Abstract
Interleukin‐7 is a cytokine with well‐established roles in lymphocyte development and more recently, an expanded role in immune function. IL‐7Rα is highly expressed by innate lymphoid cells (ILCs), but how IL‐7 directs the development or function of ILCs is not well studied. Using mice with inducible deletion of IL‐7Rα, we showed that loss of IL‐7 signaling led to impaired production of IL‐5, IL‐13 and amphiregulin in lung ST2+ group 2 innate lymphoid cells (ILC2s) following influenza/A infection. Conversely, mice treated with IL‐7 increased production of IL‐5 and IL‐13 by lung ILC2s. Moreover, we showed that IL‐7 enhanced GATA3 and CD25 expression in ILC2s and loss of IL‐7 signaling led to their reduced expression. Altogether, this study demonstrates that IL‐7 regulates the function of ILC2s during airway viral infection and induces GATA3 and CD25 expression.
Keywords: type 2 airway immune response, interleukin‐7, influenza/A, innate lymphoid cells, amphiregulin, GATA3, ILC2 function, CD25, interleukin‐5, interleukin‐13
Graphical Abstract
IL‐7 is required for optimal IL‐5, IL‐13 and amphiregulin production in ILC2s during influenza/A infection; additionally, IL‐7 regulates GATA3 in lung and bone marrow ILC2s.
Abbreviations
- AREG
amphiregulin
- BM
Bone marrow
- IL‐7
Interleukin‐7
- ILC2
Group 2 innate lymphoid cells
- TFP
Tomato fluorescent protein
- TSLP
Thymic stromal lymphopoietin
- WT
Wild type
- γc
Common gamma chain receptor
1. INTRODUCTION
Group 2 innate lymphoid cells (ILC2s) are an important population of immune cells discovered nearly a decade ago. 1 , 2 They were found to have an important role in mucosal immunity against helminths and promoting tissue repair following influenza/A virus infection, but in some contexts, lead to influenza/A induced airway hyper‐reactivity and asthma. 3 , 4 , 5 ILC2s share multiple similarities with type 2 helper T cells in developmental lineage, transcriptional programming and cytokine production properties. 6 However, ILCs lack antigen receptors and rely on cytokines produced by mucosal tissues that act as alarmins to become activated. The cytokine milieu that regulates ILC2s during an immune response varies between different mucosal tissues and is critical to direct optimal responses and tissue repair while limiting allergic responses and tissue fibrosis. ILC2s respond to multiple cytokines such as interleukin‐33 (IL‐33), thymic stromal lymphopoietin (TSLP) and gamma‐chain (γc) cytokines such as IL‐2 and IL‐7, either individually or synergistically to produce effector cytokines such as IL‐5 and IL‐13. 5
IL‐7 is a hematopoietic cytokine mainly produced by thymic epithelial cells of the cortex and medulla and stromal cells of the thymus and bone marrow (BM) where it plays an important role in the development of lymphoid cells including ILCs. 2 , 7 , 8 In the previous decade it has become apparent that IL‐7 is important for the development of ILC2s in various tissues including in the lungs. 2 Using a reporter mouse, we and others have shown that IL‐7 is produced peripherally in mucosal tissues including the lungs and is inducible in response to viral infection with influenza/A. 9 A recent study has also shown a role for IL‐7 in ILC2 function and cytokine production in the stomach in response to bacterial infection. 10 Furthermore, IL‐7 and TSLP were shown to enhance cytokine production in IL‐33‐activated ILC2s suggesting IL‐7 and TSLP play a role as co‐activators. 5 , 11 However, it is not clear if IL‐7 is important for ILC2 function in the lungs. While both IL‐7 and TSLP signal through the IL‐7Rα, IL‐7 requires heterodimerization of IL‐7Rα with the γc receptor (CD132) and TSLP signals through a heterodimer of the IL‐7Rα with TSLP receptor (TSLPR) leading to the recruitment of different JAK proteins. 12 Furthermore, IL‐7 has a stronger binding affinity to its corresponding receptor and leads to a stronger activation of STAT5 compared to TSLP. 13 , 14 IL‐7 and TSLP play multiple distinct and overlapping roles in the biology of ILC2s but these distinctions are not entirely clear. 13
GATA3 is a transcription factor known to play an important role in ILC2s. It is required for the production of the type 2 cytokines IL‐5 and IL‐13 in ILC2s. 15 In addition to inducing cytokine expression during an immune response, GATA3 is also important for the development of all helper ILCs by supporting the generation of a common precursor. 6 , 16 Both T cells and ILCs also rely on STAT5 for development, survival and homeostasis as their populations are largely reduced in STAT5A‐ and STAT5B‐ deficient mice. 17 Interestingly, GATA3 expression is reduced in intestinal ILC2s of STAT5A‐ and STAT5B‐ deficient mice. 17 IL‐2 and TSLP are cytokines that regulate GATA3 expression and are known to activate STAT5. 15 , 18 , 19 , 20
In this study, we sought to determine if IL‐7 is required for normal ILC2 function and uncover the transcriptional network it is involved in. We found that IL‐7 is required for inducing a cytokine response in lung ILC2s following influenza/A infection. We also found that IL‐7 is required for normal expression of GATA3 and CD25 in bone marrow ST2+ ILC2s. This suggests that the role of IL‐7 in ILC2 development as well as function is tied to regulation of GATA3 expression.
2. METHODS
2.1. Mice
Mice were housed and used in the Center for Disease Modelling facility (CDM) and Modified Barrier Facility (MBF) at The University of British Columbia (UBC) and all work with animals was carried out with approval and in accordance with the guidelines of the UBC Animal Care and Biosafety Committees. Il‐7rα449F mice were generated as previously described. 21 C57BL/6, Il‐7rα –/– (B6.129S7‐Il7rtm1Imx /J), Ubc‐CreERT2 (B6.Cg‐ Ndor1Tg(UBC‐cre/ERT2)1Ejb /2J), ROSA26‐tdTomato (B6.Cg‐Gt(ROSA)26Sortm14(CAG‐tdTomato)Hze /J) and rag1−/− (B6.129S7‐Rag1tm1Mom/J) mice were obtained from the Jackson Laboratory (Bar Harbour, ME, USA). Tslpr–/– mice were a gift from Dr. James Ihle (St. Jude children's research hospital, Memphis Tennessee). Il‐7eGFP/eGFP mice were a gift from Dr. J. Mike McCune (UCSF, San Francisco, CA). 22 Transgenic IL‐7 mice were a gift from Dr. Philip Leder (Harvard Medical School, Boston, MA). 23 Il‐7rαfl/fl mice were a gift from Dr. Alfred Singer (National Cancer Institute, Bethesda, MD). 24
In all cases, age‐matched and sex‐matched male and female mice between the ages of 6–12 weeks were used.
2.2. Virus
Influenza/A/PR/8/34 (PR8) was purchased from Charles River Laboratories (Wilmington, MA). Mice were sub‐lethally infected under anesthesia (isoflurane) with 500 Hemagglutinin Units (HAU) of influenza/A/PR8 intranasally in 12.5 μL of sterile PBS. In all cases, infection lasted 5 days before experimental endpoint.
2.3. Tissue preparation
Lungs were excised and processed by mincing with scissors followed by enzymatic digestion using 180 units/ml Collagenase IV and 20 ug/ml DNase I (Worthington biochemical LS004188 and LS002139) in 5 ml RPMI incubated at 37° C for 45 mins in a shaker incubator before filtering through 70 μm filters and lysing RBCs with ACK lysis buffer. Bone marrow cells we extracted from single or both femurs by flushing with 10 ml cold PBS (+10% FBS, 2 mM EDTA) and treated with ACK lysis buffer.
2.4. Antibodies and Flow cytometry
All cell surface staining was done at 4°C in the dark. Antibodies to ST2 [RMST2‐2], IL‐7Rα [ebioSB/199], CD25 [PC61], GATA3 [TWAJ], Sca‐1 [D7], CD122 [Tmb1], Ki67 [SolA15] and IL‐13 [eBio13A] were purchased from Thermo Fisher (Waltham, Massachussets). Antibodies to ST2 [DJ8] was purchased from MD Biosciences (Oakdale, MN). Antibodies to TCR‐β [H57‐597], TCR‐γδ [UC‐13D5], CD45[30]‐F11, IL‐7Rα [ebioSB/199], CD25 [PC61], KLRG1 [MAFA] and IL‐5 [TRFK5], were purchased from BioLegend (San Diego, California). CD4 [RM4‐5], CD132 [TUGm2] and Bcl‐2 [N46‐467] were purchased from BD Biosciences (Franklin Lakes, New Jersey). Antibodies to CD3ε [2C11], B220 [RA‐6B2], NK1.1 [PK136], CD11b [M1/70], CD11c [N418], Gr‐1 [RB6‐8C5] and Ter119 [Ter119] were purchased from AbLab (Vancouver, British Columbia).
Viability staining [cat# L34957 and 65‐0865‐14] and Annexin V binding protein (88‐8103‐72) (Thermo Fisher) were used according to manufacturer's instructions.
Lineage cocktail includes, CD3ε, CD4, TCR‐β, TCR‐γδ, B220, NK1.1, CD11b, CD11c, Gr‐1 and Ter119.
Samples were collected on either LSRII (BD Biosciences) or the Attune NxT (Thermo Fisher) and data were analyzed with FlowJo software Tree Star (Ashland, Oregon).
2.5. Ex vivo ILC2 stimulation
To measure IL‐5 and IL‐13, 5 × 106 lung cells were re‐stimulated for 4 hours (at 37° C 5% CO2) with either 1% BSA RPMI media alone containing Brefeldin/A (BD Biosciences); 1% BSA RPMI media containing 50 ng/ml PMA and 500 ng/ml Ionomycin from Sigma‐Aldrich (St. Louis, Missouri) and Brefeldin/A or 40 ng/ml IL‐33 from R&D systems (Minneapolis, Minnesota) and Brefeldin/A. Intracellular cytokine staining was done using the Cytofix/Cytoperm kit from BD Biosciences.
To stimulate bone marrow ILC2s with cytokines, 5–10 million bone marrow cells were stimulated for 13–16 hours (at 37° C 5% CO2) with 1% BSA RPMI, 50μM β‐mercaptoethanol and 1% pen/strep containing either 10 ng/ml recombinant murine IL‐7 from Peprotech (Cranbury, NJ) or IL‐2 (Peprotech) in combination with anti‐IL‐2 mAb [S4B6] (ThermoFisher, catalog no. 16‐7020‐85). Intracellular staining was performed using the Fixation and Cell Permeabilization kit (ThermoFisher).
2.6. pSTAT5 assay
To measure induction of pSTAT5, total dispersed lung cells were starved in 1% BSA RPMI for 2 hours then stimulated with 20 ng/ml IL‐7 (Peprotech) for 30 mins then immediately fixed with 2% PFA for 15 mins to stop reaction. Cells were further fixed and permeabilized in ice cold 90% methanol for 30 minutes in 4° C then stained with surface and pSTAT5 antibodies for 20 mins in 4° C. Cells were analyzed on the Attune NxT analyzer.
3. RESULTS AND DISCUSSION
3.1. Mice deficient in IL‐7 signaling have reduced lung ILC2s and abrogated effector responses to influenza/A
Since the IL‐7Rα is known to play an important role in ILC2 development, we assessed the relative contribution of IL‐7 and TSLP by examining the number of ST2+ ILC2s in the lungs of multiple IL‐7 signaling deficient mouse models. 2 We compared wild type (WT) mice to IL‐7Rα mutant mice where the cytoplasmic chain of IL‐7Rα has a tyrosine residue required for STAT5 phosphorylation substituted to phenylalanine (IL‐7Rα449F). 21 In addition, we used IL‐7Rα–/– mice, IL‐7eGFP/eGFP mice – which serve as IL‐7 ligand knock‐out mice due to insertion of eGFP gene in the endogenous IL‐7 locus – and TSLPR–/– mice. ST2+ lung ILC2s express other key ILC2 markers such as KLRG1, Sca‐1 and GATA3 (Supp. Fig. 1A). Flow cytometric analysis of lung cells revealed nearly a 10‐fold reduction in ST2+ ILC2s in all IL‐7/IL‐7R signaling deficient mice while TSLPR–/– mice have a modest loss (Fig. 1A and Supp. Fig. 1A, B). We sought to analyze lung ILC2s at 5 days post infection (5 dpi) with influenza/A (PR8) to assess the role of IL‐7 in ILC2 biology during inflammation. We elected to assess at 5 dpi due to the early nature of ILC2 responses and to enable assessment of ILC2s prior to accumulation of T cells. The reduction in IL‐7Rα449F and TSLPR–/– ILC2 numbers persists at 5 dpi (Supp. Fig. 1C). Interestingly however, IL‐7eGFP/eGFP and IL‐7Rα449F mice have a defect in generating IL‐5 and IL‐13 producing ILC2s while TSLPR–/– mice are not affected (Fig. 1B and Supp. Fig. 1D). This defect in cytokine production by lung ILC2s in influenza infected mice was demonstrated with re‐stimulation using phorbol 12‐myristate 13‐acetate/ ionomycin (PMA/I) or IL‐33. WT ILC2s expressing both IL‐5 and IL‐13 were only re‐activated with PMA/I re‐stimulation (Fig. 1B), while IL‐33 re‐induced IL‐5 alone in these cells. In addition, IL‐7eGFP/eGFP ILC2s have reduced amphiregulin (AREG) production (Fig. 1C). One caveat is the apparent equivalence in AREG induction by IL‐33 relative to media alone. This may reflect either Influenza‐induced endogenous IL‐33 masking any further manifestation by exogenous IL‐33 or that the time course of stimulation is not optimal. 4 These findings indicate that in addition to its role in the development of lung ILC2s, IL‐7 also plays a role in production of key cytokines and an epithelial repair effector in ILC2s. Further experiments involving re‐stimulation of ILC2s with various cytokine cocktails at different concentrations and durations would provide more insights into the role of IL‐7 in the context of the lung cytokine milieu before and after influenza infection.
FIGURE 1.
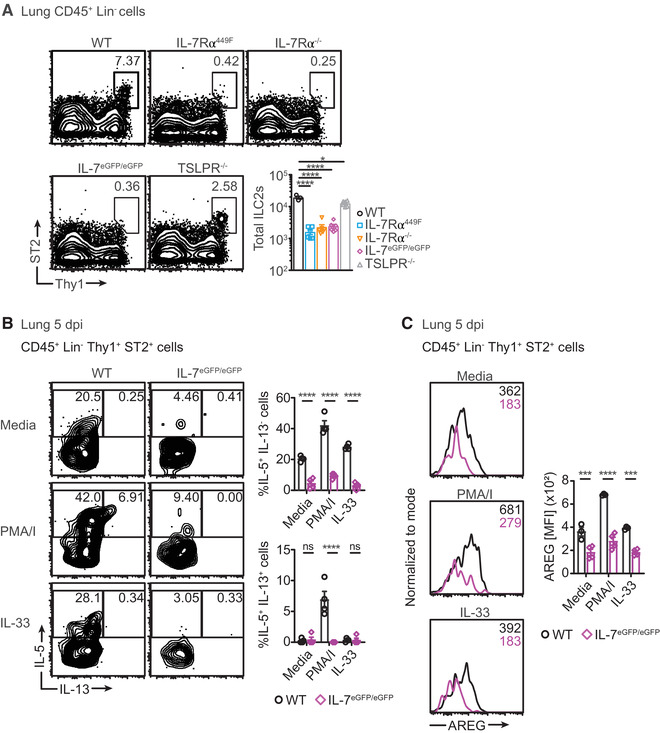
Lung ILC2s are diminished and dysfunctional in IL‐7 signaling deficient mice. (A) Scatter plots and representative bar charts of total ST2+ ILC2s in the lungs of wild type (WT), IL‐7Rα449F, IL‐7Rα–/–, IL‐7eGFP/eGFP and TSLPR–/– mice at steady‐state. Data are representative of two independent experiments, n = 3–6. (B) Frequency plots and representative bar charts of cytokine producing cells in ST2+ ILC2s in the lungs of WT and IL‐7eGFP/eGFP mice 5 days post infection (5 dpi) with influenza in resting state and after PMA/I or IL‐33 re‐stimulation. (C) Histogram plots and representative bar charts of the median fluorescence intensity (MFI) of amphiregulin (AREG) in ST2+ ILC2s in the lungs of WT and IL‐7eGFP/eGFP mice 5 dpi with influenza in resting state and after PMA/I or IL‐33 re‐stimulation. Data are representative of two independent experiments, n = 4–5. *P < 0.05, ***P < 0.001 ****P < 0.0001 and ns = not significant by one‐way and two‐way ANOVA with Tukey's post‐test. Data points represent individual mice and error bars indicate the mean ± SEM
3.2. IL‐7 induces optimal cytokine response in ILC2s independent of its role in development
Our experiments thus far have examined the role of IL‐7 in ILC2 effector responses by using models with germline disruption of IL‐7 signaling. To determine if IL‐7Rα is required by ILC2s to function properly while preserving normal development, we generated mice with inducible IL‐7Rα (CD127) deletion (i7RO).
First, we obtained mice that express Cre‐ERT2 fusion recombinase protein driven by the ubiquitin promoter (Ubc‐CreERT2, Fig. 2A). We bred them to mice that have loxP sites flanking exon 3 of the IL‐7Rα genes (Il‐7rαfl/fl ) which were then crossed to mice with a flox‐stop‐flox sequence upstream of the tdTomato fluorescent protein (TFP) driven by ROSA 26 promoter. The i7RO mice were treated with tamoxifen (as described in Fig. 2B) which binds ERT2, allowing Cre recombinase to enter the nucleus and delete the floxed Il‐7rα exon and the stop codon upstream of TFP. Therefore, cells that have experienced deletion of IL‐7Rα also express TFP. In addition to successfully deleting IL‐7Rα and expressing TFP, our tamoxifen treatment schedule results in a proportion of cells without Cre activity (TFP‐) that can serve as an internal control (Fig. 2C, D). We show that tamoxifen treatment results in IL‐7Rα expression profile similar to that of IL‐7Rα–/– mice in TFP+ cells albeit in CD8 T cells (Supp. Fig. 2A). Following influenza/A infection, TFP+ ILC2s have less IL‐5+ IL‐13+ cells compared to TFP‐ ILC2s (Fig. 2E). This indicates that ILC2s that have developed in an IL‐7 sufficient environment still require IL‐7 for optimal cytokine production. Of note, the profile of the IL‐5 and IL‐13 producing cells in the i7RO mice is varied compared to WT and other IL‐7 signaling deficient mice. This is possibly due to housing of i7RO mice in a separate animal facility with lower microbial exclusion criteria that could have a dampening effect. To further define the effector function of IL‐7, we treated rag1–/– mice with IL‐7 intraperitoneally for 4 days after influenza/A infection and assessed ILC2 cytokine production (Fig. 2F). IL‐7 treatment resulted in both increase of total number of lung ILC2s and percent of IL‐5+ IL‐13+ ILC2s (Fig. 2G, H). We note that there is no difference in percent of IL‐5 single positive cells in ILC2s. Previous research has shown that IL‐5+ IL‐13+ ILC2s play a unique role in pathology during viral lung infection. 25 Altogether, these results indicate that IL‐7 is sufficient for inducing cytokine production by ILC2s. In addition, our use of rag1–/– mice demonstrates that IL‐7 can induce cytokine production in ILC2s without T cells and B cells as intermediates.
FIGURE 2.
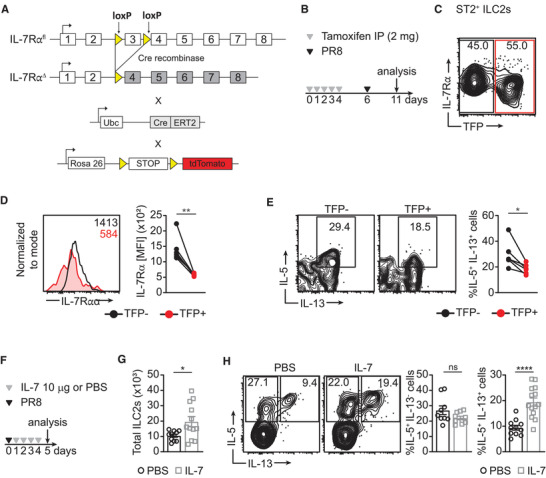
Cytokine production by ILC2s is reduced in acute IL‐7 signaling deficient mice. (A) Strategy for crossing mice to create inducible Il‐7rα knock‐out (i7RO) mice. Ubc, ubiquitin; ERT2; estrogen receptor. (B) Schematic illustrating tamoxifen treatment schedule. (C) and (D) IL‐7Rα expression in i7RO mice presented as scatter plots (C) and histogram and graph representing IL‐7Rα expression in paired samples. (E) Frequency of IL‐5+ IL‐13+ cells in tomato florescent protein (TFP) ‐ and TFP+ ILC2s of i7RO mice presented as flow cytometry plots and graph. Data are representative of 2 experiments, n = 4–5. (F) Schematic illustrating IL‐7 treatment in rag1–/– mice. (G) Total number of lung ST2+ ILC2s and (H) frequency of IL‐5+ IL‐13– and IL‐5+ IL‐13+ cells in ILC2s of rag1–/– mice treated with IL‐7 intraperitoneally. Data is pooled from two independent experiments, n = 5. *P < 0.05, **P < 0.01 and ****P < 0.0001 as determined by paired and unpaired two‐tailed Student's t‐test. Data points represent individual mice and error bars indicate the mean ± SEM
3.3. IL‐7 induces GATA3 expression in lung and BM ILC2s
GATA3 is a transcription factor important for the development of all ILCs and for cytokine production in ILC2s. 15 , 16 Considering IL‐7 is an activator of STAT5 and pSTAT5 binds the promoter regions of GATA3, we asked if IL‐7 induces GATA3 expression. 7 , 15 To determine this, we examined GATA3 expression in lung ILC2s of WT and IL‐7eGFP/eGFP mice in the course of influenza infection. We recapitulated earlier findings where ILC2 numbers are reduced in IL‐7eGFP/eGFP mice at steady‐state and after infection (5 dpi) while in WT mice, there is a marked increase in number of ILC2s following infection (Fig. 3A). GATA3 was slightly but not significantly reduced in IL‐7eGFP/eGFP lung ILC2s at steady‐state. At 5 dpi, lung ILC2s of both genotypes experienced significant downregulation of GATA3, as has been shown previously. 26 However, IL‐7eGFP/eGFP ILC2s have further reduced GATA3 expression compared to WT mice at 5 dpi (Fig. 3B). This suggests that IL‐7 contributes to maintaining the cytokine production properties of lung ILC2s during influenza infection possibly by stabilizing GATA3 expression.
FIGURE 3.
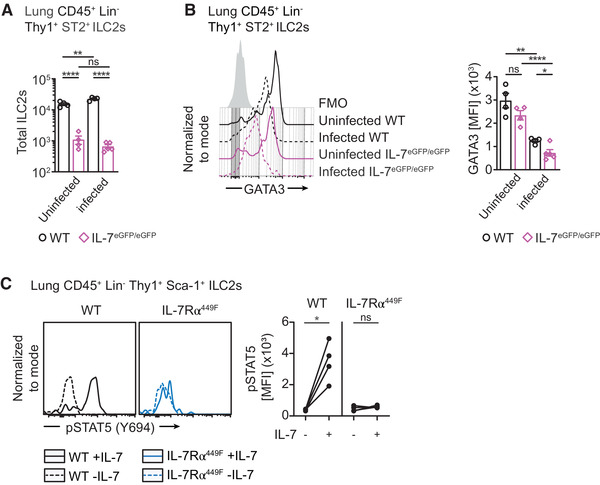
IL‐7 in GATA3 induction and STAT5 activation in lung ILC2s. (A) Bar graph of total lung ILC2s in WT and IL‐7eGFP/eGFP mice at steady‐state and 5 dpi with influenza. Representative data from two independent experiments. (B) Histogram and bar graph of GATA3 expression in lung ILC2s of WT and IL‐7eGFP/eGFP mice at steady‐state and 5 dpi with influenza. Data representative of two independent experiments, n = 4–6. (C) STAT5 activation following treatment with IL‐7 for 30 minutes in steady‐state lung ILC2s of WT and IL‐7Rα449F mice. Data representative of two experiments, n = 3–4. *P < 0.05, **P < 0.01 and ns = not significant, as determined by ANOVA with Tukey's post‐test and paired two‐tailed Student's t‐test. Data points represent individual mice and error bars indicate the mean ± SEM
STAT5 is required for the expression of GATA3 in intestinal ILC2s. 17 We assessed the activation of STAT5 in WT and IL‐7Rα449F lung ILC2s at steady‐state. We show that pulsing whole, dispersed lung cells in vitro with IL‐7 for 30 minutes induces pSTAT5 in WT but not in IL‐7Rα449F lung ILC2s (Fig. 3C). Consistent with T cells, 21 this demonstrates the importance of the Y449 residue of the IL‐7Rα for the activation of STAT5 in ILC2s and possibly, the induction of GATA3.
GATA3 is also known to be important for the development of all ILCs and maturation of ILC2s. We examined PLZF expressing ILC precursors (ILCPs) in the bone marrow (BM) and found reduced numbers of and GATA3 expression in ILCPs of IL‐7Rα449F mice compared to WT mice (Supp. Fig. 2B, C). We also investigated the expression of GATA3 in BM ST2+ ILC2s of adult WT, IL‐7Rα449F, TSLPR–/– and IL‐7eGFP/eGFP mice. We found that IL‐7Rα449F and IL‐7eGFP/eGFP mice have reduced number of ST2+ ILC2s and GATA3 expression while TSLPR–/– ILC2s present with normal GATA3 expression (Fig. 4A‐D). To confirm that IL‐7 alone is capable of inducing GATA3 in ILC2s, we used ex vivo stimulation of IL‐7eGFP/eGFP BM ILC2s with IL‐7. We found that acute (∼15 hrs) treatment with IL‐7 induces high GATA3 expression in ILC2s (Fig. 4E). Furthermore, using i7RO mice, we demonstrated that acute deletion of IL‐7Rα led to reduced GATA3 expression in ILC2s (Fig. 4F‐H). Taken together, our results show that IL‐7 is important for normal expression of GATA3 in ILC2s and acute gain or loss of function of this cytokine largely alters the expression of GATA3.
FIGURE 4.
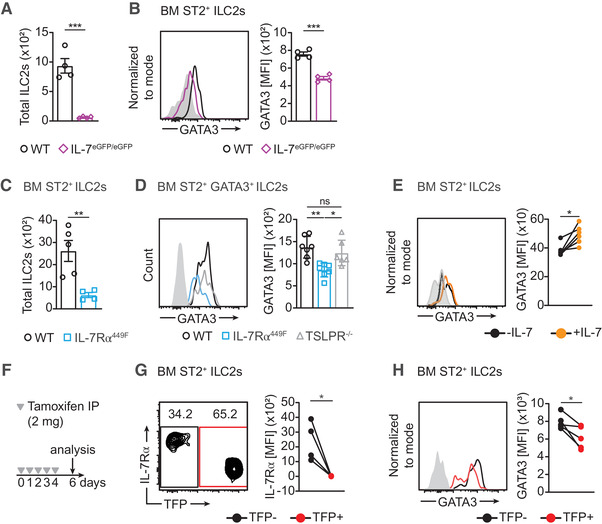
GATA3 expression in BM ILC2s requires IL‐7. (A) Bar graph of total bone marrow (BM) ILC2s in a single femur of WT and IL‐7 eGFP/eGFP mice. Representative data from two independent experiments. (B) Histogram and bar graph of GATA3 expression in BM ILC2s of WT and IL‐7 eGFP/eGFP mice. (C) Bar graph of total bone marrow (BM) ILC2s in a single femur of WT and IL‐7Rα449F mice. Representative data from two independent experiments. (D) Histogram and bar graph of GATA3 expression in BM ILC2s of WT, IL‐7Rα449F and TSLPR–/– mice. (E) GATA3 expression in BM IL‐7eGFP/eGFP ILC2s with and without ex vivo IL‐7 treatment (∼15 hrs) as paired samples represented in histogram and bar charts. Representative of two independent experiments. (F) Schematic illustrating tamoxifen treatment schedule of i7RO mice. (G) IL‐7Rα and (H) GATA3 expression in TFP‐ and TFP+ BM ILC2s of i7RO mice presented as histograms and bar charts. Grey filled histogram represents FMO (Fluorescence Minus One). Data representative of two independent experiments, n = 4–6. *P < 0.05, **P < 0.01 and ns = not significant, as determined by ANOVA with Tukey's post‐test and paired two‐tailed Student's t‐test. Data points represent individual mice and error bars indicate the mean ± SEM
3.4. IL‐7 signaling regulates the high affinity IL‐2 receptor α chain (CD25)
Since IL‐7Rα449F disruption abrogates STAT5 activation, the impaired GATA3 induction in IL‐7 signaling deficient ILC2s suggests that IL‐7 may regulate it via STAT5. 27 This prompted us to ask if IL‐7 also regulates expression of the high affinity IL‐2Rα (CD25) since STAT5 and GATA3 can induce CD25 in T cell subsets. 28 , 29 , 30 As predicted, IL‐7Rα449F and IL‐7eGFP/eGFP ILC2s have reduced CD25 expression compared to WT mice while TSLPR–/– mice are unaffected (Fig. 5A and Supp. Fig. 2D). By using i7RO mice, we also determined that, despite developing in an IL‐7 signaling sufficient environment, acute loss of IL‐7Rα in ILC2s resulted in reduced CD25 expression (Fig. 5B). We also measured CD25 expression in WT BM ILC2s treated ex vivo with IL‐7. We found that CD25 expression was enhanced in paired samples treated with IL‐7 showing that IL‐7 induced CD25 expression (Fig. 5C). Furthermore, we examined transgenic IL‐7 (TgIL‐7) mice that over‐express IL‐7 systemically including in the BM and we showed that TgIL‐7 BM ILC2s had higher surface CD25 expression compared to WT mice (Fig. 5D). To determine if reduction in CD25 expression affects GATA3 expression, we treated IL‐7eGFP/eGFP BM ILC2s with IL‐2 in combination with the anti‐IL‐2 mAb (S4B6). S4B6 has been documented to bind IL‐2 non‐competitively and facilitate binding of IL‐2 to the β chain (CD122) and γ chain (CD132) thus compensating for loss of the α chain (CD25). 31 With this technique we demonstrated that rescuing IL‐2 did not result in induction of GATA3 (Supp. Fig. 2E). Further assessment of downstream signaling in addition to STAT5 phosphorylation will be necessary to determine how IL‐7 controls CD25 expression and loss of function models are required to determine if IL‐2 signaling plays a role in GATA3 expression.
FIGURE 5.
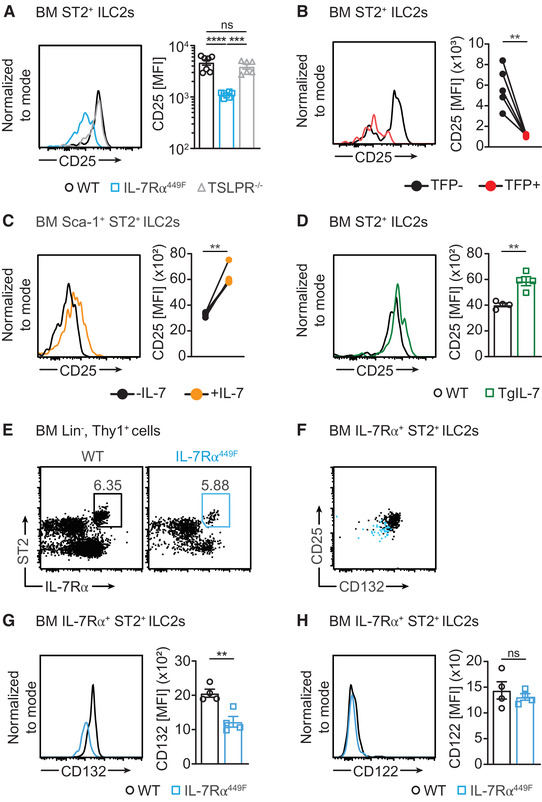
CD25 expression in ILC2s is regulated by IL‐7. (A) Histogram and bar graph of CD25 expression in BM ILC2s of WT, IL‐7Rα449F and TSLPR–/– mice. (B) CD25 expression in TFP‐ and TFP+ BM ILC2s presented as histograms and bar charts. Data representative of two independent experiments. (C) CD25 expression in WT BM ILC2s with and without in vitro IL‐7 treatment (∼15 hrs) as paired samples represented in histogram and bar charts. Representative of three independent experiments. (D) CD25 expression in WT and Tg‐IL‐7 mice presented as histograms and bar charts. Representative data from two independent experiments, n = 3–6. (E) Flow cytometry plot illustrating frequency of IL‐7Rα+ ST2+ ILC2s. (F) Flow cytometry plot showing CD132 and CD25 expression in BM ILC2s. (G) CD132 and (H) CD122 expression in WT and IL‐7Rα449F ILC2s presented as histograms and bar charts. Representative data from two independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001 and ns = not significant, as determined by ANOVA with Tukey's post‐test and paired and unpaired two‐tailed Student's t‐test. Data points represent individual mice and error bars indicate the mean ± SEM
We sought to measure the expression of the β and γ chain receptors to determine the extent to which IL‐7 influences signaling components of IL‐2 and other γ‐chain cytokines and therefore we compared BM IL‐7Rα+ ST2+ ILC2s of WT and IL‐7Rα449F mice (Fig. 5E). Interestingly, IL‐7Rα449F ILC2s that express reduced CD25 also have reduced CD132 expression suggesting IL‐7 controls signal transduction of IL‐2 and possibly also other γ‐chain cytokines (Fig. 5F and G). However, we observed no difference in CD122 expression between IL‐7Rα449F and WT ST2+ ILC2s (Fig. 5H). The reduction in CD132 expression suggests that IL‐7 controls the sensitivity of ILC2s to other γ chain cytokines.
3.5. Impaired IL‐7Rα signaling does not affect ILC2 survival and proliferation in the BM
The importance of IL‐7 in survival of lymphocytes prompted us to ask if the reduced number of ILC2s observed in the BM was due to reduced survival. We found that the expression of the anti‐apoptotic factor Bcl‐2 was similar in WT and IL‐7Rα449F steady‐state BM ST2+ ILC2s suggesting that IL‐7 does not control Bcl‐2 expression (Fig. 6A). Alternatively, Bcl‐2 expression in ILC2s may not depend on the Tyr 449 motif. To definitively measure cell death, we used Annexin V staining and found that WT and IL‐7Rα449F ILC2s were composed of similar proportions of dead and early apoptotic cells (Fig. 6B). We also measured Ki67 expression to assess proliferation and found no difference between WT and IL‐7 eGFP/eGFP BM ILC2s (Fig. 6C). Taken together, this suggests that IL‐7Rα signaling does not control survival or proliferation of BM ILC2s. Our observation is in agreement with an earlier report that complete deletion of the IL‐7Rα does not affect Bcl‐2 expression in small intestinal ILC2s. They also found that IL‐15 can compensate and support their survival. 32 How loss of IL‐7 impacts the survival of ILC2s in other tissues and if other cytokines can also compensate for the survival of ILC2s is unclear. Our finding that loss of IL‐7Rα signaling affects the expression of the common γ‐chain receptor – which is required for IL‐15 signaling – indicates that non‐γ chain cytokines in the BM may also be required to compensate for IL‐7.
FIGURE 6.
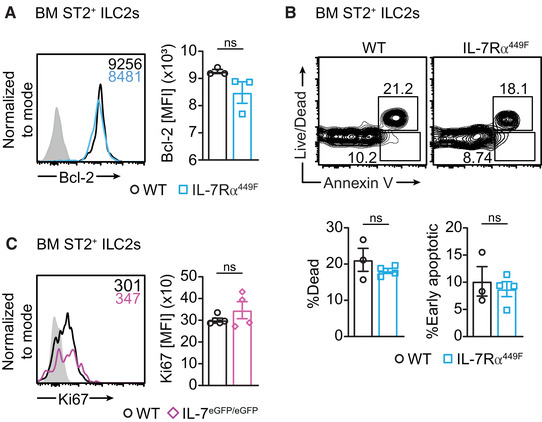
IL‐7 receptor mutant mice have normal Bcl‐2 expression, survival and proliferation. (A) Bcl‐2 expression in BM ILC2s of WT and IL‐7Rα449F mice presented as histograms and bar charts. Grey filled histogram represents isotype control. (B) Annexin V and viability staining of WT and IL‐7Rα449F mice. Amcyan (dead cell) hi and Annexin V hi staining represent dead cells and Amcyan (dead cell) low/ Annexin V hi represent early apoptotic cells. Data are representative of two independent experiments, n = 3–4. Grey filled histogram represents isotype control. (C) Ki67 expression in BM ILC2s in WT and IL‐7eGFP/eGFP mice presented as histograms and bar charts. Grey filled histogram represents FMO. ns = not significant, as determined by two‐tailed Student's t‐test. Data points represent individual mice and error bars indicate the mean ± SEM
In summary, we have shown that IL‐7 plays an integral role in inducing cytokine production by lung ST2+ ILC2s following influenza/A infection. ILC2s are known to play an important function in repairing damaged tissue following influenza/A infection and when exacerbated, this process could lead to airway hyper‐responsiveness and allergic inflammation. 3 , 4 Further analysis is necessary to understand whether ILC2 stimulation by IL‐7 supports tissue repair versus hyper‐responsivity. Considering multiple mucosal alarmins are involved in the activation and function of ILC2s in the lungs, IL‐7 may act as a co‐stimulator of ST2+ ILC2s working synergistically with IL‐33. 5 , 11 The extent of the effect of IL‐7, and whether it can independently activate and stimulate ILC2s in vivo needs further investigation. We have also shown that loss of IL‐7 signaling abrogates GATA3 expression in a BM population of ILC2s. This suggests that IL‐7 regulation of GATA3 may be important for development of ILC2s as well as function. 6 , 15 In addition, we demonstrated that CD25 is regulated by IL‐7 indicating that IL‐7 may regulate ILC2 function and development by controlling cytokine signals. IL‐2 has indeed previously been shown to enhance IL‐5 and IL‐13 production in ILC2s. 33 While IL‐7 is known to be important for lymphocyte survival, it seems to have a non‐essential role in Bcl‐2 expression in BM ILC2s and their survival. Further research is required to map the network that IL‐7 regulates and understand how IL‐7 controls ILC2 development and function and where GATA3 lies in that network.
AUTHORSHIP
AS conceived and designed the project, performed and analyzed experiments, and wrote the manuscript. EM and JL performed and analyzed experiments and reviewed the manuscript. JS performed and analyzed experiments. NA conceived and designed the project and reviewed the manuscript.
CONFLICT OF INTEREST DISCLOSURE
The authors declare no conflict of interest.
Supporting information
Supplement Material
Supplement Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Laura Mathae and Jennie Jackson at the University of British Columbia for reviewing the manuscript and providing their critical input. We would also like to thank the staff at the Centre for Disease Modeling and Modified Barrier Facility for animal husbandry and the UBC Flow Cytometry Facility. Graphical abstract was created with ©BioRender ‐ biorender.com. Work in the laboratory was supported by a grant to NA from the Canadian Institutes for Health Research (MOP‐126060 and PJT‐156119). AS was supported by the John Richard Turner Fellowship.
Sheikh A, Lu J, Melese E, Seo JH, Abraham N. IL‐7 induces type 2 cytokine response in lung ILC2s and regulates GATA3 and CD25 expression. J Leukoc Biol. 2022;112:1105–1113. 10.1002/JLB.3AB1220-819RRR
Contributor Information
Abdalla Sheikh, Email: abdalla.sheikh@uhn.ca.
Ninan Abraham, Email: ninan@mail.ubc.ca.
REFERENCES
- 1. Spits H, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145‐9. [DOI] [PubMed] [Google Scholar]
- 2. Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue‐associated c‐Kit(+)Sca‐1(+) lymphoid cells. Nature. 2010;463:540‐4. [DOI] [PubMed] [Google Scholar]
- 3. Monticelli LA, et al. Innate lymphoid cells promote lung tissue homeostasis following acute influenza virus infection. Nature immunology. 2011;12:1045‐1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang YJ, et al. Innate lymphoid cells mediate influenza‐induced airway hyper‐reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halim TimotheusYF, et al. Lung natural helper cells are a critical source of Th2 cell‐type cytokines in protease allergen‐induced airway inflammation. Immunity. 2012;36:451‐463. [DOI] [PubMed] [Google Scholar]
- 6. Zhu J, GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Frontiers in Immunology, 2017. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu W, et al. NFIL3 Orchestrates the Emergence of Common Helper Innate Lymphoid Cell Precursors. Cell Reports. 2015;10:2043‐2054. [DOI] [PubMed] [Google Scholar]
- 8. Mazzucchelli RI, et al. Visualization and identification of IL‐7 producing cells in reporter mice. PLoS ONE. 2009;4:e7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dahlgren MW, et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity. 2019;50:707‐722 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Satoh‐Takayama N, et al. Bacteria‐induced group 2 innate lymphoid cells in the stomach provide immune protection through induction of IgA. Immunity. 2020;52:635‐649 e4. [DOI] [PubMed] [Google Scholar]
- 11. Kabata H, Moro K, Koyasu S. The group 2 innate lymphoid cell (ILC2) regulatory network and its underlying mechanisms. Immunol Rev. 2018;286:37‐52. [DOI] [PubMed] [Google Scholar]
- 12. Verstraete K, et al. Structural basis of the proinflammatory signaling complex mediated by TSLP. Nat Struct Mol Biol. 2014;21:375‐382. [DOI] [PubMed] [Google Scholar]
- 13. Sheikh A, Abraham N. Interleukin‐7 receptor alpha in innate lymphoid cells: more than a marker. Front Immunol. 2019;10:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walsh ST. Structural insights into the common gamma‐chain family of cytokines and receptors from the interleukin‐7 pathway. Immunol Rev. 2012;250:303‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mjösberg J, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649‐659. [DOI] [PubMed] [Google Scholar]
- 16. Yagi R, et al. The transcription factor GATA3 is critical for the development of all IL‐7Rα‐Expressing innate lymphoid cells. Immunity. 2014;40:378‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villarino AV, et al. Subset‐ and tissue‐defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J Exp Med. 2017;214:2999‐3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoyler T, et al. The transcription factor GATA‐3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamane H, Zhu J, Paul WE. Independent roles for IL‐2 and GATA‐3 in stimulating naive CD4+ T cells to generate a Th2‐inducing cytokine environment. J Exp Med. 2005;202:793‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wan YY. GATA3: a master of many trades in immune regulation. Trends Immunol. 2014;35:233‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Osborne LC, et al. Impaired CD8 T cell memory and CD4 T cell primary responses in IL‐7R alpha mutant mice. J Exp Med. 2007;204:619‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller CN, et al. IL‐7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int Immunol. 2013;25:471‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rich BE, et al. Cutaneous lymphoproliferation and lymphomas in interleukin 7 transgenic mice. J Exp Med. 1993;177:305‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCaughtry TM, et al. Conditional deletion of cytokine receptor chains reveals that IL‐7 and IL‐15 specify CD8 cytotoxic lineage fate in the thymus. J Exp Med. 2012;209:2263‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stier MT, et al. Respiratory syncytial virus infection activates IL‐13‐producing group 2 innate lymphoid cells through thymic stromal lymphopoietin. J Allergy Clin Immunol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silver JS, et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol. 2016;17:626‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plumb AW, et al. Interleukin‐7 in the transition of bone marrow progenitors to the thymus. Immunology & Cell Biology. 2017;95:916‐924. [DOI] [PubMed] [Google Scholar]
- 28. Bitar M, et al. Evaluating STAT5 phosphorylation as a mean to assess T cell proliferation. Front Immunol. 2019;10:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sadovnik I, et al. Identification of CD25 as STAT5‐dependent growth regulator of leukemic stem cells in Ph+ CML. Clin Cancer Res. 2016;22:2051‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA‐3 for the function of regulatory T cells. Immunity. 2011;35:337‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spangler JB, et al. Antibodies to interleukin‐2 elicit selective T cell subset potentiation through distinct conformational mechanisms. Immunity. 2015;42:815‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinette ML, et al. IL‐15 sustains IL‐7R‐independent ILC2 and ILC3 development. Nat Commun. 2017;8:14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roediger B, et al. IL‐2 is a critical regulator of group 2 innate lymphoid cell function during pulmonary inflammation. J Allergy Clin Immunol. 2015;136:1653‐1663 e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Material
Supplement Material


