Summary
The emergence and spread of artemisinin-tolerant malaria parasites threatens malaria control programmes worldwide. Mutations in the propeller domain of the Kelch13 protein confer Plasmodium falciparum artemisinin resistance (ART-R). ART-R is linked to the reduced susceptibility of temporary growth-arrested ring-stage parasites, but the metabolic mechanisms remain elusive. We generated two PfKelch13 mutant lines via CRISPR-Cas9 gene editing which displayed a reduced susceptibility accompanied by an extended ring stage. The metabolome of ART-induced ring-stage growth arrest parasites carrying PfKelch13 mutations showed significant alterations in the tricarboxylic acid (TCA) cycle, glycolysis, and amino acids metabolism, pointing to altered energy and porphyrin metabolism with metabolic plasticity. The critical role of these pathways was further confirmed by altering metabolic flow or through chemical inhibition. Our findings uncover that the growth arrestment associated with ART-R is potentially attributed to the adaptative metabolic plasticity, indicating that the defined metabolic remodeling turns out to be the trigger for ART-R.
Subject areas: Multidrug resistant organisms, Parasitology
Graphical abstract

Highlights
-
•
Typical PfKelch13 mutant parasite lines confer to ART resistance have been constructed
-
•
Modified SLOPE protocol allows for the enrichment of early ring-stage parasites
-
•
Metabolomics screening points to altered energy metabolism with metabolic plasticity
-
•
Altering metabolic flow or chemical inhibition confirms the screening results
Multidrug resistant organisms; Parasitology
Introduction
Malaria remains one of the most important tropical diseases and causes significant morbidity and mortality, especially in sub-Saharan Africa. In 2020, the world health organization (WHO) reported 241 million cases of malaria worldwide, resulting in 627,000 deaths.1 Currently, the two main strategies for the control of malaria are vector control (long-lasting insecticide-treated nets and indoor residual spraying) and management of malaria cases (malaria rapid diagnostic tests and artemisinin-based combination therapies[ACTs]).2,3 First-line treatments for managing uncomplicated falciparum malaria, recommended globally by the WHO, are based on ACT, combining a potent but short-lived artemisinin (ART) derivative and a slower-acting companion drug that increases the effectiveness of treatment and reduces the risk of recrudescence and the development of resistance.4,5,6
Plasmodium falciparum parasites’ partial resistance/tolerance to ART (ART-R) has, however, emerged less than a decade ago following the use of ACT in Southeast Asia.7,8,9 Clinical and biological investigations have demonstrated that ART-R was associated with an increased number of ring stages capable of undergoing growth arrest.10,11 Mutations in the propeller domain of a kelch gene located on chromosome 13 (Pfkelch13), e.g., C580Y, Y493H, R539T, I543T, R561H, etc, are a major determinant of both in vitro and in vivo ART-R.12,13,14,15,16,17 To date, the exact mechanism by which mutations in Pfkelch13 enhance tolerance to ART remains controversial.18,19 P. falciparum susceptibility to ART is, however, closely associated with the altered temporal responses of parasites at various stages.20,21
Growth arresting or quiescent states are an effective survival mechanism in a wide range of microorganisms, plants, and insects. This response is triggered by environmental stress causing cell cycle development arrest in order to enhance survival when facing unfavorable conditions.22,23,24 So far, the clearest example of this phenomenon in malaria parasites is the hypnozoite stage of Plasmodium vivax and related species.25 Upon exposure to ART, synchronized P. falciparum parasites exhibit stage-dependent susceptibility profiles correlated with hemoglobin digestion.19,26 Interestingly, parasites at the early ring stage (2–4 h post invasion) displayed higher sensitivity in evaluation of the resistance level, which was developed as an in vitro ART assay, i.e., ring-stage survival assay (RSA0–3h).27,28
Previous studies have reported that P. falciparum early ring-stage parasites play a crucial role in the development of drug tolerance during the intraerythrocytic developmental cycle (IDC).11,29 Early ring-stage parasites are protected from dihydroartemisinin (DHA) by a temporary developmental arrest followed by a return to a normal developmental cycle when drug pressure is removed. However, the physiological state resulting from growth arrest along with the metabolic mechanism that leads to decreased ART susceptibility has not been fully characterized. We hypothesize that investigation of the metabolic profiles of these forms could aid understanding of how temporary ring-stage growth arrest is triggered and how that could drive the development of novel antimalarials. Although it has been previously demonstrated that parasites altered their transcriptomic and proteomic profiles after the DHA pulse,30 the metabolic alterations and the adaptative changes that Pfkelch13 mutant parasites undergo before developing into growth-arrested forms remain elusive.
Here, we present the results of experiments in which the metabolism-based growth-arrest profile of parasites with varying ART-R profiles were investigated. The in vitro DHA susceptibility phenotypes and IDC development profiles were evaluated in mutant parasite lines. As host erythrocyte contamination is known to significantly affect the reliability of metabolomic data, we developed a protocol for the generation of adequate early-stage parasite samples with sufficient quality and with reduced host cell contamination. Untargeted metabolomics screening was then performed, and a finely tuned adaptive metabolic mechanism was obtained for the first time by investigating the abundance of significantly changed metabolome corresponding to amino acid, energy, and porphyrin metabolism after DHA exposure. Taking together, we have identified that the growth arrestment-associated DHA tolerance is potentially attributed to the adaptative metabolic remolding for mutant parasite lines. We then assayed whether metabolic stimulation or chemical inhibition could protect parasites from the damage caused by DHA, indicating that the metabolic plasticity turns out to be critical for parasite survival when exposed to DHA pulse during IDC life cycle.
Results
In vitro DHA susceptibility of Pfkelch13 493H (3D7Y493H) and 580Y (3D7C580Y) gene-edited parasite lines
We generated two ART-R lines using CRISPR-Cas9i gene editing,31 through the introduction of two mutations, Y493H and C580Y, in the propeller domain of Pfkelch13 gene in the 3D7 strain (Figure S1A). To assess the in vitro susceptibility of Pfkelch13 493H (3D7Y493H) and 580Y (3D7C580Y) gene-edited parasite lines and control (3D7WT) to DHA, RSA0–3h was performed on three technical replicates. Survival rates of 3D7Y493H and 3D7C580Y parasite lines were significantly higher (5.8 and 10.9%, respectively, p < 10−3) compared to the 3D7WT control line (0.078%) (Figures 1A and 1B).
Figure 1.
Pfkelch13 3D7Y493H and 3D7C580Y mutant parasites exhibits reduced susceptibility to DHA and prolonged ring stage
(A) Alignment of the mutated region of Pfkelch13.
(B) 0-3 h ring-stage survival assay (RSA0–3h) determination for transfection control with the WT Pfkelch13 gene (3D7WT) and 3D7-Pfkelch13 engineered mutant lines (3D7Y493H and 3D7C580Y). Survival rates for each parasite line were calculated by dividing the number of surviving DHA-treated parasites by the number of DMSO-treated parasites and expressed in percentage. p value indicate statistical comparisons of survival rate between the control and mutant parasites by Holm-Bonferroni method adjusted two-sided t test (n = 3). Data are represented as mean ± SD.
(C) RSA0–3h50% assay was designed to evaluate the in vitro susceptibility of the 3D7WT and the mutant parasite lines against various DHA concentrations (ranging from 0.5 to 1000 nM). Tightly synchronized 0-3 h post-invasion parasites were subjected to 6 h exposure with varied DHA concentrations. The survival rates of the parasites under this treatment conditions were plotted against the drug concentration. The dose-response curve was fitted by sigmoidal model (n = 3). Data are represented as mean ± SD.
(D and E) Comparison of intraerythrocytic development patterns between Pfkelch13 WT and mutant parasites. Purified schizonts from tightly synchronized parasite lines were used to obtain early-ring-stage parasites. Blood smears were made every 6 h, and the percentage of each stage was counted in 100 parasites. Ring, trophozoite, and schizont stages are distinguished by different colors, while WT and mutant parasites were shown as continuous and dashed lines, respectively. ∗ (p value < 0.05) indicate statistical comparisons of developmental stages between WT and mutant parasites at selected time point (labeled by colored background) by Student’s t test (n = 3). Data are represented as mean ± SD.
(F) Microscopic examination of Giemsa-stained blood smears revealed the ring stage for transgenic strains was ∼6 h longer than WT, while prolonged ring stage along with shortened trophozoite stage contributed to a similar whole IDC life (Scale bar = 5 μM).
As the RSA0–3h reflects only the survival rates for parasites exposed to 700 nM DHA for 6 h, we extended the in vitro susceptibility profiles of the mutant parasite lines to various concentrations of DHA (from 0.5–1000 nM) and different exposure times (from 1–9 h). We then defined the RSA0–3h50% as the DHA concentration and the exposure time required to kill 50% of the viable parasites observed in the nonexposed cultures (DMSO). The 3D7Y493H and 3D7C580Y parasite lines displayed different dose-response curves. Upon 6 h exposure, 10-fold higher RSA0–3h50% survival rates (42.57 nM for 3D7Y493H and 53.51 nM for 3D7C580Y) were observed compared to the 3D7WT control line (4.92 nM) (Figures 1C and S1B). These DHA concentrations and exposure periods were then used in subsequent metabolomics experiments.
We next investigated the IDC profiles (from 0–3 h) of post-invasion ring-stages of 3D7WT, 3D7Y493H (Figure 1D), and 3D7C580Y (Figure 1E) in the absence of DHA. Generally, the developmental stage was based on the overall size of parasites, including the ratios of the area and diameter of the nucleus to those of the cytoplasm, and the amount of visible pigment and mature nuclei after Giemsa staining. Specifically, for late rings (∼18 h) and early trophozoites (∼24 h), we discriminate them from each other according to the appearance of pigment in brown as well as the filling degree of cytoplasm.
The total length of the IDC of each parasite line was approximately 48 h (from rings to trophozoites and schizonts). While the length of the whole IDC remained unchanged, the progression of 3D7Y493H and 3D7C580Y ring stages was delayed by ∼6 h compared to 3D7WT, and the subsequent trophozoite-stage duration was shortened (Figure 1F). The ring to trophozoite transition for 3D7Y493H and 3D7C580Y occurred at ∼24 h and ∼3 h later than 3D7WT (estimated at ∼21 h), respectively. These altered IDC profiles of the 3D7Y493H and 3D7C580Y mutant parasites were observed in three independent biological replicates in the absence of drug treatment.
We then evaluated in vitro IDC growth up to 72 h under different concentrations of DHA. The proportions of the parasite stages were monitored by microscopic examination of Giemsa-stained smears. Parasites were categorized into rings, trophozoites/schizonts, or growth-arrested/dead forms at each time point. Ring-stage development was defined during the first 18 h with cytoplasm occurring in a ring shape with little to no hemozoin visible. In the following 12 h, parasites became increasingly enlarged and dense with clearly observable hemozoin deposition and accumulation. Thereafter, with the progressive accumulation of hemozoin, divided nuclei could be observed at 36 h, and hemozoin was into a granulate form before mature merozoites developed at ∼46 h.
In addition to these morphologically normal parasites, growth-arrested parasites displayed a rounded morphology with retained cytoplasm and chromatin which differed from those of dead forms. Following administration of 700 nM DHA for 6 h, only a small portion (∼10%) of morphologically normal 3D7WT ring stages were observed up to 24 h while normal 3D7Y493H and 3D7C580Y ring and mature stages were detected up to 72 h, indicating an increased capacity to withstand DHA (Figure 2A). Likewise, following administration of 50 nM DHA for 6 h, normal 3D7Y493H and 3D7C580Y ring and mature stages were seen across the IDC development, and this was not observed in the 3D7WT control line (Figure 2B). Overall, under both conditions, the 3D7Y493H and 3D7C580Y parasites were less susceptible to DHA exposure.
Figure 2.
Delayed clearance, growth retardation, and recrudescence from DHA-induced growth arrest of Pfkelch13-mutated parasite
Percentage of parasites (n = 100) with different morphology after DHA exposure was monitored by microscopic examination. Parasites were morphologically categorized into ring, trophozoite/schizont, or growth-arrested/dead form at each time point. Stage distributions for parasites after vehicle or DHA treatment at 72 h.
(A) After treatment of 700 nM DHA for 6 h, no variable parasite in 3D7WT was observed in the second life cycle whereas both ring- and mature-stage parasites were observed for 3D7Y493H and 3D7C580Y, respectively. Data are represented as mean ± SD.
(B) Following the exposure of 50 nM DHA for 6 h, DHA-resistant lines indicated a delayed IDC with more variable parasites than 3D7WT. Data are represented as mean ± SD.
(C) Morphological analysis of prolonged IDC development for 3D7Y493H and 3D7C580Y parasites after 6-h exposure of DHA exposure, and the insert indicated the representative images of dead, growth-arrested, and active forms of parasites. (Scale bar = 5 μM).
(D) Changes in the SYTO-61 staining profile of viable parasites after drug exposure for 88h. WT and 3D7 mutant parasites were subjected to 6-h DHA pulses at 50 nM and SYTO-61 signals were measured by flow cytometry when DMSO-treated parasite had progressed into mid-late stage (∼40h) in the next cycle. Asterisks indicate an absolute increase in the number of early-stage parasites due to growth arrestment.
To further quantify drug-induced growth retardation during the ring to trophozoite transition of synchronized parasites (0–3 hpi) exposed to 50 nM DHA for 6 h, we monitored the morphology of the parasite stages at every 6 h over 60 h. Compared to the 3D7WT control line, we observed a higher proportion of 3D7Y493H and 3D7C580Y parasites with rounded cytoplasm and condensed chromatin (defined as growth-arrested parasites), in addition to pyknotic forms with collapsed nuclei (dead parasites) (Figure 2C).
For 3D7Y493H and 3D7C580Y parasite lines stained with SYTO-61, corrected SYTO-61 signals referred to the median value of the viable parasite population. Examination of the population profile of SYTO-61 staining by flow cytometry indicated an absolute increase in the number of viable parasites for mutant parasites in the subsequent IDC, which was closely associated with growth retardation upon DHA exposure. (Figure 2D).
Ring-stage parasites enrichment
Next, we have improved a protocol to enrich growth-arrested ring-stage parasites for metabolomic studies based on previously reported research.32 The protocol was based on streptolysin O (SLO), a pore-forming exotoxin that can selectively lyse uninfected erythrocytes. Lysis efficiency was assessed by measuring the proportions of infected and uninfected erythrocytes in bulk cultures using a range of SLO concentrations. Lysis of uninfected erythrocytes was effective at 875 U of SLO (95.9 ± 4.1%, 92.0 ± 2.9%, 92.4 ± 2.6% for 3D7WT, 3D7Y493H, and 3D7C580Y parasites, respectively) (Figures S2A and S2B). Next, we improved the protocol by using 70% Percoll (v/v) to separate intact and lysed erythrocytes. By using this approach (streptolysin O+Percoll, named SLOPE), we obtained parasitemia for ring-stage parasites reaching 38.1 ± 1.9%, 36.4 ± 2.4%, 36.8 ± 3.1% for the 3D7WT, 3D7Y493H, and 3D7C580Y parasite lines, respectively, resulting in a ∼7-fold increase in ring-stage parasites and reduced host cell contamination. We later confirmed the removal of ghost cells by immunofluorescence (Figures S2D–S2F). Briefly, confocal microscopy using a set of antibodies (anti-CD235a, an external surface protein; anti-spectrin, an intracell protein only accessible in lysed erythrocytes) and Hoechst dye (parasite DNA staining) was performed to detect intact erythrocytes (infected and uninfected) from ghosts. The separation was achieved using the SLOPE approach, resulting in 50% intact cells with significantly increased rings. Less than 5% of cells were ghost cells according to quantification using acquired images.
Finally, we determined the impact of enrichment on viability and metabolic activity of enriched ring-stage parasites by measuring growth curve and morphological examination. We observed no significant changes in growth rates or parasite morphology between enriched and untreated parasites, indicating the feasibility of using this method to perform metabolomics analysis, as described below (Figure S2C).
Metabolomic profile of enriched parasites
To investigate whether the SLOPE method could separate parasite-specific signals from host contamination, enriched parasites were harvested and analyzed. The metabolomics profiles of untreated control and saponin-lysed parasites were almost indistinguishable in a principal-component analysis (PCA) score plot (Figures 3B–3D), suggesting relatively poor enrichment and/or a low discriminatory power for the separation of parasite-specific signals and background host contamination. In contrast, the SLOPE-pretreated preparations showed a significantly reduced ghost cell signal, consistent with the data obtained from immunofluorescence assay. SLOPE pretreatment clearly differentiated metabolomics profiles from treated and untreated parasites. First, partial least squares discriminant analysis (PLS-DA) score plot revealed distinct separations between groups without over fitting (Figure S3) and clearly separated metabolomics profiles from ring-stage samples. Second, we observed a significant reduction in the relative intensity of three representative host-derived metabolites (cytidine 5’-diphosphoethanolamine, Valylleucine, and hypoxanthine) in SLOPE-treated parasites (Figures 3E–3G).
Figure 3.
Metabolic profile of parasites with different treatment by untargeted metabolomics strategy
(A) Schematic workflow for the metabolomics study in which sample collection, ring-stage parasite enrichment and data acquisition are illustrated. Highly synchronized Pfkelch13 WT and mutant parasite lines were subjected to either 50 nM DHA or 0.1% DMSO exposure for 6 h followed by thorough wash to remove remaining DHA. Cultures were treated with SLO to preferentially lyse uninfected erythrocytes and then layered on Percoll gradient to separate lysed ghosts from intact cells by centrifugation. Followed by metabolites quenching and extraction, the metabolic basis of artemisinin-induced growth-arrested parasites was investigated by through high-resolution MS-based untargeted metabolomics approach.
(B–D) PCA scatterplot for acquired metabolomes in 3D7WT, 3D7Y493H, and 3D7C580Y parasites. The metabolic profiles of untreated control and saponin lysed parasites were hardly distinguished because of ghost contamination, while enriched ring-stage parasites treated by streptolysin-O or constructed methods showed a clear different metabolic profile (n = 5).
(E–G) Relative signal of three representative compounds which was able to reflect the host metabolic effect on classifier accuracy. Like above-acquired metabolic profile, host-derived metabolic signal was significantly reduced after sample pretreatment, indicating the feasibility of this method to reduce host contamination as well as obtain parasite-specific metabolome which was favorable for following metabolomics analysis. p value indicates statistical comparisons of the relative abundance between the control and mutant parasites by Holm-Bonferroni method adjusted two-sided t test (n = 5). Data are represented as mean ± SD.
Untargeted metabolomic profiles of parasites following DHA treatment
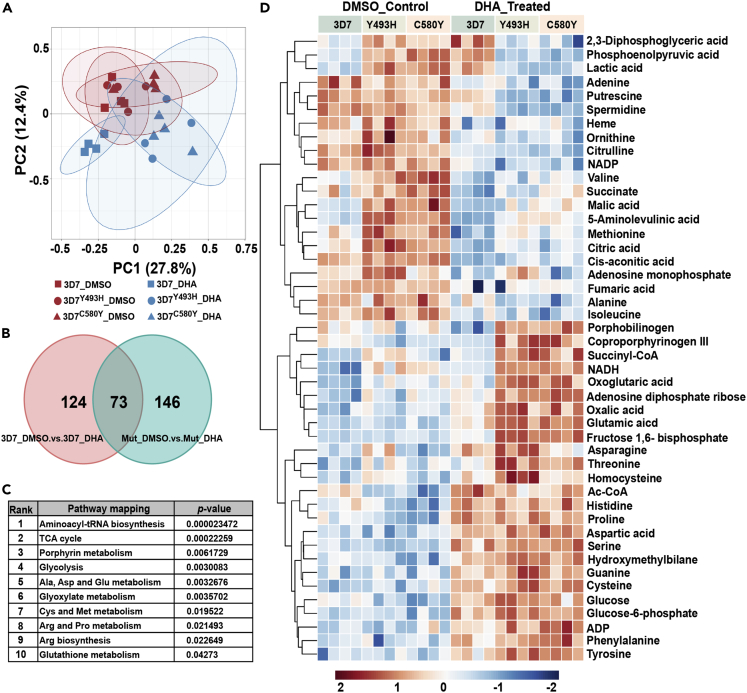
Next, we applied the SLOPE protocol to define the metabolomic profile of ring-stage parasites exposed to 50 nM DHA for 6 h according to the schematic workflow (Figure 3A). At this drug concentration, either viable subpopulation which were still positive by the RSA0–3h in contrast to the 3D7WT strain (Figure 1C) or pyknotic parasites at ring stage were enriched. The untargeted metabolomics profiles revealed a total of 1,000 metabolite features of which 920 were included in subsequent analyses. A distinct separation pattern of four clusters was noticed, indicating specific metabolomics changes induced by DHA treatment (Figure 4A). The changes induced by DHA were detected by conducting a supervised PLS-DA model including a “variable important in projection” (VIP) larger than 1.5. A comparison of the metabolomics profiles measured by univariate analysis of variance from DHA-exposed and nonexposed parasites identified 124 and 146 metabolites (adjusted p value <0.05) in the wild-type (WT, 3D7WT) and mutant parasite lines (3D7Y493H and 3D7C580Y), respectively. As shown in Table 1, the metabolomics changes induced by DHA were deduced from compounds shared between exposed and nonexposed cultures and included the annotations of the most significant metabolites (Figure 4B). Overall, overlapped metabolic profile indicated that amino acid metabolism-, porphyrin metabolism-, and energy metabolism-related pathways belonged to highly ranked pathways (Figure 4C).
Figure 4.
Untargeted metabolomics screening of Pfkelch13-associated metabolome after DHA exposure and the schematic workflow is listed in Figure 3.
PCA (A) scatterplot for acquired metabolic profile in vehicle (n = 4 for each group) or DHA-treated (n = 4 for each group)3D7WT, 3D7Y493H, and 3D7C580Y parasites. Clearly separated clusters of DHA-resistant parasite indicated distinguishable metabolic pattern compared with 3D7WT whereas that was similar at initial state without DHA treatment. Besides, permutation tests have validated the robustness of multivariate statistical model without over fitting. (B) Venn diagram showing significant overlap of 73 significantly alerted metabolites in pairwise comparisons between the WT and mutant lines after DHA pulse. (C) Pathway enrichment analysis of the DHA-related metabolome. Pathway enrichment analysis has been performed using a subset of metabolites listed in Table 1, indicating that amino acid metabolism, porphyrin metabolism, and energy metabolism related pathways belonged to high ranked pathways. Using a two-sided t test adjusted for multiple comparisons using the Holm-Bonferroni method (D) Heatmap of significantly changed metabolites from enriched metabolic pathways. Each line represents a detected metabolite and data were scaled and depicted on a log2 scale. Columns represent each biological replicate from four independent experiments with/without DHA exposure. Overall, metabolite abundances in DHA-treated parasites showed metabolic alterations in Pfkelch13 WT and mutant parasite strains.
Table 1.
Different metabolites between DHA-exposed and nonexposed parasites
| Upregulated metabolites | ||||||||
|---|---|---|---|---|---|---|---|---|
| No | Metabolite | m/z (amu) | RTa (min) | VIPb | 3D7WT DMSO vs DHA |
3D7Mut DMSO vs DHA |
||
| FCc | adjusted p value | FC | adjusted p value | |||||
| 1 | Glutamic acid | 147.0533 | 1.39 | 1.56 | 1.515 | 0.0419 | 2.067 | 1.73E-07 |
| 2 | Fructose 1,6- bisphosphate | 339.9952 | 1.11 | 1.59 | 0.647 | 0.0040 | 2.414 | 3.63E-07 |
| 3 | Serine | 105.0425 | 1.28 | 1.68 | 2.111 | 0.0279 | 2.631 | 4.81E-07 |
| 4 | N-Acetyl-D-Glucosamine 6-Phosphate | 301.0563 | 10.04 | 1.72 | 1.391 | 0.0781 | 1.668 | 6.98E-07 |
| 5 | Hydroxymethylbilane | 854.2853 | 1.27 | 1.61 | 0.771 | 0.0450 | 1.743 | 7.44E-07 |
| 6 | Tyrosine | 181.0737 | 5.16 | 1.70 | 1.469 | 0.0274 | 1.810 | 1.90E-06 |
| 7 | Glucose 6-phosphate | 260.0299 | 10.19 | 1.55 | 1.601 | 0.0422 | 1.828 | 2.09E-06 |
| 8 | Uridine diphosphate-N-acetylglucosamine | 607.0817 | 14.80 | 1.60 | 1.374 | 0.0144 | 2.131 | 2.35E-06 |
| 9 | Glycerophosphocholine | 257.1028 | 12.99 | 1.53 | 1.677 | 0.0209 | 2.126 | 4.37E-06 |
| 10 | Adenosine diphosphate ribose | 559.0717 | 1.43 | 2.01 | 1.697 | 0.0097 | 2.075 | 5.65E-06 |
| 11 | Aspartic acid | 133.0351 | 1.20 | 1.67 | 1.332 | 0.0019 | 1.532 | 5.84E-06 |
| 12 | Inosine | 268.2263 | 13.74 | 1.75 | 1.742 | 0.0143 | 2.217 | 6.78E-06 |
| 13 | Cysteine | 121.0195 | 4.52 | 1.81 | 1.307 | 0.0033 | 1.965 | 1.24E-05 |
| 14 | Glyoxylic acid | 74.0003 | 8.12 | 1.88 | 0.702 | 0.0219 | 1.916 | 1.36E-05 |
| 15 | Glucose | 180.0634 | 9.43 | 1.95 | 1.465 | 0.0294 | 2.698 | 1.56E-05 |
| 16 | 3-Methyl-2-oxovaleric acid | 130.0628 | 12.74 | 1.56 | 1.282 | 0.0354 | 1.989 | 2.19E-05 |
| 17 | α-Ketoisovaleric acid | 116.0471 | 12.48 | 1.52 | 1.153 | 0.0473 | 1.616 | 2.28E-05 |
| 18 | Proline | 115.0632 | 12.74 | 1.51 | 1.285 | 0.0180 | 1.444 | 2.48E-05 |
| 19 | Putrescine | 88.1002 | 1.39 | 1.64 | 0.768 | 0.0402 | 1.434 | 2.62E-05 |
| 20 | Adenosine monophosphate | 347.0631 | 1.41 | 1.78 | 1.472 | 0.0481 | 1.842 | 2.77E-05 |
| 21 | Pantothenic Acid | 219.1117 | 6.21 | 1.50 | 1.598 | 0.0247 | 2.123 | 3.47E-05 |
| 22 | Citrulline | 175.0955 | 2.10 | 1.64 | 1.239 | 0.0492 | 1.707 | 4.09E-05 |
| 23 | Coproporphyrinogen III | 660.3158 | 8.83 | 1.87 | 0.709 | 0.0038 | 2.484 | 4.20E-05 |
| 24 | GDP-L-fucose | 589.0818 | 1.26 | 1.77 | 1.502 | 0.0155 | 1.795 | 4.55E-05 |
| 25 | Phenylalanine | 165.0788 | 9.41 | 1.54 | 1.612 | 0.0402 | 2.151 | 5.42E-05 |
| 26 | L-methionine | 149.0512 | 8.77 | 1.64 | 0.570 | 0.0234 | 1.673 | 6.24E-05 |
| 27 | Ac-CoA | 809.1251 | 8.77 | 1.53 | 1.703 | 0.0194 | 2.379 | 6.99E-05 |
| 28 | CDP-ethanolamine | 446.0611 | 8.99 | 1.52 | 1.900 | 0.0110 | 2.565 | 7.54E-05 |
| 29 | Histidine | 155.0693 | 12.15 | 1.71 | 1.468 | 0.0537 | 1.988 | 9.35E-05 |
| 30 | Guanine | 151.0493 | 9.50 | 1.63 | 1.650 | 0.0134 | 1.970 | 1.09E-04 |
| 31 | ADP | 427.0294 | 1.43 | 1.83 | 1.251 | 0.0104 | 1.573 | 1.13E-04 |
| 32 | Oxalic acid | 89.9952 | 1.71 | 1.50 | 1.254 | 0.0324 | 1.937 | 1.17E-04 |
| 33 | Galactosylsphingosine | 461.3360 | 14.79 | 1.61 | 1.711 | 0.0240 | 3.102 | 1.42E-04 |
| 34 | NADH | 665.1244 | 14.88 | 1.65 | 2.192 | 0.0030 | 1.907 | 2.01E-04 |
| 35 | Threonine | 119.0582 | 9.98 | 1.51 | 1.289 | 0.0431 | 1.465 | 3.26E-04 |
| 36 | Malic acid | 134.0215 | 12.65 | 1.78 | 1.147 | 0.0019 | 1.569 | 3.87E-04 |
| 37 | Porphobilinogen | 226.0958 | 6.73 | 1.95 | 0.506 | 0.0109 | 1.612 | 4.11E-04 |
| 38 | dCMP | 307.0554 | 8.46 | 1.64 | 1.823 | 0.0158 | 1.818 | 8.48E-04 |
| 39 | Succinyl-CoA | 867.1294 | 14.81 | 1.87 | 1.372 | 0.0401 | 2.228 | 9.18E-04 |
| 40 | Dodecanedioic acid | 294.1859 | 13.47 | 1.60 | 1.381 | 0.0311 | 1.803 | 0.0011 |
| 41 | Succinyladenosine | 383.1076 | 6.44 | 1.77 | 1.615 | 0.0020 | 1.751 | 0.0029 |
| 42 | Homocysteine | 135.0354 | 1.17 | 1.53 | 1.510 | 0.0363 | 2.023 | 0.0067 |
| 43 | Asparagine | 132.0535 | 12.65 | 1.63 | 1.296 | 0.0068 | 1.725 | 0.0205 |
| 44 | α-Ketoglutaric acid | 146.021545 | 12.644 | 1.88 | 1.255 | 0.0361 | 1.553 | 0.0434 |
| Downregulated metabolites | ||||||||
|---|---|---|---|---|---|---|---|---|
| No | Metabolite | m/z (amu) | RTa (min) | VIPb | 3D7WT DMSO vs DHA |
3D7Mut DMSO vs DHA |
||
| FCc | adjusted p value | FC | adjusted p value | |||||
| 1 | Stearic acid | 284.2722 | 14.79 | 1.72 | 0.521 | 0.0461 | 0.336 | 3.17E-11 |
| 2 | Cis-aconitic acid | 174.0165 | 9.38 | 1.75 | 0.636 | 0.0008 | 0.394 | 3.54E-08 |
| 3 | Lactic acid | 90.0317 | 12.64 | 1.69 | 1.729 | 0.0153 | 0.473 | 8.57E-08 |
| 4 | Spermidine | 145.1577 | 1.09 | 1.56 | 0.454 | 0.0662 | 0.211 | 7.30E-08 |
| 5 | Oleic acid | 282.2560 | 14.54 | 1.67 | 0.596 | 0.0017 | 0.286 | 1.19E-06 |
| 6 | Uridine diphosphate-N-acetylmuraminate | 679.1024 | 14.67 | 1.70 | 0.679 | 0.3101 | 0.455 | 7.47E-07 |
| 7 | NADP | 743.0754 | 16.00 | 1.71 | 0.471 | 0.0267 | 0.282 | 1.74E-06 |
| 8 | Pipecolic acid | 129.0788 | 1.84 | 1.60 | 0.395 | 0.0585 | 0.278 | 2.21E-06 |
| 9 | Pyroglutamic acid | 129.0424 | 6.73 | 1.57 | 0.596 | 0.0147 | 0.286 | 3.63E-06 |
| 10 | Isoleucine | 131.0945 | 3.07 | 1.83 | 0.742 | 0.0413 | 0.550 | 5.12E-06 |
| 11 | Fumaric acid | 116.0108 | 1.98 | 1.94 | 1.208 | 0.0093 | 0.363 | 1.76E-05 |
| 12 | Valine | 117.0788 | 9.36 | 1.67 | 0.754 | 0.0466 | 0.570 | 1.89E-05 |
| 13 | 5-Aminolevulinic acid | 131.0588 | 1.35 | 1.57 | 0.520 | 0.0062 | 0.295 | 4.50E-05 |
| 14 | Citric acid | 192.0271 | 8.57 | 2.38 | 0.438 | 0.0036 | 0.317 | 5.60E-05 |
| 15 | Citrulline | 175.0957 | 2.10 | 1.64 | 0.486 | 0.0007 | 0.346 | 6.52E-05 |
| 16 | Undecanedioic acid | 216.1363 | 10.30 | 1.73 | 0.478 | 0.0074 | 0.660 | 7.54E-05 |
| 17 | Deoxycytidine | 227.0910 | 11.96 | 1.55 | 0.656 | 0.0435 | 0.572 | 1.07E-04 |
| 18 | 2,3-Diphosphoglyceric acid | 265.9596 | 1.15 | 1.58 | 1.810 | 0.0083 | 0.621 | 1.29E-04 |
| 19 | Alanine | 89.0475 | 9.40 | 1.74 | 0.750 | 0.0463 | 0.542 | 1.15E-04 |
| 20 | Creatine | 131.0693 | 1.35 | 1.83 | 0.576 | 0.0358 | 0.344 | 3.02E-04 |
| 21 | Phosphoenolpyruvic acid | 167.9826 | 1.15 | 1.55 | 2.018 | 0.0011 | 0.155 | 1.76E-04 |
| 22 | Succinic acid | 118.0264 | 12.99 | 1.80 | 0.589 | 0.0298 | 0.671 | 4.50E-04 |
| 23 | Adenine | 135.0544 | 2.77 | 1.79 | 0.496 | 0.0095 | 0.544 | 4.98E-04 |
| 24 | Pyridoxine | 129.0788 | 1.84 | 1.60 | 0.511 | 0.0005 | 0.423 | 7.36E-04 |
| 25 | Ornithine | 132.0898 | 12.74 | 1.81 | 0.796 | 0.0469 | 0.421 | 0.0011 |
| 26 | Inosinic acid | 348.0471 | 1.39 | 1.51 | 0.460 | 0.0171 | 0.387 | 0.0015 |
| 27 | Heme | 616.1761 | 9.72 | 1.82 | 0.449 | 0.0274 | 0.408 | 0.0015 |
| 28 | Isovaleric acid | 102.0680 | 1.54 | 1.60 | 0.634 | 0.0384 | 0.441 | 0.0025 |
| 29 | Mannitol 1-phosphate | 262.0454 | 15.69 | 1.85 | 0.732 | 0.0158 | 0.612 | 0.0379 |
retention time.
variable important in projection.
fold change.
We then applied a metabolome wide association strategy to investigate associations between perturbations in the metabolome and the in vitro DHA susceptibility phenotypes of the three parasite lines. As shown in the heatmap (Figure 4D), alongside an increase in the abundance of glucose, hierarchical clustering analysis revealed a major cluster with decreasing intensity for relative level for most of the intermediates canonically linked to tricarboxilic acid (TCA) cycle following DHA pulse. Of note, the main metabolic changes observed in ring-stage parasites exposed to DHA were significant decreases in the glycolysis pathway and the TCA cycle as well as an increased amino acids metabolism (Figures 5 and 6). These changes suggest that parasites exhibit a finely tuned adaptive metabolic mechanism upon DHA pulse. The accumulation of intermediates in glycolysis was able to enhance the activity pyruvate kinase, resulting the decrease amount of phosphoenolpyruvic acid which further regulates the gene expression in TCA cycle. Thus, the perturbation in glycolysis was capable to affect multiple processes during metabolic switching. Besides, phosphoenolpyruvic acid, a substrate in the peptidoglycan synthesis, affected metabolome including GlcNAc-P, UDP-GlcNAc, and UDP-MurNAc, suggesting a decreased carbon fluxes in glycolysis. Notably, the level of α-ketoglutaric acid, one derivative from amino acid oxidation, exhibited a higher abundance, suggesting a possible increase for amino acids catabolism, consistent with pathway enrich analysis. Meanwhile, as it has been proved that glutamic acid and α-ketoglutaric acid constitute a branching point for TCA cycle and amino acids, its significantly upregulated level along with its precursors (including proline, histidine, and asparagine) could suggest a metabolism shifted toward glutamic acid production. Besides, thiol-containing amino acids, known to be associated with either energy or antioxidative metabolism, were also significantly affected upon DHA exposure. As shown in Figure 6, the relative abundance for intermediates corresponding to thiol-containing amino acid metabolism (including cysteine and serine as well as their metabolic products also responsible for energy metabolism and antioxidative stress) was significantly altered. Among them, it should be noted that spermidine is responsible for cellular cycle control, strengthening our hypothesis that the IDC process is arrested.33
Figure 5.
Proposed metabolic network of glucose metabolism, TCA, and heme biosynthetic pathways after DHA exposure
Boxplot (DMSO: light brown/DHA: green) corresponds to the relative abundance for each metabolite after normalization (n = 4 and data are represented as mean ± SD). Abbreviations: Glc: glucose; G6P: glucose 6-phosphate; F1,6-BP: fructose 1,6- bisphosphate; PEP: phosphoenolpyruvic acid; Lac: lactic acid; GlcNAc-P: N-acetyl-glucosamine 6-phosphate; UDP-GlcNAc: UDP-N-acetylglucosamine; UDP-MurNAc: UDP-N-acetylmuraminate; ALA: 5-Aminolevulinic acid; PGB: porphobilinogen; HMB: hydroxymethylbilane; CPIII: coproporphyrinogen III; Mal: malic acid; OA: Oxaloacetic acid; Ac-CoA: acetyl-CoA; Cit: citric acid; Cis-CAN: cis-aconitic acid; aKG: α-ketoglutaric acid; Suc-CoA: succinyl-CoA; Suc: Succinic acid; Fum: fumaric acid; GA: glyoxylic acid; Oxa: oxalic acid
Figure 6.
Metabolic network of selected amino acids and their intermediates after DHA exposure
Boxplot (DMSO: light brown/DHA: green) correspond to the relative abundance for each metabolite after normalization (n=4 and data are represented as mean ± SD). Abbreviations: Ile: isoleucinei; M2OA: 3-methyl-2-oxovaleric acid; Ac-CoA: acetyl-CoA; TCA: tricarboxylic acid; Val: valine; aKI: α-ketoisovaleric acid; Suc-CoA: succinyl-CoA; aKG: α-ketoglutaric acid; Ser: serine; Ala: alanine; Pro: proline; Glu: glutamic acid; Asp: asparagine; Asn: Aspartic acid; Met: methionine; HomoCys: homocysteine; Cys; cysteine; GSH: Glutathione; Put: putrescine; Spe: spermidine.
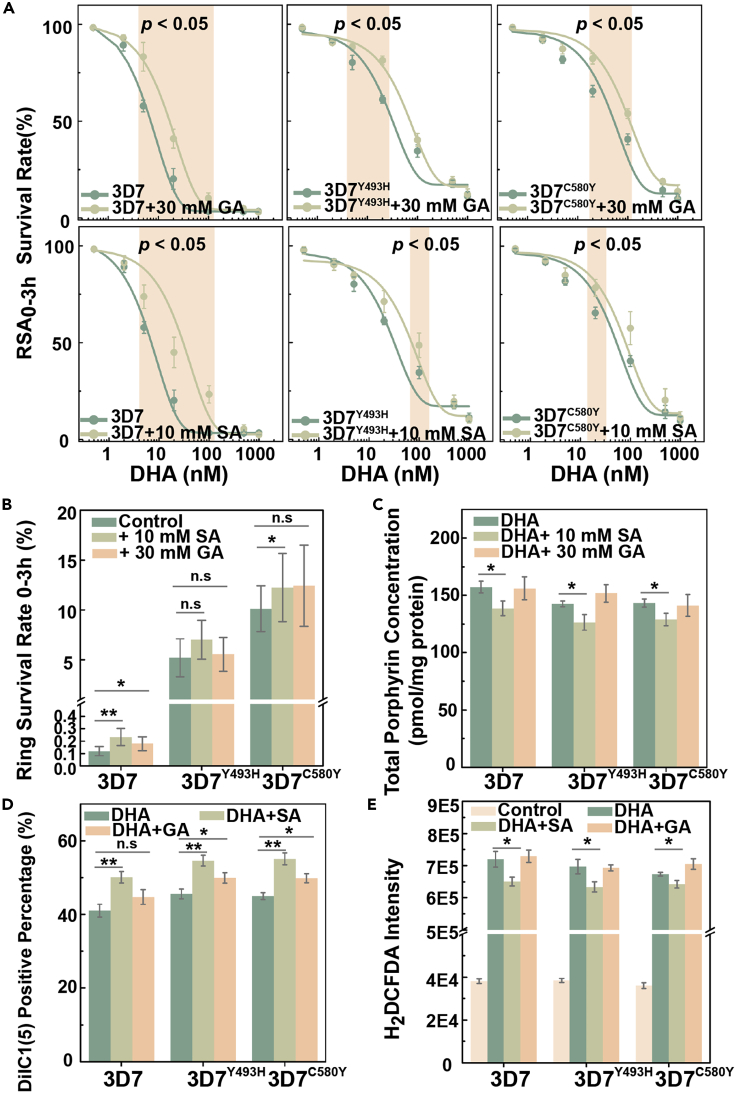
To validate the relationship between decreased TCA metabolism and the temporary growth arrest of parasites exposed to DHA, we investigated the effects of glyoxylic acid (GA) on the 3D7WT, 3D7Y493H, and 3D7C580Y lines. The effect of GA and succinyl acetone (SA) preincubation on the function of mitochondrion and generation of reactive oxygen species (ROS) as well as the growth of Plasmodium was monitored before subsequent analysis (Figures S4 and S5). We observed that the addition of 30 mM GA before DHA exposure promoted a decrease of DHA susceptibility to both the 3D7WT control line and the 3D7Y493H and 3D7C580Y lines (Figures 7A and 7B). These results demonstrate that changes in metabolic flux have an impact on parasite susceptibility to DHA, consistent with the results from the metabolomics screening assays. This finding recalls the bacterial tolerance to antimicrobial agents that can be induced by changing the TCA metabolic flux into a glyoxylate shunt and points out that pathogens susceptibility can be metabolically tuned.34 In addition, we revealed that exogenous supplementation with GA also elicits a protective effect against DHA-induced lethality, for currently unknown reasons.
Figure 7.
Affected drug susceptibility by chemical inhibition of corresponding pathway
(A) Top panel showed preincubating parasites with 30 mM for 1h GA had reduced DHA susceptibility in 3D7WT, 3D7Y493H, and 3D7C580Y parasites; bottom panel presented preincubating parasites with 10 mM SA for 7h had reduced DHA susceptibility in 3D7WT, 3D7Y493H, and 3D7C580Y parasites. p value indicates statistical comparisons between WT and mutant parasites at selected time point (labeled by colored background) by Student’s t test.
(B) RSA0–3h determination for parasites pretreated with SA or GA as a result of inhibition of TCA cycle or heme biosynthetic pathway. Changed DHA susceptibility was obtained due to the change of metabolic state.
(C) Total porphyrin levels (combination of heme and PPIX) in parasites showed a reduced porphyrin concentration due to alterations in heme biosynthetic pathways.
(D and E) DHA-dependent mitochondrial dysfunction and reactive oxygen species generation measurement. Parasites were exposed to 700 nM DHA for the evaluation of drug susceptibility with/without inhibition.
Statistical comparisons between WT and mutant parasites are conducted by Student’s t test after Bonferroni correction (∗ p < 0.05; ∗∗ p < 0.01).
To assess the role of porphyrin metabolism in parasite susceptibility to DHA, we examined whether its inhibition could affect the survival of parasites. We pretreated DHA-exposed parasites with SA, an inhibitor of 5-aminolevulinic acid dehydratas (ALAD) in porphyrin metabolism, and observed a slight decrease in parasite susceptibility to DHA, especially for concentrations around EC50 (Figures 7A and 7B). Based on assessment of the total level of porphyrins, we confirmed that heme production was associated with changes in in vitro DHA susceptibility. Indeed, porphyrin levels as evaluated by fluorescence significantly decreased after SA treatment (Figure 7C), indicating that modulation of heme metabolism leads to changes in in vitro DHA susceptibility.
Furthermore, we also observed that mitochondrial membrane potential was significantly increased following DHA treatment in SA-exposed parasites compared to untreated parasites (Figure 7D). Oxidative stress was assessed by measuring the level of ROS (through the measurement of H2DCFDA) caused by DHA treatment. A ∼10-fold increase in ROS was detected in DHA-exposed parasites compared to nonexposed controls, indicating lethality was caused by excess oxidative stress. Meanwhile, oxidative stress caused by DHA exposure was significantly higher in 3D7WT than in 3D7Y493H and 3D7C580Y (p = 0.049 and 0.036, respectively). Furthermore, ROS levels were slightly decreased, consistent with results from RSA, for parasites pretreated with inhibitor (Figure 7E). One of the most well-documented modes of action for DHA is through the binding of the endoperoxide with heme-related Fe2+ producing abundant free radicals that alkylate parasite proteins, resulting in parasite death.35,36,37 ROS is also responsible for mitochondrial depolarization which impairs normal function, and excess levels of ROS can induce DNA double-strand breaks which further cause lethal effect for parasites.
Discussion
In this study, we aimed at elucidating whether ring-stage growth arrest induced by DHA confers a survival benefit to P. falciparum parasites with mutations in the Pfkelch13 gene. Previous metabolomics P. falciparum studies mainly focused on the late blood stages of life cycle; here we explored the metabolic profile of ring stages which is closely associated with growth arrest upon DHA exposure. We compared the metabolic profiles of growth-arrested ring-stage parasites in relation with their in vitro susceptibility to DHA as measured by the RSA.38 Herein, we investigated the correlation of the relative abundance of altered metabolome with a metabolic process resulting from DHA pulse that parasites have to face during survival. Parasites intended to extend the less sensitive ring stage for the escape of lethal effect.
Based on a novel enrichment method (SLOPE), adaptive metabolic changes were evaluated to address the hypothesis that Pfkelch13 WT and mutant lines constitute two different populations upon DHA pulse. The metabolome of ring-stage parasites displayed altered energy and porphyrin metabolism profiles, including changes in the TCA cycle, glycolysis, and amino acid and pyruvate metabolic pathways. We initially hypothesized that mutant parasites were capable to orchestrate a metabolic switch corresponding to energy metabolism. Considering either glucose or amino acids as the energy supply in parasites, metabolic network revealed the metabolic switch from glucose to amino acids catabolism occurs during arrested growth. Notably, perturbed porphyrin metabolism is also a metabolic feedback for hemoglobin catabolism which has been previously reported for the mode of action of DHA.39 Meanwhile, for the corresponding antioxidative stress metabolism, previous research has reported an enhanced glutathione production by screening the biological changes, which was consistent with observed metabolic network.40 Although the altered levels of TCA cycle has been reported by measuring level of intermediates, little is known about the adaptative metabolic plasticity that mutant parasites undergo upon DHA exposure which turns out to be critical for survival.30 Based on the metabolomic screening results, we were able to alter the in vitro susceptibility to DHA by either changing metabolic flux (through exogenous supplementation with GA) or through chemical inhibition (through the addition of SA), indicating that metabolic disruption protects parasites from the damage caused by DHA exposure. Overall, this study provides metabolomics-based evidence for elucidating the role of Pfkelch13 mutations in ART-R.
In addition to the conventional resistance phenotype measured by RSA, detailed in vitro stage-specific growth rates during a single IDC were determined, indicating either a ∼6 h delayed growth for DHA-resistant engineered strains or the existence of growth-arrested forms of parasites following exposure to 50 nM DHA. Despite the extended ring-stage period, the overall IDC development time for engineered lines remained almost similar to the WT due to the shortened trophozoite stage period. We concluded that the altered development profile, especially concerning the extended ring-stage development, was closely related to the resistance phenotype, and this was consistent with previously reported transcriptome profiles in which a prolonged ring stage allows the upregulation of unfolded protein responses that protect the parasite from drug-induced protein damages.41 The altered IDC profile confers the additional advantage that the most susceptible stage in asexual development receives reduced exposure. Previous studies have indicated that the late-stage parasites (trophozoite or schizont) are more sensitive to various antimalarials, and therefore based on our findings, we consider that an extended ring stage may have evolved as a protective mechanism as it reduces the exposure time for these sensitive later stages during the IDC.20,27,42 Because ring-stage parasites are metabolically less active and less susceptible to DHA than mature stages, it is conceivable that extended ring stages would enable the parasites to better survive DHA exposure, given its relatively short in vivo half-life.42,43 Some reports have shown that growth-arrested parasites resulting from drug pressure applied to ring-stage parasites can survive for several days before returning to normal development.11,21 According to the results, we hypothesized that the extended ring stage could be an advantage in which parasites were growth-arrested with subsequent development following DHA treatment.
ART-induced arrested growth remains understudied due to the rarity of arrested parasites following drug exposure. Furthermore, most metabolomic studies focus on the parasite stages that occur late in the IDC because of the availability of efficient purification methods such as magnetic sorting which significantly reduces background noise resulting from excess uninfected erythrocytes.30,40 For the early-stage parasites, the lack of an efficient enrichment strategy has greatly hindered attempts to investigate their metabolomics. Furthermore, recent downstream omics studies have revealed that the proteome or metabolome of early stage forms are significantly affected by host contamination due to a high ratio of host to parasite-specific signals.44 To investigate the metabolome in arrested ring-stage-associated DHA resistance, we present here a novel enrichment method (SLOPE) to obtain ring-stage parasites by effective removal of uninfected host erythrocytes without affecting parasitic viability. Clearly separated metabolic profiles were obtained for SLOPE-enriched ring-stage samples, demonstrating its suitability for metabolic analysis. We found that the major shifts in the metabolome of parasites with DHA-resistance-associated mutations in the Pfkelch13 gene under growth arrest involved the TCA cycle, glycolysis, and amino acid and pyruvate metabolism. In most of free-living microorganisms, the TCA cycle acts as the central wheel for carbon metabolism in which carbon-derived metabolites like pyruvate produced from glycolysis are converted into intermediate products for energy production. Previous studies have revealed that during the asexual development stages, levels of TCA cycle intermediates change periodically, indicating that the full TCA cycle functions with mitochondrial acetyl-coenzyme A (CoA) derived from amino acid catabolism.45 It should be noted that parasites are capable to use either glucose or amino acids as the source for energy metabolism.46 Therefore, on the basis of metabolomics screening, significantly upregulated level of glucose as well as related amino acids indicated that either amino acids had been serving for synthesis of glucose or the intracellular glucose consumption had been arrested. Thus, we deduce that upon DHA pulse, there was a metabolic switch from glucose to amino acids consumption for fueling the energy supply in growth-arrested parasites.
Based on above-mentioned metabolomics findings, we have investigated whether inhibition of the metabolic pathways implicated above would affect DHA susceptibility. Parasites pretreated with GA or SA displayed reduced DHA susceptibility as measured by an increased survival in the RSA0–3h50%. RSA0–3h revealed a slight reduction in in vitro susceptibility following addition of GA or SA, demonstrating that decreased metabolism or altered metabolic flux profiles induced by growth arrest could contribute to increased DHA tolerance. Therefore, according to our metabolomics data, we hypothesize that GA may be an inducer of antibiotic tolerance through the biochemical shunting of the TCA cycle.
In conclusion, the generation of Pfkelch13 3D7493H and 3D7580Y mutants using the Cas9i system, coupled with various DHA susceptibility assays, revealed an association between Pfkelch13 mutations and growth arrest-mediated partial resistance to DHA. Furthermore, based on a novel enrichment method, we provide metabolic evidence that DHA-induced growth arrest ring stages are associated with significantly decreased energy metabolism activity. DHA susceptibility is affected by both chemical inhibition and changes in metabolic flux. These findings could help understand how mutant parasite lines undergo adaptative changes upon drug pulse and provide useful information in understanding the critical metabolic plasticity from metabolic perspective.
Limitations of the study
The here-presented study was aimed at investigating the underlying metabolic mechanisms for PfKelch13-mutation-mediated ART-R in human malaria parasites by untargeted metabolic profiling. Limitations of this study included using the mix of both growth-arrested and dead parasites resulting from proposed enrichment protocol for following metabolomics analysis. Although flow cytometric sorting was not performed due to the lack of efficiency for acquiring sufficient parasites, the metabolome of dead parasites had been taken into consideration for background subtraction during data analysis which could help circumvent this problem. Therefore, we believed the unrelated metabolic signal resulting from either dead parasites or host erythrocytes could be greatly reduced, suggesting that the acquired metabolome was closely associated with growth-arrested parasites. Meanwhile, based on the primary untargeted screening analysis, more detailed targeted profiling aimed at quantitatively investigating the alterations for metabolites involved in related pathways had not been performed. Although the biological efficacy for part of proposed metabolome and pathways has been validated by either changing metabolic flow or chemical inhibition, it still remained as a comprehensive evaluation in which detailed investigation for the alterations of related metabolic pathways has to be carried out. Moreover, in spite of the fact that PfKelch13-mutations was closely associated with ART-R, a brief validation for results obtained from this study should be considered in more filed isolates which were unavailable due to global COVID-19 situation. Ultimately, each study has its own limitations and requirements; thus additional studies are required for detailed characterization and validation of defined metabolic pathways in more filed isolates with PfKelch13 mutations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Anitbodies | ||
| PE conjugated CD235a Monoclonal Antibody | Invitrogen | Cat# MA1-19661;RRID: AB_1071750 |
| Mouse Anti-alpha 1 Spectrin monoclonal antibody | Abcam | Cat# ab11751;RRID: AB_2194328 |
| Goat Anti-Mouse IgG (Alexa Fluor® 59) antibody | Abcam | Cat# ab150116;RRID: AB_2650601 |
| Bacterial and virus strains | ||
| Plasmodium.falciparum 3D7 strain | This paper | N/A |
| Plasmodium.falciparum 3D7-Cas9i strain | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 | Gibco | Cat# 31800089 |
| HEPES | Gibco | Cat# 11344041 |
| AlbuMAX™ I | ThermoFisher | Cat# 11020088 |
| Giemsa’ stain | Sigma-Aldrich | Cat# 32884 |
| Hypoxanthine | Sigma-Aldrich | Cat# V900452 |
| DSM1 | Sigma-Aldrich | Cat# 5333040001 |
| WR99210 hydrochloride | Sigma-Aldrich | Cat# SML2976 |
| Streptolysin O | Sigma-Aldrich | Cat# S5265 |
| SYTO™ 61 | Invitrogen | Cat# S11343 |
| Methanol LC-MS grade | Merck | Cat# 1.06035 |
| Acetonitrile LC-MS grade | Merck | Cat# 1.00029 |
| Formic acid for LC-MS | Merck | Cat# 5.33002 |
| Critical commercial assays | ||
| NucleoSpin Gel and PCR Clean-up Kit | MACHEREY-NAGEL | Cat# 740609 |
| NucleoBond Xtra Midi Plus Kit | MACHEREY-NAGEL | Cat# 740410 |
| ClonExpressII One Step Cloning Kit | Vazyme Biotech | Cat# C112-02 |
| MitoProbe DilC1(5) Assay Kit | ThermoFisher | Cat# M34151 |
| H2DCFDA Assay Kit | ThermoFisher | Cat# D399 |
| Oligonucleotides | ||
| Primer for plasmid construction, see Table S1 | This paper | N/A |
| Deposited data | ||
| Raw data | This paper | Metabolights: MTBLS6388 |
| Software and algorithms | ||
| SIEVE | ThermoFisher | N/A |
| TraceFinder | ThermoFisher | N/A |
| MetaboAnalyst | Xia et al.,47 | http://www.metaboanalyst.ca |
| GraphPad Prism8 | GraphPad | https://www.graphpad.com |
| SIMCA-P 13.0 | Umetrics | http://www.umetrics.com/products/simca |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, J.C (caojun@jipd.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All P. falciparum strains were cultured with human O+ erythrocytes (provided by Shanghai Blood Center) at 2% hematocrit in RPMI 1640 media (Thermo Fisher Scientific, MA) supplemented with 0.5% (w/v) Albumax I, 50 mg/L hypoxanthine, 25 mM NaHCO3, 25 mM HEPES and 10 mg/L gentamycin as previously described.48 Parasite cultures were maintained at 37°C under 5% O2, 5% CO2 and 90% N2. Thin blood smears were prepared by methanol fixation and Giemsa’s solution stain (Sigma-Aldrich, St. Louis, MO). Parasitemia was measured by bright-field microscopy with 100× oil objective and bright field image of parasite morphology was captured on an optical microscope. The experimental protocols, including parasite culture and gene transfection were reviewed and approved by the corresponding Review Committees of Jiangsu Institute of Parasitic Diseases (approval no. IRB00008756).
Method details
Plasmid construction
For enhancing transfection efficiency, the Cas9 endonuclease was integrated into the genome of wide type P. falciparum 3D7 strain for stable expression.31 Specifically, C-terminal fragment (1219 bp) of PfP230p was amplified with primers P1/P2 (SI, Table S1) and cloned into pUF1-Cas9 plasmid at the restriction sites EcoRI and AvrII. Then the blasticidin cassette was replaced with yDHODH gene with primer P3/P4 (SI, Table S1) at the restriction sites NcoI and XbaI to obtain pUF1-DHODH-Cas9i plasmid.
Plasmids containing Pfkelch13 mutations were constructed based on the pL6cs-sgRNA plasmid (consisting of a sgRNA expression cassette and homology regions) via multiple cloning steps as previously described. Briefly, guide sequences (P5/P6) were cloned into pL6cs-sgRNA after annealing at the restriction sites AvrII and XhoI. Then two fragments of PF3D7_1343700 with desired mutations and corresponding donor DNA were amplified from genomic DNA 3D7 with the primers P7/P8, P9/P10, P11/P12, P13/P14 for Y493H, C580Y and homology regions, respectively (SI, Table S1). These fragments were cloned into the pL6cs-sgRNA plasmid using restriction sites AscI and AflII. The constructs were then transformed to XL10 competent cells (Vazyme Biotech Co., Ltd) for sequencing. Correctly sequenced plasmids were extracted by NucleoBond Xtra Midi Plus (Macherey-Nagel, Germany) for transfection.
Parasite gene transfection
Parasite were tightly synchronized with either 5% sorbitol or 40/70% Percoll enrichment according to protocol. Briefly, for sorbitol-synchronization, parasite pellet was collected by centrifugating at 2500 rpm for 5min followed by the addition of 10 mL sorbitol. After incubating at 37 °C for 10min, cultures were centrifuged at 2500 rpm and washed for 3 times with medium. For Percoll-synchronization, parasite pellet was carefully overlayed on the gradient followed by centrifugating at 3100 rpm for 20min. After discarding the top layer of gradient, parasites were washed with medium and returned to normal culture condition.
Transfections were performed by electroporating erythrocytes infected with late-stage trophozoites with 150 μg purified plasmid DNA resuspended in Cytomix (120 mM KCl, 10 mM KH2PO4, 25 mM HEPES, 2 mM EGTA, 0.15 mM CaCl2, 5 mM MgCl2, pH 7.6) with 310V pulse voltage.49 For construction of the Cas9i parental strain, the pUF1-DHODH-Cas9i plasmid was first transfected into the 3D7 strain episomally followed by 1.5 μM DSM1 drug selection to obtain integrative parental strain via single-crossover recombination.
pL6cs-sgRNA plasmids with the desired mutations were then transfected into the Cas9-integrated parental strain to obtain the corresponding Pfkelch13 mutant parasite strains. Positive-selection was conducted by 2.5 nM WR99210 at 72h post-transfection due to co-expressed hDHFR marker and culture media with selection drug was renewed daily during first week. Cultured parasites were microscopically monitored every week post electroporation until the observation of viable parasites under drug selection. Sequences of Pfkelch13 of mutant strains were validated by Sanger sequencing before experiments were initiated.
Chemical and density-based enrichment protocol
The enrichment assay was performed according to previously reported method with some modifications.32 SLO (Sigma-Aldrich, St. Louis, MO) was activated and the hemolytic activity for each batch was evaluated in triplicate monthly. Specifically, 25,000 units of SLO was resuspended in 5 mL PBS (pH 7.4) containing 0.1% BSA and then be incubated with 5 mM dithiothreitol at 37°C for 1h. Then activated SLO was divided into aliquots and stored at −20°C for long-term application. Hemolytic units of SLO were defined as the amount required for 50% lysis of 50 μL erythrocytes at 2% hematocrit at 37°C and the hemolytic activity for each batch of SLO was evaluated in triplicate monthly. Erythrocytes were pre-incubated with complete medium at 37°C for 4h and resuspended in PBS at 2% hematocrit. Then different volumes of SLO were added into an aliquot of 50 μL erythrocytes and incubated at 37°C for 30min. The hemoglobin level was spectrophotometrically determined at 412 nm on SpectraMax iD3 microplate reader. Cell density of tightly synchronized ring stage parasite culture after two rounds of 5% sorbitol treatment was firstly adjusted to 2% parasitemia and then subjected to SLO lysis analysis. Different amounts of SLO were added into (0, 250, 375, 500, 625, 750, 875, 1000, 1250U) culture followed by thorough pipetting and incubation at 25°C for 30 min. Following the addition of 5 mL PBS, the mixture was centrifuged at 2500 rpm for 5 min and cell pellets were washed twice with PBS to remove remaining SLO. Cell pellets were then resuspended in 1 mL PBS, carefully layered onto 70% Percoll and further centrifuged at 3100 rpm for 20 min to remove permeabilizated uninfected erythrocytes. The lower layer (consisting of infected cells) was collected and washed twice with PBS to obtain ring-stage parasites.
To investigate the enrichment efficiency, lysis curves were plotted to quantitatively access the proportion of infected erythrocytes. Briefly, tightly synchronized parasites were treated with varying concentrations of SLO as described above. After staining infected cells with Hoechst for 30 min, flow cytometry was applied to evaluate the percentage of lysed uninfected cells. Total erythrocytes were gated according to their forward and side scatter parameters and uninfected cells were calculated by subtracting infected cells from total cells. The lysis percentage was calculated by comparing either the infected or uninfected cells in SLO treated samples with that in untreated controls.
Immunofluorescence assays were performed using Nikon fluorescence confocal microscopy. Either intact erythrocytes or lysed ghosts were visualized by anti-CD235a antibody which is localized on the outer surface of erythrocytes. As spectrin turned out to be a cytoskeleton-localized protein with inaccessibility for anti-spectrin antibody in intact cells, therefore lysed ghost cells without intact cytosolic member were able to be captured due to the passage of corresponding antibody and visualized as both CD235a+ and Spectrin+. Samples were fixed in 4% paraformaldehyde and blocked with 1% BSA followed by the addition of mouse anti-alpha I spectrin antibody (Abcam, Cambridge, MA) at 1:100 dilution. After washing with ice-cold PBS three times, samples were incubated with Alexa Fluor 594 conjugated goat anti mouse secondary antibody (Abcam, Cambridge, MA) at 1:2000 dilution. Following thorough washing, samples were further incubated with PE-CD235a antibody (Thermo Fisher Scientific, MA) and Hoechst (Thermo Fisher Scientific, MA) at 1:1000 and 1:10,000 dilution respectively and subjected to immunofluorescence assays.
To investigate the effect of treatment on the growth and development of parasites, purified parasites were further subjected to culture for growth curve analysis. As described above, tightly synchronized early-stage parasites were split into two parts, one for untreated control while one received treatment as above. After microscopically determining the parasitemia, fresh erythrocytes were added to the culture at 2% hematocrit and the starting parasitemia was adjusted to 0.5%. The growth curve of each sample was monitored during three complete life cycles for evaluation of whether treatment impacts on parasite growth.
Phenotype validation
Ring-stage survival assays (RSA0–3h and RSA0–3h50%)
In vitro survival assays were performed as previously described to assess resistance phenotypes.28 Briefly, parasites were twice synchronized by 5% sorbitol at 40h intervals to narrow down the life cycle window, then 40/70% Percoll treatment was performed, and remaining parasites were cultured for 3 h with fresh erythrocytes to allow merozoite invasion. Cultures were then further synchronized with 5% sorbitol to eliminate any remaining late-stage parasites and to obtain 0-3h ring stages. Finally, cultures were dispensed into 24-well plates at 0.5% parasitemia, 2% hematocrit. For the RSA0–3h assay, parasites were exposed to 700 nM DHA or 0.1% DMSO and cultured for 6 h prior to washing twice in RPMI 1640 incomplete media to remove excess drug and returned to standard culture conditions for 66 h in new wells. Survival rates were microscopically assessed by counting the percentage of viable parasites (∼10,000 erythrocytes). Survival rates were calculated as the ratios of viable parasites in DHA and DMSO treated samples, and each sample was assessed in triplicate.
Since conventional RSA0–3h only measures the survival rate for parasites after single dose of DHA pulse (700 nM), RSA0–3h50% was further determined to evaluate the in vitro sensitivity as well as the required concentration and exposure period applied in metabolomics study. For RSA0–3h50%, 0-3h ring-stage parasites were adjusted to 0.5% parasitemia and 2% hematocrit followed by the exposure of serially diluted DHA ranging from 0.5 to 1000 nM (0, 0.5, 2, 5, 20, 50, 100, 200, 500, 1000 nM) or 0.1% DMSO in opaque 96-well microplate with 200 μL culture/well. Following incubation for several periods (1h, 3h, 6h, 9h), parasites were thoroughly washed four times with 200 μL RPMI 1640 incomplete media and transferred to a new plate for ensuring the complete removal of DHA. Parasites were then returned to standard culture conditions followed by assessment of variability by measuring the parasitemia. Briefly, parasites were stained with 20 μL SYTO-61 (2 μM diluted in PBS) and incubated at room temperature for 20 min before fluorescence analysis at the excitation and emission wavelength of 628 and 645 nm, respectively. Parasite viability was calculated as the ratio of the fluorescence intensity for viable parasites in DHA-treated parasites and DMSO-treated controls after background subtraction according to the following equation, in which FLcontrol and FLkill refers to the fluorescence intensity measured in samples without drug or samples treated by constant culture with 700 nM DHA to ensure 100% parasite killing, respectively. All samples were analyzed in triplicate. RSA0–3h50% values were obtained by fitting data to a non-linear regression model and the value corresponding to drug concentrations producing 50% loss of parasite viability were recorded.
| Viability= (FL- FLkill)/(FLcontrol- FLkill) |
Proportions of growth-arrested parasites
To evaluate the propagation of different parasite stage strains, parasites were highly synchronized with 0-3h window as described above. Parasite life cycle analysis was assessed by Giemsa-stained thin-blood smears in triplicate every 6 h during 50h propagation. The percentage of parasites at different stages in three replicates were averaged and plotted against time.
Parasite propagation was further quantified following drug treatment at 700 nM or 50 nM. Thin blood smears were made daily to quantify growth-arrested, dead or other viable parasite stages according to morphology. Specifically, growth-arrested parasites had condensed chromatin surrounded by a relatively small amount of cytoplasm while dead parasites had degraded chromatin or cytoplasm.
DHA-induced growth retardation assay
It is of note that there is a portion of viable parasites could enter into a growth-arrested state after drug pulse in which they are were actually alive but unable or delayed to replicate within this period. Therefore, growth effect induced by drug pulse was assessed by labeling with SYTO-61. In this assay, highly synchronized parasites were divided in parallel for either DMSO or DHA treated while SYTO-61 labeling was performed when DMSO-treated parasite had progressed into mid-late stage (∼40h) in the cycle after drug treatment (∼88h after DHA treatment). It could ensure distinguishable growth alteration resulted from viable parasites and the signal of viable parasite was accessed by flow cytometry on BD FACS Verse cytometer. For the subtraction of unviable background signal, the SYTO-61 positive events at specific concentration referred to the median of viable parasites had been normalized by reducing those from killed control to obtain the SYTO-61 frequency histogram.
Metabolomic identification for proposed approach
Parasite culture samples with at least 5×107 infected erythrocytes (including untreated control, saponin-lysed, SLO treated and SLO+Percoll treated) were collected for evaluation to define whether the SLOPE was able to distinguish erythrocyte-based background from parasite-specific metabolome signals. Briefly, infected erythrocytes were harvested and extracted in ice-cold methanol on dry ice to quench enzymatic activity. Following quenching, samples were immediately lysed and subjected to a two-step extraction method to specifically profile the parasite-derived metabolome for metabolomics analyses. After aspirating the supernatant culture medium by centrifugation at 2500 rpm for 5 min, erythrocytes were immediately harvested and subjected to various treatments as quickly as possible. Following lysis or treatment, 50 μL cell pellet was carefully added into 450 μL ice-cold 100% methanol, after immediate vortex, samples were centrifuged at 2500 rpm for 5 min and returned to dry ice to remove large-molecular-weight proteins. Supernatant was carefully collected followed by the addition of 450 μL 80% methanol. Samples were subjected to sonication on ice for 15 min and then centrifuged at 12,000 rpm for 10 min. Resulting supernatants were pooled and stored at −80°C for metabolomics analyses.
Untargeted metabolomics analysis
To obtain the metabolomes of ring-stage parasites, 0–3 hpi parasites were treated with either 50 nM DHA or 0.1% DMSO for 6 h in four independent biological replicates. Infected erythrocytes were enriched as above prior to metabolome extraction. Metabolic profiling of Pfkelch13 mutant and WT parasites were carried out in parallel for comparing metabolic states after drug treatment. Medium-based metabolites were ruled out for the subsequent data analyses. The metabolic profile of WT parasites exposed to 700 nM DHA which is composed of the signal resulted from dead parasites has been applied for background subtraction before following multivariate statistical analysis.
Untargeted metabolomics mass data was acquired on a high-resolution Q Exactive Orbitrap mass spectrometer equipped with a heated electrospray source (HESI) in both positive and negative modes simultaneously. For both positive and negative modes, the operating parameters were set as follows: a spray voltage of 3 kV, a capillary temperature of 300°C, sheath gas flow of 40 arbitrary units, auxiliary gas flow of 10 arbitrary units, sweep gas of 2 arbitrary units and S-Lens RF level of 50. In the full scan analysis (70–1050 amu), the resolution was set at 700 000 with an automatic gain control target of 1×106 charges and a maximum injection time of 120 ms. Chromatographic separation was performed on a UPLC Ultimate 3000 system equipped with a 1.9 mm Hypersil Gold C18 column (100 mm × 2.1 mm), and the column was maintained at 4°C. A Multi-step gradient consisting of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) was applied and the gradient operated at a flow rate of 0.4 mL min-1 by linearly increasing solvent B from 5% to 95% over 15 min, then the column was washed with 95% solvent B for 2 min and re-equilibrated in 5% solvent B. The UPLC autosampler temperature was set at 4°C and the injection volume for each sample was 5 μL. Both the UPLC and the Orbitrap mass spectrometer system were controlled by the Xcalibur 2.2 software. The quality control (QC) samples were prepared by pooling the same volume of supernatant from samples pretreated in the same manner as real samples and analyzed every 10 samples to ensure stability and repeatability. The mass spectrometry was calibrated every 24 h during the profiling to ensure the mass accuracy.
Biological effectiveness analysis
Porphyrin level analysis
Porphyrin levels were quantitatively analyzed by fluorescence spectrometry through external standard methods. Samples were collected from parasites exposed to 700 nM DHA with/without inhibition. Briefly, after washing with 10 mL PBS, pelleted parasites were resuspended in 50 μL water and quick-frozen in liquid nitrogen. Parasite lysate was incubated with 200 μL 1.5M oxalic acid at 99°C for 30 min, and fluorescence intensity was recorded on a microplate reader with the excitation and emission wavelength at 400 and 662 nm respectively. Calculated porphyrin levels were further normalized to protein concentrations obtained by BCA kits.
Mitochondrial membrane potential analysis
Mitochondrial membrane potential (MMP) assay was performed to further investigate the mitochondrial function of parasites. The MMP assay was evaluated using a MitoProbe DilC1 kit (Thermo Fisher Scientific, MA) according to the manufacturer’s instructions. Briefly, parasites exposed to 700 nM DHA were incubated with DilC1 at 37°C for 30 min. After co-staining with Hoechst, the percentage of MMP positive cells were calculated as the ratio of DilC1 positive cells to Hoechst positive cells on SpectraMax iD3 microplate reader using 488 and 640 nm as the excitation wavelength, respectively.
Determination of ROS generation
Intracellular ROS levels between groups was determined as previously described. Briefly, parasites exposed to 700 nM DHA were incubated with 20 μM dichlorodihydrofluorescein diacetate at 37°C for 30 min in the dark. Then parasites were harvested and washed with PBS twice followed by saponin lysis. After thorough washing and centrifugation at 2500 rpm for 5min, cell pellets were resuspended in PBS and the fluorescence of DCF was recorded on a microplate reader.
Quantification and statistical analysis
Raw peak extraction and deconvolution for metabolomics data were performed by SIEVE software and metabolic data were analyzed using TraceFinder software for relative quantitative analysis of detected features. Multivariate statistical analysis including principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were performed by SIMCA-P 13.0 software (Umetrics, Sweden). Discriminating features were further evaluated according to the variable importance (VIP) for each metabolite in the PLS-DA model. The statistical significance was calculated using the two-tailed Student’s t-test by SPSS 25.0 software (Chicago, IL, USA) and p value were further adjusted by Benjamini and Hochberg false discovery rate adjustment for multiple features comparison. Features with significant changes (q value < 0.05 and VIP >1) were selected as discriminating metabolites. Qualitative analysis was performed according to either in-house metabolite library (level 2) acquired from authentic compounds and public database including HMDB, METLIN according to the retention time and accurate mass (level 3). The mass tolerance was set as less than 5 ppm between obtained feature and database-recorded data. Pathway enrichment analysis was performed by using the website tool MetaboAnalyst 4.0 (http://www.metaboanalyst.ca).
Additional resources
This paper did not create any additional resources.
Acknowledgments
This work was supported by National Key R&D Program of China (No. 2020YFC1200105 and 2018YFA0507300), National Natural Science Foundation of China (No. 82230077, 81971967, 82102424, 81971959, and 82172302), Natural Science Foundation of Jiangsu Province (No. BK20210061), the China Postdoctoral Science Foundation (2020M671397), the Shanghai Blue Cross Brain Hospital Co. Ltd., and Shanghai Tongji University Education Development Foundation.
Author contributions
JC, QZ, DM, and GZ conceived and designed the experiments. YZ generated transgenic parasite lines and related analysis. XY, MZ, GZ, JT, CW, and LP performed the experiments and data analysis. XY and DM wrote the manuscript with contribution from JC, QZ, and RC.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105725.
Contributor Information
Guoding Zhu, Email: jipdzhu@hotmail.com.
Didier Ménard, Email: didier.menard@pasteur.fr.
Qingfeng Zhang, Email: qfzhang@tongji.edu.cn.
Jun Cao, Email: caojun@jipd.com.
Supplemental information
Data and code availability
The data supporting the current study have been deposited at Metabolights. Accession number is listed in the key resources table; This paper did not report any original code; Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.World Health Organization World malaria report 2021. 2021. https://www.who.int/publications/i/item/9789240040496
- 2.Bastiaens G.J.H., Bousema T., Leslie T. Scale-up of malaria rapid diagnostic tests and artemisinin-based combination therapy: challenges and perspectives in sub-Saharan Africa. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001590. e1001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benelli G., Beier J.C. Current vector control challenges in the fight against malaria. Acta Trop. 2017;174:91–96. doi: 10.1016/j.actatropica.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Eastman R.T., Fidock D.A. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 2009;7:864–874. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui L., Su X.z. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev. Anti Infect. Ther. 2009;7:999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Artemisinin resistance and artemisinin-based combination therapy efficacy: status report. 2018. https://apps.who.int/iris/bitstream/handle/10665/274362/WHO-CDS-GMP-2018.18-eng.pdf
- 7.Breman J.G. Resistance to artemisinin-based combination therapy. Lancet Infect. Dis. 2012;12:820–822. doi: 10.1016/S1473-3099(12)70226-8. [DOI] [PubMed] [Google Scholar]
- 8.Tilley L., Straimer J., Gnädig N.F., Ralph S.A., Fidock D.A. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 2016;32:682–696. doi: 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodrow C.J., White N.J. The clinical impact of artemisinin resistance in Southeast Asia and the potential for future spread. FEMS Microbiol. Rev. 2017;41:34–48. doi: 10.1093/femsre/fuw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., LaCrue A.N., Teuscher F., Waters N.C., Gatton M.L., Kyle D.E., Cheng Q. Fatty acid synthesis and pyruvate metabolism pathways remain active in dihydroartemisinin-induced dormant ring stages of Plasmodium falciparum. Antimicrob. Agents Chemother. 2014;58:4773–4781. doi: 10.1128/AAC.02647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hott A., Ca Sandra D., Sparks K.N., Morton L.C., Castanares G.G., Rutter A., Kyle D.E. Artemisinin-resistant Plasmodium falciparum parasites exhibit altered patterns of development in infected erythrocytes. Antimicrob. Agents Chemother. 2015;59:3156–3167. doi: 10.1128/AAC.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.-C., Khim N., Kim S., Duru V., Bouchier C., Ma L., et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straimer J., Gnädig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MalariaGEN Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. Elife. 2016;5 doi: 10.7554/eLife.08714. e08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haldar K., Bhattacharjee S., Safeukui I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018;16:156–170. doi: 10.1038/nrmicro.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stokes B.H., Dhingra S.K., Rubiano K., Mok S., Straimer J., Gnädig N.F., Deni I., Schindler K.A., Bath J.R., Ward K.E., et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife. 2021;10 doi: 10.7554/eLife.66277. e66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birnbaum J., Scharf S., Schmidt S., Jonscher E., Hoeijmakers W.A.M., Flemming S., Toenhake C.G., Schmitt M., Sabitzki R., Bergmann B., et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367:51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- 20.Murithi J.M., Owen E.S., Istvan E.S., Lee M.C.S., Ottilie S., Chibale K., Goldberg D.E., Winzeler E.A., Llinás M., Fidock D.A., Vanaerschot M. Combining stage specificity and metabolomic profiling to advance antimalarial drug discovery. Cell Chem. Biol. 2020;27:158–171.e3. doi: 10.1016/j.chembiol.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui F.A., Boonhok R., Cabrera M., Mbenda H.G.N., Wang M., Min H., Liang X., Qin J., Zhu X., Miao J., et al. Role of Plasmodium falciparum Kelch 13 protein mutations in P. falciparum populations from northeastern Myanmar in mediating artemisinin resistance. mBio. 2020;11 doi: 10.1128/mBio.01134-19. e01134-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Virgilio C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012;36:306–339. doi: 10.1111/j.1574-6976.2011.00287.x. [DOI] [PubMed] [Google Scholar]
- 23.Rittershaus E.S.C., Baek S.-H., Sassetti C.M. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greening C., Grinter R., Chiri E. Uncovering the metabolic strategies of the dormant microbial majority: towards integrative approaches. mSystems. 2019;4 doi: 10.1128/mSystems.00107-19. e00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groger M., Fischer H.S., Veletzky L., Lalremruata A., Ramharter M. A systematic review of the clinical presentation, treatment and relapse characteristics of human Plasmodium ovale malaria. Malar. J. 2017;16:112. doi: 10.1186/s12936-017-1759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klonis N., Crespo-Ortiz M.P., Bottova I., Abu-Bakar N., Kenny S., Rosenthal P.J., Tilley L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. USA. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klonis N., Xie S.C., McCaw J.M., Crespo-Ortiz M.P., Zaloumis S.G., Simpson J.A., Tilley L. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc. Natl. Acad. Sci. USA. 2013;110:5157–5162. doi: 10.1073/pnas.1217452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duvalsaint M., Kyle D.E. Phytohormones, isoprenoids, and role of the apicoplast in recovery from dihydroartemisinin-induced dormancy of Plasmodium falciparum. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01771-17. e01771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok S., Stokes B.H., Gnädig N.F., Ross L.S., Yeo T., Amaratunga C., et al. Artemisinin-resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic program to enhance survival. Nat. Commun. 2021;12:530–614. doi: 10.1038/s41467-020-20805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y., Wang F., Wang C., Zhang X., Jiang C., Ding F., Shen L., Zhang Q. Optimization of CRISPR/Cas system for improving genome editing efficiency in Plasmodium falciparum. Front. Microbiol. 2020;11:625862. doi: 10.3389/fmicb.2020.625862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown A.C., Moore C.C., Guler J.L. Cholesterol-dependent enrichment of understudied erythrocytic stages of human Plasmodium parasites. Sci. Rep. 2020;10:4591–4605. doi: 10.1038/s41598-020-61392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González N.S., Huber A., Algranati I.D. Spermidine is essential for normal proliferation of trypanosomatid protozoa. FEBS Lett. 2001;508:323–326. doi: 10.1016/s0014-5793(01)03091-5. [DOI] [PubMed] [Google Scholar]
- 34.Meylan S., Porter C.B.M., Yang J.H., Belenky P., Gutierrez A., Lobritz M.A., Park J., Kim S.H., Moskowitz S.M., Collins J.J. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert A., Benoit-Vical F., Meunier B. The key role of heme to trigger the antimalarial activity of trioxanes. Coord. Chem. Rev. 2005;249:1927–1936. doi: 10.1016/j.ccr.2004.12.022. [DOI] [Google Scholar]
- 36.Meunier B., Robert A. Heme as trigger and target for trioxane-containing antimalarial drugs. Acc. Chem. Res. 2010;43:1444–1451. doi: 10.1021/ar100070k. [DOI] [PubMed] [Google Scholar]
- 37.Cui L., Wang Z., Miao J., Miao M., Chandra R., Jiang H., Su X.z., Cui L. Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol. Microbiol. 2012;86:111–128. doi: 10.1111/j.1365-2958.2012.08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding C.R., Sidik S.M., Petrova B., Gnädig N.F., Okombo J., Herneisen A.L., Ward K.E., Markus B.M., Boydston E.A., Fidock D.A., Lourido S. Genetic screens reveal a central role for heme metabolism in artemisinin susceptibility. Nat. Commun. 2020;11:4813. doi: 10.1038/s41467-020-18624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobbold S.A., Chua H.H., Nijagal B., Creek D.J., Ralph S.A., McConville M.J. Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front-line antimalarial drugs. J. Infect. Dis. 2016;213:276–286. doi: 10.1093/infdis/jiv372. [DOI] [PubMed] [Google Scholar]
- 40.Siddiqui G., Srivastava A., Russell A.S., Creek D.J. Multi-omics based identification of specific biochemical changes associated with PfKelch13-mutant artemisinin resistant Plasmodium falciparum. J. Infect. Dis. 2017;215:1435–1444. doi: 10.1093/infdis/jix156. [DOI] [PubMed] [Google Scholar]
- 41.Mok S., Ashley E.A., Ferreira P.E., Zhu L., Lin Z., Yeo T., Chotivanich K., Imwong M., Pukrittayakamee S., Dhorda M., et al. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S., Leang R., Ringwald P., et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava A., Philip N., Hughes K.R., Georgiou K., MacRae J.I., Barrett M.P., Creek D.J., McConville M.J., Waters A.P. Stage-specific changes in Plasmodium metabolism required for differentiation and adaptation to different host and vector environments. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1006094. e1006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carey M.A., Covelli V., Brown A., Medlock G.L., Haaren M., Cooper J.G., Papin J.A., Guler J.L. Influential parameters for the analysis of intracellular parasite metabolomics. mSphere. 2018;3 doi: 10.1128/mSphere.00097-18. e00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olszewski K.L., Morrisey J.M., Wilinski D., Burns J.M., Vaidya A.B., Rabinowitz J.D., Llinás M. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar M., Skillman K., Duraisingh M.T. Linking nutrient sensing and gene expression in Plasmodium falciparum blood-stage parasites. Mol. Microbiol. 2021;115:891–900. doi: 10.1111/mmi.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang X., Shen S., Tang J., He X., Zhao Y., Wang C., He X., Guo G., Liu M., Wang L., et al. A cascade of transcriptional repression determines sexual commitment and development in Plasmodium falciparum. Nucleic Acids Res. 2021;49:9264–9279. doi: 10.1093/nar/gkab683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shang X., Wang C., Fan Y., Guo G., Wang F., Zhao Y., Sheng F., Tang J., He X., Yu X., et al. Genome-wide landscape of ApiAP2 transcription factors reveals a heterochromatin-associated regulatory network during Plasmodium falciparum blood-stage development. Nucleic Acids Res. 2022;50:3413–3431. doi: 10.1093/nar/gkac176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the current study have been deposited at Metabolights. Accession number is listed in the key resources table; This paper did not report any original code; Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.