Abstract
The Pacific cupped oyster Crassostrea gigas is one of the most ‘globalized’ marine invertebrates and its production is predominant in many parts of the world including Europe. However, it is threatened by mortality events associated with pathogenic microorganisms such as the virus OsHV‐1 and the bacteria Vibrio aestuarianus. C. gigas is also a host for protozoan parasites including haplosporidians. In contrast with Haplosporidium nelsoni previously detected in Europe, H. costale was considered exotic although its presence in French oysters was suggested in the 1980s based on ultrastructural examination. Here, a combination of light and transmission electron microscopy, PCR and sequencing allowed characterizing the presence of the parasite in the context of low mortality events which occurred in 2019 in France. Histological observation revealed the presence of uninucleated, plasmodial and spore stages within the connective tissues of some oysters. Ultrastructural features were similar to H. costale ones in particular the presence of axe‐shaped haplosporosomes in spore cytoplasms. Three fragments of the genome including partial small subunit rRNA gene, the ITS‐1, 5.8S and ITS‐2 array and part of the actin gene were successfully sequenced and grouped with H. costale homologous sequences. This is the first time that the presence of H. costale was confirmed in C. gigas in France. Furthermore, a TaqMan real‐time PCR assay was developed and validated [DSe = 92.6% (78.2–99.8) and DSp = 95.5% (92.3–98.6)] to enable the rapid and specific detection of the parasite. The application of the PCR assay on archived samples revealed that the parasite has been present in French oyster populations at least since 2008. Considering the little information available on this parasite, the newly developed TaqMan assay will be very helpful to investigate the temporal and geographic distribution and the life cycle of the parasite in France and more generally in C. gigas geographic range.
Keywords: Crassostrea gigas, Haplosporida, Haplosporidium costale, oyster, parasite, real‐time PCR
1. INTRODUCTION
Originated from the north eastern Asia, the Pacific cupped oyster Crassostrea gigas is currently produced in different parts of the world (Herbert et al., 2016) including France where its production is estimated around 85,000 t per year (FAO, 2019). This production is based on wild spat collected in the field and on hatchery‐produced spat, mainly triploids. Movements of oysters regularly occur between spat collection areas or hatcheries and oyster growing areas in France and also between France and some European countries including Ireland, Italy, Portugal, Spain or United Kingdom.
Massive mortality events of Crassostrea gigas have been reported since the 1990s in France (Samain & McCombie, 2007). Associated with the presence of the virus OsHV‐1 or with the presence of the bacteria Vibrio aestuarianus, these mortalities have strongly impacted the production of oysters in France and more widely in Europe [EFSA Panel on Animal Health and Welfare (AHAW), 2015]. Protozoan parasites such as Perkinsus marinus, Mikrocytos mackini and Haplosporidium nelsoni have been shown to infect C. gigas. Whereas the two first parasite species have only been detected in America and are exotic to Europe, H. nelsoni has also been reported in C. gigas with low prevalence in Northern America (Friedman, 1996; Friedman et al., 1991; Haskin & Andrews, 1988; Stephenson et al., 2003), Asia (Burreson et al., 2000; Chun, 1972; Kamaishi & Yoshinaga, 2002; Kang, 1980; Kern, 1976; Wang et al., 2010) and Europe (Lynch et al., 2013; Renault et al., 2000). Its presence does not seem associated with the occurrence of mortality in C. gigas.
In France, the surveillance of mollusc diseases is mainly based on the reporting of mortality events by producers. Although this passive approach might present some biases, it aims to detect emerging situations using mortality as an alert signal (Lupo et al., 2014). Investigation of Crassostrea gigas abnormal mortality is usually carried out not only by testing oysters by PCR for the detection of the virus OsHV‐1 or the bacteria Vibrio aestuarianus but also by histology. Histology allows the observation of lesions and the detection of a range of pathogens including protozoan parasites (Carnegie et al., 2016).
Haplosporidium costale belongs to the phylum Cercozoa, order Haplosporida and family Haplosporidiidae (Arzul & Carnegie, 2015). Also called SSO (Sea Side Organisms), it has been reported in the context of seasonal mortality events affecting the American oyster Crassostrea virginica in high salinity (>25‰) coastal bays on the east coast of the United States (Andrews, 1984; Andrews & Castagna, 1978; Couch & Rosenfield, 1968). Low prevalence and intensity of infection have also been reported in C. virginica on the Atlantic coast of Canada. Although the presence of parasite DNA has been detected in C. gigas in China (Wang et al., 2010), no effect has been reported in this oyster species. In C. virginica, infections with H. costale seem to be acquired in early summer. Plasmodia are generally not observed in oyster tissues before the following spring. Plasmodia rapidly multiply and sporulate in the connective tissues of the digestive gland, mantle and gonad, sporulation coinciding with host death in May to June. In contrast with H. costale, the congeneric species H. nelsoni sporulates in epithelia of the digestive tubules. Nevertheless, in the absence of spores, histology does not allow distinguishing between both parasite species (Burreson & Ford, 2004). Specific molecular assays were developed to detect H. costale in oysters including a conventional PCR targeting the 18S (Ko et al., 1995) and a multiplex PCR assay allowing the concurrent detection of H. nelsoni, H. costale and Perkinsus marinus (Penna et al., 2001, 1999; Russell et al., 2000, 2004).
Primary investigations prompted by the report of low Crassostrea gigas mortality revealed the presence of Haplosporidium‐like parasites in oyster tissues. Molecular and ultrastructural analyses were subsequently carried out to characterize the parasite species and pointed out Haplosporidium costale.
Following the report of the parasite Haplosporidium costale for the first time in France, a TaqMan™ real‐time PCR assay was developed in order to allow its rapid and specific detection. Its specificity and sensitivity were determined and it was compared with the conventional PCR assay developed by Stokes and Burreson (2001) and previously recommended by the OIE (OIE, 2003). This TaqMan™ assay was finally tested on archived samples collected in main oyster farming areas in France either in the context of mortality events or studies by the French network of surveillance of mollusc diseases (REPAMO).
2. MATERIAL AND METHODS
2.1. Oyster Crassostrea gigas samples
Between December 2018 and June 2019, low and recurrent mortalities were observed at the Ifremer (Institut français de recherche pour l'exploitation de la mer) nursery facilities in Bouin, Vendée in a batch of juvenile oysters Crassostrea gigas (coded 19–030 hereafter). Cumulative mortality during the nursery period reached 7.2%. Produced in March 2018 in the Ifremer hatchery in La Tremblade, Charente Maritime, these oysters were transferred to the Ifremer nursery in Bouin in May 2018. In March 2019, moribund oysters (17 individuals, weighting around 10 g) were collected to carry out primary investigations.
A second sampling (10 moribund oysters) was done in June 2019 at the Ifremer experimental farm located in the Seudre River in La Tremblade on a batch of adult oysters (coded 19–082 hereafter) showing 0.004%/week mortality rate. Initially produced in the Ifremer hatchery in La Tremblade in June 2016, these oysters grew in the facilities in La Tremblade except between August and October 2017, and between July 2018 and June 2019 when they grew in the facilities in Bouin.
During the production period in La Tremblade and Bouin, oysters were exposed to a 400 L/h seawater flow enriched with a cultured phytoplankton diet (Isochrysis galbana, Tetraselmis suecica and Skeletonema costatum) provided ad libitum (50,000 cells/ml).
2.2. Oyster tissue processing and diagnostic approach
At reception, moribund oysters were opened, checked for the presence of macroscopic signs and processed as described in the procedure available on the EURL for mollusc diseases website (https://www.eurl‐mollusc.eu/content/download/143184/file/SampleProcessing_Edition1.pdf).
Sections of organs including gills, mantle, gonad and digestive gland were prepared from each oyster for histology. In addition, about 50 mg of gills, adductor muscle, digestive gland and mantle were collected for bacteria isolation and remaining tissues were frozen (sample 19–030) or fixed in absolute ethanol for molecular analyses (sample 19–082).
Presumptive methods including real‐time PCR for the detection of OsHV‐1 from approximately 25 mg of mantle and gills tissues according to Martenot et al. (2010) and real‐time PCR for the detection of bacteria belonging to Vibrio Splendidus clade or Vibrio aestuarianus species from main isolated bacteria according to Saulnier et al. (2017) were carried out and did not yield positive results except the detection of bacteria belonging to the Splendidus clade in some individuals.
Consequently, histological examination of tissues was done in order to test the presence of parasites or lesions. Among oysters collected in June, gill and digestive gland imprints were also done from one individual showing abnormal black colouration of the soft tissues. The observation of Haplosporidium‐like parasites in histology and on imprints prompted to carry out molecular analyses, in situ hybridization and ultrastructural examination that are described below.
2.3. Imprints, histology and in situ hybridization
Gill and digestive gland imprints were fixed in absolute ethanol, stained with Hemacolor (Merck) and directly observed on a BX50 microscope (Olympus).
After at least 48 h in Davidson's fixative, tissue sections were maintained in 70% ethanol until they were dehydrated and embedded in paraffin for histology according to standard procedures (Howard et al., 2004). Two‐ to three‐micrometre thick tissue sections were stained with haematoxylin and eosin. Slide examination was done using a BX50 (Olympus) microscope. Infection intensity was determined according to the criteria established by da Silva and Villalba (2004). In situ hybridization protocol was adapted from (Stokes & Burreson, 2001). Three‐micrometre thick tissue sections on silane‐prep™slides (Sigma, France) were dewaxed, rehydrated and treated with proteinase K [100 μg/ml in TE buffer (Tris 50 mM, EDTA 10 mM)] at 37°C for 10 min. Slides were dehydrated by immersion in an absolute ethanol bath and air‐dried. Sections were then incubated with 100 μl of hybridization buffer [50% formamide, 10% dextran sulphate, 4× saline‐sodium citrate buffer (0·06 M Na3 citrate, 0·6 M NaCl, pH 7), 250 μg/ml yeast tRNA and 10% Denhardt's solution] containing 5 ng/μl of 3′digoxigenin‐labelled SSO1318 oligoprobe (Eurogentec).
Target DNA and digoxigenin‐labelled probe were denatured at 95°C for 5 min and hybridization was carried out overnight at 42°C. Sections were washed in 2× SSC at room temperature (RT) (2 × 5 min), in 0·4× SSC at 42°C (10 min) and in solution I (100 mM maleic acid, 0·15 M NaCl, pH 7·5) for 5 min. Tissues were then incubated in blocking reagent (Roche) (1% w/v) in solution I for 30 min at room temperature. Specifically bound probe was detected using an alkaline phosphatase‐conjugated mouse IgG antibody against digoxigenin diluted at 1·5 U/ml in solution I (1 h, RT). Excess of antibody was removed by two washes in solution I (1 min) and equilibrated in solution II (0·1 M Tris pH 8, 0·1MNaCl, 0·05 M MgCl2, pH 9·5). Slides were incubated in NBT/ BCIP, a chromogenic substrate for alkaline phosphatase, diluted in solution II (20 μl/ml) in the dark. Slides were subsequently observed using a BX 50 microscope (Olympus). Negative controls included samples without digoxigenin‐labelled probe in the hybridization mixture or without antibodies during the revelation step. Positive control consisted of Crassostrea virginica infected with Haplosporidium costale originating from the United States (kindly provided by R. Carnegie).
2.4. Molecular analyses for the detection and characterization of Haplosporidium costale
2.4.1. DNA extraction
Total DNA was extracted from approximately 25 mg of −20°C frozen gill, digestive gland and mantle tissues using the Wizard® Genomic DNA Purification Kit (Promega, Inc.) according to the manufacturer's protocol. Extracted samples were stored at 4°C until being tested by PCR.
2.4.2. DNA amplification
Different combinations of primers were used to test the presence of the parasite Haplosporidium costale and to amplify parts of its genome (Table 1). PCR tests were carried out using a T100 thermal cycler (BioRad). For primers previously described, PCR analyses were done following authors’ recommendations (Table 1). For primers designed in the present study, reactions of 20 μl contained 1× buffer (Promega), 1.5 mM MgCl2, each primer at 0.5 μM, each dNTP at 0.125 μM, 2 U goTaq polymerase (Promega) and 200–250 ng template DNA. Cycling parameters were 94°C for 5 min; 40 cycles of amplification at 94°C for 1 min; annealing temperature as described in Table 1 for 1 min. Final elongation was carried out at 72°C for 10 min.
TABLE 1.
Primers used in this study to test the presence of parasites of the genus Haplosporidium and to sequence parts of its genome
| Primer pair | Sequences | Tm | Amplicon expected size | Region amplified | Reference |
|---|---|---|---|---|---|
| HapF1 | GTTCTTTCWTGATTCTATGMA | 49 | 330 bp | 18S SSU rDNA | Renault et al., 2000 |
| HapR2 | GATGAAYAATTGCAATCAYCT | 49 | |||
| HPNF3 | CATTAGCATGGAATAATAAAACACGAC | 55 | 600 bp | 18S SSU rDNA | Catanese et al., 2018 |
| HPNR3 | GCGACGGCTATTTAGATGGCTGA | 55 | |||
| SSO‐A | CACGACTTTGGCAGTTAGTTTTG | 55 | 550 bp | 18S rRNA gene | Stokes & Burreson, 2001 |
| SSO‐B | CGAACAAGCGCTAGCAGTACAT | 55 | |||
| 16SA | AACCTGGTTGATCCTGCCAGT | 65 | ∼1400 bp | 18S rRNA gene | Medlin et al., 1988 |
| 16SB | TGATCCTTCTGCAGGTTCACCTAC | 65 | |||
| Haplo18Sl_int_1F | GAAACGGCTACCACATCCAC | 50 | 630 bp | 18S rRNA gene | This study |
| Haplo18Sl_int_1R | CCCCGGCTTTAGTTCTTGAT | 50 | |||
| ITSf | GGGATAGATGATTGCAATTRTTC | 45 | ITS | Hill et al., 2010 | |
| ITS‐B | TATGCTTAAATTCAGCGGGT | 45 | ∼800 bp | ITS | Carnegie et al., 2014 |
| ITS 2.2 | CCTGGTTAGTTTCTTTTCCTCCGC | 45 | ∼600 bp | ITS | |
| Actin_Hapl1_F | ACTGGCATTGTGCTTGACAG | 52 | ∼400 bp | Actin gene | This study |
| Actin_Hapl1_R | TTCACAGTGATCGTGGAAGG | 52 | |||
| Actin_Hapl2_F | GATGTACGTCGGGATTCAGG | 52 | ∼300 bp | ||
| Actin_Hapl2_R | TGTGCGGGTACATCGTAGTG | 52 |
Positive and negative controls were included in each PCR run. Positive controls consisted of DNA extracted from known infected samples. Negative controls consisted of bi‐distilled water used in the extraction and real‐time PCR steps.
Amplification products were resolved on 1% agarose gels stained with ethidium bromide and visualized using UV illuminator. Expected amplicon size is provided for each primer pair in Table 1.
Correct size products were excised from the gels, purified using Amicor Kit (Millipore) and sequenced.
2.4.3. Sequencing
Purified PCR products were sent to LightRun Sequencing to the company Eurofins genomics, DE. Chromatograms were analysed using FinchTV 1.4 (Geospiza).
Some purified PCR products were home sequenced using the ABI Prism Big Dye Terminator v3.1 sequencing kit following manufacturer's instructions. DNAs were sequenced using the ABI 3130xl Avant Genetic Analyses (Applied Biosystems). Analyses of the sequences were completed using Finch TV (Biospiza).
2.4.4. Phylogenetic analyses
Sequences obtained were compared with those in the GenBank database using the BLAST algorithm (Altschul et al., 1997).
2.4.5. 18S sequences
Available 18S gene sequences from Haplosporidian organisms and the Paramyxida Marteilia refringens were downloaded from GenBank and included in the phylogenetic analyses together with the sequences longer than 1000 bp obtained in the present study. Alignments were performed using ClustalW (Thompson et al., 1994) in MEGA 7 (Kumar et al., 2016) with default parameters. The analysis involved 46 nucleotide sequences and a total of 2071 characters.
Prior to the phylogenetic analysis, the program jModelTest 0.1.1 (Posada, 2008) was used to select the best fitting substitution model according to the corrected Akaike information criterion (AIC) (Hurvich & Tsai, 1993). A total of 88 candidate models including models with equal/unequal base frequencies, with/without a proportion of invariable sites (+I) and with/without rate variation among sites (+G) were tested.
The best‐fit model of nucleotide substitution was the GTR+I+G model. Tree topology was therefore inferred based on a Bayesian approach using MrBayes v 3.1.2 (Huelsenbeck & Ronquist, 2001) and implementing the GTR + I + G model of nucleotide substitution.
2.4.6. Internal transcribed spacer (ITS) regions, including the ITS 1, 5.8S rRNA gene and ITS 2
Available ITS1–5.8S–ITS2 sequences from Carnegie et al. (2014) were downloaded from GenBank and aligned with sequences obtained in this study as described above.
Evolutionary divergence was estimated between sequences obtained in this study and closest sequences found in GenBank using the Maximum Composite Likelihood model (Tamura et al., 2004). The rate variation among sites was modelled with a gamma distribution (shape parameter = 1). Differences in the composition bias among sequences were considered in evolutionary comparisons (Tamura & Kumar, 2002). Evolutionary analyses were conducted in MEGA 7 (Kumar et al., 2016).
The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated, resulting in a total 409 positions.
2.4.7. Ultrastructural examination (19–082)
Pieces (1 mm3) of mantle, gills and digestive gland of one oyster showing an abnormal black colouration and for which examination of imprints revealed the presence of parasites looking like parasites of the genus Haplosporidium were immediately fixed with 4% glutaraldehyde for 24 h at 4°C. The samples were then washed in 0.2 M cacodylate buffer (3 × 30 min) and post‐fixed for 1 h in 1% osmium tetroxide (OsO4) in 0.2 M cacodylate buffer, cleared in propylene oxide and embedded in epon resin. Ultrathin sections were obtained using copper grids and double‐stained with 5% uranyl acetate and 5% lead citrate and then examined at 80 kV on a JEOL 1110 transmission electron microscope equipped with a Morada digital camera and iTEM imaging software (Soft Imaging System, Olympus).
2.5. Real‐time TaqMan™ PCR development
2.5.1. Oyster samples
For the development of the real‐time PCR, the oyster samples coded 19–030 and 19–082 described above were used.
Diagnostic sensitivity and specificity of the new PCR assay were determined using oyster samples from three populations displaying presumably different levels of prevalence (488 oysters in total): (i) an oyster population supposed to be parasite free from a high biosecurity level area of Ifremer experimental facilities in Bouin, (ii) an oyster population with a presumably moderate parasite prevalence (10–30%) including oysters from contaminated facilities at Bouin in which the parasite was initially described and (iii) a wild oyster population from Vendée with a presumably low parasite prevalence (<10%) (Supplementary Material Table S1).
In addition, the presence of the parasite in previous years was checked using the new PCR assay in archived Crassostrea gigas DNA material obtained in different contexts: mortality, macroscopic abnormalities or field studies (263 individuals from 17 batches, information available in Table 9).
TABLE 9.
Detection frequency of Haplosporidium costale DNA in samples of farmed oysters collected in two main French oyster farming areas (Bassin de Marennes Oléron and Bassin d'Arcachon) since 2008
| Number of samples positive by real‐time PCR by range of CT values | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Geographical area (total number of batches) | Year | Sampling dates | Site/GPS coordinates | Oyster age | Context of sampling: notified mortalities, macroscopic anomalies or field study | Detection frequency (No. of positive/No. of tested individuals) | <30 | >30 and < 37 | >37 |
| Bassin de Marennes Oléron (12) | 29% (59/205) | ||||||||
| 2009 | 20/08/2009 | Agnas; 45.873056, −1.170833 | 1–2 years | field study | 25% (3/12) | 0 | 2 | 1 | |
| 2011 | 03/05/2011 | Agnas; 45.873056, −1.170833 | 1–2 years | field study | 0% (0/12) | ||||
| 18/08/2011 | Barat; 45.808806, −1.159970 | >2 years | macroscopic anomalies (black gills) | 0% (0/5) | |||||
| 2012 | 16/10/2012 | La Baudissière; 45.924259, −1.233623 | >2 years | mortality | 25% (3/12) | 0 | 0 | 3 | |
| 14/11/2012 | Agnas; 45.873056, −1.170833 | 1–2 years | field study | 10% (1/10) | 0 | 0 | 1 | ||
| 2013 | 16/05/2013 | Artouan; 45.762818, −1.088398 | 1–2 years | field study | 67% (8/12) | 0 | 4 | 4 | |
| 07/08/2013 | Lamouroux; 45.903214, −1.144970 | >2 years | mortality | 10% (2/20) | 0 | 1 | 0 | ||
| 2014 | 08/04/2014 | La Floride; 45.803243, −1.153969 | >2 years | field study | 10% (2/20) | 0 | 1 | 1 | |
| 04/09/2014 | Agnas; 45.873056, −1.170833 | >2 years | mortality | 42% (5/12) | 0 | 2 | 3 | ||
| 2016 | 01/09/2016 | La Floride; 45.803243, −1.153969 | >2 years | field study | 0% (0/30) | ||||
| 01/09/2016 | La Floride; 45.803243, −1.153969 | >2 years | field study | 53% (16/30) | 0 | 6 | 10 | ||
| 2017 | 28/08/2017 | La Floride; 45.803243, −1.153969 | >2 years | field study | 19/30 | 1 | 15 | 3 | |
| Bassin Arcachon (5) | 31% (18/58) | ||||||||
| 2008 | 17/07/2008 | Le Tès; 44.665, −1.13829 | >2 years | mortality | 60% (6/10) | 0 | 4 | 2 | |
| 2013 | 04/07/2013 | Grand Banc; 44.680440, −1.201571 | >2 years | mortality | 17% (2/12) | 0 | 1 | 1 | |
| 25/07/2013 | Marens; 44.704443, −1.204934 | 1–2 years | mortality | 33% (4/12) | 0 | 3 | 1 | ||
| 2014 | 02/04/2014 | Gujan; 44.655431, −1.067254 | >2 years | mortality | 15% (3/20) | 0 | 0 | 3 | |
| 2016 | 21/11/2016 | Courbey; 44.690785, −1.230244 | >2 years | macroscopic anomalies (adductor muscle calcification) | 75% (3/4) | 0 | 3 | 0 | |
Bold values are detection frequencies at the geographic area scale.
2.5.2. Oyster DNA extraction
DNA extraction was performed on approximately 25 mg of gill, digestive gland and mantle tissues from fresh oyster tissues or archived material frozen or conserved in ethanol as described above.
2.5.3. Primers and probe design
We used the primer set targeting the 18s gene (1358F 1507R) initially designed for conventional PCR (Ko et al., 1995). In addition, two probes were designed using Primer Express® Software v3.0 (Applied Biosystems, Foster City, CA, USA) after comparing Haplosporidium costale sequence with closely related ones: Haplosporidium lusitanicum (AY449713.1), H. pickfordi (AY452724.1), H. edule (DQ458793.1), H. raabei (HQ176468.1), H. montforti (DQ219484.1) and H. nelsoni (U19538.2). The set was validated in silico using NCBI primerBLAST (Basic Local Alignment Search Tool; www.ncbi.nlm.nih.gov/blast/). The sequences were as follows: 1358F (forward primer): 5′‐TACTGCTAGCGCTTGTTCGCAAGAT‐3′; 1507R (reverse primer): 5′‐TCGGGTCGGCCCGCTGACTGGGT‐3′; Probe1 (probe): 5′‐FAM‐ GAAGGTCTGGGCTGCACGCG‐3′‐TAMRA; Probe2: 5′‐FAM‐ AGGGACAATCTGTGCTCAGCAGATGG‐3′‐TAMRA. The amplicon size was 149 bases. The oligonucleotides were synthesized and purchased from Eurogentec.
2.5.4. Real‐time TaqMan™ assay
DNA (5 μl) was transferred into 15 μl of PCR mix, consisting of TaqMan® Supermix (Biorad, 10 μl), primers 1358F (0.3 μl, 300 nM f.c.), 1507R (0.3 μl, 300 nM f.c.) and Probe2 (0.3 μl, 300 nM f.c.). Ultrafast QPCR Mastermix® (Agilent) was also tested in similar conditions. Assays were run on Mx3000 and CFX Connect machines (Agilent, Biorad). The reaction was run at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s.
To confirm the specificity of the amplification observed in real‐time PCR on the sample 19–030, PCR products were sequenced using the primers 1358F 1507R (Ko et al., 1995). Briefly, PCR products were purified by ExoStar (GE Healthcare‐Life Sciences). Sequence reactions were performed with BigDye® Terminator V3.1 (Applied Biosystems), followed by dye purification with BigDye X‐terminator™ purification kit and sequencing with ABI3003xl (Applied Biosystems). All sequences were identical and corresponded to the targeted region, as confirmed by Blast analyses.
A sample showing a fluorescent signal exceeding the fluorescent background level was considered positive regardless of the threshold cycle (CT) obtained and a sample showing no fluorescent signal above the background level was considered negative.
2.6. Validation
2.6.1. Inclusivity and exclusivity tests
Inclusivity and exclusivity were evaluated by testing a panel of samples infected with Haplosporidium costale parasites or with closely related parasites. These samples were from the collection of the National and European Union Reference Laboratories for Mollusc Diseases and previously characterized (information available in Table 6).
TABLE 6.
Panel of tested samples and results of inclusivity and exclusivity tests
| Parasite species | Isolation source | Identification method | TaqMan real‐time PCR result (+ /–) | |
|---|---|---|---|---|
| Inclusivity | Haplosporidium costale (n = 3) | Oyster, Crassostrea virginica (USA provided by Ryan Carnegie, VIMS), Crassostrea gigas (France: Vendée) | Histology, ISH, PCR and gene sequencing | + (100%) |
| Exclusivity | Haplosporidium nelsoni (n = 1) | Oyster, Crassostrea virginica (USA, provided by Ryan Carnegie, VIMS) | Histology, PCR and gene sequencing | – (0%) |
| Haplosporidium nelsoni (n = 3) | Oyster, Crassostrea gigas (France: Thau, Cancale, Brest) | Histology, PCR and gene sequencing | ||
| Haplosporidium pinnae (n = 2) | Pinna nobilis (France: Corsica and Calanques) | Histology, PCR and gene sequencing | ||
| Haplosporidium armoricanum (n = 1) | Flat oyster, Ostrea edulis (the Netherlands provided by Marc Engeslma, WUR) | Histology, PCR and gene sequencing | ||
| Minchinia tapetis (n = 2) | Cockle, Cerastoderma edule (France: Arcachon) | Histology, PCR and gene sequencing | ||
| Minchinia mytili (n = 2) | Mussels, Mytilus edulis (France: Baie des Veys) and Mytilus galloprovincialis (France: Thau) | Histology, PCR and gene sequencing | ||
| Bonamia ostreae (n = 1) | Flat oyster, Ostrea edulis (France: Baie de Quiberon) | Histology, PCR and gene sequencing |
Note: –, negative results; +, positive results.
2.6.2. Analytical characteristics
The real‐time PCR limit of detection (LDpcr) was determined using a synthetic plasmid including 691 bp (from position 817 to 1507) of Haplosporidium costale 18S gene in pUC57 (Eurogentec) as well as naturally infected oyster samples (samples 19–030 and 19–082 detailed above).
Three independent trials were performed on three independent twofold serial dilutions (10–0.375 DNA copies per μl of template), with eight replicates of each dilution level. LDpcr was determined as the lowest number of target nucleic acid generating 95% of positive results. Standard curves were generated by linear regression analysis of the CT values versus the log10 copy number for each standard dilution. The measured quantity for each dilution level was determined in retrospect by using the formula: log x = [CT − b]/a, where a is the slope, b is the Y intercept and x is the quantity. The quantification limit of the assay was then determined according to the model proposed in the standard NF U47‐600‐2 (Agence française de normalization, 2015).
Repeatability, preliminary reproducibility and the influence of matrix DNA concentration on amplification were also estimated (Supplementary Material S1).
2.6.3. Diagnostic characteristics
Diagnostic sensitivity (DSe) and specificity (DSp) were assessed using Bayesian latent approach by comparing the conventional PCR assay developed by Stokes and Burreson (2001) and the new real‐time PCR. Parameters were estimated by Markov Chain Monte Carlo (MCMC) methods via Gibbs sampling. Analysis was performed as described by Joseph et al. (1995) by adapting the model to two methods and three populations. To consider the possible dependence between PCR assays, conditional dependence was modelled using the covariance between methods within the diseased class (Dendukuri & Joseph, 2001; Vacek, 1985) (Supplementary Material S2). Firstly, analysis was performed without informative priors (model 1); additional models were then tested by restricting the prevalence of the negative group to 0 (model 2) and by pre‐determining the conventional PCR DSe (model 3). As few information was available for the conventional PCR targeting Haplosporidium costale, DSe estimation was based on data available for a conventional PCR targeting H. nelsoni, a closely related parasite (Gagné et al., 2015). So, the beta priors (152.8758, 27.8016) were obtained using the betabuster software (downloaded from http://www.epi.ucdavis.edu/diagnostictests/betabuster.html) assuming that the DSe of the conventional PCR was above 80% with a mode set at 85% and 95% certainty. The models were implemented in JAGS within the R statistical software environment using packages rjags. For each model, four Monte–Carlo Markov chains (MCMCs) were run with 50,000 iterations with the first 10,000 iterations discarded as a ‘burn‐in’. Convergence of the MCMC chain was assessed by confirming that the history plots ran stably and did not switch to different regions.
3. RESULTS
3.1. Imprint examination, histology and in situ hybridization
In sample 19–030, 11 out of 13 oysters tested in histology showed Haplosporidium parasites (Table 2) including uninucleated (Figure 1a), plasmodial stages (Figure 1b) and sporonts in connective tissues of the digestive gland, gills, gonadal follicles, labial palps, kidney and muscle. Plasmodial stages appeared binucleated or with few nuclei. Some of them had an irregular shape. Some ‘foamy’ forms were also noticed in the connective tissues of the digestive gland (Figure 1c). Infection level was light to high and the presence of the parasite appeared associated with haemocyte infiltration (Figure 1d), necrosis, abnormal nuclear pictures and the presence of undetermined bacteria.
TABLE 2.
Summary of the results (number of positive individuals/number of tested individuals) obtained on the two samples of oysters tested by histology, imprints, in situ hybridization (ISH) and PCR regarding the presence of Haplosporidium costale
FIGURE 1.
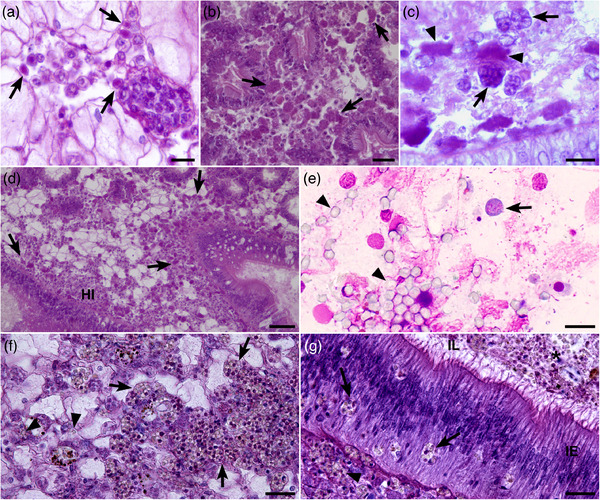
Histological haematoxylin eosin tissue sections (a–d, f and g) and Hemacolor® tissue imprints (e) showing Haplosporidium costale parasites in different tissues of Crassostrea gigas. (a) Uninucleated parasites (arrows) in a venous sinus of the mantle. (b) Plasmodial stages with an irregular form (arrows) in the connective tissue of the digestive gland. (c) Foamy form (arrow) and plasmodial stage (arrowhead) in the connective tissue of the digestive gland. (d) Haemocytic infiltration (HI) in the connective tissue of the digestive gland associated with parasite plasmodial stages (arrows). (e) Plasmodial cell (arrow) and spores (arrowhead) on digestive gland imprint. (f) Sporonts (arrows) and free spores (arrowhead) in the connective tissue around gonad follicles. (g) Sporonts and spores in different positions around the intestine: sporonts (arrowhead) in the connective tissue of intestine, sporonts (arrow) inside the intestine epithelium (IE) and spores (*) into the intestine lumen (IL). Scale bars: a, c and e = 10 μm; b, f and g = 20 μm; d = 50 μm
In sample 19–082, in situ parasites including spores were observed on gill and digestive gland imprints prepared from the oyster (Table 2; Figure 1e) showing an abnormal black colour of the soft tissues. Out of the 10 tested oysters in histology, only this oyster appeared infected with parasites, mostly spores, invading the connective tissues of the digestive gland, gills and around the gonad (Figure 1f). Some spores could be seen crossing the digestive epithelia (Figure 1g). No haemocyte infiltration was observed in this infected oyster. The other tested individuals showed haemocyte infiltration or necrosis but no parasite were detected.
In situ hybridization revealed positive labelling in all the oysters found infected with the parasite in histology. Although all parasite stages appeared recognized by the probe, spores showed less or sometimes low labelling compared to other parasite stages (not shown).
3.2. Molecular characterization
Depending on the primer pair, amplification at the expected size was obtained for 10–17 of 17 oysters from sample 19–030 and for 1–5 of 10 oysters for sample 19–082 (Table 2).
Some PCR products were sequenced (Table 3). For a same primer pair, sequences were identical. Whatever the primer pair used, these sequences showed 99%–100% identity with Haplosporidium costale (KC578010.1; AF387122.1) (Table 3).
TABLE 3.
Number of sequences obtained by genome fragment and oyster sample and results of the BlastN analysis
| BlastN results | ||||||
|---|---|---|---|---|---|---|
| Gene | Primers (expected size) | Number of obtained sequences 19–030 | Number of obtained sequences 19–082 | Identity | Closest match (reference number) | Species |
| 18S | SSOA‐SSOB (557 nt) | 7 | 2 | 100% (532 nt) | KC578010.1 | Haplosporidium costale |
| HapF1‐ HapR1 (330 nt) | 7 | – | 100% (295 nt) | AF387122.1 | Haplosporidium costale | |
| HPNF3‐HPNR3 (600 nt) | 6 | – | 99.62% (527 nt) | KC578010.1 and JX977120.1 | Haplosporidium costale | |
| Long fragment (combination of primers) | 1 | ‐ | 99.45% (1445 nt) | AF387122.1 and JX977120.1 | Haplosporidium costale and Haplosporidium sp. from Saccostrea glomerata | |
| ‐ | 1 | 98.31% (1354 nt) | AF387122.1 | Haplosporidium costale | ||
| ITS region | Combination of primers | 2 (100% identical) | ‐ | 100% (453 nt) | KF790901.1 | Haplosporidium costale |
| 1 | 99.56% (453 nt) | KF790901.1 | Haplosporidium costale | |||
| Actin | Combination of primers | 1 | 1 | 100% (597 nt) | AY450407.1 | Haplosporidium costale |
In order to extend sequences obtained on the 18S fragment and to amplify the internal transcribed spacer (ITS) region and actin gene, different primer pairs were tested and combined (Table 1).
Two sequences, 1445 and 1354 bp in size, were obtained on the 18S fragment for samples 19–030 and 19–082, respectively (GenBank references: MZ666334 and MZ666335). These sequences displayed between 98.31% and 99.45% identities with Haplosporidium costale (AF387122.1) and Haplosporidium sp. from Saccostrea glomerata (JX977120.1)
Three 453 bp sequences were obtained for the ITS region including partial small subunit ribosomal RNA gene, the ITS 1, 5.8S rRNA gene, ITS 2, and partial sequence of the large subunit ribosomal RNA gene. Two sequences obtained from oysters from sample 19–030 were 100% identical (GenBank reference: MZ666374) and the third one from 19–082 (GenBank reference: MZ666375) showed two transversions (G instead of C and A instead of C in position 267 and 272 respectively in 19–082). These sequences appeared 99.56% and 100% identical with Haplosporidium costale (KF790901.1).
Finally, two identical sequences (598 nt), one from each oyster sample, were obtained for the actin gene (GenBank reference: MZ673037). These sequences were 100% identical with H. costale (AY450407.1).
Phylogenetic analysis carried out on 18S long sequences from Haplosporidian organisms and using Marteilia refringens as outgroup showed that Haplosporidian species were included in a paraphyletic group (Figure 2). Species of the genus Haplosporidium clustered in a main clade separated from species of the genera Bonamia and Minchinia. Whereas Bonamia species formed a monophyletic group well supported by Bayesian posterior probabilities, the relationships between Minchinia species were not clear. Urosporidium species clustered together within the Haplosporidium clade. Among Haplosporidium species, H. nelsoni, H. diporeirae, H. pinnae and H. carcini grouped together whereas apart from H. paragon which grouped with Haplosporidium sp. in Syllis nipponica and Urosporidium species, other species were distributed in two clusters, one including H. edule, H. raabei, H. pickfordi, H. tuxtlensis, H. lusitanicum, H. montforti and a second one with Haplosporidium sp. MYE from Ostrea edulis characterized as H. armoricanum (Engelsma pers comm) and the group of very closely related H. costale and Haplosporidium sp. from Saccostrea glomerata. Both sequences obtained from Crassostrea gigas from France were included in this monophyletic group well supported by Bayesian posterior probability (0.92).
FIGURE 2.
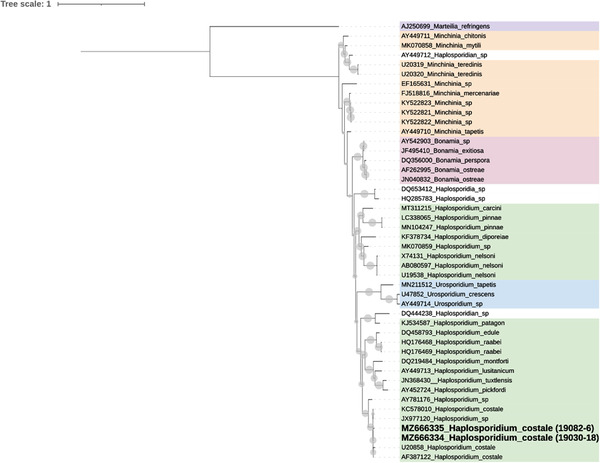
Phylogenetic tree (50% majority‐rule consensus) using Bayesian Inference (MrBayes 3.1.2) based on the small subunit ribosomal gene of Haplosporidian. Circles at the node correspond to Bayesian probabilities superior to 50%. Marteilia refringens was used as the outgroup. Sequences obtained in this study are indicated in bold. Affiliations at a genus level are specified using coloured ranges. The analysis involved 46 nucleotide sequences and a total of 2071 characters. The 46 nucleotide sequences included available 18S gene sequences from Haplosporidian organisms and the Paramyxida Marteilia refringens downloaded from GenBank and 18S sequences longer than 1000 bp obtained in the present study
The alignment of 18S–ITS1–5.8S–ITS2–28S sequences from Haplosporidium costale (KF790901–2) and Haplosporidium sp. from Saccostrea glomerata (KF790894–900) allowed identifying two groups: the first one including two sequences obtained from Crassostrea virginica in North America (Nova Scotia‐Canada and Maine and Virginia, USA) and the second one including the seven sequences from S. glomerata from New South Wales, Australia. Both sequences obtained from Crassostrea gigas from France appeared closer to North American (0.12%) than Australian ones (5.74%) (Table 4).
TABLE 4.
18S–ITS1–5.8S–ITS2–28S sequences: estimates of evolutionary divergence over sequence pairs between groups
| Distance | St. error | ||
|---|---|---|---|
| French seq | Australian seq | 0.0574 | 0.0128 |
| French seq | N. American seq | 0.0012 | 0.0012 |
| Australian seq | N. American seq | 0.0559 | 0.0126 |
Note: The numbers of base substitutions per site from averaging over all sequence pairs between groups are shown. Standard error estimates are shown in the last column. French Seq corresponds to sequences obtained in the present study. Australian seq corresponds to sequences of Haplosporidium sp. from Saccostrea glomerata (KF790894–900). N. American seq corresponds to sequences of H. costale from Nova Scotia‐Canada and Maine and Virginia, USA (KF790901–2).
Among the Northern American and French sequences, polymorphism consisted of 2 transversions for one of the French sequences and a deletion of 29 nucleotides in one Northern American.
The alignment of French, Northern American and Australian sequences showed one variable region framed by two conserved ones. This variable region which probably contains the ITS‐1 appears distinct between French–Northern American and Australian sequences.
3.3. Ultrastructural description
Both plasmodia and sporonts were observed in the connective tissue of gills and digestive gland (Figure 3a). Dimensions and numbers of organelles are reported for each parasite stage in Table 5.
FIGURE 3.
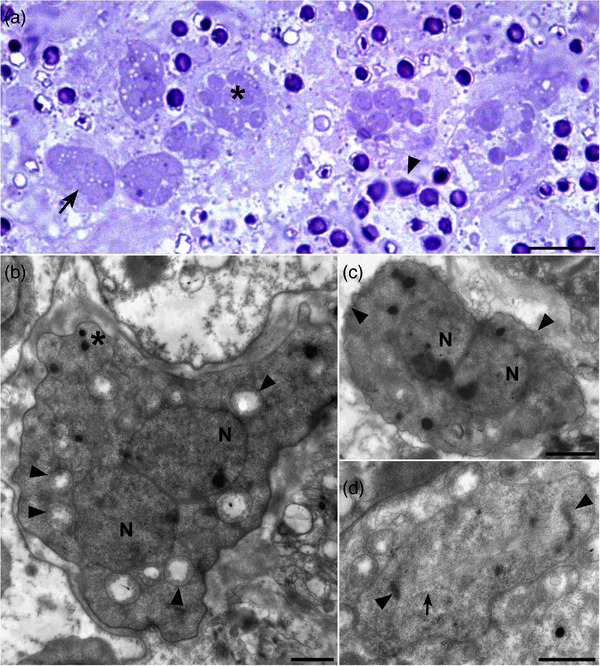
Semithin section (a) and ultrathin sections (b–d) of Haplosporidium costale plamodia infecting different tissues of Crassostrea gigas. (a) Plasmodial (arrow) and sporont (arrowhead) stages in the connective tissue of gills. Note the presence of intermediary stages (*). (b) Plasmodial stage with two nuclei (N). Note the presence of several mitochondria (arrowheads) and some lipid droplets (*) inside the cytoplasm. (c) Plasmodial stage with two nuclei (N) presenting a fibrillar ribbed layer (arrowheads). (d) Nucleus in division presenting intranuclear microtubules (arrow) and centrosomes (arrowheads). Scale bars: a = 10 μm; b, c and d = 1 μm
TABLE 5.
Haplosporidium costale observed in Crassostrea gigas samples in the current study. Main ultrastructural features: dimensions and numbers of organelles for plasmodia, sporonts and spores
| Feature | Mean ± SD | Range | n |
|---|---|---|---|
| Plasmodia | |||
| Length (μm) | 7.1 ± 1.9 | 5–10.2 | 7 |
| Width (μm) | 5.0 ± 1.6 | 3.1–7.1 | 7 |
| Number of nuclei | 1.8 ± 0.9 | 0–3 | 7 |
| Nucleus diameter (μm) | 2.3 ± 0.9 | 1.6–4.7 | 13 |
| Number of lipid droplets | 2.9 ± 1.6 | 1–5 | 7 |
| Number of mitochondria | 5.1 ± 4.3 | 1–15 | 7 |
| Sporont | |||
| Length (μm) | 11.5 ± 2.2 | 7.3–15.0 | 13 |
| Width (μm) | 8.6 ± 1.5 | 6.4–11.9 | 13 |
| Number of spores | 6.8 ± 3 | 2–13 | 13 |
| Spore | |||
| Length (μm) | 2.8 ± 0.6 | 1.6–4.9 | 38 |
| Width (μm) | 2.3 ± 0.4 | 1.4–2.9 | 38 |
| Length endosporoplasm (μm) | 2.6 ± 0.6 | 1.1–4.6 | 38 |
| Width endosporoplasm (μm) | 2.2 ± 0.4 | 1.0–2.9 | 38 |
| Nucleus diameter (μm) | 1.36 ± 0.2 | 0.85–1.63 | 13 |
| Number of endospore lipid droplets | 0.1 ± 0.3 | 0–1 | 38 |
| Number of epispore lipid droplets | 0.1 ± 0.3 | 0–1 | 38 |
| Number of endospore mitochondria | 1.4 ± 1.3 | 0–5 | 38 |
| Number of epispore mitochondria | 1.1 ± 1.3 | 0–5 | 38 |
| Number of endospore dense bodies | 0.4 ± 0.7 | 0–2 | 38 |
| Number of epispore dense bodies | 0.4 ± 0.7 | 0–2 | 38 |
| Number of haplosporosomes | 2.5 ± 4.2 | 0–19 | 38 |
| Number of spherical haplosporosomes | 1.7 ± 3.4 | 0–17 | 38 |
| Haplosporosome spherical diameter (nm) | 120.5 ± 35 | 75–277 | 65 |
| Number of axe‐shaped haplosporosomes | 0.8 ± 1.3 | 0–4 | 38 |
| Axe haplosporosome length (nm) | 343.3 ± 135.3 | 169.3–708.2 | 31 |
| Axe haplosporosome shaft width (nm) | 106.5 ± 20.8 | 67.9–153.3 | 30 |
| Axe haplosporosome head width (nm) | 176.4 ± 37.9 | 100.8–247.9 | 16 |
| Operculum width (μm) | 1.51 ± 04 | 0.9–2.1 | 8 |
| Spore wall thickness (nm) | 82.7 ± 13.4 | 59.5–100.8 | 12 |
3.3.1. Plasmodia
Plasmodia including up to three nuclei appeared amoeboid in shape and measured 7.1 ± 1.9 μm in the long dimension (Figure 3b). A fibrillar ribbed layer was observed around some plasmodial stages (Figure 3c).
Nuclei, 2.3 μm in mean diameter, were often paired suggesting division by fission and usually showed large nucleolus. Intranuclear microtubules were noticed in one plasmod (Figure 3d).
The cytoplasm included up to 15 mitochondria with tubular cristae generally located around the nucleus as well as few lipid droplets (up to 5) (Figure 3b). Smooth and anastomosing endoplasmic reticulum could be observed but not Golgi apparatus.
3.3.2. Sporonts
Sporonts, 11.5 ± 2.2 μm in length and 8.6 ± 1.5 μm in width, with up to 13 uninucleated sporoblasts were observed close to plasmodial stages in the connective tissue of gills and digestive gland (Figure 4a). Young sporonts with sporoblasts showing dotted walls suggesting spore wall formation could be seen (Figure 4b). Sporoblasts measuring 2.3 ± 0.4 μm by 2.8 ± 0.6 μm consisted of endosporoplasms surrounded with a dense episporoplasm which included sometimes one lipid droplet, dense bodies and up to 5 mitochondria (Figure 4c).
FIGURE 4.
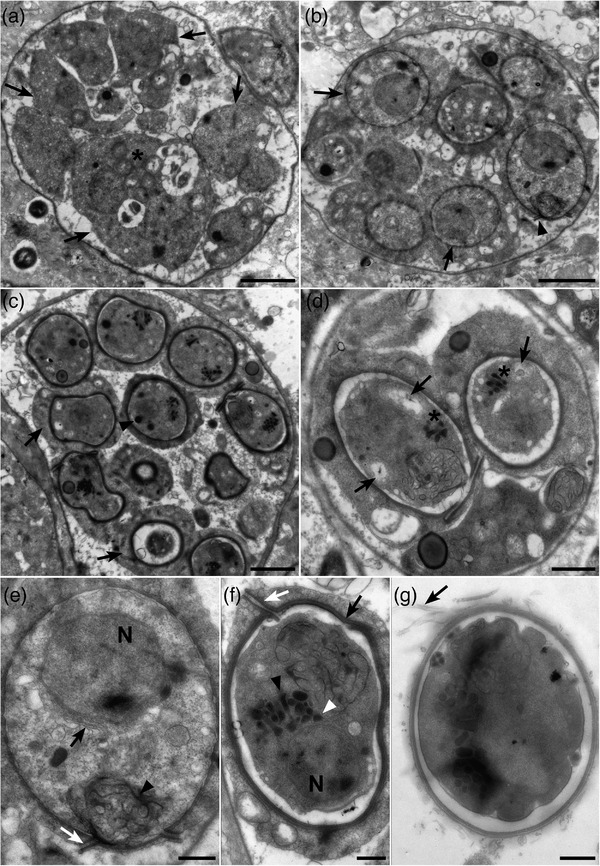
Ultrathin sections of Haplosporidium costale sporonts infecting different tissues of Crassostrea gigas. (a) Early sporont showing different endosporoplasms (arrows) including nucleus, mitochondria (*) but no haplosporosome or spore wall. (b) Sporont with spore wall formation in progress which delimits sporoblast (arrows). Note the presence of an operculum (arrowhead) in a young spore. (c) Sporont with sporoblasts composed of endosporoplasms surrounded with an episporoplasm (arrows). Some mitochondria and lipid droplets can be seen in the episporoplasm. Endosporoplasms include nucleus, mitochondria, lipid droplets (arrowheads) and haplosporosomes. (d) Sporont with two spores containing haplosporosomes (*) and mitochondria (arrows). (e) Spore with smooth endoplasmic reticulum (arrowhead) beneath the operculum (white arrow) and Golgi apparatus (black arrow) near the nucleus (N). (f) Mature spore with a wall consisting in three layers (black arrow) and with an operculum attached to the spore wall by a hinge (white arrow). Note the presence of spherical (white arrowhead) or axehead‐shaped (black arrowhead) haplosporosomes near nucleus (N). (g) Spore presenting epispore cytoplasmic extensions (arrow). Scale bars: a, b and c = 2 μm; d = 1 μm; e, f and g = 0.5 μm
Endosporoblasm length ranged between 1.1 and 4.6 μm and contained one nucleus (1.35 μm in diameter), sometimes one lipid droplet, dense bodies and up to 5 mitochondria. Lipid droplets could be seen in young sporoblasts and rarely in mature ones. On the contrary, numerous haplosporomes (up to 19) located in the endosporoplasm occurred in mature sporoblasts and less in young ones (Figure 4d). Nuclear Membrane Bound Golgi (NM‐BG) (Figure 4e) could be observed. The spherule, consisting of long cisternae of smooth endoplasmic reticulum, was observed more frequently below the operculum in mature spores (Figure 4e). Haplosporosomes appeared spherical or axehead‐shaped (Figure 4f).
The mature spore wall, 82.7 ± 13.4 μm across, included a thick external electron dense layer, a lighter layer and finally an inner denser layer (Figure 4f).
In addition, spores showed an operculum, 0.9–2.1 μm long, attached by a hinge to the spore wall (Figure 4f). The operculum was more often observed in mature spores but sometimes in less mature ones (Figure 4d). No structure looking like filament was clearly observed although some pictures suggest the presence of epispore cytoplasmic extensions (Figure 4g).
3.4. Method development and validation
To allow a large evaluation of Haplosporidium costale DNA presence in French Pacific oyster populations, a real‐time TaqMan PCR method was developed. Two DNA probes were designed and tested at different concentrations in a TaqMan assay amplifying 149 bp of the 18S region (Figure 5). Earlier amplifications and a better efficiency (estimated at 98.5%) were observed with probe 2 at a final concentration of 300 nM. This mix was thus selected.
FIGURE 5.
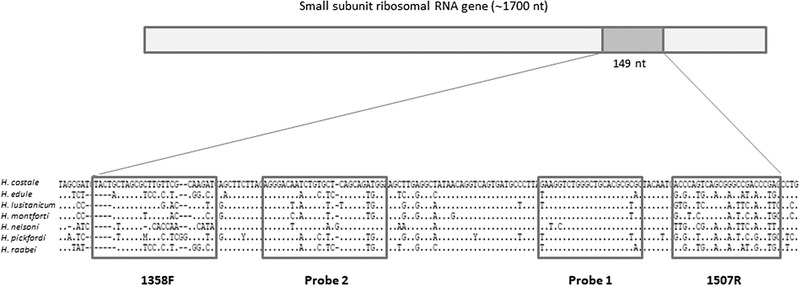
Alignment of 18S sequences of Haplosporidium costale and closely related parasites. Position of primers and probes is represented by boxes
Inclusivity and exclusivity tests yielded expected results with only Haplosporidium costale DNA detected positive whereas DNA from closely related parasites was negative (Table 6). Real‐time PCR detection limit (LDPCR) was evaluated on both infected oyster DNA and plasmidic DNA. LDPCR 95% was estimated at 4.25 copies/μl (Supplementary material Table S2). Moreover, repeatability and preliminary reproducibility of the assay were evaluated and considered as excellent (variations < 5%) (Supplementary material Table S3). The influence of Crassostrea gigas DNA matrix was also tested and no PCR inhibition was observed for DNA concentration ranging between 5 and 250 ng/μl (Supplementary material Table S4).
Finally, real‐time and conventional PCR assays were compared using a panel of field samples (Table 7). No positive results were obtained with both PCR assays in samples from the Haplosporidium costale‐free oyster population. However, the real‐time PCR detected more positive samples than the conventional PCR in populations with low and moderate parasite prevalence.
TABLE 7.
Results obtained with the new real‐time PCR and the conventional PCR (Stokes & Burreson, 2001) on oysters collected from three populations of Crassostrea gigas displaying different prevalence of Haplosporidium costale
| Conventional PCR | Real‐time PCR | Number of individuals | |
|---|---|---|---|
| Parasite‐free population | – | – | 170 |
| – | + | 0 | |
| + | – | 0 | |
| + | + | 0 | |
| Population with low parasite prevalence | – | – | 152 |
| – | + | 19 | |
| + | – | 1 | |
| + | + | 9 | |
| Population with moderate parasite prevalence | – | – | 98 |
| – | + | 7 | |
| + | – | 0 | |
| + | + | 32 |
Note: –, negative results (no amplification by PCR); +, positive results (amplification by PCR).
Diagnostic sensitivity (DSe) and specificity (DSp) were estimated using Bayesian latent class analysis with conditional dependence and different models were evaluated (Table 8). In the three models, the Markov Chain Monte Carlo appeared to converge. All history plots and quantile plots were stable. DSp of both the real‐time and conventional PCR assays was very high in these three models, ranging from 95.5% to 99.5%. In contrast, DSe differed between models and between PCR assays (Table 8). Real‐time PCR DSe was always higher than conventional PCR DSe. The use of priors in conditional dependence (model 3) improved the estimation of parameters specially the DSe which reached 83.6% and 92.6% for the conventional and the real‐time PCR, respectively.
TABLE 8.
Prevalence, diagnostic sensitivity (DSe) and specificity (DSp) of the new real‐time PCR and the conventional PCR (Stokes & Burreson, 2001) assays estimated by Bayesian latent class analysis in conditional dependence with the 95% probability intervals (in parentheses)
| Parameters | Model 1 | Model 2 | Model 3 |
|---|---|---|---|
| Real‐time PCR | |||
| DSe | 57.7 (27–97.9) | 67.4 (30–98.6) | 92.6 (78.2–99.8) |
| DSp | 98.6 (94.8–100) | 98.7 (95.1–100) | 95.5 (92.3–98.6) |
| PCR | |||
| DSe | 38.5 (17–71.1) | 44.8 (18.9–73.7) | 83.6 (77.8–88.7) |
| DSp | 99.4 (98.1–100) | 99.4 (98.2–100) | 99.5 (98.5–100) |
| Prevalence | |||
| Free | 1.2 (0–5.1) | 0 | 0.6 (0–2.3) |
| Low | 27.5 (9.5–59.1) | 23.3 (8.6–52.4) | 8.2 (3.7–14.7) |
| Moderate | 54.5 (25–96.4) | 46.1 (24.2–92.2) | 27.2 (19.6–35.7) |
Note: Three populations of Crassostrea gigas displaying different prevalence of Haplosporidium costale were used in this analysis: Parasite‐free population (Free), population with low parasite prevalence (Low) and population with moderate parasite prevalence (Moderate). Models include model without informative prior (1), model with an estimation of the prevalence of the free population (model 2) and model with prior for the DSe PCR (model 3)
Moreover, depending on the model, the parasite prevalence in the different populations fluctuated. The prevalence was close to zero for the negative population in the models 1 and 3, ranged from 8.2% to 27.5% for the low‐infected population and was between 27.2% and 54.5% for the moderate‐infected population. The lowest values in the different groups were obtained with model 3 using prior (Table 8).
3.5. Application on field samples
Using this real‐time PCR assay, Haplosporidium costale DNA presence was screened on 263 archived samples collected from two main French farming areas located in the Atlantic coast (Marennes Oléron and Arcachon bay) to investigate if the parasite was present before 2019. On average, 44% and 27% of tested oysters were positive in Arcachon bay and in Marennes Oléron bay, respectively. Detection frequency ranged from 0% to 75% in Atlantic samples, depending on the year. Notably, first detection corresponded to oysters sampled in 2008 in Arcachon (Table 9). In this sample collection, H. costale DNA was detected in oysters quasi‐continuously from 2008 to 2017 in France. Indeed, despite the low number of individuals available some years and the lack of samples in 2010 and 2015, positive results were obtained in 2008, 2009, 2012, 2013, 2014, 2016 and 2017 (Table 9).
Finally, parasite DNA could be amplified in oysters sampled during reported mortality events (8 batches), but also in the context of field studies outside any mortality event (6 batches).
4. DISCUSSION
The parasite Haplosporidium costale was first reported to cause high mortalities in the American oyster Crassostrea virginica along the Atlantic coast of Virginia in the 1960s (Andrews, 1984; Wood & Andrews, 1962). The parasite was later reported in C. virginica on the east coast of the United States and on the Atlantic coast of Canada as well as in the Pacific oyster C. gigas in China (Wang et al., 2010).
In contrast with Haplosporidium nelsoni previously detected in Europe (Lynch et al., 2013; Renault et al., 2000), H. costale was considered exotic. Although a parasite looking like H. costale was described in transmission electron microscopy in Crassostrea gigas in France in 1988/1989, the lack of molecular information did not allow concluding about its formal identification (Comps & Pichot, 1991).
In the present study, a combination of microscopy and molecular tools has allowed characterizing the presence of the parasite Haplosporidium costale in the context of low mortality events in Pacific oysters in France.
In histology, plasmodia and spore stages were observed in the connective tissues of the digestive gland, mantle and gonad. A diversity of plasmodial stage shapes was noticed and notably the presence of foamy forms generally found during infection with Haplosporidium costale (Ryan Carnegie, pers com). Haemocytic infiltration was also reported in infected oysters. The parasite distribution in oyster tissues and the association with intense haemocyte infiltration are concordant with H. costale tropism and impact in C. virginica (Burreson & Stokes, 2006).
Ultrastructural features of the parasite detected in France resemble Haplosporidian ones (Hine et al., 2009; Rosenfield et al., 1969) and are closer to Haplosporidium costale and H. armoricanum (Hine et al., 2009; Perkins, 1969; Rosenfield et al., 1969) than other members of the genus. Plasmodia usually show paired nuclei surrounded by mitochondria as described in H. costale in C. virginica (Perkins, 1969). Sporulation appears similar to the spore maturation described for H. costale including the formation of spore wall with an operculum, the presence of a complex membranous organelle, the spherule, as well as the appearance of axe‐headed haplosporosomes (Perkins, 1969). Similarly to H. costale, no filament were observed on spores in our study (Perkins, 1969; Rosenfield et al., 1969). However, some pictures could suggest the presence of epispore cytoplasmic extensions (Figure 4g) as it was reported for H. armoricanum in Ostrea edulis in Galicia (Azevedo et al., 1999).
The presence of Haplosporidian parasites looking like Haplosporidium costale in transmission electron microscopy was earlier reported in three oysters Crassostrea gigas collected from Thau lagoon, South of France (Comps & Pichot, 1991). Interestingly, these parasitized oysters showed a grey colouration as observed for one oyster collected in June 2019 in our study. Sporocyst and spore size as well as the number of spores in sporocysts were smaller in our study compared to the data available in Comps and Pichot (1991) and compared to data available for H. armoricanum (Hine et al., 2007). These features could vary with host species and sites as previously suggested (Perkins & Van Banning, 1981). The resemblance between H. armoricanum and H. costale spores was already highlighted based on ultrastructural investigations (Hine et al., 2009, 2007, 2020). H. armoricanum was characterized based on several examinations of flat oysters from France, Spain and the Netherlands (Azevedo et al., 1999; Bachère & Grizel, 1983; Cahour et al., 1980; Hine et al., 2007; Perkins & van Banning, 1981; van Banning, 1977). The similarity of spores from H. costale and H. armoricanum raises questions regarding their relative taxonomic position. Although no molecular information is currently available for H. armoricanum, Hine et al. (2007) previously reported that their molecular phylogenies were closely related.
As with many non‐cultivable micro eukaryotes, Haplosporidium costale genome is poorly represented in public databases. However, three fragments of the genome including part of the small subunit rRNA gene, the ITS‐1, 5.8S and ITS‐2 array and part of the actin gene were successfully sequenced. The actin gene sequence showed 100% identities with the sequence obtained from Crassostrea virginica from Virginia (USA). Two long 18S sequences (>1360 bp) were obtained and were between 98.31% and 99.45% similar to H. costale from Crassostrea virginica from Virginia (USA) and Haplosporidium sp. from Saccostrea glomerata from New South Wales (Australia). Interestingly, the ITS1, 5.8S and ITS‐2 sequence appeared more discriminating and revealed a higher distance between the French and the Australian isolates than with the North American ones. Our results confirm that the parasite observed in histology and transmission electron microscopy in C. gigas juveniles and adults in France is H. costale. However, more sequence data would be required in order to better evaluate the diversity between these different isolates and between H. costale and the closely related species H. armoricanum.
Disease control requires different diagnostic tools, some generic and others more specific, the choice depending on the surveillance objective. For targeted surveillance, the availability of a rapid and specific diagnostic tool is an advantage because it makes possible to diagnose a large number of individuals in a short time for a given infectious agent. A real‐time PCR assay was thus developed allowing the specific detection of Haplosporidium costale and no other closely related mollusc parasites belonging to the genera Haplosporidium, Minchinia and Bonamia. Primers developed by Ko et al. (1995) were shown to be specific and this specificity was reinforced by the use of a probe. The probe showed no sequence homology with other Haplosporidium species except with H. orchestiae but the primers did not present homology with this species. Several conventional (Ko et al., 1995; Stokes & Burreson, 2001) and multiplex (Penna et al., 2001; Russell et al., 2004) PCR detecting this parasite exist but the development of a real‐time PCR provides several benefits such as a reduced time of analysis, an increased sensitivity and a decreased cross‐contamination risk. This real‐time PCR presented a detection limit between 2.5 and 5 copies/μl which is similar to other real‐time assays developed for other pathogens of molluscs (Canier et al., 2020; Polinski et al., 2015). Furthermore, the sensitivity of this real‐time PCR was not affected by the presence of host genomic DNA even with high DNA load (250 ng/μl host gDNA), contrary to the sensitivity of the real‐time PCR detecting Mikrocytos mackini which was reduced by 20% in presence of 80 ng/μl host gDNA (Polinski et al., 2015). The robustness of this method is particularly interesting when speaking about method transfer to routine diagnostic laboratories.
This new real‐time PCR was compared with the conventional PCR from Stokes and Burreson (2001) on a panel of field samples categorized in three prevalence groups. Diagnostic sensitivity and specificity were estimated using Bayesian analyses. Whatever the model used, real‐time and conventional PCR specificity was high. Meanwhile, the sensitivity varied according to the model and the PCR and was particularly low when no prior was used which was not in agreement with data available in the literature for PCR assays (Aranguren & Figueras, 2016; Caraguel et al., 2012; Ramilo et al., 2013). The addition of prior in the model gave sensitivity values comparable to other PCR or real‐time PCR targeting mollusc pathogens (Canier et al., 2020; Gagné et al., 2015; Polinski et al., 2021). The use of prior is generally recommended in Bayesian methods if prior determination is based on independent data and/or based on expert scientific knowledge (Johnson et al., 2018; Cheung et al., 2021). Finally, this new real‐time PCR assay was used in a survey to screen H. costale DNA presence in France on archived samples collected between 2008 and 2017, partly in the context of mortality events, in two main French oyster producing areas. Parasite DNA has been detected in France at least since 2008. Although no parasite was observed in histology, parasite DNA was detected in 15%–75% of tested animals. Detection frequency of the parasite DNA was not higher in samples collected in the context of mortality events suggesting that the presence of the parasite may not have an impact on the oyster health.
As far as we know, this is the first time that the presence of Haplosporidium costale is confirmed in Crassostrea gigas in France and a TaqMan real‐time PCR assay is developed to enable the rapid and specific detection of the parasite. The application of the PCR assay on archived samples revealed that the parasite has been present in French oyster populations at least since 2008. The lack of impact of the parasite on C. gigas, which seems less susceptible than other species such as C. virginica, certainly explains why it has not been previously reported. The parasite may have been present in France for many years as suggested by the description of Haplosporidium like parasites similar to H. costale in Pacific oysters collected in 1988 and 1989 in Thau lagoon in France (Comps & Pichot, 1991).
Our results raise questions regarding the factors having contributed to the development of the infection and the occurrence of low mortality in experimental batches of Crassostrea gigas (Lupo et al. in preparation). Considering the little information available on this parasite, the newly developed TaqMan assay will be very helpful to investigate the temporal and geographic distribution and the life cycle of the parasite in France and more generally in C. gigas geographic range.
CONFLICT OF INTEREST
The authors declare no conflicts of interests in relation to this work.
ETHICAL APPROVAL
No ethical approval was required.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
Authors are very grateful to Ryan Carnegie (VIMS, USA) and Marc Engelsma (WUR, the Netherlands) for having provided reference material for Haplosporidium nelsoni and H. armoricanum, respectively.
This work was funded by the French Institute for Exploitation of the Sea (Ifremer) and by DGAL (French National Services of Food and Agriculture) through the National Reference Laboratory for Mollusc Diseases, Ifremer, La Tremblade.
Arzul, I. , Garcia, C. , Chollet, B. , Serpin, D. , Lupo, C. , Noyer, M. , Tourbiez, D. , Berland, C. , Dégremont, L. , & Travers, M.‐A. (2022). First characterization of the parasite Haplosporidium costale in France and development of a real‐time PCR assay for its rapid detection in the Pacific oyster, Crassostrea gigas . Transboundary and Emerging Diseases, 69, e2041–e2058. 10.1111/tbed.14541
DATA AVAILABILITY STATEMENT
The sequence data have been submitted to the GenBank databases under accession numbers MZ666334, MZ666335, MZ666374, MZ673037.
REFERENCES
- Agence française de normalisation . (2015). Méthodes d'analyse en santé animale ‐ PCR (réaction de polymérisation en chaîne) – Partie 2: Exigences et recommandations pour le développement et la validation de la PCR en santé animale. Standard NF U47‐600‐2, Février. 51 p.
- Altschul, S. , Madden, T. L. , Schäffer, A. A. , Zhang, J. , Zhang, Z. , Miller, W. , & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, J. D. (1984). Epizootiology of diseases of oysters (Crassostrea virginica), and parasites of associated organisms in eastern North America. Helgoländer Meeresuntersuchungen, 37(14), 149–166. 10.1007/BF01989300 [DOI] [Google Scholar]
- Andrews, J. D. , & Castagna, M. (1978). Epizootiology of Minchinia costalis in susceptible oysters in Seaside Bays of Virginia's Eastern Shore, 1959–1976. Journal of Invertebrate Pathology, 32(2), 124–138. 10.1016/0022-2011(78)90022-8 [DOI] [Google Scholar]
- Aranguren, R. , & Figueras, A. (2016). Moving from histopathology to molecular tools in the diagnosis of molluscs diseases of concern under EU legislation. Frontiers in Physiology, 7, 538. 10.3389/fphys.2016.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzul, I. , & Carnegie, R. B. (2015). New perspective on the haplosporidian parasites of molluscs. Journal of Invertebrate Pathology, 131, 3242. 10.1016/j.jip.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Azevedo, C. , Montes, J. , & Corral, L. (1999). A revised description of Haplosporidium armoricanum, parasite of Ostrea edulis L. from Galicia, northwestern Spain, with special reference to the spore‐wall filaments. Parasitology Research, 85(12), 977–983. 10.1007/s004360050669 [DOI] [PubMed] [Google Scholar]
- Bachère, E. , & Grizel, H. (1983). Mise en évidence d’Haplosporidium sp. (Haplosporida–Haplosporidiidae) parasite de l'huître plate Ostrea edulis . Revue des Travaux de l'Institut des Pêches Maritimes, 46, 226–232.
- Burreson, E. M. , & Ford, S. E. (2004). A review of recent information on the Haplosporidia, with special reference to Haplosporidium nelsoni (MSX disease). Aquatic Living Resources, 17, 499–517. [Google Scholar]
- Burreson, E. M. , & Stokes, N. A. (2006). 5.2.2 Haplosporidiosis of oysters. In Executive Committee (ed.), Suggested procedures for the detection and identification of certain finfish and shellfish pathogens. Diseases of Molluscan Shellfish. American Fisheries Society's Fish Health Section. Chapter 5. [Google Scholar]
- Burreson, E. M. , Stokes, N. A. , & Friedman, C. S. (2000). Increased virulence in an introduced pathogen: Haplosporidium nelsoni (MSX) in the Eastern Oyster Crassostrea virginica . Journal of Aquatic Animal Health, 12(1), 18. 10.1577/1548-8667(2000)012<0001:IVIAIP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cahour, A. , Poder, M. , & Balouet, G. (1980). Présence de Minchinia armoricana (Haplosporea, Haplosporida) chez Ostrea edulis d'origine francaise. Compte Rendu des Séances de la Société de Biologie, 174, 359–368. [Google Scholar]
- Canier, L. , Dubreuil, C. , Noyer, M. , Serpin, D. , Chollet, B. , Garcia, C. , & Arzul, I. (2020). A new multiplex real‐time PCR assay to improve the diagnosis of shellfish regulated parasites of the genus Marteilia and Bonamia . Preventive Veterinary Medicine, 183, 105–126. 10.1016/j.prevetmed.2020.105126 [DOI] [PubMed] [Google Scholar]
- Caraguel, C. , Stryhn, H. , Gagné, N. , Dohoo, I. , & Hammell, L. (2012). Use of a third class in latent class modelling for the diagnostic evaluation of five infectious salmon anaemia virus detection tests. Preventive Veterinary Medicine, 104(12), 165–173. 10.1016/j.prevetmed.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Carnegie, R. B. , Arzul, I. , & Bushek, D. (2016). Managing marine mollusc diseases in the context of regional and international commerce: Policy issues and emerging concerns. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1689), 2015.0215. 10.1098/rstb.2015.0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnegie, R. B. , Hill, K. M. , Stokes, N. A. , & Burreson, E. M. (2014). The haplosporidian Bonamia exitiosa is present in Australia, but the identity of the parasite described as Bonamia (formerly Mikrocytos) roughleyi is uncertain. Journal of Invertebrate Pathology, 115, 3340. 10.1016/j.jip.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Catanese, G. , Grau, A. , Valencia, J. M. , Garcia‐March, J. R. , Vázquez‐Luis, M. , Alvarez, E. , Deudero, S. , Darriba, S. , Carballal, M. J. , & Villalba, A. (2018). Haplosporidium pinnae sp. Nov., a haplosporidan parasite associated with mass mortalities of the fan mussel, Pinna nobilis, in the Western Mediterranean Sea. Journal of Invertebrate Pathology, 157, 924. 10.1016/j.jip.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Cheung, A. , Dufour, S. , Jones, G. , Kostoulas, P. , Stevenson, M. A. , Singanallur, N. B. , & Firestone, S. M. (2021). Bayesian latent class analysis when the reference test is imperfect. Revue Scientifique et Technique, 40, 271–286. 10.20506/rst.40.1.3224 [DOI] [PubMed] [Google Scholar]
- Chun, S. K. (1972). Preliminary studies on the sporozoan parasites in oysters on the southern coast of Korea. Bulletin of the Korean Fisheries Society, 7680. [Google Scholar]
- Comps, M. , & Pichot, Y. (1991). Fine spore structure of a haplosporidan parasitizing Crassostrea gigas: Taxonomic implications. Diseases of Aquatic Organisms, 11, 7377. 10.3354/dao011073 [DOI] [Google Scholar]
- Couch, J. A. , & Rosenfield, A. (1968). Epizootiology of Minchinia costalis and Minchinia nelsoni in oysters introduced into Chincoteague Bay. Proceedings of the National Shellfisheries Association, Virginica. 5159.
- da Silva, P. M. , & Villalba, A. (2004). Comparison of light microscopic techniques for the diagnosis of the infection of the European flat oyster Ostrea edulis by the protozoan Bonamia ostreae . Journal of Invertebrate Pathology, 85, 97–104. 10.1016/j.jip.2003.12.010 [DOI] [PubMed] [Google Scholar]
- Dendukuri, N. , & Joseph, L. (2001). Bayesian approaches to modeling the conditional dependence between multiple diagnostic tests. Biometrics, 57(1), 158–167. 10.1111/j.0006-341x.2001.00158.x [DOI] [PubMed] [Google Scholar]
- EFSA Panel on Animal Health and Welfare (AHAW) . (2015). Oyster mortality. EFSA Journal, 13(6), 4122. 10.2903/j.efsa.2015.4122 [DOI] [Google Scholar]
- FAO . (2019). Aquaculture production. Fishstat Plus. Available online at https://www.fao.org/fishery/statistics/software/fishstat/en
- Friedman, C. S. (1996). Haplosporidian infection of the Pacific oyster, Crassostrea gigas (Thunberg), in California and Japan. Journal of Shellfish Research, 15, 597–600. [Google Scholar]
- Friedman, C. S. , Cloney, D. F. , Manzer, D. , & Hedrick, R. P. (1991). Haplosporidiosis of the Pacific oyster, Crassostrea gigas . Journal of Invertebrate Pathology, 58(3), 367–372. 10.1016/0022-2011(91)90182-P [DOI] [PubMed] [Google Scholar]
- Gagné, N. , Veniot, A. , Stephenson, M. , & McClure, C. (2015). Performance characteristics of polymerase chain reaction and histological methods for the detection of Haplosporidium nelsoni in the eastern oyster (Crassostrea virginica). Journal of Veterinary Diagnostic Investigation, 27(4), 476–488. 10.1177/1040638715592666 [DOI] [PubMed] [Google Scholar]
- Haskin, H. H. , & Andrews, J. D. (1988). Uncertainties and speculations about the life cycle of the eastern oyster pathogen Haplosporidium nelsoni (MSX). American Fisheries Society Special Publication, 18, 522. [Google Scholar]
- Herbert, R. J. H. , Humphreys, J. , Davies, C. J. , Roberts, C. , Fletcher, S. , & Crowe, T. P. (2016). Ecological impacts of non‐native Pacific oysters (Crassostrea gigas) and management measures for protected areas in Europe. Biodiversity and Conservation, 25(14), 2835–2865. 10.1007/s10531-016-1209-4 [DOI] [Google Scholar]
- Hill, K. M. , Carnegie, R. B. , Aloui‐Bejaoui, N. , Gharsalli, R. E. , White, D. M. , Stokes, N. A. , & Burreson, E. M. (2010). Observation of a Bonamia sp. Infecting the oyster Ostrea stentina in Tunisia, and a consideration of its phylogenetic affinities. Journal of Invertebrate Pathology, 103(3), 179–185. 10.1016/j.jip.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Hine, P. , Carnegie, R. , Burreson, E. , & Engelsma, M. (2009). Inter‐relationships of haplosporidians deduced from ultrastructural studies. Diseases of Aquatic Organisms, 83, 247–256. 10.3354/dao02016 [DOI] [PubMed] [Google Scholar]
- Hine, P. , Engelsma, M. , & Wakefield, S. (2007). Ultrastructure of sporulation in Haplosporidium armoricanum . Diseases of Aquatic Organisms, 77, 225–233. 10.3354/dao01822 [DOI] [PubMed] [Google Scholar]
- Hine, P. , Morris, D. J. , Azevedo, C. , Feist, S. W. , & Casal, G. (2020). Haplosporosomes, sporoplasmosomes and their putative taxonomic relationships in rhizarians and myxozoans. Parasitology, 147(14), 1614–1628. 10.1017/S0031182020001717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, D. H. , Lewis, J. L. , Keller, B. J. , & Smith, C. S. (2004). Histological Techniques for Marine Bivalve Mollusks and Crustaceans, NOAA Technical Memorandum NOS NCCOS 5, 218 p.
- Huelsenbeck, J. P. , & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England), 17(8), 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hurvich, C. M. , & Tsai, C. ‐L. (1993). A corrected Akaike information criterion for vector autoregressive model selection. Journal of Time Series Analysis, 14(3), 271–279. 10.1111/j.1467-9892.1993.tb00144.x [DOI] [Google Scholar]
- Johnson, W. O. , Jones, G. , & Gardner, I. A. (2018). Gold standards are out and Bayes is in: Implementing the cure for imperfect reference tests in diagnostic accuracy studies. Preventive Veterinary Medicine, 167, 113–127. 10.1016/j.prevetmed.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Joseph, L. , Gyorkos, T. W. , & Coupal, L. (1995). Bayesian estimation of disease prevalence and the parameters of diagnostic tests in the absence of a gold standard. American Journal of Epidemiology, 141, 263–272. [DOI] [PubMed] [Google Scholar]
- Kamaishi, T. , & Yoshinaga, T. (2002). Detection of Haplosporidium nelsoni in Pacific oyster Crassostrea gigas in Japan. Fish Pathology, 37(4), 193–195. 10.3147/jsfp.37.193 [DOI] [Google Scholar]
- Kang, P. A. (1980). On the Minchinia sp. infection in the oysters from Chungmu area. Bulletin of the Fisheries Research Development Agency, 25–28. [Google Scholar]
- Kern, F. G. (1976). Sporulation of Minchinia sp. (Haplosporida, Haplosporidiidae) in the Pacific oyster Crassostrea gigas (Thunberg) from the Republic of Korea. The Journal of Protozoology, 23(4), 498–500. 10.1111/j.1550-7408.1976.tb03826.x [DOI] [Google Scholar]
- Ko, Y. T. , Ford, S. E. , & Fong, D. (1995). Characterization of the small subunit ribosomal RNA gene of the oyster parasite Haplosporidium costale . Molecular Marine Biology and Biotechnology, 4(3), 236240. [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo, C. , Osta Amigo, A. , Mandard, Y. V. , Peroz, C. , & Renault, T. (2014). Improving early detection of exotic or emergent oyster diseases in France: Identifying factors associated with shellfish farmer reporting behaviour of oyster mortality. Preventive Veterinary Medicine, 116(12), 168–182. 10.1016/j.prevetmed.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Lynch, S. A. , Villalba, A. , Abollo, E. , Engelsma, M. , Stokes, N. A. , & Culloty, S. C. (2013). The occurrence of haplosporidian parasites, Haplosporidium nelsoni and Haplosporidium sp., in oysters in Ireland. Journal of Invertebrate Pathology, 112(3), 208–212. 10.1016/j.jip.2012.11.013 [DOI] [PubMed] [Google Scholar]
- Martenot, C. , Oden, E. , Travaillé, E. , Malas, J. P. , & Houssin, M. (2010). Comparison of two real‐time PCR methods for detection of ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas . Journal of Virological Methods, 170(12), 86–89. 10.1016/j.jviromet.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Medlin, L. , Elwood, H. J. , Stickel, S. , & Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S‐like rRNA‐coding regions. Gene, 71(2), 491–499. 10.1016/0378-1119(88)90066-2 [DOI] [PubMed] [Google Scholar]
- OIE . (2003). SSO disease (Haplospordium costale). In Manual of diagnostic tests for aquatic animals (Chapter 3.1.6, 2003e éd., p. 255259).
- Penna, M. ‐S. , Khan, M. , & French, R. A. (2001). Development of a multiplex PCR for the detection of Haplosporidium nelsoni, Haplosporidium costale and Perkinsus marinus in the eastern oyster (Crassostrea virginica, Gmelin, 1971). Molecular and Cellular Probes, 15(6), 385–390. 10.1006/mcpr.2001.0386 [DOI] [PubMed] [Google Scholar]
- Penna, S. , French, R. A. , Volk, J. , Karolus, J. , Sunila, I. , & Smolowitz, R. (1999). Diagnostic screening of oyster pathogens: Preliminary field trials of multiplex PCR. Journal of Shellfish Research, 18, 319–320. [Google Scholar]
- Perkins, F. O. (1969). Electron microscope studies of sporulation in the oyster pathogen, Minchinia costalis (Sporozoa: Haplosporida). The Journal of Parasitology, 55(5), 897–920. 10.2307/3277152 [DOI] [Google Scholar]
- Perkins, F. O. , & van Banning, P. (1981). Surface ultrastructure of spores in three genera of Balanosporida, particularly Minchinia armoricana van Banning, 1977—The taxonomic significance of spore wall ornamentation in the Balanosporida. Journal of Parasitology, 67, 866–874. [Google Scholar]
- Polinski, M. P. , Laurin, E. , Delphino, M. K. V. C. , Lowe, G. J. , Meyer, G. R. , & Abbott, C. L. (2021). Evaluation of histopathology, PCR, and qPCR to detect Mikrocytos mackini in oysters Crassostrea gigas using Bayesian latent class analysis. Disease of Aquatic Organisms, 144, 21–31. 10.3354/dao03566 [DOI] [PubMed] [Google Scholar]
- Polinski, M. , Lowe, G. , Meyer, G. , Corbeil, S. , Colling, A. , Caraguel, C. , & Abbott, C. L. (2015). Molecular detection of Mikrocytos mackini in Pacific oysters using quantitative PCR. Molecular and Biochemical Parasitology, 200(12), 19–24. 10.1016/j.molbiopara.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Posada, D. (2008). jModelTest : Phylogenetic model averaging. Molecular Biology and Evolution, 25(7), 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Ramilo, A. , Navas, J. , Villalba, A. , & Abollo, E. (2013). Species‐specific diagnostic assays for Bonamia ostreae and B. exitiosa in European flat oyster Ostrea edulis: Conventional, real‐time and multiplex PCR. Diseases of Aquatic Organisms, 104(2), 149–161. 10.3354/dao02597 [DOI] [PubMed] [Google Scholar]
- Renault, T. , Stokes, N. , Chollet, B. , Cochennec, N. , Berthe, F. , Gérard, A. , & Burreson, E. (2000). Haplosporidiosis in the Pacific oyster Crassostrea gigas from the French Atlantic coast. Diseases of Aquatic Organisms, 42, 207–214. 10.3354/dao042207 [DOI] [PubMed] [Google Scholar]
- Rosenfield, A. , Buchanan, L. , & Chapman, G. B. (1969). Comparison of the fine structure of spores of three species of minchinia (Haplosporida, Haplosporidiidae). The Journal of Parasitology, 55(5), 921–941. 10.2307/3277153 [DOI] [Google Scholar]
- Russell, S. S. , Penna, S. , & French, R. A. (2000). Comparative evaluation of the multiplex PCR with conventional detection methods for Haplosporidium nelsoni (MSX), Haplosporidium costale (SSO), and Perkinsus marinus (Dermo) in the eastern oyster, Crassostrea virginica . Journal of Shellfish Research, 19, 580–581. [Google Scholar]
- Russell, S. , Frasca, S. , Sunila, I. , & French, R. (2004). Application of a multiplex PCR for the detection of protozoan pathogens of the eastern oyster Crassostrea virginica in field samples. Diseases of Aquatic Organisms, 59, 85–91. 10.3354/dao059085 [DOI] [PubMed] [Google Scholar]
- Samain, J. ‐F. , & McCombie, H. (2007). Summer mortality of Pacific oyster Crassostrea gigas. The Morest project. Éditions Quae/Ifremer. [Google Scholar]
- Saulnier, D. , De Decker, S. , Tourbiez, D. , & Travers, M. A. (2017). Development of a duplex Taqman real‐time PCR assay for rapid identification of Vibrio splendidus‐related and V. aestuarianus strains from bacterial cultures. Journal of Microbiological Methods, 140, 67–69. 10.1016/j.mimet.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Stephenson, M. F. , McGladdery, S. E. , Maillet, M. , & Veniot, A. (2003). First reported occurrence of MSX in Canada. Journal of Shellfish Research, 355, Abstract. [Google Scholar]
- Stokes, N. , & Burreson, E. (2001). Differential diagnosis of mixed Haplosporidium costale and Haplosporidium nelsoni infections in the eastern oyster, Crassostrea virginica, using DNA probes. Journal of Shellfish Research, 20, 207–213. [Google Scholar]
- Tamura, K. , & Kumar, S. (2002). Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Molecular Biology and Evolution, 19(10), 1727–1736. 10.1093/oxfordjournals.molbev.a003995 [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Nei, M. , & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor‐joining method. Proceedings of the National Academy of Sciences of the United States of America, 101(30), 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacek, P. M. (1985). The effect of conditional dependence on the evaluation of diagnostic tests. Biometrics, 41(4), 959–968. [PubMed] [Google Scholar]
- van Banning, P. (1977). Minchinia armoricana sp. Nov. (Haplosporida), a parasite of the European flat oyster, Ostrea edulis . Journal of Invertebrate Pathology, 30, 199–206. [Google Scholar]
- Wang, Z. , Lu, X. , Liang, Y. , & Wang, C. (2010). Haplosporidium nelsoni and H. costale in the Pacific oyster Crassostrea gigas from China's coasts. Diseases of Aquatic Organisms, 89, 223–228. 10.3354/dao02196 [DOI] [PubMed] [Google Scholar]
- Wood, J. L. , & Andrews, J. D. (1962). Haplosporidium costale (Sporozoa) associated with a disease of Virginia oysters. Science, 136, 710–711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The sequence data have been submitted to the GenBank databases under accession numbers MZ666334, MZ666335, MZ666374, MZ673037.


