Abstract
The emergence of a novel class of infectious agent composed exclusively of a misfolded protein (termed prions) has been a challenge in modern biomedicine. Despite similarities on the behavior of prions with respect to conventional pathogens, the many uncertainties regarding the biology and virulence of prions make them a worrisome paradigm. Since prions do not contain nucleic acids and rely on a very different way of replication and spreading, it was necessary to invent a novel technology to study them. In this article, we provide an overview of such a technology, termed protein misfolding cyclic amplification (PMCA), and summarize its many applications to detect prions and understand prion biology.
Keywords: Prions, Protein misfolding cyclic amplification, Biomedicine
Introduction
Prion diseases, also called transmissible spongiform encephalopathies (TSEs), affect both animals and humans (Prusiner 1998). Animal prion diseases include scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) or “mad cow” disease in cattle, chronic wasting disease (CWD) in cervids, transmissible mink encephalopathy (TME) in mink, feline spongiform encephalopathy (FSE) in felines, and camel prion disease (CPD) in dromedaries (Collinge 2001; Babelhadj et al. 2018). Human prion diseases can be classified into three categories depending on their origin: sporadic, genetic, and acquired forms (Collinge 2001). Sporadic Creutzfeldt-Jacob disease (sCJD) is the most common human prion disease, and other sporadic human diseases include fatal insomnia (sFI) and variably protease-sensitive prionopathy (VPSPr) (Wadsworth and Collinge 2010). Familial or genetic CJD, fatal familial insomnia (FFI), and Gerstmann-Sträussler-Scheinker syndrome (GSS) are the genetic forms of human prion diseases (Mead et al. 2019). Acquired human prion diseases are caused by accidental infections. Kuru was caused by ritual ingestion of prion-infected human brains. Iatrogenic CJD is transmitted through medical and neurosurgical procedures using prion-containing biological materials and prion-contaminated surgical instruments, respectively (Collinge 2001). The new variant form of CJD (vCJD) was contracted by the consumption of meat products from BSE-affected cows (Will et al. 1996; Bruce et al. 1997).
Prion diseases are 100% fatal disorders characterized by neuronal loss, vacuolation in the neuropil, deposition of protein aggregates, and atypical inflammatory responses (Soto and Satani 2011). The disease-associated protein aggregates were later found to be the infectious agent that can cause prion diseases in animals and humans (Prusiner 1998). Such infectious agent was named “prion” for being both proteinaceous and infectious (Prusiner 1982). Prions are composed of a misfolded and aggregated version of the prion protein, termed PrPSc (Prusiner 1991). The normal cellular form of the protein, or PrPC, is abundantly expressed in the central nervous system of all mammals (Weissmann et al. 1993). It was hypothesized that prion diseases develop when PrPC is misfolded to PrPSc, through either the spontaneous conversion or the seeded conversion induced by exogenous PrPSc (Prusiner 1991). Despite a plethora of evidence supporting the “prion hypothesis,” it took the prion research community more than 3 decades to reach a unanimous verdict for it, which was achieved only after the in vitro generation of genuinely infectious prions (Castilla et al. 2005a) by the groundbreaking invention of the protein misfolding cyclic amplification technology, or PMCA (Saborio et al. 2001).
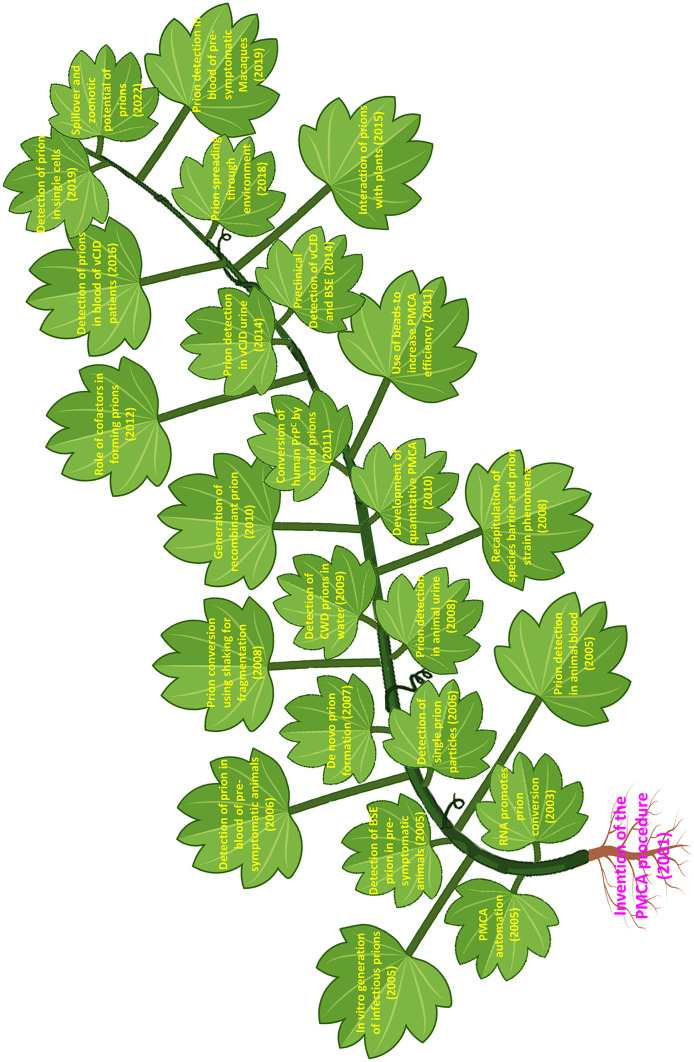
In this article, we will provide a brief overview of the invention of the PMCA technology as well as the working principle behind it. We will discuss the application of PMCA to detect prions in body fluids, with a focus on its use for diagnostic purposes. In addition, we will describe the impact of PMCA on advancing prion research, with respect to the prion conversion mechanism, the prion strain and species barrier phenomena, and the zoonotic potential of animal prion diseases. It is clear that the invention of PMCA opened a new area of research in prion diseases. Soon after the first publication, multiple groups achieved a series of important milestones in prion research using the PMCA technology (Fig. 1).
Fig. 1.
Schematic representation of the main milestones in prion research accomplished using the PMCA technology. For space constrains, we could not include many other studies which have been also very important. We apologize to our colleagues for this
The invention of PMCA
According to the “prion hypothesis,” the infectious prion is mostly or solely composed of PrPSc, which causes prion disease by converting PrPC to PrPSc through a templating mechanism (Prusiner 1991). It was first demonstrated by Caughey and colleagues that by mixing partially purified PrPSc and 35S-methionine-labeled recombinant PrPC in test tubes, PrPSc is capable of converting PrPC to a protease-resistant form with a similar band pattern to that of the original PrPSc seed (Kocisko et al. 1994). However, the low efficiency of the first cell-free conversion system hampered the chance of evaluating the prion infectivity associated with the newly formed 35S-labeled protease-resistant PrP species. In prion disease models, inoculation of minute amounts of PrPSc leads to the accumulation of a huge quantity of PrPSc in the brain at the clinical phase of the disease. Thus, it is clear that from the time infectious prions are inoculated to the time the animals develop the disease, there is a massive amplification of PrPSc at the expense of PrPC(Prusiner 1998). PMCA was developed by Soto and colleagues to recapitulate the efficient in vivo conversion process in the test tube (Saborio et al. 2001). Conceptually analogous to the amplification of DNA fragments by the polymerase chain reaction (PCR), the amplification of PrPSc by PMCA is achieved by subjecting the PrPSc seeds and the PrPC substrates to a cyclical process of incubation and fragmentation (Soto et al. 2002; Castilla et al. 2006; Morales et al. 2012). During the incubation phase, similar to the extension/elongation step of PCR, PrPSc fibrils serve as templates for the PrPC building blocks and the incoming PrPC molecules adapt to the conformation of PrPSc and incorporate into the growing ends of PrPSc. During the fragmentation phase, the mechanical/physical forces break the elongated PrPSc fibrils into shorter fragments, providing more templates for the next cycle of elongation. Conventional PMCA typically uses sonication as a fragmentation force, but adjusted versions include other forms of fragmentation, such as shaking in a modified PMCA reaction, often referred to as QuIC (Wilham et al. 2010). With cycles of incubation and fragmentation, minimal amounts of PrPSc can be replicated and amplified to a level that can be easily detected by conventional biochemical and biophysical means (Saa et al. 2006a).
The first report of the PMCA procedure was published in 2001 (Saborio et al. 2001) using brain homogenates from a prion-affected hamster diluted to the level where PrPSc was barely detectable by immunoblotting. Simply mixing and incubating the diluted PrPSc with the brain homogenate of a healthy hamster, which contains a large amount of fresh PrPC, led to the detection of an increased amount of PrPSc. Following cycles of incubation-sonication, the quantity of PrPSc increased dramatically, validating the in vitro amplification of protein conformation by the PMCA technique (Saborio et al. 2001). It was further demonstrated that the amounts of amplified PrPSc increase with the number of PMCA cycles, and PrPSc seed, as low as 6–12 picograms, can be readily detected in just 10 PMCA cycles. While this original report established the principle behind PMCA, subsequent studies enabled it to automatize the reaction and improve dramatically the efficiency of prion amplification (Fig. 1). Indeed, the PMCA technology was automated by utilizing a programmable microplate horn sonicating system (Castilla et al. 2005a). The automated PMCA can be carried out in a serial manner and allows highly effective amplification of minute amounts of PrPSc. Optimized PMCA was shown to be able to amplify PrPSc from diseased animal brain homogenate that is diluted down to 10−12 (w/v), which theoretically contains the equivalent of a single particle of PrPSc (Saá et al. 2006a). Such a level of detection represents a 3 billion-times and 4000 times higher detection sensitivity as compared to standard immunoblotting techniques and animal bioassays, respectively (Saá et al. 2006a). It was also shown that PMCA can be used to amplify all different types of prions present in distinct animal species and humans, even at pre-symptomatic stages of the disease (Soto et al. 2005). Other improvements of the technology included the introduction of beads from diverse materials (Pritzkow et al 2011; Gonzalez-Montalban et al. 2011) as well as other reagents including EDTA or digitonin (Moda et al. 2014).
Use of PMCA for prion detection and diagnosis of prion diseases
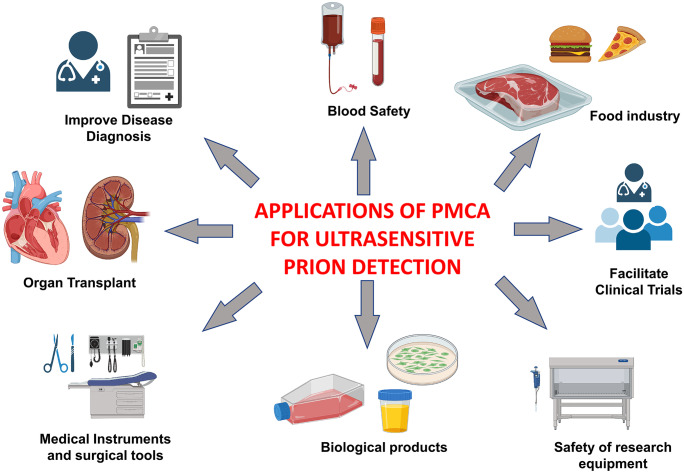
Currently, there is no accurate antemortem diagnosis for human prion diseases (Soto 2004; Zerr 2022). The clinical diagnosis for a probable or possible case is based on a series of clinical tests in combination with the rate of disease progression, CSF biomarkers, and brain imaging techniques (Soto 2004). Definite diagnosis requires neuropathological examinations and/or biochemical detection of PrPSc (Soto 2004; Zerr 2022). Although a definite diagnosis can be made using biopsy samples from a patient brain, such a procedure is very intrusive and poses a great risk to patients’ lives. Almost all definite diagnoses for CJD are made with autopsy samples. Although human prion diseases are rare, in the case of variant Creutzfeldt-Jakob disease (vCJD) caused by exposure to cattle products contaminated with BSE prions, a few vCJD cases have been linked to transfusion of blood donated by preclinical vCJD carriers (Llewelyn et al. 2004; Peden et al. 2004; Wroe et al. 2006). Such secondary transmissions of prions are supported by blood transfusion studies in animal models (Houston et al. 2000; Bons et al. 2002; Andréoletti et al. 2012; Lacroux et al. 2012). Therefore, it would be important to have a technique to detect prions in the blood of people incubating vCJD or sCJD to ensure the safety of blood transfusion and other blood products. Such a technique for ultrasensitive detection of prions will have multiple applications in various fields, including disease diagnosis, blood safety, clinical trials, food industry, biological products, and safety of research and medical equipment (Fig. 2). Table 1 highlights some of the articles reporting the use of PMCA to detect prions in human biological samples.
Fig. 2.
The applications of PMCA to detect prions span many diverse fields including disease diagnosis, clinical trials, food industry, and various applications in enhancing biosafety of diverse products
Table 1.
Application of PMCA for ultrasensitive prion detection in body fluids of patients affected by vCJD
| Sample | Disease group Npositive/Ntotal | Control group Npositive/Ntotal | Reported sensitivity | Reported specificity | Reference |
|---|---|---|---|---|---|
| Blood | 3/4 | 0/141 | 75% | 100% | Lacroux et al (2014) |
| Blood | 18/18 | 1/238 | 100% |
99.2% (analytical) 100% (diagnostic) |
Bougard et al (2016) |
| Blood | 14/14 | 0/153 | 100% | 100% | Concha-Marambio et al (2016) |
| Urine | 13/14 | 0/224 | 92.9% | 100% | Moda et al (2014) |
| CSF | 40/41 | 0/57 | 97.6% | 100% | Bougard et al (2018) |
| CSF | 15/15 | 0/41 | 100% | 100% | Barria et al (2018) |
Detection of prions in blood by PMCA
Using animal bioassays studies, it has been estimated that the median lethal dose (LD50) of prions in blood is millions fold lower than what is in diseased animal brains (Brown et al. 2001), making it a great challenge to directly detect PrPSc in blood samples. PMCA has the established ability to amplify and detect trace amounts of PrPSc (Soto et al. 2005; Saá et al. 2006a; Morales et al. 2012), and its amplification efficiency and detection sensitivity can be enhanced exponentially by increasing the number of PMCA cycles (Saborio et al. 2001). In a pilot study, PMCA achieved 89% sensitivity and 100% specificity in detecting PrPSc in blood samples from experimentally infected and diseased hamsters (Castilla et al. 2005b). A follow-up study with blood samples collected at different time points during the pre-symptomatic phase of the disease from intraperitoneally infected hamsters showed PrPSc detection in blood as early as 20 days post-infection (Saá et al. 2006b). This study also showed a unique temporal distribution pattern of PrPSc in the blood (Saá et al. 2006b). These promising studies demonstrated the great potential of PMCA to detect prions in blood samples.
The first report of PMCA detection of blood vCJD prion was published in 2014 (Lacroux et al. 2014). In this study, Andreoletti and colleagues were able to amplify and detect prions in blood samples from 3 out of 4 confirmed vCJD patients by using ovine PrPC as substrate, which was selected because it outperformed other substrate candidates, including bovine, murine, and human PrPC. Although this particular PMCA system achieved 100% specificity in detecting PrPSc, its sensitivity only improved marginally compared to previous reports using other techniques (Edgeworth et al. 2011), which might be due to the fact that its detection limit was only equivalent to 10−7 vCJD brain homogenate (Lacroux et al. 2014). Two independent studies published in 2016, when PMCA conditions were optimized to detect PrPSc in dilutions as low as 10−9 (Bougard et al. 2016) or 10−10 (Concha-Marambio et al. 2016) vCJD brain homogenates, both groups reported 100% sensitivity and specificity in detecting prions in vCJD blood samples (Bougard et al. 2016; Concha-Marambio et al. 2016). Coste group used plasminogen-coated magnetic nanobeads to capture PrPSc from blood samples and then subjected the enriched PrPSc to serial PMCA procedures that contain brain homogenate of transgenic mice overexpressing human PrP with methionine at amino acid 129. Under this PMCA condition, 18 out of 18 blood samples were identified to contain vCJD prions, and, importantly, preclinical blood samples from two donors who later developed vCJD were also tested positive for vCJD prions (Bougard et al. 2016). In the other report, Soto’s group first removed from the blood samples soluble proteins and other components that might interfere with PMCA efficacy. The pre-treated samples were then amplified by PMCA using brain homogenate from humanized mice with Met/Met genotype at codon 129. In addition to being able to detect PrPSc in 14 out of 14 vCJD blood samples of any fractions, this highly sensitive PMCA system could amplify PrPSc from just a few microliters, or even sub-microliter, of whole blood samples (Concha-Marambio et al. 2016).
Due to the scarcity of preclinical vCJD blood samples, in order to investigate the efficacy of PMCA for preclinical detection of prions in vCJD blood, a study was done in the blood of nonhuman primates that were peripherally infected with macaque-adapted vCJD prions (Concha-Marambio et al. 2020). These blood samples were collected longitudinally from 3 infected macaques that developed clinical signs 25–27 months post-infection. The PMCA assay correctly detected prions from all blood samples collected during the whole preclinical stage without obtaining any PrPSc signal from uninfected animal blood. Strikingly, prions could be detected in blood as early as 65 days post-infection, more than 2 years before the clinical onset of the disease (Concha-Marambio et al. 2020). Another very important implication of this study is the observation that the amount of PrPSc in blood tended to increase along the progression of the disease. Since PMCA can be used for the semi-quantitation of prions in a sample (Chen et al. 2010), the technique might be useful in monitoring disease progression, especially the response of patients to therapies.
Detecting vCJD prions in urine
Prion infectivity has been found in urine samples in prion-infected rodents, although PrPSc was not detected in these studies (Seeger et al. 2005; Kariv-Inbal et al. 2006; Murayama et al. 2007; Gregori et al. 2008), which is most likely due to the extremely low quantity of PrPSc present in urine. To test whether PrPSc could be amplified and detected by PMCA from the urine of prion-infected animals, Gonzalez-Romero et al. infected hamsters by the intraperitoneal route and collected urine samples during the symptomatic stage of the disease. With a limited number of animals, 80% sensitivity and 100% specificity were achieved after extended rounds of PMCA (Gonzalez-Romero et al. 2008). It was estimated, semi-quantitatively, that the amount of PrPSc present in urine is at least 10 times less than that in blood. A study in urine samples from vCJD patients reported the identification of 13 out of 14 vCJD urine samples and none out of the 224 controls, achieving a sensitivity of 92.9% and a specificity of 100% (Moda et al. 2014).
Detecting prions in CSF
The first study to detect BSE prions from CSF was published in 2010 (Murayama et al. 2010). In this study, CSF samples were collected from 3 BSE-infected cows at terminal stages, but only one CSF specimen tested positive for PMCA. In a 2014 report, 3 macaques were intracerebrally infected with BSE prions and CSF, and blood samples were longitudinally collected during both the disease incubation periods and the clinical phases (Murayama et al. 2014). Surprisingly, mouse PrPC was used as the PMCA substrate to amplify PrPSc in these fluids. No prions were detected in the preclinical samples, CSF, or blood. For the samples collected after disease onset, prions were consistently amplified from CSF collected at multiple time points during the symptomatic period from all 3 macaques. Conversely, only one blood sample collected from one animal at its terminal stage could seed the PrPSc amplification (Murayama et al. 2014). From this study, it seems that prions appear at earlier time points and/or in higher amounts in CSF than in blood, although it is also possible that blood contains stronger PMCA inhibitors that prevent amplification.
For detection of vCJD prions in human CSF, Barria et al. applied an enhanced, highly sensitive PMCA procedure that consists of 1 or 2 rounds (48 or 96 h) of amplification. Fifteen out of 15 vCJD CSF samples tested positive for prions, and none of the controls contained PrPSc, presenting 100% sensitivity and specificity (Barria et al. 2018). One of the CSF samples was from the only confirmed vCJD patient who bore the Met/Val genotype at codon 129. Similar findings from a larger cohort analyzed blindly using PMCA technology were reported with 97.6% (40/41) sensitivity and 100% specificity (0/57) in the same year by another team (Bougard et al. 2018). The same 129MV vCJD CSF sample was among the 40 positively identified cases. Twelve confirmed 129MV sCJD CSF samples were also subjected to the same PMCA procedure, and none of them showed PrPSc seeding capability. Therefore, it is noteworthy that PMCA might represent a specific diagnostic tool for 129MV vCJD, because it was very challenging to clinically differentiate 129MV vCJD from sCJD in terms of neuroimaging and clinical symptoms and signs (Mok et al. 2017). It will also be interesting to apply PMCA to test the seeding ability of the blood sample from this particular 129MV vCJD patient. Nevertheless, the lack of detection in sCJD CSF and blood is likely due in part to the usually poor amplification of sCJD prions by PMCA. The set of PMCA conditions for human prion amplification has mostly been optimized for efficient detection of vCJD prions. More recent studies have shown that modifications of some of the experimental conditions of the PMCA assay may lead to good amplification of sCJD prions (Bélondrade et al. 2021; Cazzaniga et al. 2022). Thus, the detection of sCJD prions in biological fluids needs to be re-attempted with the optimized conditions.
In summary, the consistently high sensitivity of PMCA in detecting extremely small quantities of PrPSc present in different tissues and body fluids demonstrates its great potential to be used as a live diagnostic tool for human prion diseases. The ability to achieve a pre-clinical diagnosis of prion infection might be crucial for early intervention and effective treatment of the disease.
Applications of PMCA to understand prion biology
PMCA successfully recapitulates in the laboratory the efficient replication process of prions. Thus, in addition to its use for highly sensitive prion detection, PMCA has contributed enormously to the study of the intricate biology of this unconventional infectious agent (Fig. 1). In the sections below, we will briefly summarize the many studies published to understand diverse aspects of prion diseases using PMCA. Table 2 highlights some of the main published studies using PMCA to understand the unique biology of prions.
Table 2.
PMCA studies of prion biology
| Prion biology | Study | PMCA seeds | PMCA substrates | Summary |
|---|---|---|---|---|
| Prion hypothesis | Castilla et al. (2005a) | 263 K | Normal hamster brain homogenate | PMCA-amplified PrPres induces bona fide prion disease in wild-type hamsters |
| Deleault et al. (2007) | Purified Sc237, purified 139H, or no seed | Purified hamster PrPC with co-purified lipids, plus RNA cofactor | Generation of infectious PrPSc from purified PrPC requires RNA cofactor in either seeded or non-seeded PMCA reactions | |
| Wang et al. (2010, 2015), Pan et al. (2020) | No seed | Bacterially expressed and purified recombinant mouse PrP, plus lipid and RNA cofactors | De novo formation of infectious recombinant prion by PMCA requires lipid and RNA cofactors | |
| Prion strain | Castilla et al. (2008b) | 5 murine prion strains, 4 human prion strains | Normal mouse brain homogenate, brain homogenate from transgenic mice expressing human PrP | PMCA-amplified prions faithfully recapitulate the strain-specific biochemical and biological properties of the seed prion strains |
| Wang et al. (2010), Kim et al. (2010), Deleault et al. (2012b) | No seed, mouse prion, or hamster prion | Recombinant mouse or hamster PrP | PMCA generated infectious recombinant prions manifest novel strain characteristics, which demonstrates the critical role of in vivo cofactors in maintaining strain properties | |
| Prion species barrier | Castilla et al. (2008a) | Mouse and hamster prions | Normal mouse and hamster brain homogenates | PMCA not only recapitulates the species barrier phenomenon in vitro, but also demonstrates its great potential to investigate strain adaptation and stabilization |
| Green et al. (2008) | Mouse prion | Brain homogenate from transgenic mice expressing deer PrP | PMCA-facilitated strain adaptation recapitulates the in vivo cross-species transmission that requires hundreds of days of adaptation and stabilization | |
| Pritzkow (2022) | North American CWD and Norwegian CWD prions | Normal mouse brain homogenate, brain homogenates from transgenic mice expressing PrP of various species | Inter-species PMCA studies suggest North American and Norwegian CWD prions represent different strains with distinct cross-species conversion potentials | |
| Zoonotic potential of animal prion disease | Jones et al. (2009) | BSE, scrapie | Brain homogenate from transgenic mice expressing human PrP | First PMCA study to show the conversion susceptibility of human PrP to BSE but not scrapie prions |
| Barria et al. (2011) | CWD | Brain homogenate from transgenic mice expressing human PrP | CWD, after either in vivo passages in animals or in vitro passages by PMCA, is able to convert human PrPC to PrPSc, indicating the potential of CWD prions to infect humans | |
| Wang et al. (2021) | CWD | Non-CJD human brain homogenate | CWD-seeded, PMCA-amplified human PrPSc is able to induce bona fide prion disease in humanized transgenic mice, demonstrating the ability of CWD prions to cross the species barrier and infect humans | |
| Pritzkow (2022) | North American CWD and Norwegian CWD prions | Brain homogenate from transgenic mice expressing human PrP | Cross-species PMCA results indicate North American CWD prions have more zoonotic potential than Norwegian CWD prions |
Using PMCA to study the prion hypothesis
Although a great deal of studies supports the prion hypothesis (Gabizon et al. 1988; Hsiao et al. 1990; Biieler et al. 1993; Telling et al. 1996; Sigurdson et al. 2009; Jackson et al. 2009), the general thinking was that the definitive evidence requires the in vitro generation of infectious prions, with defined components, which are capable of causing bona fide prion diseases in vivo (Soto and Castilla 2004). The generation of synthetic mammalian prions in the laboratory was first reported by Prusiner’s group in 2004 (Legname et al. 2004). Amyloid fibrils were formed in vitro using recombinant PrP and inoculated into transgenic mice overexpressing truncated mouse PrP. After prolonged incubation periods, the animals developed neurological diseases accompanied by the accumulation of protease-resistant PrP (termed PrPres), which was transmissible to wild-type mice with a shortened incubation time. Neuropathological characterization of these mice suggested a novel prion strain. However, these in vitro-generated amyloid fibrils were unable to infect wild-type animals, which indicate that low prion infectivity is associated with this material (Legname et al. 2004). In addition, it is well established that transgenic mice overexpressing mutated forms of the prion protein gene are prone to the spontaneous generation of some disease-related features (Chiesa et al. 1998; Westaway et al. 1994). Thus, since synthetic prions were not capable to infect wild-type animals, it remains possible that the results observed corresponded to spontaneous pathological changes in these mice.
In 2005, Soto and colleagues reported that using PMCA, they were able to generate PrPSc that preserved many biochemical and biophysical properties of the original PrPSc seed. More importantly, when inoculated into wild-type animals, the PMCA amplified PrPSc induced a deadly transmissible disease that was similar to the prion disease caused by the original PrPSc seed (Castilla et al. 2005a). This work not only provided the strongest support for the prion hypothesis, but also demonstrated the great value of PMCA in studying prions.
To further understand the mechanism of prion formation, several groups carried out PMCA studies with crude brain homogenate or purified PrPSc from diseased animals as seeds and purified PrPC, either from healthy animals or from bacteria, as substrates. Using PMCA, Supattapone and colleagues generated infectious prions from a mixture of purified PrPSc and PrPC plus an auxiliary cofactor, synthetic polyA RNA (Deleault et al. 2007). Interestingly, in unseeded PMCA reactions containing only PrPC with co-purified lipids and the cofactor molecules, PrPres could form spontaneously in test tubes and cause wild-type animals to develop prion disease, which was transmissible on the second passage (Deleault et al. 2007). Subsequently, Wang et al. reported that bona fide prions can be generated by PMCA using just bacterially expressed recombinant mouse PrP, RNA, and lipids (Wang et al. 2010). The PMCA-generated recombinant prion induced prion diseases in wild-type mice with a highly synchronized incubation period comparable to that of some native mouse prions. Follow-up animal infection assays by intraperitoneal and oral routes further demonstrate that recombinant prion behaves like naturally occurring prions in all respects (Wang et al. 2015; Pan et al. 2020). Collectively, these PMCA studies provided unambiguous evidence in support of the prion hypothesis.
Understanding prion strains and species barriers by PMCA
Like conventional infectious diseases, prion disorders can manifest in the same host species with various distinct clinical and pathological phenotypes, a property referred to as strain diversity (Weissmann 1991; Bruce 1993). For transmissible diseases caused by viruses, bacteria, or fungi, different strains are determined by distinct genetic variations of the pathogens. For prions, it has been proposed that each strain represents a unique self-replicating conformation of the same prion protein (Morales et al. 2007; Aguzzi et al. 2007; Collinge 2010; Morales 2017; Soto and Pritzkow 2018). Although atomistic level high-resolution structural studies of various strains are required to prove this hypothesis, a series of PMCA studies have provided valuable insights into this perplexing issue (Deleault et al. 2007, 2012b; Castilla et al. 2008b; Wang et al. 2010; Kim et al. 2010). Using 5 murine prion strains and 4 human prion strains, it was reported that PMCA can faithfully replicate the biochemical properties of all strains, such as electrophoretic mobility and glycosylation pattern after protease digestion (Castilla et al. 2008b). Critically, all 5 PMCA-amplified murine prions caused transmissible diseases in wild-type mice, which manifested strain-specific characteristics that are indistinguishable from the diseases induced by the brain-derived native strains (Castilla et al. 2008b). These experiments demonstrated that crude brain homogenates contain all the cellular factors necessary for faithful in vitro strain replication by PMCA. Indeed, the first highly infectious recombinant prion generated by PMCA in the presence of synthetic phospholipids and mouse liver RNA produced prions with certain novel strain characteristics, most likely because the cofactors used in vitro are different from the in vivo conversion factors (Wang et al. 2010). Similarly, in another recombinant prion study, where only recombinant hamster PrP and no cellular cofactors were used, the PMCA-generated recombinant prions elicited different neuropathological changes from the native 263 K prion that was used to seed the formation of recombinant prions (Kim et al. 2010). These studies suggested the important roles of cellular cofactors in forming and maintaining prion strains. In an attempt to isolate the brain molecules that facilitate the in vivo prion conversion, Supattapone’s group identified the endogenous phosphatidylethanolamine (PE) as the sole cofactor needed to produce prions (designated as recPrPSc−PE) by PMCA (Deleault et al. 2012a). When used to propagate different mouse prion strains, including the first recombinant prion, ME7, and 301C, the PE cofactor induced stain convergence: all three distinct strains were converted into a single new strain (Deleault et al. 2012b).
Similar to classical infectious agents, the interspecies transmission of prions often leads to longer incubation time, incomplete attack rate, subclinical infection, or even complete blockage, a phenomenon called species barrier (Prusiner 1993; Bruce et al. 1994; Priola 1999; Clarke et al. 2001; Collinge and Clarke 2007). Within the prion hypothesis framework, the species barrier is thought to be due to the mismatch of the PrP primary sequences of the two species. In 2008, we showed that PMCA can recapitulate the species barrier phenomenon (Castilla et al. 2008a). A clear barrier to prion conversion was observed between hamster and mouse, showing no amplification of mouse RML prion with hamster brain homogenate substrate, and vice versa. Because of the extremely powerful amplification ability of PMCA, when a higher amount of RML murine prions or 263 K hamster prions was applied as seed, hamster PrPC or mouse PrPC was converted to PrPSc, both of which could be propagated indefinitely (Castilla et al. 2008a). Interestingly, with successive rounds of prion replication, the strain characteristics were progressively adapted to the new PrPC host sequence (Castilla et al. 2008a). This study indicated that PMCA, in addition to recapitulating the specie barrier phenomenon in vitro, has the great potential to investigate the cross-species transmission characterized by strain adaptation and stabilization that often require a series of in vivo transmissions. Similarly, Telling and colleagues showed that PMCA amplification of mouse RML prions at expense of cervid PrPC generated PrPSc infectious to cervidized transgenic mice (Green et al. 2008). Thus, serial PMCA abrogated a transmission barrier that required several hundred days of adaptation and subsequent stabilization in transgenic mice. In a recent study (Pritzkow et al. 2022), we used PMCA to study the transmission of various CWD isolates to various animal species. We were able to identify specific profiles of inter-species transmission of distinct CWD prion strains (Pritzkow et al. 2022).
Application of PMCA to investigate the zoonotic potential of animal prion diseases
The causal relationship between BSE and vCJD (Collinge and Rossor 1996; Will et al. 1996; Collinge et al. 1996; Bruce et al. 1997; Hill et al. 1997; Collinge 1997) and the observation that no human prion diseases have been linked to natural ovine scrapie (EFSA Panel on Biological Hazards (BIOHAZ) 2011) have offered a great opportunity to evaluate the applicability of PMCA to study the zoonotic potential of animal prion diseases. In 2009, Jones et al. reported that using PMCA, they were able to reproduce the conversion susceptibility of human PrPC to BSE prions but not scrapie prions (Jones et al. 2009). Moreover, interspecies conversion using different polymorphic versions of human PrPC followed the expected pattern based on epidemiological data and studies in animal models.
Currently, one of the biggest zoonotic concerns comes from the CWD epidemic in North America (Houston and Andréoletti 2019; Pritzkow 2022). CWD was initially recognized and limited to northern Colorado in 1967 but has since been found in 30 US states and 4 Canadian provinces of North America (USGA Map 2022), with recently reported cases in Norway, Sweden, and Finland of Europe (Tranulis et al. 2021) and South Korea of Asia (Lee et al. 2013). Thus, answering the question of whether or not CWD prions can infect humans is highly relevant. Conventional rodent infection studies from independent labs have reported failed attempts to transmit CWD prions to transgenic mice that overexpress human PrP (Kong et al. 2005; Tamgüney et al. 2006; Sandberg et al. 2010; Wilson et al. 2012; Kurt et al. 2015; Race et al. 2019; Wadsworth et al. 2021) with the exception of a recent study (Hannaoui et al. 2022). Using non-human primates, the Bartz group reported two squirrel monkeys that were intracerebrally challenged with CWD prions developed prion diseases and reached terminal stages in 31 and 34 months after infection (Marsh et al. 2005). The transmission of CWD prions to squirrel monkeys was confirmed later in a study from the Chesebro group (Race et al. 2009). However, the same CWD inocula did not induce prion disease in another non-human primate, Cynomolgus macaques (Race et al. 2009). Two follow-up studies from the same group reported the same results that squirrel monkeys are highly susceptible to CWD infection but Cynomolgus macaques are resistant to CWD transmission (Race et al. 2014, 2018). Since the PrP sequences of both squirrel monkeys and Cynomolgus macaques differ from human PrP at various positions, it is complicated to draw definitive conclusions in terms of cross-species transmission of CWD to humans. To this end, PMCA offers an efficient platform to address such a significant yet challenging issue.
In a PMCA study, the first attempt to convert human PrP by CWD prions was unsuccessful (Barria et al. 2011), which was in agreement with the transgenic animal studies that there is a strong species barrier between humans and cervids. However, after serial passaging of CWD PrPSc in transgenic mice expressing cervid PrP, the in vivo adapted CWD prions were able to convert human PrPC to PrPSc (Barria et al. 2011). Owing to the efficient strain adaptation and stabilization ability of PMCA (Green et al. 2008; Castilla et al. 2008a), after one or two rounds of amplification, the in vitro-adapted CWD prions could efficiently convert human PrPC to its scrapie form. Noteworthy, the CWD-seeded PMCA converted human PrPSc displayed a unique protease-resistant banding pattern that is different from those of known CJD PrPSc but reminiscent of that of CWD prion (Barria et al. 2011). Using the PMCA technology, we recently compared North America CWD prions with the newly discovered Norwegian CWD prions, in terms of their potential to transmit to other animals and their zoonotic potential to transmit to humans (Pritzkow et al. 2022). Interestingly, while both North America and Norwegian CWD prions were estimated to possess the potential to transmit to other animals, only North America CWD prions were found to be able to convert human PrPC (Pritzkow et al. 2022). This zoonotic potential difference might be due to the fact that North American CWD prions have existed for a longer time, which has allowed them to become more stabilized and virulent after many rounds of natural infection. In the latest study combining PMCA conversion and animal bioassay, the Zou group showed that in vitro-converted CWD-human PrPSc could induce bona fide prion diseases in transgenic mice expressing human PrPC (Wang et al. 2021). These PMCA studies highlighted the possibility, albeit low so far, that CWD prions can cross the species barrier to infect humans.
Using PMCA to study the interaction of prions with various materials
Shedding of CWD prions through secretion and excretion and their interaction with various elements of the surrounding environment has likely helped the fast transmission of CWD among cervids (Pritzkow et al. 2021). Miller et al. reported that under experimental conditions, naïve mule deer became CWD infected after living in paddocks where either infected deer had resided or infected deer carcasses have decomposed a couple of years earlier (Miller et al. 2004). Later, Johnson et al. demonstrated that purified hamster prions could bind to clay soil without diminishing or losing their infectivity (Johnson et al. 2006). Instead, the soil-bound PrPSc displayed higher infectious titer than the unbound PrPSc in orally infected hamsters (Johnson et al. 2007).
While soil is considered the major environmental reservoir for prions, other natural and man-made common environmental materials, such as wood, rocks, plastic, glass, cement, and various metallic surfaces, might also be able to bind prions and act as vectors for disease transmission. To test this possibility, we exposed the surfaces of these materials to various prions, including 263 K, RML, vCJD, and CWD, and studied their attachment and retention of prion infectivity by both in vitro PMCA assay and in vivo animal assay (Pritzkow et al. 2018). PMCA studies identified distinct prion binding capacities associated with different prions, and rodent assays confirmed the ability of various material surfaces to disseminate prion diseases (Pritzkow et al. 2018). Since plants are the most common food for animals that can be naturally affected by prion diseases, these herbal organisms might also play a role in transmitting prion diseases. In an experimental setting, Pritzkow et al. demonstrated that prions from diverse origins, including CWD in urine and feces, effectively bind to wheat grass roots and leaves and can be readily detected by PMCA (Pritzkow et al. 2015). When attached to living plants, prions could be detected even weeks later, showing their persistence. Moreover, when naïve grass was planted in prion-contaminated soil, prions could be absorbed from the soil to the grass roots and further transported to stems and leaves that are not in direct contact with contaminated soil (Pritzkow et al. 2015). Finally, detection of CWD prions by PMCA in one of two environmental water samples from a CWD endemic area (Nichols et al. 2009) and natural mineral licks (Plummer et al. 2018) demonstrated the persistence of prions in the natural environment.
Application of PMCA to screen for prions in biological products and medical equipment
Biological products derived from living organisms have been increasingly used for treating human diseases. For example, stem cells are considered a great option to treat aging and other aging-associated diseases, such as Alzheimer’s disease and Parkinson’s disease (Bali et al. 2016; Yasuhara et al. 2017). Lessons learned from the accidental transmission of the human immunodeficiency virus (HIV) and hepatitis C virus (HCV) through blood transfusions have driven the standardization of blood donation procedures to include routine screening tests for HIV and HCV (di Minno et al. 2016). Iatrogenic CJD arises from incidental transmission of CJD via the use of contaminated biological materials (e.g., human growth hormone, dura mater, and corneal grafts) or neurosurgical instruments. Because prions can lead to incurable fatal diseases and are inert to the conventional inactivation methods used to eliminate viral, bacterial, and fungi contaminations, it is particularly important to screen cells and other biological materials for prion contamination. As a proof-of-principle study, we reported that using the PMCA technology, a single prion-infected cell can be detected in a group of over one million or more non-infected cells (Lyon et al. 2019). A cell line that has been used to generate anti-cancer vaccines was tested free of vCJD and BSE prions using PMCA. For reusable surgical instruments contaminated with prions, although there are already decontamination protocols in place (Belay et al. 2010, 2013), PMCA tests following the decontamination procedures will ensure these valuable medical devices are truly free of prions.
Final conclusions and remarks
One of the main technologies used for high sensitive detection of conventional infectious agents (e.g., SARS-CoV-2, HIV, Hepatitis) is the amplification of nucleic acids by PCR. Prions are unconventional infectious agents composed exclusively of a protein that is responsible for a group of fatal neurodegenerative diseases in humans and animals. The unorthodox nature of prion requires an unconventional tool to detect and study them. The PMCA technology was invented and developed to meet this challenge. Analogous to the detection of viruses by PCR through amplification of the viral genetic materials, prions can be detected by PMCA via the propagation of the infectious protein. Powered by its cyclic nature of amplification, the ultrasensitive PMCA technique is capable of amplifying prions from the equivalent of single molecules of the infectious agent (Saá et al. 2006a). This enabled the detection of minute amounts of prions in various body fluids and peripheral tissues, even from pre-symptomatic individuals (Moda et al. 2014; Bougard et al. 2016; Concha-Marambio et al. 2016; Barria et al. 2018). The 100% sensitivity and specificity demonstrated by PMCA in the blood studies (Bougard et al. 2016; Concha-Marambio et al. 2016) bode well for its application as a standard diagnostic tool for prion diseases.
In addition to its use for prion detection and disease diagnosis, PMCA has also had a great impact on studies to understand the complex biology of prions (Soto 2011). PMCA allows the faithful in vitro propagation of prions under proper conditions, which enabled the in vitro generation of infectious material in the laboratory (Castilla et al. 2005a; Deleault et al. 2007; Wang et al. 2010), representing the most conclusive evidence for the prion hypothesis. Such feature of PMCA also makes possible the in-depth studies of the mechanisms underlying prion conversion and propagation as well as to shed light on the complex phenomenon of strain diversity and species barrier (Castilla et al. 2008a, b; Deleault et al. 2012b). Beyond, PMCA has been utilized for drug repurposing to identify therapeutic leads for treating prion diseases (Barret et al. 2003).
Finally, since the process of protein misfolding and aggregation in prion disease has remarkable similarities with those undergone by several other proteins associated with various neurological disorders (Soto and Pritzkow 2018), the principles behind PMCA have recently been used to develop cyclic amplification procedures to diagnose these diseases. Several reports have shown the use of this technology to detect misfolded protein aggregates implicated in highly prevalent diseases, including Alzheimer’s and Parkinson’s diseases (Salvadores et al. 2014; Shahnawaz et al. 2017, 2020).
Funding
This work was partially supported by grant P01 AI077774 from the NIH to CS.
Declarations
Ethical approval
Not applicable.
Informed consent
Not applicable.
Conflict of interest
Claudio Soto is a Founder, Chief Scientific Officer, and Member of the Board of Directors of Amprion Inc., a biotechnology company that focuses on the commercial use of PMCA for high-sensitivity detection of misfolded protein aggregates implicated in various neurodegenerative diseases. Sandra Pritzkow also has a conflict of interest with Amprion. The University of Texas Health Science Center at Houston has licensed patents and patent applications to Amprion.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fei Wang and Sandra Pritzkow contributed equally to this article.
References
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- Andréoletti O, Litaise C, Simmons H, Corbière F, Lugan S, Costes P, Schelcher F, Vilette D, Grassi J, Lacroux C. Highly efficient prion transmission by blood transfusion. PLoS Pathog. 2012;8:e1002782. doi: 10.1371/JOURNAL.PPAT.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babelhadj B, di Bari MA, Pirisinu L, Chiappini B, Gaouar SBS, Riccardi G, Marcon S, Agrimi U, Nonno R, Vaccari G. Prion disease in dromedary camels in Algeria. Emerg Infect Dis. 2018;24:1029–1036. doi: 10.3201/EID2406.172007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali P, Lahiri D, Banik A, Nehru B, Anand A. Potential for stem cells therapy in Alzheimer’s disease: do neurotrophic factors play critical role? Curr Alzheimer Res. 2016;14:208–220. doi: 10.2174/1567205013666160314145347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret A, Tagliavini F, Forloni G, Bate C, Salmona M, Colombo L, de Luigi A, Limido L, Suardi S, Rossi G, Auvré F, Adjou KT, Salès N, Williams A, Lasmézas C, Deslys JP. Evaluation of quinacrine treatment for prion diseases. J Virol. 2003;77:8462. doi: 10.1128/JVI.77.15.8462-8469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria MA, Lee A, Green AJE, Knight R, Head MW. Rapid amplification of prions from variant Creutzfeldt-Jakob disease cerebrospinal fluid. J Pathol Clin Res. 2018;4:86–92. doi: 10.1002/CJP2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria MA, Telling GC, Gambetti P, Mastrianni JA, Soto C. Generation of a new form of human PrPScin vitro by interspecies transmission from cervid prions. J Biol Chem. 2011;286:7490–7495. doi: 10.1074/jbc.M110.198465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay ED, Blase J, Sehulster LM, Maddox RA, Schonberger LB. Management of neurosurgical instruments and patients exposed to Creutzfeldt-Jakob disease. Infect Control Hosp Epidemiol. 2013;34:1272–1280. doi: 10.1086/673986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay ED, Schonberger LB, Brown P, Priola SA, Chesebro B, Will RG, Asher DM. Disinfection and sterilization of prion-contaminated medical instruments. Infect Control Hosp Epidemiol. 2010;31:1304–1306. doi: 10.1086/657579. [DOI] [PubMed] [Google Scholar]
- Bélondrade M, Nicot S, Mayran C, Bruyere-Ostells L, Almela F, di Bari MA, Levavasseur E, Watts JC, Fournier-Wirth C, Lehmann S, Haïk S, Nonno R, Bougard D. Sensitive protein misfolding cyclic amplification of sporadic Creutzfeldt-Jakob disease prions is strongly seed and substrate dependent. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-83630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biieler H, Aguui A, Sailer A, Greiner R-A, Autenried P, Ague M, Weissmann C (1993) Mice devoid of PrP are resistant to scrapie. Cell 73:1339–1347 [DOI] [PubMed]
- Bons N, Lehmann S, Mestre-Francès N, Dormont D, Brown P. Brain and buffy coat transmission of bovine spongiform encephalopathy to the primate Microcebus murinus. Transfusion. 2002;42:513–516. doi: 10.1046/J.1537-2995.2002.00098.X. [DOI] [PubMed] [Google Scholar]
- Bougard D, Bélondrade M, Mayran C, Bruyère-Ostells L, Lehmann S, Fournier-Wirth C, Knight RS, Will RG, Green AJE. Diagnosis of methionine/valine variant Creutzfeldt-Jakob disease by protein misfolding cyclic amplification. Emerg Infect Dis. 2018;24:1364–1366. doi: 10.3201/EID2407.172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougard D, Brandel JP, Bélondrade M, Béringue V, Segarra C, Fleury H, Laplanche JL, Mayran C, Nicot S, Green A, Welaratne A, Narbey D, Fournier-Wirth C, Knight R, Will R, Tiberghien P, Haïk S, Coste J (2016) Detection of prions in the plasma of presymptomatic and symptomatic patients with variant Creutzfeldt-Jakob disease. Sci Transl Med 8. 10.1126/SCITRANSLMED.AAG1257/SUPPL_FILE/8-370RA182_SM.PDF [DOI] [PubMed]
- Brown P, Cervenáková L, Diringer H. Blood infectivity and the prospects for a diagnostic screening test in Creutzfeldt-Jakob disease. J Lab Clin Med. 2001;137(1):5–13. doi: 10.1067/mlc.2001.111951. [DOI] [PubMed] [Google Scholar]
- Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond B Biol Sci. 1994;343:405–411. doi: 10.1098/RSTB.1994.0036. [DOI] [PubMed] [Google Scholar]
- Bruce ME. Scrapie strain variation and mutation. Br Med Bull. 1993;49:822–838. doi: 10.1093/OXFORDJOURNALS.BMB.A072649. [DOI] [PubMed] [Google Scholar]
- Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock CJ. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Castilla J, Saá P, Soto C. Detection of prions in blood. Nat Med. 2005;11:982–985. doi: 10.1038/nm1286. [DOI] [PubMed] [Google Scholar]
- Castilla J, Saá P, Morales R, Abid K, Maundrell K, Soto C. Protein misfolding cyclic amplification for diagnosis and prion propagation studies. Methods Enzymol. 2006;412:3–21. doi: 10.1016/S0076-6879(06)12001-7. [DOI] [PubMed] [Google Scholar]
- Castilla J, Gonzalez-Romero D, Saá P, Morales R, de Castro J, Soto C. Crossing the species barrier by PrPSc replication in vitro generates unique infectious prions. Cell. 2008;134:757–768. doi: 10.1016/j.cell.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla J, Morales R, Saá P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J. 2008;27:2557–2566. doi: 10.1038/emboj.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga, FA, Bistaffa E, De Luca CMG (2022) PMCA-based detection of prions in the olfactory mucosa of patients with sporadic Creutzfeldt-Jakob disease. Front Aging Neurosci 14. 10.3389/FNAGI.2022.848991/FULL [DOI] [PMC free article] [PubMed]
- Chen B, Morales R, Barria MA, Soto C. Estimating prion concentration in fluids and tissues by quantitative PMCA. Nat Meth. 2010;7:519–520. doi: 10.1038/nmeth.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Ghetti B, Harris DA. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21(6):1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Jackson GS, Collinge J, Pepys MB, Barron LD, Masel J, Tahari-Alaoui A, Lansbury P, Dobson CM, Exley C, Feizi T. The molecular biology of prion propagation. Philos Trans R Soc Lond B Biol Sci. 2001;356:185–195. doi: 10.1098/RSTB.2000.0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J. Human prion diseases and bovine spongiform encephalopathy (BSE) Hum Mol Genet. 1997;6:1699–1705. doi: 10.1093/HMG/6.10.1699. [DOI] [PubMed] [Google Scholar]
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/ANNUREV.NEURO.24.1.519. [DOI] [PubMed] [Google Scholar]
- Collinge J. Prion strain mutation and selection. Science. 2010;328:1111–1112. doi: 10.1126/SCIENCE.1190815/ASSET/740BD400-3CBA-4867-8FF0-7E8975A40B65/ASSETS/GRAPHIC/328_1111_F1.JPEG. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/SCIENCE.1138718/SUPPL_FILE/COLLINGE.SOM.PDF. [DOI] [PubMed] [Google Scholar]
- Collinge J, Rossor M. A new variant of prion disease. Lancet. 1996;347:916–917. doi: 10.1016/S0140-6736(96)91407-5. [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle KCL, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of “new variant” CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- Concha-Marambio L, Chacon MA, Soto C. Preclinical detection of prions in blood of nonhuman primates infected with variant Creutzfeldt-Jakob disease. Emerg Infect Dis. 2020;26:34–43. doi: 10.3201/EID2601.181423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha-Marambio L, Pritzkow S, Moda F, Tagliavini F, Ironside JW, Schulz PE, Soto C (2016) Detection of prions in blood from patients with variant Creutzfeldt-Jakob disease. Sci Transl Med 8. 10.1126/SCITRANSLMED.AAF6188/SUPPL_FILE/8-370RA183_SM.PDF [DOI] [PMC free article] [PubMed]
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A. 2007;104:9741–9746. doi: 10.1073/PNAS.0702662104/SUPPL_FILE/02662FIG13.JPG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Piro JR, Walsh DJ, Wang F, Jiyan M, Geoghegan JC, Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A. 2012;109:8546–8551. doi: 10.1073/PNAS.1204498109/SUPPL_FILE/PNAS.201204498SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A. 2012;109:E1938–E1946. doi: 10.1073/PNAS.1206999109/SUPPL_FILE/PNAS.201206999SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Minno G, Perno CF, Tiede A, Navarro D, Canaro M, Güertler L, Ironside JW. Current concepts in the prevention of pathogen transmission via blood/plasma-derived products for bleeding disorders. Blood Rev. 2016;30:35–48. doi: 10.1016/J.BLRE.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgeworth JA, Farmer M, Sicilia A, Tavares P, Beck J, Campbell T, Lowe J, Mead S, Rudge P, Collinge J, Jackson GS. Detection of prion infection in variant Creutzfeldt-Jakob disease: a blood-based assay. The Lancet. 2011;377:487–493. doi: 10.1016/S0140-6736(10)62308-2. [DOI] [PubMed] [Google Scholar]
- EFSA Panel on Biological Hazards (BIOHAZ) (2011) Joint Scientific Opinion on any possible epidemiological or molecular association between TSEs in animals and humans EFSA J 9. 10.2903/J.EFSA.2011.1945
- Gabizon R, McKinley MP, Groth D, Prusiner SB (1988) Immunoaffinity purification and neutralization of scrapie prion infectivity. Proc Natl Acad Sci 85:6617–6621 10.1073/PNAS.85.18.6617 [DOI] [PMC free article] [PubMed]
- Gonzalez-Montalban N, Makarava N, Ostapchenko VG, Savtchenk R, Alexeeva I, Rohwer RG, Baskakov IV. Highly efficient protein misfolding cyclic amplification. PLoS Pathog. 2011;7:e1001277. doi: 10.1371/journal.ppat.1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS Lett. 2008;582:3161–3166. doi: 10.1016/j.febslet.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, Telling GC. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog. 2008;4:e1000139. doi: 10.1371/JOURNAL.PPAT.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori L, Kovacs GG, Alexeeva I, Budka H, Rohwer RG. Excretion of transmissible spongiform encephalopathy infectivity in urine. Emerg Infect Dis. 2008;14:1406–1412. doi: 10.3201/EID1409.080259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannaoui S, Zemlyankina I, Chang SC, Arifin MI, Béringue V, McKenzie D, Schatzl HM, Gilch S. Transmission of cervid prions to humanized mice demonstrates the zoonotic potential of CWD. Acta Neuropathol. 2022 doi: 10.1007/s00401-022-02482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AF, Desbruslais M, Joiner S, Sidle KCL, Gowland I, Collinge J, Doey LJ, Lantos P. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- Houston F, Andréoletti O. Animal prion diseases: the risks to human health. Brain Pathol. 2019;29:248–262. doi: 10.1111/BPA.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston F, Foster JD, Chong A, Hunter N, Bostock CJ. Transmission of BSE by blood transfusion in sheep. Lancet. 2000;356:999–1000. doi: 10.1016/S0140-6736(00)02719-7. [DOI] [PubMed] [Google Scholar]
- Hsiao KK, Scott M, Foster D, Groth DF, Dearmond SJ, Prusiner SB. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science. 1990;250:1587–1590. doi: 10.1126/SCIENCE.1980379. [DOI] [PubMed] [Google Scholar]
- Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, Jasanoff A, Lindquist S. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63:438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CJ, Pedersen JA, Chappell RJ, McKenzie D, Aiken JM. Oral transmissibility of prion disease is enhanced by binding to soil particles. PLoS Pathog. 2007;3:e93. doi: 10.1371/JOURNAL.PPAT.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CJ, Phillips KE, Schramm PT, McKenzie D, Aiken JM, Pedersen JA. Prions adhere to soil minerals and remain infectious. PLoS Pathog. 2006;2:e32. doi: 10.1371/JOURNAL.PPAT.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Wight D, Barron R, Jeffrey M, Manson J, Prowse C, Ironside JW, Head MW. Molecular model of prion transmission to humans. Emerg Infect Dis. 2009;15:2013–2016. doi: 10.3201/EID1512.090194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariv-Inbal Z, Ben-Hur T, Grigoriadis NC, Engelstein R, Gabizon R. Urine from scrapie-infected hamsters comprises low levels of prion infectivity. Neurodegener Dis. 2006;3:123–128. doi: 10.1159/000094770. [DOI] [PubMed] [Google Scholar]
- Kim J, il, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, Surewicz WK Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285:14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B (1994) Cell-free formation of protease-resistant prion protein. Nature 1994 370:6489 370:471–474. 10.1038/370471a0 [DOI] [PubMed]
- Kong Q, Huang S, Zou W, et al. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosci. 2005;25:7944–7949. doi: 10.1523/JNEUROSCI.2467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt TD, Jiang L, Fernández-Borges N, et al. Human prion protein sequence elements impede cross-species chronic wasting disease transmission. J Clin Invest. 2015;125:2548–2548. doi: 10.1172/JCI82647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C, Bougard D, Litaise C, et al. Impact of leucocyte depletion and prion reduction filters on TSE blood borne transmission. PLoS ONE. 2012;7:e42019. doi: 10.1371/JOURNAL.PONE.0042019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C, Comoy E, Moudjou M, et al. Preclinical detection of variant CJD and BSE prions in blood. PLoS Pathog. 2014;10:e1004202. doi: 10.1371/JOURNAL.PPAT.1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Sohn HJ, Kim MJ, Kim HJ, Lee WY, Yun EI, Tark DS, Cho IS, Balachandran A. Strain characterization of the Korean CWD cases in 2001 and 2004. J Vet Med Sci. 2013;75:95–98. doi: 10.1292/JVMS.12-0077. [DOI] [PubMed] [Google Scholar]
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB (2004) Synthetic mammalian prions. Science 305:673–676. 10.1126/SCIENCE.1100195/SUPPL_FILE/LEGNAME.SOM.PDF [DOI] [PubMed]
- Llewelyn CA, Hewitt PE, Knight RSG, Amar K, Cousens S, MacKenzie J, Will RG. Possible transmission of variant Creutzfeldt-Jakob disease by blood transfusion. Lancet. 2004;363:417–421. doi: 10.1016/S0140-6736(04)15486-X. [DOI] [PubMed] [Google Scholar]
- Lyon A, Mays CE, Borriello F, Telling GC, Soto C, Pritzkow S. Application of PMCA to screen for prion infection in a human cell line used to produce biological therapeutics. Sci Rep. 2019;9:1–6. doi: 10.1038/s41598-019-41055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh RF, Kincaid AE, Bessen RA, Bartz JC. Interspecies transmission of chronic wasting disease prions to squirrel monkeys (Saimiri sciureus) J Virol. 2005;79:13794–13796. doi: 10.1128/JVI.79.21.13794-13796.2005/ASSET/3E3A1645-08E2-4E2B-91A1-7A0172ACE8B8/ASSETS/GRAPHIC/ZJV0210569610002.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, Lloyd S, Collinge J (2019)Genetic factors in mammalian prion diseases. Annu Rev Genet 3;53(1):117-47 [DOI] [PubMed]
- Miller MW, Williams ES, Hobbs NT, Wolfe LL. Environmental sources of prion transmission in mule deer. Emerg Infect Dis. 2004;10:1003–1006. doi: 10.3201/EID1006.040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moda F, Gambetti P, Notari S, Concha-Marambio L, Catania M, Park K-W, Maderna E, Suardi S, Haïk S, Brandel J-P, Ironside J, Knight R, Tagliavini F, Soto C. Prions in the urine of patients with variant Creutzfeldt-Jakob disease. New Engl J Med. 2014;371:530–539. doi: 10.1056/NEJMOA1404401/SUPPL_FILE/NEJMOA1404401_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok T, Jaunmuktane Z, Joiner S, Campbell T, Morgan C, Wakerley B, Golestani F, Rudge P, Mead S, Jäger HR, Wadsworth JDF, Brandner S, Collinge J. Variant Creutzfeldt-Jakob disease in a patient with heterozygosity at PRNP codon 129. New Engl J Med. 2017;376:292–294. doi: 10.1056/NEJMC1610003/SUPPL_FILE/NEJMC1610003_DISCLOSURES.PDF. [DOI] [PubMed] [Google Scholar]
- Morales R (2017) Prion strains in mammals: different conformations leading to disease. PLoS Pathog 13. 10.1371/JOURNAL.PPAT.1006323 [DOI] [PMC free article] [PubMed]
- Morales R, Abid K, Soto C (2007) The prion strain phenomenon: molecular basis and unprecedented features. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1772:681–691. 10.1016/J.BBADIS.2006.12.006 [DOI] [PMC free article] [PubMed]
- Morales R, Duran-Aniotz C, Diaz-Espinoza R, Camacho M, Soto C. Protein misfolding cyclic amplification of infectious prions. Nat Prot. 2012;7:1397–1409. doi: 10.1038/nprot.2012.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Masujin K, Imamura M, Ono F, Shibata H, Tobiume M, Yamamura T, Shimozaki N, Terao K, Yamakawa Y, Sata T. Ultrasensitive detection of PrPSc in the cerebrospinal fluid and blood of macaques infected with bovine spongiform encephalopathy prion. J Gen Virol. 2014;95:2576–2588. doi: 10.1099/VIR.0.066225-0/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- Murayama Y, Yoshioka M, Masujin K, Okada H, Iwamaru Y, Imamura M, Matsuura Y, Fukuda S, Onoe S, Yokoyama T, Mohri S. Sulfated dextrans enhance in vitro amplification of bovine spongiform encephalopathy PrPSc and enable ultrasensitive detection of bovine PrPSc. PLoS ONE. 2010;5:e13152. doi: 10.1371/JOURNAL.PONE.0013152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Yoshioka M, Okada H, Takata M, Yokoyama T, Mohri S. Urinary excretion and blood level of prions in scrapie-infected hamsters. J Gen Virol. 2007;88:2890–2898. doi: 10.1099/VIR.0.82786-0/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- Nichols TA, Pulford B, Wyckoff AC, Meyerett C, Michel B, Gertig K, Hoover EA, Jewell JE, Telling GC, Zabel MD. Detection of protease-resistant cervid prion protein in water from a CWD-endemic area. Prion. 2009;3:171–183. doi: 10.4161/PRI.3.3.9819/SUPPL_FILE/KPRN_A_10909819_SM0001.ZIP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Yang J, Zhang X, Chen Y, Wei S, Yu G, Pan YH, Ma J, Yuan C. Oral ingestion of synthetically generated recombinant prion is sufficient to cause prion disease in wild-type mice. Pathogens. 2020;9:653. doi: 10.3390/PATHOGENS9080653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AH, Head MW, Ritchie DL, Bell PJE, Ironside PJW. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet. 2004;364:527–529. doi: 10.1016/S0140-6736(04)16811-6. [DOI] [PubMed] [Google Scholar]
- Plummer IH, Johnson CJ, Chesney AR, Pedersen JA, Samuel MD. Mineral licks as environmental reservoirs of chronic wasting disease prions. PLoS ONE. 2018;13:e0196745. doi: 10.1371/JOURNAL.PONE.0196745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priola SA. Prion protein and species barriers in the transmissible spongiform encephalopathies. Biomed Pharmacoth. 1999;53:27–33. doi: 10.1016/S0753-3322(99)80057-2. [DOI] [PubMed] [Google Scholar]
- Pritzkow S, Wagenführ K, Daus ML, Boerner S, Lemmer K, Thomzig A, Mielke M, Beekes M. Quantitative detection and biological propagation of scrapie seeding activity in vitro facilitate use of prions as model pathogens for disinfection. PLoS ONE. 2011;6(5):e20384. doi: 10.1371/journal.pone.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritzkow S. Transmission, strain diversity, and zoonotic potential of chronic wasting disease. Viruses. 2022;14:1390. doi: 10.3390/V14071390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritzkow S, Gorski D, Ramirez F, Soto C (2021) Prion dissemination through the environment and medical practices: facts and risks for human health Clin Microbiol Rev 34. 10.1128/CMR.00059-19 [DOI] [PMC free article] [PubMed]
- Pritzkow S, Gorski D, Ramirez F, Telling GC, Benestad SL, Soto C. North American and Norwegian chronic wasting disease prions exhibit different potential for interspecies transmission and zoonotic risk. J Infect Dis. 2022;225:542–551. doi: 10.1093/INFDIS/JIAB385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritzkow S, Morales R, Lyon A, Concha-Marambio L, Urayama A, Soto C. Efficient prion disease transmission through common environmental materials. J Biol Chem. 2018;293:3363–3373. doi: 10.1074/jbc.M117.810747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritzkow S, Morales R, Moda F, Khan U, Telling GC, Hoover E, Soto C. Grass plants bind, retain, uptake, and transport infectious prions. Cell Rep. 2015;11:1168–1175. doi: 10.1016/j.celrep.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/SCIENCE.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/SCIENCE.1675487. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Genetic and infectious prion diseases. Arch Neurol. 1993;50:1129–1153. doi: 10.1001/ARCHNEUR.1993.00540110011002. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/PNAS.95.23.13363/ASSET/784B140D-D7D4-4C97-B807-3147C0C74B30/ASSETS/GRAPHIC/PQ2183010010.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade-White KD, Miller MW, et al. Susceptibilities of nonhuman primates to chronic wasting disease. Emerg Infect Dis. 2009;15:1366–1376. doi: 10.3201/EID1509.090253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade-White KD, Phillips K, Striebel J, Race R, Chesebro B. Chronic wasting disease agents in nonhuman primates. Emerg Infect Dis. 2014;20:833–837. doi: 10.3201/EID2005.130778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Williams K, Chesebro B. Transmission studies of chronic wasting disease to transgenic mice overexpressing human prion protein using the RT-QuIC assay. Vet Res. 2019;50:1–14. doi: 10.1186/S13567-019-0626-2/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race b, Williams k, Orrú CD, Hughson AG, Lubke L, Chesebro B (2018) Lack of transmission of chronic wasting disease to cynomolgus macaques. J Virol 92. 10.1128/JVI.00550-18/ASSET/64DC9F0A-AA34-4632-BB9D-7D25A6287056/ASSETS/GRAPHIC/ZJV0141836620009.JPEG [DOI] [PMC free article] [PubMed]
- Saá P, Castilla J, Soto C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J Biol Chem. 2006;281:35245–35252. doi: 10.1074/jbc.M603964200. [DOI] [PubMed] [Google Scholar]
- Saá P, Castilla J, Soto C. Presymptomatic detection of prions in blood. Science. 2006;313:92–94. doi: 10.1126/SCIENCE.1129051/SUPPL_FILE/SAA.SOM.PDF. [DOI] [PubMed] [Google Scholar]
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- Salvadores N, Shahnawaz M, Scarpini E, Tagliavini F, Soto C. Detection of misfolded Aβ oligomers for sensitive biochemical diagnosis of Alzheimer’s disease. Cell Rep. 2014;7:261–268. doi: 10.1016/j.celrep.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Sandberg MK, Al-Doujaily H, Sigurdson CJ, Glatzel M, O’Malley C, Powell C, Asante EA, Linehan JM, Brandner S, Wadsworth JDF, Collinge J. Chronic wasting disease prions are not transmissible to transgenic mice overexpressing human prion protein. J Gen Virol. 2010;91:2651–2657. doi: 10.1099/VIR.0.024380-0/CITE/REFWORKS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger H, Heikenwalder M, Zeller N, Kranich J, Schwarz P, Gaspert A, Seifert B, Miele G, Aguzzi A. Medicine: coincident scrapie infection and nephritis lead to urinary prion excretion. Science. 2005;310:324–326. doi: 10.1126/SCIENCE.1118829/SUPPL_FILE/SEEGER.SOM.PDF. [DOI] [PubMed] [Google Scholar]
- Shahnawaz M, Mukherjee A, Pritzkow S, Mendez N, Rabadia P, Liu X, Hu B, Schmeichel A, Singer W, Wu G, Tsai AL, Shirani H, Nilsson KPR, Low PA, Soto C. Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature. 2020;578:273–277. doi: 10.1038/s41586-020-1984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnawaz M, Tokuda T, Waraga M, Mendez N, Ishii R, Trenkwalder C, Mollenhauer B, Soto C. Development of a biochemical diagnosis of Parkinson disease by detection of α-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol. 2017;74:163–172. doi: 10.1001/JAMANEUROL.2016.4547. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KPR, Hornemann S, Heikenwalder M, Manco G, Schwarz P, Ott D, Rülicke T, Liberski PP, Julius C, Falsig J, Stitz L, Wüthrich K, Aguzzi A. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci U S A. 2009;106:304–309. doi: 10.1073/PNAS.0810680105/SUPPL_FILE/SM1.WMV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Diagnosing prion diseases: needs, challenges and hopes. Nature Rev Microbiol. 2004;2:809–819. doi: 10.1038/nrmicro1003. [DOI] [PubMed] [Google Scholar]
- Soto C. Prion hypothesis: the end of the controversy? Trends Biochem Sci. 2011;36:151–158. doi: 10.1016/j.tibs.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C, Anderes L, Suardi S, Cardone F, Castilla J, Frossard MJ, Peano S, Saa P, Limido L, Carbonatto M, Ironside J, Torres JM, Pocchiari M, Tagliavini F. Pre-symptomatic detection of prions by cyclic amplification of protein misfolding. FEBS Lett. 2005;579:638–642. doi: 10.1016/J.FEBSLET.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Soto C, Castilla J. The controversial protein-only hypothesis of prion propagation. Nat Med. 2004;10:S63. doi: 10.1038/nm1069. [DOI] [PubMed] [Google Scholar]
- Soto C, Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C, Saborio GP, Anderes L. Cyclic amplification of protein misfolding: application to prion-related disorders and beyond. Trends Neurosci. 2002;25(8):390–394. doi: 10.1016/s0166-2236(02)02195-1. [DOI] [PubMed] [Google Scholar]
- Soto C, Satani N. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol Med. 2011;17:14–24. doi: 10.1016/J.MOLMED.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamgüney G, Giles K, Bouzamondo-Bernstein E, Bosque PJ, Miller MW, Safar J, DeArmond SJ, Prusiner SB. Transmission of elk and deer prions to transgenic mice. J Virol. 2006;80:9104–9114. doi: 10.1128/JVI.00098-06/SUPPL_FILE/TAMGUNEY_JVIROL_SUPPLEMENT.DOC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling GC, Haga T, Torchia M, Tremblay P, DeArmond SJ, Prusiner SB. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10:1736–1750. doi: 10.1101/GAD.10.14.1736. [DOI] [PubMed] [Google Scholar]
- Tranulis MA, Gavier-Widén D, Våge J, Nöremark M, Korpenfelt SL, Hautaniemi M, Pirisinu L, Nonno R, Benestad SL (2021) Chronic wasting disease in Europe: new strains on the horizon Acta Vet Scand 63. 10.1186/S13028-021-00606-X [DOI] [PMC free article] [PubMed]
- USGA Map (2022) Distribution of chronic wasting disease in North America | U.S. Geological Survey. https://www.usgs.gov/media/images/distribution-chronic-wasting-disease-north-america-0. Accessed 27 Aug 2022
- Wadsworth JDF, Collinge J. Molecular pathology of human prion disease. Acta Neuropathol. 2010;121:69–77. doi: 10.1007/S00401-010-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth JDF, Joiner S, Linehan JM, Jack K, Al-Doujaily H, Costa H, Ingold T, Taema M, Zhang F, Sandberg MK, Brandner S, Tran L, Vikøren T, Våge J, Madslien K, Ytrehus B, Benestad SL, Asante EA, Collinge J. Humanized transgenic mice are resistant to chronic wasting disease prions from Norwegian reindeer and moose. J Infect Dis. 2021 doi: 10.1093/INFDIS/JIAB033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/SCIENCE.1183748/SUPPL_FILE/WANG.SOM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McGovern G, Zhang Y, Wang F, Zha L, Jeffrey M, Ma J. Intraperitoneal infection of wild-type mice with synthetically generated mammalian prion. PLoS Pathog. 2015;11:e1004958. doi: 10.1371/JOURNAL.PPAT.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Qin K, Camacho MV, Cali I, Yuan J, Shen P, Greenlee J, Kong Q, Mastrianni JA, Zou WQ. Generation of human chronic wasting disease in transgenic mice. Acta Neuropathol Commun. 2021;9(1):158. doi: 10.1186/s40478-021-01262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C. A “unified theory” of prion propagation. Nature. 1991;352:679–683. doi: 10.1038/352679a0. [DOI] [PubMed] [Google Scholar]
- Weissmann C, Bueler H, Fischer M, Aguet M. Role of the PrP gene in transmissible spongiform encephalopathies. Intervirology. 1993;35:164–175. doi: 10.1159/000150307. [DOI] [PubMed] [Google Scholar]
- Westaway D, DeArmond SJ, Cayetano-Canlas J, Groth D, Foster D, Yang SL, Torchia M, Carlson GA, Prusiner SB. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76(1):117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/JOURNAL.PPAT.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will RG, Ironside JW, Zeidler M, Cousens SN, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofmar A, Smith PG. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–925. doi: 10.1016/S0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- Wilson R, Plinston C, Hunter N, et al. Chronic wasting disease and atypical forms of bovine spongiform encephalopathy and scrapie are not transmissible to mice expressing wild-type levels of human prion protein. J Gen Virol. 2012;93:1624–1629. doi: 10.1099/VIR.0.042507-0/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- Wroe SJ, Pal S, Siddique D, Hyare H, Macfarlane R, Joiner S, Linehan JM, Brandner S, Wadsworth JD, Hewitt P, Collinge J. Clinical presentation and pre-mortem diagnosis of variant Creutzfeldt-Jakob disease associated with blood transfusion: a case report. Lancet. 2006;368:2061–2067. doi: 10.1016/S0140-6736(06)69835-8. [DOI] [PubMed] [Google Scholar]
- Yasuhara T, Kameda M, Sasaki T, Tajiri N, Date I. Cell therapy for Parkinson’s disease. Cell Transplant. 2017;26:1551–1559. doi: 10.1177/0963689717735411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr I. Laboratory diagnosis of Creutzfeldt-Jakob disease. New Engl J Med. 2022;386:1345–1350. doi: 10.1056/nejmra2119323. [DOI] [PubMed] [Google Scholar]