Abstract
Background and Aims
Innate lymphoid cells (ILCs) are tissue‐resident lymphocytes that play critical roles in cytokine‐mediated regulation of homeostasis and inflammation. However, relationships between their immune phenotypic characteristics and HCC remain largely unexplored.
Approach and Results
We performed single‐cell RNA sequencing analysis on sorted hepatic ILC cells from human patients with HCC and validated using flow cytometry, multiplex immunofluorescence staining, and functional experiments. Moreover, we applied selection strategies to enrich ILC populations in HCC samples to investigate the effects of B cells on the immune reaction of inducible T cell costimulator (ICOS)+ ILC2 cells. Dysregulation of ILCs was manifested by the changes in cell numbers or subset proportions in HCC. Seven subsets of 3433 ILCs were identified with unique properties, of which ICOS+ ILC2a were preferentially enriched in HCC and correlated with poor prognosis. Mechanistically, we report that B cells, particularly resting naïve B cells, have a previously unrecognized function that is involved in inflammatory differentiation of ILC2 cells. B cell–derived ICOSL signaling was responsible for exacerbating inflammation through the increased production of IL‐13 in ICOS+ ILC2a cells. Heat shock protein 70 (HSP70) genes Heat Shock Protein Family A Member 1A (HSPA1A) and Heat Shock Protein Family A Member 1B (HSPA1B) were highly expressed in ILC2s in late‐stage HCC, and targeting to ICOS and its downstream effector HSP70 in ILC2s suppressed tumor growth and remodeled the immunosuppressive tumor microenvironment.
Conclusions
This in‐depth understanding sheds light on B cell‐driven innate type 2 inflammation and provides a potential strategy for HCC immunotherapy.

Abbreviations
- Abs
antibodies
- CFSE
carboxyfluorescein succinimidyl ester
- CM
conditioned medium
- CX3CR1
C‐X3‐C Motif Chemokine Receptor 1
- EOMES
Eomesodermin
- GATA3
GATA Binding Protein 3
- HAVCR2
HAV Cellular Receptor 2
- HSP70
heat shock protein 70
- HSPA1A
Heat Shock Protein Family A Member 1A
- HSPA1B
Heat Shock Protein Family A Member 1B
- IFN‐γ
interferon‐gamma
- ILC1
group‐1 innate lymphoid cell
- ILC2
group‐2 innate lymphoid cell
- ILC3
group‐3 innate lymphoid cell
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KIR2DL4
KIR, Two Ig Domains And Long Cytoplasmic Tail 4
- KLR
Killer Cell Lectin Like Receptor
- LAG3
Lymphocyte Activating 3
- Lin
lineage
- MDSC
myeloid‐derived suppressor cells
- PB
peripheral blood
- PBMC
peripheral blood mononuclear cell
- PD‐1
programmed cell death 1
- scRNA‐seq
single‐cell RNA sequencing
- TBX21
T‐Box Transcription Factor 21
- TCGA
The Cancer Genome Atlas
- TCF7
Transcription Factor 7
- TLS
tertiary lymphoid structures
- t‐SNE
t‐distributed stochastic neighbor embedding
- XCL
X‐C Motif Chemokine Ligand
INTRODUCTION
HCC is the most common primary liver malignancy and the third leading cause of cancer‐related death worldwide.[ 1 ] This malignancy usually develops in chronic inflammatory liver disease (e.g., fibrosis or cirrhosis).[ 2 ] Accumulating evidence support that these inflammatory disorders have a promotion effect on the initiation and progression of HCC.[ 1 , 3 ]
Chronic inflammation also disturbs the hepatic immune system, allowing cancer cells to easily evade immune surveillance.[ 4 ] During the past decade, cancer immunotherapies have dramatically altered the oncological treatment landscape, with fewer off‐target effects than other agents.[ 5 ] The recent success of immune checkpoint inhibitors such as nivolumab and pembrolizumab that block the programmed cell death 1 (PD‐1)–PD‐L1 pathway, has ushered in a new era for immune therapy in HCC.[ 6 ] However, only a minority of patients with HCC benefit from the PD‐1 checkpoint inhibition, so there is an urgent need to overcome the low response rate through exploration of antitumor immune mechanisms in HCC.[ 7 ]
Innate lymphoid cells (ILCs) consisting of cytotoxic natural killer (NK) cells and ‘‘helper‐like’’ ILCs are key players in innate immunity.[ 8 ] The latter express the α‐chain of the IL‐7 receptor (CD127) and are categorized into three major groups, ILC1, ILC2, and ILC3, according to their specific transcription factor and cytokine signatures.[ 9 ] Recent research into ILCs has resulted in an unprecedented appreciation of the profound involvements of distinct tumor promoting or inhibiting activities.[ 10 ] Whether these findings represent a general feature of ILC subsets or different developmental states and how ILC subsets participate in the complex network with other cells and molecules in the tumor microenvironment remain largely unknown.
Advances in single‐cell analysis of lymphocytes have allowed for a comprehensive understanding of the immune system.[ 11 ] In tumors, the targeted single‐cell RNA sequencing (scRNA‐seq) using an immune panel of 405 transcripts has been applied to analyze infiltrating ILCs in HCC, which identified transcriptomic and phenotypic transition of nontumor ILCs toward ILC2s and noncytotoxic ILC1s.[ 12 ] Nevertheless, in‐depth investigations of ILCs transcriptomic profiles and their pathological roles remain limited. Here, we use high‐throughput sequencing technologies of polyA‐captured scRNA‐seq to profile tumor‐infiltrating ILCs from patients with HCC and identify seven immune clusters with unique functionality. A distinct ICOS+ ILC2a subset is significantly associated with adverse patient outcomes. Furthermore, we identify ICOSL derived from B cells as an important signaling to induce and sustain inflammation in ICOS+ ILC2a cells. Blockade of heat shock protein 70 (HSP70) specifically in ILC2s decreases type 2 cytokine production and suppress tumor progression. These studies describe a B cell‐driven ICOSL–ICOS pathway in ILC2 cells linking innate and adaptive immunity under inflammation conditions in HCC.
MATERIALS AND METHODS
Single‐cell RNA sequencing and data analysis
All procedures that involved human sample collection were approved by the ethics commitee of Nanjing Medical University with the presence of informed consent and adhered to the principles of the Declararion of Helsinki.
HCC‐derived single cells libraries were prepared with the BD Rhapsody Whole Transcriptome Analysis Amplification Kit (BD Biosciences, cat. no. 633801) according to the manufacturer’s instructions and sequenced on Illumina Novaseq (Illumina, San Diego, CA). We applied fastp (version 0.17.0) with default parameter to obtain qualified clean data. Unique molecular identifiers (UMI)‐tools (version 1.0.0) was used to identify the cell barcode whitelist. The UMI‐based clean data were aligned to the human hg19 using STAR aligner (version 2.7) to obtain UMI counts. Cells with <200 detectable genes and >20% mitochondrial rate were filtered out, and CD127− cells were excluded stringently before the initial analysis. The Seurat package (version 3.2.2) was used for normalization and regression based on the expression table according to the UMI counts of each sample and percentage of mitochondria to achieve scaled data. Principle component analysis (PCA) was constructed based on the top 5000 highly variable genes, and the top 20 principal components were applied for t‐distributed stochastic neighbor embedding (t‐SNE) construction. Using the graph‐based cluster method (resolution = 0.8), we acquired the unsupervised cell cluster result and further calculated marker genes using the “FindAllMarkers” function under the following criteria:
log2FC > 0.25.
p ≤ 0.05.
min.pct > 0.1.
To identify possible cell subtypes, clusters of the same cell type were selected for re‐t‐SNE analysis and graph‐based clustering. To identify differentially expressed genes among samples, “FindMarkers” with a Wilcoxon’s rank‐sum test algorithm was used under the following criteria:
log2FC > 0.25.
p ≤ 0.05.
min.pct > 0.1.
For further details regarding the materials and methods, please refer to the Supporting Information (Extended Materials and Methods).
RESULTS
The frequency of ILCs is significantly altered in patients with HCC
Human ILCs are defined by expression of CD45+lineage (Lin)−CD127+ (Figure S1A). To investigate the ILCs compartment in patients with HCC, we first determined the frequency of ILCs using flow cytometry. We observed that the overall percentage of ILCs among all live cells (CD45+) was either comparable between the peripheral blood (PB) of healthy donors and patients with HCC [median, 0.61 vs. 0.44, respectively; p > 0.05 (Figure 1A and Table S2)] or between the nontumor liver tissues and tumors from patients with HCC [median, 0.55 vs. 0.50; p > 0.05 (Figure 1B)]. ILC2s were the markedly elevated ILCs population in the PB of patients with HCC, as characterized by robust enrichment, whereas ILC1s were decreased in patients with HCC compared with healthy donors [ILC1 median: 0.65 in healthy donors vs. 0.63 in patients with HCC; p < 0.05; ILC2 median, 0.12 vs. 0.35; p < 0.0001, ILC3 median, 0.11 vs. 0.03; p < 0.05 (Figure 1C)]. Consistently, the frequency of ILC2s within total ILCs population was significantly increased, whereas ILC1s were decreased in HCC tumor tissues compared with nontumor tissues [nontumor vs. tumor tissues; ILC1 median, 0.54 vs. 0.35; p < 0.001; ILC2 median, 0.36 vs. 0.54; p < 0.01; ILC3 median, 0.04 vs. 0.05; p > 0.05 (Figure 1D)]. Furthermore, the analysis of paired tissues and PB HCC samples revealed a selective increase in ILC2s in tumor microenvironment [nontumor vs. tumor tissues; ILC1 median, 0.55 vs. 0.24; p < 0.0001; ILC2 median, 0.36 vs. 0.57; p < 0.001 (Figure 1E)]. Univariate analysis showed that ILC2 accumulation was associated with tumor size (p = 0.016), whereas there was no significant association of ILC2s with age, sex, metastasis, HBV status, liver fibrosis, tumor numbers or tumor stage in patients with HCC [p > 0.05 (Table S3)].
FIGURE 1.
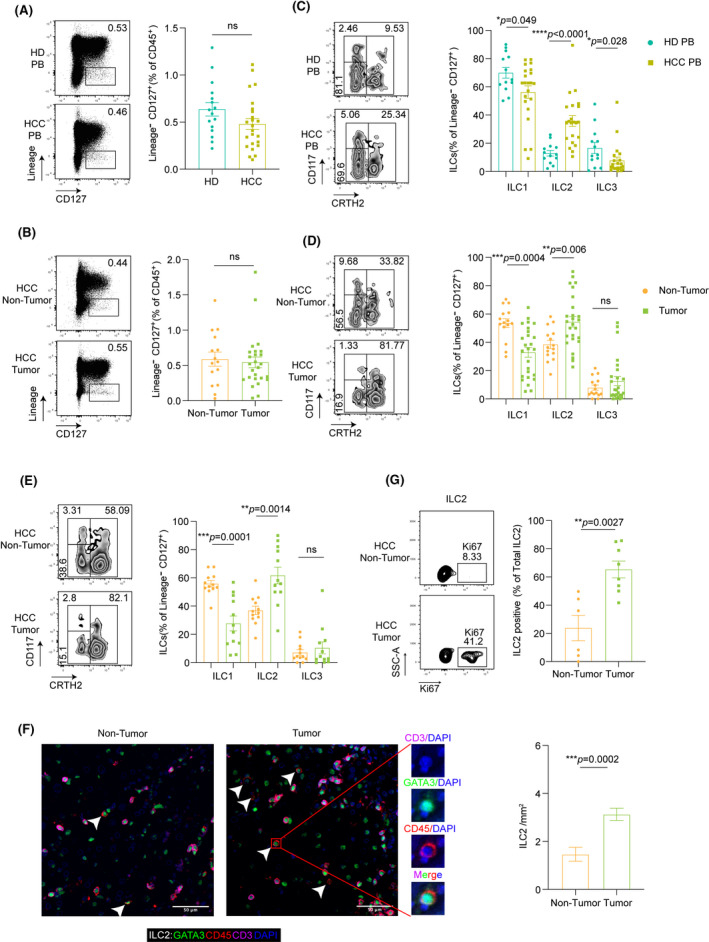
ILC2s are dramatically increased in human HCC. (A) Flow cytometry analysis of representative cases of CD45+LIN−CD127+ ILCs in the PB of healthy donors (HD) and patients with HCC. Frequencies of total ILCs in HD (n = 16) and HCC (n = 24) relative to the CD45+ lymphocytes (p = 0.1). (B) Flow cytometry analysis of representative cases of ILCs in nontumor tissues (N), tumor (T) of HCC. Frequencies of total ILCs in N (n = 15), and T (n = 26) relative to the CD45+ lymphocytes (p = 0.72). (C) Flow cytometry analysis of representative cases of ILC subsets (ILC1, ILC2, and ILC3) in the PB of HD and HCC. Relative frequencies of CD161+CD117−CRTH2− ILC1, CD161+CD117+/− CRTH2+ ILC2, CD161+ CD117+ CRTH2− ILC3 in HD (n = 13) and HCC (n = 22) relative to the total ILCs. (D) Flow cytometry analysis of representative cases of ILC subsets (ILC1, ILC2, and ILC3) in N and T tissues of HCC. Frequencies of ILC subsets in tumor (n = 26), nontumor tissues (n = 15) relative to the total ILCs. (E) Flow cytometry analysis of representative cases of ILC subsets in paired tissue samples of patients with HCC. Frequencies of ILC subsets in tumor, paired nontumor tissues (n = 12) relative to the total ILCs. (F) Percentages of Ki‐67–expressing ILC2s in tumor (n = 8), nontumor tissues (n = 6) of HCC. (G) Representative example of ILC2s in HCC tissues (n = 12) stained by fluorescent multiplex immunohistochemistry showing expression of CD3 (purple), CD45 (red), GATA3 (green), and DAPI (blue) (left panel). Scale bars: 50 μm. Summary of density information for CD3−CD45+GATA3+ ILC2s in paired nontumor and tumor tissues (right panel). Lines and error bars are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, ****p < 0.0001. All statistical analyses were performed with the Mann‐Whitney U test for (A–D, G) and Wilcoxon rank‐sum test for (E, F)
Next, multiplex IHC confirmed our findings with an increased proportion of ILC2s in tumor tissues compared with nontumor tissues [nontumor vs. tumor tissues; median, 0.5 vs. 2.5 cells/mm2; p < 0.0001 (Figure 1F)]. In addition, ILC2s from HCC tissues showed increased proliferation capacity compared with ILC2s from nontumor tissues without affecting the ILC2s’ viability [nontumor vs. tumor tissues; median, 0.22 vs. 0.64; p < 0.01 (Figure 1G and Figure S1B)]. Taken together, these results indicate that HCC represents an alteration in the ILCs composition with the expansion of ILC2s as previously reported.[ 13 , 14 ]
scRNA‐seq census of hepatic ILCs identified seven distinct cell populations in HCC
To explore the complexity of tumor‐infiltrating ILCs, we sorted CD45+Lin−CD 127+ ILCs and CD45+Lin−CD127−CD56+ NK cells by flow cytometry (Figure S2A) and performed scRNA‐seq through the BD Rhapsody platform using tumor and paired nontumor tissues from five patients with treatment‐naïve HCC (Table S4). We excluded contaminating lineage cells and low‐quality cells and finally obtained 3433 CD127+ ILCs (Figure S2B and S2C). Each cell contained an average of approximately 825 distinct genes (Figure S2D). Two‐dimensional projection by t‐SNE sorted these cells distinctly into seven groups inside HCC tumors identified by DBSCAN, designated as clusters of ILC1s (termed as ILC1‐a, b, c, and d), ILC2s (termed as ILC2a, b), and ILC3s based on marker genes representative of each cluster (Figure 2A, Figure S2E and S2F, Table S5).
FIGURE 2.
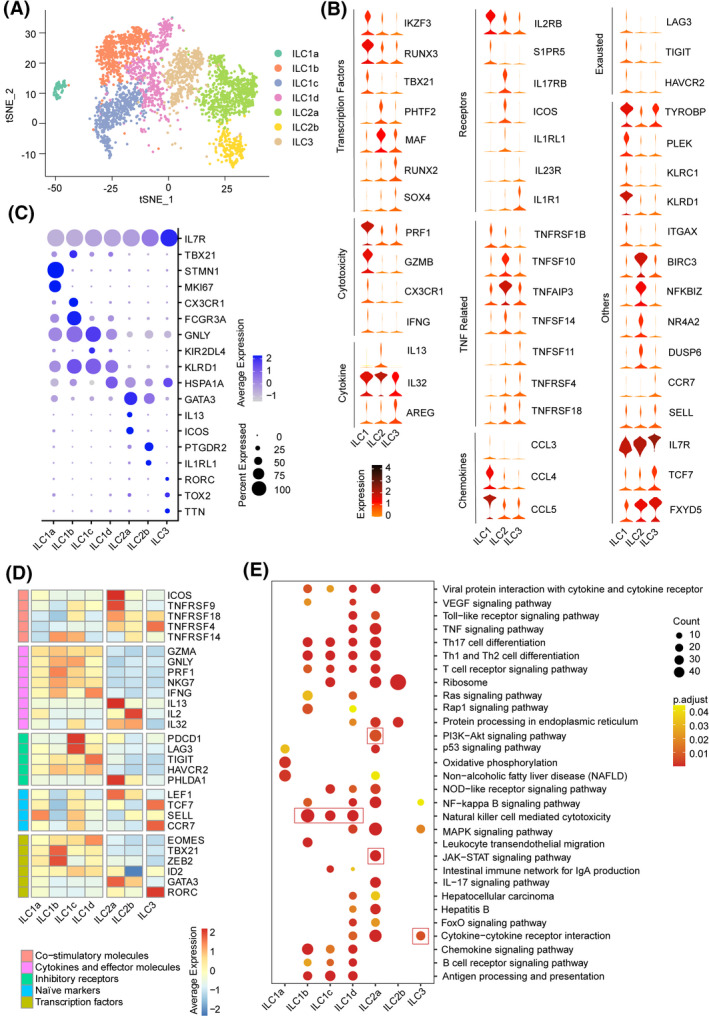
ScRNA‐seq profiling of hepatic ILC cell subtypes in HCC tumor environments. (A) The t‐SNE plot of 3343 single ILC cells from five patients, demonstrating the formation of seven distinct clusters, including four for ILC1 cells (ILC1a‐d clusters), two for ILC2 cells (ILC2a and ILC2b clusters), and one for ILC3 cells. Each dot corresponds to one single cell, colored according to cell clusters. (B) Violin plots showing the expression profile of our selection for ILCs subset (ILC1, ILC2, and ILC3 cells)‐associated genes, as grouped into functional categories (left margin) important for ILCs lineage development and the immune response. The expression is measured as read count for each gene. (C) Dot plot showing average and percent expression of the two most defining marker genes across seven ILC clusters. (D) The average expression of selected ILC cell function‐associated genes of co‐stimulatory molecules, cytokines and effector molecules, inhibitory receptors, naïve markers, and transcript factors in each cell cluster. Red boxes highlight the prominent patterns defining known ILC cell subtypes. (E) KEGG pathway analysis of enriched genes identified among the different seven ILC cell clusters. Red boxes show the major distinct pathway enriched in ILC subsets. Significant KEGG terms for the seven ILC cell clusters are shown (p ≤ 0.05)
To identify molecules unique to the three conventional CD127+ ILC cell types, we first explored differential gene expression profiles up‐regulated in each of the ILC subset. All of the identified ILC subsets expressed the common ILCs signature genes Il‐7R, Transcription Factor 7 (TCF7), and ID2 that are critical for ILCs development. Notably, a set of significantly elevated transcription factors of ILC2s were identified, including MAF involved in IL‐4 production and BIRC3 involved in apoptosis inhibition by binding to TNF receptor‐associated factor (TRAF). However, AREG transcripts were mainly detected in ILC3s, whereas only 156 of 1044 ILC2 cells expressed AREG, and IL‐5, IL‐4, IL‐9, and ARG1 transcripts were not found (Figure 2B, Table S5). We next assessed the expression of signature genes for seven ILC subpopulations and defined (Figure 2C, Table S6). Four ILC1 subsets were characterized by high expression of the t‐box transcription factor 21 gene (TBX21) as well as genes associated with cytotoxicity, including GZMA, GNLY, and PRF1. Of note, ILC1s showed higher expression of Eomesodermin (EOMES), paralleling published reports of human intraepithelial ILC1s expressing both T‐bet and EOMES (Figure 2D, Table S7).[ 15 ] ILC2 subsets shared common ILC2s markers [Prostaglandin D2 Receptor 2 (PTGDR2) and GATA Binding Protein 3 (GATA3)],[ 16 ] in which ILC2a cells showed significantly elevated expression of co‐stimulatory molecules, including ICOS and TNF/TNFR superfamilies. Analyses of ILC3s showed that transcripts encoding the archetypical cytokines (IL‐17A and IL‐22) as well as a crucial activating receptor, IL‐23R, were barely expressed. In addition, early differentiation markers, including C‐C Motif Chemokine Receptor 7 (CCR7), Selectin L (SELL), IL‐7R, and TCF7, were expressed differentially in ILC3s that appeared to be in a more resting immunological or quiescent state (Figure 2B and 2D, Table S7).
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that more than 50% of subset‐specific genes are involved in different immune signaling pathways, such as NK cell‐mediated cytotoxicity in ILC1s; the PI3K‐AKT/JAK‐STAT pathway, protein synthesis, and processing in ILC2s; and cytokine−cytokine receptor interaction in ILC3s (Figure 2E). Interestingly, ILC subsets shared a number of common transcripts with each other, including the immune signaling adaptor TYROBP between ILC1s and ILC3s, transcription factor KIT between ILC2s and ILC3s, and the immunosuppressive receptor subunits Il‐10RA as well as the chemokines CCL4 between ILC1s and ILC2s, supporting the notion that these subsets share a functional relationship and flexibility (Figure S2G and Table S8). Our findings indicate a differential immune profile coupled with immune functions in each type of ILCs, with highly variable transcriptomes and different gene expression profiles inside HCC tumors.
Differential RNA expression profiles of distinct ILCs subtypes in HCC compared with adjacent normal liver tissues
Consistent with previous studies, our data demonstrate the prevalence of ILCs in liver tissue (Figure S3A).[ 8 , 17 ] According to the scRNA‐seq data, we observed significant differences in ILC clusters in the tumor versus adjacent normal tissues (Figure 3A). ILC1s were identified predominantly in normal tissues but were reduced in tumors, in agreement with our previous findings (Figure 1D and 1E). Cluster ILC1b mainly existed in adjacent normal samples, whereas ILC1c was prevalent in tumor tissues. Of note, the two ILC2 clusters (ILC2a and ILC2b) mainly populated the cells in tumors. ILC3s represented a minor ILC population with slight elevation in tumors compared with adjacent normal tissues, whereas regulatory innate lymphoid cells (ILCregs) were not detected in tumor tissues of patients with HCC (Figure S3B). And MKI67 expression of ILC2s in tumor tissues is higher than that in adjacent tissues (Figure S3C).
FIGURE 3.
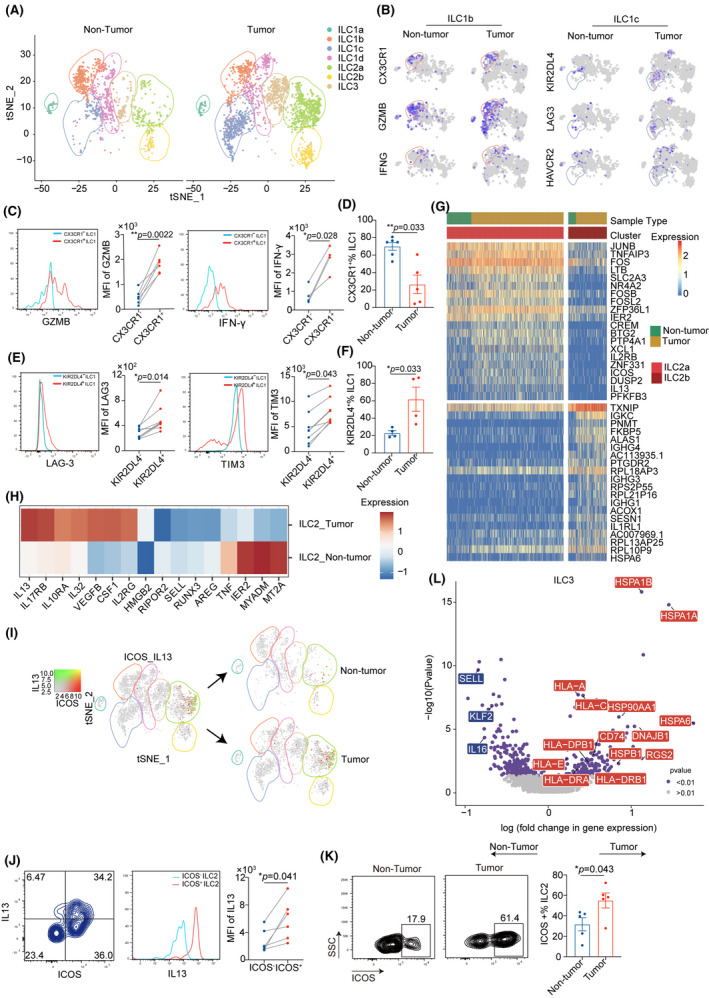
Comparison of the single‐cell landscape in hepatic ILC subsets between tumor and nontumor tissue. (A) ILCs in human tumor (n = 2313 cells) and nontumor samples (n = 1121 cells) of HCC were shown by t‐SNE plot, as gated (with color‐coding) on seven distinct clusters. (B) Feature plots showing differential expression of selected cytotoxic molecules (CX3CR1, GZMB, and IFNG) in ILC1b and inhibitory genes (KIR2DL4, LAG3, and HAVCR2) in ILC1c derived from tumor versus nontumor on the t‐SNE plot. The color scale represents normalized expression for each cell for a given gene with the “LogNormalize” method in R package Seurat 3.2.2. (C) Representative flow cytometry histogram for Granzyme B/IFN‐γ‐expressing CX3CR1+ ILC1b cells in tumors (left panel). (D) Percentage of CX3CR1+ ILC1b from tumor (n = 5) and nontumor tissues (n = 6) of patients with HCC (right panel). (E) Flow cytometry validation of immune checkpoints (LAG3 and HIVCR2) expression in KIR2DL4+ ILC1c from one representative patient (left panel). (F) Percentage of KIR2DL4+ ILC1c from tumor and nontumor liver tissues (right panel) (n = 4 per group). (G) Heatmap showing differentially expressed genes of ILC2 subsets derived from HCC tissues compared with nontumor tissues. The color density indicates the average expression of a given gene, with each row normalized by z‐score transformation. (H) Heatmap revealing the scaled expression of differentially expressed genes between cluster ILC2a and ILC2b from tumor and nontumor tissue. (I) Co‐expression of ICOS and IL‐13 in ILC2a cells illustrated in t‐SNE plots. The color scale represents normalized expression with the “LogNormalize” method for each cell for a given gene. (J) Flow cytometry analysis of IL‐13 in ICOS+ ILC2s subsets after additional 5‐h stimulation with phorbol 12‐myristate 13‐acetate (PMA) and ionomycin from HCC (left panel). (K) Percentage of ICOS+ ILC2s from tumor and paired nontumor samples (right panel) (n = 5 per group). (L) Volcano plot showing differentially expressed genes identified from ILC3 subsets isolated from HCC compared with adjacent normal tissues. Each red or blue dot denotes an individual gene with p ≤ 0.05. *p < 0.05, **p < 0.01, and ***p < 0.001, ****p < 0.0001. All statistical analyses were performed with paired‐sample t test for (C, E, and J) and the Mann‐Whitney U test for (D, F, and K)
We next examined whether these cells had unique transcriptomic profiles or functions between tumor‐normal paired tissues (Table S9). Intriguingly, ILC1s in tumors expressed high levels of inhibitory receptor genes Killer Cell Lectin Like Receptor (KLR)C1 and KIR, Two Ig Domains And Long Cytoplasmic Tail 4 (KIR2DL4), whereas normal liver tissues expressed activating receptor genes KLRG1 and KLRF1 (Figure S3D). Notably, tumor‐infiltrating ILC1s exhibited significantly higher expression of immune checkpoint receptors genes HAV Cellular Receptor 2 [(HAVCR2), encoding TIM‐3] and Lymphocyte Activating 3 (LAG3), suggesting an exhausted phenotype of ILC1s within the tumor (Figure 3B). To explore the characteristics of each transcriptional state by the clustering approach, we identified ILC1a‐d featuring a distinct pattern of TBX21 expression; ILC1b levels were highest, dominating in normal adjacent tissues and expressing the highest levels of cytolytic effector genes [FGF Binding Protein 2 (FGFBP2), Fc Fragment Of IgG Receptor IIIa (FCGR3A), C‐X3‐C Motif Chemokine Receptor 1 (CX3CR1), Granzyme B (GZMB), GZMH, and PRF1]. ILC1c, predominantly composed of cells from tumor tissues, highly expressed the exhaustion‐related gene LAG3 (Figure 3B, Figure S3F, and Table S10). Flow cytometry analysis confirmed the CX3CR1+ ILC1b, the preferential accumulation in nontumor tissues, expressed the highest levels of GZMB and interferon‐gamma (IFN‐γ) among all ILC1s, indicating that this subset might represent the important population for tumor killing [CX3CR1− ILC1b vs. CX3CR1+ ILC1b; GZMB mean, 483 vs. 1832.5; p < 0.0001; IFN‐γ mean, 803.5 vs. 2751.5; p < 0.01 (Figure 3C); nontumor vs. tumor; median, 0.75 vs. 0.21; p < 0.01 (Figure 3D)]. An increased proportion of KIR2DL4+ ILC1c in HCC tissues expressed higher levels of exhaustion markers HAVCR2 and LAG3 than in KIR2DL4− ILC1s, representing exhausted cells [KIR2DL4− ILC1 vs. KIR2DL4+ ILC1; LAG3 mean, 284 vs. 509.5; p < 0.001; TIM3 mean, 4737.25 vs. 8097.125; p < 0.001; (Figure 3E) nontumor vs. tumor; median, 0.64 vs. 0.22; p < 0.05 (Figure 3F)].
Intratumoral ILC2s highly expressed the cytokine receptors IL‐17RB, IL‐10RA, and IL‐2RG; the proinflammatory cytokines IL‐32, IL‐13, Colony Stimulating Factor 1 (CSF1), and High Mobility Group Box 2 (HMGB2); and the angiogenesis factor VEGFB, which were previously reported to be important for their contribution to cancer progression (Figure 3H). HMGB2, Heat Shock Protein Family A Member 1A (HSPA1A), and IL‐13 are associated with poor prognosis in HCC (Table S11), consistent with published reports in several other cancers.[ 18 , 19 , 20 ] A higher degree of heterogeneity was also observed between ILC2a and ILC2b subsets that were mainly enriched in tumor tissues (Figure 3G and 3I). Notably, ICOS was only expressed in ILC2a, suggesting that human ILC2s can be segregated based on their expression of ICOS, and this correlated with other markers such as IL‐13 and IL‐2RB (Figures 3J and S3G). In addition, ILC2a showed up‐regulated expression of various transcripts, including proinflammatory genes [e.g., Lymphotoxin beta (LTB), TNF Alpha Induced Protein 3 (TNFAIP3), IL‐2RB, and IL‐13], chemokine genes [e.g., Immediate Early Response 2 (IER2), X‐C Motif Chemokine Ligand (XCL) 1, C‐X‐C Motif Chemokine Ligand (CXCL) 8, XCL2, Immediate Early Response 5 (IER5)], and transcription factors GATA3, KIT, RAR‐Related Orphan Receptor A (RORA), Aryl Hydrocarbon receptor (AHR), and TCF7. It is unclear how the expression of these inflammatory genes is regulated in tumors, but we note that ILC2a expressed substantially higher levels of JUNB, FOS, FOSB, FOSL2, and JUND compared with ILC2b, promoting the formation of the transcription factor complex AP‐1 (Figure S3I and Table S12). Flow cytometry confirmed that a higher proportion of ICOS+ ILC2a from HCC tissues expressed higher amounts of IL‐13 protein than did ICOS− ILC2b (Figure 3J and 3K).
ILC3s in HCC tissues were highly enriched in transcripts involved in antigen processing and presentation, such as HSPA1A, Heat Shock Protein Family A Member 1B (HSPA1B), Major Histocompatibility Complex, Class I, A, Major Histocompatibility Complex, Class I, C, Heat Shock Protein Family A (Hsp70) Member 6, and CD74 (Figure 3L). The presence of ILC3s was further validated in HCC by multiplexed immunofluorescence (Figure S3J). Collectively, these findings revealed a more immunosuppressive phenotype of ILC clusters from tumors compared with nontumor tissues in HCC.
Single‐cell trajectory branch points correspond to ILC differentiation
ILC trajectories and dynamics were analyzed using the Monocle 2 algorithm for pseudotime to predict the future state of single cells. The trajectory evinced a clear directional flow and placed individual ILCs into different states containing two termini corresponding to two distinct cell fates. The root of the trajectory was mainly populated by replicating MKI67+ cells in the ILC1a cluster, which indicated that these proliferating cells could be precursors of ILC1s. The two termini of the tree were populated mainly by ILC2b for fate 1 and ILC2a for fate 2 (Figure 4A). Next, we analyzed differentially expressed genes along with cell fate progression to explore the plasticity of ILC subsets in HCC. The ILC1‐associated genes TBX21, EOMES, TYROBP, and PRF1 displayed progressive down‐regulation from the root to both fates, suggesting that ILC1s might confer early protection against cancer cells. In contrast, expression of ILC2s signature genes GATA3, PTGDR2, IL1RL1, and IL17RB was up‐regulated in cells differentiating into both fates, indicating that ILC2s may correlate with the late stage of HCC progression. ILC3‐associated genes KIT and RAR‐Related Orphan Receptor C (RORC) were mostly expressed in the intermediate state, supporting the fact that ILC3s facilitate plasticity with conversions to ILC1s or ILC2s (Figure S4B).[ 21 , 22 ] We observed prebranching with the ILC1 component was positively associated with cytotoxicity, whereas both fates with the ILC2 component showed a low score for the cytotoxicity signature (Figure 4B).[ 23 ] As cancer progressed, the percentage of intratumoral ILC2s increased significantly (Figure 1D), with notable accumulation from stage III and thereafter in patients with HCC [I‐II vs. III‐IV; ILC1 median, 0.39 vs. 0.15; p < 0.001; ILC2 median, 0.41 vs. 0.72; p < 0.001 (Figure 4C)]. Altogether, these trajectory data suggest that within ILC1−ILC3−ILC2 subsets, ILC subsets display a certain degree of plasticity and aberrant cellular state during HCC progression.
FIGURE 4.
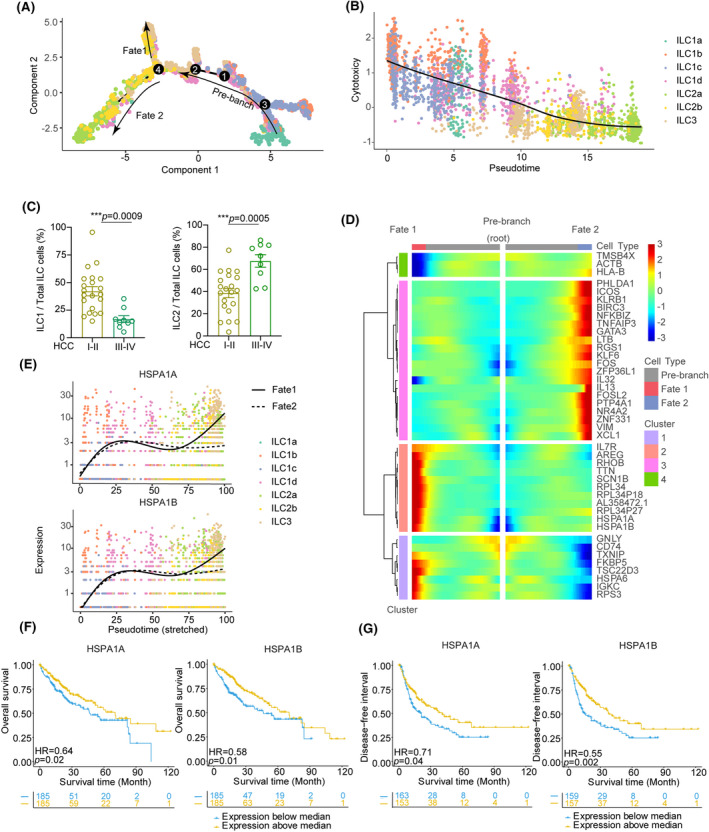
Tracing the gene expression patterns associated with ILC cell differentiation for HCC development. (A) Pseudotime trajectory of all ILCs revealing the progression of ILC1 to ILC3 to ILC2 population. Each dot corresponds to a single cell, as colored according to cell type. (B) Pseudotime for each cell was correlated with cytotoxicity score calculated by the mean expression of 12 cytotoxicity‐associated genes related to these ILC cell status. The solid line represented the relationship between cytotoxicity score with pseudotime fitting by LOESS regression. Spearman’s rank correlation coefficient was also calculated. (C) Frequencies of ILC1s and ILC2s among CD127+ ILC cells from tumor tissues of patients with early‐stage HCC (stages I and II, n = 21) or in late stage (stages III and IV, n = 9). (D) Heatmap showing genes in a branch‐dependent manner for branch point 4. Each row represents the kinetic expression of a gene. Cells are mirrored along the central axis before the trajectory branch point. (E) Pseudotime kinetics of indicated genes (HSPA1A and HSPA1B) from the root of the trajectory to fate 1 (solid line) and the cells up to fate 2 (dashed line). Each point corresponds to a single cell, and each color represents one type of ILC cell cluster. (F) Kaplan‐Meier survival curves of the overall survival of TCGA patients with HCC stratified by HSPA1A and HSPA1B mRNA expression above the median level or below the median level. (G) Kaplan‐Meier survival curves for the disease‐free interval of patients with HCC, stratified by HSPA1A and HSPA1B expression levels. HR and the corresponding p value were calculated by the Cox regression model
Furthermore, we focused on differential gene expression in cells of fates from the trajectory, in which expression of GNLY and GZMB is markedly up‐regulated at the root in ILCs early differentiation and reduced in cells of both fates. This was in contrast to genes, such as TNFAIP3 and Regulator Of G Protein Signaling 1 (RGS1), that are highly expressed along the root through to fate 2, although HSPA1A and HSPA1B (encoding HSP70) were highly expressed at the end of the two fates, despite a slight reduction occurring at point 4 (Figure 4D and 4E). HSPA1A and HSPA1B are mainly enriched in ILC1d, ILC2a, and part of ILC3 and ILC2b (Figure S4A). Moreover, NFKBIZ and FOSL2 were almost absent at early‐stage differentiation and up‐regulated exclusively in cells at fate 2. A similar trend of AREG was observed in cells derived from fate 1 (Figure S4B). Notably, Kaplan‐Meier analysis of a data set from The Cancer Genome Atlas (TCGA) revealed that at both fates, these high‐level genes are associated with poor survival in HCC (Figure 4F and 4G, Figure S5, and Table S11). Overall, our results suggest that these aberrantly expressed genes might be an important mechanism of HCC initiation and progression caused by ILCs functional plasticity and potent targets for cancer therapy.
Potential involvement of ICOS/ICOSL signaling in ILC2‐mediated type 2 inflammation triggered by B cells
To further investigate the role of the distinct tumor‐derived ICOS+ ILC2a subset in HCC, we identified 1499 differentially expressed genes in ICOS+ compared with ICOS− ILC2a cells (p ≤ 0.05) and annotated these genes using Gene Ontology (GO). Of note, pathways that related to cytokine signaling, B cell activation, and metabolism of lipids were significantly enriched (Figure 5A and Figure S6A). Gene Set Enrichment Analysis further revealed a highly enriched signature of genes associated with IL‐13 production, B cell activation, and lymphocyte differentiation which was reflected in the up‐regulation of IL‐13, HSPB1, ICOS, CD40LG, XCL1, CXCL3, and exhaustion markers CD274 and TIGIT in ICOS+ ILC2a cells (Figure S6B and S6C, Table S13). In addition, the genes involved in the catabolism of cholesterol, fatty acid, and triacylglyceride were markedly up‐regulated in ICOS+ ILC2a cells, indicating that lipid metabolism might be critical for ILC2a differentiation and/or functions in innate immunity (Figure S6D and S6E).
FIGURE 5.
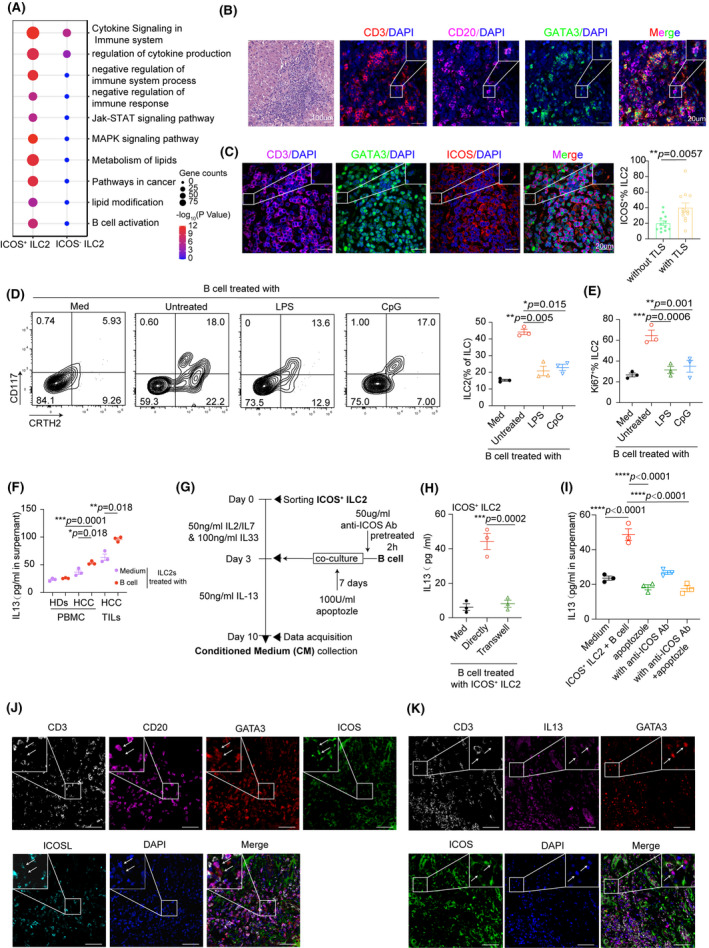
ICOS–ICOSL axis–enhanced IL‐13 production in ILC2 cells elicited by B cells. (A) GO Biological Process enrichment plot for clusters of ICOS‐positive versus ICOS‐negative among ILC2a cells. (B) H&E and immunofluorescence analysis of CD3+ T cells (red), CD20+ B cells (purple), and CD3−GATA3+ ILC2 cells (green) in TLS. Results represent three independent experiments (n = 8). Scale bar, 20 μm. (C) Representative immunofluorescence images of ICOS+ ILC2 cells in and out of TLS. Scale bars, 20 μm. Percentage of ICOS+ ILC2 cells in HCC tissues with or without TLS (n = 12 per group). (D) Purified naïve B cells were left untreated or were stimulated with LPS or CpG‐DNA ODN (CpG) for 18 h and then cocultured with autologous ILC cells at a ratio of 1:1 for 7 days. Quantitation of the percentage of ILC2 cells in the total CD127+ ILC cells before and after B cell treatment. (E) Expression of Ki‐67 of ILC2 cells with indicated treatments were detected by FACS. (F) IL‐13 concentration in CM of ILC2 cells or plus B cells from PBMCs and TILs in patients with HCC was measured by ELISA assay. PBMCs of healthy donors were served as control. (G) B cells were pretreated with ICOS‐neutralizing antibody (50 μg/ml) or isotype and then cultured alone or with autologous ICOS+ ILC2 cells from HCC PBMCs in the presence or absence of apoptozole (100 U/ml) as indicated in the workflow. (H) Quantitative analysis of IL‐13 levels in ICOS+ ILC2 subsets after cultured for 7 days in medium or with B cells directly or in a Transwell chamber by ELISA. (I) Analysis of IL‐13 levels in ICOS+ ILC2 subsets cultured alone or stimulated with B cells, ICOS Abs, apoptozole, or both from HCC PBMCs for 7 days. Data are presented as means ± SEM (C to I). (J) Representative immunofluorescence images of ICOS+ILC2 cells and ICOSL+B cell. Scale bars, 50 μm (n = 4). (K) Representative immunofluorescence images of ICOS+ILC2 cells secreting IL‐13. Scale bars, 50 μm (n = 4). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Student t test for C; rank‐transformed one‐way ANOVA combined with multiple comparison for (D, F); one‐way ANOVA combined with multiple comparison for (E, H, I)
In general, ICOSL is expressed in antigen presenting cells, and its receptor expressed on ILC2a cell populations might broaden the possibilities to establish cell‐to‐cell interactions (Figure S7A).[ 24 ] Previously, we reported ICOSL was substantially expressed by all B cell populations.[ 24 ] Again, we detected the ICOSL expression by flow cytometry and noted that the ICOSL intensities were much higher on naïve B cells than on memory B cells and plasma cells (Figure S7B). However, its functional role with respect to the effect on ILC cell properties was not investigated. Using multiplexed immunofluorescent staining, we confirmed that B cells were in close contact with ILC2s within tertiary lymphoid structures (TLS), suggesting that direct interactions may exist between B and ILC2 cells (Figure 5B). Interestingly, a higher number of ICOS+ ILC2a cells accumulated in HCC with TLS compared with those without TLS [without TLS vs. with TLS; median, 0.20 vs. 0.40; p < 0.01 (Figure 5C)].
Because it has been reported that several well‐known ILC2 activators, like IL‐33, IL‐25, and TSLP, are diminished in HCC samples,[ 25 ] we hypothesized that B cells might contribute to the proliferation of hepatic ILC2s through the ICOSL–ICOS axis. To test this, freshly isolated blood ILCs were incubated with autologous B cells or such cells stimulated with lipopolysaccharide (LPS) or CpG‐DNA ODN in the presence of IL‐2, IL‐7, and IL‐33. ILC–specific transcription factors were measured and the results revealed that naïve B cells had a greater capacity to increase gene expression of GATA3 and reduce T‐bet expression in ILC cells (Figure S7C). We also found that B cells, especially the naïve B cells, actively enhanced the cell proliferation of ILC2 and IL‐13 secretion (Figure 5D and 5E, Figure S7D). We then evaluated the propensity of ILC2 to produce IL‐13 in the context of naïve B cell stimulation in vitro. Flow cytometry showed that the percentage of IL‐13+ ILC2 cells isolated from peripheral blood mononuclear cells (PBMCs) of patients with HCC following B cell treatment was significantly increased compared with those from healthy donors (Figure S7E and S7F). In addition, HCC‐infiltrating ICOS+ ILC2 showed the greatest potential to generate IL‐13 in response to B cells by ELISA compared with those isolated from PBMC of patients with HCC or healthy donors (Figure 5F). We next purified B cells from the cocultures and observed that ICOS+ ILC2s, but not B cells, had a significant increase in IL‐13 transcript levels by real‐time PCR (Figure S7G). To further explore the potential role of B cells in facilitation of ICOS‐dependent production of IL‐13 in ILC2 cells, human IL‐7R+CRTH2+ICOS+ were sorted at high purity (>96%) from HCC PBMCs (Figure S7H and S7I), cultured for four days in the presence of IL‐2, IL‐7, and IL‐33 to attain selective activation of ICOS+ ILC2a cells, and then cocultured with B cells to detect the concentration of IL‐13 as indicated in the workflow (Figure 5G). Cell‐cell contacts between B cells and ICOS+ ILC2a cells were required for IL‐13 secretion because this process was completely abrogated when ILC2a were cultured indirectly with B cells in different chambers or directly with B cells in the presence of anti‐ICOS antibodies (Abs) (Figure 5H). Furthermore, ICOS Abs in combination with the Hsp70 inhibitor apoptozole presented stronger inhibition of IL‐13 secretion than ICOS Abs or apoptozole alone (Figure 5I). Consistent with this, transfecting ILC2 cells with small interfering RNAs (siRNAs) for ICOS also effectively suppressed B cell–elicited inflammatory ILC2 subset for IL‐13 production (Figure S7J). Also, we confirmed that B cells were most often in contact with IL‐13–producing ICOS+ ILC2 cells using multiple immunofluorescent staining (Figure 5J and 5K). Taken together, our results indicate that B cell–derived ICOSL signaling may be responsible for exacerbating inflammation through the increased production of IL‐13 in ILC2a cells.
HSP70 blockade inhibits IL‐13 secretion in ILC2 cells and suppresses tumor progression
HSP70 is reportedly correlated with the malignant behavior of HCC cancer cells and is critical for the regulation of T helper 2 (Th2) cytokine production, including IL‐4, IL‐5, and IL‐13 in airway inflammation.[ 26 ] We next explore whether HSP70 molecules are responsible for IL‐13 production in ICOS+ ILC2a cells, resulting in cell dysfunction in shaping the interaction network with their surrounding microenvironment. We observed that conditioned medium (CM) from the ICOS+ ILC2a and B cell co‐culture system augmented the proliferation and antiapoptosis of IL13Ra1+ HCC cell lines Hep3B and HuH7, whereas apoptozole resulted in a significant reduction in IL‐13 levels and attenuated tumor cell growth without affecting ILC2a cell numbers (Figure S7K‐M). Also, we synchronized the proliferation status of tumor cells by treatment with CM containing HSP70 inhibitor apoptozole as well as IL‐13. The increased presence of an additional cytokine IL‐13 could counteract the effect of apoptozole and partially rescue the defects in cellular proliferation of HCC cells (Figure 6A). Moreover, the blockade of the ICOS/ICOSL interactions between ILC2a and B cells also suppressed the proliferation of CM‐treated tumor cells. Notably, combined treatment with apoptozole and anti‐ICOS monotherapy exhibited the most effectively inhibitory effect on tumor growth (Figure 6B).
FIGURE 6.
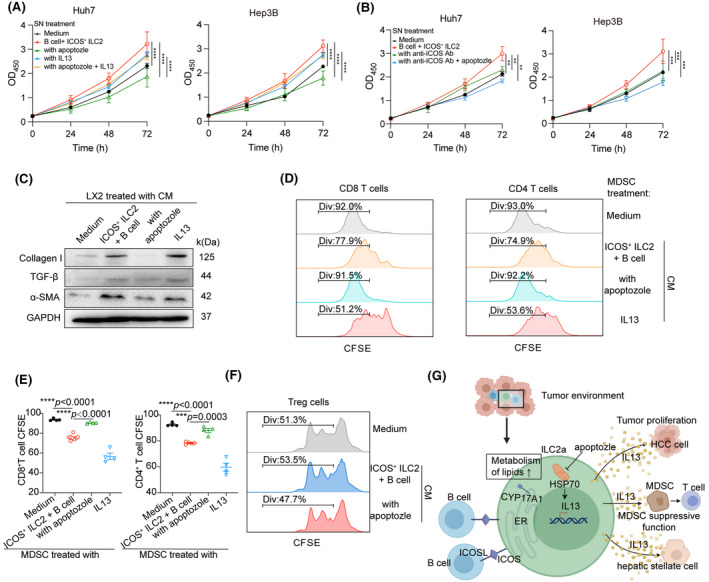
ICOS+ ILC2 cells promote HCC by remodeling an immunosuppressive microenvironment through IL‐13 secretion. (A) ICOS+ ILC2 cells were cocultured with autologous B in the presence or absence of apoptozole and then the filtered CM was harvested after 7 days. Proliferation of Hep3B and Huh7 cells treated with medium, IL‐13, or CM from co‐culture of ILC2 and B cells for 3 days were measured by CCK‐8 (shown as OD450). (B) Blocking ICOS in B cells or plus apoptozole abrogated ICOS+ ILC2 cell‐mediated IL‐13 production on day 7. Proliferation of Hep3B and Huh7 cells treated with medium or CM from co‐culture of ILC2 and B cells for 3 days were measured by CCK‐8 (shown as OD450). (C) Analysis the activation of HSC LX‐2 cultured alone or stimulated with CM as indicated in (A) for 3 days. Protein levels of collagen Ⅰ, TGF‐β, and α‐SMA in LX‐2 cells were determined by western blotting (n = 3). (D) Representative FACS histograms of CFSE‐labeled activated T cells cultured alone or with purified CD11b+CD14+CD15−HLA‐DRlowCD33+ M‐MDSC cells pretreated with medium or CM as indicated in (A). Cell proliferation of T cells was indicated by reduction of CFSE intensity as a result of cell division. (E) Quantification of the proliferation index in CD8+ and CD4+ T cells for (D). (F) CD4+CD25+ nTreg cells were isolated from PBMC of patients with HCC by magnetic cell sorting in the presence of plate‐bound anti‐CD3 (2 μg/ml) for 3 days and cultured alone or stimulated with CM as indicated in (A) for 3 days. The proliferation index of CFSE‐labeled nTreg cells was analyzed by flow cytometry (n = 3). (G) Schematic model depicting the hepatic ICOS+ ILC2 cells inducing a suppressive immune microenvironment in HCC. B cells have increased production of IL‐13 associated with lipid metabolism in inflammatory ILC2a cells through ICOS/ICOSL axis, which finally form an immunosuppressive microenvironment. Data are presented as mean ± SEM of 3 to 5 different independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Rank‐transformed two‐way ANOVA combined with multiple comparison for (A, B); Rank‐transformed one‐way ANOVA combined with multiple comparison for (E)
The ILC2/IL‐13 axis has been reported to be involved in immune evasion, thus pointing to a detrimental role for ILC2 in the context of cancer immunotherapy.[ 14 , 27 ] As we observed that HSC (hHSC) LX‐2 expressed IL‐13Rα1 (Figure S7N), we further treated LX‐2 with CM from ICOS+ ILC2a and B cell co‐culture system and showed that CM could activate hHSC by elevating the expression of α‐smooth muscle actin (α‐SMA) and collagen I, as well as proinflammatory cytokine TGF‐β (Figure 6C). A higher IL‐13Rα1 density was also detected on M‐myeloid‐derived suppressor cells (MDSC)s compared with CD14+ monocytes (Figure S7O), as suggested in a previous study.[ 28 ] Flow analysis of carboxyfluorescein succinimidyl ester (CFSE) dilution in T cells stimulated by anti‐CD3/CD28 showed that IL‐13 or co‐culture CM‐treated M‐MDSCs were able to significantly inhibit the proliferation of both CD8+ and CD4+ T cells, in contrast to untreated M‐MDSCs (Figure 6D and 6E). However, treatment with CM containing apoptozole not only inhibited the features of hHSC activation but also attenuated M‐MDSC‐inhibited autologous T cells to be proliferative in vitro. In addition, we observed that co‐culture CM had no impact on the proliferation of Treg cells treated with or without apoptozole, suggesting that IL‐13 were not involved in the ILC2‐mediated effects on Tregs (Figure 6F, Figure S7P). These studies confirm that ILC2a cells foster an immunosuppressive microenvironment, which indicated the potential utility of targeting Hsp70 to eradicate HCC (Figure 6G).
ILC2a cells are enriched in HCC and can be used as an independent indicator for poor prognosis
Because our data suggest that the ILC2a subset is a highly prevalent effector population in the microenvironment of HCC, we predicted that the single‐cell–derived gene signature from the ILC2a cluster would provide important prognostic information. Using available gene expression data from TCGA, we found that ILC2a were negatively associated with relapse‐free and overall survival in HCC (Figure 7A‐D). The 25 selected informative genes were used to construct the ILC2a signature (Table S14). We found that the ILC2a signature was significantly associated with worse prognosis, and the HRs were larger than any of those for CD127, CD294, ICOS, or GATA3 single‐gene expression, as well as the published gene signatures representing TH2 and ILC2’s processes. Both univariate and multivariate analyses suggested that the ILC2a signature has a superior ability to discriminate HCC prognosis (Figure 7E and Figure S8). As expected, a higher expression of the ILC2a signature was observed in patients with HCC and advanced disease (Figure 7F). The predictive ILC2a signatures were also verified using an independent data set deposited in LIRI‐JP HCC cohort (Figure S9). Our findings suggest that ILC2a may be a critical mediator for improved clinical outcomes observed in HCC.
FIGURE 7.
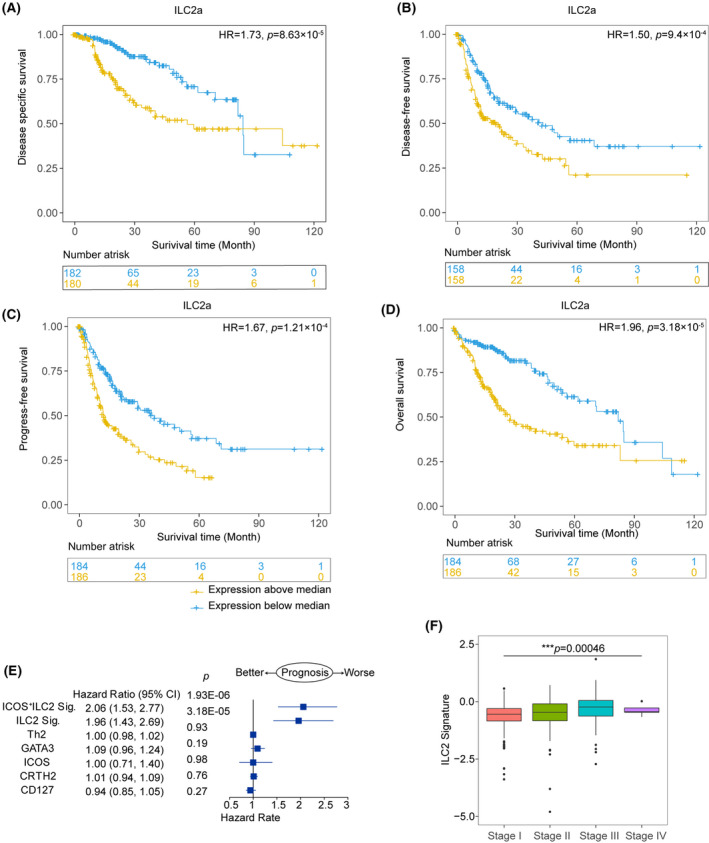
Prognostic significance of the ILC2a gene signature developed from single‐cell data from human HCCs. Kaplan‐Meier survival curves for disease‐specific survival (A), disease‐free survival, (B) progression‐free survival (C), and overall survival (D) from 371 liver HCC (LIHC) samples from the TCGA data set, showing significant prognostic separation according to the ILC2a signature derived from single‐cell data. (E) Prognostic effect of the ILC2a signature compared with other ILC2s cell signatures (indicated on the y axis; see Materials and Methods) as well as single‐gene expression of CD127, CRTH2, ICOS, and GATA3 as continuous variables in HCC. Forest plot shows the HRs (blue squares) and confidence intervals (horizontal ranges) for disease‐free survival derived from multivariable cox regression models adjusted for age, sex, and tumor stage. (F) Box plots show the distributions of ILC2a related gene signature among different tumor stages in HCC (n = 1775 cases), which was tested by one‐way ANOVA combined with multiple comparison. Results were derived from the publicly available TCGA‐LIHC data set. Box plots show the median (center bar), the third and first quartiles (upper and lower edge of box, respectively), and the largest and smallest value ≤1.5 times the interquartile range (limits of upper and lower whiskers, respectively)
DISCUSSION
HCC is considered an immunogenic tumor that arises in a chronically inflamed liver due to hepatitis virus infections, toxins, and fatty liver disease, among others.[ 29 ] Innate immunity directly or indirectly participates in tumor control, and ILCs are preferentially enriched in the human liver, indicating their pivotal role in liver immunology.[ 30 ] Our single‐cell analysis of the transcriptomes of diverse ILC population was not in line with previous studies using human tonsils and mice, which might represent species or different pathophysiological states.[ 31 ] Importantly, our current study on the interactions of ILCs with B cells reveals the unique ability of ILCs to bridge innate and adaptive immunity while responding to tumors. Given the detrimental effect of tumor immunosurveillance, ILC2a is therefore well placed as an antitumoral drug target.
It has been reported that ILC1‐like cells in patients with cancer may promote immune surveillance by secreting IFN‐γ and TNF‐α, whereas the TGF‐β–driven conversion of NK cells to ILC1s leads to tumor escape.[ 32 ] Herein, we paint a rather detailed picture of the features of four distinct transcriptional states among ILC1s. Remarkably, the majority of tumor‐infiltrating ILC1s (~80%) were represented with exhaustion in their transcriptional states, especially by clusters of ILC1c and ILC1d. In contrast, CX3CR1+ effector cells (ILC1b) enriched in adjacent normal tissues exhibited a well‐defined cytotoxicity gene expression program. The dynamic behavior of the branch points to the pseudotrajectory of cellular development: ILC1b expressed activating receptors at an early stage and ILC1c‐d expressed exhausted inhibitory receptors at a late stage, suggesting that ILC1s undergo functional conversion during HCC progression. It should be noted that inconsistency with the findings in mice from ILC precursors and the colorectal cancer model, we did not detect the expression of PD‐1/PD‐L1 in all ILCs in HCC tissues.[ 33 ] This dissection of immune checkpoint expression in ILCs is thus of interest, as ILCs may also be targeted by suitable checkpoint inhibitors or associated with the efficacy of such treatment.
Our analysis based on patients with HCC also revealed two distinct ILC2 subsets differing in their gene expression profiles: ICOS/IL‐13 (ILC2a)– and IL1RL1/CD25 (ILC2b)–expressing subsets. This gave rise to the identification of a homogeneous and distinct cluster of cells that matched the previous definitions of two subtypes of ILC2s: the Lin− KLRG1+ IL‐25R− ST2+ ILC2 subset (termed ILC2NAT) present in naïve mice and a second population of Lin− KLRG1+ cells that were IL‐25R+ and ST2− (termed ILC2INFLAM) on helminth infection.[ 34 ] Notably, ILC2b express IL‐17RB and IL‐1RL1, as well as CD25, the nonsignaling IL‐2Rα subunit that increases the affinity of IL‐2R (CD122 and γc) for IL‐2, which is in contrast to murine ILC2NAT cells (Figure S3H). Our results led to the identification of additional ILC2 subsets, suggesting that ILC2s might be more diverse than initially proposed.
Interaction between different types of immune cells is essential for immune system regulation. Of note, we revealed a previously unrecognized mechanism involving in B cell–mediated ILC2a dysfunction through the ICOS–ICOSL axis. Our data support the finding that ILC2‐involving in innate type 2 inflammation can promote tumor development but provide additional evidence for the role of B cell in mediating ICOS–ICOSL signaling regulating ILC2a function. Given that the ILC2a identified in HCC expressed higher levels of IL‐13, ICOS blockade or HSP70 inhibitor might diminish the ILC2a suppressive phenotype. Importantly, our results provide important insights into the connection between innate and adaptive immune systems, suggesting that ILC2a may be reprogrammed by B cells within the tumor microenvironment and contribute to HCC development and progression.
While exploring the transcriptional profile of ILC3s, we only identified one cluster characterized by expression of SELL but not NCRs and CCR6. Most surprisingly was the absence of the ILC3s signature cytokine IL‐22. These findings suggest that only NKp44− ILC3s are present in HCC tissues, which might represent a set of transcriptionally less active, naïve ILC3s. In general, further investigation of whether ILC3s produce all effector molecules or whether other functionally distinct ILC3 subsets are present in HCC tissues is warranted.
In summary, this study highlights the potential characteristics of infiltrating ILCs to enhance our understanding of innate‐adaptive immune crosstalk in HCC, opening up the possibility of developing targeted drugs or biomarkers of clinical significance to orchestrate the immune system to eradicate cancers.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
Conception and design: Yun Chen and Juncheng Dai; development of methodology: Juncheng Dai, Jiajing Luo, Yuanlin He; acquisition of data (managed patients, provided facilities, provided bioinformatics supervision, performed experiments, and so on): Yun Chen, Juncheng Dai, Jiajing Luo, Changxian Li, Guannan Zhang, Yu Jin and Jinying Lu; analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Juncheng Dai, Yuanlin He, Na Qin, Nanxi Wang; graphics preparation: Juncheng Dai, Jiajing Luo, Yuanlin He; writing of the manuscript: Yun Chen, Jiajing Luo and Xiaohuan Guo, with input and final approval of all authors.
ETHICS STATEMENT
All samples were anonymously coded in accordance with local ethical guidelines (as stipulated by the Declaration of Helsinki). Written informed consent was obtained from the patients, and the protocol was approved by the Review Board of Nanjing Medical University (EC206/09). All experiments were performed in accordance with relevant guidelines and regulations.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Dongming Kuang from Sun Yat‐sen University and Yuting Ma from Center of Systems Medicine Chinese Academy of Medical Sciences for providing professional advice. We appreciate Dr. Yao Yao and Zhongyi Yan for their technical support.
He Y, Luo J, Zhang G, Jin Y, Wang N, Lu J, et al. Single‐cell profiling of human CD127+ innate lymphoid cells reveals diverse immune phenotypes in hepatocellular carcinoma. Hepatology. 2022;76:1013–1029. 10.1002/hep.32444
Yuanlin He, Jiajing Luo, and Guannan Zhang contributed equally to this work.
Funding information
This work was supported by project grants from The National Natural Science Foundation of China (82071767 and 81772602), Jiangsu Provincial key research development program of China (BE2018750), Key Laboratory of Emergency and Trauma, Ministry of Education (KLET‐201913), and Jiangsu provincial higher educational key research development program (19KJA360002)
SEE EDITORIAL ON PAGE 903
Contributor Information
Juncheng Dai, Email: djc@njmu.edu.cn.
Yun Chen, Email: chenyun@njmu.edu.cn.
DATA AVAILABILITY STATEMENT
The single cell RNA‐seq data that support the findings of this study are available from the corresponding author Juncheng Dai on reasonable request. Juncheng Dai, djc@njmu.edu.cn.
REFERENCES
- 1. Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–62. [DOI] [PubMed] [Google Scholar]
- 2. Koyama Y, Brenner DA. Liver inflammation and fibrosis. J Clin Invest. 2017;127(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66(6):1300–12. [DOI] [PubMed] [Google Scholar]
- 4. Zheng C, Zheng L, Yoo J‐K, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single‐cell sequencing. Cell. 2017;169(7):1342–56.e16. [DOI] [PubMed] [Google Scholar]
- 5. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52(1):17–35. [DOI] [PubMed] [Google Scholar]
- 6. Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24(7):1518–24. [DOI] [PubMed] [Google Scholar]
- 8. Lim AI, Li Y, Lopez‐Lastra S, Stadhouders R, Paul F, Casrouge A, et al. Systemic Human ILC precursors provide a substrate for tissue ILC differentiation. Cell. 2017;168(6):1086–100.e10. [DOI] [PubMed] [Google Scholar]
- 9. Luther C, Warner K, Takei F. Unique progenitors in mouse lymph node develop into CD127+ NK cells: thymus‐dependent and thymus‐independent pathways. Blood. 2011;117(15):4012–21. [DOI] [PubMed] [Google Scholar]
- 10. Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. 2018;18(11):671–88. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–45.e20. [DOI] [PubMed] [Google Scholar]
- 12. Heinrich B, Gertz EM, Schäffer AA, Craig A, Ruf B, Subramanyam V, et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut. Published online August 2, 2021. 10.1136/gutjnl-2021-325288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poposki JA, Klingler AI, Tan BK, Soroosh P, Banie H, Lewis G, et al. Group 2 innate lymphoid cells are elevated and activated in chronic rhinosinusitis with nasal polyps. Immun Inflamm Dis. 2017;5(3):233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chevalier MF, Trabanelli S, Racle J, Salomé B, Cesson V, Gharbi D, et al. ILC2‐modulated T cell‐to‐MDSC balance is associated with bladder cancer recurrence. J Clin Invest. 2017;127(8):2916–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simoni Y, Fehlings M, Kløverpris HN, McGovern N, Koo SL, Loh CY, et al. Human innate lymphoid cell subsets possess tissue‐type based heterogeneity in phenotype and frequency. Immunity. 2017;46(1):148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. 2018;174(5):1054–66. [DOI] [PubMed] [Google Scholar]
- 17. Jeffery HC, McDowell P, Lutz P, Wawman RE, Roberts S, Bagnall C, et al. Human intrahepatic ILC2 are IL‐13positive amphiregulinpositive and their frequency correlates with model of end stage liver disease score. PLoS One. 2017;12(12):e0188649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han Q, Xu L, Lin W, Yao X, Jiang M, Zhou R, et al. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene. 2019;38(16):3019–32. [DOI] [PubMed] [Google Scholar]
- 19. Sojka DR, Gogler‐Pigłowska A, Vydra N, Cortez AJ, Filipczak PT, Krawczyk Z, et al. Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non‐small cell lung carcinoma (NSCLC); an implication for NSCLC treatment. Sci Rep. 2019;9(1):14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barderas R, Bartolomé RA, Fernandez‐Aceñero MJ, Torres S, Casal JI. High expression of IL‐13 receptor alpha2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res. 2012;72(11):2780–90. [DOI] [PubMed] [Google Scholar]
- 21. Cella M, Gamini R, Sécca C, Collins PL, Zhao S, Peng V, et al. Subsets of ILC3‐ILC1‐like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol. 2019;20(8):980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, Rogge L, et al. IL‐12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med. 2016;213(4):569–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non‐small‐cell lung cancer by single‐cell sequencing. Nat Med. 2018;24(7):978–85. [DOI] [PubMed] [Google Scholar]
- 24. Zeng QH, Wei Y, Lao XM, Chen DP, Huang CX, Lin QY, et al. B cells polarize pathogenic inflammatory T helper subsets through ICOSL‐dependent glycolysis. Sci Adv. 2020;6(37):eabb6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X, Ye L, Zhang Q, Shen H, Li S, Zhang X, et al. Group‐2 innate lymphoid cells promote hepatocellular carcinoma progression through CXCL2‐neutrophil induced immunosuppression. Hepatology. 2021;74(5):2526–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yombo DJK, Mentink‐Kane MM, Wilson MS, Wynn TA, Madala SK. Heat shock protein 70 is a positive regulator of airway inflammation and goblet cell hyperplasia in a mouse model of allergic airway inflammation. J Biol Chem. 2019;294(41):15082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mchedlidze T, Waldner M, Zopf S, Walker J, Rankin A, Schuchmann M, et al. Interleukin‐33‐dependent innate lymphoid cells mediate hepatic fibrosis. Immunity. 2013;39(2):357–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jovanovic IP, Pejnovic NN, Radosavljevic GD, Pantic JM, Milovanovic MZ, Arsenijevic NN, et al. Interleukin‐33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer. 2014;134(7):1669–82. [DOI] [PubMed] [Google Scholar]
- 29. Younes R, Bugianesi E. Should we undertake surveillance for HCC in patients with NAFLD? J Hepatol. 2018;68(2):326–34. [DOI] [PubMed] [Google Scholar]
- 30. Heymann F, Tacke F. Immunology in the liver–from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13(2):88–110. [DOI] [PubMed] [Google Scholar]
- 31. Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, et al. An Id2(RFP)‐reporter mouse redefines innate lymphoid cell precursor potentials. Immunity. 2019;50(4):1054–68.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao Y, Souza‐Fonseca‐Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. 2017;18(9):1004–15. [DOI] [PubMed] [Google Scholar]
- 33. Moral JA, Leung J, Rojas LA, Ruan J, Zhao J, Sethna Z, et al. ILC2s amplify PD‐1 blockade by activating tissue‐specific cancer immunity. Nature. 2020;579(7797):130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai T, Qiu J, Ji Y, Li W, Ding Z, Suo C, et al. IL‐17‐producing ST2(+) group 2 innate lymphoid cells play a pathogenic role in lung inflammation. J Allergy Clin Immunol. 2019;143(1):229–44.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The single cell RNA‐seq data that support the findings of this study are available from the corresponding author Juncheng Dai on reasonable request. Juncheng Dai, djc@njmu.edu.cn.


