Abstract
Adenoviruses (AdVs) are widespread in vertebrates. They infect the respiratory and gastrointestinal tracts, the eyes, heart, liver, and kidney, and are lethal to immunosuppressed people. Mastadenoviruses infecting mammals comprise several hundred different types, and many specifically infect humans. Human adenoviruses are the most widely used vectors in clinical applications, including cancer treatment and COVID‐19 vaccination. AdV vectors are physically and genetically stable and generally safe in humans. The particles have an icosahedral coat and a nucleoprotein core with a DNA genome. We describe the concept of AdV cell entry and highlight recent advances in cytoplasmic transport, uncoating, and nuclear import of the viral DNA. We highlight a recently discovered “linchpin” function of the virion protein V ensuring cytoplasmic particle stability, which is relaxed at the nuclear pore complex by cues from the E3 ubiquitin ligase Mind bomb 1 (MIB1) and the proteasome triggering disruption. Capsid disruption by kinesin motor proteins and microtubules exposes the linchpin and renders protein V a target for MIB1 ubiquitination, which dissociates V from viral DNA and enhances DNA nuclear import. These advances uncover mechanisms controlling capsid stability and premature uncoating and provide insight into nuclear transport of nucleic acids.
Keywords: DNA nuclear transport, Mind bomb 1 (MIB1) E3 ubiquitin ligase, nuclear pore complex (NPC), ubiquitin proteasome system, virus entry and disassembly
Viruses invade organisms and cells, cause disease, persist for a lifetime, or are eliminated by host defense. Viral vectors are beneficial to clinical therapy or preventive vaccination. Adenoviruses are widespread human pathogens and the most widely used vectors in gene therapy. Here we provide an updated coherent concept for nuclear entry of a human virus, a key process in infection and immunity.
1. INTRODUCTION
Adenoviruses (AdVs) comprise more than 300 different types of pathogens, including more than 100 from humans (Greber, 2020; Harrach et al., 2019). Human AdVs (HAdV) are classified into species A to G, as curated by the Human Adenovirus Working Group (http://hadvwg.gmu.edu/). They not only cause self‐limiting respiratory, ocular, blood‐borne, or intestinal infections, but also severe infections in immunocompromised individuals, possibly involving reactivation of persistent viruses (Ghebremedhin, 2014; Lion, 2019; Matthes‐Martin et al., 2013; Mohamed Ismail et al., 2019; Prasad & Greber, 2021).
Their large genomic capacity, the high stability, and ease of production have made HAdVs important vectors for gene therapy, oncolytic applications, and vaccinations (Brucher et al., 2021; Gao et al., 2019; Li, 2019; Schmid et al., 2018; Waehler et al., 2007). For example, AdVs can be grown to high titers and purified to near homogeneity under good manufacturing practice (Raty et al., 2008; Verma & Weitzman, 2005). This has recently been achieved also with helper‐dependent, so‐called gutless HAdV vectors lacking any viral sequences except the encapsidation signal and the inverted terminal repeats (Brucher et al., 2021; Parks et al., 1996). Such vectors express no viral proteins and allow transgene expression over prolonged periods of time, weeks, and months (Alonso‐Padilla et al., 2016; Sakhuja et al., 2003).
AdVs are widely used gene delivery agents because they efficiently transduce both dividing and quiescent cells and have high physical and genetic stability. A large number of variants is available for tailored applications and can bypass preexisting immunity, which is a key issue in many therapeutic vector applications (Fausther‐Bovendo & Kobinger, 2014). Notably, infection pathways of AdVs have been elucidated in sufficient detail by mechanistic studies and omics approaches to grant clinical applications to AdV vectors, such as HAdV types of the species B and C and simian AdV (Capone et al., 2013; Mendonca et al., 2021; Zhao et al., 2019). HAdV‐E4 and HAdV‐B7 were successfully used as a vaccine to suppress acute respiratory diseases caused by adenoviruses in recruits (Hoke et al., 2012; Top et al., 1971).
Despite these achievements, major challenges for the field have remained, including the immunogenic features of AdVs, mechanisms limiting vector efficacies in therapies, or the nature of cell‐to‐cell infection variability (Allen & Byrnes, 2019; Appaiahgari & Vrati, 2015; Atasheva et al., 2019; Sohn & Hearing, 2019; Suomalainen & Greber, 2021). Here, we provide an update on the biology of AdV cell entry into cells with a focus on virion stability, uncoating, and nuclear import of the viral genome.
2. THE VIRION
AdV particles have an icosahedral symmetry of pseudo T = 25, where each of the 20 facets harbors 12 trimers of the major coat protein hexon (Benevento et al., 2014; van Oostrum & Burnett, 1985). The hexon protein (also referred to as protein II) contains hypervariable regions (HVRs) exposed to the virion outside. The HVRs are subject to immune recognition by both neutralizing antibodies and virus‐specific T‐cell responses (Deal et al., 2013). Each of the 12 vertices is made up of 5 copies of penton base (III), which anchor the trimeric fiber protein (IV) protruding away from the capsid (Cheneau & Kremer, 2020; Stasiak & Stehle, 2020). The HAdV particles are held together by cementing proteins, as indicated by high resolution X‐ray and cryo‐EM structure analyses (Dai et al., 2017; Liu et al., 2010; Perez‐Illana et al., 2021; Rafie et al., 2021; Reddy & Nemerow, 2014; Yu et al., 2017, 2022). Initial disputes about the location of minor proteins have now been settled (reviewed in 41). For example, five copies of protein IIIa are located inside the virion beneath each vertex and they link the pentons to the peripentonal hexons. IIIa also links to the minor capsid protein VIII, but the location of the entire IIIa is unknown since the C‐terminal 40% of the protein are not icosahedrally ordered and remain unresolved. The roles of IIIa and VIII in virus entry are unknown, aside from the observation that they are released from the incoming HAdV‐C2 particles before or during endosomal escape (Greber et al., 1993).
The minor capsid stabilizing protein IX is located on the virion surface. IX increases the thermoresistance of virions, as indicated by HAdV‐C5 knockout mutants (Colby & Shenk, 1981). It has 140 amino acids in case of HAdV‐C5, and the virion harbors 240 copies of IX. Like protein V (see below), IX is only found in human and nonhuman Mastadenoviruses. The N‐terminal domains of three IX proteins form a triskelion in the valleys between hexons, four per facet, one in the center, and three toward each of the edges. The N terminus of IX is separated by an unstructured rope‐like stretch of amino acids, which extend to the C‐terminal alpha helical domain. Toward the edge of the facets, three α‐helices (each originating from a distinct triskelion of the same facet) bundle up to a trimeric parallel coiled‐coil structure, which projects toward the outside of the virion. This bundle is joined by a protein IX helix from a neighboring facet, and thereby forms a tetrameric bundle of three parallel and one antiparallel α‐helices (Dai et al., 2017; Yu et al., 2017). In this way, there are 20 protein IX molecules per facet arranged into three tetrameric helical bundles and four trimeric triskelions. While the triskelions of IX mediate thermoresistance of the virion (Vellinga et al., 2005), the helical bundles help the virion to orchestrate the capsid disassembly at the nuclear pore complex (NPC) (Strunze et al., 2011).
Another small virion protein has a well‐described function, the internal protein VI. VI disrupts membranes (Luisoni et al., 2015; Moyer et al., 2011; Wiethoff et al., 2005). There are 360 copies of VI per virion. Some of the cryo‐EM density of VI could be localized near the cavities of hexon trimers in HAdV‐C5 and D26, although not in all cavities, of which there are 240 in the virion (Dai et al., 2017; Yu et al., 2017). The full protein VI has not been localized suggesting that large parts of it are icosahedrally disordered and positioned in various intravirion locations. It remains to be elucidated how the competition for hexon cavity binding by another internal protein, VII, impinges on the localization of VI in the virion (Dai et al., 2017; Hernando‐Perez et al., 2020). Intriguingly, there are no traces of VII in the cavities of hexon in the membrane disruption‐defective TS1 virions, which lack sufficient activity of the viral protease p23 to process a range of minor virion proteins, including the precursors of IIIa, VI, VII, VIII, X, terminal protein and also the scaffold protein L1‐52/55 K (Mangel & San, 2014; Yu et al., 2022). Likewise, virions lacking VII are defective at endosomal lysis (Ostapchuk et al., 2017), presumably because they fail to expose protein VI for membrane disruption (Burckhardt et al., 2011; Hernando‐Perez et al., 2020; Pied & Wodrich, 2019).
Within the capsid, AdV particles harbor a single copy of a linear double‐stranded viral DNA attached at both ends to a single copy of the terminal protein. The genome is condensed with about 800 copies of the major DNA‐associated protein VII (Martin‐Gonzalez et al., 2019). It also contains about 150 copies of the minor DNA‐binding protein V, which link the DNA to the capsid by binding to penton base and protein VI (Perez‐Berna et al., 2015; Perez‐Vargas et al., 2014). Another basic protein of the virion, protein X, also known as μ, has about the same abundance as V, but a mass of only 9 kDa, four time less than V. Proteins VII, V, and μ make crosslinkable contacts and are part of the DNA core (Chatterjee et al., 1985). It has been proposed that VII wraps and bundles DNA segments by interacting with μ, and V has DNA decondensing functions (Gallardo et al., 2021).
In sum, the structure, location, and function of proteins in the HAdV capsid are well understood thanks to cryo‐EM analyses, cell biological and immunological studies in model organisms, and applications of vectors in preclinical and clinical settings, although the molecular structure of the DNA core is much less understood. In turn, cryo‐EM structures of nonhuman AdVs from dog, cattle, bats, or lizard are emerging, and report interesting similarities, but also differences to HAdV capsids. Differences affect the location and interconnection of stabilizing proteins (Cheng et al., 2014; Hackenbrack et al., 2017; Marabini et al., 2021; Schoehn et al., 2008). It will be important to annotate the structural differences between human and animal AdVs and assign them to functions in entry and morphogenesis. This may then pave the way for systematic considerations of nonhuman AdVs in gene therapy and vaccinations (Bots & Hoeben, 2020; Greber & Gomez‐Gonzalez, 2021).
3. STEPS IN AdV ENTRY UPSTREAM OF THE CELL NUCLEUS
AdV entry is best characterized in epithelial cells, although entry into immune cells is of emerging importance (for reviews, see [Barry et al., 2020; Greber & Flatt, 2019]). Here we carve out the principles of AdV entry, as derived mostly from studies with HAdV‐C, the best characterized AdV. Entry occurs in sequential steps, starting with virion attachment (Arnberg, 2012; Stewart & Nemerow, 2007), cell signaling (Wolfrum & Greber, 2013), endocytosis (Meier & Greber, 2003), endosomal rupture and removal of disrupted endosomes (Luisoni et al., 2016; Montespan et al., 2017; Suomalainen et al., 2013), transport through the cytosol (Greber & Way, 2006; Scherer et al., 2020), separation of the genome from the capsid, and genome nuclear import (see Figure 1; [Flatt & Greber, 2017]).
FIGURE 1.
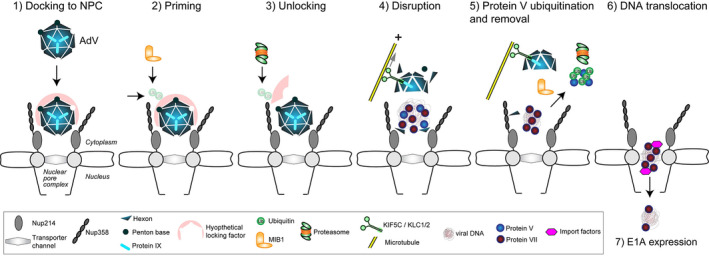
An inclusive model for adenovirus disassembly at the NPC and nuclear import of viral DNA. The HAdV‐C5 capsid with pseudo T = 25 icosahedral symmetries effectively protects and efficiently delivers its DNA cargo into host cells. During infectious cell entry, virions are dismantled in a coordinated and well‐controlled manner (Greber et al., 1994). At the plasma membrane, the virion sheds the fibers and some of the penton base proteins and, subsequently, also the internal stabilizing proteins IIIa and VIII (Greber et al., 1993; Nakano et al., 2000). It exposes the membrane lytic protein VI (Burckhardt et al., 2011; Wodrich et al., 2010), and upon rupture of the endosomal membrane, releases the protein VI, as well as a small fraction of the internal DNA decondensing protein V (Burckhardt et al., 2011; Puntener et al., 2011). The leaky virion attaches to microtubule motors and traffics toward the cell nucleus (Scherer et al., 2020; Wang et al., 2018), where it detaches from the microtubules and binds to the NPC protein Nup214 (Wang et al., 2017) (1). This binding occurs through the major capsid protein hexon (Cassany et al., 2015; Trotman et al., 2001). A locking factor, or the lack of critical ubiquitination providing a positive signal for dismantling, precludes the NPC‐docked virion from disassembly (Bauer et al., 2021). The E3 ubiquitin ligase MIB1 primes the virion for disassembly at the NPC by recruiting the disassembly machinery (2), or targeting the locking factor for proteasomal degradation (3). The disruption of the capsid is mediated by the conventional kinesin KIF5C and its light chain KLC1/2 pulling on microtubules near the NPC against the holding force of the nucleus (Joseph & Dasso, 2008; Strunze et al., 2011) (4). The TPR domain of KLC1/2 attaches the motor complex to the facet‐stabilizing protein IX (de Vrij et al., 2011; Strunze et al., 2011). Kinesin activation might be mediated by binding of Nup358 to the stalk of KIF5C, as shown by an unrelated in vitro study demonstrating allosteric activation of the ATPase activity (Cho et al., 2009). When the capsid is sufficiently disrupted, MIB1‐mediated ubiquitination of protein V leads to the detachment of V from the capsid and the viral DNA (5). This reaction renders the viral DNA in complex with the condensing protein VII a nuclear import substrate recognized by nuclear import factors (6). Remarkably, insufficient ubiquitination of protein V increases the misdelivery of viral DNA to the cytoplasm, instead of the nucleus, as shown by the all lysine to arginine HAdV‐C5 mutant (Bauer et al., 2021)
3.1. Receptors, attachment factors, and facilitators
All viruses interact with extracellular proteins, sugars, lipids, and solutes before binding to target cells and initiating infection (Atasheva et al., 2019; Bremner et al., 2009; Doronin et al., 2012; Eichholz et al., 2016; Hendrickx et al., 2014; Kelkar et al., 2006; Khare et al., 2012). We use the terms “receptor, attachment factor, and infection facilitator” to refer to host molecules located outside of the cell and influencing the course of infection, as defined earlier (Yamauchi & Greber, 2016). Accordingly, receptors are cell surface molecules that directly bind to a component of the virus particle. This binding reaction leads to infection of a susceptible cell. Attachment factors are host molecules that directly bind to the virion but do not necessarily lead to cell infection, whereas facilitators are molecules that enhance the infection without directly contacting the virion.
HAdVs typically initiate infection by high affinity or high avidity binding of their fiber knob proteins. This has been widely demonstrated with cells lacking fiber receptors and retargeted AdV particles (for conceptual reviews, see [Arnberg, 2012; Baker, Greenshields‐Watson, et al., 2019; Barry et al., 2020; Excoffon, 2020; Luisoni & Greber, 2016; Nemerow & Stewart, 2016; Wolfrum & Greber, 2013]). The importance of fiber knob binding to receptors was first demonstrated with coxsackievirus AdV receptor (CAR), a high affinity attachment factor for most HAdVs, including canine AdV‐2 (CAdV‐2) and avian AdV CELO (Soudais et al., 2000), but not for the B, D, and G types (Bergelson et al., 1997; Freimuth et al., 1999; Kirby et al., 2000; Roelvink et al., 1998; Tomko et al., 1997). Species B and D use CD46 (Cupelli et al., 2010; Fleischli et al., 2005, 2007; Gaggar et al., 2003; Gustafsson et al., 2010; Pache et al., 2008; Persson et al., 2009; Russell, 2004; Sakurai et al., 2006, 2007; Sirena et al., 2004, 2005; Tuve et al., 2006; Wang, Li, Yumul, et al., 2011; Wang, Yumul, et al., 2013) or desmoglein (DSG)‐2 (Hemsath et al., 2022; Trinh et al., 2012; Vassal‐Stermann et al., 2019; Wang et al., 2015; Wang, Li, Liu, et al., 2011; Wang, Li, Yumul, et al., 2011; Wang, Yumul, et al., 2013). AdV‐D26 has been reported to use CD46 or sialic acid as a receptor through fiber knob or hexon as ligands (Baker, Mundy, et al., 2019; Hemsath et al., 2022; Persson et al., 2021).
Hexon was initially shown to be an AdV‐C2, C5, D26, and B35 ligand for virion binding to the scavenger receptor SR‐A6 (MARCO) (Maler et al., 2017; Stichling et al., 2018). Scavenger receptors constitute a large family of structurally diverse cell surface receptors. Besides viruses, they interact with and mediate the uptake of a wide range of ligands into immune cells. Ligands include modified and nonmodified self‐molecules, nonopsonized particles, and microbial ligands. Regarding viruses, human and murine SR‐A6 have been implicated in infection of epithelial cells with herpes simplex virus type 1, and SR‐A1 and SR‐F1/2 (SREC‐1) are surface receptor candidates for HAdV‐C5 on Kupffer cells and liver sinusoidal endothelial cells (Khare et al., 2012; MacLeod et al., 2013).
HAdV‐C2/C5 hexon interactions with SR‐A6 lead to virus entry, transduction, and cytokine responses in murine macrophages in vitro and in vivo, but not in human epithelial A549 cells, where CAR and integrins function as entry receptors (Khare et al., 2012; Maler et al., 2017; Stichling et al., 2018). Notably, the HAdV‐C5 hexon has a negatively charged hypervariable region 1 binding to SR‐A6 (Stichling et al., 2018). Another hypervariable region of hexon, HVR7 is, however, likely not involved in the SR‐A6 entry pathway, as the replacement of the HVR7 INTETL sequence with GNNSTY (ablating factor X binding) does not affect macrophage entry (Ma et al., 2015; Schmid et al., 2018). These results are in agreement with human mononuclear phagocyte data, where coagulation factor X (FX) in complex with HAdV‐C5 did not lead to enhancement of an innate immune response via a Toll‐like receptor pathway compared to HAdV‐C5 alone (Eichholz et al., 2015).
3.2. Following receptor binding
Upon binding to a fiber receptor, HAdVs typically engage a secondary receptor for endocytic uptake, usually an active state αvβ3/αvβ5 integrin through an arginine–glycine–aspartate (RGD) sequence in the penton base protein. This elicits cell survival signals and triggers endocytosis and eventually also endosomal lysis (Luisoni & Greber, 2016; Nemerow, 2000). The latter is triggered by upstream actomyosin‐dependent drifting motions of virus particles bound to CAR, which leads to exposure of the membrane lytic protein VI, and sphingolipid conversion to ceramide in the plasma membrane (Burckhardt et al., 2011; Luisoni et al., 2015; Wiethoff & Nemerow, 2015). Although an early paper had reported that the function of incoming protein VI required intact microtubules (Wodrich et al., 2010), it is now accepted that endosomal escape is independent of microtubules (Lagadec et al., 2021; Suomalainen et al., 1999). Protein VI readily separates from capsid upon rupture of endosomes, as shown by immunostainings and galectin‐3/8 reporter protein expression to monitor broken endosomes (Burckhardt et al., 2011; Luisoni et al., 2016; Maier et al., 2012).
In contrast to HAdVs, mouse AdV (MAdV) types 1 and 3 bind with their fiber knobs to integrins αvβ6 and αvβ8 at both high and low affinity and thereby trigger infection (Bieri et al., 2021). Interestingly, MAdV‐1 and ‐3 evolved a dual integrin binding motif, RGD, and LXXL (L for leucine and X for any amino acid). This motif has higher affinity for extended open integrins engaged in cytoskeletal interaction, and lower affinity for conformationally closed integrins that are not constrained by the cytoskeleton (Bieri et al., 2021; Campbell & Humphries, 2011). The former integrins are immobile, and the latter ones are mobile in the plasma membrane, arguing that virion interactions with extended open and closed integrins could be involved in the exposure of protein VI from the MAdVs. Such mechanism would be akin to HAdV‐C2/5 interactions with mobile CAR binding to fiber, and immobile αvβ3/5 integrins binding to RGD of penton base. This situation gives rise to mechanical stress on the virion and the dissociation of fibers and some of the pentons, the activation of lysosomal secretion, the exposure of protein VI, endosomal lysis, and infection (Greber, 2016). Interestingly, HAdV‐C5 containing a quadruple cysteine‐constrained RGD in its fiber knob had a lower specific infectivity in CAR‐negative cells than wild‐type HAdV‐C5 in the corresponding CAR‐positive cells, indicating that both mobile and immobile receptors are together enhancing infection (Burckhardt et al., 2011; Nagel et al., 2003).
3.3. Cytoplasmic trafficking
Long‐range cytoplasmic transport of virus particles is required for infection, particularly if the virus replicates in the cell nucleus (Dohner & Sodeik, 2005; Greber & Way, 2006; Scherer et al., 2020; Wang et al., 2018; Welte, 2004; Witte et al., 2018). HAdVs use motor proteins for bidirectional trafficking on microtubules to the nucleus after endosomal escape (Engelke et al., 2011; Gazzola et al., 2009). In nonpolarized cells, dynein/dynactin‐based transport prevails over kinesin‐based transport and leads to virion enrichment near the centrosome proximal to the nucleus (Raynaud‐Messina & Merdes, 2007; Suomalainen et al., 1999). In polarized epithelial cells, kinesin‐mediated transport may be required to bring incoming particles from the apical plasma membrane to the nucleus, as the microtubule minus ends are located predominantly near the apical plasma membrane (Müsch, 2004).
While the ligand for intermediate chain (IC) and light IC of cytoplasmic dynein was identified to be hexon (Bremner et al., 2009), the nature of the trigger rendering hexon competent to bind dynein has remained controversial. Bremner et al. suggested that low pH was the trigger, and that the hexon HVR1 was involved in dynein recruitment (Bremner et al., 2009; Scherer & Vallee, 2015). This was based on IC binding experiments with pH 4.4‐treated hexon or virions, and dispase treatment of HAdV‐C5, which cleaves in HVR1 and abrogates nuclear transport of incoming virus. However, infectious incoming HAdV‐C penetrates from early endosomes and is not known to pass through acidic endosomes (Gastaldelli et al., 2008; Maier et al., 2012; Suomalainen et al., 2013). This argues that another cue than low pH might render hexon competent to bind dynein. Likewise, the genetic ablation of HVR1 had no effects on HAdV‐C5 transduction of human lung epithelial cells, although it strongly affected the binding of the particles to SR‐A6 of murine macrophage MPI‐2 cells (Stichling et al., 2018). It thus remains unknown how dynein precisely binds to HAdV‐C5.
An opposing cytoplasmic motor to dynein for HAdV‐C5 transport is conventional kinesin, which occurs in three isoforms, kinesin‐1A, 1B, and 1C heavy chains (formerly known as KIF5A, 5B, and 5C), and KLC1 and KLC2 light chains. The knockdown of KIF5B led to the enrichment of incoming particles near the centrosome suggesting that this motor was involved in transporting virions away from the centrosome to the nuclear membrane (Zhou et al., 2018). Biochemical interaction studies further suggested that KIF5B binds to penton base via the stalk domain. Interestingly, stochastic simulations of HAdV‐C motion bursts gave rise to a model, according to which a small number of 1–2 kinesin and dynein motors were attached to motile virions in order to accommodate the motion bursts and the rapid directional switches (Gazzola et al., 2009). Notably, microtubule‐dependent HAdV burst are short, and cytosolic particles remain mostly inactive in the cytoplasm (Strunze et al., 2005; Suomalainen et al., 1999; Zhou et al., 2018). The stochastic simulations also suggested that minus‐ and plus end‐directed motors compete with each other for a common binding site on the HAdV‐C particle. This notion is important in light of the observation that the incoming particles detach from microtubules preferentially very close to the nuclear membrane (Wang et al., 2017). This detachment depends on nuclear export mediated by the export factor CRM1 (Lagadec et al., 2021; Smith et al., 2008; Strunze et al., 2005), as indicated by the CRM1‐specific inhibitor leptomycin B (LMB) and the rescue of infection by LMB‐resistant CRM1 in LMB‐treated cells (Wang et al., 2017). It is still unclear, if CRM1 directly binds to the viral capsids.
Interestingly, it was recently reported that the genetic knockout of the human noncoding RNA nc886, also known as pre‐miR‐886 or CBL3, inhibited adenoviral gene expression and replication (Saruuldalai et al., 2022). Nc886 is transcribed by RNA polymerase III and known to inhibit the activation of protein kinase R (PKR) (Kunkeaw et al., 2013). Its knockout (KO) in SV40 immortalized, primary thyroid follicular epithelial cells (Nthy‐ori3‐1) reduced the HAdV‐C5 DNA delivery to the nucleus as measured by protein VII immunofluorescence, but it did not affect the entry of the virus into the cytoplasm. This effect was independent of PKR. The nc886 KO cells showed increased mRNA levels of four kinesin genes, namely KIF5C, KIF20A, KIF22, and KIF23. This might hint to the possibility that one or several of these kinesin heavy chains have antiviral functions when overexpressed, for example by dysregulating virion transport or uncoating. However, alternative interpretations of the nc886 KO effects are feasible, such as increased type 1 interferon levels (Lee et al., 2011, 2021).
4. CAPSID DISASSEMBLY AT THE NPC TRIGGERED BY MIB1
Recently, the Charlie Rice laboratory and the Urs Greber laboratory independently discovered the importance of the cellular E3 ubiquitin ligase MIB1 for HAdV‐C infection. They showed that MIB1 triggers the onset of capsid disassembly at the NPC. MIB1 has well‐described regulatory roles in endocytic pathways, for example controlling WNT/β‐catenin and DELTA/NOTCH signaling (Berndt et al., 2011; Luxan et al., 2013). MIB1 is a ring finger E3 ubiquitin ligase with a catalytic cysteine residue in the C‐terminal RING3 domain (Guo et al., 2016). The Greber laboratory initially identified MIB1 through an arrayed, genome‐wide RNA interference screen against the replication defective HAdV‐C5_dE1‐GFP lacking E1, scoring the expression of GFP (Bauer et al., 2019). The Rice laboratory used a gene‐trap screen in haploid cells infected with replication defective or competent HAdV‐C5 reporter viruses (Sarbanes et al., 2021). Besides blocking HAdV‐C, the KO of MIB1 also blocked HAdV‐A31, B3, and D8 infections, but did not affect a large panel of unrelated DNA and RNA viruses, indicating that MIB1 is rather specifically involved in HAdV infections (Bauer et al., 2019; Sarbanes et al., 2021).
The KO of MIB1 by both laboratories uncovered an unprecedented virus entry phenotype, namely a very strong arrest of incoming virions at the cytoplasmic side of the NPC. The block occurred at the stage of DNA uncoating and release, as shown by transmission electron microscopy, and fluorescence microscopy employing either direct DNA labeling using copper(I)‐catalyzed azide‐alkyne cyclo‐addition (click) chemistry (Bauer et al., 2019; Wang, Suomalainen, et al., 2013), or immunostaining of protein VII (Sarbanes et al., 2021). Protein VII condenses the viral DNA, helps to protect the genome in the leaky cytoplasmic capsid, and accompanies the genome into the cell nucleus (Martin‐Gonzalez et al., 2019; Puntener et al., 2011).
Both laboratories excluded an involvement of MIB1 in steps upstream of the NPC by assessing viral endocytosis, protein VI exposure (a surrogate for endosomal rupture), and transport to the nucleus (Bauer et al., 2019). They also used chemical inhibitors, such as LMB to block the attachment of virions to the NPC (Strunze et al., 2005), and showed that LMB was effective also in the absence of MIB1 (Bauer et al., 2019, 2021; Sarbanes et al., 2021). Furthermore, both laboratories ectopically expressed catalytically inactive MIB1 in MIB1‐KO HeLa or Hap1 cells and demonstrated that the ubiquitination activity of MIB1 was crucial for virion uncoating at the NPC. Sarbanes et al. also showed that the proteasome inhibitor MG132 blocked the separation of the viral DNA from the capsid but did not affect the targeting of the virus particles to the nucleus (Sarbanes et al., 2021). This suggested that the ubiquitination activity of MIB1 promoted HAdV infection by triggering the proteasomal degradation of factor(s) that impair capsid disassembly at the NPC. Based on proximity ligation and proteomics data, Sarbanes et al. speculated that one of these factors might be ribonucleoprotein particles directly or indirectly ubiquitinated by MIB1. It remains to be explored how this relates to an earlier observation that ribonuclease‐sensitive factors blocked the binding of HAdV‐C2 in reconstituted in vitro systems using nuclear envelopes from rat liver (Trotman et al., 2001).
Overall, the convergence of the RNAi and the Hap1 screens is remarkable. MIB1 was among the strongest RNAi hits and one among just a few that could be validated (Bauer et al., 2019). Likewise, MIB1 was the sole host factor identified in the haploid screen, besides the viral receptors (Sarbanes et al., 2021). These notions illustrate the considerable noise in RNAi screens on one hand, and a possible bias of insertional mutagenesis screens toward entry factors in viral infection phenotypes, on the other hand. Regardless, the results discussed here highlight the power of genetic screens to elucidate host factors in viral infections, as shown earlier by pioneering RNA interference screens (Cherry et al., 2005; Coyne et al., 2011; Karlas et al., 2010; Panda et al., 2011; Snijder et al., 2012) and more recently insertional mutageneses, and CRISPR KO screens (for a review, see [Puschnik et al., 2017]).
5. EXPLORING PERINUCLEAR INTERRELATIONS
Live‐cell imaging was used to explore how MIB1 interacted with NPC‐docked HAdV‐C5 particles (Bauer et al., 2019). Tetracycline‐induced ectopic expression of GFP‐MIB1 in MIB1‐KO cells revealed not only a transient interaction of GFP‐MIB1 with NPC‐docked virions but visualized also a dissociation of the virion DNA from the capsid. Together with the notion that catalytically active MIB1 is required for DNA release to the nucleus and infection (Bauer et al., 2019; Sarbanes et al., 2021), these results very strongly argue that crucial proviral ubiquitination events occur at the virion/NPC docking stage in MIB1 normal cells.
To identify viral targets of MIB1 ubiquitination, Bauer, Gomez‐Gonzalez et al. used differential ubiquitin pulldown assays with a diglycine‐specific antibody from normal and MIB1‐KO cells, followed by quantitative mass spectrometry (Bauer et al., 2021). They found that four lysine residues of protein V were specifically ubiquitinated in normal, but not in MIB1‐KO, cells. Two other incoming virion proteins were also ubiquitinated, protein VI and penton base, but independently of MIB1. Notably, purified HAdV‐C5 particles contained no detectable ubiquitinated proteins, as concluded from mass spectrometry experiments with good peptide coverage larger than 50%, except for the low abundant terminal protein, IVa2, fiber, and protein X where coverage was between 15% and 20% (Bauer et al., 2019).
Protein V occurs in about 150 copies per virion (Benevento et al., 2014). It links the inner DNA core with the capsid wall and a fraction of it associates with proteins VI and VIII (Perez‐Vargas et al., 2014; Reddy & Nemerow, 2014). Both protein VI and VIII are quantitatively released at early steps of HAdV‐C2 entry (Burckhardt et al., 2011; Greber et al., 1993). In good accordance with these data, about one third of the protein V‐GFP fusion proteins was rapidly released from incoming HAdV‐C2 within the first 30 min, while the rest was released upon disassembly and DNA release at the NPC (Bauer et al., 2021; Puntener et al., 2011). Protein V has 368 amino acids, including 26 lysine residues, and strongly and unspecifically attaches to DNA through multiple binding sites (Perez‐Vargas et al., 2014).
Interesting phenotypes were observed when cell entry of HAdV‐C5 virions lacking protein V was compared to that of particles in which all lysines of V were changed to arginines (V‐KR). The absence of protein V reduced the thermostability of the virion and nuclear import of the viral DNA. It also strongly increased the fraction of prematurely released virion DNA in the cytosol, before virus attachment to the NPC, as observed in cells treated with LMB. This gave rise to enhanced cytokine production in macrophages, depending on the cytosolic DNA sensor cGAS (Bauer et al., 2021). Importantly, the KO of protein V rendered the infection independent of MIB1 but required more inoculum than infection of normal cells with wild‐type HAdV‐C5. These data indicate that protein V of incoming HAdV‐C5 is a direct or indirect ubiquitination target of MIB1. Remarkably, the V‐KR mutant was defective of DNA nuclear import in normal cells but was not impaired at DNA release from the NPC‐docked virions. Instead, the uncoated viral DNA was mistargeted to the cytosol to a large extent, whereas in LMB‐treated cells the incoming V‐KR particles were stable, in contrast to virions lacking V. Similar to wild‐type virions, uncoating of V‐KR virions was impaired at the NPC in MIB1‐KO cells. Reverting 2 of the 26 mutated arginine residues to lysines restored the nuclear import phenotype and infection. These data strongly argue that (1) ubiquitination and degradation of protein V are key for viral DNA nuclear import, and (2) MIB1 has another ubiquitination target besides protein V to license the uncoating process of the NPC‐docked virion.
6. AN INTEGRATED VIEW ON HAdV CYTOPLASMIC TRAFFICKING AND DISASSEMBLY AT THE NPC
Based on the data discussed above, we suggest the following model for the cytoplasmic events in HAdV‐C cell entry downstream of endosomal escape. Cytoplasmic virions are destabilized by the detachment of proteins IIIa, IV, VI, and VIII and lack a fraction of penton base and protein V. These particles engage with dynein/dynactin and kinesin motors and traffic on microtubules toward the nucleus. Although the particles are leaky and their DNA is accessible to staining by click chemistry reagents (Wang, Suomalainen, et al., 2013), their protein V is inaccessible to proximity ligation by APEX‐MIB1 (Sarbanes et al., 2021). The resistance of protein V to MIB1 ubiquitination allows V to function as a linchpin between the viral capsid and the DNA core. This greatly enhances the stability of the virions in the cytoplasm and protects them against premature disassembly. In subsequent steps, the virion detaches from the microtubules near the nuclear envelope, depending on a CRM1‐mediated signal from the nucleus (Wang et al., 2017).
As depicted in Figure 1, the virion docks to the NPC by direct hexon binding to Nup214, as suggested by chemical cross‐linking (Trotman et al., 2001) as well as depletion of Nup214 and restoration of NPC binding by expression of the N‐terminal 137 amino acids of Nup214 (Cassany et al., 2015). The NPC‐docked HAdV‐C5 is in a locked‐in state and prevented from disassembly by a factor(s) that require MIB1‐dependent ubiquitination and proteasomal degradation (Bauer et al., 2021; Sarbanes et al., 2021). Once the inhibitory factor(s) are released or the rupture process licensed, the virion is disrupted by KIF5C/TPR1/2‐mediated pulling forces on protein IX, which work against tethering forces of the NPC (Strunze et al., 2011). This process may be assisted in yet unknown ways by the W142‐P143 domain of CRM1 (Lagadec et al., 2021). Capsid rupture also disrupts the NPC, as shown by increased influx of fluorescent dextrans into the nucleus (Strunze et al., 2011). The viral DNA condensed with protein VII and linked to the terminal protein is then imported into the nucleus, depending on a range of host factors, including histone H1, heat shock cognate protein 70, and importins α, β1, β2, and 7 (Hindley et al., 2007; Saphire et al., 2000; Trotman et al., 2001; Wodrich et al., 2006). Further studies of viral DNA import into host cell nuclei may take advantage of live‐cell imaging chemistry with sufficient signal to noise ratios combined with super‐resolution microscopy. Such studies are required to provide mechanistic insights into the apparently stochastic nature of viral DNA nuclear import and misdelivery to the cytosol (Flatt & Greber, 2015; Wang, Suomalainen, et al., 2013).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENT
We acknowledge the present and past members of the Greber group for discussions, in particular Alfonso Gomez‐Gonzalez and Michael Bauer. Open Access Funding provided by Universitat Zurich. [Correction added on 20 May 2022, after first online publication: CSAL funding statement has been added.]
Greber, U. F. & Suomalainen, M. (2022). Adenovirus entry: Stability, uncoating, and nuclear import. Molecular Microbiology, 118, 309–320. 10.1111/mmi.14909
DATA AVAILABILITY STATEMENT
Data sharing not applicable ‐ no new data generated.
REFERENCES
- Allen, R.J. & Byrnes, A.P. (2019) Interaction of adenovirus with antibodies, complement, and coagulation factors. FEBS Letters, 593(24), 3449–3460. [DOI] [PubMed] [Google Scholar]
- Alonso‐Padilla, J. , Papp, T. , Kajan, G.L. , Benko, M. , Havenga, M. , Lemckert, A. et al. (2016) Development of novel adenoviral vectors to overcome challenges observed with HAdV‐5‐based constructs. Molecular Therapy, 24(1), 6–16. 10.1038/mt.2015.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaiahgari, M.B. & Vrati, S. (2015) Adenoviruses as gene/vaccine delivery vectors: promises and pitfalls. Expert Opinion on Biological Therapy, 15(3), 337–351. [DOI] [PubMed] [Google Scholar]
- Arnberg, N. (2012) Adenovirus receptors: implications for targeting of viral vectors. Trends in Pharmacological Sciences, 33(8), 442–448. [DOI] [PubMed] [Google Scholar]
- Atasheva, S. , Yao, J. & Shayakhmetov, D.M. (2019) Innate immunity to adenovirus: lessons from mice. FEBS Letters, 593(24), 3461–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A.T. , Greenshields‐Watson, A. , Coughlan, L. , Davies, J.A. , Uusi‐Kerttula, H. , Cole, D.K. et al. (2019) Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions. Nature Communications, 10(1), 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A.T. , Mundy, R.M. , Davies, J.A. , Rizkallah, P.J. & Parker, A.L. (2019) Human adenovirus type 26 uses sialic acid‐bearing glycans as a primary cell entry receptor. Science Advances, 5(9), eaax3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, M.A. , Rubin, J.D. & Lu, S.C. (2020) Retargeting adenoviruses for therapeutic applications and vaccines. FEBS Letters, 594(12), 1918–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, M. , Flatt, J.W. , Seiler, D. , Cardel, B. , Emmenlauer, M. , Boucke, K. et al. (2019) The E3 ubiquitin ligase mind bomb 1 controls adenovirus genome release at the nuclear pore complex. Cell Reports, 29(12), 3785–3795 e8. [DOI] [PubMed] [Google Scholar]
- Bauer, M. , Gomez‐Gonzalez, A. , Suomalainen, M. , Schilling, N. , Hemmi, S. & Greber, U.F. (2021) A viral ubiquitination switch attenuates innate immunity and triggers nuclear import of virion DNA and infection. Science Advances, 7(51), eabl7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento, M. , Di Palma, S. , Snijder, J. , Moyer, C.L. , Reddy, V.S. , Nemerow, G.R. et al. (2014) Adenovirus composition, proteolysis, and disassembly studied by in‐depth qualitative and quantitative proteomics. The Journal of Biological Chemistry, 289(16), 11421–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J.M. , Cunningham, J.A. , Droguett, G. , Kurt‐Jones, E.A. , Krithivas, A. , Hong, J.S. et al. (1997) Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science, 275(5304), 1320–1323. [DOI] [PubMed] [Google Scholar]
- Berndt, J.D. , Aoyagi, A. , Yang, P. , Anastas, J.N. , Tang, L. & Moon, R.T. (2011) Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/beta‐catenin signaling. The Journal of Cell Biology, 194(5), 737–750. 10.1083/jcb.201107021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri, M. , Hendrickx, R. , Bauer, M. , Yu, B. , Jetzer, T. , Dreier, B. et al. (2021) The RGD‐binding integrins alphavbeta6 and alphavbeta8 are receptors for mouse adenovirus‐1 and ‐3 infection. PLoS Pathogens, 17(12), e1010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bots, S.T.F. & Hoeben, R.C. (2020) Non‐human primate‐derived adenoviruses for future use as oncolytic agents? International Journal of Molecular Sciences, 21(14). 10.3390/ijms21144821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner, K.H. , Scherer, J. , Yi, J. , Vershinin, M. , Gross, S.P. & Vallee, R.B. (2009) Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host & Microbe, 6(6), 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucher, D. , Kirchhammer, N. , Smith, S.N. , Schumacher, J. , Schumacher, N. , Kolibius, J. et al. (2021) iMATCH: an integrated modular assembly system for therapeutic combination high‐capacity adenovirus gene therapy. Molecular Therapy – Methods & Clinical Development, 20, 572–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt, C.J. , Suomalainen, M. , Schoenenberger, P. , Boucke, K. , Hemmi, S. & Greber, U.F. (2011) Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host & Microbe, 10(2), 105–117. [DOI] [PubMed] [Google Scholar]
- Campbell, I.D. & Humphries, M.J. (2011) Integrin structure, activation, and interactions. Cold Spring Harbor Perspectives in Biology, 3(3), a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone, S. , D'Alise, A.M. , Ammendola, V. , Colloca, S. , Cortese, R. , Nicosia, A. et al. (2013) Development of chimpanzee adenoviruses as vaccine vectors: challenges and successes emerging from clinical trials. Expert Review of Vaccines, 12(4), 379–393. [DOI] [PubMed] [Google Scholar]
- Cassany, A. , Ragues, J. , Guan, T. , Begu, D. , Wodrich, H. , Kann, M. et al. (2015) Nuclear import of adenovirus DNA involves direct interaction of hexon with an N‐terminal domain of the nucleoporin Nup214. Journal of Virology, 89(3), 1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, P.K. , Vayda, M.E. & Flint, S.J. (1985) Interactions among the three adenovirus core proteins. Journal of Virology, 55(2), 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheneau, C. & Kremer, E.J. (2020) Adenovirus‐Extracellular Protein Interactions and Their Impact on Innate Immune Responses by Human Mononuclear Phagocytes. Viruses, 12(12). 10.3390/v12121351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Huang, X. , Li, X. , Xiong, W. , Sun, W. , Yang, C. et al. (2014) Cryo‐EM structures of two bovine adenovirus type 3 intermediates. Virology, 450‐451, 174–181. [DOI] [PubMed] [Google Scholar]
- Cherry, S. , Doukas, T. , Armknecht, S. , Whelan, S. , Wang, H. , Sarnow, P. et al. (2005) Genome‐wide RNAi screen reveals a specific sensitivity of IRES‐containing RNA viruses to host translation inhibition. Genes & Development, 19(4), 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K.I. , Yi, H. , Desai, R. , Hand, A.R. , Haas, A.L. & Ferreira, P.A. (2009) RANBP2 is an allosteric activator of the conventional kinesin‐1 motor protein, KIF5B, in a minimal cell‐free system. EMBO Reports, 10(5), 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby, W.W. & Shenk, T. (1981) Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. Journal of Virology, 39(3), 977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, C.B. , Bozym, R. , Morosky, S.A. , Hanna, S.L. , Mukherjee, A. , Tudor, M. et al. (2011) Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host & Microbe, 9(1), 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupelli, K. , Muller, S. , Persson, B.D. , Jost, M. , Arnberg, N. & Stehle, T. (2010) Structure of adenovirus type 21 knob in complex with CD46 reveals key differences in receptor contacts among species B adenoviruses. Journal of Virology, 84(7), 3189–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , Wu, L. , Sun, R. & Zhou, Z.H. (2017) Atomic structures of minor proteins VI and VII in the human adenovirus. Journal of Virology, 91(24), e00850‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, C. , Pekosz, A. & Ketner, G. (2013) Prospects for oral replicating adenovirus‐vectored vaccines. Vaccine, 31(32), 3236–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner, K. & Sodeik, B. (2005) The role of the cytoskeleton during viral infection. Current Topics in Microbiology and Immunology, 285, 67–108. 10.1016/j.tim.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Doronin, K. , Flatt, J.W. , Di Paolo, N.C. , Khare, R. , Kalyuzhniy, O. , Acchione, M. et al. (2012) Coagulation factor X activates innate immunity to human species C adenovirus. Science, 338(6108), 795–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichholz, K. , Bru, T. , Tran, T.T. , Fernandes, P. , Welles, H. , Mennechet, F.J. et al. (2016) Immune‐complexed adenovirus induce AIM2‐mediated pyroptosis in human dendritic cells. PLoS Pathogens, 12(9), e1005871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichholz, K. , Mennechet, F.J. & Kremer, E.J. (2015) Human coagulation factor x‐adenovirus type 5 complexes poorly stimulate an innate immune response in human mononuclear phagocytes. Journal of Virology, 89(5), 2884–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke, M.F. , Burckhardt, C.J. , Morf, M.K. & Greber, U.F. (2011) The dynactin complex enhances the speed of microtubule‐dependent motions of adenovirus both towards and away from the nucleus. Viruses, 3(3), 233–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon, K.J.D.A. (2020) The coxsackievirus and adenovirus receptor: virological and biological beauty. FEBS Letters, 594, 1828–1837. [DOI] [PubMed] [Google Scholar]
- Fausther‐Bovendo, H. & Kobinger, G.P. (2014) Pre‐existing immunity against Ad vectors: humoral, cellular, and innate response, what's important? Human Vaccines & Immunotherapeutics, 10(10), 2875–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt, J.W. & Greber, U.F. (2015) Misdelivery at the nuclear pore complex‐stopping a virus dead in its tracks. Cell, 4(3), 277–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt, J.W. & Greber, U.F. (2017) Viral mechanisms for docking and delivering at nuclear pore complexes. Seminars in Cell & Developmental Biology, 68, 59–71. [DOI] [PubMed] [Google Scholar]
- Fleischli, C. , Sirena, D. , Lesage, G. , Havenga, M.J. , Cattaneo, R. , Greber, U.F. et al. (2007) Species B adenovirus serotypes 3, 7, 11 and 35 share similar binding sites on the membrane cofactor protein CD46 receptor. The Journal of General Virology, 88(Pt 11), 2925–2934. [DOI] [PubMed] [Google Scholar]
- Fleischli, C. , Verhaagh, S. , Havenga, M. , Sirena, D. , Schaffner, W. , Cattaneo, R. et al. (2005) The distal short consensus repeats 1 and 2 of the membrane cofactor protein CD46 and their distance from the cell membrane determine productive entry of species B adenovirus serotype 35. Journal of Virology, 79(15), 10013–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth, P. , Springer, K. , Berard, C. , Hainfeld, J. , Bewley, M. & Flanagan, J. (1999) Coxsackievirus and adenovirus receptor amino‐terminal immunoglobulin V‐related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. Journal of Virology, 73(2), 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggar, A. , Shayakhmetov, D.M. & Lieber, A. (2003) CD46 is a cellular receptor for group B adenoviruses. Nature Medicine, 9(11), 1408–1412. [DOI] [PubMed] [Google Scholar]
- Gallardo, J. , Perez‐Illana, M. , Martin‐Gonzalez, N. & San, M.C. (2021) Adenovirus structure: what is new? International Journal of Molecular Sciences, 22(10). 10.3390/ijms22105240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Mese, K. , Bunz, O. & Ehrhardt, A. (2019) State‐of‐the‐art human adenovirus vectorology for therapeutic approaches. FEBS Letters, 593(24), 3609–3622. [DOI] [PubMed] [Google Scholar]
- Gastaldelli, M. , Imelli, N. , Boucke, K. , Amstutz, B. , Meier, O. & Greber, U.F. (2008) Infectious adenovirus type 2 transport through early but not late endosomes. Traffic, 9(12), 2265–2278. [DOI] [PubMed] [Google Scholar]
- Gazzola, M. , Burckhardt, C.J. , Bayati, B. , Engelke, M. , Greber, U.F. & Koumoutsakos, P. (2009) A stochastic model for microtubule motors describes the in vivo cytoplasmic transport of human adenovirus. PLoS Computational Biology, 5(12), e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebremedhin, B. (2014) Human adenovirus: viral pathogen with increasing importance. European Journal of Microbiology and Immunology, 4(1), 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber, U.F. (2016) Virus and host mechanics support membrane penetration and cell entry. Journal of Virology, 90(8), 3802–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber, U.F. (2020) Adenoviruses—infection, pathogenesis and therapy. FEBS Letters, 594(12), 1818–1827. [DOI] [PubMed] [Google Scholar]
- Greber, U.F. & Flatt, J.W. (2019) adenovirus entry: from infection to immunity. Annual Review of Virology, 6(1), 177–197. 10.1146/annurev-virology-092818-015550 [DOI] [PubMed] [Google Scholar]
- Greber, U.F. & Gomez‐Gonzalez, A. (2021) Adenovirus—a blueprint for gene delivery. Current Opinion in Virology, 48, 49–56. 10.1016/j.coviro.2021.03.006 [DOI] [PubMed] [Google Scholar]
- Greber, U.F. , Singh, I. & Helenius, A. (1994) Mechanisms of virus uncoating. Trends in Microbiology, 2(2), 52–56. 10.1016/0966-842x(94)90126-0 [DOI] [PubMed] [Google Scholar]
- Greber, U.F. & Way, M. (2006) A superhighway to virus infection. Cell, 124(4), 741–754. [DOI] [PubMed] [Google Scholar]
- Greber, U.F. , Willetts, M. , Webster, P. & Helenius, A. (1993) Stepwise dismantling of adenovirus 2 during entry into cells. Cell, 75(3), 477–486. [DOI] [PubMed] [Google Scholar]
- Guo, B. , McMillan, B.J. & Blacklow, S.C. (2016) Structure and function of the Mind bomb E3 ligase in the context of Notch signal transduction. Current Opinion in Structural Biology, 41, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, D.J. , Andersson, E.K. , Hu, Y.L. , Marttila, M. , Lindman, K. , Strand, M. et al. (2010) Adenovirus 11p downregulates CD46 early in infection. Virology, 405(2), 474–482. [DOI] [PubMed] [Google Scholar]
- Hackenbrack, N. , Rogers, M.B. , Ashley, R.E. , Keel, M.K. , Kubiski, S.V. , Bryan, J.A. et al. (2017) Evolution and cryo‐electron microscopy capsid structure of a north American bat adenovirus and its relationship to other mastadenoviruses. Journal of Virology, 91(2). 10.1128/JVI.01504-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach, B. , Tarjan, Z.L. & Benko, M. (2019) Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Letters, 593(24), 3660–3673. [DOI] [PubMed] [Google Scholar]
- Hemsath, J.R. , Liaci, A.M. , Rubin, J.D. , Parrett, B.J. , Lu, S.C. , Nguyen, T.V. et al. (2022) Ex vivo and in vivo CD46 receptor utilization by species D human adenovirus serotype 26 (HAdV26). Journal of Virology, 96(3), e0082621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx, R. , Stichling, N. , Koelen, J. , Kuryk, L. , Lipiec, A. & Greber, U.F. (2014) Innate immunity to adenovirus. Human Gene Therapy, 25, 265–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando‐Perez, M. , Martin‐Gonzalez, N. , Perez‐Illana, M. , Suomalainen, M. , Condezo, G.N. , Ostapchuk, P. et al. (2020) Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proceedings of the National Academy of Sciences of the United States of America, 117(24), 13699–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley, C.E. , Lawrence, F.J. & Matthews, D.A. (2007) A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic, 8(10), 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke, C.H., Jr. , Hawksworth, A. & Snyder, C.E., Jr. (2012) Initial assessment of impact of adenovirus type 4 and type 7 vaccine on febrile respiratory illness and virus transmission in military basic trainees, March 2012. MSMR, 19(3), 2–4. [PubMed] [Google Scholar]
- Joseph, J. & Dasso, M. (2008) The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Letters, 582(2), 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas, A. , Machuy, N. , Shin, Y. , Pleissner, K.P. , Artarini, A. , Heuer, D. et al. (2010) Genome‐wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature, 463(7282), 818–822. [DOI] [PubMed] [Google Scholar]
- Kelkar, S. , De, B.P. , Gao, G. , Wilson, J.M. , Crystal, R.G. & Leopold, P.L. (2006) A common mechanism for cytoplasmic dynein‐dependent microtubule binding shared among adeno‐associated virus and adenovirus serotypes. Journal of Virology, 80(15), 7781–7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare, R. , Reddy, V.S. , Nemerow, G.R. & Barry, M.A. (2012) Identification of adenovirus serotype 5 hexon regions that interact with scavenger receptors. Journal of Virology, 86(4), 2293–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, I. , Davison, E. , Beavil, A.J. , Soh, C.P. , Wickham, T.J. , Roelvink, P.W. et al. (2000) Identification of contact residues and definition of the CAR‐binding site of adenovirus type 5 fiber protein. Journal of Virology, 74(6), 2804–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkeaw, N. , Jeon, S.H. , Lee, K. , Johnson, B.H. , Tanasanvimon, S. , Javle, M. et al. (2013) Cell death/proliferation roles for nc886, a non‐coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene, 32(32), 3722–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadec, F. , Carlon‐Andres, I. , Ragues, J. , Port, S. , Wodrich, H. & Kehlenbach, R.H. (2021) CRM1 promotes capsid disassembly and nuclear envelope translocation of adenovirus independently of its export function. Journal of Virology, 96, JVI0127321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S. , Bao, X. , Lee, H.H. , Jang, J.J. , Saruuldalai, E. , Park, G. et al. (2021) Nc886, a Novel suppressor of the type I interferon response upon pathogen intrusion. International Journal of Molecular Sciences, 22(4). 10.3390/ijms22042003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , Kunkeaw, N. , Jeon, S.H. , Lee, I. , Johnson, B.H. , Kang, G.Y. et al. (2011) Precursor miR‐886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA, 17(6), 1076–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. & Lieber, A. (2019) Adenovirus vectors in hematopoietic stem cell genome editing. FEBS Letters, 593(24), 3623–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion, T. (2019) Adenovirus persistence, reactivation, and clinical management. FEBS Letters, 593(24), 3571–3582. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Jin, L. , Koh, S.B. , Atanasov, I. , Schein, S. , Wu, L. et al. (2010) Atomic structure of human adenovirus by cryo‐EM reveals interactions among protein networks. Science, 329(5995), 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisoni, S. , Bauer, M. , Prasad, V. , Boucke, K. , Papadopoulos, C. , Meyer, H. et al. (2016) Endosomophagy clears disrupted early endosomes but not virus particles during virus entry into cells. Matters, 6, 13–22. 10.19185/matters.201606000013 [DOI] [Google Scholar]
- Luisoni, S. & Greber, U.F. (2016) Biology of adenovirus cell entry—receptors, pathways, mechanisms. In: Curiel, D. (Ed.) Adenoviral vectors for gene therapy, 2nd edition. London, UK: Academic Press, Elsevier, pp. 27–58. [Google Scholar]
- Luisoni, S. , Suomalainen, M. , Boucke, K. , Tanner, L.B. , Wenk, M.R. , Guan, X.L. et al. (2015) Co‐option of membrane wounding enables virus penetration into cells. Cell Host & Microbe, 18(1), 75–85. [DOI] [PubMed] [Google Scholar]
- Luxan, G. , Casanova, J.C. , Martinez‐Poveda, B. , Prados, B. , D'Amato, G. , MacGrogan, D. et al. (2013) Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nature Medicine, 19(2), 193–201. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Duffy, M.R. , Deng, L. , Dakin, R.S. , Uil, T. , Custers, J. et al. (2015) Manipulating adenovirus hexon hypervariable loops dictates immune neutralisation and coagulation factor X‐dependent cell interaction in vitro and in vivo. PLoS Pathogens, 11(2), e1004673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod, D.T. , Nakatsuji, T. , Yamasaki, K. , Kobzik, L. & Gallo, R.L. (2013) HSV‐1 exploits the innate immune scavenger receptor MARCO to enhance epithelial adsorption and infection. Nature Communications, 4, 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, O. , Marvin, S.A. , Wodrich, H. , Campbell, E.M. & Wiethoff, C.M. (2012) Spatiotemporal dynamics of adenovirus membrane rupture and endosomal escape. Journal of Virology, 86(19), 10821–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maler, M.D. , Nielsen, P.J. , Stichling, N. , Cohen, I. , Ruzsics, Z. , Wood, C. et al. (2017) Key role of the scavenger receptor MARCO in mediating adenovirus infection and subsequent innate responses of macrophages. mBio, 8(4). 10.1128/mBio.00670-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel, W.F. & San, M.C. (2014) Structure, function and dynamics in adenovirus maturation. Viruses, 6(11), 4536–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini, R. , Condezo, G.N. , Krupovic, M. , Menendez‐Conejero, R. , Gomez‐Blanco, J. & San, M.C. (2021) Near‐atomic structure of an atadenovirus reveals a conserved capsid‐binding motif and intergenera variations in cementing proteins. Science Advances, 7(14). 10.1126/sciadv.abe6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Gonzalez, N. , Hernando‐Perez, M. , Condezo, G.N. , Perez‐Illana, M. , Siber, A. , Reguera, D. et al. (2019) Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure. Nucleic Acids Research, 47(17), 9231–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes‐Martin, S. , Boztug, H. & Lion, T. (2013) Diagnosis and treatment of adenovirus infection in immunocompromised patients. Expert Review of Anti‐Infective Therapy, 11(10), 1017–1028. [DOI] [PubMed] [Google Scholar]
- Meier, O. & Greber, U.F. (2003) Adenovirus endocytosis. The Journal of Gene Medicine, 5(6), 451–462. 10.1002/jgm.409 [DOI] [PubMed] [Google Scholar]
- Mendonca, S.A. , Lorincz, R. , Boucher, P. & Curiel, D.T. (2021) Adenoviral vector vaccine platforms in the SARS‐CoV‐2 pandemic. NPJ Vaccines, 6(1), 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Ismail, A. , Zhou, X. , Dyer, D.W. , Seto, D. , Rajaiya, J. & Chodosh, J. (2019) Genomic foundations of evolution and ocular pathogenesis in human adenovirus species D. FEBS Letters, 593(24), 3583–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montespan, C. , Marvin, S.A. , Austin, S. , Burrage, A.M. , Roger, B. , Rayne, F. et al. (2017) Multi‐layered control of Galectin‐8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathogens, 13(2), e1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, C.L. , Wiethoff, C.M. , Maier, O. , Smith, J.G. & Nemerow, G.R. (2011) Functional genetic and biophysical analyses of membrane disruption by human adenovirus. Journal of Virology, 85(6), 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsch, A. (2004) Microtubule organization and function in epithelial cells. Traffic, 5(1), 1–9. [DOI] [PubMed] [Google Scholar]
- Nagel, H. , Maag, S. , Tassis, A. , Nestle, F.O. , Greber, U.F. & Hemmi, S. (2003) The alphavbeta5 integrin of hematopoietic and nonhematopoietic cells is a transduction receptor of RGD‐4C fiber‐modified adenoviruses. Gene Therapy, 10(19), 1643–1653. [DOI] [PubMed] [Google Scholar]
- Nakano, M.Y. , Boucke, K. , Suomalainen, M. , Stidwill, R.P. & Greber, U.F. (2000) The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. Journal of Virology, 74(15), 7085–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow, G.R. (2000) Cell receptors involved in adenovirus entry. Virology, 274(1), 1–4. [DOI] [PubMed] [Google Scholar]
- Nemerow, G.R. & Stewart, P.L. (2016) Insights into adenovirus uncoating from interactions with integrins and mediators of host immunity. Viruses, 8(12). 10.3390/v8120337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oostrum, J. & Burnett, R.M. (1985) Molecular composition of the adenovirus type 2 virion. Journal of Virology, 56(2), 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapchuk, P. , Suomalainen, M. , Zheng, Y. , Boucke, K. , Greber, U.F. & Hearing, P. (2017) The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathogens, 13(6), e1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pache, L. , Venkataraman, S. , Reddy, V.S. & Nemerow, G.R. (2008) Structural variations in species B adenovirus fibers impact CD46 association. Journal of Virology, 82(16), 7923–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, D. , Das, A. , Dinh, P.X. , Subramaniam, S. , Nayak, D. , Barrows, N.J. et al. (2011) RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative‐strand RNA viruses. Proceedings of the National Academy of Sciences of the United States of America, 108(47), 19036–19041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, R.J. , Chen, L. , Anton, M. , Sankar, U. , Rudnicki, M.A. & Graham, F.L. (1996) A helper‐dependent adenovirus vector system: removal of helper virus by Cre‐mediated excision of the viral packaging signal. Proceedings of the National Academy of Sciences of the United States of America, 93(24), 13565–13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Berna, A.J. , Marion, S. , Chichon, F.J. , Fernandez, J.J. , Winkler, D.C. , Carrascosa, J.L. et al. (2015) Distribution of DNA‐condensing protein complexes in the adenovirus core. Nucleic Acids Research, 43, 4274–4283. 10.1093/nar/gkv187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Illana, M. , Martinez, M. , Condezo, G.N. , Hernando‐Perez, M. , Mangroo, C. , Brown, M. et al. (2021) Cryo‐EM structure of enteric adenovirus HAdV‐F41 s structural variations among human adenoviruses. Science Advances, 7(9). 10.1126/sciadv.abd9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Vargas, J. , Vaughan, R.C. , Houser, C. , Hastie, K.M. , Kao, C.C. & Nemerow, G.R. (2014) Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. Journal of Virology, 88(16), 9287–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B.D. , John, L. , Rafie, K. , Strebl, M. , Frangsmyr, L. , Ballmann, M.Z. et al. (2021) Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proceedings of the National Academy of Sciences of the United States of America, 118(3). 10.1073/pnas.2020732118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B.D. , Muller, S. , Reiter, D.M. , Schmitt, B.B. , Marttila, M. , Sumowski, C.V. et al. (2009) An arginine switch in the species B adenovirus knob determines high‐affinity engagement of cellular receptor CD46. Journal of Virology, 83(2), 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pied, N. & Wodrich, H. (2019) Imaging the adenovirus infection cycle. FEBS Letters, 593(24), 3419–3448. [DOI] [PubMed] [Google Scholar]
- Prasad, V. & Greber, U.F. (2021) The endoplasmic reticulum unfolded protein response—homeostasis, cell death and evolution in virus infections. FEMS Microbiology Reviews, 45(5). 10.1093/femsre/fuab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntener, D. , Engelke, M.F. , Ruzsics, Z. , Strunze, S. , Wilhelm, C. & Greber, U.F. (2011) Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. Journal of Virology, 85(1), 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschnik, A.S. , Majzoub, K. , Ooi, Y.S. & Carette, J.E. (2017) A CRISPR toolbox to study virus‐host interactions. Nature Reviews Microbiology, 15(6), 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafie, K. , Lenman, A. , Fuchs, J. , Rajan, A. , Arnberg, N. & Carlson, L.A. (2021) The structure of enteric human adenovirus 41‐A leading cause of diarrhea in children. Science Advances, 7(2). 10.1126/sciadv.abe0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raty, J.K. , Lesch, H.P. , Wirth, T. & Yla‐Herttuala, S. (2008) Improving safety of gene therapy. Current Drug Safety, 3(1), 46–53. 10.2174/157488608783333925 [DOI] [PubMed] [Google Scholar]
- Raynaud‐Messina, B. & Merdes, A. (2007) Gamma‐tubulin complexes and microtubule organization. Current Opinion in Cell Biology, 19(1), 24–30. 10.1016/j.ceb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Reddy, V.S. & Nemerow, G.R. (2014) Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proceedings of the National Academy of Sciences of the United States of America, 111(32), 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelvink, P.W. , Lizonova, A. , Lee, J.G. , Li, Y. , Bergelson, J.M. , Finberg, R.W. et al. (1998) The coxsackievirus‐adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. Journal of Virology, 72(10), 7909–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, S. (2004) CD46: a complement regulator and pathogen receptor that mediates links between innate and acquired immune function. Tissue Antigens, 64(2), 111–118. [DOI] [PubMed] [Google Scholar]
- Sakhuja, K. , Reddy, P.S. , Ganesh, S. , Cantaniag, F. , Pattison, S. , Limbach, P. et al. (2003) Optimization of the generation and propagation of gutless adenoviral vectors. Human Gene Therapy, 14(3), 243–254. [DOI] [PubMed] [Google Scholar]
- Sakurai, F. , Akitomo, K. , Kawabata, K. , Hayakawa, T. & Mizuguchi, H. (2007) Downregulation of human CD46 by adenovirus serotype 35 vectors. Gene Therapy, 14(11), 912–919. [DOI] [PubMed] [Google Scholar]
- Sakurai, F. , Murakami, S. , Kawabata, K. , Okada, N. , Yamamoto, A. , Seya, T. et al. (2006) The short consensus repeats 1 and 2, not the cytoplasmic domain, of human CD46 are crucial for infection of subgroup B adenovirus serotype 35. Journal of Controlled Release, 113(3), 271–278. [DOI] [PubMed] [Google Scholar]
- Saphire, A.C. , Guan, T. , Schirmer, E.C. , Nemerow, G.R. & Gerace, L. (2000) Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. The Journal of Biological Chemistry, 275(6), 4298–4304. [DOI] [PubMed] [Google Scholar]
- Sarbanes, S.L. , Blomen, V.A. , Lam, E. , Heissel, S. , Luna, J.M. , Brummelkamp, T.R. et al. (2021) E3 ubiquitin ligase Mindbomb 1 facilitates nuclear delivery of adenovirus genomes. Proceedings of the National Academy of Sciences of the United States of America, 118(1). 10.1073/pnas.2015794118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruuldalai, E. , Park, J. , Kang, D. , Shin, S.‐P. , Im, W.R. , Lee, H.‐H. et al. (2022) A host non‐coding RNA, nc886, plays a pro‐viral role by promoting virus trafficking to the nucleus. Molecular Therapy—Oncolytics, 24, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, J. & Vallee, R.B. (2015) Conformational changes in the adenovirus hexon subunit responsible for regulating cytoplasmic Dynein recruitment. Journal of Virology, 89(2), 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, J. , Yi, J. & Vallee, R.B. (2020) Role of cytoplasmic dynein and kinesins in adenovirus transport. FEBS Letters, 594, 1838–1847. [DOI] [PubMed] [Google Scholar]
- Schmid, M. , Ernst, P. , Honegger, A. , Suomalainen, M. , Zimmermann, M. , Braun, L. et al. (2018) Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control. Nature Communications, 9(1), 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoehn, G. , El Bakkouri, M. , Fabry, C.M. , Billet, O. , Estrozi, L.F. , Le, L. et al. (2008) Three‐dimensional structure of canine adenovirus serotype 2 capsid. Journal of Virology, 82(7), 3192–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirena, D. , Lilienfeld, B. , Eisenhut, M. , Kalin, S. , Boucke, K. , Beerli, R.R. et al. (2004) The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. Journal of Virology, 78(9), 4454–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirena, D. , Ruzsics, Z. , Schaffner, W. , Greber, U.F. & Hemmi, S. (2005) The nucleotide sequence and a first generation gene transfer vector of species B human adenovirus serotype 3. Virology, 343(2), 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.G. , Cassany, A. , Gerace, L. , Ralston, R. & Nemerow, G.R. (2008) A neutralizing antibody blocks adenovirus infection by arresting microtubule‐dependent cytoplasmic transport. Journal of Virology, 82(13), 6492–6500. 10.1128/JVI.00557-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, B. , Sacher, R. , Ramo, P. , Liberali, P. , Mench, K. , Wolfrum, N. et al. (2012) Single‐cell analysis of population context advances RNAi screening at multiple levels. Molecular Systems Biology, 8, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, S.Y. & Hearing, P. (2019) Adenoviral strategies to overcome innate cellular responses to infection. FEBS Letters, 593(24), 3484–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudais, C. , Boutin, S. , Hong, S.S. , Chillon, M. , Danos, O. , Bergelson, J.M. et al. (2000) Canine adenovirus type 2 attachment and internalization: coxsackievirus‐adenovirus receptor, alternative receptors, and an RGD‐independent pathway. Journal of Virology, 74(22), 10639–10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak, A.C. & Stehle, T. (2020) Human adenovirus binding to host cell receptors: a structural view. Medical Microbiology and Immunology, 209(3), 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, P.L. & Nemerow, G.R. (2007) Cell integrins: commonly used receptors for diverse viral pathogens. Trends in Microbiology, 15(11), 500–507. [DOI] [PubMed] [Google Scholar]
- Stichling, N. , Suomalainen, M. , Flatt, J.W. , Schmid, M. , Pacesa, M. , Hemmi, S. et al. (2018) Lung macrophage scavenger receptor SR‐A6 (MARCO) is an adenovirus type‐specific virus entry receptor. PLoS Pathogens, 14(3), e1006914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunze, S. , Engelke, M.F. , Wang, I.H. , Puntener, D. , Boucke, K. , Schleich, S. et al. (2011) Kinesin‐1‐mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host & Microbe, 10(3), 210–223. [DOI] [PubMed] [Google Scholar]
- Strunze, S. , Trotman, L.C. , Boucke, K. & Greber, U.F. (2005) Nuclear targeting of adenovirus type 2 requires CRM1‐mediated nuclear export. Molecular Biology of the Cell, 16(6), 2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M. & Greber, U.F. (2021) Virus infection variability by single‐cell profiling. Viruses, 13(8). 10.3390/v13081568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M. , Luisoni, S. , Boucke, K. , Bianchi, S. , Engel, D.A. & Greber, U.F. (2013) A direct and versatile assay measuring membrane penetration of adenovirus in single cells. Journal of Virology, 87(22), 12367–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M. , Nakano, M.Y. , Keller, S. , Boucke, K. , Stidwill, R.P. & Greber, U.F. (1999) Microtubule‐dependent plus‐ and minus end‐directed motilities are competing processes for nuclear targeting of adenovirus. The Journal of Cell Biology, 144(4), 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko, R.P. , Xu, R. & Philipson, L. (1997) HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proceedings of the National Academy of Sciences of the United States of America, 94(7), 3352–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top, F.H., Jr. , Dudding, B.A. , Russell, P.K. & Buescher, E.L. (1971) Control of respiratory disease in recruits with types 4 and 7 adenovirus vaccines. American Journal of Epidemiology, 94(2), 142–146. [DOI] [PubMed] [Google Scholar]
- Trinh, H.V. , Lesage, G. , Chennamparampil, V. , Vollenweider, B. , Burckhardt, C.J. , Schauer, S. et al. (2012) Avidity binding of human adenovirus serotypes 3 and 7 to the membrane cofactor CD46 triggers infection. Journal of Virology, 86, 1623–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman, L.C. , Mosberger, N. , Fornerod, M. , Stidwill, R.P. & Greber, U.F. (2001) Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nature Cell Biology, 3(12), 1092–1100. [DOI] [PubMed] [Google Scholar]
- Tuve, S. , Wang, H. , Ware, C. , Liu, Y. , Gaggar, A. , Bernt, K. et al. (2006) A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. Journal of Virology, 80(24), 12109–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassal‐Stermann, E. , Effantin, G. , Zubieta, C. , Burmeister, W. , Iseni, F. , Wang, H. et al. (2019) CryoEM structure of adenovirus type 3 fibre with desmoglein 2 shows an unusual mode of receptor engagement. Nature Communications, 10(1), 1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellinga, J. , Van der Heijdt, S. & Hoeben, R.C. (2005) The adenovirus capsid: major progress in minor proteins. The Journal of General Virology, 86(Pt 6), 1581–1588. [DOI] [PubMed] [Google Scholar]
- Verma, I.M. & Weitzman, M.D. (2005) Gene therapy: twenty‐first century medicine. Annual Review of Biochemistry, 74, 711–738. [DOI] [PubMed] [Google Scholar]
- de Vrij, J. , van den Hengel, S.K. , Uil, T.G. , Koppers‐Lalic, D. , Dautzenberg, I.J. , Stassen, O.M. et al. (2011) Enhanced transduction of CAR‐negative cells by protein IX‐gene deleted adenovirus 5 vectors. Virology, 410(1), 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehler, R. , Russell, S.J. & Curiel, D.T. (2007) Engineering targeted viral vectors for gene therapy. Nature Reviews. Genetics, 8(8), 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.H. , Burckhardt, C.J. , Yakimovich, A. & Greber, U.F. (2018) Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses, 10(4). 10.3390/v10040166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.H. , Burckhardt, C.J. , Yakimovich, A. , Morf, M.K. & Greber, U.F. (2017) The nuclear export factor CRM1 controls juxta‐nuclear microtubule‐dependent virus transport. Journal of Cell Science, 130(13), 2185–2195. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Ducournau, C. , Saydaminova, K. , Richter, M. , Yumul, R. , Ho, M. et al. (2015) Intracellular signaling and desmoglein 2 shedding triggered by human adenoviruses Ad3, Ad14, and Ad14P1. Journal of Virology, 89(21), 10841–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Li, Z.Y. , Liu, Y. , Persson, J. , Beyer, I. , Moller, T. et al. (2011) Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nature Medicine, 17(1), 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Li, Z. , Yumul, R. , Lara, S. , Hemminki, A. , Fender, P. et al. (2011) Multimerization of adenovirus serotype 3 fiber knob domains is required for efficient binding of virus to desmoglein 2 and subsequent opening of epithelial junctions. Journal of Virology, 85(13), 6390–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.H. , Suomalainen, M. , Andriasyan, V. , Kilcher, S. , Mercer, J. , Neef, A. et al. (2013) Tracking viral genomes in host cells at single‐molecule resolution. Cell Host & Microbe, 14(4), 468–480. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Yumul, R. , Cao, H. , Ran, L. , Fan, X. , Richter, M. et al. (2013) Structural and functional studies on the interaction of adenovirus fiber knobs and desmoglein 2. Journal of Virology, 87(21), 11346–11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M.A. (2004) Bidirectional transport along microtubules. Current Biology, 14(13), R525–R537. [DOI] [PubMed] [Google Scholar]
- Wiethoff, C.M. & Nemerow, G.R. (2015) Adenovirus membrane penetration: tickling the tail of a sleeping dragon. Virology, 479–480C, 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoff, C.M. , Wodrich, H. , Gerace, L. & Nemerow, G.R. (2005) Adenovirus protein VI mediates membrane disruption following capsid disassembly. Journal of Virology, 79(4), 1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte, R. , Andriasyan, V. , Georgi, F. , Yakimovich, A. & Greber, U.F. (2018) Concepts in light microscopy of viruses. Viruses, 10(4). 10.3390/v10040202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodrich, H. , Cassany, A. , D'Angelo, M.A. , Guan, T. , Nemerow, G. & Gerace, L. (2006) Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. Journal of Virology, 80(19), 9608–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodrich, H. , Henaff, D. , Jammart, B. , Segura‐Morales, C. , Seelmeir, S. , Coux, O. et al. (2010) A capsid‐encoded PPxY‐motif facilitates adenovirus entry. PLoS Pathogens, 6(3), e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum, N. & Greber, U.F. (2013) Adenovirus signalling in entry. Cellular Microbiology, 15(1), 53–62. 10.1111/cmi.12053 [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y. & Greber, U.F. (2016) Principles of virus uncoating: cues and the snooker ball. Traffic, 17(6), 569–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Mullen, T.M. , Abrishami, V. , Huiskonen, J.T. , Nemerow, G.R. & Reddy, V.S. (2022) Structure of a cell entry defective human adenovirus provides insights into precursor proteins and capsid maturation. Journal of Molecular Biology, 434(2), 167350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Veesler, D. , Campbell, M.G. , Barry, M.E. , Asturias, F.J. , Barry, M.A. et al. (2017) Cryo‐EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Science Advances, 3(5), e1602670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Punga, T. & Pettersson, U. (2019) Adenovirus in the omics era—a multipronged strategy. FEBS Letters, 594, 1879–1890. 10.1002/1873-3468.13710 [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Scherer, J. , Yi, J. & Vallee, R.B. (2018) Role of kinesins in directed adenovirus transport and cytoplasmic exploration. PLoS Pathogens, 14(5), e1007055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable ‐ no new data generated.


