. Q. , & Passamaneck, Y. J. (2012). Development of the larval anterior neurogenic domains of Terebratalia transversa (Brachiopoda) provides insights into the diversification of larval apical organs and the spiralian nervous system. EvoDevo, 3, 3. 10.1186/2041-9139-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp, E. , Kragl, M. , Rubin, L. , & Tanaka, E. M. (2005). Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development, 132, 3243–3253. 10.1242/dev.01906 [DOI] [PubMed] [Google Scholar]
- Shao, Y. , Wang, X. B. , Zhang, J. J. , Li, M. L. , Wu, S. S. , Ma, X. Y. , Wang, X. , Zhao, H. F. , Li, Y. , Zhu, H. H. , & Wu, D. D. (2020). Genome and single‐cell RNA‐sequencing of the earthworm Eisenia andrei identifies cellular mechanisms underlying regeneration. Nature Communications, 11, 1–15. 10.1038/s41467-020-16454-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, J. , Yang, S. , Xu, L. , Wu, C. , Wu, X. , Li, A. , Li, A. , Yu, Y. , Ni, H. , Fukuda, M. , & Zhou, J. (2004). Bystin as a novel marker for reactive astrocytes in the adult rat brain following injury. European Journal of Neuroscience, 20, 873–884. 10.1111/j.1460-9568.2004.03567.x [DOI] [PubMed] [Google Scholar]
- Singh, B. N. , Weaver, C. V. , Garry, M. G. , & Garry, D. J. (2018). Hedgehog and Wnt signaling pathways regulate tail regeneration. Stem Cells and Development, 27, 1426–1437. 10.1089/scd.2018.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F. (1896). Notes on species of North American Oligochaeta: II. Illinois, Natural History Survey, Bulletin, 4, 397–413. [Google Scholar]
- Spassov, D. S. , & Jurecic, R. (2003). Mouse Pum1 and Pum2 genes, members of the Pumilio family of RNA‐binding proteins, show differential expression in fetal and adult hematopoietic stem cells and progenitors. Blood Cells, Molecules, and Diseases, 30, 55–69. 10.1016/S1079-9796(03)00003-2 [DOI] [PubMed] [Google Scholar]
- Stratton, J. A. , Holmes, A. , Rosin, N. L. , Sinha, S. , Vohra, M. , Burma, N. E. , Trang, T. , Midha, R. , & Biernaskie, J. (2018). Macrophages regulate Schwann cell maturation after nerve injury. Cell Reports, 24, 2561–2572. 10.1016/j.celrep.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Sugio, M. , Yoshida‐Noro, C. , Ozawa, K. , & Tochinai, S. (2012). Stem cells in asexual reproduction of Enchytraeus japonensis (Oligochaeta, Annelid): Proliferation and migration of neoblasts. Development, Growth & Differentiation, 54, 439–450. 10.1111/j.1440-169X.2012.01328.x [DOI] [PubMed] [Google Scholar]
- Sun, F. , Palmer, K. , & Handel, M. A. (2010). Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development, 137, 1699–1707. 10.1242/dev.043125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Zhong, Y. , Qiu, W. , Guo, J. , Gui, L. , & Li, M. (2020). MiR‐26 regulates ddx3x expression in medaka (Oryzias latipes) gonads. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 246, 110456. 10.1016/j.cbpb.2020.110456 [DOI] [PubMed] [Google Scholar]
- Tadokoro, R. , Sugio, M. , Kutsuna, J. , Tochinai, S. , & Takahashi, Y. (2006). Early segregation of germ and somatic lineages during gonadal regeneration in the annelid Enchytraeus japonensis . Current Biology, 16, 1012–1017. 10.1016/j.cub.2006.04.036 [DOI] [PubMed] [Google Scholar]
- Tao, J. , Han, Q. , Zhou, H. , & Diao, X. (2019). Transcriptomic responses of regenerating earthworms (Eisenia foetida) to retinoic acid reveals the role of pluripotency genes. Chemosphere, 226, 47–59. 10.1016/j.chemosphere.2019.03.111 [DOI] [PubMed] [Google Scholar]
- Tao, T. , Sun, J. , & Zhu, M. S. (2020). The triple functional domain protein Trio with multiple functions in the nervous system. Journal of Neurology & Neuromedicine, 5, 22–30. [Google Scholar]
- Tauran, Y. , Poulain, S. , Lereau‐Bernier, M. , Danoy, M. , Shinohara, M. , Segard, B. D. , Kato, S. , Kido, T. , Miyajima, A. , Sakai, Y. , Plessy, C. , & Leclerc, E. (2019). Analysis of the transcription factors and their regulatory roles during a step‐by‐step differentiation of induced pluripotent stem cells into hepatocyte‐like cells. Molecular Omics, 15, 383–398. 10.1039/C9MO00122K [DOI] [PubMed] [Google Scholar]
- Tellez‐Garcia, A. A. , Álvarez‐Martínez, R. , López‐Martínez, J. M. , & Arellano‐Carbajal, F. (2021). Transcriptome analysis during early regeneration of Lumbriculus variegatus . Gene Reports, 23, 101050. 10.1016/j.genrep.2021.101050 [DOI] [Google Scholar]
- Teves, M. E. , Roldan, E. R. , Krapf, D. , Strauss, J. F., III , Bhagat, V. , & Sapao, P. (2020). Sperm differentiation: The role of trafficking of proteins. International Journal of Molecular Sciences, 21, 3702. 10.3390/ijms21103702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium . (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research, 49, D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant‐Klun, I. , Knez, K. , Tomazevic, T. , & Skutella, T. (2013). Gene expression profiling of human oocytes developed and matured in vivo or in vitro. BioMed Research International, 2013, e879489. 10.1155/2013/879489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizziano‐Cantonnet, D. , Lasalle, A. , Di Landro, S. , Klopp, C. , & Genthon, C. (2018). De novo transcriptome analysis to search for sex‐differentiation genes in the Siberian sturgeon. General and Comparative Endocrinology, 268, 96–109. 10.1016/j.ygcen.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Vladislavovich, A. A. (2021). Экспрессия ParaHox‐генов при паратомии олигохеты Nais communis [ParaHox gene expression during paratomy in the oligochaete Nais communis] (BSc Thesis). Saint Petersburg State University. https://dspace.spbu.ru/bitstream/11701/32426/1/Diplom_AmosovA.V.pdf
- Weber, G. (1983). Enzymes of purine metabolism in cancer. Clinical Biochemistry, 16, 57–63. 10.1016/s0009-9120(83)94432-6 [DOI] [PubMed] [Google Scholar]
- Yi, Y. J. , Zimmerman, S. W. , Manandhar, G. , Odhiambo, J. F. , Jonakova, V. , Sutovsky, M. , Park, C. S. , & Sutovsky, P. (2010). The activity of ubiquitin activating enzyme UBA1 is required for sperm capacitation, acrosomal exocytosis, and sperm‐egg coat penetration during porcine fertilization. Biology of Reproduction, 83, 555–555. 10.1093/biolreprod/83.s1.555 [DOI] [PubMed] [Google Scholar]
- Yin, C. , Jia, X. , Zhao, Q. , Zhao, Z. , Wang, J. , Zhang, Y. , Li, Z. , Sun, H. , & Li, Z. (2020). Transcription factor 7‐like 2 promotes osteogenic differentiation and boron‐induced bone repair via lipocalin 2. Materials Science and Engineering: C, 110, 110671. 10.1016/j.msec.2020.110671 [DOI] [PubMed] [Google Scholar]
- Zattara, E. E. , & Bely, A. E. (2011). Evolution of a novel developmental trajectory: Fission is distinct from regeneration in the annelid Pristina leidyi . Evolution & Development, 13, 80–95. 10.1111/j.1525-142X.2010.00458.x [DOI] [PubMed] [Google Scholar]
- Zattara, E. E. , & Bely, A. E. (2016). Phylogenetic distribution of regeneration and asexual reproduction in Annelida: Regeneration is ancestral and fission evolves in regenerative clades. Invertebrate Biology, 135, 400–414. 10.1111/ivb.12151 [DOI] [Google Scholar]
- Zattara, E. E. , Turlington, K. W. , & Bely, A. E. (2016). Long‐term time‐lapse live imaging reveals extensive cell migration during annelid regeneration. BMC Developmental Biology, 16, 1–21. 10.1186/s12861-016-0104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Cai, C. , Li, S. , Wang, W. , Li, Y. , Chen, J. , Zhu, X. , & Zeng, Y. A. (2016). Essential roles of cyclin Y‐like 1 and cyclin Y in dividing Wnt‐responsive mammary stem/progenitor cells. PLoS Genetics, 12, e1006055. 10.1371/journal.pgen.1006055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Rotgans, B. , Wang, T. , & Cummins, S. F. (2016). REGene: A literature‐based knowledgebase of animal regeneration that bridge tissue regeneration and cancer. Scientific Reports, 6, 1–11. 10.1038/srep23167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, P. , Patel, B. , McMenamin, M. , Moran, E. , Paprocki, A. M. , Kihara, M. , Schramm, R. D. , & Latham, K. E. (2005). Effects of follicle size and oocyte maturation conditions on maternal messenger RNA regulation and gene expression in rhesus monkey oocytes and embryos. Biology of Reproduction, 72, 890–897. 10.1095/biolreprod.104.035881 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
Raw sequence files are available at NCBI SRA under BioProject PRJNA830434.


