Abstract
Purpose
Endometriosis, a gynecological disease, is difficult to be cured. Currently, to identify more potential biomarkers for the early diagnosis of endometriosis is urgently needed. Insulin like growth factor 2 (IGF2) has been revealed to correlate with endometriosis. This research aimed to further explore the role of IGF2 and its up-stream mechanism in endometriosis.
Methods
Primary ectopic endometrial stromal cells (EESCs) were extracted from ectopic endometrial tissues which were pathological endometrial tissues resected from three patients with II-III endometriosis. Primary normal endometrial stromal cells (NESCs) were extracted from normal endometrial tissues of two patients with grade III cervical dysplasia and one patient with uterine leiomyoma III. Four endometriotic cell lines (EEC145T, hEM15A, hEM5B2, and 12Z) and normal human endometrial epithelial cells (hEECs) were purchased. Cell proliferation, migration, and invasion were evaluated through functional assays. The molecular interaction between RNAs was investigated through mechanistic analyses.
Results
We discovered that IGF2 was upregulated in purchased endometriotic cells and primary EESC. Suppression of IGF2 hampered cell proliferation, migration, and invasion. Furthermore, insulin-like growth factor 2 antisense RNA (IGF2-AS) was uncovered to positively regulate IGF2 expression and enhanced proliferative, migratory, and invasive abilities of endometriotic cells. Mechanistically, miR-370-3p was found to bind with IGF2-AS and IGF2. IGF2-AS competitively bind with miR-370-3p to upregulate IGF2. Furthermore, IGF2-AS was revealed to activate the PI3K/AKT/mTOR signaling pathway through targeting miR-370-3p/IGF2 axis.
Conclusion
IGF2-AS promotes endometriotic cell growth via regulating IGF2/miR-370-3p axis and further activating PI3K/AKT/mTOR signaling pathway.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-022-02638-2.
Keywords: IGF2-AS, IGF2, miR-370-3p, Endometriosis, Proliferation
Introduction
Endometriosis is a frequently diagnosed inflammatory disease featured with the existence of endometrium-like tissues outside the uterus [1, 2]. The underlying pathologic mechanism is associated with abnormally programmed endometrial progenitor/stem cells [3, 4]. Endometriosis affects approximately one-tenth of women at reproductive age, leading to pelvic pain, dysmenorrhea, even infertility, or ovarian cancer [5]. Medical and surgical treatments have been adopted to treat endometriosis [6], while increased invasiveness and high metastasis of endometriotic cells made it difficult to be cured [7, 8]. Thus, a thorough exploration of pathogenesis and development of endometriosis is urgently needed. Kobayashi H et al. have pointed out that dysregulation of insulin-like growth factor 2 (IGF2) might facilitate endometriosis predisposition [9]. Kim H et al. have revealed that IGF-II 820G > A polymorphism is closely associated with endometriosis development in Korean women [10]. Our study targeted at exploring the function and underlying mechanism of IGF2 in endometriotic cells.
Long non-coding RNAs (lncRNAs) are molecules with no less than 200 nucleotides which are crucial participators in various biological processes [11]. Their function has a close association with the subcellular location. In cell nucleus, lncRNAs affect gene expression at the epigenetic and transcriptional levels; in cell cytoplasm, they modulate gene expression at the post-transcriptional and translational levels. LncRNAs with aberrant expression have been confirmed to act as tumor suppressor or promoter and play crucial parts in the development of tumors [12, 13]. Moreover, it has been uncovered that lncRNAs play a significant role in endometriosis development. SNP rs710886 A > G reduces lncRNA PCAT1 expression, thus decreasing the risk of endometriosis [14]. LncRNA MALAT1 mediates endometriosis pathogenesis via modulation on MMP-9 expression to facilitate endometrial cell apoptosis [15]. LncRNA CHL1-AS2 level is remarkably higher in ectopic endometrium tissues than in eutopic endometrium tissues or normal endometrium tissues [16]. LncRNA LINC00261 suppresses endometriotic cell growth and migration [17]. LncRNA AC002454.1 is highly expressed in endometriosis and synergistically works with CDK6 to promote endometrial cell migration and invasion [18]. Exosome-transmitted lncRNA aHIF is upregulated in serum of endometriosis patients and promotes angiogenesis in endometriosis [19].
The competitive endogenous RNA (ceRNA) network is a typical post-transcriptional mechanism in various biological processes. Accumulating evidence has suggested that lncRNAs are able to serve as the ceRNA to bind to microRNAs (miRNAs) to antagonize the suppression of miRNAs on mRNAs expression. Xu Z et al. have disclosed that lncRNA H19 axis mediates stromal cell invasion and migration via sponging miR-216a-5p to elevate ACTA2 expression in endometriosis [20]. Shi Y et al. have pointed out that lncRNA CHL1-AS1 sponges miR-6076 to upregulate CHL1 expression, thus promoting cell proliferative and migratory capabilities in endometrial cancer [21]. Wang Y et al. have revealed that LINC00958 regulates DOLPP1 expression via serving as an endogenous sponge of miR-761 [22].
In our research, we sought to delve into the detailed function of IGF2 in endometriotic cells. Furthermore, the regulatory mechanism of IGF2 with other RNAs was also deeply explored by a series of mechanism assays. We hope that this research could be informative and instructive for scholars to understand endometriosis and develop effective therapeutic targets.
Materials and methods
Clinical specimens
Ectopic endometrial tissues were pathological endometrial tissues resected from three patients with II-III endometriosis. The average age was 35, and the menstrual cycle range was between 22 and 30 days. Normal endometrial tissues were collected from two patients with grade 3 cervical dysplasia and one patient with uterine leiomyoma III via hysterectomy. The intermural myomas centered completely within the muscular wall of the uterine attached to the mucosa. The average age was 30, and the menstrual cycle range was between 28 and 32 days. The pathological diagnosis was done preoperatively through color Doppler ultrasound and verified after the surgery. The stage of endometriosis was determined based on lesion size and position, adhesion between ovaries and fallopian tubes, and the closure of rectouterine fossa. Considering specific symptoms, these patients who had given birth were encouraged to undergo surgical treatment to cure endometriosis, and the surgery was approved by the patients as well as their families. All clinical tissue samples were acquired from the First People’s Hospital of Wenling. Patients who received hormone treatment or suffered from concurrent malignancies were excluded from this experiment [23]. Clinical sample study was approved by the First People’s Hospital of Wenling.
Isolation procedures of primary endometrial stromal cells and cell culture
Isolation procedures of primary normal endometrial stromal cells (NESCs) and primary ectopic endometrial stromal cells (EESCs) followed the protocols mentioned in the previous study [23]. Briefly, normal or ectopic endometrial tissues were first cut up and then treated with phosphate-buffered saline (PBS) containing collagenase (1 mg/ml, 15 U/mg) and 1% penicillin/streptomycin in an orbital shaker for digestion at 37 °C. The acquired suspension of primary NESC (1.5 × 105) and EESC (1.5 × 105) was set in T25 cell culture flasks (Corning) and cultivated in Dulbecco’s modified Eagle medium (DMEM)/Ham’s F12 (1:1). The medium was mixed with 10% fetal bovine serum (FBS; Gibco) and antibiotics (1% penicillin/streptomycin). Cell purity was tested by immunofluorescence staining utilizing vimentin (ab8978, Abcam) and cytokeratin (ab76126, Abcam) antibodies. Two primary cell lines (CC cell line and corresponding normal cell line) with high purity and good viability were chosen in the subsequent experiments.
Cell lines
Endometriotic cells of EEC145T, hEM15A, hEM5B2, and 12Z were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Human endometrial epithelial cells (hEECs) were purchased from Procell company (Cat No.: CP-H058, China). Endometriotic cells (each cell line: 3 × 106) were cultivated in DMEM with 10% fetal calf serum (FCS) (PAA by GE Healthcare Life Sciences, Chalfont St Giles, UK), 1% glutamine (2 mM), and 1% penicillin–streptomycin. Meanwhile, 3 × 106 hEEC cells grew in 90% EMEM (MEM + NEAA) and 10% FBS at 37 °C with 95% air and 5% CO2.
Cell transfection
This assay was done to knock down or overexpress RNAs in primary NESC and EESC as well as hEM15A and hEM5B2 cells. The expression of IGF2 was downregulated by short hairpin RNAs (shRNAs) targeting IGF2 (sh-IGF2#1/sh-IGF2#2) with sh-NC as negative control. Similarly, sh-IGF2-AS#1/sh-IGF2-AS#2 was utilized for silencing IGF2-AS expression. MiR-370-3p mimics were adopted to overexpress miR-370-3p, with NC mimics as negative control. Moreover, miR-370-3p inhibitor was employed to inhibit miR-370-3p. All vectors used in our study were bought from Sigma Aldrich (Shanghai, China) and transfected into cells with Lipofectamine 3000 (Invitrogen, Carlsbad, CA).
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
This assay was done to test the expression of RNAs in primary NESC and EESC as well as five purchased endometriotic cell lines (each cell line: 2 × 106). First, 1 ml Trizol reagent (Invitrogen) was adopted to extract total RNA from the ectopic endometrium cells under manufacturer’s instruction. In the process of reverse transcription, the first strand cDNA was synthesized in a First Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA) by utilizing gene specific primers. Then, SYBR® Green qPCR Master Mix (Applied Biosystems, Foster City, CA, USA) in ABI 7500 Real-Time PCR Systems was used for qPCR analysis. The PCR conditions were as follows: denaturation for 30 s at 95 °C, annealing treatment for 45 s at 55 °C, and amplification for 30 s at 72 °C. The cycle was repeated 30 times. The microvolume spectrophotometer was employed to detect the absorbance at 260 nm to calculate RNA quantity. RNA purity was evaluated based on the ratio of absorbance at 260 nm/230 nm. RNA integrity number (RIN) was between 6 and 8, which meant extracted RNAs were in good quality. The 2−ΔΔCt method was utilized to measure the expression of target genes. GAPDH or U6 acted as the internal controls. The experiment was performed in triplicate.
Cell counting kit 8 (CCK-8) assay
Cell-counting kit 8 (Dojingdo Molecular Technologies, Rockville, Japan) was used to test cell proliferation. Primary NESC and EESC as well as hEM15A and hEM5B2 cells were transfected with different plasmids. Sh-NC acted as control. Cell viability was monitored at 0, 24, 48, and 72 h after transfection. Then, former medium was replaced by 10 µl of CCK-8 solution to cultivate 1 × 105 endometriotic cells or 3 × 103 primary cells for another 4 h. At last, the cell proliferative ability was assessed by microplate reader (Bio-Tek, Winooski, VT) at the length of 450 nm. The experiment was performed in triplicate.
5-Ethynyl-2′-deoxyuridine (EdU) assay
EdU assay was applied to evaluate proliferation of hEM15A and hEM5B2 cells. Cells were transfected with different plasmids. Sh-NC acted as control. To begin with, 2.5 × 105 cells were cultured in 100 μl EdU solution for 2 h. Later, cells were fixed by means of 4% paraformaldehyde. Finally, cells were fixed in 70% ethanol, and then stained by Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (RiboBio, China). Fluorescence microscopy was utilized to observe cell growth. ImageJ software was applied to count the number of cells stained by EdU/DAPI. The rate of EdU positive cells was calculated according to the formula: (EdU positive cells/DAPI-stained cells) × 100%. The experiment was performed in triplicate.
Colony formation assay
This assay was employed to examine cell proliferative ability. At first, hEM15A and hEM5B2 cells were transfected with different plasmids. Sh-NC acted as control. Then, cell samples were planted into 6-well plates (500 cells/each well) at 37 °C for 14 days. Colonies were dyed with the application of 0.5% crystal violet (0.9 ml) in 4% paraformaldehyde, followed by manual observation. The experiment was performed in triplicate.
Wound healing assay
This assay was utilized to assess cell migration. Primary NESC and EESC as well as hEM15A and hEM5B2 cells (each cell line: 1.2 × 105) were transfected with different plasmids. Sh-NC acted as control. Cells were seeded in six-well plates and cultivated. When cell confluence reached 70–80%, cell-free lane was obtained by using 200 μl pipette tip. A straight line was assured using ruler as a guide. Culture medium was discarded to remove detached cells. Then, 1 ml PBS was used for washing cells for 2 times. Subsequently, cells were continuously cultured for 24 h at 37 °C with 5% CO2. ImageJ software was employed to capture and analyze the images of wound. By comparing the changes of wound width at different time points, the speed of cell migration could be evaluated. If the cell migration capability was strong, the gap of wound would narrow down within certain time intervals. The experiment was performed in triplicate.
Transwell migration and invasion assay
This assay was done to evaluate cell migratory and invasive properties. Cells were transfected with different plasmids. Sh-NC acted as control. Before the invasion assay, the top insert was covered with Matrigel (Catalog No. 356234, EMD Millipore). Apart from this step, the migration assays were under the same protocol. The top compartment was filled with 0.1 ml single cell suspension. The lower compartment was added with medium supplemented with 20% FBS. After cells were cultured for 24 h, 0.5% crystal violet was used to stain membrane for 25 min at room temperature. Eventually, a light microscope (magnification, 40 ×) was used to observe invaded and migrated cells, and ImageJ software was used for cell counting. Cell migratory and invasive capabilities were evaluated by observing and comparing the number of cells that successfully migrated and invaded into the lower chamber. Primary NESC and EESC as well as hEM15A and hEM5B2 cells were involved in transwell invasion assay, while hEM15A and hEM5B2 cells were also involved in transwell migration assay (each cell line: 1 × 106). The experiment was performed in triplicate.
Western blot assay
This assay was performed to analyze protein levels in primary NESC and EESC as well as hEM15A and hEM5B2 cells (each cell line: 2 × 106). Cells were first dissolved with the assistance of RIPA buffer mixed with protease and phosphatase inhibitor (Roche, Shanghai China). Next, proteins were isolated by 10% SDS-PAGE. Afterwards, proteins were moved to PVDF membrane. The membrane sealed by 5% nonfat milk was cultured with primary antibodies for a whole night at 4 °C, and then incubated with secondary antibodies at room temperature. In the end, western blots were developed by the ECL luminescent liquid (Thermo Fisher Scientific, Rochester, NY). GAPDH acted as internal control. The experiment was performed in triplicate.
Subcellular fraction assay
This assay was carried out to localize IGF2-AS in cells. The cytoplasmic and nuclear RNAs in hEM15A and hEM5B2 cells (each cell line: 1 × 107) were acquired and purified as per the instruction of Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA). The extracted RNAs (IGF2-AS, GAPDH as reference gene for cytoplasm, U6 as reference gene for nucleus) were analyzed by qRT-PCR. The experiment was performed in triplicate.
Luciferase reporter assay
This assay was applied to verify the binding of miR-370-3p and IGF2-AS/IGF2. Wild-type or mutated IGF2 (IGF2 WT/Mut) or IGF2-AS WT/Mut was sub-cloned into pmirGLO dual-luciferase vector (Promega, Madison, WI, USA). These constructed plasmids were co-transfected with miR-370-3p mimics or their corresponding NC mimics into primary NESC and EESC as well as hEM15A and hEM5B2 cells (each cell line: 1 × 104). After incubation for 48 h, luciferase reporter assay system (Promega, Madison WI) was employed to measure the relative luciferase activities. Renilla luciferase activity was regarded as internal control. The experiment was performed in triplicate.
RNA pull down assay
This assay was applied to test the binding of miR-370-3p and IGF2-AS. The protein extracts of hEM15A and hEM5B2 cells (each cell line: 2 × 107) were collected from RIPA lysis buffer. Then they were mixed with the biotin-tagged probes of IGF2-AS. IGF2-AS no biotin probe was used as control. Magnetic beads were added after digestion. At last, the RNA–protein mixture was treated with proteinase K and then qRT-PCR was utilized for analysis. The experiment was performed in triplicate.
RNA immunoprecipitation (RIP)
This assay was conducted to indirectly detect the binding affinity of miR-370-3p and IGF2-AS/IGF2. A Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was applied to complete RIP assay. At first, 2 × 107 endometriotic cells (primary NESC and EESC as well as hEM15A and hEM5B2 cells) were lysed in RIP lysis buffer and then precipitated by antibody against Argonaute 2 (Ago2), with immunoglobulin G (IgG) as control. The final precipitates were eluted and subjected to qRT-PCR. The experiment was performed in triplicate.
Statistical analysis
Quantitative data was shown as the mean ± standard deviation (SD). Differences among multiple groups or between two groups were respectively analyzed by means of the one-way/two-way analysis of variance (ANOVA) or Student’s t-test. SPSS 20.0 software (Chicago, IL, USA) was utilized for analyzing statistical data. Each independent experiment performed in primary cell lines and purchased endometriotic cell lines were repeated in triplicate. P value < 0.05 was considered statistically significant. When the results had a remarkable difference between the experimental group and the NC group, or among different experimental groups, the bars were marked with asterisks. If P value was less than 0.05, one asterisk was marked. If P value was less than 0.01, two asterisks were marked accordingly.
Results
IGF2 knockdown restricts the proliferative, migratory, and invasive capabilities of primary NESC and EESC
First, we performed a series of assays in primary NESC and EESC. Based on the collected data of qRT-PCR, IGF2 was highly expressed in EESC (Fig. 1A). The knockdown efficacy of sh-IGF2#1/2 was proved to be high through qRT-PCR assay (Fig. 1B). CCK-8 was performed, and the results indicated that cell viability was restrained after IGF2 knockdown (Fig. 1C). As shown in wound healing assays, migration of NESC and EESC was impeded due to IGF2 knockdown (Fig. 1D). Data of transwell invasion assays reflected that cell invasion was hindered by IGF2 depletion (Fig. 1E). Moreover, collected western blots demonstrated that MMP2 and MMP9 protein levels were reduced by IGF2 downregulation (Fig. 1F). In short, IGF2 accelerated proliferation, migration, and invasion of primary NESC and EESC.
Fig. 1.
The influence of insulin like growth factor 2 (IGF2) knockdown on biological behaviors of primary normal and ectopic endometrial stromal cells was examined by CCK-8, wound healing, and transwell assays. A The expression of IGF2 in primary normal endometrial stromal cells (NESC) and primary ectopic endometrial stromal cells (EESC) was tested by qRT-PCR. B The knockdown efficiency of sh-IGF2#1/2 was examined by qRT-PCR in primary NESC and EESC. (sh-IGF2#1 and sh-IGF2#2 were two constructed plasmids containing different sequences for IGF2 knockdown, while sh-NC served as negative control.) C–E CCK-8, wound healing, and transwell assays were performed to evaluate the proliferation, migration, and invasion capabilities of primary NESC and EESC cells. F The levels of invasion-related proteins upon IGF2 downregulation in primary NESC and EESC were detected through western blot. *P < 0.05, **P < 0.01
IGF2 knockdown inhibits the proliferative, migratory, and invasive capabilities of transformed endometriotic cells
In this section, we tried to figure out the role of IGF2 in endometriotic cells. Expression of IGF2 in transformed endometriotic cells (EEC145T, hEM15A, hEM5B2, and 12Z) and control hEEC cells was first examined. As shown in qRT-PCR assay, IGF2 expression in endometriotic cells was higher compared to control cells (Fig. S1A). Since hEM15A and hEM5B2 cells had the most abundant expression of IGF2, they were used for the following assays. Next, the knockdown of IGF2 by sh-IGF2#1/2 was verified to be efficient (Fig. S1B). Furthermore, functional assays were performed to explore the influence of IGF2 depletion on hEM15A and hEM5B2 cell malignant behaviors. CCK-8 assay revealed that the OD value was decreased in cells transfected with sh-IGF2#1/2, suggesting cell viability was inhibited by IGF2 depletion (Fig. S1C). Moreover, EdU assay indicated that cell proliferation could be repressed by IGF2 knockdown since we observed that the ratio of EdU positive cells was significantly reduced after IGF2 was silenced (Fig. S1D). In addition, results of colony formation assays demonstrated that the number of cell colonies obviously dropped in sh-IGF2 groups, which further proved that cell proliferation could be repressed by IGF2 knockdown (Fig. S1E).
As displayed in wound healing assay, wound closure was slower in sh-IGF2#1/2 groups, suggesting the cell migration was inhibited by IGF2 downregulation (Fig. S2A). In addition, transwell assays were carried out to further explore the function of IGF2 on cell migration and invasion. It was shown in Fig. S2B and C that depletion of IGF2 caused a decrease in the number of migrated and invaded cells. Furthermore, invasion-associated protein levels (MMP2 and MMP9) were evaluated in western blot assay. The results demonstrated that MMP2 and MMP9 protein level was diminished by IGF2 knockdown (Fig. S2D). Taken together, IGF2 depletion restrained proliferation, migration, and invasion of transformed endometriotic cells.
Fig. 2.
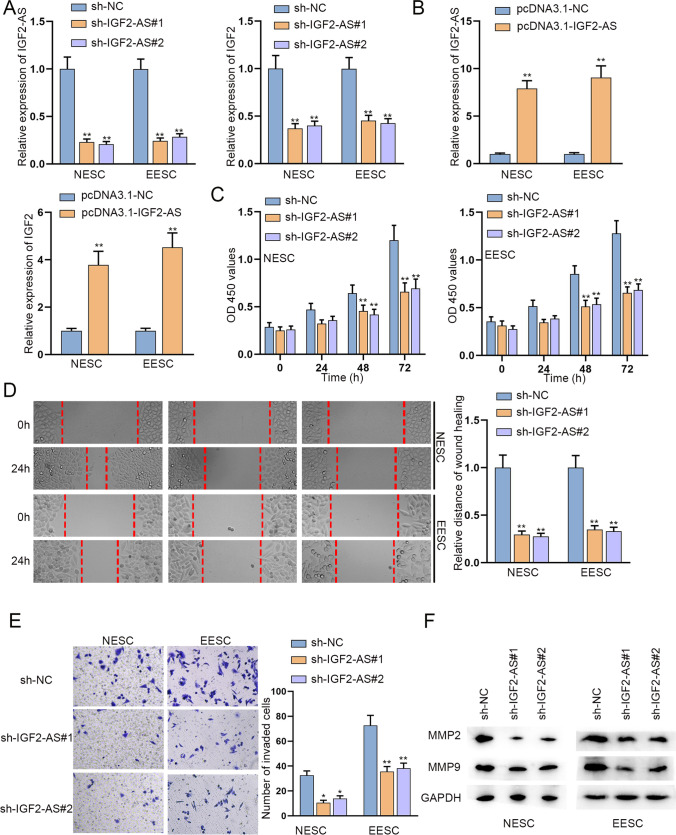
The influence of insulin like growth factor 2 antisense RNA (IGF2-AS) depletion on biological behaviors of primary NESC and EESC was examined by CCK-8, wound healing, and transwell assays. A IGF2-AS expression in primary NESC and EESC was quantified by qRT-PCR and the expression of IGF2 was detected before and after IGF2-AS depletion. B The overexpression efficiency of pcDNA3.1-IGF2-AS was examined by qRT-PCR in primary NESC and EESC. C–E Proliferation, migration, and invasion abilities of primary NESC and EESC in response to IGF2-AS depletion were confirmed via CCK-8, wound healing, and transwell assays. F Invasion-associated protein levels upon IGF2-AS deficiency were measured with the help of western blot. *P < 0.05, **P < 0.01
IGF2-AS positively regulates IGF2 and IGF2-AS knockdown suppresses proliferation, migration, and invasion of primary NESC and EESC
Since the nearby antisense RNAs of mRNAs can exert regulatory effects on mRNAs [24], we wondered if IGF2-AS (insulin-like growth factor 2 antisense RNA) could regulate IGF2 to further influence the malignant processes of endometriotic cells. Firstly, it was found that knockdown efficiency of sh-IGF2-AS#1/2 was high and IGF2-AS depletion diminished IGF2 expression in primary NESC and EESC (Fig. 2A). Meanwhile, the successful transfection of pcDNA3.1-IGF2-AS led to the contrary results (Fig. 2B). Furthermore, the data of CCK-8 indicated that IGF2-AS downregulation diminished primary NESC and EESC viability (Fig. 2C). Subsequently, wound healing assays were conducted in primary NESC and EESC. As shown in Fig. 2D, the wound closure was slower after IGF2-AS knockdown. In transwell assays, we could notice that the invasion ability of primary NESC and EESC was weakened due to IGF2-AS knockdown (Fig. 2E). Moreover, the collected western blots demonstrated that IGF2-AS downregulation restrained cell migratory and invasive abilities (Fig. 2F). Thus, we concluded that IGF2-AS upregulated IGF2 and IGF2-AS downregulation restricted the malignant behaviors of primary NESC and EESC.
IGF2-AS positively regulates IGF2 and IGF2-AS knockdown hampers proliferation, migration, and invasion of transformed endometriotic cells
In this part, we firstly downregulated IGF2-AS in hEM15A and hEM5B2 cells and the knockdown efficiency was verified by qRT-PCR. Then, we found that knockdown of IGF2-AS significantly reduced the expression of IGF2 (Fig. S3A). Similarly, IGF2-AS upregulation enhanced IGF2 expression (Fig. S3B). Next, loss-of-function assays were conducted to examine the function of IGF2-AS in hEM15A and hEM5B2 cells. As shown in CCK-8 assay, IGF2-AS depletion hampered cell viability (Fig. S3C). EdU assay results suggested that cell proliferation was repressed by IGF2-AS depletion since the number of EdU positive cells declined in sh-IGF2-AS-transfected cells (Fig. S3D). Moreover, colony formation assays indicated that the number of colonies was reduced by down-regulation of IGF2-AS (Fig. S3E). In wound healing assays, we observed that the wound closure was slower in sh-IGF2-AS#1/2 groups (Fig. S4A). In addition, transwell assays indicated that the number of migrated or invaded cells significantly declined after IGF2-AS knockdown (Fig. S4B and C). Furthermore, it was revealed in western blot assays that the protein level of MMP2 and MMP9 was reduced after the transfection of sh-IGF2-AS#1/2 (Fig. S4D). To summarize, IGF2-AS positively regulates IGF2 and IGF2-AS depletion inhibits proliferation, migration, and invasion of transformed endometriotic cells.
MiR-370-3p binds with both IGF2-AS and IGF2 in transformed endometriotic cells
In this part, we explored the mechanism for IGF2-AS to regulate IGF2. The subcellular location of IGF2-AS was detected at first. According to the data from subcellular fraction assay, IGF2-AS was predominantly distributed in the cytoplasm of hEM15A and hEM5B2 cells (Fig. S5A). The cytoplasmic role of IGF2-AS indicated the post-transcriptional regulatory mechanism of IGF2-AS. CeRNA pattern is a typical post-transcriptional network that lncRNAs can elevate the expression of mRNAs through sponging miRNAs [25]. Hence, we wondered if IGF2-AS could compete with IGF2 to bind with certain miRNAs to form the ceRNA pattern in hEM15A and hEM5B2 cells. Based on starBase database [26] (screened by 6 cancer types in pan-Cancer), 10 miRNAs which bound to both IGF2-AS and IGF2 were identified (Fig. S5B). Then, RNA pull down assay was conducted to verify the binding of these 10 miRNAs and IGF2-AS. The results revealed that miR-491-5p and miR-370-3p were significantly pulled down by IGF2-AS (Fig. S5C). Furthermore, miR-370-3p and miR-491-5p expression was enhanced in cells and the overexpression efficiency was verified by qRT-PCR assay (Fig. S5D and E). Then we detected IGF2 expression in cells transfected with miR-491-5p mimics and found that IGF2 expression was almost unchanged after miR-491-5p overexpression (Fig. S5F). However, we detected IGF2 expression in cells transfected with miR-370-3p mimics and noticed that IGF2 expression significantly declined after miR-370-3p augment. Meanwhile, the protein level of IGF2 was also decreased by miR-370-3p overexpression (Fig. S6A). Thus, we selected miR-370-3p to conduct further assays. The binding sequences of IGF2-AS/IGF2 and miR-370-3p were displayed (Fig. S6B). Based on the following luciferase reporter assay, luciferase activity of IGF2-AS/IGF2-WT reduced when miR-370-3p was upregulated, while IGF2-AS/IGF2-MUT was not impacted (Fig. S6C). Furthermore, RIP assay results showed that IGF2-AS, IGF2, and miR-370-3p were largely enriched in anti-Ago2 (Fig. S6D). Based on the data, IGF2-AS could regulate IGF2 via binding with miR-370-3p in transformed endometriotic cells.
IGF2-AS exacerbates the malignant behaviors of endometriotic cells through binding with miR-370-3p and upregulating IGF2
After verifying the ceRNA mechanism in endometriotic cells, we performed rescue assays in hEM15A and hEM5B2 cells to explore the role of IGF2-AS/miR-370-3p/IGF2 axis in cell biological functions. It was revealed in CCK-8, EdU, and colony formation assays that the inhibiting impact of IGF2-AS depletion on cell proliferation was completely offset by miR-370-3p inhibitor or pcDNA3.1-IGF2 (Fig. S7A–C). Furthermore, wound healing, transwell, and western blot assays uncovered that miR-370-3p inhibition or IGF2 augment fully counteracted the suppressive effects of IGF2-AS knockdown on cell migration and invasion (Fig. S7D–G). To sum up, IGF2-AS bound with miR-370-3p to upregulate IGF2, finally strengthening proliferation, migration, and invasion abilities of endometriotic cells.
IGF2-AS regulates IGF2/miR-370-3p axis in primary NESC and EESC to promote cell malignant behaviors
We also tested the ceRNA mechanism in primary NESC and EESC. At first, data of qRT-PCR and western blot showed IGF2 expression dropped upon miR-370-3p overexpression (Fig. 3A). The results of luciferase reporter assay revealed that miR-370-3p could bind to IGF2-AS and IGF2 as the luciferase activity of IGF2-AS/IGF2-WT was weakened due to miR-370-3p augment (Fig. 3B–C). Moreover, we conducted RIP assay and further proved the binding affinity between miR-370-3p and IGF2-AS/IGF2 (Fig. 3D). Functional assays were then performed in primary NESC and EESC. Based on the results of CCK-8 assays, the inhibitory effects of IGF2-AS knockdown on cell proliferation were completely offset by miR-370-3p inhibition or IGF2 upregulation (Fig. 3E). In addition, the wound width and western blots displayed in Fig. 3F and G manifested IGF2-AS depletion restricted cell migration and invasion, while miR-370-3p inhibition or IGF2 augment fully reversed the results. In summary, IGF2-AS could promote proliferation, migration, and invasion of primary NESC and EESC by regulating miR-370-3p/IGF2 axis.
Fig. 3.
Luciferase reporter, RIP, and rescue experiments were performed to confirm the mechanism of IGF2-AS/miR-370-3p/IGF2 network. A Relative expression of IGF2 upon miR-370-3p overexpression in primary NESC and EESC was detected by qRT-PCR and western blot. B–C Luciferase activity of wild type and mutant IGF2-AS/IGF2 upon miR-370-3p overexpression was tested by luciferase reporter assays. D The correlation of IGF2-AS, miR-370-3p and IGF2 was proven by RIP assay. E–G Cell proliferation, migration, and invasion of NESC and EESC transfected with indicated plasmids (sh-NC, sh-IGF2-AS#1, sh-IGF2-AS#1 + miR-370-3p inhibitor, or sh-IGF2-AS#1 + pcDNA3.1-IGF2) were tested by CCK-8, wound healing, and western blot assays. **P < 0.01
IGF2-AS activates PI3K/AKT/mTOR signaling pathway through targeting miR-370-3p/IGF2 axis
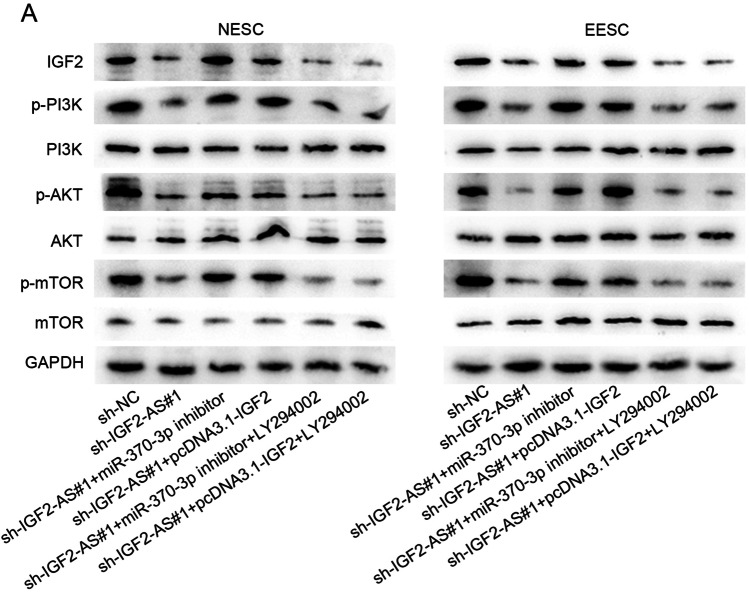
IGF2 has been reported to activate PI3K/AKT/mTOR signaling pathway in some diseases and cancers [27–29]. Thus, we wondered whether IGF2-AS could activate PI3K/AKT/mTOR signaling pathway through targeting miR-370-3p/IGF2 axis. LY294002 is known as a PI3K inhibitor [30]. By western blot assay, we discovered that the protein level of IGF2, phosphorylated (p)-PI3K, p-AKT, and p-mTOR was inhibited by IGF2-AS depletion in hEM15A and hEM5B2 cells, which was recovered totally by the co-transfection of miR-370-3p inhibitor or pcDNA3.1-IGF2, while the results were reversed by LY294002 (Fig. S8A). The similar changes of western blots could be observed in primary NESC and EESC (Fig. 4A). These findings reflected that IGF2-AS activated PI3K/AKT/mTOR signaling pathway through targeting miR-370-3p/IGF2 axis.
Fig. 4.
Western blot assays were conducted to measure the key protein levels of PI3K/AKT/mTOR signaling pathway. The primary NESC and EESC were divided into six groups with different treatments, including sh-NC, sh-IGF2-AS#1, sh-IGF2-AS#1 + miR-370-3p inhibitor, and sh-IGF2-AS#1 + pcDNA3.1-IGF2, sh-IGF2-AS#1 + miR-370-3p inhibitor + LY294002 and sh-IGF2-AS#1 + pcDNA3.1-IGF2 + LY294002. A The protein levels of IGF2, p-PI3K, PI3K, p-AKT, AKT, p-mTOR, and mTOR were tested by western blot assays under indicated conditions
Discussion
Endometriosis has similar characteristics of tumors in cell proliferation, distant migration, and invasion [31]. The main symptoms of endometriosis are dysmenorrhea and intercourse pain [32]. The morbidity of endometriosis presents a rising trend [33]. The current study focused on the role and mechanisms of IGF2. We firstly verified that IGF2 expressed at a higher level in endometriotic cells relative to the normal cells. Then, functional assays revealed that IGF2 exerted the pro-proliferation, pro-migration, and pro-invasion effects on endometriotic cells.
Previous researchers have demonstrated the regulatory network between mRNA and their nearby lncRNAs in human diseases. For instance, it is suggested that FOXD3-AS1 might promote glioma cell proliferative, migratory, and invasive abilities via upregulating FOXD3 [34]. TPT1-AS1 induces tumor growth and metastasis of epithelial ovarian cancer through elevating TPT1 expression [24]. Herein, we found that IGF2-AS positively regulated IGF2 expression. IGF2-AS has been reported to inhibit apoptosis and neuronal loss in damaged mouse neural stem cell derived neurons [35]. Role of IGF2-AS in endometriosis has not been studied before. Our study firstly uncovered that IGF2-AS contributed to proliferation, invasion, and migration of endometriotic cells.
Subsequently, how IGF2-AS modulated IGF2 expression was in exploration. SinceAs reported, the subcellular location of lncRNAs determines how they play the regulatory roles [13], so we detected subcellular location of IGF2-AS. The result showed that IGF2-AS was a cytoplasmic lncRNA, indicating the ceRNA potential of IGF2-AS. Based on bioinformatics analysis and experimental results, IGF2-AS served as the sponge of miR-370-3p to elevate IGF2 expression. Hu Z et al. have pointed out that miR-370-3p suppresses proliferation of endometriotic cells [36]. Additionally, miR-370-3p expression is increased in high responders to resistance training in breast cancer survivors [37]. MiR-370-3p along with 5 other miRNAs is differentially expressed in gestational trophoblastic neoplasia vs. complete hydatidiform mole [38]. Moreover, lncRNA H19 prompts TGF-β-induced EMT via functioning as an endogenous sponge of miR-370-3p in ovarian cancer cells [39]. The results of our study demonstrated that IGF2-AS regulated miR-370-3p/IGF2 axis to promote the malignant processes in endometriotic cells. The ceRNA network in endometriosis has been verified in multiple reports. Liu S et al. have pointed out that H19 upregulates ITGB3 through modulating miR-124-3p to promote ectopic endometrial cell proliferation and invasion [40]. Qiu J et al. have revealed that H19 relieves endometriosis through regulating miR-342-3p/IER3 axis [41]. Furthermore, IGF2 has been reported to activate PI3K/AKT/mTOR signaling pathway in some diseases and cancers [27–29]. Similarly, we also proved that IGF2-AS could activate PI3K/AKT/mTOR signaling pathway through miR-370-3p/IGF2 axis.
In summary, our study uncovered a novel ceRNA mechanism of IGF2-AS/miR-370-3p/IGF2 in endometriotic cells. IGF2-AS positively regulated IGF2 expression via competitively binding with miR-370-3p to actuate PI3K/AKT/mTOR signaling pathway, thus facilitating proliferative, migratory, and invasive capabilities of endometriotic cells. This study verified the crucial role of IGF2-AS in endometriosis, which might contribute to endometriosis diagnosis or treatment.
Supplementary Information
Acknowledgements
We appreciate the support of all lab members.
Abbreviations
- lncRNA
Long non-coding RNA
- IGF2-AS
Insulin like growth factor 2 antisense RNA
- ceRNA
Competing endogenous RNA
- miRNA
MicroRNA
- IGF2
Insulin like growth factor 2
- qRT-PCR
Quantitative real-time PCR
- CCK-8
Cell counting kit-8
- EdU
5-Ethynyl-2′-deoxyuridine
- RIP
RNA immunoprecipitation
- shRNA
Short hairpin RNA
- FBS
Fetal bovine serum
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- PVDF
Polyvinylidene fluoride
- DMEM
Dulbecco’s modified Eagle’s medium
- SD
Standard deviation
- NC
Negative control
- ANOVA
Analysis of variance
- ATCC
American Type Culture Collection
- OD
Optical density
Data availability
Research data are not shared.
Declarations
Ethics approval and consent to participate
All clinical samples were acquired from the First People’s Hospital of Wenling following the ethical and legal standards. All patients have signed a statement of consent.
Consent for publication
The paper has been proofread by all authors enrolled in this study, and they have permitted the submission and publication of this paper.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jingjing Feng, Email: fenxie45558@163.com.
Xiao Cheng, Email: chengtui1572500830@163.com.
References
- 1.Zondervan KT, Becker CM, Koga K, Missmer SA, Taylor RN, Viganò P. Endometriosis. Nat Rev Dis Primers. 2018;4(1):9. doi: 10.1038/s41572-018-0008-5. [DOI] [PubMed] [Google Scholar]
- 2.Laganà AS, Salmeri FM, Ban Frangež H, Ghezzi F, Vrtačnik-Bokal E, Granese R. Evaluation of M1 and M2 macrophages in ovarian endometriomas from women affected by endometriosis at different stages of the disease. Gynecol Endocrinol. 2020;36(5):441–444. doi: 10.1080/09513590.2019.1683821. [DOI] [PubMed] [Google Scholar]
- 3.Bulun SE, Yilmaz BD, Sison C, Miyazaki K, Bernardi L, Liu S, et al. Endometriosis. Endocr Rev. 2019;40(4):1048–1079. doi: 10.1210/er.2018-00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laganà AS, Salmeri FM, Vitale SG, Triolo O, Götte M. Stem cell trafficking during endometriosis: may epigenetics play a pivotal role? Reproductive sciences (Thousand Oaks, Calif) 2018;25(7):978–979. doi: 10.1177/1933719116687661. [DOI] [PubMed] [Google Scholar]
- 5.Giudice LC, Kao LC. Endometriosis. Lancet (London, England) 2004;364(9447):1789–1799. doi: 10.1016/s0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 6.Benagiano G, Guo SW, Puttemans P, Gordts S, Brosens I. Progress in the diagnosis and management of adolescent endometriosis: an opinion. Reprod Biomed Online. 2018;36(1):102–114. doi: 10.1016/j.rbmo.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Vercellini P, Vigano P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–275. doi: 10.1038/nrendo.2013.255. [DOI] [PubMed] [Google Scholar]
- 8.Czyzyk A, Podfigurna A, Szeliga A, Meczekalski B. Update on endometriosis pathogenesis. Minerva Ginecol. 2017;69(5):447–61. doi: 10.23736/s0026-4784.17.04048-5. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi H. Imprinting genes associated with endometriosis. EXCLI J. 2014;13:252–264. [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Park JH, Ku SY, Kim SH, Choi YM, Kim JG. Association between endometriosis and polymorphisms in insulin-like growth factors (IGFs) and IGF-I receptor genes in Korean women. Eur J Obstet Gynecol Reprod Biol. 2011;156(1):87–90. doi: 10.1016/j.ejogrb.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–33. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22(12):5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing C, Sun SG, Yue ZQ, Bai F. Role of lncRNA LUCAT1 in cancer. Biomed Pharmacother. 2021;134:111158. doi: 10.1016/j.biopha.2020.111158. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Xing Q, Feng T, He M, Yu W, Chen H. SNP rs710886 A>G in long noncoding RNA PCAT1 is associated with the risk of endometriosis by modulating expression of multiple stemness-related genes via microRNA-145 signaling pathway. J Cell Biochem. 2019 doi: 10.1002/jcb.29406. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Chen LH, Zhang B, Zheng QM. The modulation of endometriosis by lncRNA MALAT1 via NF-kappaB/iNOS. Eur Rev Med Pharmacol Sci. 2019;23(10):4073–80. doi: 10.26355/eurrev_201905_17908. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Wu W, Ye X, Ma R, Luo J, Zhu H, et al. Aberrant expression of CHL1 gene and long non-coding RNA CHL1-AS1, CHL1-AS2 in ovarian endometriosis. Eur J Obstet Gynecol Reprod Biol. 2019;236:177–182. doi: 10.1016/j.ejogrb.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Sha L, Huang L, Luo X, Bao J, Gao L, Pan Q, et al. Long non-coding RNA LINC00261 inhibits cell growth and migration in endometriosis. J Obstet Gynaecol Res. 2017;43(10):1563–1569. doi: 10.1111/jog.13427. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Wang Y, Chen P, Ma Y, Wang S, Tian Y et al. AC002454.1 and CDK6 synergistically promote endometrial cell migration and invasion in endometriosis. Reproduction (Cambridge, England). 2019;157(6):535–43. 10.1530/rep-19-0005. [DOI] [PubMed]
- 19.Qiu JJ, Lin XJ, Zheng TT, Tang XY, Zhang Y, Hua KQ. The exosomal long noncoding RNA aHIF is upregulated in serum from patients with endometriosis and promotes angiogenesis in endometriosis. Reproductive sciences (Thousand Oaks, Calif). 2019;26(12):1590–1602. 10.1177/1933719119831775. [DOI] [PubMed]
- 20.Xu Z, Zhang L, Yu Q, Zhang Y, Yan L, Chen ZJ. The estrogen-regulated lncRNA H19/miR-216a-5p axis alters stromal cell invasion and migration via ACTA2 in endometriosis. Mol Hum Reprod. 2019;25(9):550–561. doi: 10.1093/molehr/gaz040. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Zha J, Zuo M, Yan Q, Song H. Long noncoding RNA CHL1-AS1 promotes cell proliferation and migration by sponging miR-6076 to regulate CHL1 expression in endometrial cancer. J Cell Biochem. 2019 doi: 10.1002/jcb.29486. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Huang T, Sun X, Wang Y. Identification of a potential prognostic lncRNA-miRNA-mRNA signature in endometrial cancer based on the competing endogenous RNA network. J Cell Biochem. 2019;120(11):18845–18853. doi: 10.1002/jcb.29200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou CF, Liu MJ, Wang W, Wu S, Huang YX, Chen GB, et al. miR-205-5p inhibits human endometriosis progression by targeting ANGPT2 in endometrial stromal cells. Stem Cell Res Ther. 2019;10(1):287. doi: 10.1186/s13287-019-1388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Gao H, Li X, Zhu Y, Peng S, Yu J, et al. LncRNA TPT1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Sci. 2019;110(5):1587–1598. doi: 10.1111/cas.14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, et al. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi: 10.1186/s12935-019-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian B, Zhao Y, Liang T, Ye X, Li Z, Yan D, et al. Curcumin inhibits urothelial tumor development by suppressing IGF2 and IGF2-mediated PI3K/AKT/mTOR signaling pathway. J Drug Target. 2017;25(7):626–636. doi: 10.1080/1061186x.2017.1306535. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2019;38(1):497. doi: 10.1186/s13046-019-1470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu Q, Wang L, Yu F, Gao H, Lei T, Li P, et al. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther. 2015;16(4):623–633. doi: 10.1080/15384047.2015.1019185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Kuramitsu Y, Baron B, Kitagawa T, Tokuda K, Akada J, et al. PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. Int J Oncol. 2017;50(2):606–612. doi: 10.3892/ijo.2016.3804. [DOI] [PubMed] [Google Scholar]
- 31.Angioni S. New insights on endometriosis. Minerva Ginecol. 2017;69(5):438–9. doi: 10.23736/s0026-4784.17.04089-8. [DOI] [PubMed] [Google Scholar]
- 32.Borghese B, Santulli P, Marcellin L, Chapron C. Definition, description, clinicopathological features, pathogenesis and natural history of endometriosis: CNGOF-HAS Endometriosis Guidelines. Gynecologie, Obstetrique, Fertilite & Senologie. 2018;46(3):156–167. doi: 10.1016/j.gofs.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Imesch P, Fink D. Endometriosis Update 2016. Praxis. 2016;105(5):253–257. doi: 10.1024/1661-8157/a002295. [DOI] [PubMed] [Google Scholar]
- 34.Chen ZH, Hu HK, Zhang CR, Lu CY, Bao Y, Cai Z, et al. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am J Transl Res. 2016;8(10):4106–4119. [PMC free article] [PubMed] [Google Scholar]
- 35.Song C, Song C, Chen K, Zhang X. Inhibition of long non-coding RNA IGF2AS protects apoptosis and neuronal loss in anesthetic-damaged mouse neural stem cell derived neurons. Biomed Pharmacother. 2017;85:218–24. doi: 10.1016/j.biopha.2016.10.094. [DOI] [PubMed] [Google Scholar]
- 36.Hu Z, Mamillapalli R, Taylor HS. Increased circulating miR-370-3p regulates steroidogenic factor 1 in endometriosis. Am J Physiol Endocrinol Metab. 2019;316(3):E373–E382. doi: 10.1152/ajpendo.00244.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagstrom AD, Denham J. microRNAs in high and low responders to resistance training in breast cancer survivors. Int J Sports Med. 2018;39(6):482–489. doi: 10.1055/a-0592-7691. [DOI] [PubMed] [Google Scholar]
- 38.Zhao JR, Cheng WW, Wang YX, Cai M, Wu WB, Zhang HJ. Identification of microRNA signature in the progression of gestational trophoblastic disease. Cell Death Dis. 2018;9(2):94. doi: 10.1038/s41419-017-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Huang Y, Deng X, Luo M, Wang X, Hu H, et al. Long noncoding RNA H19 promotes transforming growth factor-beta-induced epithelial-mesenchymal transition by acting as a competing endogenous RNA of miR-370-3p in ovarian cancer cells. Onco Targets Ther. 2018;11:427–440. doi: 10.2147/ott.S149908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Qiu J, Tang X, Cui H, Zhang Q, Yang Q. LncRNA-H19 regulates cell proliferation and invasion of ectopic endometrium by targeting ITGB3 via modulating miR-124-3p. Exp Cell Res. 2019;381(2):215–222. doi: 10.1016/j.yexcr.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Liu L, Zhong Y, Cai M, Gao J, Tan C, et al. LncRNA H19 over-expression inhibited Th17 cell differentiation to relieve endometriosis through miR-342-3p/IER3 pathway. Cell Biosci. 2019;9:84. doi: 10.1186/s13578-019-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data are not shared.