Abstract
Background
PR domain containing 16 (PRDM16) is a key transcriptional regulator in the development of craniofacial, adipose, and neural tissues. Our lab identified PRDM16 expression in the epithelial cells of the Kölliker's organ (KO) that starts at ~E13.5 and is maintained until KO disappearance. A transgenic mouse model that carries a gene trap null allele of Prdm16 (Prdm16 cGT ) was used to characterize the impact of Prdm16 loss on cochlear development.
Results
At P0 Prdm16 cGT null cochlea exhibited hypoplastic KO, shortened cochlear duct, increased density of hair cells (HCs) and supporting cells (SCs) in the apical turn as well as multiple isolated ectopic HCs within the KO domain. KO epithelial cells proliferation rate was reduced in the apical turn of the developing Prdm16 cGT null cochlea vs controls. Bulk RNA sequencing of cochlear duct cells at E14.5 followed by quantitative real time PCR and mRNA Fluorescence in‐situ hybridization (FISH) validation identified differentially expressed genes in Prdm16 cGT null vs littermate control cochleae. Upregulated genes at E14.5 included Fgf20, as well as several Notch pathway genes (Lfng, Hes1, and Jag1).
Conclusions
This study characterizes Prdm16 expression during cochlear development and establishes its requirement for KO development.
Keywords: Cochlear development, Kölliker's organ, PRDM16
Key Findings
Prdm16 is a novel marker for Kölliker's organ in the developing mammalian cochlea
Prdm16 is required for Kölliker's organ cell proliferation and is involved in spiral limbus and tectorial membrane development
Prdm16 inhibit sensory fates in Kölliker's organ cells during development
1. INTRODUCTION
Mammalian cochlear development involves a precisely orchestrated series of events that convert a simple thickened epithelium (otic placode) to a complex structure connected to the central nervous system. In mice, the cochlear duct arises from the otocyst around embryonic day 11 (E11). 1 As the cochlear duct extends and coils, a subset of cells within its floor begins to develop as the prosensory domain that will give rise to the organ of Corti. 2 This domain is localized to a narrow strip extending along the cochlear duct floor. 1 On the neural side of the developing organ of Corti, a group of epithelial cells constitutes Kölliker's organ (KO). This organ is a transient epithelial structure that undergoes remodeling during the embryonic and early postnatal stages. As the cochlea matures, KO columnar cells are replaced by cuboidal cells lining the mature inner sulcus and represent approximately 12% of the original cell count. 3 Thyroid hormone has been shown to regulate this process as its deficiency leads to prolonged survival of KO cells and malformed tectorial membrane. 4 , 5 The role of KO is still under investigation and is thought to be involved in tectorial membrane formation 6 and generating intrinsic spontaneous activity that drives primary afferent auditory neurons. 7 , 8 Previous studies have shown that KO epithelial cells have some capacity to generate sensory hair cells upon forced expression of genes involved in hair cell differentiation such as Atoh1. 9 , 10 , 11 The mechanism underlying the regulation of KO cells' capacity to generate hair cells is still unclear. It has been proposed that the Notch signaling pathway restricts the sensory fate at the neural boundary of the developing organ of Corti, 12 while Hedgehog signaling represses the sensory competence of the KO cells. 13 Molecular mechanisms governing KO development, sensory competence, and the ultimate function of KO in the mature cochlea are still under investigation.
Using single‐cell transcriptomic analysis and immunostaining, we identified that PR domain‐containing 16 (Prdm16) is expressed within KO during cochlear development. This gene encodes a protein that belongs to the PRDM family which consists of 17 members and is characterized by the combination of a PR‐SET domain and a number of Zn‐finger domains. 14 The PR domain exhibits Histone‐H3 monomethylase activity, while zinc‐finger domains are capable of sequence‐specific DNA binding. 15 Prdm16 has been identified as a key regulator in the development of multiple diverse cell types, including neuronal stem cells, 16 hematopoietic stem cells, 17 craniofacial, 18 , 19 and adipose tissues. 20 A few molecular mechanisms were identified downstream of Prdm16, including repression of transforming growth factor beta (TGF‐β) superfamily signaling during craniofacial development 18 , 21 and chromatin remodeling activity during brown adipose tissue development. 22 The role of Prdm16 in mammalian cochlea development, and ultimately in hearing function, has not been investigated. PRDM16 is located at 1p36.32 in the human genome (hg38), and deletion of genetic material from the short (p) arm of chromosome 1 lead to 1p36 deletion syndrome. 23 Sensorineural hearing impairment was reported in 82% of patients with 1p36 deletion syndrome, 24 yet the underlying gene responsible for hearing impairment in 1p36 deletion syndrome has not been identified. This study describes the expression of Prdm16 during mammalian cochlear development and the developmental consequences of its loss.
2. RESULTS
To identify novel genes required for mouse cochlear development, we utilized the 10× genomics platform and bioinformatics analysis to capture and analyze individual single‐cell transcriptome profiles from WT cochlear duct cells at E14.5. Graph‐based clustering was used to identify clusters of cells with similar transcriptional profiles. We utilized previously identified markers for different populations of cells including mesenchyme markers Tbx18 & Pou3f4, 25 , 26 non‐sensory roof epithelial markers Fgf9, Otx1 & Otx2, 27 , 28 prosensory domain markers Sox2, Jag1 & Hey2, 29 , 30 , 31 KO markers Fgf10 & tecta 32 and future outer sulcus markers Bmp4 & Lmx1a 1 to cluster different population of cells within the developing cochlear duct (Figure 1A). To uncover novel genes expressed within each population, we performed differential gene expression analysis across known populations. A list of top 20 differentially expressed genes within each population is shown in Table 1 and the whole data set is shown in Table S1. Analysis of differentially expressed genes within the KO population at E14.5 identified Prdm16 as a novel marker for this population (Figure 1A). Next, we characterized the expression of Prdm16 in WT cochleae through immunostaining using anti‐PRDM16 antibody 33 concomitantly with anti‐SOX2 staining as established marker for the prosensory domain and supporting cells later in development. Our data confirmed PRDM16 expression within the nuclei of KO cells as early as E13.5, and its expression is maintained in the KO domain throughout development until postnatal day 7 (P7) (Figure 1B). Expression appeared first in the basal and middle regions of E13.5 cochlear duct then extended to the cochlea's apical turn around E15.5. We observed partial overlap between the PRDM16 and SOX2 domain at all time points in the region of inner supporting cells. PRDM16 staining shows moderate level of expression within the interdental cells (IDCs) and low level of expression within the stria vascularis (SV) (Figure 1B). Such expression is maintained until P0 when it rapidly declines. The KO start diminishing in size at P0 while PRDM16 expression is maintained, and the whole organ disappears by ~P10, leaving only inner sulcus cells. Since JAG1 is known to be restricted to the prosensory domain around E14.5, we performed co‐staining with PRDM16 and confirmed that PRDM16 expression is limited to the KO, since there is no co‐localization of PRDM16 and JAG1 (Figure 1C).
FIGURE 1.

Prdm16 expression marks the KO during mouse cochlear development. (A) t‐SNE plot representing graph‐based clustering of single cell gene expression in WT E14.5 isolated cochlear duct cells showing different cell clusters (color‐coded) and enriched gene sets per cluster. (B) Immunostaining of WT cochlear ducts for PRDM16 (green) and SOX2 (red) at multiple time points (E13.5‐P7) showing nuclear expression of Prdm16 within KO epithelial cells. White boxes indicate magnified middle turn at each time point. (C) Immunostaining of E14.5 WT cochlear section (middle turn) showing a clear demarcation between JAG1 and PRDM16 expression (white arrowheads). IDCs, interdental cells; KO, Kölliker's organ; OC, organ of Corti; PD, prosensory domain; SG, spiral ganglion; SV, stria vascularis (scale bar = 100 μm).
TABLE 1.
Top 20 enriched genes within each cluster at E14.5 in WT mouse cochlea.
| Kölliker's organ Genes | L2FC | P value | Prosensory domain Genes | L2FC | P value | Outer sulcus Genes | L2FC | P value | Roof Genes | L2FC | P value | Mesen‐chyme Genes | L2FC | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 150 001‐5O10Rik | 4.50 | 2.18E‐50 | Fgf20 | 4.31 | 1.35E‐64 | Rubie | 4.62 | 1.46E‐67 | Kctd8 | 6.35 | 5E‐124 | AI593442 | 9.83 | 2.07E‐42 |
| Snap91 | 4.20 | 8.49E‐61 | Bdnf | 3.82 | 1.36E‐45 | Bmp4 | 4.32 | 4.70E‐83 | Otx2 | 6.14 | 2.4E‐137 | Pou3f4 | 8.99 | 8.64E‐96 |
| Efhd1 | 3.64 | 1.58E‐51 | Gng8 | 3.47 | 1.38E‐39 | Lmo3 | 4.18 | 7.90E‐63 | Slc6a15 | 5.91 | 3.98E‐95 | Col3a1 | 8.99 | 1.47E‐92 |
| Clic6 | 3.26 | 3.86E‐40 | Hey2 | 3.42 | 2.65E‐47 | Tac1 | 3.40 | 1.67E‐37 | Lrp2 | 5.91 | 2.69E‐70 | Tbx18 | 8.67 | 7.12E‐123 |
| Rgcc | 2.70 | 6.94E‐30 | Sox2 | 3.37 | 3.08E‐49 | Hs3st1 | 2.68 | 2.43E‐35 | Oc90 | 5.58 | 7.2E‐142 | Otor | 8.50 | 1.71E‐97 |
| Nrxn3 | 2.60 | 1.78E‐23 | Runx1t1 | 3.27 | 8.18E‐32 | Pcdh17 | 2.47 | 6.37E‐23 | Meis2 | 5.53 | 8.3E‐99 | Dnm3os | 8.32 | 1.88E‐67 |
| Cryab | 2.50 | 3.07E‐23 | Cyp26b1 | 3.13 | 1.41E‐34 | Rasgef1b | 2.38 | 1.66E‐26 | Gsc | 5.49 | 5.85E‐93 | Prrx1 | 8.00 | 2.23E‐78 |
| Tmod1 | 2.49 | 7.62E‐26 | Tm4sf1 | 2.94 | 8.14E‐31 | Gata2 | 2.36 | 4.27E‐29 | Ecel1 | 5.20 | 1.12E‐83 | Bgn | 7.55 | 1.63E‐69 |
| Spock1 | 2.48 | 6.5E‐22 | Ntf3 | 2.80 | 4.26E‐34 | Fst | 2.29 | 1.80E‐25 | Cacna2d1 | 4.95 | 9.64E‐84 | Cped1 | 7.23 | 5.27E‐72 |
| Ebf1 | 2.47 | 6.32E‐27 | Fat3 | 2.73 | 1.18E‐29 | C1qtnf12 | 2.10 | 7.49E‐23 | Fibin | 4.85 | 1.8E‐82 | Tgfbi | 7.08 | 7.47E‐60 |
| Tecta | 2.46 | 4.3E‐26 | Prr15 | 2.60 | 8.08E‐23 | Sgk1 | 2.01 | 6.88E‐19 | Meg3 | 4.35 | 1.35E‐89 | Apod | 7.07 | 1.71E‐35 |
| Fgf10 | 2.35 | 6.02E‐22 | Lor | 2.58 | 2.25E‐24 | Itih5 | 1.99 | 5.81E‐20 | Fut9 | 4.34 | 2.53E‐73 | Pdgfra | 6.76 | 5.80E‐57 |
| Prdm16 | 2.30 | 8.07E‐19 | Cdkn1b | 2.54 | 3.01E‐26 | Rprm | 1.80 | 3.20E‐16 | Tbx1 | 4.30 | 2.43E‐78 | Col26a1 | 6.08 | 6.31E‐65 |
| Dcn | 1.97 | 1.33E‐13 | Tectb | 2.47 | 6.48E‐24 | Gata3 | 1.77 | 1.57E‐17 | Mme | 4.19 | 1.21E‐72 | Col1a2 | 6.05 | 4.84E‐57 |
| Chst15 | 1.95 | 2.7E‐15 | S100a1 | 2.46 | 1.13E‐24 | Unc5b | 1.76 | 1.23E‐15 | Krt19 | 4.07 | 9.85E‐65 | Eva1b | 5.92 | 2.52E‐60 |
| Cenpf | 1.86 | 1.79E‐11 | Lgr5 | 2.40 | 4.56E‐23 | Smad7 | 1.73 | 6.66E‐16 | Ap1s2 | 3.91 | 2.26E‐62 | Clmp | 5.39 | 7.04E‐45 |
| Top2a | 1.78 | 1.03E‐11 | Sox21 | 2.40 | 5.58E‐24 | Ppfibp1 | 1.73 | 3.10E‐15 | Pid1 | 3.78 | 8.18E‐64 | Car3 | 4.94 | 1.62E‐24 |
| Mia | 1.78 | 1.58E‐13 | Rassf9 | 2.38 | 1.14E‐23 | Sfrp1 | 1.68 | 1.00E‐15 | Ehbp1 | 3.60 | 6.29E‐56 | Atp1a2 | 4.69 | 1.14E‐33 |
| Tectb | 1.76 | 1.91E‐11 | Ntm | 2.25 | 3.59E‐20 | 923 010‐2O04Rik | 1.65 | 2.16E‐14 | Lurap1l | 3.54 | 6.64E‐59 | Foxc1 | 4.51 | 6.65E‐35 |
| Cdh4 | 1.74 | 1.46E‐11 | Hs6st2 | 2.24 | 4.43E‐20 | Angpt1 | 1.58 | 4.14E‐12 | Phldb2 | 3.36 | 4.07E‐49 | Dab2 | 4.37 | 2.74E‐22 |
To understand the role of Prdm16 in cochlear development, we used the Prdm16 cGT gene trap null mutant mouse strain 33 and validated the loss of PRDM16 in the cochlea by immunohistochemistry (Figure 2A). Since Prdm16 cGT null mutants die shortly after birth due in part to complications related to presence of cleft secondary palate, we characterized the cochlear phenotype at P0. H&E and immunostaining of cochlear sections at P0 showed hypoplastic KO in Prdm16 cGT null cochlea compared with heterozygote littermate controls (Prdm16 cGT/+ ) (Figure 2B,C). The thickness of KO was significantly reduced across the whole length of the cochlear duct (Student's t‐test P value <0.001, n = 5) (Figure 2B‐D). We also observed failed development of the spiral limbus (SL), along with detachment and fragmentation of tectorial membrane (TM) across all cochlear turns in Prdm16 cGT null mice (Figure 2B‐D). Additionally, SV thickness was significantly reduced across all cochlear turns in Prdm16 cGT null cochlea (Student's t‐test P‐value <0.001, n = 5) (Figure 2D). We did not observe any difference between Prdm16 heterozygotes and WT cochlea.
FIGURE 2.
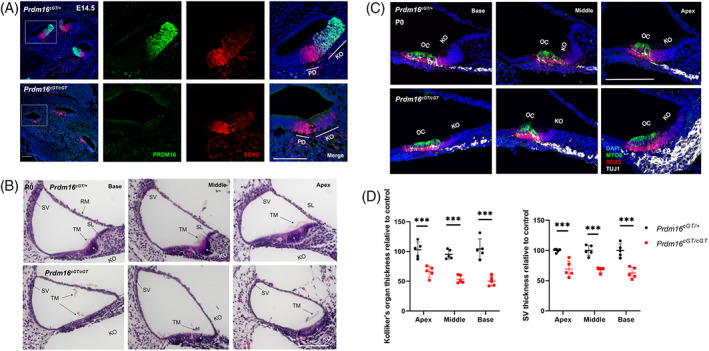
Prdm16 is required for normal development of multiple cochlear structures including KO, spiral limbus, stria vascularis and tectorial membrane. (A) Validation of Prdm16 deletion in cochlear duct of Prdm16 cGT mouse model by immunostaining for PRDM16 (green) and SOX2 (red) at E14.5 showing lack of PRDM16 signal in Prdm16 cGT null cochlear sections. White boxes indicate magnified turns. (B) H&E staining of cochlear duct sections from Prdm16 cGT null (Prdm16 cGT/cGT ) and littermate heterozygote controls (Prdm16 cGT/+ ) pups at P0 showing hypoplastic KO & spiral limbus with detached tectorial membrane and thin stria vascularis. (C) Immunostaining of cochlear duct sections from Prdm16 cGT null and littermate heterozygote controls at P0 showing DAPI, MYO6 (hair cell marker), SOX2 (supporting cell marker), TUJ1 (neuronal marker) staining showing high HC density in the apical turn of Prdm16 cGT null cochlea. (D) Graphs showing average KO thickness and SV thickness at P0 in Prdm16 cGT null relative to heterozygous controls at each turn (n = 5 per group, Mean ± SD, multiple Student's t‐tests, P‐value *** < 0.001). KO, Kölliker's organ; OC, organ of Corti; PD, prosensory domain; RM, Reisner's membrane; SV, stria vascularis; SL, spiral limbus; TM, tectorial membrane (scale bar = 100 μm).
To test if the organ of Corti (OC) is impacted by Prdm16 loss we performed whole mount immunostaining with markers of HCs (MYO6) and SCs (SOX2). Prdm16 cGT null mutants exhibit shortened cochlear ducts (62% compared with heterozygote control cochleae) (Student's t‐test P value <0.001, n = 5) (Figure 3A‐C), and the density of HCs and SCs per 100 μm of cochlear length was significantly increased in the apical turn (IHC and SC density around 150% relative to controls, and OHC around 140% relative to controls) (Student's t‐test P value <0.001, n = 5) (Figure 3B‐C). Taken together, shortening of the cochlear duct and increased density of HCs and SCs in the apical turn point to a defect in the extension of the apical cochlear turn in Prdm16 cGT null cochlea. Interestingly, we also identified ectopic HCs within the KO in Prdm16 cGT null cochlea that are MYO6‐positive and exhibit stereocilia bundles, as evident by phalloidin staining (Figure 4A). The number of ectopic HCs in the whole cochlea ranged from 6 to 32 cells/cochlea (N = 5) and they occurred either in clusters of 4 to 5 cells or as individual cells within the KO (Figure 4A). Immunostaining for the neuronal marker, TUJ1, revealed that 100% of the ectopic HCs are innervated with nerve fibers running through the KO (Figure 4A). The distance between ectopic HCs and native IHCs ranged from 38 to 98 μm. Ectopic HCs appeared to have pear‐shaped cell body, with basal nucleus, apical stereocilia bundle and a calyx afferent surrounding the basolateral membrane as evident by TUJ1 staining (Figure 4A). Scanning electron microscopy of the stereocilia bundles of ectopic HCs shows cylindrical arrangement of multiple rows of stereocilia compared with the U or V‐like arrangement of stereocilia in native HCs at P0 (Figure 4B). Within the Prdm16 cGT null cochlea, some native IHCs and OHCs showed stereocilia bundles with rotated axes at P0 (Figure 4B).
FIGURE 3.

Prdm16 cGT Null mice exhibit short cochlear duct and high density HCs & SCs in the apical turn at P0. Whole‐mount immunostaining of cochlear epithelium from P0 Prdm16 cGT/cGT and littermate heterozygote controls stained with hair cell marker (MYO6) and F‐actin enriched stereocilia bundles marker (phalloidin) in (A) and supporting cell marker (SOX2) in (B). Arrowheads show the beginning and the end of cochlear length measurement. Scale bar = 200 μm. (C) Quantification of cochlear length, IHC, OHC, and SC density in different cochlear turns at P0 (n = 5 per group, Mean ± SD, multiple Student's t tests, P value *** < 0.001). IHC, Inner hair cells; OHC, outer hair cells, SC: Supporting cells.
FIGURE 4.

Prdm16 cGT null cochlea shows ectopic HCs within the KO. (A) Whole‐mount immunostaining of cochlear epithelium from P0 Prdm16 cGT/cGT and littermate heterozygote controls stained with hair cell marker (MYO6), F‐actin enriched stereocilia bundles marker (phalloidin), neuron marker (TUJ1) showing no ectopic HCs outside the organ of Corti in controls (top panel), while Prdm16 cGT null cochlea shows either cluster of ectopic HCs or individual ectopic HC in the KO region. Ectopic HCs are innervated as shown with TUJ1 staining. (B) Scanning electron microscopy of cochlear epithelium from P0 Prdm16 cGT/cGT and littermate heterozygote controls showing stereocilia bundles of IHCs, OHCs and ectopic HC. Within the Prdm16 cGT null cochlea, some native IHCs and OHCs showed rotated axis of stereocilia bundles (yellow arrows). Blue boxes mark the magnified regions. Scale bar = 200 μm unless otherwise specified.
To understand the possible mechanism underlying the hypoplastic KO and short cochlear duct in Prdm16 cGT null cochlea, we assessed the proliferation of KO cells during cochlear development at E14.5 and E16.5 in Prdm16 cGT null mutant compared with heterozygote littermates using EdU proliferation marker. The percentage of KO cell incorporating EdU proliferation marker was significantly reduced in apical turns of Prdm16 cGT null cochlea vs controls at both E14.5 (Student's t‐test P < 0.05, n = 4) and E16.5 (Student's t‐test P < 0.001, n = 4) (Figure 5A‐C). We noticed that by E16.5, KO cells in the base have already stopped proliferating.
FIGURE 5.

Prdm16 regulates KO cell proliferation during development. Immunostaining of cochlear sections from Prdm16 cGT/cGT and littermate heterozygote controls showing EdU (proliferation marker), DAPI and SOX2 (prosensory domain marker) staining in the apical turn at E14.5 (A) and E16.5 (B). Scale bar = 100 μm. White boxes mark the magnified regions (KO, Kölliker's organ; PD, prosensory domain). (C) Percentage of KO cells incorporating EdU at E14.5 and E16.5 for Prdm16 cGT/cGT and littermate heterozygous controls at each turn (n = 4 per group, Mean ± SD, multiple Student's t tests, P value * < 0.05, *** < 0.001).
To investigate the molecular mechanisms underlying the role of Prdm16 during cochlear development, we collected bulk RNA from three Prdm16 cGT null, three heterozygote, and three WT E14.5 cochlear ducts. Analysis of RNA sequencing revealed downregulation of 277 genes and upregulation of 218 genes in Prdm16 cGT null cochleae compared with WT (Log2 fold change >0.5, FDR adjusted P value <0.05, n = 3) (Figure 6A‐C). The top 50 differentially expressed genes are shown in Table 2, and the whole data set is shown in Table S2. We cross‐referenced differentially expressed genes to domain‐specific genes from our scRNA sequencing data at E14.5 (Figure 1A and Table S1). Interestingly, upregulated genes included multiple prosensory‐specific genes such as Fgf20, S100a1, Lor, Lfng, Jag1, and p27Kip1 (Cdkn1b). We also noticed upregulation of two Prdm family genes in response to Prdm16 loss; Mecom (Prdm3) and Prdm1 indicating a compensatory response which has been shown previously. 19 Among the downregulated genes were KO‐specific genes, including Efhd1, Itga8, Gsn, Clic6, Fgf10, and Tecta. We utilized the Database of Annotation, Visualization and Integrated Discovery (DAVID) 34 , 35 to uncover enriched gene ontologies within upregulated and downregulated genes. We found multiple Notch signaling pathway genes enriched within the upregulated gene set including Lfng, Postn, Jag1, and Hes1 (Figure 6D). Enriched gene ontologies within downregulated genes included positive regulation of proliferation and chloride transport (Figure 6E).
FIGURE 6.

RNA sequencing detects differentially expressed genes (DEGs) in Prdm16 cGT null cochlear duct at P0. (A) Volcano plot showing DEGs with statistical significance in red. (B) Heatmap of top 50 DEGs that show upregulation in Prdm16 cGT null vs WT. (C) Heatmap of top 50 DEGs that show downregulation in Prdm16 cGT null vs WT (D, E) Graphs representing enriched gene ontologies in DEGs (P values are shown on the x‐axis on top). The number of genes within each category is shown on the right side of each graph. (n = 3 per group, Waldic Statistics, FDR P value <0.05).
TABLE 2.
Top 50 up‐and downregulated genes in Prdm16 cGT null cochlear duct compared to WT control at E14.5 (RNA sequencing data).
| Downregulated gene symbol | Log2 fold change | FDR adj P value | Upregulated gene symbol | Log2 fold change | FDR adj P value |
|---|---|---|---|---|---|
| Upp1 | −3.03095 | 8.65E‐64 | Chst5 | 2.208205 | 1.88E‐42 |
| Calb1 | −2.91107 | 1.21E‐48 | Chrdl1 | 1.892087 | 2.88E‐88 |
| Prdm16 | −2.86781 | 1.24E‐139 | Cpm | 1.887978 | 1.72E‐43 |
| 1500015O10Rik | −2.63236 | 5.64E‐170 | Mdga1 | 1.550947 | 3.40E‐15 |
| Stmn2 | −2.1146 | 1.22E‐27 | Fam163a | 1.49125 | 5.46E‐27 |
| Pla2r1 | −1.96931 | 5.29E‐58 | S100b | 1.475811 | 1.60E‐12 |
| Abcg5 | −1.91204 | 4.17E‐22 | Postn | 1.462296 | 1.41E‐32 |
| Gucy1b3 | −1.87374 | 9.84E‐24 | Scnn1b | 1.384264 | 4.12E‐26 |
| Efhd1 | −1.79445 | 3.69E‐48 | Gna14 | 1.332706 | 2.95E‐21 |
| Scn9a | −1.73202 | 1.31E‐14 | P2rx2 | 1.305185 | 2.03E‐16 |
| Gucy1a3 | −1.68744 | 7.69E‐16 | Ddc | 1.300187 | 3.01E‐26 |
| Pou3f2 | −1.66321 | 4.48E‐21 | Lfng | 1.252448 | 5.57E‐17 |
| Clic6 | −1.61144 | 3.08E‐40 | Vstm2b | 1.239903 | 6.31E‐22 |
| E030013I19Rik | −1.60824 | 1.03E‐14 | Enpp1 | 1.217822 | 1.22E‐27 |
| Itga8 | −1.5675 | 4.40E‐15 | Prox1 | 1.187037 | 5.91E‐20 |
| Gsn | −1.54794 | 2.85E‐14 | Pdk4 | 1.174113 | 3.36E‐11 |
| Sh3bgrl2 | −1.52764 | 7.75E‐32 | Dlx1 | 1.171678 | 2.46E‐20 |
| Snap91 | −1.51172 | 2.81E‐53 | Fgf20 | 1.148081 | 1.63E‐12 |
| Tspan8 | −1.42692 | 1.32E‐23 | S100a13 | 1.122683 | 6.38E‐09 |
| Pappa2 | −1.41921 | 1.87E‐30 | Gpld1 | 1.113356 | 2.17E‐21 |
| Car2 | −1.29245 | 1.66E‐22 | Smpx | 1.098375 | 2.32E‐08 |
| Esrrb | −1.27153 | 1.93E‐18 | Lsamp | 1.098119 | 2.34E‐17 |
| Serpinb5 | −1.26163 | 3.17E‐08 | Fermt1 | 1.097341 | 5.40E‐19 |
| Ptk2b | −1.25885 | 2.54E‐13 | Anxa5 | 1.087628 | 4.01E‐24 |
| Pcdh19 | −1.24398 | 1.66E‐09 | Lhfp | 1.087242 | 5.07E‐16 |
| Rgcc | −1.18559 | 4.31E‐25 | Mecom | 1.082385 | 9.35E‐11 |
| Pcdh11x | −1.17733 | 2.20E‐17 | Tulp2 | 1.073672 | 7.04E‐11 |
| Nr1h5 | −1.16182 | 3.13E‐08 | Aldh1a1 | 1.0728 | 3.31E‐17 |
| Tac1 | −1.14629 | 3.57E‐18 | Clrn1 | 1.069625 | 2.14E‐14 |
| Tmprss6 | −1.14532 | 6.83E‐12 | Prss33 | 1.068649 | 6.17E‐13 |
| Itgb6 | −1.13618 | 1.23E‐12 | Prdm1 | 1.052381 | 9.71E‐09 |
| Nr2e1 | −1.11374 | 3.46E‐07 | Zfp385c | 1.045068 | 2.07E‐14 |
| Lrrtm3 | −1.11234 | 2.41E‐07 | Jade2 | 1.033174 | 3.02E‐07 |
| Vstm2a | −1.10418 | 9.18E‐07 | Cthrc1 | 1.016233 | 2.51E‐13 |
| Prr15l | −1.10112 | 1.03E‐14 | Fam196a | 0.995588 | 8.63E‐15 |
| Spock1 | −1.09519 | 4.39E‐35 | Tmem59l | 0.986324 | 1.21E‐07 |
| Cnga2 | −1.09179 | 1.42E‐12 | S100a1 | 0.980682 | 1.00E‐05 |
| Ehf | −1.07324 | 4.44E‐06 | Prss23 | 0.97987 | 4.43E‐11 |
| B3galt2 | −1.02835 | 1.32E‐06 | Nnat | 0.972767 | 8.53E‐21 |
| Frzb | −1.01417 | 1.60E‐23 | Tlr2 | 0.965138 | 1.31E‐14 |
| Ltbp4 | −1.00191 | 4.25E‐12 | Myo7a | 0.93987 | 1.00E‐09 |
| Cyp26c1 | −0.99843 | 2.76E‐14 | Rims2 | 0.928665 | 6.96E‐10 |
| Pcolce2 | −0.99366 | 5.07E‐05 | Thsd7b | 0.928559 | 2.17E‐05 |
| Vwc2 | −0.9856 | 8.68E‐11 | Dlx2 | 0.919608 | 2.72E‐12 |
| D930020B18Rik | −0.97348 | 1.01E‐06 | Mafb | 0.919096 | 2.53E‐07 |
| Gpr37 | −0.95309 | 1.77E‐05 | Cdh6 | 0.918861 | 9.76E‐08 |
| Zdhhc14 | −0.95026 | 4.12E‐16 | Sgk1 | 0.91884 | 0.000133 |
| Ntng1 | −0.93912 | 1.72E‐08 | Dapl1 | 0.897578 | 2.01E‐05 |
| Frk | −0.93653 | 7.63E‐17 | Cbln1 | 0.889931 | 1.67E‐07 |
| Dennd4a | −0.93638 | 8.38E‐12 | Rora | 0.889867 | 5.98E‐10 |
To validate the differentially expressed genes in Prdm16 cGT null cochlea, quantitative real time PCR (qPCR) was performed using quadruplicate biological replicates per genotype (null, heterozygous and WT) to validate a subset of differentially expressed genes identified from RNA sequencing. We validated the upregulation of S100a1, Lfng, Fgf20, Sall1, Jag1, Tectb, and Hes1 and downregulation of Pou3f2, Itga8, Ethd1, Clic6, Sporck1, Rgcc, tecta, Gsn, Clic5, Thrb, and Fgf10 (N = 4, Student's t‐test P value * < 0.05, ** < 0.01, and *** < 0.001) (Figure 7A,B). We extended the validation of these genes by analyzing their domains of expression using mRNA‐FISH and immunostaining on three independent samples per genotype group. Jag1, Fgf20, p27Kip1, and Lfng showed expanded expression domains beyond the prosensory region to include parts of the KO (Figure 7C‐G). We also observed downregulation of Clic6 and Tecta expression within the KO in Prdm16 cGT null cochlea (Figure 7C,D). Taken together, Prdm16 loss results in the downregulation of KO‐specific genes and upregulation of prosensory markers within the KO domain.
FIGURE 7.
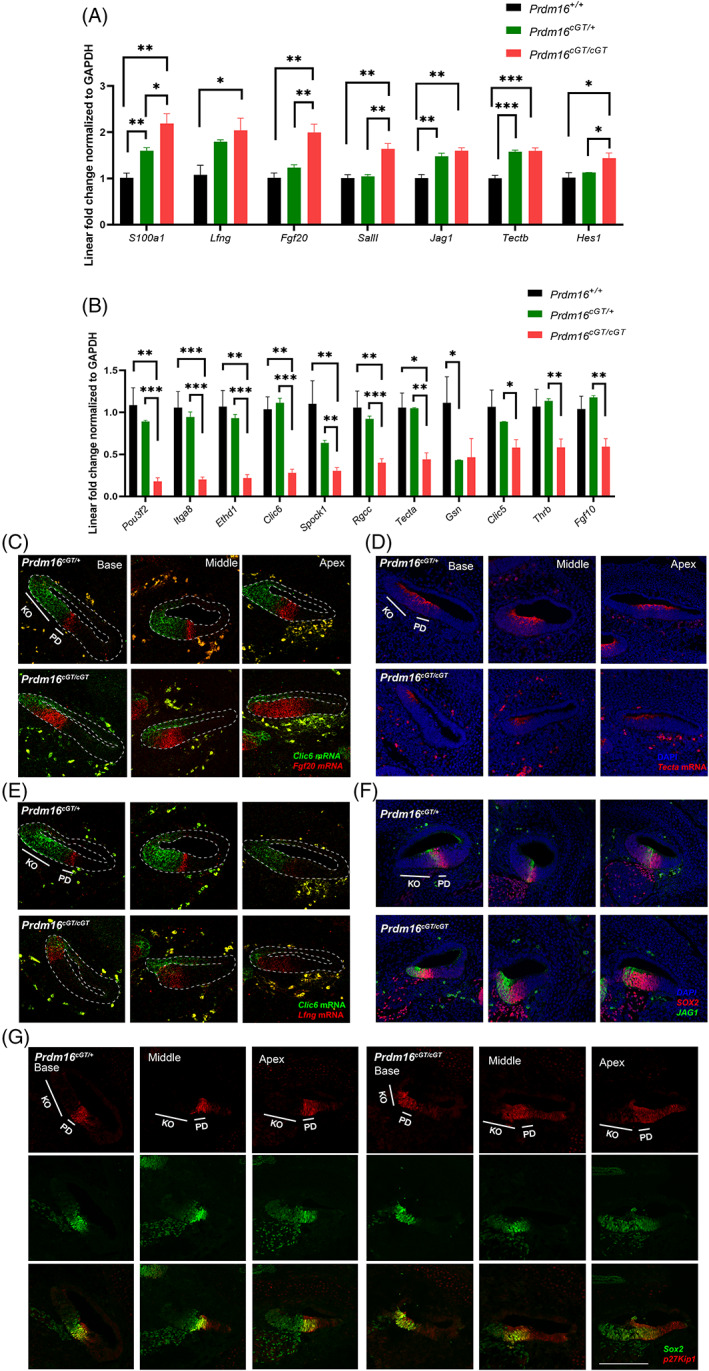
Validation of DEGs in Prdm16 cGT null cochleae vs controls. Real‐time qRT‐PCR showing linear fold change in gene expression of select upregulated genes (A) and downregulated genes (B) normalized to GAPDH expression (n = 4 per group, Mean ± SD, multiple Student's ttests, P value * < 0.05, ** < 0.01, *** < 0.001). (C–E) RNA‐FISH of E14.5 cochlear sections showing DAPI, Clic6 mRNA (KO marker), Fgf20 mRNA (prosensory domain marker), Tecta mRNA (marker of both KO and PD) and Lfng mRNA (marker of the boundary between PD and KO) showing expanded domain of prosensory gene expression and downregulation of KO‐specific genes in the KO of Prdm16 cGT null cochlea. (F) Immunostaining of E14.5 cochlear sections for JAG1 (green), SOX2 (red) and DAPI showing expanded JAG1 expression within the KO in Prdm16 cGT null cochleae. (G) Immunostaining of E14.5 cochlear sections for p27Kip1 (red), SOX2 (green) showing expanded p27Kip1 expression within the KO in Prdm16 cGT null cochleae. KO, Kölliker's organ; PD, prosensory domain (scale bar = 100 μm).
3. DISCUSSION
This work identifies Prdm16 as a novel marker of KO during mouse cochlear development. Its expression is consistent within KO epithelial cells throughout cochlear development and early postnatal development; therefore, it can be utilized as a reliable marker of this cell population that improves upon currently used KO markers, Fgf10 or Tecta 32 that are also expressed within the developing prosensory domain during early cochlear development, whereas Prdm16 expression domain stops at the border of the prosensory domain. Additionally, we identified Prdm16 expression within the stria vascularis and interdental cells during cochlear development. Prdm16 cGT null cochleae are shorter, exhibit decreased proliferation of KO cells and increased density of both HCs and SCs within the apical turn. This demonstrates that Prdm16 is necessary for the proliferation of KO epithelial cells. Proliferation of these cells is subsequently required for normal lengthening of the apical turn and to achieve normal HC and SC density within the organ of Corti. Cochlear lengthening during development is dependent upon multiple mechanisms, including convergent extension, 36 radial intercalation and cellular growth. 37 Due to KO location on the modiolar side of the developing cochlear duct, we demonstrate that KO lengthening is permissive to cochlear lengthening during development.
The absence of spiral limbus and the subsequent loss of modiolar anchorage of the TM in P0 Prdm16 cGT null cochlea provide new evidence for the involvement of Prdm16 in spiral limbus development and thereby correct TM anchorage. Additionally, we observed fragmentation of TM in Prdm16 cGT null cochlea. Given previous evidence of KO involvement in TM formation, 3 , 38 , 39 and the hypoplastic KO phenotype observed in Prdm16 cGT null cochlea, we suggest that Prdm16 loss impacts TM formation. Most of the histological preparations described in our study used dehydrated/rehydrated specimens, which makes it hard to analyze the morphology of TM. Future studies utilizing fresh cochlear preparations and subsequent μCT scanning are needed to better study TM morphology in Prdm16 cGT null cochlea. Proper development of TM, assembly of its components, and its maintenance are essential for hearing, as evident by loss of function gene mutations in different genes that code for TM proteins (eg, α‐tectorin) and result in hearing impairment in mice and human. 40 , 41 , 42 Given the loss of modiolar anchorage of TM and its fragmentation at birth, we anticipate hearing deficits in Prdm16 cGT null mutants if they were able to survive postnatally. To this end, we are currently generating cochlear‐specific Prdm16 conditional mutants to test this hypothesis.
The presence of ectopic HCs within the KO in Prdm16 cGT null cochlea is indicative of a role of Prdm16 in repressing HC fate in KO. The morphology of ectopic HCs is consistent with type I vestibular HCs, which are characterized by pear‐shaped cell bodies innervated by a calyx afferent surrounding the basolateral membrane 43 and cylindrically arranged stereocilia bundles. 44 These ectopic HCs were scarce and showed variability across samples, yet all ectopic HCs were innervated from the spiral ganglion. Our findings are in line with multiple previous studies that showed KO epithelial cell capacity to generate sensory HCs. 9 , 10 , 11 , 45 Since few KO cells turned into ectopic HCs in Prdm16 cGT null cochlea, we project that Prdm16 is not the sole regulator of KO sensory competence. Other members of the PRDM family, including Prdm1 and Mecom, showed compensatory upregulation in Prdm16 cGT null cochleae, and this may explain why only a few KO cells changed fates to form ectopic HCs.
To further understand the molecular mechanisms underlying the role for Prdm16 during cochlea development, we performed gene expression analysis in Prdm16 cGT null cochlea and identified that multiple sensory domain specific genes are upregulated in KO cells, including Fgf20, Lfng, p27Kip1, and Jag1. 29 , 30 , 31 In‐situ and immuno‐expression analyses showed expanded domains of expression of Fgf20, Lfng, p27Kip1, and Jag1 to include parts of the KO at E14.5 in Prdm16 cGT null cochlea vs controls. Jag1 and Lfng have been shown to be initially expressed within KO 46 before becoming restricted to the differentiating prosensory domain around E14.5. 47 In Prdm16 cGT null cochlea, such restriction was not evident, indicating a role for Prdm16 in regulating their expression. Additionally, KO‐specific genes are downregulated in Prdm16 cGT null cochlea, including Clic5, Clic6, Gsn and Fgf10. Taken together, Prdm16 is required to inhibit prosensory‐specific genes and maintain the KO‐specific genes during KO development. Our observation of expanded prosensory domain at the expense of KO along with ectopic expression of prosensory genes, including several Notch signaling pathway genes, in the KO demonstrates a role of Prdm16 in repressing these genes in KO epithelial cells to define the boundary between the KO and the prosensory domain of the cochlea. Since Notch signaling is responsible for specifying the prosensory domain through lateral induction during cochlear development, 48 upregulation of Notch pathway genes secondary to Prdm16 loss, and subsequently the production of ectopic HCs in the KO, points to a role for Prdm16 in repressing Notch signaling within the KO. Previous studies found an interplay between Prdm16 and Notch signaling in different systems. Prdm16 modulates Notch signaling during arterial specification. 49 , 50 Loss of the Notch target genes, Hes1, 3 & 5 cause downregulation of Prdm16 expression in the ventricular zone of the developing telencephalon. 51 Prdm16 has also been demonstrated to be a key regulator in the cell fate choice between skeletal muscle and brown adipose in common progenitors during embryogenesis and also influences beige adipose function and its metabolic function postnatally. 20 , 52 , 53 The established role for Prdm16 and Notch signaling in these systems and our data pertaining to sensory specification of the inner ear suggest that Prdm16 is involved in the modulation of Notch signaling in a variety of cellular contexts.
Additionally, gene expression analysis shows downregulation of Tecta and upregulation of Tectb in Prdm16 cGT null cochlea vs control. α‐tectorin (TECTA) and β‐tectorin (TECTB) are essential proteins in the structure of the TM, 54 and mouse mutants for α‐tectorin exhibit abnormal TM and hearing loss. 40 , 55 Our scRNA sequencing data as well as mRNA FISH data shows that Tecta mRNA is expressed by KO cells (Figures 1A and 7C), and the TM phenotype in Prdm16 cGT null cochlea phenocopies that observed in α‐tectorin mutant cochlea. 55 Taken together, we predict that the loss of KO in Prdm16 cGT null cochlea results in dysregulation of tectorin subunit expression that results in abnormal TM morphology. Postnatal studies in Prdm16 conditional knockouts to evaluate TM defects will provide insight into the impact of tectorin dysregulation.
Few studies have examined the development of KO and its derivatives in mammalian cochlea, 4 , 5 , 6 , 7 , 56 and we believe that utilizing Prdm16 as a novel KO marker and exploring its role as a regulator of this cell population will provide new insight into the development and function of KO in mammalian cochlea. Our work sets the stage for follow up studies that will further investigate a potential role for Prdm16 in human hearing, including the possibility of providing insight into its contribution to the underlying etiology of hearing impairment that is a common component of the 1p36 deletion syndrome phenotype.
4. EXPERIMENTAL PROCEDURES
4.1. Animals
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Midwestern University Institutional Animal Care and Use Committee. All efforts were made to minimize animal suffering. For PRDM16 expression analysis, wildtype (WT) C57BL/6J (Jax: 000664) mice were timed mated. Breeding females were checked daily for presence of post‐copulatory vaginal plugs, and if present, the developmental stage of the litter was considered embryonic day (E) 0.5. At the time points of interest, pregnant female mice were euthanized, and embryos were collected. The Prdm16 cGT mutant mouse strain was previously reported. 33 Prdm16 cGT/+ male and female mice were timed mated to generate Prdm16 cGT null embryos (Prdm16 cGT/cGT ), heterozygote (Prdm16 cGT/+ ), and WT (Prdm16 +/+) littermate controls. The Prdm16 cGT/+ strain was maintained on an FVB/NJ inbred strain background (Jax: 001800).
4.2. Single cell RNA sequencing (ScRNA seq)
One WT pregnant female was euthanized at E14.5, and cochleae were dissected from three pups of either sex in sterile cold HBSS, then placed in dispase (1 U/mL) in DMEM/F‐12 (Stem cell Technologies) for 15 min at 37°C. Following the incubation period, the lateral wall, and mesenchymal cells were dissected away to isolate the cochlear duct. Cochlear ducts from three pups were combined in a single tube and incubated in 0.25% trypsin‐EDTA for 15 min at 37°C with gentle trituration every 5 min, then trypsin was inactivated by adding an equal volume of DMEM/F‐12. Dissociated cells were then passed through a 30 μm strainer to exclude cell aggregates, pelleted at 300×g and then resuspended in 100 μl of cold PBS supplemented with 1% fetal bovine serum (FBS, Gibco: 26140079). Single cells were captured and lysed, and mRNAs were reverse transcribed into cDNAs using a 10X Genomics Chromium Controller at University of Nebraska Medical Center Sequencing core. cDNA libraries were prepared using Chromium Single Cell 3′ Reagents according to the manufacturer's instructions. Libraries were sequenced on an Illumina NextSeq to generate 60 bp of sequence to identify transcript identity. Sequences were aligned to the Ensembl mouse MM10 assembly using Cell Ranger 2.1.1 analysis software (10× Genomics). Processing of the Cell Ranger output data was done with Loupe Browser v.5 (10× Genomics). Gene expression‐based clustering information for the cells, including t‐SNE and UMAP projections and differential gene expression was done utilizing Loupe Browser v.5. The raw data from the ScRNA sequencing of E14.5 cochlear duct is deposited in gEAR database 57 and can be accessed through the following link: https://umgear.org/p?s=ba412295&g=sox2
4.3. Immunostaining
WT C57BL/6J embryos, Prdm16 cGT null embryos (Prdm16 cGT/cGT ), and littermate heterozygote controls (Prdm16 cGT/+ ) at different developmental time points were collected in cold PBS, then dissected to remove skin, cranial vault, brain tissue and mandible. The skull base was then fixed with 4% paraformaldehyde overnight at 4°C then washed with PBS three times. For cryosection immunostaining, samples were washed in 10%, 20%, and 30% sucrose solutions at 4°C then embedded in OCT, and frozen on dry ice then stored at −80°C until further processing. OCT‐embedded samples were serially sectioned horizontally (10 μm thick) at −20°C using a cryostat (Leica CM1950). The sections were mounted on positively charged microscope slides (Globe Scientific Inc) and left to dry at room temperature overnight. For paraffin section immunostaining, fixed skull bases were paraffin processed using a Sakura Tissue‐Tek VIP 5 Tissue Processor overnight, then embedded in paraffin using the Sakura Tissue‐Tek TEC Paraffin Embedding Station and 7 μm sections were collected onto slides using a Thermo Scientific Microm HM325 Rotary Microtome. Slides were then placed on 40°C plate to dry out, then washed three times in Xylene, descending grades (100%, 95%, and 70%) of ethanol for 5 min each. Antigen retrieval was performed by boiling slides in Citrate Buffer pH = 6 for 8 min at 95 to 100°C. Paraffin or cryosections were then washed in PBS, permeabilized with PBS containing 0.1% Tween 20, then blocked with PBS containing 0.1% Tween 20 and 5% normal donkey serum (Southern Biotech 0030‐01). Samples were incubated in primary antibody overnight at 4°C. Samples were then washed with PBS and incubated with a secondary antibody for 2 hours at room temperature then washed, placed on a glass microscope slide in DAPI mounting media, cover‐slipped, and photographed using a Nikon A1R confocal microscope. For whole mount immunostaining, P0 pup heads were collected, inner ears were dissected in cold PBS, fixed with 4% paraformaldehyde overnight at 4°C then washed with PBS three times. Cochlear basilar membrane was micro‐dissected, permeabilized with PBS containing 0.5% Triton, blocked with PBS containing 0.5% Triton and 5% normal donkey serum. Samples were incubated in primary antibody overnight at 4°C. Samples were then washed with PBS and incubated with a secondary antibody for 2 hours at room temperature then washed, placed on a glass microscope slide in DAPI mounting media, cover‐slipped, and photographed using a Nikon A1R confocal microscope. Primary antibodies/stains used: Phalloidin (R&D Systems, 1:100), MYO6 (Proteus, 1:200), SOX2 (R&D Systems, 1:200), JAG1 (DHSB, 1:50), PRDM16, 33 and p27Kip1 (Invitrogen 1:200).
4.4. EdU proliferation assay
Pregnant females were injected with EdU (400 μg/g body weight) 2 hours before collecting the embryos. Cryosections were obtained as stated previously. Staining for incorporated EdU along with SOX2 antibody and DAPI was performed according to Click‐iT EdU Cell Proliferation Kit (Invitrogen C10340) manufacturer protocol. The stained cryosection slides were then imaged on a Nikon A1R confocal microscope.
4.5. Bulk RNA extraction and sequencing
RNAqueous RNA isolation kit (Invitrogen, AM1931) was used for bulk RNA extraction as per the manufacturer protocol. Briefly, E14.5 cochlear ducts from both inner ears of one embryo were microdissected in DEPC‐treated cold PBS, collected in RNA extraction buffer and homogenized (Bel‐Art 650 000 000). RNA extraction and subsequent removal of genomic DNA were performed according to the manufacturer's protocol. Three littermates per genotype were used. The yield and integrity of total RNA from microdissected samples were measured using a 2100 Bioanalyzer (Agilent Technologies). Next generation sequencing and bioinformatics analysis were performed at Northwestern Sequencing core. Briefly, TruSeq mRNA‐Seq Library Prep was used to create cDNA libraries according to manufacturer's protocol. Each library was sequenced to generate 50 base pair single reads on the Illumina HiSeq Sequencing (Illumina). The sequence reads were aligned to the mouse reference genome sequence (USCS mm10) using STAR aligner. 58 Alignments were assembled and annotated using Cufflinks. 59 DESeq2 60 was used to detect differentially expressed gene transcripts. The raw data from bulk RNA sequencing of E14.5 cochlear duct is deposited in Gene expression Omnibus (GSE193046) and can be accessed through the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193046
4.6. Pathway analysis
The database DAVID (the Database of Annotation, Visualization and Integrated Discovery) 34 , 35 was used for pathway analysis. The differentially expressed gene transcripts (q < 0.05) identified from the RNA‐Seq data were input into DAVID, which identified enriched biological pathways.
4.7. RNA fluorescence in situ hybridization (FISH)
Cryosections (10 μm) from E14.5 embryos were used for RNA FISH analysis following the manufacturer's protocol (Molecular instruments, HCR RNA‐FISH protocol for fresh frozen or fixed frozen tissue sections https://files.molecularinstruments.com/MI‐Protocol‐RNAFISH‐FrozenTissue‐Rev2.pdf). 61 Probes used are included in Table S3).
4.8. Quantitative real time PCR
Reverse transcription of total RNA was performed using GoScript Reverse Transcription System (Promega A5001), and quantitative real time PCR (qPCR) was performed using PowerUp SYBR Green (Applied Biosytems, A25742), each according to manufacturer's protocols. Data analysis was performed using the comparative CT method, 62 and data were normalized to detection of GAPDH RNA. Primers used are included in Table S4.
4.9. Data analysis and statistics
Data analysis was done using Image J software Version 1.53n. 63 Cochlear length was measured from the tip of the apex to the base using MYO6 immunostaining as a marker for the hair cells. To measure HC and SC density, at least 400 μm regions of the base, middle, and apex of whole‐mount immunostained cochleae were counted and normalized to 100 μm. KO and stria vascularis thickness was measured using H&E‐stained sections. The thickness was measured at three different points at 25%, 50%, and 75% of the length of the structure in five consecutive 7 μm thick sections then averaged per each biological sample. For EdU proliferation assay, the percentage of KO cells incorporating EdU marker was calculated by counting all KO cells on the neural side of SOX2‐positive cells using DAPI staining, then counting the number of EdU‐positive cells within this population and calculating the percentage using five consecutive 10 μm thick sections then averaged per each biological sample. For each experiment, the numbers of samples (n) is indicated. The P value for difference between samples was calculated using either multiple‐testing adjusted P value (for differential expression using DESeq2 60 ) or a multiple unpaired two‐tailed Student's t‐test (for length, thickness, cell density, percentage of proliferation quantification, and qRT‐PCR), and P < 0.05 was considered as significant.
AUTHOR CONTRIBUTIONS
Michael Ebeid: Conceptualization (lead); data curation (lead); formal analysis (lead); funding acquisition (lead); investigation (lead); methodology (equal); project administration (lead); resources (lead); software (lead); supervision (lead); validation (lead); visualization (equal); writing – original draft (lead); writing – review and editing (lead). Kathy Barnas: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); visualization (equal); writing – review and editing (equal). Hongji Zhang: Investigation (equal); methodology (equal); project administration (equal); validation (equal); writing – review and editing (equal). Amal Yaghmour: Formal analysis (equal); investigation (equal); methodology (equal); writing – review and editing (equal). Gabriele Noreikaite: Formal analysis (equal); investigation (equal); methodology (equal); writing – review and editing (equal). Bryan C. Bjork: Conceptualization (equal); methodology (equal); resources (equal); supervision (equal); writing – review and editing (equal).
Supporting information
TABLE S1 Complete list of differentially expressed genes within each population of cochlear duct cells at E14.5 from Wilde‐type mouse using single cell RNA sequencing.
TABLE S2 Complete list of differentially expressed genes in Prdm16 cGT null cochlear duct cells at E14.5 compared to littermate WT controls using bulk RNA sequencing.
TABLE S3 List of RNA FISH probes name, size, accession numbers and linked fluorophores.
TABLE S4 List of quantitative real time PCR primer sequences.
ACKNOWLEDGMENTS
This study has been funded by Midwestern University startup fund (ME) and American Hearing Research foundation (AHRF, ME). Confocal imaging, tissue histology and scanning electron microscopy was performed at Midwestern University Core Facility, Downers Grove, IL. RNA sequencing was performed at Northwestern University sequencing core and was supported by an award from the Midwestern University Core Facility Outsourcing Fund, Downers Grove, IL. scRNA sequencing was performed at University of Nebraska Medical Center. We would like to thank Dr. Sung‐Ho Huh at University of Nebraska Medical Center, Dr. Suzanne Mansour at University of Utah and Dr. Claus‐Peter Richter at Northwestern University for their valuable input and support.
Ebeid M, Barnas K, Zhang H, Yaghmour A, Noreikaite G, Bjork BC. PRDM16 expression and function in mammalian cochlear development. Developmental Dynamics. 2022;251(10):1666‐1683. doi: 10.1002/dvdy.480
Funding information American Hearing Research Foundation, Grant/Award Number: ME 2018; Midwestern University; Startup fund, Core facility outsourcing fund
REFERENCES
- 1. Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18(9):3327‐3335. doi: 10.1523/jneurosci.18-09-03327.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kelley MW, Xu XM, Wagner MA, Warchol ME, Corwin JT. The developing organ of Corti contains retinoic acid and forms supernumerary hair cells in response to exogenous retinoic acid in culture. Development Dec 1993;119(4):1041–53. [DOI] [PubMed] [Google Scholar]
- 3. Hinojosa R. A note on development of Corti's organ. Acta Otolaryngol Sep‐Oct 1977;84(3–4):238–51. doi: 10.3109/00016487709123963 [DOI] [PubMed] [Google Scholar]
- 4. Uziel A, Gabrion J, Ohresser M, Legrand C. Effects of hypothyroidism on the structural development of the organ of Corti in the rat. Acta Otolaryngol Nov‐Dec 1981;92(5–6):469–80. doi: 10.3109/00016488109133286 [DOI] [PubMed] [Google Scholar]
- 5. Legrand C, Bréhier A, Clavel MC, Thomasset M, Rabié A. Cholecalcin (28‐kDa CaBP) in the rat cochlea. Development in normal and hypothyroid animals. An immunocytochemical study. Brain Res 1988;466(1):121–9. doi: 10.1016/0165-3806(88)90090-9 [DOI] [PubMed] [Google Scholar]
- 6. Anniko M. Embryogenesis of the mammalian inner ear. III. Formation of the tectorial membrane of the CBA/CBA mouse in vivo and in vitro. Anat Embryol (Berl). 1980;160(3):301‐313. doi: 10.1007/bf00305110 [DOI] [PubMed] [Google Scholar]
- 7. Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE. The origin of spontaneous activity in the developing auditory system. Nature. 2007;450(7166):50‐55. doi: 10.1038/nature06233 [DOI] [PubMed] [Google Scholar]
- 8. Babola TA, Li S, Wang Z, Kersbergen CJ, Elgoyhen AB, Coate TM, Bergles DE Purinergic signaling controls spontaneous activity in the auditory system throughout early development. J Neurosci Jan 27 2021;41(4):594–612. doi: 10.1523/jneurosci.2178-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci Jun 2000;3(6):580–6. doi: 10.1038/75753 [DOI] [PubMed] [Google Scholar]
- 10. Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32(19):6699‐6710. doi: 10.1523/jneurosci.5420-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puligilla C, Kelley MW. Dual role for Sox2 in specification of sensory competence and regulation of Atoh1 function. Dev Neurobiol Jan 2017;77(1):3–13. doi: 10.1002/dneu.22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Basch ML, Brown RM 2nd, Jen H‐I, et al. Fine‐tuning of notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. Elife. 2016;5:e19921. doi: 10.7554/eLife.19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Driver EC, Pryor SP, Hill P, et al. Hedgehog signaling regulates sensory cell formation and auditory function in mice and humans. J Neurosci. 2008;28(29):7350‐7358. doi: 10.1523/JNEUROSCI.0312-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007;7(1):187. doi: 10.1186/1471-2148-7-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishibashi J, Seale P. Functions of Prdm16 in thermogenic fat cells. Temperature (Austin). 2015;2(1):65‐72. doi: 10.4161/23328940.2014.974444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol Oct 2010;12(10):999–1006. doi: 10.1038/ncb2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aguilo F, Avagyan S, Labar A, Sevilla A, Lee DF, Kumar P, Lemischka IR, Zhou BY, Snoeck HW Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood May 12, 2011;117(19):5057–66. doi: 10.1182/blood-2010-08-300145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bjork BC, Turbe‐Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19(5):774‐789. doi: 10.1093/hmg/ddp543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shull LC, Sen R, Menzel J, Goyama S, Kurokawa M, Artinger KB. The conserved and divergent roles of Prdm3 and Prdm16 in zebrafish and mouse craniofacial development. Dev Biol. 2020;461(2):132‐144. doi: 10.1016/j.ydbio.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seale P, Kajimura S, Yang W, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38‐54. doi: 10.1016/j.cmet.2007.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warner DR, Wells JP, Greene RM, Pisano MM. Gene expression changes in the secondary palate and mandible of Prdm16(−/−) mice. Cell Tissue Res. 2013;351(3):445‐452. doi: 10.1007/s00441-012-1525-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harms MJ, Lim HW, Ho Y, et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 2015;29(3):298‐307. doi: 10.1101/gad.252734.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shaffer LG, Heilstedt HA. Terminal deletion of 1p36. The Lancet. 2001;358:S9. doi: 10.1016/S0140-6736(01)07022-2 [DOI] [PubMed] [Google Scholar]
- 24. Heilstedt HA, Ballif BC, Howard LA, Kashork CD, Shaffer LG. Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Genet. 2003;64(4):310‐316. doi: 10.1034/j.1399-0004.2003.00126.x [DOI] [PubMed] [Google Scholar]
- 25. Trowe MO, Maier H, Schweizer M, Kispert A. Deafness in mice lacking the T‐box transcription factor Tbx18 in otic fibrocytes. Development. 2008;135(9):1725‐1734. doi: 10.1242/dev.014043 [DOI] [PubMed] [Google Scholar]
- 26. Phippard D, Heydemann A, Lechner M, et al. Changes in the subcellular localization of the Brn4 gene product precede mesenchymal remodeling of the otic capsule. Hear Res. 1998;120(1–2):77‐85. doi: 10.1016/s0378-5955(98)00059-8 [DOI] [PubMed] [Google Scholar]
- 27. Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial–mesenchymal interactions. Dev Biol. 2004;273(2):350‐360. doi: 10.1016/j.ydbio.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 28. Morsli H, Tuorto F, Choo D, Postiglione MP, Simeone A, Wu DK. Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development. 1999;126(11):2335‐2343. [DOI] [PubMed] [Google Scholar]
- 29. Morrison A, Hodgetts C, Gossler A, Hrabé de Angelis M, Lewis J. Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mech Dev. 1999;84(1–2):169‐172. doi: 10.1016/s0925-4773(99)00066-0 [DOI] [PubMed] [Google Scholar]
- 30. Hume CR, Bratt DL, Oesterle EC. Expression of LHX3 and SOX2 during mouse inner ear development. Gene Expr Patterns. 2007;7(7):798‐807. doi: 10.1016/j.modgep.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham‐McDonogh O. Hesr1 and Hesr2 may act as early effectors of notch signaling in the developing cochlea. Dev Biol. 2008;316(1):87‐99. doi: 10.1016/j.ydbio.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn Jun 2003;227(2):203–15. doi: 10.1002/dvdy.10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strassman A, Schnütgen F, Dai Q, et al. Generation of a multipurpose Prdm16 mouse allele by targeted gene trapping. Dis Model Mech. 2017;10(7):909‐922. doi: 10.1242/dmm.029561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44‐57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 35. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1‐13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129(10):2495‐2505. [DOI] [PubMed] [Google Scholar]
- 37. Driver EC, Northrop A, Kelley MW. Cell migration, intercalation and growth regulate mammalian cochlear extension. Development. 2017;144(20):3766‐3776. doi: 10.1242/dev.151761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim DJ, Anniko M. Developmental morphology of the mouse inner ear. A scanning electron microscopic observation. Acta Otolaryngol Suppl. 1985;422:1‐69. [PubMed] [Google Scholar]
- 39. Zine A, Romand R. Development of the auditory receptors of the rat: a SEM study. Brain Res. 1996;721(1–2):49‐58. doi: 10.1016/0006-8993(96)00147-3 [DOI] [PubMed] [Google Scholar]
- 40. Legan PK, Goodyear RJ, Morín M, et al. Three deaf mice: mouse models for TECTA‐based human hereditary deafness reveal domain‐specific structural phenotypes in the tectorial membrane. Hum Mol Genet. 2014;23(10):2551‐2568. doi: 10.1093/hmg/ddt646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia A, Gao SS, Yuan T, et al. Deficient forward transduction and enhanced reverse transduction in the alpha tectorin C1509G human hearing loss mutation. Dis Model Mech. 2010;3(3–4):209‐223. doi: 10.1242/dmm.004135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verhoeven K, Laer LV, Kirschhofer K, et al. Mutations in the human α‐tectorin gene cause autosomal dominant non‐syndromic hearing impairment. Nat Genet. 1998;9(1):60‐62. doi: 10.1038/ng0598-60 [DOI] [PubMed] [Google Scholar]
- 43. Meredith FL, Rennie KJ. Channeling your inner ear potassium: K(+) channels in vestibular hair cells. Hear Res. 2016;338:40‐51. doi: 10.1016/j.heares.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 44. Assad JA, Corey DP. An active motor model for adaptation by vertebrate hair cells. J Neurosci. 1992;12(9):3291‐3309. doi: 10.1523/JNEUROSCI.12-09-03291.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1‐Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22(2):377‐390. doi: 10.1016/j.devcel.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2010;30(45):15044‐15051. doi: 10.1523/jneurosci.3547-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kiernan AE, Xu J, Gridley T. The notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genetics. 2006;2(1):e4. doi: 10.1371/journal.pgen.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hartman Byron H, Reh Thomas A, Bermingham‐McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proceedings of the National Academy of Sciences. 2010;107(36):15792‐15797. doi: 10.1073/pnas.1002827107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beerens M, Van Wauwe J, Craps S, et al. Prdm16 and notch functionally and physically interact during artery development. bioRxiv. 2021;471275. doi: 10.1101/2021.12.05.471275 [DOI] [Google Scholar]
- 50. Aranguren XL, Agirre X, Beerens M, et al. Unraveling a novel transcription factor code determining the human arterial‐specific endothelial cell signature. Blood. 2013;122(24):3982‐3992. doi: 10.1182/blood-2013-02-483255 [DOI] [PubMed] [Google Scholar]
- 51. Kinameri E, Inoue T, Aruga J, et al. Prdm proto‐oncogene transcription factor family expression and interaction with the Notch‐Hes pathway in mouse neurogenesis. PLoS One. 2008;3(12):e3859. doi: 10.1371/journal.pone.0003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961‐967. doi: 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cohen P, Levy JD, Zhang Y, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156(1–2):304‐316. doi: 10.1016/j.cell.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Richardson GP, Lukashkin AN, Russell IJ. The tectorial membrane: one slice of a complex cochlear sandwich. Curr Opin Otolaryngol Head Neck Surg. 2008;16(5):458‐464. doi: 10.1097/MOO.0b013e32830e20c4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim D‐K, Kim JA, Park J, Niazi A, Almishaal A, Park S. The release of surface‐anchored α‐tectorin, an apical extracellular matrix protein, mediates tectorial membrane organization. Sci Adv. 2019;5(11):eaay6300. doi: 10.1126/sciadv.aay6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dayaratne MWN, Vlajkovic SM, Lipski J, Thorne PR. Kölliker's organ and the development of spontaneous activity in the auditory system: implications for hearing dysfunction. Biomed Res Int. 2014;2014:367939. doi: 10.1155/2014/367939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Orvis J, Gottfried B, Kancherla J, et al. gEAR: gene expression analysis resource portal for community‐driven, multi‐omic data exploration. Nat Methods. 2021;18(8):843‐844. doi: 10.1038/s41592-021-01200-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29(1):15‐21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511‐515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choi HMT, Schwarzkopf M, Fornace ME, et al. Third‐generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145(12):dev165753. doi: 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101‐1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 63. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9(7):676‐682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Complete list of differentially expressed genes within each population of cochlear duct cells at E14.5 from Wilde‐type mouse using single cell RNA sequencing.
TABLE S2 Complete list of differentially expressed genes in Prdm16 cGT null cochlear duct cells at E14.5 compared to littermate WT controls using bulk RNA sequencing.
TABLE S3 List of RNA FISH probes name, size, accession numbers and linked fluorophores.
TABLE S4 List of quantitative real time PCR primer sequences.


