Abstract
Embryonic stem cells (ESCs) exhibit an unusual cell cycle profile containing a short G1 phase. Whether this feature is required to maintain pluripotency is a matter of debate. Here, we report that the short G1 phase is a consequence of MEK1/2 kinase-mediated promotion of G1/S transition, but not necessarily coupled with pluripotency maintenance. We find that compared to primed ESCs, naïve ESCs exhibit a significantly longer G1 phase due to the inhibition of MEK1/2 kinases. MEK1/2 inhibition increases intracellular level of reactive oxygen species (ROS), leading to the stabilization of p53 protein. The genetic ablation of p53 largely converts the cell cycle profile of naïve ESCs to that of primed ESCs. These results demonstrate that pluripotency and proliferation are separable cellular events, and the short G1 phase of primed ESCs is a manifestation of the intricate interplay between classical oncogenes MEK1/2 and tumor suppressor gene TP53 to promote G1/S transition.
Keywords: Cell cycle, Embryonic stem cell, P53, ROS
Cell cycle; Embryonic stem cell; P53; ROS.
1. Introduction
Embryonic stem cells (ESCs) are self-renewing pluripotent cells, defined by their capability to differentiate into all somatic cell types (Evans, 2011). The regulatory network to maintain pluripotency is well characterized (Hackett and Surani, 2014). Core pluripotency transcription factors, such as POU class 5 homeobox 1 (POU5F1, commonly known as OCT4) and SRY-box transcription factor 2 (SOX2), orchestrate multiple cellular processes to enforce epigenetic and metabolic programs to maintain ESCs in an undifferentiated state (Young, 2011). Recent studies demonstrate that ESCs can exist in different pluripotent states dependent on cell culture conditions (Nichols and Smith, 2009). Ground-state pluripotency or naïve ESCs (resembling inner cell mass in blastocyst) can be induced in murine ESC culture by chemical inhibition of mitogen-activated protein kinase and glycogen synthase kinase 3 (termed “2i”) (Ying et al., 2008). On the other hand, murine ESCs grown in serum plus leukemia inhibitory factor (LIF) are more heterogeneous, containing primed ESCs resembling post-implantation epiblast (Kolodziejczyk et al., 2015). Naïve and primed ESCs share many similarities in vitro. Both are capable of self-renewal, express OCT4 and SOX2 at similar levels, and can differentiate into three germ layers (Martello and Smith, 2014). However, without 2i, ESCs express many lineage-specific transcripts and are prone to spontaneous differentiation (Marks et al., 2012).
Murine ESCs are fast-proliferating cells, with a doubling time around 12 h as well as an unusually short G1 phase (Li et al., 2012; Wang et al., 2011). This property of ESCs is not well understood at the mechanistic level. Nevertheless, fast proliferation is thought to promote pluripotency maintenance, because differentiating ESCs exhibit longer doubling time as well as the G1 phase (Boward et al., 2016). However, recent studies demonstrate that ESCs can enter a non-proliferative dormant state, albeit transiently, both in vitro and in vivo (Bulut-Karslioglu et al., 2016; Scognamiglio et al., 2016). In addition, during the course of this study, Ter Huurne et al. demonstrated that the G1 phase of ESCs is longer in 2i than serum conditions in a p53-dependent manner (Ter Huurne et al., 2017, 2020). How 2i condition activates p53 pathway remains to elucidated. These observations suggest that fast proliferative rate may not be a prerequisite for pluripotency maintenance.
Previously, we showed that Tp53, encoding the tumor suppressor p53 protein, is required for acute DNA damage-induced apoptosis, but dispensable for the G2/M arrest in murine ESCs (He et al., 2016). Here, we demonstrate that 2i induces cytosolic reactive oxygen species to activate p53, resulting in different cell cycle structures in naïve and primed ESCs. We estimate that p53-dependent regulation of the cell cycle accounts for approximately 13% of differentially expressed genes between these two cellular states.
2. Results
2.1. ESCs grown in 2i condition exhibit a longer G1 phase
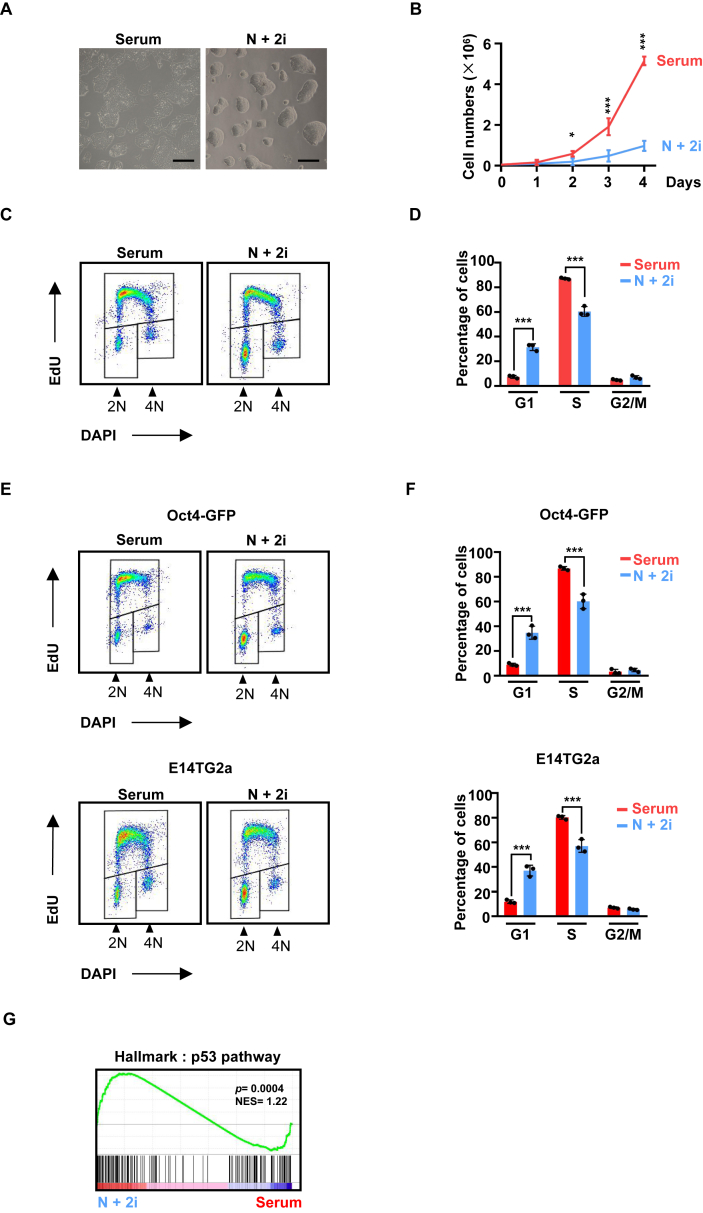
ESCs can self-renew in different culture conditions. For following experiments, we mostly used R1/E (R1) ES cell line, and key observations were also validated using additional ES cell lines, ES-E14TG2a (E14TG2a) and Oct4-GFP (Ohbo et al., 2003). As expected, compared to serum condition (Serum), murine ESCs routinely cultured in serum-free containing N2B27 medium with 2i/LIF condition (N + 2i) (Ying et al., 2008), containing small-molecule inhibitors of mitogen-activated protein kinase (MEKi) and glycogen synthase kinase 3 (GSKi), were more morphologically uniform (Figure 1A). However, we noticed that ESCs in 2i condition proliferated at a slower rate than ones in serum (Figure 1B). This difference was not due to accelerated cell death, but due to extended G1 phase in 2i condition (Figure 1C). Concomitant with lengthened G1 phase, S phase was shortened in 2i condition without obvious change in G2/M (Figure 1D). These results indicate that 2i condition induces certain regulators of the G1/S transition of the cell cycle. Similarly, two other ESC cell lines also exhibited longer G1 phases in 2i (Figures 1E, 1F). To understand the underlying mechanism, we carried out transcriptome analysis by RNA sequencing (RNA-seq). Gene set enrichment analysis (GSEA) was performed using the hallmark gene set collection in the Molecular Signatures Database (MSigDB) (Liberzon et al., 2015). Transcripts per kilobase of exon model per million mapped reads (TPM>1) and false discovery rate (FDR<0.01) were set as cutoff. Interestingly, hallmark gene set “p53 pathway” was upregulated in 2i condition (Figure 1G).
Figure 1.
ESCs grown in 2i condition exhibit a longer G1 phase. (A) Microscopic morphology of R1/E ESCs grown in serum (Serum) and 2i condition (N+2i). Scale bar, 200 μm. (B) Proliferative rate of ESCs in serum and 2i conditions. Equal number of cells were seeded at day 0, and cell numbers were counted every day for 4 days (n = 3). ∗, P < 0.05; ∗∗∗, P < 0.001 (unpaired t test). (C) Cell cycle profiling by fluorescence-activated cell sorting (FACS) analysis of ESCs grown in serum and 2i. Representative result was shown. (D) Quantification of FACS analysis as in (C) (n = 3). The bar plot represents mean ± SD. ∗∗∗, P < 0.001 (unpaired t test). (E) Cell cycle profiling by FACS analysis of additional ESC cell lines, Oct4-GFP and ES-E14TG2a (E14). (F) Quantification of FACS analysis as in (E) (n = 3). The bar plot represents mean ± SD. ∗∗∗, P < 0.001 (unpaired t test).(G) Hallmark analysis of differentially expressed genes between serum and 2i.
2.2. Prolonged G1 phase induced by 2i depends on Tp53
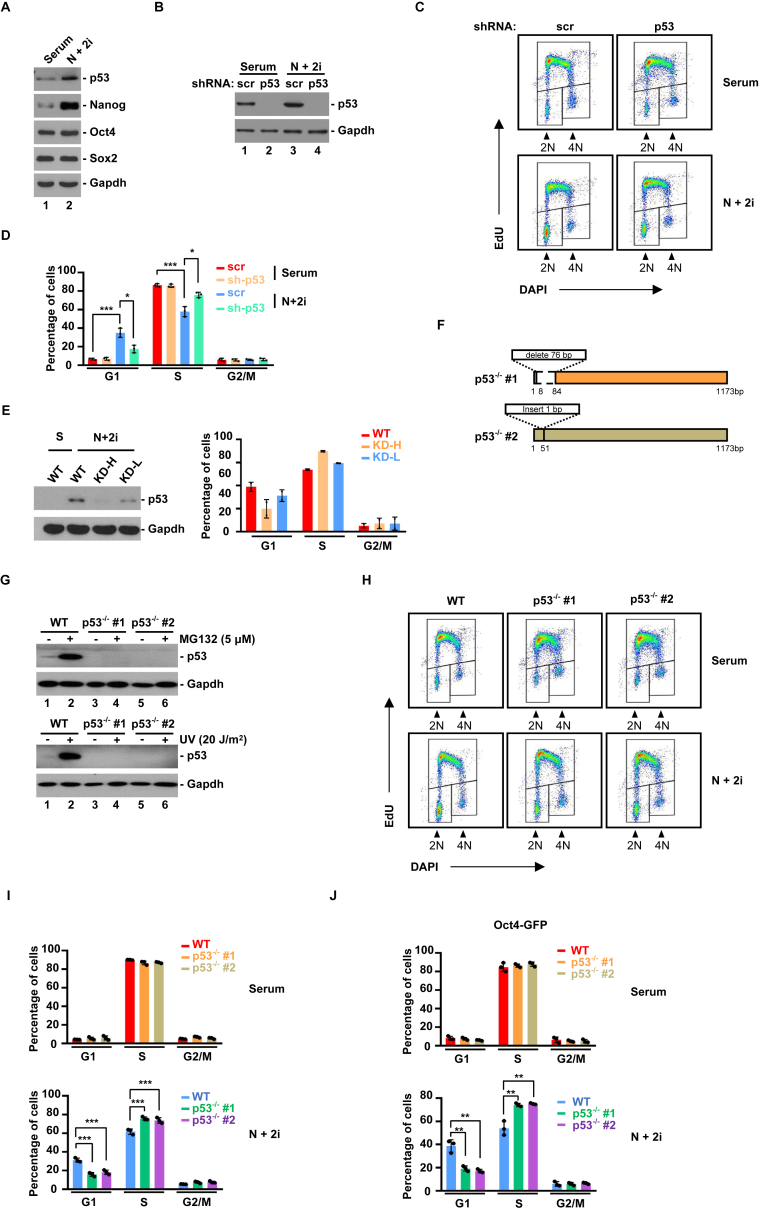
Tp53, encoding tumor suppressor p53 protein, is a well-established master regulator of the G1/S transition in various types of somatic cells (Levine, 2020). Its role in ESCs is less defined. Previously, we found that p53 is dispensable for acute DNA damage-induced cell cycle arrest in ESCs (He et al., 2016). We decided to investigate whether it was involved under homeostatic conditions. As expected, protein levels of Oct4 and Sox2 were comparable between serum and 2i condition, consistent with the fact that pluripotency are maintained in both conditions (Figure 2A). Nanog protein level was significantly elevated in 2i condition (Figure 2A), because unlike in serum, the majority of ESCs in 2i homogenously express Nanog (Munoz Descalzo et al., 2012). Of note, Nanog is not essential for pluripotency maintenance, but can reduce the chance of spontaneous differentiation (Chambers et al., 2007). This is consistent with the fact that ESCs in 2i are more uniform compared to those in serum (Figure 1A). In agreement with GSEA analysis (Figure 1G), we found that p53 protein was induced by 2i condition (Figure 2A). To determine whether this increase in p53 protein was biologically relevant, we analyzed the cell cycle profiles in ESCs with or without stable Tp53 knockdown. These cell lines were previously used to characterize the role of Tp53 in acute DNA damage response in ESCs (He et al., 2016). Stable knockdown of Tp53 efficiently eliminated 2i-induced increase in p53 protein (Figure 2B). FACS analysis revealed that knockdown of Tp53 largely restored the cell cycle profile of ESCs in 2i back to that in serum (Figures 2C, 2D). Meanwhile, knockout efficiency was positively correlated with the degree of cell cycle recovery (Figure 2E). In contrast, knockdown of Tp53 did not alter the cell cycle structure of ESCs in serum (Figures 2C, 2D). This is because p53 protein level is usually low in serum condition, or this could indicate other mechanisms of G1/S transition at play. The G2/M phase was similar between serum and 2i conditions (Figure 1D), and not altered by Tp53 knockdown (Figure 2D). These results indicate that Tp53 may be responsible for lengthened G1 phase in 2i. To confirm, we generated Tp53 knockout cell lines by CRISPR/Cas9 genomic editing technology. Two independent Tp53 knockout cell lines were subjected to further analyses (p53−/− #1 and #2). Knockout was confirmed by PCR analysis of targeted region (Figure 2F), and the absence of p53 protein by either MG132 or ultraviolet (UV) treatment (Figure 2G). FACS analysis revealed that cell cycle profile in 2i was converted to that in serum in Tp53 knockout cell lines (Figures 2H, 2I). In addition, knockout of Tp53 in another ESC cell line had similar effect (Figure 2J). Thus, 2i condition stabilizes p53 protein, resulting in a longer G1 phase.
Figure 2.
2i condition activates p53 to prolong G1 phase (A) Protein blot analysis of indicated proteins.(B) Knockdown of Tp53 expression by shRNA. Scrambled (scr) shRNA was used as control.(C) FACS analysis of ESCs with or without Tp53 knockdown in serum or 2i conditions.(D) Quantification of FACS analysis as in (C) (n = 3). The bar plot represents mean ± SD. ∗, P < 0.05; ∗∗∗, P < 0.001 (unpaired t test).(E) Quantification of FACS analysis and protein blot analysis of ESCs with Tp53 knockdown in high efficiency and in low efficiency. The bar plot represents mean ± SD.(F) Diagram of two independent p53-knockout cell lines.(G) Confirmation of knockout efficiency by MG132 or UV treatment. WT, wild-type.(H) FACS analysis of ESCs with or without Tp53 knockout in serum or 2i conditions.(I) Quantification of FACS analysis as in (G) (n = 3). The bar plot represents mean ± SD. ∗∗∗, P < 0.001 (unpaired t test). (J) Quantification of FACS analysis of another ESC cell line (Oct4-GFP) with or without Tp53 knockout. The bar plot represents mean ± SD. ∗∗, P < 0.01 (unpaired t test).
2.3. Knockout of Tp53 does not affect pluripotency in vitro
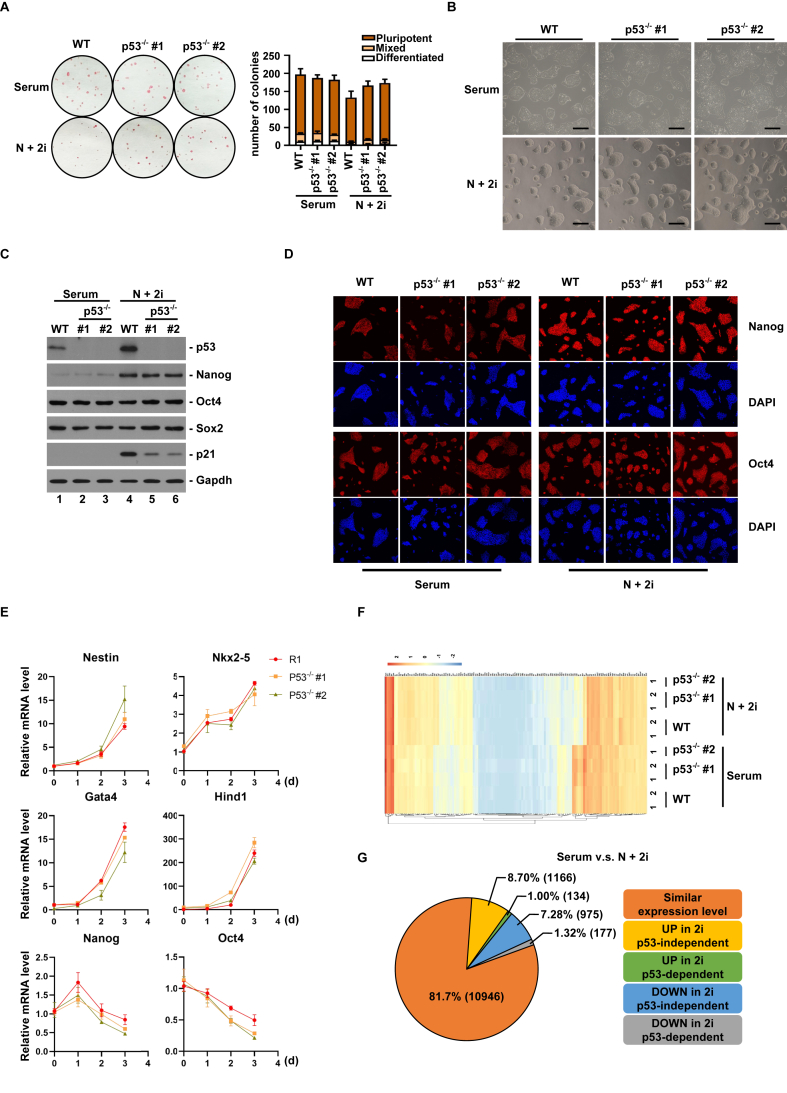
We analyzed whether Tp53 had any impact on pluripotency maintenance in vitro. In both serum and 2i conditions, Tp53 knockout cell lines were positive for alkaline phosphatase staining, a maker of pluripotent ESCs (Figure 3A), had comparable microscopic cell morphology as parental cells (Figure 3B), and expressed similar level of pluripotency factors Oct4, Sox2 and Nanog (Figure 3C,D). Differentiation experiments showed that p53 deletion did not affect the ability of ESCs to differentiate into different layers (Figure 3E). These observations indicate that Tp53 may not be required for pluripotency maintenance in both culture conditions. To confirm, we carried out transcriptome analysis by RNA-seq of wild-type and two Tp53 knockout cell lines grown in serum and 2i conditions. Focusing on 140 genes in the category of “signaling pathways regulating pluripotency of stem cells - Mus musculus (mouse)” (KEGG pathway: mmu04550), we found that these cell lines clustered on the basis of culture conditions rather than the status of Tp53 (Figure 3F), indicating that Tp53 is not required for pluripotency. Overall, among 13,398 genes well expressed in either serum or 2i condition (TPM>1), 18.3% (2,452 genes) exhibited a 2-fold difference between serum and 2i conditions. Among these differentially expressed genes, 134 upregulated and 177 downregulated genes in 2i condition were dependent on Tp53, accounting for approximately 13% of differentially expressed genes between serum and 2i conditions (Figure 3G). Thus, the difference in transcriptome between two culture conditions is partially caused by Tp53-regulated cell cycle structure.
Figure 3.
Tp53 is dispensable for pluripotency maintenance in vitro. (A) Alkaline phosphatase (AP) staining of ESCs with or without Tp53 knockout, grown in serum or 2i condition. Colonies with uniform AP staining, mixed positive and negative staining, or negative AP staining were assigned as pluripotent, mixed and differentiated ESC colonies, respectively (n = 3). (B) Microscopic examination of ESCs with or without Tp53 knockout, grown in serum or 2i condition. Scale bar, 200 μm.(C) Protein blot analysis of indicated proteins.(D) Immunofluorescence analysis of Oct4 and Nanog. Scale bar, 200μm. (E) qPCR analysis of linage-specific transcripts during the course of ESCs differentiation with or without Tp53 knockout. The bar plot represents mean ± SD. (F) Cluster analysis of expression levels of 140 genes in the category of “signaling pathways regulating pluripotency of stem cells - Mus musculus (mouse)” (KEGG pathway: mmu04550).(G) Transcriptomic differences between ESCs grown in serum and 2i, and their dependency on p53.
2.4. Inhibition of MEK pathway leads to p53 activation
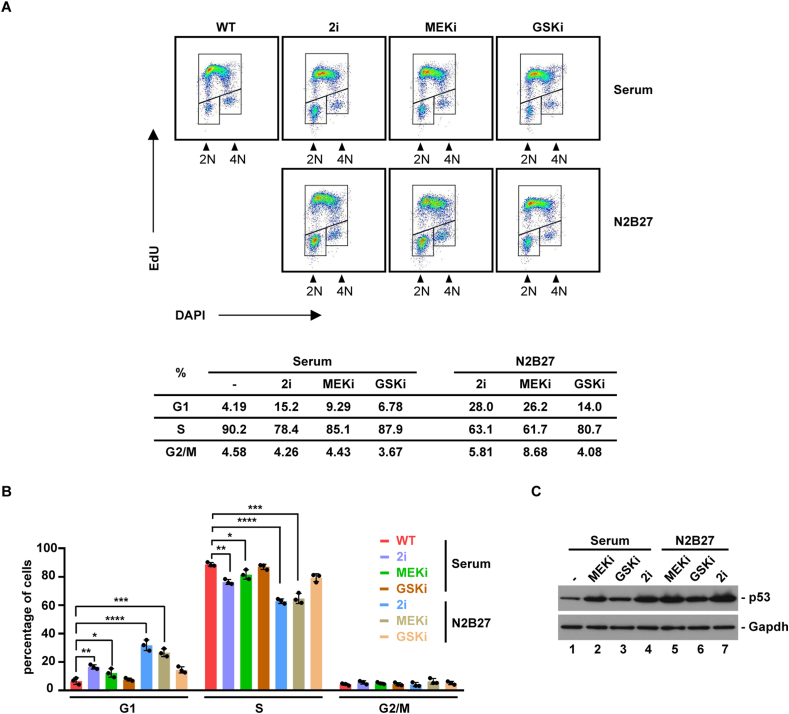
To understand the mechanism of p53 activation in 2i condition, we examined the effect of individual small-molecule inhibitor, that is, small-molecule inhibitors of mitogen-activated protein kinase (MEKi) and glycogen synthase kinase 3 (GSKi). Original 2i culture system, as used in all experiments above, is free of serum and contains N2B27 medium (N) supplemented with MEKi/GSKi (2i) (Ying et al., 2008). Similar to N+2i, the addition of 2i to serum condition lengthened the G1 phase, albeit to a lesser degree. Prolonged G1 phase was largely caused by MEKi (Figure 4A, B). Accordingly, p53 levels were increased primarily by MEKi (Figure 4C). Thus, the inhibition of oncogenic MEK signaling in ESCs upregulate tumor suppressor protein p53 to regulate the G1/S transition.
Figure 4.
Inhibition of MEK pathway leads to p53 activation. (A) Effect of small-molecule inhibitors of MEK and GSK (MEKi and GSKi) on ESC cell cycle profile. 2i denotes MEKi plus GSKi. (B) Quantification of FACS analysis as in (A) (n = 3). The bar plot represents mean ± SD. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001 (unpaired t test).(C) Effects of MEKi, GSKi or 2i on p53 protein.
2.5. MEKi induces reactive oxygen species to activate p53
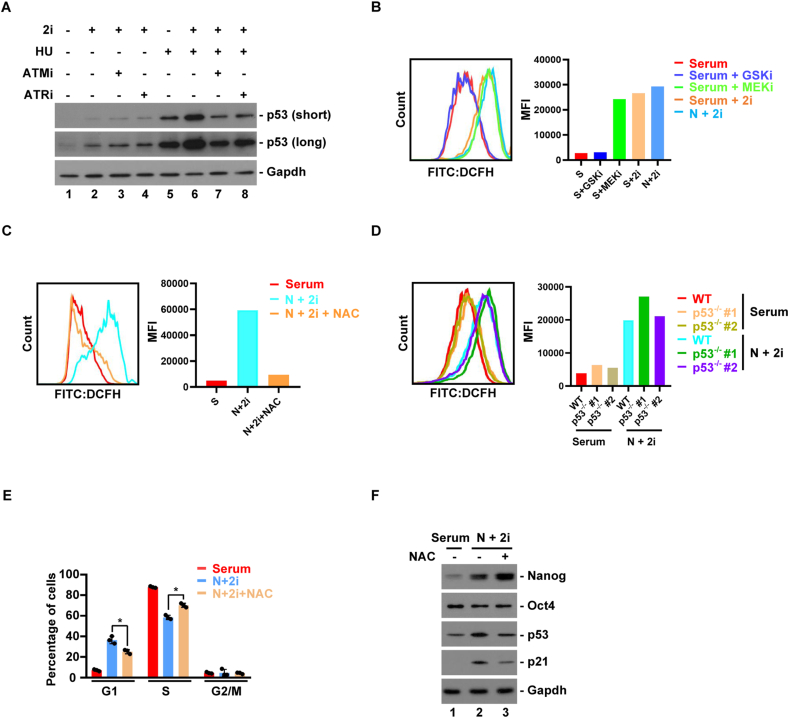
We investigated how 2i promoted p53 activities in ESCs. p53 protein can be stabilized by ATM/ATR-mediated DNA damage response pathway (Zhou and Elledge, 2000). We reasoned that this pathway was unlikely to contribute 2i-induced p53 activities, because we previously found that Tp53 is dispensable for cell cycle control under acute DNA damage condition (He et al., 2016). Indeed, p53 protein was induced to a much higher level by hydroxyurea (HU) than 2i (Figure 5A, compare lanes 1, 2 and 5). Small-molecule inhibitors of ATM and ATR (ATMi and ATRi) could inhibit HU-induced increase in p53 protein level, but not that induced by 2i (Figure 5A). Thus, 2i-induced p53 is not activated by ATM/ATR-mediated canonical DNA damage response pathway. ESCs grown in 2i proliferated at a slower rate than in serum, but still at a comparable rate as most cancer cell lines. We thus considered more physiological signaling pathways that can mildly activate p53 (Kruiswijk et al., 2015). In this regard, we found that reactive oxygen species (ROS) was accumulated to a higher level in 2i than in serum (Figure 5B). Interestingly, higher level of ROS was induced by MEKi but not GSKi (Figure 5B). Treatment of ESCs in 2i with antioxidant N-acetylcysteine (NAC) reduced ROS level to that in serum (Figure 5C). The ROS level was indistinguishable in Tp53 knockout and parental cell lines in both serum and 2i culture conditions (Figure 5D), indicating that activation of p53 may be a downstream event of elevated ROS. Indeed, treatment with NAC partially restored the G1 phase (Figure 5E), and reduced p53 protein level in 2i (Figure 5F). Of note, we found that antioxidants can only reduce ROS levels in 2i condition within a short time window (Supplementary Figure 1). p53 protein level in 2i condition rose to its original level at 3 h after NAC treatment (Supplementary Figure 1 A). Similarly, glutathione monoethyl ester (GSH-MEE), a cell-permeable derivative of glutathione, could reduce p53 level in 2i condition at 1 h but quickly lost its potency (Supplementary Figure 1B). This technical issue might explain why G1 phase was only partially restored by antioxidant treatment. Alternatively, this could also indicate that additional signaling pathways may be induced by 2i to fully activate p53.
Figure 5.
Reactive oxygen species is induced by MEKi to promote p53 activity. (A) Effects of small-molecule inhibitors of ATM and ATR (ATMi and ATRi) on 2i- or hydroxyurea (HU)-induced p53 protein expression.(B) FACS analysis and MFI quantification of cellular reactive oxygen species (ROS) level in ESCs grown in indicated conditions.(C) Effect of antioxidant, N-acetylcysteine (NAC), on 2i-induced ROS.(D) FACS analysis and MFI quantification of ROS levels in ESCs with or without Tp53 knockout, grown in serum or 2i condition.(E) Effect of NAC on 2i-induced increase in the G1 duration. The bar plot represents mean ± SD. ∗, P < 0.05 (unpaired t test).(F) Effect of NAC on 2i-induced p53 protein expression.
We attempted to identify other factors to account for 2i-induced p53 activation. Based on our transcriptomic analysis (Supplementary Figure 2A), we selected several factors for further examination including Hoxc12, p19ARF and Dusp6 (Supplementary Figure 2B). We confirmed by qPCR analysis that 2i induced the expression of p19ARF, encoded by Cdkn2a (Supplementary Figure 2D). In multiple somatic cell types, p19ARF increases p53 protein level via its ability to neutralize E3 ligase Mdm2 (Sherr, 2006). We generated two independent p19ARF knockout cell lines (Supplementary Figure 2C). Surprisingly, knockout of p19ARF failed to prevent 2i-induced p53 protein accumulation (Supplementary Figure 2E), 2i-induced increase in ROS level (Supplementary Figure 2F) or the G1 phase (Supplementary Figure 2J). Hoxc12 is an uncharacterized member of the Hox gene family. Members in this family are best known for their function in body pattern specification (Pearson et al., 2005). We confirmed by qPCR that Hoxc12 expression was highly induced by 2i (Supplementary Figure 2G). However, genetic knockout of Hoxc12 did not prevent 2i-induced p53 protein accumulation (Supplementary Figure 2H), 2i-induced increase in ROS level (Supplementary Figure 2I) or the G1 phase (Supplementary Figure 2J). Lastly, we investigated the role of Dusp6. Dusp6 was recently shown to decrease p53 protein level in somatic cell lines (Wu et al., 2018). We confirmed by qPCR that Dusp6 expression was repressed by 2i (Supplementary Figure 2K). We ectopically expressed Dusp6 in ESCs, and found that it did not prevent 2i-induced increase in p53 protein level. In fact, overexpression of Dusp6 further increased p53 protein level and that of p21, a p53 target gene (Supplementary Figure 2L). Because Dusp6 is known to inhibit MEK activity (Lin et al., 2020), this result is consistent with our findings that MEKi increased p53 protein level. These observations highlight a complex, cell type-specific regulation of p53 activity.
2.6. 2i promotes mitochondria-independent production of reactive oxygen species
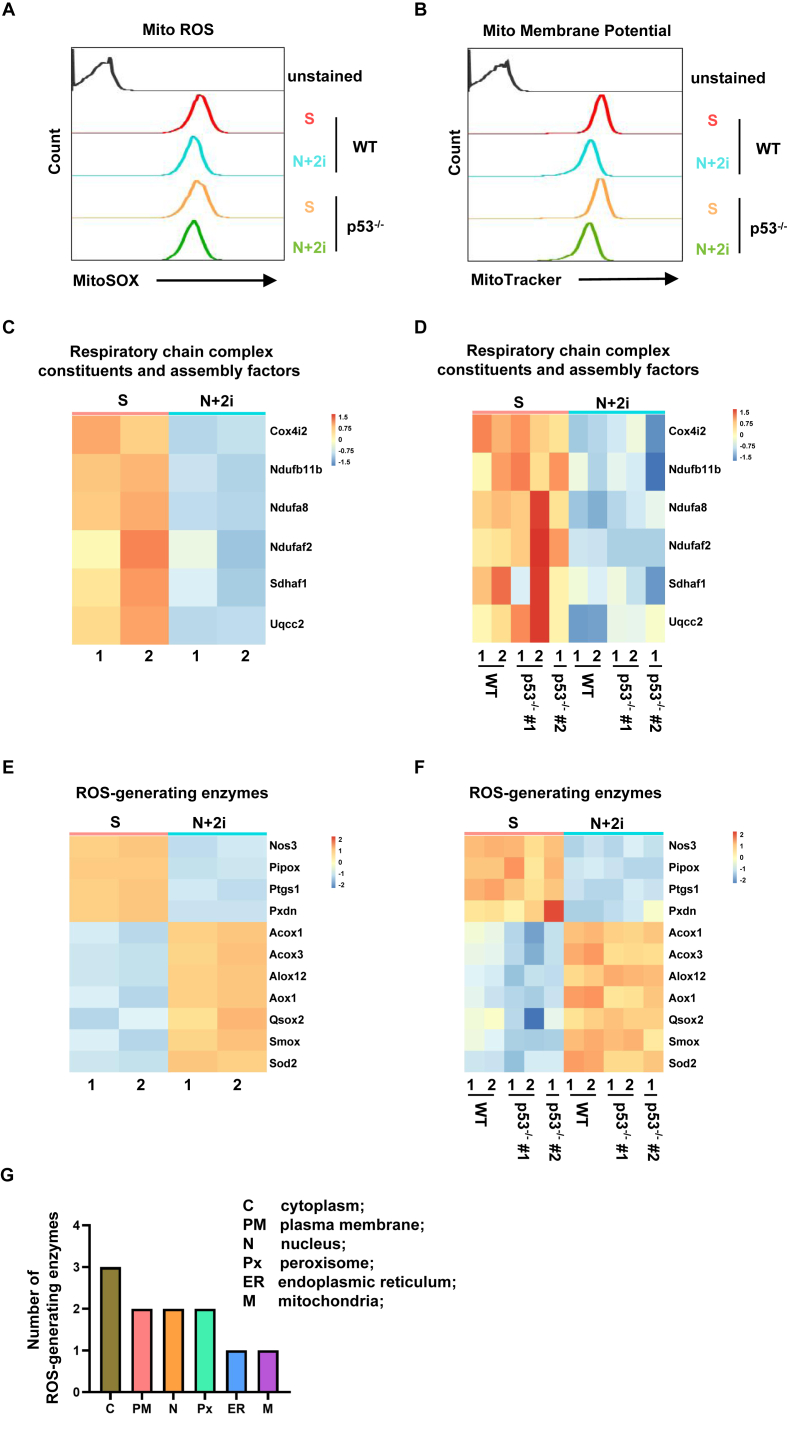
We investigated the cellular sources of 2i-induced ROS production. Mitochondrial electron transfer chain (ETC) complexes as well as over 50 oxidases localized in various cellular compartments can generate ROS (Forrester et al., 2018; Sies and Jones, 2020). To monitor mitochondrial ROS, wild-type and p53 knockout cells were stained with MitoSOX Red, followed by FACS analysis. Although total cellular ROS is increased in 2i condition (Figure 5), mitochondrial ROS level was slightly reduced in 2i compared to serum condition (Figure 6A). Concomitantly, mitochondrial membrane potential was moderately reduced in 2i condition (Figure 6B), indicating attenuated ETC activities. Consistently, transcriptomic analysis by RNA-seq revealed that multiple ETC constituents and assembly factors were downregulated in 2i condition (Figure 6C). These 2i-induced effects were independent of p53 (Figure 6A, B and D). These observations demonstrate that 2i-induced ROS production is independent of mitochondrial activity. We further queried the expression level of other oxidases known to generate ROS (Sies and Jones, 2020). Among more than 50 oxidases, 11 were differentially expressed in serum and 2i conditions. 7 out of 11 oxidases were upregulated in 2i (Figure 6E). 2i-induced differential expression of these oxidases was independent of p53 (Figure 6F), consistent with the notion that p53 activation is downstream of enhanced ROS production. Based on a recent analysis of ROS-generating enzymes (Sies and Jones, 2020), these 11 oxidases were distributed on various intracellular organelles and only 1 was on mitochondria (Figure 6G). Taken together, we concluded that 2i-induces mitochondria-independent ROS production by promoting the expression of cytosolic oxidases.
Figure 6.
2i promotes mitochondria-independent production of reactive oxygen species. (A) FACS analysis of mitochondrial ROS level in wild-type (WT) and p53 knockout cells (p53−/−) in serum (S) or 2i condition. (B) FACS analysis of mitochondrial membrane potential in wild-type (WT) and p53 knockout cells (p53−/−) in serum (S) or 2i condition. (C) Heatmap representation of differentially expressed genes, coding mitochondrial respiratory chain complex constituents and assembly factors, in serum and 2i conditions. (D) The effect of 2i-induced p53 on the expression of mitochondrial respiratory chain complex constituents and assembly factors as in (C). (E) Heatmap representation of differentially expressed genes, coding cellular oxidases capable of generation ROS, in serum and 2i conditions. (F) The effect of 2i-induced p53 on the expression of cellular oxidases as in (E). (G) Cellular localization of differentially expressed oxidases as in (E).
3. Discussion
ESCs are fast-proliferating cells, characterized by an extremely short G1 phase of the cell cycle, a feature long thought to be required for pluripotency maintenance. Nevertheless, the molecular basis for truncated G1 is not clear, and whether short G1 is coupled with pluripotency is under constant debate. On one hand, several studies show that differentiation signals primarily act in the G1 phase, support the notion that a short G1 phase is beneficial for pluripotency maintenance (Borghese et al., 2010; Pauklin and Vallier, 2013). On the other hand, ESCs can enter non-proliferating, dormant state without compromising pluripotency (Bulut-Karslioglu et al., 2016; Scognamiglio et al., 2016). Consistent with our results, other studies also find that the G1 phase is prolonged in 2i condition (Ter Huurne et al., 2017, 2020), and can be manipulated without compromising pluripotency (Li et al., 2012). Because 2i condition is regarded as the ground-state pluripotency of ESCs (Ying and Smith, 2017), the length of G1 phase seems to be an independent cellular event uncoupled with pluripotency. To reconcile these observations, we propose that the relationship between the length of G1 phase and ESC pluripotency maintenance is contextual dependent on the induction signal. When ESCs are shielded from differentiation signals such as in 2i condition, the length of G1 phase and pluripotency are separable cellular events. On the other hand, under strong differentiation signals, increased G1 phase renders ESCs more susceptible for differentiation. Under 2i, we find that increased G1 phase is caused by p53 activation, which in turn caused by the inhibition of MEK1/2 kinases (MEKi). p53 is a master regulator of the G1/S transition (Levine, 2020). Thus, the short G1 phase of primed ESCs is a manifestation of the intricate interplay between classic oncogenes MEK1/2 and tumor suppressor gene TP53 to promote the G1/S transition.
Optimal culturing systems of ESCs are prerequisites to fulfill their potential in regenerative medicine (Halliwell et al., 2020). An ideal growth medium should maintain pluripotency without compromising other aspects of ESCs. The discovery of “2i” culture condition, using chemical inhibitors of oncogenic MEK signaling to capture pluripotency in murine ESCs, is a landmark on this front (Ying et al., 2008). Subsequently, numerous studies have investigated chemical inhibitors of various signaling pathways in order to optimize pluripotency maintenance for both murine and human ESCs (Gafni et al., 2013; Ying and Smith, 2017). However, recent studies also demonstrate several pitfalls of this culture condition. Long-term culturing of ESCs in 2i paradoxically compromises their developmental potentials in vivo (Choi et al., 2017; Yagi et al., 2017). Our results showing that 2i activates p53 pathway may offer a potential explanation. The function of p53 in mammalian development has long been overshadowed by its prominent role as a tumor suppressor (Manfredi, 2019). Activated p53 pathway, when ESCs cultured in 2i/LIF conditions, may protect ESCs from DNA damage induced by Reactive Oxygen Species (ROS), and maintained genome integrity and stability. On the other hand, however, recent studies have demonstrated that even moderate activation of p53 activities during embryogenesis leads to a variety of developmental defects in mice (Bowen et al., 2019). Strikingly, p53 is activated in human diseases such as Treacher Collins syndrome, and genetic inactivation of p53 fully rescues the craniofacial phenotypes in murine models (Calo et al., 2018). Thus “2i” culture condition shields ESCs from differentiation, but at the same time seems to stress ESCs. In fact, several human ESC lines spontaneously inactivate TP53 to gain growth advantage (Merkle et al., 2017). We propose that the activation status of p53 should be taken into account for efforts to optimize ESC culturing medium. It will also be of interest to disentangle effectors downstream of MEK1/2 inhibited by 2i to identify how differentiation is blocked. Manipulation of these effectors may avoid the activation of p53.
4. Experimental procedures
4.1. ESC culture
R1/E (SCRC-1036) and ES-E14TG2a (CRL-1821) ESC cell lines were purchased from ATCC. Oct4-GFP ES cell line was previously described (Ohbo et al., 2003). For serum culture condition, all feeder-free murine ESC cell lines were cultured in Dulbecco’s modified Eagle’s medium, containing 15% ESC-grade fetal bovine serum (Gemini Bio-Products), supplemented with penicillin/streptomycin, 103 U/ml LIF (Millipore), 2 mM L-Glutamine, 100 μM 2-mercaptoethanol, and 100 μM nonessential amino acids (Invitrogen) in a 37 °C humidified incubator with 5% CO2. For 2i culture condition,ESCs were cultured in serum-free N2B27 supplemented with MEK inhibitor PD0325901 (1 μM), GSK3 inhibitor CH99021 (3 μM) and 103 U/ml LIF (Millipore), as previously described (Ying et al., 2008). Routine maintenance and alkaline phosphatase staining of ESCs were carried out as previously described (Dai et al., 2014). For cell differentiation assay, LIF was removed and 0.1 mM retinoic acid (Sigma) was added, cells were harvested at the indicated time points for the following analyses(Dai et al., 2014).
4.2. CRISPR/Cas9 knockout and shRNA knockdown cell lines
ESCs with stable knockdown of Tp53 was characterized previously (He et al., 2016). CRISPR/Cas9 was used to generate Tp53, p19ARF and Hoxc12 knockout cell lines. gRNAs were designed using the online tool (horizondiscovery.com), and listed in Supplementary Table. Cells were split at clonal density, and colonies were picked for expansion. Genomic DNA from individual colony was extracted using the GeneJET Genomic DNA Purification Kit (Thermo Scientific). The targeted region was PCR amplified and sequenced. Two independent p19ARF and Hoxc12 knockout cell lines were generated.
4.3. qPCR analysis
Total RNA was extracted by Trizol (Invitrogen) following the manufacturer’s instructions, treated with DNase I (Invitrogen), and reverse transcribed by random hexamers or oligo (dT) using M-MLV Reverse Transcriptase (Invitrogen). qPCR was performed by using EvaGreen Supermix (Bio-Rad) on a Bio-Rad CFX96 Real-Time PCR Detection System. Primer sequences were listed in the Supplementary Table. Expression levels of each gene were normalized to Gapdh mRNA and quantified as previously described (Peirson et al., 2003).
4.4. Immunoblotting
Cells were trypsinized and washed with PBS. Cell pellet were lysed in ice cold buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM KCl, 20 mM Tris [pH7.9], 0.5 mM EDTA, 10% glycerol, 1 mM DTT, 0.1% PMSF, EDTA-free complete protease inhibitor mixture (Roche) and 0.5% Nonidet P-40) for 15 min. Protein concentration was measured using BCA Protein assay (Thermo Scientific). Total cell lysates were mixed with SDS-PAGE sample buffer, followed by brief sonication and centrifugation at 13,000 g for 1 min. The antibodies used were listed in Supplementary Table.
4.5. Immunofluorescence
Cells were grown on glass slide, fixed with 4% paraformaldehyde for 15min, permeabilized with 0.3% Triton X-100 in PBS for 10 min, blocked in 3% BSA for 1hr, then incubated with anti-Oct4 and anti-Nanog overnight at 4 °C. After washing in PBS for 20min, cells were incubated with Alexa Fluor 568 goat anti-mouse IgG secondary antibodies for 1 h. For DNA visualization, DAPI was incubated with 0.5 mg/ml for 5min. Cells were observed by Olympus FV3000 confocal microscopy.
4.6. FACS analysis
Cells were incubated with EdU using BD Pharmingen EdU click proliferation kit (BD Biosciences), following manufacturer’s instruction, then stained with DAPI to analyze cell cycle profile. To detect intracellular ROS, mitochondrial ROS and mitochondrial membrane potential, 10 μM DCFH-DA, MitoSOX Red (Thermo Scientific) and MitoTracker (Thermo Scientific), respectively, were added in medium and incubated for 30 min at 37 °C, cells were trypsinized and washed 2 times with PBS. At least 10,000 events were collected by BD FACSCelesta, and analyzed by FlowJo V10 software.
4.7. RNA sequencing and data analysis
Three biological replicas for each condition were subjected to transcriptomic analysis. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. The resulting fastq file was quality controlled using FastQC v0.11.8, and adapters were trimmed using Trimmomatic v0.39. Paired-end clean reads were aligned to the reference genome (GRCm38.p6) using Hisat2 v2.0.5. featureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. Transcripts per kilobase of exon model per million mapped reads (TPM) was calculated based on the length of the gene and reads count mapped to this gene. Pearson correlation was >0.98 among biological replicas. Downstream analysis for differential gene expression of the raw count table was performed using the DESeq2 R package (1.16.1). The volcano plots of each group comparison were generated by R. Gene Set Enrichment Analysis (GSEA) was performed using the lists of hallmark gene sets from the Molecular Signature Database (MsigDB) using default parameters. Differential gene expression analysis was performed using the DESeq2 R package (1.16.1). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed. False discovery rate (FDR) < 0.05 and log2 fold change >1 were set as cutoff for differentially expressed genes.
4.8. Statistical analysis
Growth curve of ESCs were constructed by GraphPad Prism version 8.0.1. All data were shown as means ± SD, derived from at least three independent experiments. For all statistical tests (Student’s t test), P < 0.05 was considered statistically significant. In all figures, ∗ denotes P < 0.05, ∗∗ denotes P < 0.01, ∗∗∗ denotes P < 0.001 and ∗∗∗∗ denotes P < 0.0001.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Declarations
Author contribution statement
Peixuan Zhang and Ling Qin: Conceived and designed the experiments, Analyzed and interpreted the data; Wrote the paper.
Jinfeng Jiang and Tong Qiu: Performed the experiments.
Chao Yang: Analyzed and interpreted the data.
Yuan Yuan: Contributed reagents, materials, and analysis the data.
Funding statement
Dr Zhang Peixuan was supported by Sichuan Provincial Postdoctoral Science Foundation [0082204153037].
This work was supported by Key Technologies Research and Development Program [2017YFA0104200], Department of Science and Technology of Sichuan Province [2021YJ0012, 2020YFS0460, 2019YFH0081], National Natural Science Foundation of China [31971141, 81671115, 31800690], National Major Science and Technology Projects of China [2018ZX09733001], Sichuan Province Science and Technology Support Program [2019YFH0081].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This work was supported by Chinese Postdoctoral Science Foundation (0082204153037), National Natural Science Foundation of China (31971141, 81671115 and 31800690), National Key Research and Development Program (2017YFA0104200), National Major Scientific and Technological Special Project for Significant New Drugs Development (2018ZX09733001), Science and Technology Department of Sichuan Province (2021YJ0012, 2020YFS0460 and 2019YFH0081), Sichuan Science and Technology Program(2019YFH0081).
Appendix A. Supplementary data
The following are the supplementary data related to this article:
.
References
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A., Brustle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- Boward B., Wu T., Dalton S. Concise review: control of cell fate through cell cycle and pluripotency networks. Stem Cells. 2016;34:1427–1436. doi: 10.1002/stem.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen M.E., McClendon J., Long H.K., Sorayya A., Van Nostrand J.L., Wysocka J., Attardi L.D. The spatiotemporal pattern and intensity of p53 activation dictates phenotypic diversity in p53-driven developmental syndromes. Dev. Cell. 2019;50:212–228. doi: 10.1016/j.devcel.2019.05.015. e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., Biechele S., Jin H., Macrae T.A., Hejna M., Gertsenstein M., Song J.S., Ramalho-Santos M. Inhibition of mTOR induces a paused pluripotent state. Nature. 2016;540:119–123. doi: 10.1038/nature20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Gu B., Bowen M.E., Aryan F., Zalc A., Liang J., Flynn R.A., Swigut T., Chang H.Y., Attardi L.D., et al. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature. 2018;554:112–117. doi: 10.1038/nature25449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Choi J., Huebner A.J., Clement K., Walsh R.M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S.Y., et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Luan G., Deng L., Lei T., Kang H., Song X., Zhang Y., Xiao Z.X., Li Q. Primordial dwarfism gene maintains Lin28 expression to safeguard embryonic stem cells from premature differentiation. Cell Rep. 2014;7:735–746. doi: 10.1016/j.celrep.2014.03.053. [DOI] [PubMed] [Google Scholar]
- Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat. Rev. Mol. Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- Forrester S.J., Kikuchi D.S., Hernandes M.S., Xu Q., Griendling K.K. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018;122:877–902. doi: 10.1161/CIRCRESAHA.117.311401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A., et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Halliwell J., Barbaric I., Andrews P.W. Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat. Rev. Mol. Cell Biol. 2020;21:715–728. doi: 10.1038/s41580-020-00292-z. [DOI] [PubMed] [Google Scholar]
- He H., Wang C., Dai Q., Li F., Bergholz J., Li Z., Li Q., Xiao Z.X. p53 and p73 regulate apoptosis but not cell-cycle progression in mouse embryonic stem cells upon DNA damage and differentiation. Stem Cell Rep. 2016;7:1087–1098. doi: 10.1016/j.stemcr.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk A.A., Kim J.K., Tsang J.C., Ilicic T., Henriksson J., Natarajan K.N., Tuck A.C., Gao X., Buhler M., Liu P., et al. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- Levine A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer. 2020;20:471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- Li V.C., Ballabeni A., Kirschner M.W. Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12550–12555. doi: 10.1073/pnas.1206740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.K., Wu W., Ponce R.K., Kim J.W., Okimoto R.A. Negative MAPK-ERK regulation sustains CIC-DUX4 oncoprotein expression in undifferentiated sarcoma. Proc. Natl. Acad. Sci. U.S.A. 2020;117:20776–20784. doi: 10.1073/pnas.2009137117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi J.J. p53 and development: shedding light on an evolutionary enigma. Dev. Cell. 2019;50:128–129. doi: 10.1016/j.devcel.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Merkle F.T., Ghosh S., Kamitaki N., Mitchell J., Avior Y., Mello C., Kashin S., Mekhoubad S., Ilic D., Charlton M., et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature. 2017;545:229–233. doi: 10.1038/nature22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Descalzo S., Rue P., Garcia-Ojalvo J., Martinez Arias A. Correlations between the levels of Oct4 and Nanog as a signature for naive pluripotency in mouse embryonic stem cells. Stem Cells. 2012;30:2683–2691. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Ohbo K., Yoshida S., Ohmura M., Ohneda O., Ogawa T., Tsuchiya H., Kuwana T., Kehler J., Abe K., Scholer H.R., et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 2003;258:209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Peirson S.N., Butler J.N., Foster R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast A.M., Baumgartner D., Carnevalli L.S., Atzberger A., Haas S., et al. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell. 2016;164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- Ter Huurne M., Chappell J., Dalton S., Stunnenberg H.G. Distinct cell-cycle control in two different states of mouse pluripotency. Cell Stem Cell. 2017;21:449–455 e444. doi: 10.1016/j.stem.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Huurne M., Peng T., Yi G., van Mierlo G., Marks H., Stunnenberg H.G. Critical role for P53 in regulating the cell cycle of ground state embryonic stem cells. Stem Cell Rep. 2020;14:175–183. doi: 10.1016/j.stemcr.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Alexander P., McKnight S.L. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harbor Symp. Quant. Biol. 2011;76:183–193. doi: 10.1101/sqb.2011.76.010835. [DOI] [PubMed] [Google Scholar]
- Wu C.E., Koay T.S., Esfandiari A., Ho Y.H., Lovat P., Lunec J. ATM dependent DUSP6 modulation of p53 involved in synergistic targeting of MAPK and p53 pathways with trametinib and MDM2 inhibitors in cutaneous melanoma. Cancers. 2018;11 doi: 10.3390/cancers11010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Kishigami S., Tanaka A., Semi K., Mizutani E., Wakayama S., Wakayama T., Yamamoto T., Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Smith A. The art of capturing pluripotency: creating the right culture. Stem Cell Rep. 2017;8:1457–1464. doi: 10.1016/j.stemcr.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
Data Availability Statement
Data will be made available on request.