Abstract
The serotype M6 group A streptococcal RofA regulator was previously shown to exert a direct positive control of protein F1 expression and, concomitantly, fibronectin binding. Using a serotype M6 rofA mutant, we demonstrate here that this regulator has a potentially indirect negative influence on the expression of the mga, emm6, pel-sagA, and speA virulence genes. Additionally, the rofA mutant exhibited reduced eukaryotic cell internalization rates in combination with decreased host cell viability.
Binding to the intercellular matrix protein fibronectin is a crucial step in the pathogenesis of infections by group A streptococci (GAS) (9). Such binding promotes specific and firm bacterial adherence to human epithelial tissues and, eventually, the internalization into human epithelial cells as well. There are several fibronectin-binding molecules described to be expressed on the GAS surface, such as proteins F1 and SfbI (7, 30), F2 (11), SOF and SfbII (13, 28), FBP54 (1), PFBP (29), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (25).
Among these molecules, protein F1 was found to be present in 50 to 70% of all clinical isolates tested (5, 19, 33) and to be the predominant fibronectin-binding factor in serotype M6 GAS isolates. The structure and function of protein F1 in this specific serotype was well studied. Fibronectin-binding activity was located in two conserved domains of the protein (24, 31). Due to a bridging effect of the bound fibronectin to α5β1- and αvβ3-type integrins, the streptococci attached to buccal epithelial cells and various epithelial cell lines (8, 17, 22, 23, 34). As one consequence, the bacteria can internalize into the epithelial cells (10, 18, 21). This feature has been associated with persistent colonization of asymptomatic human carriers by GAS (20), as well as with the capability to cause invasive diseases (16).
Although considerable N-terminal variability of protein F and/or SfbI sequences has been demonstrated (12), the constitutive expression of these proteins could be disadvantageous for the bacteria because of the constant exposure to the immune system. There are also environmental conditions such as nutrient limitation under which attachment to an anatomical compartment is not necessarily beneficial for GAS. Thus, it seemed logical that the expression of protein F1 would respond to environmental conditions. A few stimuli such as the oxygen partial pressure (35) and superoxide levels (4) have been identified. So far, only one regulatory gene, the rofA gene, has been found to be involved in the response to these environmental conditions (2, 3). The corresponding RofA protein was recently described to exert its regulatory activity by directly binding to two conserved boxes in the promoter region of the protein F1-encoding prtF1 gene (6). Quantitative determination of fibronectin binding by serotype M6 GAS mutants harboring an insertional rofA inactivation indicated that RofA acted as a positive regulator of prtF1, and its own gene under decreased O2 partial pressure (3). However, some of its regulatory activity depended on the genomic background (2).
Analysis of the serotype M1 genome sequence revealed the presence of a rofA gene and, simultaneously, the presence of two genes that were 30 to 50% homologous to rofA (6, 27). In a serotype M49 GAS strain, the nra gene was identified as exhibiting 60% homology to rofA (27). These findings prompted Granok et al. (6) to describe a RofA-like family of proteins (RALPs). Among these genes, the function of nra was determined as that of a negative regulatory factor for several virulence genes (27) and thus as controlling properties necessary for prolonged intracellular persistence (G. Molinari, M. Rohde, S. R. Talay, G. S. Chhatwal, S. Beckert, and A. Podbielski, submitted for publication).
In the present study, the effect of a serotype M6 rofA gene on virulence genes other than prtF1 as well as on eukaryotic cell adherence and internalization was investigated. The results suggest that RALPs are global regulators with a predominantly negative activity and are involved in cell attachment and invasion.
Generation of a serotype M6 GAS rofA insertional mutant.
A fragment of the rofA gene from serotype M6 isolate 2/66 of the GAS strain collection from the Prague WHO reference laboratory was amplified by PCR. The fragment corresponding to positions 31 to 980 of the published M6 rofA sequence (3) was cloned into the plasmid vector pSF152 (32). Integration of the plasmid by single cross-over resulted in disruption of the 1,491-bp rofA gene at bp 798.
Effects of rofA inactivation on prtF1 expression and fibronectin binding.
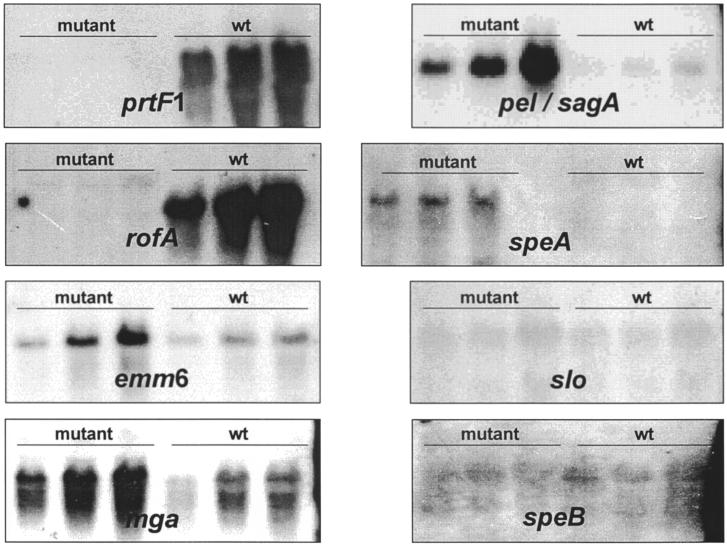
Experiments were done to confirm that this mutation of rofA was affecting the protein F1 transcription and fibronectin binding as reported elsewhere (3). Total RNA was prepared according to an established protocol (27) from wild-type bacteria, and the isogenic rofA mutant was grown to mid-log growth phase in a standing culture under a 20% O2–5% CO2 atmosphere. A prtF1-specific probe (19) was used for a semiquantitative Northern blot hybridization analysis (Fig. 1). The prtF1 message was found to be decreased at least eightfold in the rofA mutant.
FIG. 1.
Quantitation of virulence gene mRNAs in a serotype M6 GAS strain (wt) and its isogenic rofA regulator mutant. Total RNA was isolated from both strains grown to mid-log phase, was serially diluted in twofold increments, separated by denaturing gel electrophoresis, and subjected to Northern blot hybridizations with digoxigenin-labeled virulence gene-specific probes. CSPD (Roche Molecular Biochemicals, Mannheim, Germany) and autoradiography were used to detect hybridization. The amounts of total RNA used in the assays were 20 μg (mga and rofA), 10 μg (prtF1, slo, and speB), and 5 μg (emm6, pel-sagA, and speA).
Binding of soluble 125I-labeled fibronectin was measured according to the protocol of Kreikemeyer et al. (13) using commercially available matrix proteins (Gibco-BRL, Eggenstein, Germany). For the assays, the two strains were grown to early-log, early-stationary, and stationary growth phases as standing cultures under a 20% O2–5% CO2 atmosphere. The results are the average of three independent experiments. By as early as 3 h of growth in the CO2-enriched atmosphere, the rofA mutant bound 30% less fibronectin. In the stationary phase, the fibronectin-binding of the mutant was further reduced to 61% of the wild-type levels (Table 1).
TABLE 1.
Intercellular matrix protein binding by a serotype M6 GAS strain and its isogenic rofA regulator mutanta
| Protein and strain type | Mean % dpm ± SD
|
||
|---|---|---|---|
| 3 h | 8 h | Overnight | |
| Fibronectin | |||
| Wild type | 54 ± 5 | 58 ± 8 | 54 ± 9 |
| Mutant | 38 ± 2 | 37 ± 6 | 33 ± 4 |
| Fibrinogen | |||
| Wild type | 52 ± 4 | 51 ± 3 | 53 ± 3 |
| Mutant | 45 ± 2 | 40 ± 3 | 44 ± 3 |
| Collagen I | |||
| Wild type | 24 ± 2 | 25 ± 1 | 31 ± 2 |
| Mutant | 24 ± 1 | 24 ± 1 | 31 ± 3 |
Soluble matrix proteins were 125I labeled and employed for the binding assays as described by Kreikemeyer et al. (13). The bacteria were grown in Todd Hewitt-yeast extract broth to the early-logarithmic (3 h), early-stationary (8 h), and late-stationary (overnight) growth phases prior to the assays. The amount of radioactive material, measured in decays per minute (dpm), used for the assays was assigned a value of 100%; the amount of bound radioactive material (measured in dpm) was in turn related to these values. The table contains results from three independent assays.
Effects of rofA inactivation on expression of virulence genes and functions.
The rofA homologue nra was found to control the expression of at least a dozen GAS genes and the collagen-binding activity in a serotype M49 GAS strain ((27); Molinari et al., submitted). Our next objective was to determine whether a functional rofA gene has a comparably broad regulatory activity.
Binding experiments with 125I-labeled fibrinogen and collagen were performed under conditions similar to those of the fibronectin-binding assays. The mutant exhibited a slightly reduced fibrinogen binding (Table 1). The differences increased from exponential to stationary growth phase. Since protein F1 was demonstrated to bind fibrinogen in addition to fibronectin (12), the decreased fibrinogen binding of the rofA mutant could be explained by the reduction in prtF1 gene transcription. Collagen-binding was unaltered in the rofA mutant, indicating that this feature does not depend on a functional rofA gene in serotype M6 strains. Thus, different RALP members are involved in control of GAS binding to distinct matrix proteins (3, 27; this study).
The expression of several known virulence genes was studied in the rofA mutant by semiquantitative Northern blot analysis. The genes of the two secreted factors streptolysin O and cysteine protease SpeB appeared to be unaffected by the rofA mutation (Fig. 1). As found for nra mutants (Molinari et al., submitted), the expression of the genes encoding the superantigen SpeA and the streptolysin S (Pel-SagA), as well as the global positive regulator Mga, were increased 8- to 32-fold in the mutant (Fig. 1). These results suggest that RALPs such as RofA and Nra are involved in different regulatory circuits. Both act on the speA gene transcription, for which no other regulator has been determined. Moreover, both act on the mga regulator gene transcription, which in turn could affect the expression of the pel-sagA gene expression.
Increased transcription of M6 protein gene (emm6) in the rofA mutant as opposed to negligible changes in the emm49 expression in an nra mutant (Molinari et al., submitted) indicated that RALP control of the emm gene expression is possibly indirectly exerted and could be additionally modulated by superimposed serotype-specific regulatory circuits. Granok et al. (6) described a RofA consensus binding box which they located upstream of the rofA sequences in the genomes of serotypes M6 and M1, as well as upstream of the serotype M1 clpX stress-induced gene. When an analysis was done on published sequences, the conserved binding box could not be detected in the promoter regions of mga, emm6, pel-sagA, or speA. This finding could indicate that RofA binds to sequences other than the identified binding box or that changes in transcription of all these genes are due to an indirect effect of RofA.
The rofA-specific transcript in the mutant was larger than that of the wild-type strain due to a read-through transcription into the integrated pSF152 plasmid. Such read-through transcription has also been observed in other pSF152-derived mutants (26). The intensity of the rofA-specific band in the mutant's RNA analysis suggested an at least eightfold-less-abundant message in the mutant, thus confirming the positive autoregulatory effect of a functional RofA mutant described by Fogg et al. (2).
Impact of rofA mutation on eukaryotic cell attachment and internalization.
In protein F1-expressing GAS strains, this protein was found to play a major role in bacterial interactions with eukaryotic cells. Thus, our rofA mutant was predicted to be less adhesive and invasive than the corresponding wild type.
To test this assumption, adhesion and internalization experiments were performed using bacteria from overnight cultures and HEp-2 cells following the protocol of Molinari et al. (submitted). Adherent and internalized bacteria were quantitated by plate viability counts. The reported results are the average of six independent experiments. For a better comparison, the results were normalized by arbitrarily setting the wild-type adherence as 100% and relating the other viability counts to this value.
The percent adhesion and percent internalization values were 100 and 2.7 ± 1.6 for the wild type and 59.9 ± 33.1 and 0.21 ± 0.20 for the mutant, respectively.
Eukaryotic cell attachment of the rofA mutant was therefore decreased by 40% compared to the parental strain. This reduction correlated well to measurements of decreased fibronectin binding by the mutant, suggesting that the altered matrix protein binding was sufficient to explain the changes in cell adhesion.
In contrast, the internalization rate of the mutant was only 7% of the wild-type value, indicating that superimposed additional mechanisms besides fibronectin-binding influenced the uptake of the rofA mutant into HEp-2 cells.
Eukaryotic cell viability in presence of the rofA mutant.
A decreased host cell viability could lead to a reduction of bacterial viability counts when using culture media containing antibiotics.
Therefore, the host cell viability in the attachment and internalization assays was controlled by using the fluorescent LIVE/DEAD stain according to the manufacturer's instructions (Molecular Probes/Mobitec, Göttingen, Germany). After 4 h of coincubation, ca. 20 to 30% of the HEp-2 cells showed typical red fluorescence with the DEAD stain in the presence of the mutant, while <5% of the eukaryotic cells did so in the presence of the parental strain (data not shown).
An even stronger reduction in HEp-2 cell viability was recorded in the presence of nra mutant bacteria (Molinari et al., submitted). In both cases, the decreased eukaryotic cell viability could potentially be explained by an overproduction of secreted harmful factors like SpeA or streptolysin S (Pel-SagA) by the mutants.
Conclusion.
In this study, we investigated the impact of a mutation in a member of the RALP regulator family, rofA, on virulence gene expression and the concomitant biological behavior of a serotype M6 GAS strain. In agreement with previous publications (3), expression of the protein F1-encoding prtF1 gene and fibronectin binding as well as the truncated rofA gene was decreased in the mutant, confirming the status of RofA as a positive regulator for these virulence traits. However, a functional rofA gene also appeared to act as a negative regulator for the expression of several other virulence genes. These results agreed with many of those obtained for another RALP member, the nra gene (27). Since the promoters of the controlled genes lack the recently identified RofA binding box (6), it needs to be determined whether RofA regulation is indirect for these virulence genes. Because a functional rofA decreased the expression of mga, the global positive virulence regulator in the serotype M6 strain (14), rofA could be the superimposed negative regulator, whose presence was recently predicted in a study on Mga functions in a serotype M6 strain (15). Like the homologous Nra regulator (Molinari et al., submitted), RofA influences the host cell viability once the bacteria are internalized into the eukaryotic cells. This property is obviously associated with control mechanisms exceeding the known regulation of protein F1 expression. Since other RALP members have a similar function (Molinari et al., submitted), these regulators could have an important general role in stabilizing the intracellular status of GAS.
Acknowledgments
We thank A. Flosdorff for expert technical assistance and B. A. Buttaro, Philadelphia, Pa., for critical reading.
S.B. and A.P. were supported by grant A09.22 of the IZKF, Ulm, Germany; B.K. and A.P. were supported by DFG grant Po391/6-2; and A.P. was also supported by DFG grant Po391/3-4.
REFERENCES
- 1.Courtney H S, Li Y, Dale J B, Hasty D L. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect Immun. 1994;62:3937–3946. doi: 10.1128/iai.62.9.3937-3946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogg G C, Caparon M G. Constitutive expression of fibronectin-binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J Bacteriol. 1997;179:6172–6180. doi: 10.1128/jb.179.19.6172-6180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an μγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 4.Gibson C M, Caparon M G. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J Bacteriol. 1996;178:4688–4695. doi: 10.1128/jb.178.15.4688-4695.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodfellow A M, Hibble M, Talay S R, Kreikemeyer B, Currie B J, Sriprakash K S, Chhatwal G S. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J Clin Microbiol. 2000;38:389–392. doi: 10.1128/jcm.38.1.389-392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granok A B, Parsonage D, Ross R P, Caparon M G. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J Bacteriol. 2000;182:1529–1540. doi: 10.1128/jb.182.6.1529-1540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanski E, Caparon M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus (Streptococcus pyogenes) Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanski E, Horwitz P A, Caparon M G. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect Immun. 1992;60:5119–5125. doi: 10.1128/iai.60.12.5119-5125.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasty D L, Ofek I, Courtney H S, Doyle R J. Multiple adhesins of streptococci. Infect Immun. 1992;60:2147–2152. doi: 10.1128/iai.60.6.2147-2152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jadoun J, Ozeri V, Burstein E, Skutelsky E, Hanski E, Sela S. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J Infect Dis. 1998;178:147–158. doi: 10.1086/515589. [DOI] [PubMed] [Google Scholar]
- 11.Jaffe J, Natanson-Yaron S, Caparon M C, Hanski E. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol Microbiol. 1996;21:373–384. doi: 10.1046/j.1365-2958.1996.6331356.x. [DOI] [PubMed] [Google Scholar]
- 12.Katerov V, Andreev A, Schalen C, Totolian A A. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology. 1998;144:119–126. doi: 10.1099/00221287-144-1-119. [DOI] [PubMed] [Google Scholar]
- 13.Kreikemeyer B, Talay S R, Chhatwal G S. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol Microbiol. 1995;17:137–145. doi: 10.1111/j.1365-2958.1995.mmi_17010137.x. [DOI] [PubMed] [Google Scholar]
- 14.McIver K S, Heath A S, Grenn B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A Streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver K S, Thurman A S, Scott J R. Regulation of mga transcription in the group A Streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J Bacteriol. 1999;181:5373–5383. doi: 10.1128/jb.181.17.5373-5383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molinari G, Chhatwal G S. Invasion and survival of Streptococcus pyogenes in eukaryotic cells correlates with the source of the clinical isolates. J Infect Dis. 1998;177:1600–1607. doi: 10.1086/515310. [DOI] [PubMed] [Google Scholar]
- 17.Molinari G, Rohde M, Guzman C A, Chhatwal G S. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell Microbiol. 2000;2:145–154. doi: 10.1046/j.1462-5822.2000.00040.x. [DOI] [PubMed] [Google Scholar]
- 18.Molinari G, Talay S R, Valentin-Weigand P, Rohde M, Chhatwal G S. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natanson S, Sela S, Moses A E, Musser J M, Caparon M G, Hanski E. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J Infect Dis. 1995;171:871–878. doi: 10.1093/infdis/171.4.871. [DOI] [PubMed] [Google Scholar]
- 20.Neeman R, Keller N, Barzilai A, Korenman Z, Sela S. Prevalence of internalization-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet. 1998;352:1974–1977. doi: 10.1016/S0140-6736(97)12452-7. [DOI] [PubMed] [Google Scholar]
- 21.Okada N, Tatsuno I, Hanski E, Caparon M G, Sasakawa C. Streptococcus pyogenes protein F promotes invasion of HeLa cells. Microbiology. 1998;144:3079–3086. doi: 10.1099/00221287-144-11-3079. [DOI] [PubMed] [Google Scholar]
- 22.Okada N, Watarai M, Ozeri V, Hanski E, Caparon M, Sasakawa C. A matrix form of fibronectin mediates enhanced binding of Streptococcus pyogenes to host tissue. J Biol Chem. 1997;272:26978–26984. doi: 10.1074/jbc.272.43.26978. [DOI] [PubMed] [Google Scholar]
- 23.Ozeri V, Rosenshine I, Mosher D F, Fässler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 24.Ozeri V, Tovi A, Burstein I, Natanson-Yaron S, Caparon M G, Yamada K M, Akiyama S K, Vlodavsky I, Hanski E. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 1996;15:989–998. [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podbielski A, Woischnik M, Leonard B A B, Schmidt K H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 28.Rakonjac J V, Robbins J C, Fischetti V A. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect Immun. 1995;63:622–631. doi: 10.1128/iai.63.2.622-631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocha C L, Fischetti V A. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect Immun. 1999;67:2720–2728. doi: 10.1128/iai.67.6.2720-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talay S R, Valentin-Weigand P, Jerlström P G, Timmis K N, Chhatwal G S. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect Immun. 1992;60:3837–3844. doi: 10.1128/iai.60.9.3837-3844.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talay S R, Valentin-Weigand P, Timmis K N, Chhatwal G S. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol Microbiol. 1994;13:531–539. doi: 10.1111/j.1365-2958.1994.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 32.Tao L, LeBlanc D J, Ferretti J J. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene. 1992;120:105–110. doi: 10.1016/0378-1119(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 33.Valentin-Weigand P, Talay S R, Kaufhold A, Timmis K N, Chhatwal G S. The fibronectin binding domain of the Sfb protein adhesin of Streptococcus pyogenes occurs in many group A streptococci and does not cross-react with heart myosin. Microb Pathog Res. 1994;17:111–120. doi: 10.1006/mpat.1994.1057. [DOI] [PubMed] [Google Scholar]
- 34.Valentin-Weigand P, Talay S R, Timmis K N, Chhatwal G S. Identification of a fibronectin-binding protein as adhesin of Streptococcus pyogenes. Zentbl Bakteriol. 1993;278:238–245. doi: 10.1016/s0934-8840(11)80841-8. [DOI] [PubMed] [Google Scholar]
- 35.VanHeyningen T, Fogg G, Yates D, Hanski E, Caparon M. Adherence and fibronectin binding are environmentally regulated in the group A streptococci. Mol Microbiol. 1993;9:1213–1222. doi: 10.1111/j.1365-2958.1993.tb01250.x. [DOI] [PubMed] [Google Scholar]