Abstract
Dihydrocapsaicin is the main bioactive component in Capsicum plants, which is widely used in China and India as a food drug and additive. In this study, the biotransformation of dihydrocapsaicin was performed using four cultivated human intestinal fungal strains in vitro. Eight metabolites, including seven previously undescribed metabolites (1 and 3−8) and one known analog (2), were obtained. Numerous spectroscopic data, such as NMR and HRESIMS, were collected to determine their structures. Based on the structures of the dihydrocapsaicin metabolites, the main biotransformation reactions were revealed to be hydroxylation, alcohol oxidation, and lactylation. In particular, the lactylation of hydroxyl groups is mainly mediated by Rhizopus oryzae R2701. In addition, metabolite 1 showed significant inhibitory effect on lysine-specific demethylase 1 (LSD1) (IC50 1.99 μM). Therefore, the biotransformation of dihydrocapsaicin by intestinal fungi afforded various derivatives, which were important resources for developing LSD1 inhibitors and potential application in cancer treatment.
Keywords: Dihydrocapsaicin, Biotransformation, Intestinal fungi, LSD1
Dihydrocapsaicin; Biotransformation; Intestinal fungi; LSD1.
1. Introduction
Lysine-specific demethylase 1 (LSD1) contributes significantly to mediate the genes expression involved in diseases, such as cancer, adipogenesis, inflammation, and cardiovascular diseases [1, 2, 3, 4, 5, 6, 7, 8]. Many tumors, such as glioblastoma [9], breast [3,10], prostate [11], lung [12], and gastric cancers [13], have been found to overexpress LSD1. To date, most developed LSD1 inhibitors are chemically synthesized, whereas few natural LSD1 inhibitors have been identified. However, bioactive natural products play a crucial role in developing novel drugs.
Dihydrocapsaicin and capsaicin are both known as the major constitutes with various biological activities in Capsicum plants, such as hot peppers [14]. Dihydrocapsaicin exerts anticancer activities against various malignant tumors, such as breast cancer, colorectal cancer, and glioma [15]. Recently, capsaicin and its metabolites were identified as an efficient LSD1 inhibitor [16,17]. In this study, we found that dihydrocapsaicin is also a natural LSD1 inhibitor with an inhibition of 76.5% at 10 μM, and the new discovery has attracted our interest because dihydrocapsaicin derived from natural product can be used for designing new LSD1 inhibitors.
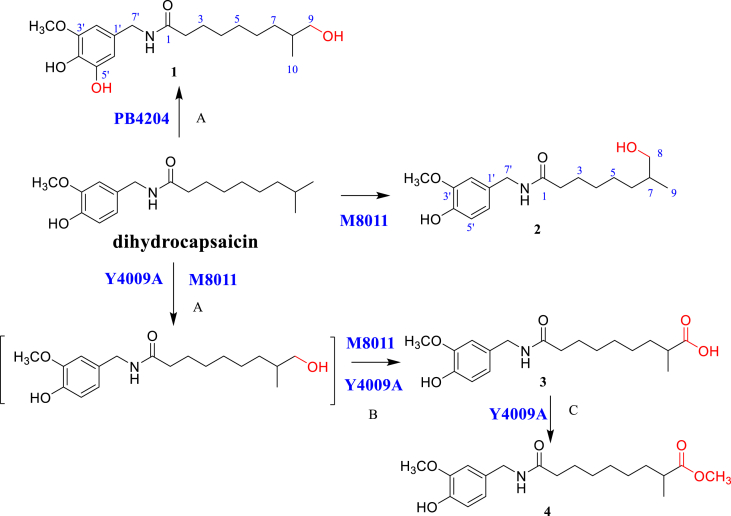
Biotransformation has been extensively studied because it has been identified as an effective technique for converting plentiful or inexpensive organic compounds into otherwise scarce or costly analogs [18]. Previous studies reported that biotransformation products of the capsaicin side chain have similar biological activities as those of capsaicin by reason of similar structures [14,19]. Therefore, to search for more bioactive LSD1 inhibitors, dihydrocapsaicin was biotransformed by human intestinal fungi, which led to the identification of eight metabolites, including 7 new metabolites (1 and 3−8) and one known analog (2) (Figures 1 and 2). In addition, metabolites (1−8) were evaluated for their inhibitory effect on LSD1. Notably, metabolite 1 showed significant inhibitory activity against LSD1, with an inhibition rate of 80.7%, which was stronger than that of dihydrocapsaicin. Furthermore, metabolite 1 showed strongest inhibitory effect on LSD1 (IC50 1.99 μM). Therefore, metabolite 1 could be used to develop biological agent to combat cancer cell migration and invasion. In this study, LSD1 was identified for the first time as a target of metabolite 1, which can provide a novel scaffold for further optimization of LSD1 inhibitor. Herein, we report details of the biotransformation procedures and hypothetical biotransformation pathway of dihydrocapsaicin and the isolation, structure elucidation, and inhibitory effect on LSD1 of the metabolites.
Figure 1.
Hypothetical biotransformation pathway of dihydrocapsaicin by Candida parapsilosis M8011, Aspergillus japonicus Y4009A, and Aspergillus fumigatus PB4204: (A) hydroxylation; (B) alcohol oxidation; (C) esterification.
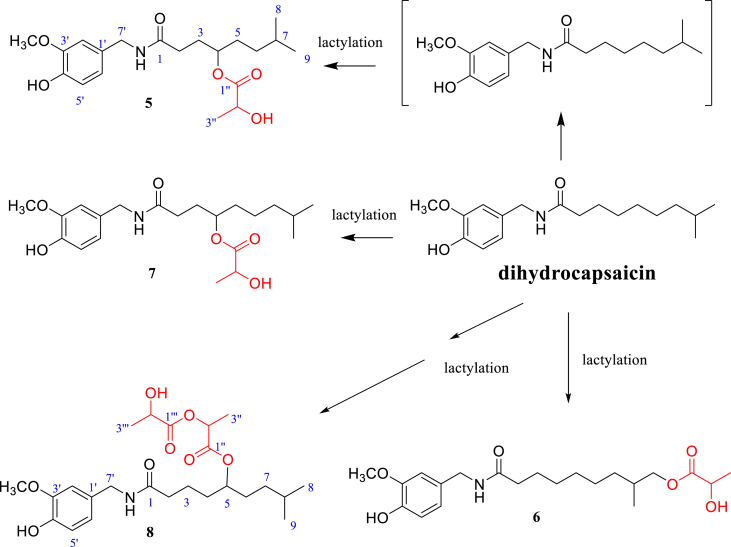
Figure 2.
Hypothetical biotransformation pathway of dihydrocapsaicin by Rhizopus oryzae R2701.
2. Materials and methods
2.1. Apparatus
Bruker 600 MHz spectrometer was used to acquire NMR spectra of the isolated metabolites. AB SciexX500r TOF mass spectrometer was used to acquire HRESIMS data. UltiMate 3000 instrument equipped with a DAD detector was used to collect analytical HPLC data. A liquid chromatography instrument equipped with an YMC RP C18 (250 × 20 mm, 5 μm) preparative column was used for the purification of metabolites. The separation of metabolites was performed in column chromatography using 200−300 mesh silica gel. TLC was carried out for the detection and analysis of metabolites using silica gel GF254 plates (2.5 × 7.5 cm, 5 × 10 cm). Acetonitrile and methanol (chromatographic-grade) were obtained from Sigma–Aldrich, and chemical grade solvents for all other solvents (Kemiou Chemical Reagent Co., Ltd.). Spots on silica gel plates were observed by spraying with a mixture of a 10% sulfuric acid and 95% ethanol solution followed by heating or the naked eye under UV light.
2.2. Microorganisms and culture media
In this research project, all human intestinal fungi were gifted by Professor Xiaochi Ma's laboratory (Dalian Medical University), which was approved by the Ethics Committee of Xinhua Hospital affiliated with Dalian University (XH2020A008).
Further, the gut fungal strains in this study were cultivated from human feces and identified as Candida parapsilosis (ID: M8011), Aspergillus fumigatus (ID: PB4204), Aspergillus japonicus (ID: Y4009A), and Rhizopus oryzae (ID: R2701), which were deposited at the Dalian Medical University. We performed all cultures and metabolism experiments on human intestinal fungi in potato medium. In general, the potato medium was prepared as the followed protocol: Peeled potato (200 g) was boiled in water for 30 min, filtered the solution, added 20 g of glucose, and then diluted to 1 L with water. Sterilization of potato media at 121 °C for 20 min was performed.
2.3. Biotransformation of dihydrocapsaicin by fungi
Four strains of the intestinal fungi were screened in order to evaluate their metabolic capability for dihydrocapsaicin.
The fungi were cultured in the potato medium (50 mL) was stored in Erlenmeyer flasks (100 mL), then the incubation was continued at 32 °C, 170 rpm for 48 h. The dihydrocapsaicin (2.0 mg) dissolved in DMSO (20 μL) was transferred to each flask, and then incubated for 5 days. The culture of fungi without dihydrocapsaicin was used as the blank group. After the co-incubation, the corresponding metabolites were extracted from the fungal fermentation broths with ethyl acetate, which was detected for the metabolites by HPLC in order to evaluate the metabolism of dihydrocapsaicin. On the basis of the screening experiments, the preparative experiments for the metabolites of dihydrocapsaicin were performed in 400 mL potato medium. The 15 mg of dihydrocapsaicin dissolved in 0.1 mL DMSO was added to the flasks, which were the pre-culture of fungi for 48 h. We extracted fermentation broths with ethyl acetate after coincubation for 5 days, and separated metabolites from the ethyl acetate extract.
2.4. Extraction and isolation of metabolites
In general, the fermentation broths were filtered and extracted by ethyl acetate (the same volume, three times). The ethyl acetate fraction was evaporated under vacuum at 45 °C.
Dihydrocapsaicin (200 mg) was biotransformed by Aspergillus fumigatus PB4204: The ethyl acetate extract (1.1 g) of fermentation was fractionated into 18 fractions by MPLC (ODS) using a gradient eluent consisting of MeOH–H2O (10:90 to 100:0) mixtures containing the trifluoroacetic acid (0.3‰, v/v). Fraction 12 was further separated by preparative HPLC to obtain 1 (83.0 mg) and the mobile phase was constituted by MeOH/H2O (45:55) containing the trifluoroacetic acid (0.3‰, v/v).
Dihydrocapsaicin (100 mg) was biotransformed by Candida parapsilosis M8011: The total ethyl acetate extract (0.5 g) of fermentation was separated into 15 fractions by MPLC (ODS) using a gradient eluent consisting of MeOH–H2O (10:90 to 100:0) mixtures containing the trifluoroacetic acid (0.3‰, v/v). Fraction 12 was separated by preparative HPLC to obtain 3 (8.0 mg) and the mobile phase was constituted by MeOH/H2O (45:55) containing the trifluoroacetic acid (0.3‰, v/v). Fraction 13 was separated by preparative HPLC to obtain 2 (10.5 mg) and the mobile phase was constituted by MeOH/H2O (65:35) containing the trifluoroacetic acid (0.3‰, v/v).
Dihydrocapsaicin (80 mg) was biotransformed by Aspergillus japonicus Y4009A: The ethyl acetate extract (0.6 g) of fermentation was fractionated into 22 fractions by MPLC (ODS) using a gradient eluent consisting of MeOH–H2O (10:90 to 100:0) mixtures containing the trifluoroacetic acid (0.3‰, v/v). Metabolite 3 (7.5 mg) was obtained from fraction 14 by the preparative HPLC, with the mobile phase as MeOH/H2O (44:55, 0.03% TFA v/v). Metabolite 4 (6.5 mg) was obtained from fraction 18 by the preparative HPLC, with the mobile phase as ACN/H2O (40:60, 0.03% TFA v/v).
Dihydrocapsaicin (300 mg) was biotransformed by Rhizopus oryzae R2701: The ethyl acetate extract (7.0 g) of the cultures of fungi was fractionated into 10 fractions by silica gel column chromatography using a gradient eluent consisting of petroleum ether−ethyl acetate (7:1 to 0:1) mixtures. Fraction 10 was separated by preparative HPLC, with mobile phase: ACN/H2O (60:40) containing the trifluoroacetic acid (0.3‰, v/v) to obtain 5 (4.4 mg), 6 (12.5 mg), 7 (11.0 mg), and 8 (3.5 mg).
2.4.1. 9,5′-dihydroxydihydrocapsaicin (1)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 338.1973 [M – H]– (calcd. for C18H28NO5–, 338.1973).
Table 1.
The 1H NMR (600 MHz) data in DMSO-d6 for metabolites 1 and 3−8. (δ in ppm, J in Hz).
| No. | 1 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|
| 2 | 2.10 t (7.8) | 2.10 t (7.8) | 2.09 t (7.2) | 2.12 m | 2.10 t (7.2) | 2.12 m | 2.11 t (6.6) |
| 3 | 1.51 m | 1.50 m | 1.50 m | 1.82 m 1.71 m |
1.51 m | 1.81 m 1.72 m |
1.51 m |
| 4 | 1.24 overlap | 1.23 overlap | 1.23 overlap | 4.77 m | 1.24 overlap | 4.81 m | 1.49 m |
| 5 | 1.24 overlap | 1.23 overlap | 1.23 overlap | 1.52 m | 1.24 overlap | 1.48 m | 4.79 m |
| 6 | 1.23 overlap | 1.22 overlap | 1.21 overlap | 1.12 m | 1.24 overlap | 1.24 m | 1.47 m |
| 7 | 1.34 m 0.98 m |
1.50 m 1.31 m |
1.51 m 1.35 m |
1.47 m | 1.30 m 1.09 m |
1.12 m | 1.09 m |
| 8 | 1.45 m | 2.29 m | 2.41 m | 0.85 d (6.6) | 1.72 m | 1.48m | 1.46 m |
| 9 | 3.24 dd (10.8,7.8) 3.15 dd (10.8, 7.8) |
0.86 d (6.6) | 3.95 dd (10.2, 6.0) 3.82 dd (10.2, 6.0) |
0.83 d (6.6) | 0.83 d (6.6) | ||
| 10 | 0.81 d (6.6) | 1.03 d (7.2) | 1.06 d (7.2) | 0.87 d (6.6) | 0.84 d (6.6) | 0.84 d (6.6) | |
| 2′ | 6.32 d (1.8) | 6.79 d (1.8) | 6.79 d (1.8) | 6.80 d (1.8) | 6.80 d (1.8) | 6.80 d (1.8) | 6.79 d (1.8) |
| 5′ | 6.69 d (8.4) | 6.69 d (7.8) | 6.69 d (8.4) | 6.69 d (7.8) | 6.69 d (7.8) | 6.69 d (7.8) | |
| 6′ | 6.31 d (1.8) | 6.62 dd (8.4,1.8) | 6.62 dd (7.8,1.8) | 6.62 dd (8.4,1.8) | 6.63 dd (7.8,1.8) | 6.63 dd (7.8,1.8) | 6.62 dd (7.8,1.8) |
| 7′ | 4.07 d (6.0) | 4.13 d (6.0) | 4.13 d (6.0) | 4.13 d (6.0) | 4.14 d (6.0) | 4.13 d (6.0) | 4.13 d (6.0) |
| 2″ | 4.10 q (7.2) | 4.14 q (6.6) | 4.10 q (6.6) | 4.99 q (6.6) | |||
| 3″ | 1.25 d (7.2) | 1.25 d (6.6) | 1.24 d (6.6) | 1.41 d (6.6) | |||
| 2‴ | 4.20 q (7.2) | ||||||
| 3‴ | 1.29 d (7.2) | ||||||
| OCH3-9 | 3.59 s | ||||||
| OCH3-3′ | 3.71 s | 3.73 s | 3.73 s | 3.74 s | 3.73 s | 3.74 s | 3.73 s |
| OH-4′ | 8.78 s | 8.80 s | 8.80 s | 8.80 s | 8.80 s | 8.80 s | 8.80 s |
| NH | 8.12 t (6.0) | 8.15 t (6.0) | 8.15 t (6.0) | 8.15 t (6.0) | 8.15 t (6.0) | 8.18 t (6.0) | 8.17 t (6.0) |
| COOH | 12.0 s |
Table 2.
The13C NMR (150 MHz) data in DMSO-d6 for metabolites 1 and 3−8. (δ in ppm).
| No. | 1 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|
| 1 | 171.9 | 171.9 | 171.9 | 171.2 | 171.9 | 171.2 | 171.5 |
| 2 | 35.4 | 35.3 | 35.3 | 31.1 | 35.4 | 31.1 | 34.9 |
| 3 | 25.4 | 25.3 | 25.3 | 29.5 | 25.3 | 29.6 | 21.1 |
| 4 | 28.8 | 28.7 | 28.6 | 73.5 | 28.6 | 73.1 | 33.0 |
| 5 | 29.2 | 28.6 | 28.5 | 31.2 | 29.0 | 33.5 | 74.8 |
| 6 | 26.4 | 26.6 | 26.5 | 33.8 | 26.1 | 22.4 | 31.2 |
| 7 | 32.9 | 33.2 | 33.2 | 27.3 | 32.6 | 38.1 | 33.6 |
| 8 | 35.3 | 38.7 | 38.6 | 22.3 | 32.1 | 27.3 | 27.3 |
| 9 | 66.3 | 177.5 | 176.3 | 22.5 | 68.5 | 22.4 | 22.2 |
| 10 | 16.8 | 17.0 | 16.9 | 16.5 | 22.4 | 22.4 | |
| 1′ | 129.7 | 130.5 | 130.5 | 130.3 | 130.5 | 130.3 | 130.4 |
| 2′ | 102.7 | 111.6 | 111.6 | 111.7 | 111.6 | 111.7 | 111.7 |
| 3′ | 148.2 | 147.4 | 147.4 | 147.4 | 147.4 | 147.4 | 147.4 |
| 4′ | 132.8 | 145.3 | 145.3 | 145.4 | 145.4 | 145.4 | 145.4 |
| 5′ | 145.6 | 115.2 | 115.1 | 115.2 | 115.2 | 115.2 | 115.1 |
| 6′ | 108.1 | 119.6 | 119.6 | 119.7 | 119.7 | 119.7 | 119.7 |
| 7′ | 42.0 | 41.8 | 41.8 | 41.9 | 41.8 | 41.9 | 41.8 |
| 1″ | 174.4 | 174.6 | 174.4 | 170.1 | |||
| 2 ″ | 66.0 | 65.9 | 66.0 | 68.5 | |||
| 3″ | 20.5 | 20.5 | 20.5 | 16.8 | |||
| 1‴ | 174.1 | ||||||
| 2‴ | 65.5 | ||||||
| 3‴ | 20.3 | ||||||
| OCH3-9 | 51.3 | ||||||
| OCH3-3′ | 55.7 | 55.5 | 55.5 | 55.5 | 55.5 | 55.5 | 55.5 |
2.4.2. 8-methyl N-vanillylcarbamoyloctanoic acid (3)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 336.1820 [M – H]– (calcd. for C18H26NO5–, 336.1816).
2.4.3. 8-methyl N-vanillylcarbamoyloctanoic acid methyl ester (4)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 350.1975 [M – H]– (calcd. for C19H28NO5–, 350.1973) and 396.2030 [M + COOH]– (calcd. for C20H30NO7–, 396.2028).
2.4.4. 4-lactyloxy-nordihydrocapsaicin (5)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 380.2078 [M – H]– (calcd. for C20H30NO6–, 380.2079) and 426.2140 [M + COOH]– (calcd. for C21H32NO8–, 426.2133).
2.4.5. 9-lactyloxy-dihydrocapsaicin (6)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 394.2242 [M – H]– (calcd. for C21H32NO6–, 394.2235).
2.4.6. 4-lactyloxy-dihydrocapsaicin (7)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 394.2232 [M – H]– (calcd. for C21H32NO6–, 394.2235) and 440.2303 [M + COOH]– (calcd. for C22H34NO8–, 440.2290).
2.4.7. 5-[2-(2-hydroxypropanoyloxy) propanoyloxy]-dihydrocapsaicin (8)
Colorless solid; 1H, and 13C NMR data were listed in Tables 1 and 2, respectively; HRESIMS, ion peak m/z 466.2453 [M – H]– (calcd. for C24H36NO8–, 466.2446).
2.5. Inhibitory effects on LSD1
The assay kit of LSD1 (Cayman Chemical) was used to evaluate the inhibitory effects of dihydrocapsaicin (DHC) and metabolites on LSD1. Generally, using fluorescence method to detect the production of H2O2 about the enzymatic reaction mediated LSD1. Resorufin (highly fluorescent compound) is produced by the reaction of H2O2 and ADHP (10-acety-3,7-dihydroxyphenoxazine) in the presence of horseradish peroxidase (HRP).
A solution of DMSO was used for dissolving all compounds, and their inhibition effects were measured at the final concentration of 10 μM. According to the operation method of assay kit, adding assay buffer (120 μL), LSD1 (20 μL), HRP (20 μL), fluorometic substance (10 μL) and inhibitor or DMSO (initial activity well) (10 μL) to three wells, after addition of peptide (10 μL), the reaction was performed at 37 °C for 30 min. The resorufin fluorescence was measured for the fluorescence intensity at emission wavelength (585−595 nm) and excitation wavelength (530−540 nm).
The percent inhibition was calculated as [(initial activity-sample)/initial activity] ×100. All experiments were carried out three times. Data are expressed as mean ± SD. Statistical analyses were performed by Student’s t-test.
Active compound solutions were suitably diluted (0.5, 1.0, 2.0, 5.0 and 10 μM) and their inhibition studies were determined. IC50 value of compound 1 was calculated using the CurveExpert 1.3 software.
3. Results and discussion
3.1. Identification of metabolites 1–8
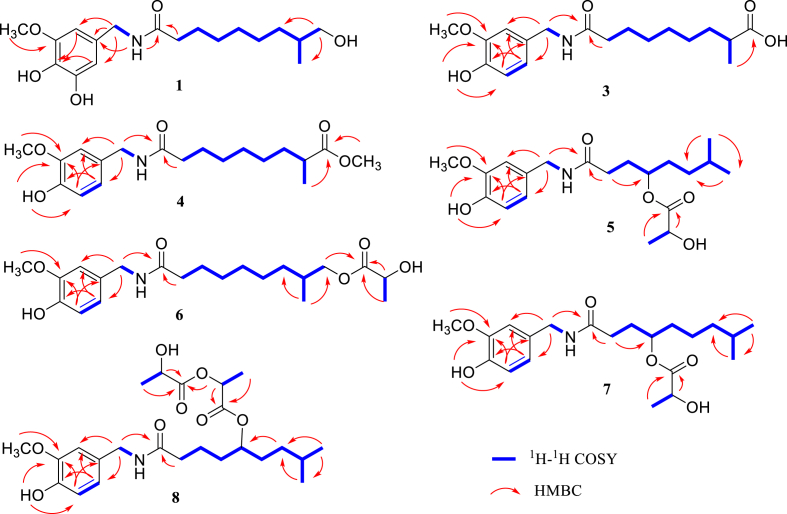
The molecular formula of 1 was found to be C18H29NO5 with 5 degrees of unsaturation by HRESIMS with the [M – H]– ion peak at m/z 338.1973 (calcd. for C18H28NO5–, 338.1973) in combination with the NMR data (Tables 1 and 2). The 1H NMR spectrum displayed signals attributable to one 3,4,5-trisubstituted phenyl moiety [δH 6.32 (d, J = 1.8 Hz, H-2′) and 6.31 (d, J = 1.8 Hz, H-6′)], one nitrogen-bearing methylene group [δH 4.07 (d, J = 6.0 Hz, H-7′)], one methoxy group [δH 3.71 (s, OCH3-3′)], one oxygen-bearing methylene group [δH 3.24 (dd, J = 10.8, 7.8 Hz, H-9a) and 3.15 (dd, J = 10.8, 7.8 Hz, H-9b)], one methylene group [δH 2.10 (t, J = 7.8 Hz, H-2)], and one methyl group [δH 0.81 (d, J = 6.6 Hz, H3-10)] (Table 1). Two exchangeable protons at δH 8.12 (t, J = 6.0 Hz, NH) and 8.78 (s, OH-4′) could be found in the 1H NMR spectrum (Table 1). In addition to carbon resonances corresponding to the above units, the 13 C NMR spectra displayed the presence of one nitrogen-bearing carbonyl carbon (δC 171.9, C-1) (Table 2). The NMR data showed that 1 was an analog of dihydrocapsaicin. Analysis of NMR spectra of 1 and dihydrocapsaicin demonstrated that the 4-hydroxy-3-methoxybenzyl moiety and methyl group (C-9) in dihydrocapsaicin were replaced by the 3,4-dihydroxy-5-methoxybenzyl unit and hydroxymethyl group (δC 66.3, C-9) in 1, respectively. This was confirmed by the HMBCs for OCH3-3′/C-3′, H-2′, H-6′/C-4′, H2-7′/C-1′, C-2′, and C-6′, and H2-9/C-7 and C-10 (Figure 3). Thus, 1 was determined to be 9,5′-dihydroxydihydrocapsaicin.
Figure 3.
Key HMBC (from H to C) and 1H–1H COSY correlations of metabolites 1 and 3−8.
Metabolite 3 was determined as molecular formula (C18H27NO5) by NMR and HRESIMS data. Comparison of the NMR data between 3 and dihydrocapsaicin suggested the difference was the substitution of the methyl unit (C-9) in dihydrocapsaicin by a carboxyl unit in 3. This was verified by 2D NMR data (Figure 3). The HMBCs for H3-10 and H-8/C-9, together with the chemical shift (δC 177.5, C-9), confirmed the above supposition (Figure 3). Finally, 3 was assigned as 8-methyl N-vanillylcarbamoyloctanoic acid.
Metabolite 4 was assigned molecular formula (C19H29NO5) by HRESIMS, which showed fourteen more mass units than 3. NMR spectra of 4 closely resembled those of 3, except for the presence of signals for a methoxy group. Comparison of the NMR data of 4 and 3 indicated that carboxyl carbon (C-9) was shielded by ΔδC −1.2 ppm. The other carbon resonances were shifted by ΔδC≤ ±0.2 ppm. These data suggested 4 was the methyl analog of 3. The key HMBCs for OCH3-9/C-9 proved that the methoxy group was connected to C-9 (Figure 3). Therefore, 4 was identified as 8-methyl N-vanillylcarbamoyloctanoic acid methyl ester.
The molecular formula (C20H31NO6) of metabolite 5 was found by the HRESIMS. Its NMR data were similar to nordihydrocapsaicin, except for the presence of signals for a carbonyl carbon (δC 174.4, C-1″), an oxygenated carbon (δC 66.0, C-2″), and a methyl carbon at (δC 20.5, C-3″). This indicated that 5 was a lactyloxy derivative of nordihydrocapsaicin, which was confirmed by the HMBCs for H-2″ and H3-3″/C-1″ and the 1H–1H COSY correlation of H-2″/H3-3″ (Figure 3). Although the HMBCs for H-5/C-1″ was not observed in 5, the molecular formula and chemical shift of C-4 (δC 73.5) suggested that the lactyloxy group must be connected to C-4. Therefore, 5 was identified as 4-lactyloxy-nordihydrocapsaicin.
The molecular formula (C21H33NO6) of metabolite 6 was found by HRESIMS. NMR data showed that 6 was a lactyloxy derivative of dihydrocapsaicin. This was verified based on the 2D NMR data, especially by the HMBCs for H2-9 and C-1″ (Figure 3). Thus, 6 was determined as 9-lactyloxy-dihydrocapsaicin.
The spectroscopic data of metabolite 7 showed that it was a lactyloxy derivative of dihydrocapsaicin with the molecular formula C21H33NO6. The HMBCs for H-4/C-1″ showed the lactyloxy unit was located at C-4 (Figure 3). Thus, 7 was identified as 4-lactyloxy-dihydrocapsaicin.
Metabolite 8 was assigned molecular formula (C24H37NO8) by HRESIMS. The HMBCs for H-2‴ and H3-3‴ /C-1‴, H-2″ and H3-3″ /C-1″, and H-2″/C-1‴, in combination with chemical shifts of these proton and carbon resonances, determined the presence of the 2-(2-hydroxypropanoyloxy) propanoyloxy group (Figure 3). Analysis of NMR spectra of 8 and dihydrocapsaicin demonstrated that 8 was a 2-(2-hydroxypropanoyloxy) propanoyloxy derivative of dihydrocapsaicin. Although the HMBC correlation of H-5/C-1″ of 8 was not observed, the molecular formula and chemical shift (C-5) confirmed the 2-(2-hydroxypropanoyloxy) propanoyloxy unit must be connected to C-5. Finally, 8 was determined to be 5-[2-(2-hydroxypropanoyloxy) propanoyloxy]-dihydrocapsaicin.
The remaining metabolite was identified as N-vanillyl-9-hydroxy-8-methyloctanamide (2) which had been reported [14].
3.2. LSD1 inhibition activity assay
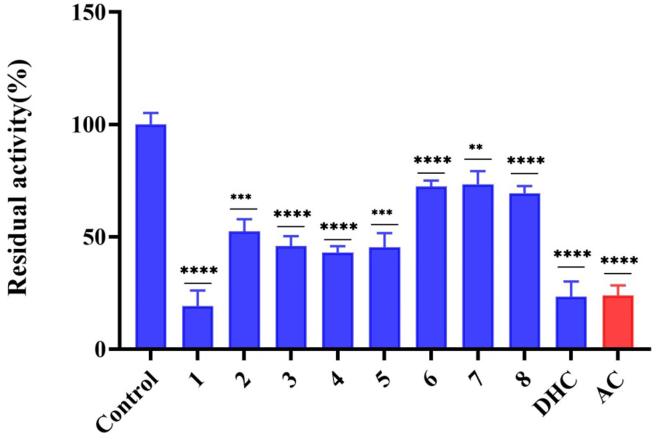
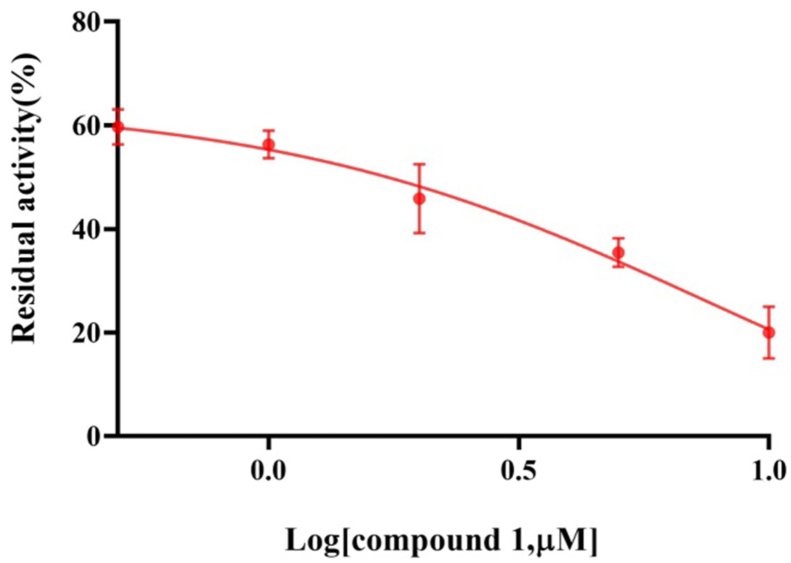
In this work, we evaluated the inhibitory effects of dihydrocapsaicin and metabolites 1−8 on LSD1 (Figure 4). As a result, the inhibitory effects of most of the metabolites against LSD1 were found to be in the range of 25–60% at 10 μM. Notably, metabolite 1 and dihydrocapsaicin displayed significant inhibitory effect on LSD1, with inhibition of 80.7% and 76.5% at 10 μM, respectively. Furthermore, metabolite 1 as the LSD1 inhibitor showed an IC50 value of 1.99 μM (Figure 5). Interestingly, metabolite 1 showed a higher LSD1 inhibition rate than dihydrocapsaicin, although they differed from each other; only C-9 and C-5′ in dihydrocapsaicin were both replaced by hydroxyl groups. However, it is unclear for the detailed mechanism of this result and need to be further studied.
Figure 4.
The inhibitory effects of metabolites 1−8, dihydrocapsaicin (DHC), and capsaicin (AC) on LSD1 at 10 μM. All experiments were carried out three times, and data were shown as mean ± standard deviation. ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001.
Figure 5.
The concentration-dependent inhibitory effect of metabolite 1 against LSD1.
As now, many reversible and irreversible LSD1inhibitors have been reported, but only a few of them are natural. For instance, baicalin was found to be the first LSD1 inhibitor (IC50 3.01 μM) [20], and α-mangostin was regarded as the first xanthone-based LSD1 inhibitor (IC50 2.81 μM) [21]. Notably, it has been reported capsaicin (IC50 0.6 μM) and capsaicin analogue (9,5′-dihydroxycapsaicin) (IC50 1.52 μM) both showed significant inhibitory effect on LSD1 [17]. In our study, the metabolite 1 showed similar inhibitory effect against LSD1 as 9,5′-dihydroxycapsaicin. These results suggest metabolite 1 may serve as a potential backbone for further development as an inhibitor of LSD1.
4. Conclusions
In the biotransformation of dihydrocapsaicin by four intestinal fungi (Aspergillus japonicus Y4009A, Rhizopus oryzae R2701, Candida parapsilosis M8011, and Aspergillus fumigatus PB4204), seven new metabolites (1 and 3−8) and one known analog (2) were afforded. The structures were unambiguously determined using NMR and HRESIMS spectra. Based on the structures of the dihydrocapsaicin metabolites, the main biotransformation reactions were revealed to be hydroxylation, alcohol oxidation, and lactylation. Metabolite 1 displayed strongest inhibitory effect on LSD1 (IC50 1.99 μM) in an in vitro bioassay. The above results indicated that biotransformation of dihydrocapsaicin by intestinal fungi was an effective method to produce LSD1 inhibitors for potential use as therapeutics in cancerous disease (glioblastoma, et al.).
Declarations
Author contribution statement
Xin He: Performed the experiments; Wrote the paper.
Baojing Zhang: Analyzed and interpreted the data.
Peng Cao; Honglei Wang; Shan Wu; Gang Wang; Fangyu Yang: Performed the experiments.
Aijing Leng; Guobiao Liang: Contributed reagents, materials, analysis tools or data.
Dawei Li: Conceived and designed the experiments.
Funding statement
Dr Dawei Li was supported by Natural Science Foundation of Liaoning Province; [2021-MS-283], Dalian Young Star of Science and Technology [2021RQ013].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Guobiao Liang, Email: liangguobiao6708@163.com.
Dawei Li, Email: lidw87@163.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Stazi G., Zwergel C., Valente S., Mai A. LSD1 inhibitors: a patent review (2010-2015) Expert Opin. Ther. Pat. 2016;26:565–580. doi: 10.1517/13543776.2016.1165209. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y., Vasilatos S.N., Boric L., Shaw P.G., Davidson N.E. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res. Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim S., Janzer A., Becker A., Zimmer A., Schüle R., Buettner R., Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 4.Liang Y., Vogel J.L., Narayanan A., Peng H., Kristie T.M. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakane N., Kwon H.S., Pagans S., Kaehlcke K., Mizusawa Y., Kamada M., Lassen K.G., Chan J., Greene W.C., Schnoelzer M., Ott M. Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1) PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musri M.M., Carmona M.C., Hanzu F.A., Kaliman P., Gomis R., Párrizas M. Histone demethylase LSD1 regulates adipogenesis. J. Biol. Chem. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janzer A., Lim S., Fronhoffs F., Niazy N., Buettner R., Kirfel J. Lysine-specific demethylase 1 (LSD1) and histone deacetylase 1 (HDAC1) synergistically repress proinflammatory cytokines and classical complement pathway components. Biochem. Biophys. Res. Commun. 2012;421:665–670. doi: 10.1016/j.bbrc.2012.04.057. [DOI] [PubMed] [Google Scholar]
- 8.Libby P., Ridker P.M., Hansson G.K. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 9.Mitra R., Ayyannan S.R. Role of lysine-specific demethylase 1 and its small molecule inhibitors in glioblastoma multiforme therapy. Anti Cancer Agents Med. Chem. 2022;22:3062–3085. doi: 10.2174/1871520622666220421092414. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 11.Cai C., He H.H., Chen S., Coleman I., Wang H., Fang Z., Chen S., Nelson P.S., Liu X.S., Brown M., Balk S.P. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:451–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi S., Ishikawa Y., Mizutani A., Iwasaki S., Matsumoto S., Kamada Y., Nomura T., Nakamura K. LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res. 2017;77:4652–4662. doi: 10.1158/0008-5472.CAN-16-3502. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Y.C., Duan Y.C., Ma J.L., Xu R.M., Zi X., Lv W.L., Wang M.M., Ye X.W., Zhu S., Mobley D., Zhu Y.Y., Wang J.W., Li J.F., Wang Z.R., Zhao W., Liu H.M. Triazole-dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J. Med. Chem. 2013;56 doi: 10.1021/jm401002r. 8543−8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M., Cho J.Y., Yu G.L., Lee H.J., Moon J.H. Bioconversion of capsaicin by Aspergillus oryzae. J. Agric. Food Chem. 2015;63:6102–6108. doi: 10.1021/acs.jafc.5b01730. [DOI] [PubMed] [Google Scholar]
- 15.Shi S., Li C., Zhang Y., Deng C., Liu W., Du J., Li Q., Ji Y., Guo L., Liu L., Hu H., Liu Y., Cui H. Dihydrocapsaicin inhibits cell proliferation and metastasis in melanoma via down-regulating β-catenin pathway. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.648052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Y., Wang Y., Huo X., Deng S., Jin L., Zhang H., Yu Z., Ning J., Ma X., Wang C. Microbial transformation of capsaicin by several human intestinal fungi and their inhibitory effects against lysine-specific demethylase 1. Phytochemistry. 2022;202 doi: 10.1016/j.phytochem.2022.113365. [DOI] [PubMed] [Google Scholar]
- 17.Jia G., Cang S., Ma P., Song Z. Capsaicin: a "hot" KDM1A/LSD1 inhibitor from peppers. Bioorg. Chem. 2020;103 doi: 10.1016/j.bioorg.2020.104161. [DOI] [PubMed] [Google Scholar]
- 18.Shimoda K., Ono T., Hamada H. Regioselective hydroxylation and dehydrogenation of capsaicin and dihydrocapsaicin by cultured cells of Phytolacca Americana. Biosci. Biotechnol. Biochem. 2021;85:103–107. doi: 10.1093/bbb/zbaa004. [DOI] [PubMed] [Google Scholar]
- 19.Onozaki H., Sasaoka K., Ezaki H. Hydrolytic degradation of capsaicin by Aspergillus niger and Aspergillus oryzae. J. Ferment. Technol. 1976;54:297–301. [Google Scholar]
- 20.Zheng Y.C., Shen D.D., Ren M., Liu X.Q., Wang Z.R., Liu Y., Zhang Q.N., Zhao L.J., Ma J.L., Yu B., Liu H.M. Baicalin, a natural LSD1 inhibitor. Bioorg. Chem. 2016;69:129–131. doi: 10.1016/j.bioorg.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Han C., Li Z., Hou J., Wang Z., Xu D., Xue G., Kong L. Bioactivity evaluation of natural product a-mangostin as a novel xanthone-based lysine-specific demethylase 1 inhibitor to against tumor metastasis. Bioorg. Chem. 2018;76:415–419. doi: 10.1016/j.bioorg.2017.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.