Abstract
The leap of retroviruses and coronaviruses from animal hosts to humans has led to two ongoing pandemics and tens of millions of deaths worldwide. Retrovirus and coronavirus nucleocapsid proteins have been studied extensively as potential drug targets due to their central roles in virus replication, among which is their capacity to bind their respective genomic RNAs for packaging into nascent virions. This review focuses on fundamental studies of these nucleocapsid proteins and how their intrinsic abilities to condense through liquid-liquid phase separation (LLPS) contribute to viral replication. Therapeutic targeting of these condensates and methodological advances are also described to address future questions on how phase separation contributes to viral replication.
Keywords: SARS-CoV-2, HIV-1, virus replication, nucleocapsid, SARS-CoV-2 N, HIV-1 NC, biomolecular condensates, BMC, liquid-liquid phase separation, LLPS
Pandemic viruses SARS-CoV-2 and HIV-1 encode nucleocapsid proteins that possess intrinsic disorder that program liquid-liquid phase separation and condensation during viral replication. Chau et al. review recent literature showing that this is critical for virus replication, including gene expression, assembly, and genome packaging, and evading host anti-viral innate responses to infection.
Introduction: Molecular and cellular aspects of protein and RNA condensation
Several membrane-bound organelles exist in the cell with well-defined functions, including the nucleus, endoplasmic reticulum (ER), and Golgi apparatus. However, recent investigations have refocused attention on the key biological functions that occur in membraneless organelles (MLOs) implicated in a growing number of cell functions from transcription to metabolism in both the nuclear and cytoplasmic compartments.1 MLOs, also known as biomolecular condensates (BMCs), include the nucleolus; Cajal bodies; nuclear speckles (nuclear subcompartments that contain pre-mRNA splicing factors); neuronal and RNA trafficking granules; stress granules; and processing bodies (P-bodies), among others.2 , 3 , 4
Believed to be fundamental to life and cellular organization,5 , 6 BMCs often form due to the presence of intrinsically disordered regions (IDRs) in proteins and multivalent macromolecular interactions between proteins and nucleic acids, leading to liquid de-mixing. Their formation mostly relies on intrinsically disordered proteins (IDPs), although multivalence and IDRs are also present in globular proteins.7 As one of the key mechanisms used in cells to control spatiotemporal organization, liquid-liquid phase separation (LLPS) relies on the coordinated condensation of proteins and nucleic acids into BMCs to concentrate select molecules while excluding others to compartmentalize, spatially organize, and coordinate cellular processes.1 While protein misfolding can form irreversible protein aggregates that exhibit solid-like properties, LLPS displays liquid-like qualities that allow for diffusion of molecular constituents within the condensate.8 Phase transitions are elicited via interactions between multivalent proteins and RNAs for normal cell function as well as pathological phase transitions, as discussed in a recent review.9
BMCs play a role in many cellular processes including chromatin organization,10 cell division,11 transcription initiation and elongation,12 mRNA translation,13 DNA damage repair,14 autophagy,15 innate immune signaling,16 and enzyme catalysis.17 These processes share a common feature: the need for multiple components to be concentrated to generate the close proximity required for function. Aberrant or dysregulated BMCs also arise due to pathological gene mutations, contributing to neurodegenerative diseases18 and to tumorigenesis.19 Finally, the ability of chemicals to permeate cellular BMCs influences their anti-cancer efficacy (e.g., cisplatin and tamoxifen).20
Condensation of proteins and RNAs through phase separation and its role in pandemic virus replication
Certain motifs within proteins contribute to the formation of BMCs: our meta-analyses of viral proteins as well as studies from other groups indicate that the presence of RNA-binding domains and zinc-coordinating domains drive condensation.13 , 21 Other folded domains in proteins such as RNA-binding motifs and protein-protein oligomerization domains contribute to BMC formation, and these folded domains may be required along with IDRs to mediate LLPS in viral and cellular complexes.1 Amino acid composition is equally important, with BMC formation arising from interactions driven by electrostatic, π-π, and hydrophobic forces, hydrogen bonds, and post-translational modifications such as phosphorylation and acetylation. In addition, the arrangement of specific sequence motifs contributes to BMCs by forming multiple weak interactions, which are described as “stickers and spacers” in polymer chemistry.22
Viruses, as obligate intracellular pathogens, utilize properties of BMCs for genome replication and interaction with host cell pathways.23 BMCs appear to be fundamental for certain viruses to assemble, such as rabies, measles, and vesicular stomatitis to produce “viral factories,” which are membraneless assemblies.24 , 25 , 26 , 27 Influenza A, Hendra, and hepatitis C viruses have been reported to generate liquid organelles for genome assembly and replication (reviewed in Etibor et al.,23 Nikolic et al.,25 Heinrich et al.,26 Alenquer et al.,28 and Wubben et al.29). Recent studies have begun to outline the importance of BMCs in the HIV-1 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication cycles (e.g., Monette et al.21 , 30 and Wang et al.31). In the case of HIV-1, BMCs appear to be involved in the entry and assembly stages of the viral replication cycle, with the primary determinant of phase separation being the nucleocapsid (NC) domain of the Gag structural protein. Similarly, the N (nucleocapsid) protein of SARS-CoV-2 forms BMCs in an RNA-dependent fashion.31, 32, 33, 34, 35, 36, 37
Despite observations that condensates contribute to the replication cycles of both HIV-1 and SARS-CoV-2, significant fundamental questions remain about the mechanisms underlying BMC formation by these two pandemic viruses and how these condensates contribute to efficient virus replication (Figure 1 ). Further understanding of the biophysical and chemical properties of viral BMCs will be essential to determine whether their disruption is a viable, effective antiviral strategy. This review will examine the available data for virus-induced phase separation in retroviruses and coronaviruses using HIV-1 and SARS-CoV-2 as exemplars, as well as the role condensates play in engaging host machineries for virus propagation.
Figure 1.
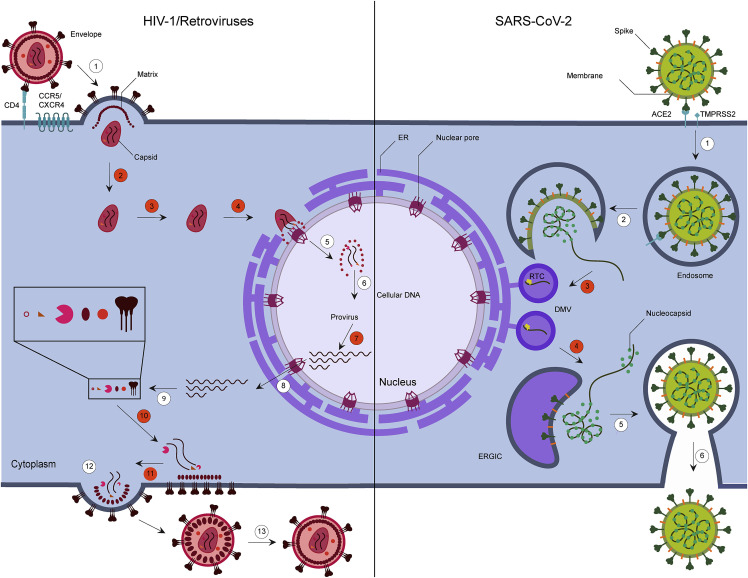
Viral replication cycles of HIV-1 and SARS-CoV-2 and implications of condensates generated by phase separation
Left: HIV-1 enters through the viral envelope glycoprotein binding with CD4 receptor and binding to chemokine coreceptor CCR5 or CXCR4, leading to fusion and entry of the virus into the cell (1). Following entry, the capsid traffics in the cytoplasm (2) toward the nuclear envelope, where uncoating likely begins (3), and then enters the nucleus via the nuclear pore (4). Capsid uncoating continues in the nucleus, where viral genomic RNA is reverse transcribed into double-stranded DNA and released into the nucleoplasm (5) for integration into the host genome (6). The integrated viral DNA is transcribed to generate a 9kb, unspliced viral genomic mRNA that can be spliced to generate mRNAs of 4 and 2kb sizes (7). The mRNAs are exported from the nucleus through the nuclear pore (8) and translated in the cytoplasm (9), and the unspliced mRNA is either translated and/or packaged with viral proteins (10) into progeny virus particles. Budding occurs (11) and is (12) followed by maturation (13) of the virus. Right: SARS-CoV-2 spike protein binds to the host receptor angiotensin-converting enzyme 2 (ACE2) and with host factor cell surface serine protease TMPRSS2 to promote viral entry (1). The RNA genome is released into the cytosol through uncoating, which is then translated into polyproteins pp1a and pp1ab and cleaved into individual non-structural proteins (nsps) (2). The nsps form the replication transcription complex (RTC), and viral genomic replication and subgenomic mRNA transcription commence in double-membrane vesicles (DMVs) (3). Subgenomic RNA translation results in structural and accessory proteins such as nucleocapsid (N), spike, membrane, and envelope proteins. The nucleocapsid protein is mostly distributed to the cytoplasm to package the genomic viral genome into hexon- or tetrahedron-shaped ribonucleoprotein complexes. The remaining proteins transit through the ER-to-Golgi intermediate compartment (ERGIC) (4), acquiring a lipid bilayer (5), and assembly and release of the virion occur (6). The stages of the viral cycle that are highlighted in orange numbered circles likely involve LLPS. In HIV-1, condensates could form at sites of maturation and assembly of the reverse transcription complexes in progeny virions, viral entry and uncoating, nuclear entry, completion of reverse transcription and integration, and later at sites of HIV-1 DNA transcription and RNA processing and following RNA nucleocytoplasmic trafficking during assembly and RNA packaging (10–11). SARS-CoV-2 N protein accumulates at RTCs (3) and forms phase-separated condensates with genomic RNA at the DMV and the ERGIC.
Why compare and contrast HIV-1 and SARS-CoV-2?
The most recent and ongoing pandemics of the 20th and 21st centuries are acquired immunodeficiency syndrome (AIDS), caused by HIV-1 and HIV-2, and COVID-19, initiated by a novel coronavirus SARS-CoV-2. The origin of HIV-1, the scourge of the early 1980s, traces back to approximately 1920.38 Simian immunodeficiency virus (SIV) was initially transmitted to humans, likely from non-human primates (chimpanzees and sooty mangabeys) via infected blood and body fluids during the butchering of bushmeat (Figure 2 ). It has been reported that numerous independent transmissions of the SIV from non-human primates to humans led to the establishment of a variety of separate lineages, some of which became established in humans as HIV-1 and HIV-2. Further diversification of HIV-1 genomic sequences was driven by the introduction of mutations arising from the low fidelity of RT in addition to recombination of viral sequences during replication. These factors led to the presence of a diverse population of viral sequences within individuals infected by HIV that then spread throughout the population, complicating the treatment and cure of HIV-1 infection due to drug-resistant mutants.39 In the case of SARS-CoV-2, numerous variants have arisen that have become increasingly capable of subverting immunity from vaccination and prior infection, making it imperative that novel, effective vaccines and antiviral drugs are developed.40 In addition, even though the replication cycles of HIV-1 and SARS-CoV-2 are substantially different, the process of viral genomic RNA packaging and virion release involves nucleocapsid proteins that have similar features. HIV-1 NC and SARS-CoV-2 N proteins undergo LLPS, form BMCs, facilitate viral RNA condensation, selectively package full-length viral genomes, and facilitate the release of virions from infected cells (Figure 3 ). Thus, comparing and contrasting the role of LLPS in the replication of these significant pandemic agents is informative and may lead to innovative therapies.
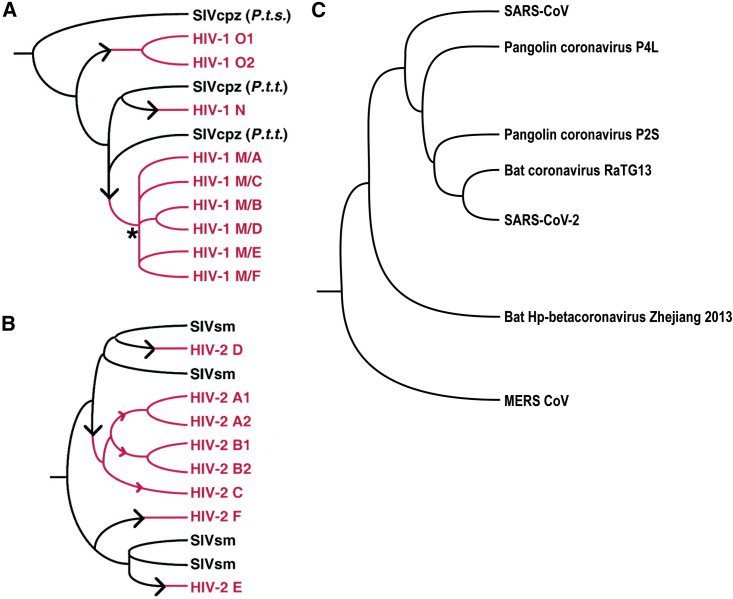
Figure 2.
Zoonotic transmission of SIV to humans and simplified evolution of betacoronaviruses
(A and B) Schematic trees illustrating multiple independent zoonotic transmissions of Simian immunodeficiency virus chimpanzee (SIVcpz) and SIV sootey mangabey (SIVsm) to humans. Branches in black indicate evolution of SIV within its natural hosts, black arrows indicate points of cross-species transmission, and branches in red indicate subsequent evolution within human hosts.
(A) SIVcpz and HIV-1. The three groups of HIV-1 (M, N, and O) are interspersed among SIVcpz strains from P.t. troglodytes (P.t.t.) and P.t. schweinfurthii (P.t.s.). The multiple subtypes of group M derive from a common ancestor indicated by a black asterisk.
(B) SIVsm and HIV-2. The six subtypes of HIV-2 (A–F) are interspersed among SIVsm lineages. Further characterization of SIVsm diversity may reveal that subtypes A, B, and C also arose through separate cross-species transmissions (indicated by the red arrows). For HIV-2, multiple isolates have been found only for subtypes A and B. Reproduced with permission and additional information can be found in Hahn et al.41
(C) Simplified phylogenetic tree showing the evolution of betacoronaviruses. SARS-CoV-1 and SARS-CoV-2 share their closest common ancestor in the sarbecovirus subgenus and have 79% sequence similarity.42 Middle Eastern respiratory syndrome (MERS)-CoV is from a different subgenus (merbecovirus) and shows a more distant relation with SARS-CoV-2, sharing 50% sequence similarity.43 The closest CoV relative of SARS-CoV-2 is bat CoV RaTG13, sharing 96% similarity, and the second closest is pangolin CoV, sharing 90% similarity.42 This suggests that bats and pangolins are intermediate hosts for SARS-CoV-2.
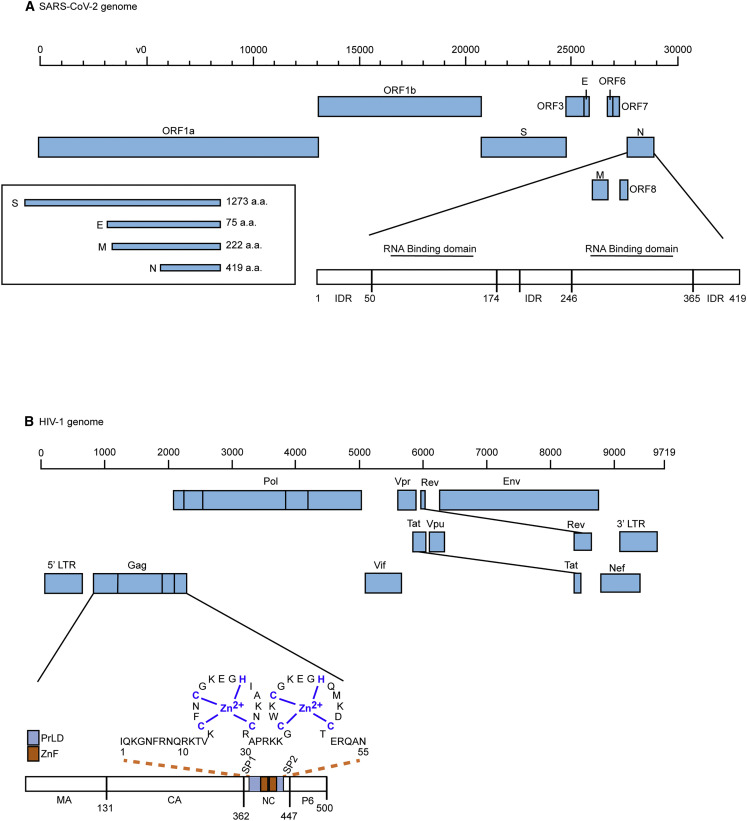
Figure 3.
Schematic diagrams of SARS-COV-2 and HIV-1 genomes, gene products, and nucleocapsid proteins
(A) Full-length viral genomic RNA (29,903 nt) for SARS-CoV-2 is illustrated including major subgenomic RNAs. ORF1a and ORF1b encode for nsps. In addition, the genome encodes four major structural proteins like spike protein (S), membrane protein (M), nucleocapsid protein (N), and envelope protein (E), as well as a number of accessory proteins. The amino acid number of each structural protein is indicated. Further, the SARS-CoV-2 N (nucleocapsid) protein domain organizations shows the RNA-binding domains in the N-terminal domain (NTD; aa 45–181); C-terminal domain (CTD; aa 248–365); dimerization domain, intrinsically disordered region (IDR; aa 1–44, 182–247, and 366–422); serine-arginine (SR) motif; nuclear localization signal (NLS); and linker region (LKR; aa 182–247). The RNA-binding domain of the NTD is essential for the packaging of viral genomic RNA, while the dimerization domain of the CTD participates in the formation of viral RNPs.32,44,45,46 Figure adapted from McBride et al.44
(B) Full-length viral genomic RNA (9,719 nt) for HIV-1 is illustrated showing 5′ LTR, Gag, Pol, Env, Vif, Vpr, Vpu, Tat, Rev, Nef, and 3′ LTR. Gag’s associated matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains are indicated. Coordinating zinc fingers are shown in the NC domain. SIV and all retroviral Gag proteins harbor one or two zinc coordinating fingers in their respective NC domains.21
SARS-CoV-2 is the cause of the devastating coronavirus infection that emerged in December 2019 (COVID-19) and rapidly spread around the world, resulting in over 5.8 million deaths worldwide as of summer 2022, although some estimates suggest much greater mortality47 (Figure 4 ). Lessons learned during the long-standing research programs to fight HIV-1 and influenza are being applied to the COVID-19 pandemic including eliciting neutralizing antibodies, vaccinology, epidemiology, and socioeconomic factors influencing infection.48 , 49 , 50 Since its discovery, SARS-CoV-2 has continued to evolve due to mutations arising during replication of the RNA genome, with selective pressure resulting in emergence of variants with increased capacity to escape immunity and spread.51 , 52 Similarities in the challenges of preventing and treating both pandemic RNA viruses HIV-1 and SARS-CoV-2 suggest that common features among these viruses could be exploited to treat these infections. Research toward SARS-CoV-2 therapies and prevention strategies will likely influence therapies, vaccines, and cures for HIV-1 and other pathogens with pandemic potential. Similar to HIV-1 and SARS-CoV-2, other viruses with pandemic potential are likely lurking in animal reservoirs and may emerge in the future by zoonotic transmission from animal to humans53 followed by adaptation to humans.
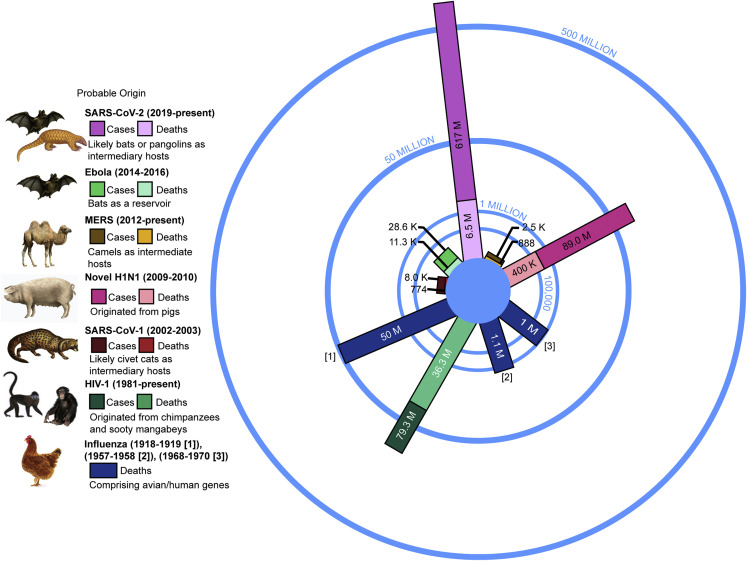
Figure 4.
Global cases and deaths of pandemics throughout history
Pandemic infections, deaths, and the probable origins of the viruses are shown for comparison as of June 2022.54 The bars display the cumulative cases, including the number of deaths. MERS, Middle East respiratory syndrome coronavirus; SARS-CoV-1/2, severe acute respiratory syndrome coronavirus 1/2; HIV-1, human immunodeficiency virus-type 1. Figure adapted from Montogomery and Macdonald55 with permission.
Potential roles of N protein condensates in SARS-CoV-2 replication
In response to the current pandemic, recent studies have focused on elucidating the function of the SARS-CoV-2 structural and non-structural proteins. Coronavirus virions encode four structural proteins that form the virion: nucleocapsid (N), envelope (E), membrane (M), and spike (S). The N protein is the most highly expressed protein in infected cells and is strongly immunogenic.44 , 56 , 57 Although coronaviruses were initially discovered over 50 years ago, many questions about fundamental aspects of the SARS-CoV-2 N protein in the replication cycle remain, which, once answered, will accelerate the development of additional effective antiviral therapies for the treatment of COVID-19 (Figure 1).
SARS-CoV-2 N protein consists of 419 amino acids with the following domain organization: N-terminal domain (NTD; amino acids 1–40); N-terminal RNA-binding domain (amino acids 41–173); serine-arginine (SR)-rich linker region, known as the LKR (amino acids 174–249); C-terminal domain that promotes dimerization (amino acids 250–364); and the C-terminal intrinsically disordered domain (IDR) (amino acids 365–419) (see Figure 3, Cubuk et al.,32 Peng et al.,45 Kang et al.,46 Sarkar et al.,58 and Yang et al.59). Large segments of the N protein are disordered, including the NTD, the SR-rich linker, and the C-terminal domain (CTD).32 Structural studies have revealed the folding of N domains using nuclear magnetic resonance (NMR), X-ray crystallography, and cryoelectron microscopy. Although the structure of the full-length protein has not yet been solved at high resolution, its structure has been predicted using computational methodology.45 , 46 , 58 , 60 , 61 , 62 , 63 Based on studies of other coronaviruses as well as SARS-CoV-2, the interaction of the NTD with the viral genomic RNA packaging sequence is essential for incorporation of genomic RNA into the virion, whereas the CTD appears to make protein-protein and protein-RNA interactions that form a multimerization interface involved in viral ribonucleoprotein (RNP) complex formation.60 , 64 , 65 , 66 , 67 , 68
In infected cells, the N proteins from SARS-CoV-2 and other CoVs localize to viral replication transcription complexes (RTCs) at membranes derived from the ER-Golgi intermediate complex (ERGIC) to form double-membrane vesicles (DMVs).35 , 69 , 70 , 71 , 72 , 73 , 74 , 75 These cellular membranes are remodeled upon infection by SARS-CoV-2 to produce DMVs that form RTCs or viral replication organelles.74 , 76 , 77 , 78 Viral RNA replication occurs inside the DMVs, and a pore has been observed in the membrane that could serve as a channel for extrusion of newly synthesized viral RNA into the cytoplasm.74 , 78 , 79 The SARS-CoV-2 N protein, which packages the viral genomic RNA, accumulates in cytoplasmic complexes56 , 71 , 80 , 81 along the membrane of the Golgi and other vesicles to initiate the process of virion assembly in conjunction with the M protein.37 , 74 The DMVs, Golgi membrane, and vesicular structures appear to be clustered near one another, presumably to coordinate RNA synthesis and packaging.74 , 82 Virus budding occurs at the ERGIC, which appear as single-membrane vesicles in cryoelectron tomograms,74 , 79 , 82 and virions accumulate primarily in large viral-containing vesicles (LVCVs). Whether formation of DMVs or other replication-related membrane compartments involve LLPS is an active area of investigation. Recently, it was shown that betacoronaviruses use the lysosomal pathway for release of virions.81 , 83 After virion release, the N protein binds to the 30 kb RNA genome to form viral RNPs, heterogeneous structures with N protein-RNA complexes tightly packed in a cylindrical or “bucket-like arrangement” when visualized using high-resolution imaging.76
Based on the presence of several IDRs in the SARS-CoV-2 N protein (Figure 3), its biophysical properties contributing to BMC formation have been investigated. Although the N protein was found to be dispersed in the cytoplasm when expressed alone in HeLa cells, it formed condensates upon arsenite treatment, a mimic of the cellular stress of viral infection.31 The N-terminal IDR (amino acids 1–39) was shown to be necessary for optimal condensate formation.31 Condensation of N-terminal IDR is dependent on G3BP1 and RNA.31 Similarly, in vitro, recombinant N protein does not form condensates alone but undergoes LLPS upon addition of RNA isolated from HeLa cells. The central linker IDR containing the SR-rich sequence also undergoes LLPS when bound to RNA, and these condensates slowly recover after photobleaching,37 suggesting that the individual IDRs within the N protein have distinct biophysical properties. Biophysical studies demonstrated that transient intramolecular interactions occur between the N-terminal NTD and RNA-binding domain and between the C-terminal IDR and the RNA-binding/dimerization domain, supporting the premise that multivalent protein interactions of the NTD, linker, and CTD underlie the ability of N to undergo LLPS when bound to RNA.32
The contribution of RNA to the formation of SARS-CoV-2 N condensates was studied in more detail by several research groups.31 , 32 , 33 , 34 , 35 , 36 , 37 , 84 Although non-viral RNA sequences can promote LLPS formation of N protein complexes, different genomic viral RNA segments appear to possess features that modulate the propensity of N to form droplets of varying sizes and with different biophysical properties.33 The authors of this study proposed that the RNA elements at the 5′ and 3′ ends of the genome promote LLPS, which may be important for the selection of full-length viral genomic RNA for packaging into virions. In contrast, other regions of the viral RNA, including the region involved in ribosome frameshifting and the predicted packaging signal, can dissolve N aggregates,33 suggesting that LLPS potentially serves a regulatory role to promote fluidity and prevent further maturation of the N-RNA complexes during viral RNA synthesis.
In cells, the N protein of beta coronaviruses localizes at RTCs, sites of viral genomic RNA replication.72 , 75 , 80 SARS-CoV-2 N appears to facilitate synthesis of subgenomic viral RNA by interacting with the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) complex, which is composed of non-structural proteins nsp12, nsp7, and nsp8.35 To determine whether LLPS of N plays a role in viral RNA synthesis, Savastano et al. used an in vitro assay to demonstrate that nsp12 formed cocondensates with N protein and RNA. Furthermore, the RdRp-RNA complex of nsp12, nsp7, and nsp8 formed phase separated droplets with the N protein and viral RNA. For betacoronavirus mouse hepatitis virus, the N protein was found to associate with RTCs dynamically, which is a characteristic feature of BMCs.75 Taken together, these studies suggest that N forms phase-separated condensates with viral genomic RNA along with the RdRp polymerase complex, potentially concentrating protein-RNA complexes that promote genome packaging and viral RNA replication while sequestering these viral components from detection by the host innate immune system. These studies support the possibility that phase separation of N and replication machinery play an important role in the RNA-dependent SARS-CoV-2 replication cycle. Importantly, data have emerged that SARS-CoV-2 N protein forms BMCs with the nsp3 protein,73 which is required for genome transcription and replication (reviewed in Lei et al.85) and is necessary to form DMVs in cells infected with a related betacoronavirus.86 SARS-CoV-2 Nsp3 contains an IDR in the N-terminal ubiquitin-like domain (Ubl1) that colocalizes in liquid droplets formed by N and RNA.73 In SARS-CoV-1, Nsp3 is implicated in the recruitment of N to sites of budding in the ERGIC and DMVs.72
Phosphorylation of N plays an important regulatory role by altering the properties of the BMCs formed.35 , 73 A major site of phosphorylation is the arginine-serine (RS)-rich sequence in the central linker region, which is modified by host kinases shortly post-infection.87 Phosphorylation of serine residues in N affect phase separation and diminish the association of N with the viral RdRp polymerase and nsp12.35 Studies by Carlson et al.73 determined that phosphorylation of N by kinases Cdk1 and GSK-3 alters the properties of the condensates formed: they were more dynamic and mobile compared with the unmodified protein. A recent preprint by the same group88 raised the intriguing possibility that the phosphorylated N protein is involved in viral RNA synthesis, whereas the unmodified N protein has stronger RNA binding and protein-protein interactions to promote viral RNP (vRNP) formation during assembly and within virions. Given these results, the role of N phosphorylation in LLPS and its influence on the interaction of N with viral and host factors may serve as a structural “switch” to segregate the two distinct functions of N during virus replication and RNA assembly. Using a phosphomimetic mutant in which the ten serines and threonines in the SR region of N were replaced with aspartate (10 Da mutant), this group determined that, in the presence of RNA, the 10 Da mutant generated condensates with a spherical droplet morphology (more liquid in nature), while the wild-type (unphosphorylated) N protein formed filamentous structures.88
In addition to vRNA, SARS-CoV-2 N protein also form condensates with the viral M protein.37 This study revealed that the M protein promotes LLPS of N, and interestingly, when N and M are mixed with RNA, mutually exclusive condensates are formed with N-RNA located in the center and N-M condensates arranged in a shell along the outer edge of the RNP core. Based on the arrangement of N-RNA and N-M phase-separated condensates, the authors posit that N coats RNA and condenses it into a vRNP first, and then M proteins located on the cytoplasmic face of the membrane assembly compartment recruit the RNPs into the virion.37 , 89 , 90 An alternative model posits that the properties of the N protein that drive LLPS formation function to compact the genomic RNA, and this viral condensate recruits the M protein through multivalent interactions with N for virus assembly.32 The N protein also partitions in vitro into LLPS droplets along with two cellular proteins known for forming condensates, hnRNP A2 and TDP-43, although the biological significance of these interactions during viral infection remains unclear.34
SARS-CoV-2 N colocalizes with G3BP1, a stress-granule-associated RNA-binding protein that forms phase-separated condensates, potentially to avoid degradation of the vRNA and triggering of the innate immune response.31 , 37 , 91 In cells, N reduces the formation of stress granules by recruiting G3BP1 and G3BP2, as well as cellular mRNAs.31 , 91 N could also interfere with the innate immune response by inhibiting signaling through the type 1 interferon-I (IFN-1) pathway.92 The N protein coimmunoprecipitates with the mitochondrial antiviral-signaling (MAVS) protein, a mediator of innate immunity against RNA viruses. MAVS forms aggregates in vivo and prion-like fibrils in vitro 93 and induces IRF3 and nuclear factor κB (NF-κB) to activate IFN-1. The N protein binds to and disrupts MAVS phase-separated droplets, interfering with MAVS-mediated IFN-1 signaling.92 This finding provides a novel mechanism involving LLPS perturbation by the N protein that may enhance the pathogenicity of SARS-CoV-2 through suppression of the innate immune response.
In conclusion, phase-separated condensates of the SARS-CoV-2 N protein appear to play a significant role in several stages of infection. The findings summarized above suggest that N phase-separated droplets are involved in vRNA replication, genome packaging, RNP condensation, and virion assembly via interactions with genomic RNA, M, vRNA replication enzymes, and nsp3, providing a potential linkage of N condensates to the DMV viral assembly compartment. The complex interplay of N with viral and cellular components will need to be sorted out in future studies; given that disruption of BMC formation could be an important therapeutic tool, further understanding the role of N protein condensates is a very worthwhile pursuit.92 , 94 , 95 The localization of N condensates appearing as aggregates have been identified by immunofluorescence in infected cells.81 The expression levels and localization of viral proteins, including N, were found to be temporal in nature, and the choice of detergent was a major determinant in identifying N in situ by immunofluorescence, for instance. Some consistency in localization between cell types was observed, but considering the divergent tropism of SARS-CoV-2, further work will be needed to assess general features of the nature of N protein localization and function in multiple cell types.96 The conservation of the N sequence makes targeting this structural protein advantageous, in contrast with the S protein, which is highly variable and has generated many variants with significant degrees of immune escape.
Potential roles of BMCs in HIV-1 replication
The etiologic retrovirus for AIDS is HIV-1. Approximately 32–42 million people have succumbed to AIDS and AIDS-related illnesses since the beginning of the epidemic, and ∼40 million people are living with this virus in 2022 (Figure 4). For those who can access treatment and respond well to potent anti-retroviral therapy, HIV-1 infection is considered a chronic and manageable disease.
The major structural proteins of HIV-1 are Gag and Gag/Pol, both precursor proteins encoded by the HIV-1 9 kb viral genomic RNA. Upon proteolytic cleavage of Gag and GagPol, mature proteins are released: matrix (MA), capsid (CA), nucleocapsid (NC), p6 and enzymes reverse transcriptase (RT), integrase (IN), and protease (PR) (Figures 1 and 3). During HIV-1 replication, Gag orchestrates virus assembly via the recruitment of the vRNA in concert with multiple host factors to initiate virion formation and release from the cell via budding at the plasma membrane. The importance of BMCs in HIV-1 replication remains incompletely characterized despite recent seminal observations. HIV-1 Gag condensate-like structures exhibit fluid-like behavior characterized by fluid movement, fusion, and fission.97 Moreover, many retroviral Gag domains possess intrinsically disordered domains juxtaposed with RNA-binding domains (RBDs).21 While intrinsically disordered domains may promote sequestration of protein partners, RBDs confer liquid-like behavior98 comparable to that observed for HIV-1 NC. An earlier systemic virtual analysis of HIV-1-encoded proteins revealed many viral components possessing intrinsic disorder.99
HIV-1 has also evolved mechanisms to block the assembly of BMCs, namely stress granules (SGs), that form in response to environmental stress, such as viral infection. Initial observations showed that HIV-1 imposed a blockade to SG assembly in many cell types.100 , 101 Subsequent detailed studies identified several virus-host interactions that mediate this blockade including interactions between the C-terminal CA domain of Gag and host factors, eukaryotic elongation factor 2 (eEF2), G3BP1, cyclophilin A, eIF4E,101 , 102 and the host RNA-binding protein Staufen1.103 HIV-1 blocks the assembly of TIAR-containing SGs under normal conditions, although in Staufen1 −/− CRISPR knockout cells, HIV-1 vRNA repositions and accumulates in TIAR+ SGs/BMCs during oxidative stress.103
Recent evidence that Gag’s C-terminal zinc-coordinating NC domain induces compositionally and functionally distinct BMCs indicates that the regulation of BMC assembly in HIV-1-expressing and -infected cells is complex.21 It has been demonstrated that HIV-1 NC has intrinsically disordered prion-like domains that contribute to zinc finger (ZnF)-dependent and zinc-chelation-sensitive condensation via LLPS regulated by cytosolic factors.21 NC-mediated condensate assemblies reposition the vRNA genome and other factors to promote virus production.21 Interestingly, while the HIV-1 CA domain mediates an SG blockade, overexpression of HIV-1 NC promotes SG assembly.104 More recently, Monette et al. demonstrated that HIV-1 condensates assemble following entry into cells with NC serving as the scaffolding condensate, whereas other members of the HIV-1 reverse transcription complex (RT, IN, and the vRNA genome, as well as the structural protein CA) defined themselves as client condensates, the condensation of which relied primarily on NC.21 , 30 Cumulatively, an equilibrium must exist between HIV-1-mediated BMC dissolution and assembly, suggesting that virus assembly relies on the continuous—yet overlapping—fluid states that would allow for consecutive virus replication steps, mediated by several viral factors. This concept, echoed by Lopez et al.,105 would allow for an active inclusion and exclusion of host and viral factors in viral condensates during the replication cycle. Although the biochemistry of Gag, NC, and Gag/Pol activities have been studied for 30–40 years, investigations have only recently revealed that HIV-1-engineered BMCs are important for pan-retrovirus replication including HIV-1 genomic RNA positioning during assembly and early events following entry.21 A recent study also provided evidence for a dynamic HIV-1 condensate (“RNP”) that is formed due to several types of NC-viral vRNA genome interactions.106 Interestingly, the viral PR was recruited to this condensate closer to the completion of virus budding at the plasma membrane, suggesting that condensation is important for correct viral maturation.
Recent work has also identified a role for phase separation through condensation early in HIV-1 infection during the integration of the proviral DNA into host cell chromatin. Di Nunzio’s group elegantly showed how IN cooperated with cleavage and polyadenylation specificity factor 6 (CPSF6), required for CA import into the nucleus early in infection,107 via condensation and phase separation.108 Additional work in 2021 led to observations that LLPS is also a key factor in the reactivation of HIV-1 proviral DNA transcription reactivation from latency, implicating components of both the host histone chaperone chromatin assembly factor 1 and polycomb repressive complex 1 complexes at the HIV-1 promoter.109 , 110 Thus, evolving evidence is demonstrating that HIV-1 regulates several gene regulatory circuits by eliciting condensation of viral and host factors, supporting the notion that compartmentalization of host processes is achieved via LLPS.
In summary, many functional similarities exist between the SARS-CoV-2 N protein and the HIV-1 Gag protein and its NC domain: (1) these proteins contain IDRs able to form BMCs and undergo LLPS through multivalent interactions; (2) they are nucleic acid-binding proteins and promote condensates containing nucleic acids, including vRNA; (3) they dimerize and form higher-order multimers; (4) they mediate the specific packaging of the viral genomic RNA; (5) they form RNPs in cells and in the virion, tightly packing the viral genome into a condensed core; and (6) in cells, they interact with SGs and form cocondensates with the SG protein G3BP1. One hypothesis to explain why SARS-CoV-2 N and HIV-1 Gag each interact with SG machinery posits that this association could serve to compartmentalize viral mRNAs undergoing translation from genomic RNAs destined for packaging into virions.
Targeting LLPS: A therapeutic strategy
The important contribution of LLPS to function offers the potential for development of novel therapeutic approaches for conditions involving these complexes such as cancer, neurodegenerative diseases, and viral infections.111 Two components of LLPS formation could be targeted: “drivers” and “controllers.” Drivers encompass elements essential for the initiation of LLPS such as IDRs in proteins and nucleic acids.112 Controllers are proteins that regulate and catalyze LLPS, including regulators of protein biosynthesis, folding, degradation, transport, and post-translational covalent modification (phosphorylation, methylation, acetylation, etc.).113
Neurodegenerative diseases can occur when protein misfolding causes the native proteins to transition into pathological states, sometimes manifesting at first as LLPS condensates.18 BMC dissolving agents may alleviate or slow progressive neurodegenerative disease and prevent irreversible plaque formation commonly associated with Alzheimer’s and other slowly progressive degenerative diseases. One therapeutic strategy is to use small molecules to modulate BMC formation. To date, compounds have targeted neurodegenerative diseases, like Alzheimer’s, amyotrophic lateral sclerosis (ALS), Nieman Pick’s disease, progressive supranuclear palsy, and frontotemporal dementia associated with the formation of aggregates involving amyloid-ꞵ or tau aggregates,114 , 115 , 116 but they could be expanded to include viruses and cancer by modulating SG assembly.117 However, the dynamic nature of the IDPs that contribute to LLPS may complicate their targeting for therapeutics.118 Alternatively, the role of nucleic acid (either RNA or DNA) in the formation and stability of BMCs suggests their potential use as therapeutics. One possible modality is aptamers, single-stranded oligonucleotides that fold into defined shapes capable of binding with high affinity and specificity to proteins/complexes to modulate their activity, a potential extensively explored in the context of prion and amyloid diseases.119 , 120 Aptamers are currently used as therapies or are undergoing clinical evaluation for macular degeneration (pegaptanib) and chronic hepatitis B virus (DCR-HBVS).121 In the context of HIV-1, aptamers have been extensively studied as decoys for Tat or Rev to prevent their interaction with TAR and RRE regulatory elements, respectively, within vRNA to inhibit HIV-1 gene expression and replication, although aptamers affecting the function of Gag, RT, and PR have also been developed.122 Aptamers against SARS-CoV-2 have been developed, but mostly for use in diagnostics, and are capable of achieving detection limits equivalent to most antibody-based detection systems.123 The therapeutic potential of aptamers for SARS-CoV-2 has largely focused on interfering with virus entry by blocking binding of the SARS-CoV-2 S trimer to the ACE2 receptor.124 Given the great specificity and affinity achievable with aptamers, their potential to alter condensate formation is worth further exploration if various issues limiting their use (metabolic instability, rapid renal clearance, intracellular delivery, and non-specific immune activation) can be overcome.125
Another approach to regulate LLPS formation is through modulation of the innate proteostasis machinery, like protein disaggregases, to prevent aberrant protein state diseases.126 Overexpression of chaperones and proteins disaggregases could counter the effect of misfolded proteins. Overall, understanding how molecules interact with BMCs and affect their stability, composition, dissolution, or rates of formation could lead to therapies for diseases that are currently considered incurable.
Given the extensive research that has highlighted the role of nucleocapsid protein LLPS in SARS-CoV-2 pathogenesis and HIV-1 replication,21 , 30 , 33 , 36 , 84 , 106 viral infections could also be termed “phase-separation-associated diseases”' as they not only produce BMCs but also interfere with host condensates.127 In the case of SARS-CoV-2 N protein, its ability to form BMCs may not only have a role in virus genome replication and assembly but also in modulation of the innate immune response to the infection.92 SARS-CoV-2 N condenses and facilitates the interaction of IKKꞵ and TAK1, leading to an aggressive inflammatory cytokine storm that causes airway damage.128 Consequently, altering the LLPS-forming capacity of nucleocapsid proteins may be a novel approach for developing antiviral drugs.
The properties of the targeted BMCs must be considered in any strategy to alter their function as the ability to partition into LLPS can also increase or decrease a drug’s therapeutic effect. For example, the effectiveness of cisplatin for the treatment of various cancers is increased 600-fold by its accumulation in aggregates where it platinates super-enhancer DNA.20 The efficacy of tamoxifen for the treatment of breast cancer is reduced upon overexpression of the gene activator MED1, which reduces the drug’s local concentration due to a MED1 increase in the volume of the transcription condensate targeted.20
Kinase inhibitors and RS proteins involved in N-mediated LLPS
Post-translational modification (PTM) of proteins can play an important role in modulating LLPS formation.113 Phosphorylation of serine/threonine residues on TIAR-2 and TAU promote LLPS, while phosphorylation of the prion-like domain (PrLD) of the fused-in-sarcoma (FUS) protein diminishes LLPS.129 , 130 , 131 Another example is the nuclear speckle, whose stability is severely altered by overexpression of kinases that modify the SR-rich domains present in several of its components.132 Consequently, identification of compounds able to increase or decrease the extent of viral protein PTMs represent another possible therapeutic approach.114
The significance of N phosphorylation to coronavirus replication became evident with the demonstration that depletion or inhibition of kinases modifying the viral N protein reduced replication. As detailed earlier, phosphorylation of a central RS-rich region of the SARS N protein, a motif conserved across coronaviruses, did not affect the protein’s ability to interact with RNA but altered the nature of the BMC formed as well as its ability to suppress host protein synthesis.87 In addition, hypophosphorylated variants of SARS-CoV-1 N protein have increased association with SGs. Overexpression of SRPK1 (a kinase able to phosphorylate the N protein RS-rich region) altered the interaction of wild-type N protein with SGs. Either overexpression of SRPK1 or its inhibition with SRPIN340 resulted in changes in N phosphorylation and a marked reduction in SARS-CoV-2 replication. Of note, SRPIN340 also inhibits replication of several unrelated viruses (hepatitis C virus, cytomegalovirus, Sindbis virus),133 , 134 suggesting that phosphorylation mediated by the affected kinase has important roles for several pathogens. In other studies, addition of GSK-3 inhibitors kenpaullone or LiCl also reduced SARS-CoV2/JMHV nucleocapsid phosphorylation and virus replication135 by altering the extent of phosphorylation of the N protein RS domain. Other compounds able to impair nucleocapsid-RNA binding in the context of other viruses were also examined for their effects on SARS-CoV-2 replication. (−)-Gallocatechin gallate (GCG), a polyphenol from green tea, disrupted N protein LLPS and inhibited virus replication.136 Two other small molecules, CVL218 and PJ34, increased LLPS of the N-vRNA-nsp12 complex, hypothesized to increase the accessibility of remdesivir to its viral target (i.e., nsp12).137 Finally, steroidal alkaloid cyclopamine compounds hardened respiratory syncytial virus condensates and blocked replication.94 Although multiple PTMs (myristylation, phosphorylation, ubiquitination, sumoylation, etc.) of HIV-1 Gag have been detected, there has been limited study of their impact on Gag’s formation of BMCs and the various stages of virus replication.138 In light of the observation that several human diseases depend on condensation and phase separation or aberrant phase transitions, there is great promise for the treatment of human pathologies involving phase separation or biomolecular condensation.
Emerging methods to evaluate BMCs
Investigators in the field have emphasized the need for rigorous approaches to identify the mechanisms driving phase separation of BMC; to develop quantitative methods to measure dynamics of BMC formation and dissolution; to understand the properties of amino acids and nucleic acids that facilitate BMCs; and to attempt to unequivocally demonstrate fluid-like nature of these complexes both in vitro and in living cells.8 There is general agreement that the following four characteristics define a phase-contrasted BMCs: the condensates can be visualized using phase-contrast microscopy; they undergo fusion and fission, demonstrating liquid-like properties; they exchange rapidly with the molecules in the surrounding milieu with a large mobile fraction and short half-time of recovery; and they are dissolved by phase-disrupting compounds such as 1,6-hexanediol, with some limitations (e.g., Düster et al.139). The Brangwynne laboratory has developed photo-activated techniques to elucidate the biophysical nature of interactions that drive the formation of BMCs in living cells, including so-called “optoDroplets”140 and Core scaffolds to promote droplets, termed “Corelets.”141 These innovative studies allow sensitive examination of light-induced condensate formation driven by IDRs or other motifs to examine the increased valency required to assemble phase-separated BMCs. Fluorescence recovery after photobleaching (FRAP) remains a useful method to examine the fluidity and macromolecule exchange dynamics of BMCs, although concerns have arisen, including the validity of comparing FRAP data from different proteins.142 Fluorescence correlative spectroscopy (FCS) and fluorescence lifetime imaging (FLIM) are additional techniques that quantitatively assess biophysical properties of condensates and their interface with the environment.8 , 143 FCS was used recently to evaluate a causal association of a SARS-CoV-2 N/α-synuclein interaction in Parkinson’s disease.144 The application of these techniques to the study of pandemic HIV-1 and SARS-CoV-2 BMCs will enhance our understanding of the functional role of phase-separated BMCs in viral replication.
Perspectives on LLPS in human viral disease (as well as in other human disease)
While there are many remaining questions concerning the biological roles of BMCs that arise from phase separation, accumulating evidence supports the critical roles for BMCs in nuclear and cytoplasmic compartments, ranging from viral transcription to virus assembly (for recent reviews, please see Etibor et al.,23 Li et al.,145 and Wu et al.146). HIV-1 and SARS-CoV-2 encode proteins with intrinsically disordered domains that mediate condensation of proteins and vRNAs in relevant viral replication complexes. Clearly, preemptive targeting of aberrant BMCs in human viral and neurodegenerative diseases would represent additional strategies that will add to the treatment armamentarium.147 Such strategies would complement the successful vaccines for SARS-CoV-2 and a potential vaccine for HIV-1,148 would enhance immune responses to viral infections,149 or could complement the new antiviral drugs (PR inhibitor nirmatrelvir combined with ritonavir, as well as the ribonucleoside analogue molnupiravir150) from major pharmaceutical companies, permitting the “phasing out” of these two viral pandemics earlier rather than later in the 21st century.
Acknowledgments
The authors apologize to authors of publications that were not included in this review due to space constraints. This work was funded by grants from the Canadian Institutes of Health Research (FRN-162447 to A.J.M. and PJT-178165 to A.W.C.), the National Institutes of Health (R21DA053689 to L.J.P. and a subcontract to A.J.M.), and a COVID-19 Innovation Fund Award (to L.J.P.) from Penn State College of Medicine, Department of Medicine.
Author contributions
A.J.M. conceived of the content and organization of this review. B.-A.C. and V.C. contributed equally to the composition and preparation of this review. All authors drafted, prepared figures, revised, and approved the final version of this manuscript; A.W.C., L.J.P., and A.J.M. secured funding.
Declaration of interests
All authors do not have any conflicts of interest.
References
- 1.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 3.Gall J.G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- 4.Sawyer I.A., Bartek J., Dundr M. Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin. Cell Dev. Biol. 2019;90:94–103. doi: 10.1016/j.semcdb.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Alberti S. Phase separation in biology. Curr. Biol. 2017;27:R1097–R1102. doi: 10.1016/j.cub.2017.08.069. [DOI] [PubMed] [Google Scholar]
- 6.Keating C.D. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res. 2012;45:2114–2124. doi: 10.1021/ar200294y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teilum K., Olsen J.G., Kragelund B.B. On the specificity of protein-protein interactions in the context of disorder. Biochem. J. 2021;478:2035–2050. doi: 10.1042/BCJ20200828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathieu C., Pappu R.V., Taylor J.P. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stortz M., Pecci A., Presman D.M., Levi V. Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol. 2020;18:59. doi: 10.1186/s12915-020-00788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong J.Y., Torres J.Z. Phase separation in cell division. Mol. Cell. 2020;80:9–20. doi: 10.1016/j.molcel.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei M.T., Chang Y.C., Shimobayashi S.F., Shin Y., Strom A.R., Brangwynne C.P. Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 2020;22:1187–1196. doi: 10.1038/s41556-020-00578-6. [DOI] [PubMed] [Google Scholar]
- 13.Roden C., Gladfelter A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spegg V., Altmeyer M. Biomolecular condensates at sites of DNA damage: more than just a phase. DNA Repair. 2021;106:103179. doi: 10.1016/j.dnarep.2021.103179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda N.N., Wang Z., Zhang H. Liquid-liquid phase separation in autophagy. J. Cell Biol. 2020;219:e202004062. doi: 10.1083/jcb.202004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibble R.W., Depaix A., Kowalska J., Jemielity J., Gross J.D. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat. Chem. Biol. 2021;17:615–623. doi: 10.1038/s41589-021-00774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 19.Song M.S., Grabocka E. Stress granules in cancer. Rev. Physiol. Biochem. Pharmacol. 2020 doi: 10.1007/112_2020_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein I.A., Boija A., Afeyan L.K., Hawken S.W., Fan M., Dall'Agnese A., Oksuz O., Henninger J.E., Shrinivas K., Sabari B.R., et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monette A., Niu M., Chen L., Rao S., Gorelick R.J., Mouland A.J. Pan-retroviral nucleocapsid-mediated phase separation regulates genomic RNA positioning and trafficking. Cell Rep. 2020;31:107520. doi: 10.1016/j.celrep.2020.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J.M., Holehouse A.S., Pappu R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Etibor T.A., Yamauchi Y., Amorim M.J. Liquid biomolecular condensates and viral lifecycles: review and perspectives. Viruses. 2021;13:366. doi: 10.3390/v13030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su J.M., Wilson M.Z., Samuel C.E., Ma D. Formation and function of liquid-like viral factories in negative-sense single-stranded RNA virus infections. Viruses. 2021;13:126. doi: 10.3390/v13010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudrière-Gesbert C., Gaudin Y., Blondel D. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 2017;8:58. doi: 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich B.S., Maliga Z., Stein D.A., Hyman A.A., Whelan S.P.J. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio. 2018;9:02290-17. doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Su J.M., Samuel C.E., Ma D. Measles virus forms inclusion bodies with properties of liquid organelles. J. Virol. 2019;93:00948-19. doi: 10.1128/JVI.00948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alenquer M., Vale-Costa S., Etibor T.A., Ferreira F., Sousa A.L., Amorim M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 2019;10:1629. doi: 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wubben J.M., Atkinson S.C., Borg N.A. The role of protein disorder in nuclear transport and in its subversion by viruses. Cells. 2020;9:2654. doi: 10.3390/cells9122654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monette A., Niu M., Nijhoff Asser M., Gorelick R.J., Mouland A.J. Scaffolding viral protein NC nucleates phase separation of the HIV-1 biomolecular condensate. Cell Rep. 2022;40:111251. doi: 10.1016/j.celrep.2022.111251. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Shi C., Xu Q., Yin H. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation into stress granules through its N-terminal intrinsically disordered region. Cell Discov. 2021;7:5. doi: 10.1038/s41421-020-00240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A., et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80:1078–1091.e6. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39:e106478. doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang M., Li Y., Song J. ATP biphasically modulates LLPS of SARS-CoV-2 nucleocapsid protein and specifically binds its RNA-binding domain. Biochem. Biophys. Res. Commun. 2021;541:50–55. doi: 10.1016/j.bbrc.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., 3rd, Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faria N.R., Rambaut A., Suchard M.A., Baele G., Bedford T., Ward M.J., Tatem A.J., Sousa J.D., Arinaminpathy N., Pépin J., et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 2012;18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 40.DeGrace M.M., Ghedin E., Frieman M.B., Krammer F., Grifoni A., Alisoltani A., Alter G., Amara R.R., Baric R.S., Barouch D.H., et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature. 2022;605:640–652. doi: 10.1038/s41586-022-04690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn B.H., Shaw G.M., De Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Boon S.S., Wang M.H., Chan R.W.Y., Chan P.K.S. Genomic and evolutionary comparison between SARS-CoV-2 and other human coronaviruses. J. Virol. Methods. 2021;289:114032. doi: 10.1016/j.jviromet.2020.114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39:e105938. doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adam D. The effort to count the pandemic's global death toll. Nature. 2022;601:312–315. doi: 10.1038/d41586-022-00104-8. [DOI] [PubMed] [Google Scholar]
- 48.Mendoza P., Lorenzi J.C.C., Gaebler C. COVID-19 antibody development fueled by HIV-1 broadly neutralizing antibody research. Curr. Opin. HIV AIDS. 2021;16:25–35. doi: 10.1097/COH.0000000000000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castel A.D., Wilbourn B., Magnus M., Greenberg A.E. SARS-CoV-2 and HIV: epidemiology, treatment, and lessons learned from HIV. AIDS Rev. 2020;22:133–142. doi: 10.24875/AIDSRev.20000070. [DOI] [PubMed] [Google Scholar]
- 50.Vasan S., Pitisuttithum P. Vaccine development lessons between HIV and COVID-19. Lancet Infect. Dis. 2021;21:759–761. doi: 10.1016/S1473-3099(21)00274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Front. Immunol. 2021;12:742167. doi: 10.3389/fimmu.2021.742167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu J., Peng P., Cao X., Wu K., Chen J., Wang K., Tang N., Huang A.L. Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. Cell. Mol. Immunol. 2022;19:293–295. doi: 10.1038/s41423-021-00836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao F., Bailes E., Robertson D.L., Chen Y., Rodenburg C.M., Michael S.F., Cummins L.B., Arthur L.O., Peeters M., Shaw G.M., et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 54.WHO 2022. https://covid19.who.int WHO Dashboard.
- 55.Montgomery R.A., Macdonald D.W. COVID-19, health, conservation, and shared wellbeing: details matter. Trends Ecol. Evol. 2020;35:748–750. doi: 10.1016/j.tree.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong Y., Ulasli M., Schepers H., Mauthe M., V'Kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94:01925-19. doi: 10.1128/jvi.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S., Lin L., Wang H., Yin J., Ren Y., Zhao Z., Wen J., Zhou C., Zhang X., Li X., et al. The epitope study on the SARS-CoV nucleocapsid protein. Dev. Reprod. Biol. 2003;1:198–206. doi: 10.1016/s1672-0229(03)01025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar S., Runge B., Russell R.W., Movellan K.T., Calero D., Zeinalilathori S., Quinn C.M., Lu M., Calero G., Gronenborn A.M., Polenova T. Atomic-resolution structure of SARS-CoV-2 nucleocapsid protein N-terminal domain. J. Am. Chem. Soc. 2022;144:10543–10555. doi: 10.1021/jacs.2c03320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang M., He S., Chen X., Huang Z., Zhou Z., Zhou Z., Chen Q., Chen S., Kang S. Structural insight into the SARS-CoV-2 nucleocapsid protein C-terminal domain reveals a novel recognition mechanism for viral transcriptional regulatory sequences. Front. Chem. 2020;8:624765. doi: 10.3389/fchem.2020.624765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ribeiro-Filho H.V., Jara G.E., Batista F.A.H., Schleder G.R., Costa Tonoli C.C., Soprano A.S., Guimarães S.L., Borges A.C., Cassago A., Bajgelman M.C., et al. Structural dynamics of SARS-CoV-2 nucleocapsid protein induced by RNA binding. PLoS Comput. Biol. 2022;18:e1010121. doi: 10.1371/journal.pcbi.1010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casasanta M., Jonaid G.M., Kaylor L., Luqiu W., Solares M., Schroen M., Dearnaley W., Wilson J., Dukes M., Kelly D. Cryo-EM structural analysis of the SARS-CoV-2 Nucleocapsid protein. Microsc. Microanal. 2021;27:1378–1380. doi: 10.1017/S1431927621005134. [DOI] [Google Scholar]
- 62.Jia Z., Liu C., Chen Y., Jiang H., Wang Z., Yao J., Yang J., Zhu J., Zhang B., Yuchi Z. Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies. FEBS J. 2022;289:3813–3825. doi: 10.1111/febs.16239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye Q., Lu S., Corbett K.D. Structural basis for SARS-CoV-2 nucleocapsid protein recognition by single-domain antibodies. Front. Immunol. 2021;12:719037. doi: 10.3389/fimmu.2021.719037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masters P.S. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 1992;125:141–160. doi: 10.1007/bf01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 66.Surjit M., Liu B., Kumar P., Chow V.T.K., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.den Boon J.A., Diaz A., Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knoops K., Kikkert M., Worm S.H.E.v.d., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80:1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cortese M., Lee J.Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J.M., Masters P.S., de Haan C.A.M. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/jvi.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolff G., Bárcena M. Multiscale electron microscopy for the study of viral replication organelles. Viruses. 2021;13:197. doi: 10.3390/v13020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mendonça L., Howe A., Gilchrist J.B., Sheng Y., Sun D., Knight M.L., Zanetti-Domingues L.C., Bateman B., Krebs A.S., Chen L., et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021;12:4629. doi: 10.1038/s41467-021-24887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.V'Kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife. 2019;8:e42037. doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scherer K.M., Mascheroni L., Carnell G.W., Wunderlich L.C.S., Makarchuk S., Brockhoff M., Mela I., Fernandez-Villegas A., Barysevich M., Stewart H., et al. SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress. Sci. Adv. 2022;8:eabl4895. doi: 10.1126/sciadv.abl4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Haan C.A.M., Rottier P.J.M. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/s0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jack A., Ferro L.S., Trnka M.J., Wehri E., Nadgir A., Nguyenla X., Fox D., Costa K., Stanley S., Schaletzky J., Yildiz A. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. PLoS Biol. 2021;19:e3001425. doi: 10.1371/journal.pbio.3001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8:01658-17. doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carlson C.R., Adly A.N., Bi M., Cheng Y., Morgan D.O. Reconstitution of the SARS-CoV-2 ribonucleosome provides insights into genomic RNA packaging and regulation by phosphorylation. bioRxiv. 2022 doi: 10.1101/2022.05.23.493138. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dinesh D.C., Chalupska D., Silhan J., Koutna E., Nencka R., Veverka V., Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020;16:e1009100. doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khan A., Tahir Khan M., Saleem S., Junaid M., Ali A., Shujait Ali S., Khan M., Wei D.Q. Structural insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020;18:2174–2184. doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nabeel-Shah S., Lee H., Ahmed N., Burke G.L., Farhangmehr S., Ashraf K., Pu S., Braunschweig U., Zhong G., Wei H., et al. SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience. 2022;25:103562. doi: 10.1016/j.isci.2021.103562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S., Dai T., Qin Z., Pan T., Chu F., Lou L., Zhang L., Yang B., Huang H., Lu H., Zhou F. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021;23:718–732. doi: 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 93.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Risso-Ballester J., Galloux M., Cao J., Le Goffic R., Hontonnou F., Jobart-Malfait A., Desquesnes A., Sake S.M., Haid S., Du M., et al. A condensate-hardening drug blocks RSV replication in vivo. Nature. 2021;595:596–599. doi: 10.1038/s41586-021-03703-z. [DOI] [PubMed] [Google Scholar]
- 95.York A. Targeting viral liquid-liquid phase separation. Nat. Rev. Microbiol. 2021;19:550. doi: 10.1038/s41579-021-00608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cascarina S.M., Ross E.D. Phase separation by the SARS-CoV-2 nucleocapsid protein: consensus and open questions. J. Biol. Chem. 2022;298:101677. doi: 10.1016/j.jbc.2022.101677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milev M.P., Brown C.M., Mouland A.J. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gotor N.L., Armaos A., Calloni G., Torrent Burgas M., Vabulas R.M., De Groot N.S., Tartaglia G.G. RNA-binding and prion domains: the Yin and Yang of phase separation. Nucleic Acids Res. 2020;48:9491–9504. doi: 10.1093/nar/gkaa681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goh G.K.M., Dunker A.K., Uversky V.N. Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genom. 2008;9:S4. doi: 10.1186/1471-2164-9-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abrahamyan L.G., Chatel-Chaix L., Ajamian L., Milev M.P., Monette A., Clément J.F., Song R., Lehmann M., DesGroseillers L., Laughrea M., et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 101.Cinti A., Le Sage V., Ghanem M., Mouland A.J. HIV-1 gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. mBio. 2016;7:e00329. doi: 10.1128/mBio.00329-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valiente-Echeverría F., Melnychuk L., Vyboh K., Ajamian L., Gallouzi I.E., Bernard N., Mouland A.J. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat. Commun. 2014;5:4819. doi: 10.1038/ncomms5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rao S., Hassine S., Monette A., Amorim R., DesGroseillers L., Mouland A.J. HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles. RNA. 2019;25:727–736. doi: 10.1261/rna.069351.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rao S., Cinti A., Temzi A., Amorim R., You J.C., Mouland A.J. HIV-1 NC-induced stress granule assembly and translation arrest are inhibited by the dsRNA binding protein Staufen1. RNA. 2018;24:219–236. doi: 10.1261/rna.064618.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lopez N., Camporeale G., Salgueiro M., Borkosky S.S., Visentín A., Peralta-Martinez R., Loureiro M.E., de Prat-Gay G. Deconstructing virus condensation. PLoS Pathog. 2021;17:e1009926. doi: 10.1371/journal.ppat.1009926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lyonnais S., Sadiq S.K., Lorca-Oro C., Dufau L., Nieto-Marquez S., Escriba T., Gabrielli N., Tan X., Ouizougun-Oubari M., Okoronkwo J., et al. The HIV-1 nucleocapsid regulates its own condensation by phase-separated activity-enhancing sequestration of the viral protease during maturation. Viruses. 2021;13:2312. doi: 10.3390/v13112312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rensen E., Mueller F., Scoca V., Parmar J.J., Souque P., Zimmer C., Di Nunzio F. Clustering and reverse transcription of HIV-1 genomes in nuclear niches of macrophages. EMBO J. 2021;40:e105247. doi: 10.15252/embj.2020105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scoca V., Di Nunzio F. Membraneless organelles restructured and built by pandemic viruses: HIV-1 and SARS-CoV-2. J. Mol. Cell Biol. 2021;13:259–268. doi: 10.1093/jmcb/mjab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma X., Chen T., Peng Z., Wang Z., Liu J., Yang T., Wu L., Liu G., Zhou M., Tong M., et al. Histone chaperone CAF-1 promotes HIV-1 latency by leading the formation of phase-separated suppressive nuclear bodies. EMBO J. 2021;40:e106632. doi: 10.15252/embj.2020106632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu L., Pan T., Zhou M., Chen T., Wu S., Lv X., Liu J., Yu F., Guan Y., Liu B., et al. CBX4 contributes to HIV-1 latency by forming phase-separated nuclear bodies and SUMOylating EZH2. EMBO Rep. 2022;23:e53855. doi: 10.15252/embr.202153855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wheeler R.J. Therapeutics—how to treat phase separation-associated diseases. Emerg. Top. Life Sci. 2020;4:307–318. doi: 10.1042/etls20190176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malinovska L., Kroschwald S., Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Owen I., Shewmaker F. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int. J. Mol. Sci. 2019;20:5501. doi: 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Darling A.L., Shorter J. Combating deleterious phase transitions in neurodegenerative disease. Biochim. Biophys. Acta. Mol. Cell Res. 2021;1868:118984. doi: 10.1016/j.bbamcr.2021.118984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pradhan A., Mishra S., Surolia A., Panda D. C1 inhibits liquid-liquid phase separation and oligomerization of tau and protects neuroblastoma cells against toxic tau oligomers. ACS Chem. Neurosci. 2021;12:1989–2002. doi: 10.1021/acschemneuro.1c00098. [DOI] [PubMed] [Google Scholar]
- 116.Zbinden A., Pérez-Berlanga M., De Rossi P., Polymenidou M. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev. Cell. 2020;55:45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 117.Babinchak W.M., Dumm B.K., Venus S., Boyko S., Putnam A.A., Jankowsky E., Surewicz W.K. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat. Commun. 2020;11:5574. doi: 10.1038/s41467-020-19211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ruan H., Sun Q., Zhang W., Liu Y., Lai L. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov. Today. 2019;24:217–227. doi: 10.1016/j.drudis.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 119.Matos C.O., Passos Y.M., do Amaral M.J., Macedo B., Tempone M.H., Bezerra O.C.L., Moraes M.O., Almeida M.S., Weber G., Missailidis S., et al. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 2020;34:365–385. doi: 10.1096/fj.201901897R. [DOI] [PubMed] [Google Scholar]
- 120.Macedo B., Cordeiro Y. Unraveling prion protein interactions with aptamers and other PrP-binding nucleic acids. Int. J. Mol. Sci. 2017;18:1023. doi: 10.3390/ijms18051023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ni S., Zhuo Z., Pan Y., Yu Y., Li F., Liu J., Wang L., Wu X., Li D., Wan Y., et al. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mater. Interfaces. 2021;13:9500–9519. doi: 10.1021/acsami.0c05750. [DOI] [PubMed] [Google Scholar]
- 122.Sullenger B.A., Gallardo H.F., Ungers G.E., Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 123.Shola David M., Kanayeva D. Enzyme linked oligonucleotide assay for the sensitive detection of SARS-CoV-2 variants. Front. Cell. Infect. Microbiol. 2022;12:1017542. doi: 10.3389/fcimb.2022.1017542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y., Juhas M., Kwok C.K. Aptamers targeting SARS-COV-2: a promising tool to fight against COVID-19. Trends Biotechnol. 2022 doi: 10.1016/j.tibtech.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fare C.A.-O., Shorter J.A.-O. (Dis)Solving the problem of aberrant protein states. Dis. Model Mech. 2021;14:dmm048983. doi: 10.1242/dmm048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brocca S.A.-O., Grandori R., Longhi S.A.-O., Uversky V.A.-O. Liquid-liquid phase separation by intrinsically disordered protein regions of viruses: roles in viral life cycle and control of virus-host interactions. Int. J. Mol. Sci. 2020;21:9045. doi: 10.3390/ijms21239045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu Y., Ma L., Cai S., Zhuang Z., Zhao Z., Jin S., Xie W., Zhou L., Zhang L., Zhao J., Cui J. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-kappaB hyper-activation and inflammation. Signal Transduct. Target. Ther. 2021;6:167. doi: 10.1038/s41392-021-00575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andrusiak M.G., Sharifnia P., Lyu X., Wang Z., Dickey A.M., Wu Z., Chisholm A.D., Jin Y. Inhibition of axon regeneration by liquid-like TIAR-2 granules. Neuron. 2019;104:290–304.e8. doi: 10.1016/j.neuron.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., Laskowski P.R., MacKenzie D., Kamath T., Commins C., et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H., O'Meally R., Dignon G.L., Conicella A.E., Zheng W., et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Duncan P.I., Stojdl D.F., Marius R.M., Scheit K.H., Bell J.C. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res. 1998;241:300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- 133.Karakama Y., Sakamoto N., Itsui Y., Nakagawa M., Tasaka-Fujita M., Nishimura-Sakurai Y., Kakinuma S., Oooka M., Azuma S., Tsuchiya K., et al. Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob. Agents Chemother. 2010;54:3179–3186. doi: 10.1128/aac.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fukuhara T., Hosoya T., Shimizu S., Sumi K., Oshiro T., Yoshinaka Y., Suzuki M., Yamamoto N., Herzenberg L.A., Herzenberg L.A., Hagiwara M. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA. 2006;103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao M., Yu Y., Sun L.-M., Xing J.-Q., Li T., Zhu Y., Wang M., Yu Y., Xue W., Xia T., et al. GCG inhibits SARS-CoV-2 replication by disrupting the liquid phase condensation of its nucleocapsid protein. Nat. Commun. 2021;12:2114. doi: 10.1038/s41467-021-22297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao D., Xu W., Zhang X., Wang X., Ge Y., Yuan E., Xiong Y., Wu S., Li S., Wu N., et al. Understanding the phase separation characteristics of nucleocapsid protein provides a new therapeutic opportunity against SARS-CoV-2. Protein Cell. 2021;12:734–740. doi: 10.1007/s13238-021-00832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bussienne C., Marquet R., Paillart J.C., Bernacchi S. Post-translational modifications of retroviral HIV-1 gag precursors: an overview of their biological role. Int. J. Mol. Sci. 2021;22:2871. doi: 10.3390/ijms22062871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Düster R., Kaltheuner I.H., Schmitz M., Geyer M. 1, 6-Hexanediol, commonly used to dissolve liquid-liquid phase separated condensates, directly impairs kinase and phosphatase activities. J. Biol. Chem. 2021;296:100260. doi: 10.1016/j.jbc.2021.100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shin Y., Berry J., Pannucci N., Haataja M.P., Toettcher J.E., Brangwynne C.P. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017;168:159–171.e14. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bracha D., Walls M.T., Wei M.T., Zhu L., Kurian M., Avalos J.L., Toettcher J.E., Brangwynne C.P. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell. 2018;175:1467–1480.e13. doi: 10.1016/j.cell.2018.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soranno A. The trap in the FRAP: a cautionary tale about transport measurements in biomolecular condensates. Biophys. J. 2019;117:2041–2042. doi: 10.1016/j.bpj.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ishikawa-Ankerhold H.C., Ankerhold R., Drummen G.P.C. Advanced fluorescence microscopy techniques--FRAP, FLIP, FLAP, FRET and FLIM. Molecules. 2012;17:4047–4132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Semerdzhiev S.A., Fakhree M.A.A., Segers-Nolten I., Blum C., Claessens M.M.A.E. Interactions between SARS-CoV-2 N-protein and alpha-synuclein accelerate amyloid formation. ACS Chem. Neurosci. 2022;13:143–150. doi: 10.1021/acschemneuro.1c00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li H., Ernst C., Kolonko-Adamska M., Greb-Markiewicz B., Man J., Parissi V., Ng B.W.L. Phase separation in viral infections. Trends Microbiol. 2022;30:1217–1231. doi: 10.1016/j.tim.2022.06.005. [DOI] [PubMed] [Google Scholar]
- 146.Wu C., Holehouse A.S., Leung D.W., Amarasinghe G.K., Dutch R.E. Liquid phase partitioning in virus replication: observations and opportunities. Annu. Rev. Virol. 2022;9:285–306. doi: 10.1146/annurev-virology-093020-013659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vendruscolo M., Fuxreiter M. Protein condensation diseases: therapeutic opportunities. Nat. Commun. 2022;13:5550. doi: 10.1038/s41467-022-32940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang P., Narayanan E., Liu Q., Tsybovsky Y., Boswell K., Ding S., Hu Z., Follmann D., Lin Y., Miao H., et al. A multiclade env-gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat. Med. 2021;27:2234–2245. doi: 10.1038/s41591-021-01574-5. [DOI] [PubMed] [Google Scholar]
- 149.Xiao Q., McAtee C.K., Su X. Phase separation in immune signalling. Nat. Rev. Immunol. 2022;22:188–199. doi: 10.1038/s41577-021-00572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Couzin-Frankel J. Antiviral pills could change pandemic's course. Science. 2021;374:799–800. doi: 10.1126/science.acx9605. [DOI] [PubMed] [Google Scholar]