Abstract
Macrophomina root rot disease (MRRD) caused by Macrophomina phaseolina is an emerging threat to the profitable cowpea production in northern Ghana.
Recommended control methods including the use of fungicides are ineffective and expensive for resource poor farmers whilst biocontrol options are not commercially available. An integrated method based on host plant resistance is considered the cheapest and most effective method of managing the disease. This study sought to confirm and characterize previously identified MRRD isolates from Northern Ghana using molecular technology, and to identify cowpea with potential sources of resistance to the MRRD. A PCR assay of ten isolates of the cowpea root rot pathogen revealed all isolates belonged to the species M. phaseolina, whilst a nucleotide BLAST of eight isolates showed 98% similarity with the sequences of Macrophomina isolates from other host available in GenBank. A sick pot method evaluation of 49 cowpea lines found 10 lines resistant to MRRD on a 1–9 disease severity scale (disease score, less than 5). A selection of eight resistant lines (Suvita 2, Abagbaala, IT97K573-1-1, IT93K-503-1-1, Hewale, AV2 3224, Nhyira and T2T4), and a susceptible check (Songotra) were evaluated against 10 isolates of M. phaseolina using a sick pot method. All the genotypes except for the susceptible check were resistant to MRRD. Thus, these genotypes could be used in cowpea MRRD resistance breeding programs.
Keywords: Macrophmina phaseolina, Cowpea lines, Root rot disease, Disease resistance, Northern Ghana
Macrophmina phaseolina; Cowpea lines; Root rot disease; Disease resistance; Northern Ghana.
1. Introduction
Cowpea (Vigna unguiculata (L.) Walp.), cultivation is an essential source of income and food security for resource-poor smallholder farmers in sub-Saharan Africa (Boukar et al., 2013). Its added ability to improve soil through fixation of atmospheric nitrogen, surviving and producing reasonable yield, and weed suppression makes it attractive for women farmers (Wiggins and Sharda, 2013; Alemu et al., 2016; Owusu et al., 2018). The crop is drought tolerant, and primarily produced for its protein-rich edible seed, with a protein content ranging from 20 to 25% of its dry weight. The leaves may also be eaten or sold as feed for farm animals (Boukar et al., 2019). An estimated 6.5 million metric tons of cowpea is produced annually from 14.5 million hectares worldwide (Boukar et al., 2019). Of this, 83.4% is produced in Africa, where 80% is produced in West Africa, with Nigeria and Niger as the leading producers (Kebede and Bekeko, 2020). In Ghana, it is the second most important legume crop. Ghana’s total cowpea production was 273,000 metric tonnes produced from 157,000 ha in 2018 most of which was cultivated in Northern Ghana (MoFA, 2019). Despite the great importance of the crop, generally, African farmers' cowpea yields have usually lag far behind their best varieties' potential yields of 2 tonnes per hectare (Kamara et al., 2018; Boukar et al., 2019; Kebede and Bekeko, 2020). These low yields are due to several factors, including biotic constraints of viral, bacterial and fungal origin, of which fungal diseases are considered the most important (Horn and Shimelis, 2020). The crop is affected by up to 40 fungal species, resulting in yield losses of 20%–100% (Horn and Shimelis, 2020).
Macrophomina phaseolina, an unimportant plant pathogen of cowpea in many areas, has emerged as a threat to cowpea production in sub-Saharan Africa (Ndiaye, 2007; Muchero et al., 2011; Kaur et al., 2012; Lamini et al., 2020). The pathogen is a soil and seed-borne polyphagous fungus, with a host range of over 500 plants (Kaur et al., 2012; Pandey et al., 2020). Macrophomina phaseolina infects plants by clogging the vascular bundle, thereby reducing the plants' ability to uptake water and nutrients, resulting in wilting and death, leading to severe yield losses (Muchero et al., 2011; Singh et al., 2012; Bodah, 2017). Common symptoms include seedling blights and root rots on infected plants (Kaur et al., 2012; Farr and Rossman, 2018; Negreiros et al., 2019). Predisposing factors associated with its occurrence include high temperature and drought conditions (Muchero et al., 2011; Kaur et al., 2012). In Sub-Saharan Africa, where cowpea is produced extensively, studies revealed increasing temperature and drought conditions which has a negative impact on crop production (Adejuwon et al., 2008; Ayanwuyi and Akintonde, 2012). A survey conducted on cultivated cowpea fields in northern Ghana in 2016 and 2017 revealed a 100% prevalence of the disease under rainfed and irrigated conditions (Lamini et al., 2020). Yield loss associated with the disease in cowpea is to be up to 10% (Ndiaye et al., 2010) under severe infestation.
Successful control of M. phaseolina is challenging due to its persistence as sclerotia in the soil and plant debris (Islam et al., 2012; Wagan et al., 2019). Recommended control of the pathogen involves a combination of strategies and methods to reduce the propagules in the soil, such as cultural, biological and chemical, which have primarily yielded less desirable results (Naseri, 2014; Lodha and Mawar, 2020). The most practical and effective management of the disease is host plant resistance, which offers the simplest, affordable and measurable disease control (Mbong et al., 2012; Khan et al., 2016; Bedawy and Moharm, 2019; Pandey et al., 2020). Cowpea sources of resistance to the MRRD have been identified and reported with most of them reporting varying levels of resistance (Muchero et al., 2011; Oladimeji et al., 2012; Ouédraogo et al., 2021). A combination of disease resistant cowpea with synthetic fungicides and microbial control agents can significantly reduce occurrence and severity of the MRRD in Ghana. Currently however, there are no synthetic fungicides or microbiolical control agents registerd in Ghana for the control of the MRRD of cowpea (EPA-Ghana, 2021). The challenge in finding cowpea as sources of resistance lies in the identification of cowpea genotypes with robust resistance to M. phaseolina (Ndiaye et al., 2010; Muchero et al., 2011). These efforts at getting sources of resistance to MRRD in cowpea have yielded few genotypes expressing resistance to the pathogens (Muchero et al., 2011; Oladimeji et al., 2012). A study by Muchero et al. (2011) reported three cowpea genotypes as highly resistant out of fourteen genotypes screened. A similar study by Oladimeji et al. (2012) found one out of five cowpea genotypes screened as resistant. In a report by Ouédraogo et al. (2021), out of 80 cowpea lines screened, five lines were completely resistant to the disease.
Although the MRRD has been reported and described in Ghana based on disease symptoms, morpho-cultural characteristics, and Koch’s postulates (Lamini et al., 2020), molecular confirmation and characterisation of the pathogen will enable the development of effective management strategies. Identifying cowpea sources of resistance will result in the timely field improvement of farmer preferred cultivars through introgression, which will boost cowpea production, resulting in improved livelihoods of resource-poor farmers. Therefore, the objectives of the present study were: (i) to identify sources of resistance against MRRD from 49 cowpea lines, and (ii) to characterize and compare the ten isolates of M. phaseolina from diseased samples of cowpea grown in different agro-climatic regions.
2. Materials and methods
2.1. Profile of experimental locations
The experiment was conducted from 2017 to 2019 at the Manga Station of the CSIR-Savanna Agricultural Research Institute (CSIR-SARI) located in the Upper East of Region of Ghana (Latitude: 10° 51′52″ Longitude: 0° 16′29″). The area falls within the Sudan Savannah agro-climatic zone characterised by a unimodal rainfall pattern. The rainy season starts from May to October, with the dry season starting from November to April. The average annual rainfall ranges from 800 to 1200 mm. The annual mean temperature during the rainy season is 28 °C whiles that of the dry season is 38 °C.
2.2. Cowpea lines
A total of 49 cowpea lines (Table 1) were assembled, consisting of cowpea breeding lines and cultivars developed by CSIR-SARI, CSIR-Crop Research Institute (CSIR-CRI), Institut Sénégalais de Recherches Agricoles (ISRA) in Senegal, International Institute of Tropical Agriculture (IITA) in Nigeria, Institut de l’Environnement et Recherches Agricoles (INERA) in Burkina Faso and the University of California, Riverside in the United States of America.
Table 1.
Cowpea lines screened for resistance to MRRD of cowpea in Northern Ghana.
| Cowpea line | Origin | Cowpea line | Origin |
|---|---|---|---|
| 1793K-503-1 | IITA/Nigeria | F4 (Sanzi × 499) (4) | CSIR-SARI/Ghana |
| 374 × Apag | CSIR-SARI/Ghana | F4(IT97K-499–35 × Sanzi) (5) | CSIR-SARI/Ghana |
| 374 × Padi | CSIR-SARI/Ghana | F4(Sanzi × 499) (2) | CSIR-SARI/Ghana |
| 499 × Apag | CSIR-SARI/Ghana | Hewale (CRI) | CSIR-CRI/Ghana |
| 499 × Omon | CSIR-SARI/Ghana | IT89D-374-57 | IITA/Nigeria |
| 503 × Apag | CSIR-SARI/Ghana | IT97K-499-35 (Songotra) | IITA/Nigeria |
| 503 × omon | CSIR-SARI/Ghana | IT99K 573-3-2-1 | IITA/Nigeria |
| 568 × Padi | CSIR-SARI/Ghana | IT99K-566-6 | IITA/Nigeria |
| Apagbaala | CSIR-SARI/Ghana | IT99K573-1-1 | IITA/Nigeria |
| Asetenapa (CRI) | CSIR-CRI/Ghana | KVX295-2-124-99 | INRA/Burkina faso |
| Asomdwie (CRI) | CSIR-CRI/Ghana | Marfotuya | CSIR-SARI/Ghana |
| AV1 3144 | CSIR-SARI/Ghana | Nhyria (CRI) | CSIR-CRI/Ghana |
| Av1 × 3161 | CSIR-SARI/Ghana | Padituya | CSIR-SARI/Ghana |
| AV2 3181 | CSIR-SARI/Ghana | Sanzi | CSIR-SARI/Ghana |
| AV2 3223 | CSIR-SARI/Ghana | SARC1-57-2 | CSIR-SARI/Ghana |
| AV2 3224 | CSIR-SARI/Ghana | SARC1-91-1 | CSIR-SARI/Ghana |
| Av2 3227 | CSIR-SARI/Ghana | Songotra | CSIR-SARI/Ghana |
| Bautawuta | CSIR-SARI/Ghana | Suvita 2 | INRA/Burkina faso |
| BC1 (Sanzi×499) Sanzi | CSIR-SARI/Ghana | T2T1 | CSIR-SARI/Ghana |
| BC1F1 (499×Sanzi) 499 (2) | CSIR-SARI/Ghana | T2T4 | CSIR-SARI/Ghana |
| BC1F4 (Sanzi × SARC) Sanzi | CSIR-SARI/Ghana | V2 3274 | CSIR-SARI/Ghana |
| Brown eye | CSIR-SARI/Ghana | Videza (CRI) | CSIR-CRI/Ghana |
| CB 27 | UCR-USA | Vita 7 | ISRA/Senegal |
| F4 (499 × Sanzi) (1) | CSIR-SARI/Ghana | Yacine | ISRA/Senegal |
2.3. Isolation of pathogen
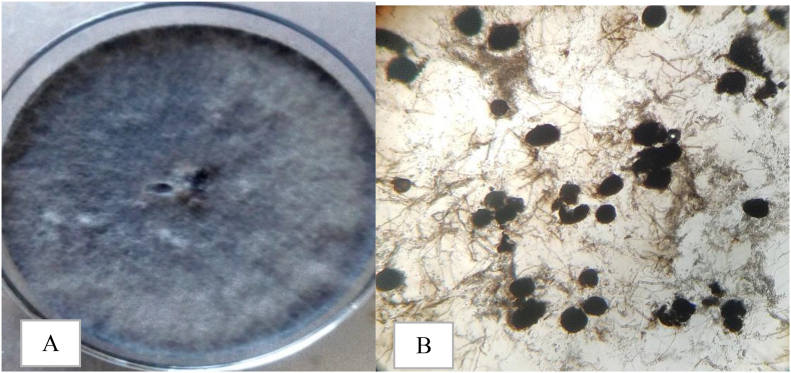
Isolates of M. phaseolina previously isolated from infected cowpea from fields across Northern Ghana were used for the studies (Table 2) (Lamini et al., 2020). The isolates were previously determined to be M. phaseolina based on disease symptoms [Figure 1(A–F)], morpho-cultural characteristics of fungus grown on potato dextrose agar (PDA) growth medium (Figure 2A), microscopic examination of the fungus under a compound microscope at ×20 magnification (Figure 2B) and pathogenicity of the fungus on cowpea [Figure 3(A and B)] (Lamini et al., 2020). Eleven isolates were retrieved from McCartney bottles containing PDA (Biolab Diagnostics Laboratory Incorporated, Hungary) stored at 4 °C, and sub cultured onto PDA (39 g/L distilled water autoclaved at 121 °C for 20 min) modified with streptomycin sulphate at 1.5 g/L.
Table 2.
Origin and characteristics of M. phaseolina collected from cowpea fields in Northern Ghana.
| Isolate | Origin | Mycelial growth | Sclerotial pattern |
|---|---|---|---|
| Arigu | Upper East | Greyish with profuse areal growth | Submerged |
| Feo | Upper East | Greyish with profuse areal growth | Submerged |
| Manga | Upper East | Greyish with profuse areal growth | Submerged |
| Asumsapelega | Upper East | Greyish profuse areal growth | Submerged |
| Sakpari | Upper East | Greyish with profuse areal growth | Submerged |
| Nafkluga | Upper East | Greyish with profuse areal growth | Submerged |
| Chinchan | Upper West | Greyish with profuse areal growth | Submerged |
| Silbelle | Upper West | Greyish with profuse areal growth | Submerged |
| Kpasenkpe | Northern | Greyish with profuse areal growth | Submerged |
| Botanga | Northern | Greyish with profuse areal growth | Submerged |
| Nyankpala | Northern | Greyish with profuse areal growth | Submerged |
Figure 1.
Symptoms of cowpea infected with M. phaseolina. (A) Cowpea field showing infected and non-infected plants, (B) cowpea plant showing chlorosis and wilting, (C) cowpea plants showing dark greyish necrosis on the stem, (D) cowpea plant showing sclerotia at the crown, (E) a comparison of root formation of noninfected versus and infected cowpea plants, (F) micro sclerotia on an infected cowpea stem.
Figure 2.
Morpho-cultural characteristics of M. phaseolina. (A) Growth of the fungus on PDA, (B) micro sclerotia of fungus (× 20 magnification).
Figure 3.
Pathogenicity of M. phaseolina on cowpea. (A) Non inoculated cowpea showing no symptoms, (B) inoculated cowpea showing symptoms of M. phaseolina infection.
3. Molecular analysis
3.1. Polymerase chain reaction
Molecular confirmation and characterisation of M. phaseolina isolates was conducted in the Biotechnology Laboratory of CSIR-SARI in Tamale, Ghana. Fungal mycelia of 11 isolates were scraped off 12-day-old cultures using a spatula, and ground in a mortar with a small amount of polyvinylpyrrolidone (PVP) as an antioxidant to prevent browning and degradation of the DNA. The ground mycelia are transferred into a 2 ml Eppendorf tube for genomic DNA extraction using a Cetyl Trimethyl Ammonium Bromide (CTAB) procedure described by Umesha et al. (2016). Extracted DNA was dissolved in 1X TE buffer, RNAse A (2 μl) was added to each tube and incubated at 37 °C for 1 h to eliminate RNA in the DNA. The quality of the genomic DNA was tested using 2% agarose gel stained with ethidium bromide. The ITS primer MpK1 [forward: CCGCCAGAGGACTATCAAAC, reverse: CGTCCGAAGCGAGGTGTAT] specific for M. phaseolina was used to identify and confirm the isolates as described by Babu et al. (2007). The polymerase chain reaction (PCR) conditions were denaturation at 94 °C for 30 s, annealing at 61 °C for 30 s and extension at 72 °C for 30 s. The PCR reaction mix used included 3 μl of ddH2O, 1 μl of DNA, 1 μl of primer and 5 μl of premix, making a total of 10 μl. Amplified products were subjected to band separation using a horizontal PAGE system. The gel used was a 6% polyacrylamide gel. The gel was run for 3 h at a voltage of 120 v. The bands were stained in an ethidium bromide solution and visualised using a UV transilluminator.
3.2. Phylogenetic test
PCR product of eight isolates with positive amplification for MPK1 (Arigu, Feo, Manga, Asumsapiliga, Botanga, Chinchan, Nafkluga and Nyankpala) out of the 11 isolates genotyped, were sequenced in South Africa, using the service of the biotechnology company Iquaba (inqaba biotec, Ghana) employing the Sanger sequencing method. Sequences received were extracted and trimmed using Bioedit software version 7.2. The sequences were nucleotide blast on National Center for Biotechnology Information (NCBI) platform. Blast results for the first 20 with high percentage identity (80–100) were selected, and transferred to Editplus software, where it was saved together with the sequence from the isolates as a FASTA file. Multiple sequence alignment was undertaken with the MEGA software version X (Kumar et al., 2008, 2018). The ClustalW alignment method (a general-purpose alignment for proteins and DNA sequences) was used with default settings for alignment parameters (gap opening penalty of 15, gap extension penalty of 6.65) and matrix parameters. The alignment file was exported as a MEGA file for phylogenic analysis. A phylogram was generated using the MEGA software version X (Kumar et al., 2008, 2018). The tree was rooted in Phytophthora palmivora. The Neighbour Joining (NJ) method was used with a Bootstrap value of 10,000 (Hall, 2013). Gaps and missing data were deleted whilst all other parameters were set to default. The final file generated was then saved for interpretation.
3.3. Spectrum of disease resistance against M. phaseolina
Two screening experiments were conducted to identify cowpea lines with putative resistance to MRRD. All experiments were undertaken under screen house conditions (30–40 °C and 70–80% RH) using the sick pot method in a CRD experimental design with five replications.
3.4. Fungi culture and inoculum preparation
A previously determined aggressive isolate of M. phaseolina (Manga) was used in the first screening experiment. For the second experiment however, ten isolates were used (Kpasenkpe isolate excluded) (Table 2). All the isolates were retrieved from PDA slants in McCartney bottles stored at 4 °C in the Plant Pathology Laboratory of CSIR-SARI. For each isolate, mycelial plugs of the fungal isolate were sub-cultured onto 90 mm Petri dishes containing 20 ml of freshly prepared acidified PDA (acidified with 1 ml/L of 85% lactic acid) following the manufacturer’s instruction. The mycelia produced was used in preparation of the inoculum.
Rice inoculum substrate prepared based on a modified protocol described by Abawi and Pastor-Corrales (1990) was used for the two screening experiments. In the modified protocol, the inoculum was prepared by weighing 50 g of rice grains in into a 250 ml conical flask, after which 20 ml distilled water was added to the grains. The contents were agitated for 15 min to allow for the grains to soak evenly. The contents were the allowed to stay for 30 min following which the conical flask was plugged with a ball of cotton wool and wrapped with aluminium foil. The contents were then autoclaved for 20 min at 103.4 kPa and 121 °C. The contents were then inoculated six mycelial discs (5 mm) obtained from 7-day old culture of M. phaseolina after it had been allowed to cool sufficiently (20–25 °C). The inoculum was incubated at room temperature (28–30 °C) for 15 days in the dark, following which they were air-dried in a 70% alcohol sterilised plastic tray at a temperature of 28–30 °C for 24 h. The substrates were then stored in Ziplock bags at 4 °C until needed.
3.5. Disease screening in screen house
In the first experiment, the 49 cowpea lines (Table 1) were screened against the Manga isolate of M. phaseolina. The cowpea lines were planted in plastic buckets (25 cm × 20 cm × 25 cm) containing heat sterilised soil. Each cowpea was inoculated with three grains of rice inoculum. The plants were irrigated as and when required. Data on disease severity was taken ten days after sowing (DAS) and at weekly intervals for five (5) weeks, following a 1–9 disease severity scale (Table 3) as described by Abawi and Pastor-Corrales (1990).
Table 3.
Disease severity key for MRRD disease of cowpea.
| Disease scale | Interpretation | Resistance rating |
|---|---|---|
| 1 | No visible symptoms on plants. | Highly resistant (HR) |
| 3 | Lesions are limited to cotyledonary tissue or hypocotyl. | Resistant (R) |
| 5 | Lesions have progressed from cotyledons to about 2 cm of stem tissues. | Tolerant (T) |
| 7 | Lesions are extensive on stem and branches. | Susceptible (S) |
| 9 | Most of the stem and growing points are affected by the development of sclerotia. | Highly susceptible (HS) |
Source: Modified after Abawi and Pastor-Corrales (1990).
Based on the 1–9 disease severity scale, cowpea genotypes with a disease severity rating of 1–4 were considered resistant whilst genotypes with a disease severity rating of 5–9 were considered susceptible in this study (Table 3).
In the second experiment, eight cowpea lines (Suvita 2, Apagbaala, T2T4, IT93K-503-1, IT99K-573-1-1, AV2 3224, Nhyira and Hewale) identified to be resistant and a susceptible check (Songotra) in the first screening were used. This was to ascertain the stability of the resistance in each cowpea line used and also to determine which had a robust resistance. Each of the cowpea line was screened against ten isolates of M. phaseolina (Table 2) collected from different locations across Northern Ghana (Kpasenkpe isolate excluded) in a 9 × 10 factorial experiment. The seeds were planted in plastic buckets (25 cm × 20 cm × 25 cm) containing heat sterilised soils. Each cowpea line was inoculated with three grains of rice containing inoculum. Data on disease severity was taken ten days after sowing (DAS) and at weekly intervals for five (5) weeks, following a 1–9 disease severity scale (Table 3).
3.6. Data analysis
The disease severity recorded at five weeks after planting (flowering stage) was used to determine the resistance levels of the 49 cowpea lines, whilst the mean disease severity scores over the period were estimated and used to calculate area under the disease progress curve (AUDPC) for each of the cowpea lines using a formula described by Campbell and Madden (1990):
where yi is the disease incidence (or severity) of the ith evaluation and yi+1 is the disease severity of the i + 1th evaluation. (ti+1−ti) is the number of days between two evaluations.
Genstat (version 12) was used for analysis of variance (ANOVA) for disease severity and AUDPC. Tukey’s HSD test (P ≤ 0.05) was used for mean separation.
4. Results
Polymerase chain reaction analysis of the 11 isolates of the M. phaseolina revealed an approximate band size of ∼340 base pairs (Figure 4). Out of the 11 isolates however, eight amplified and were used for phylogenetic studies.
Figure 4.
Amplified products obtained with primers MPK1 F&R (∼340 bp), Lane 1–11 show bands (arrowed) of DNA isolates of Macrophomina phaseolina from different locations. And L is the ladder.
The phylogeny tree (Figure 5) showed a clustering of eight isolates collected from the different locations in Northern Ghana.
Figure 5.
ClustalW multiple sequence cluster analysis of eight isolates associated with root rot disease of cowpea sampled from locations across northern Ghana.
Two major clusters were obtained. Cluster one (1) consisted of the rooted pathogen (Phytophthora palmivora) whiles cluster two (2) consisted of the sequences of the eight isolates and those obtained from the NCBI which revealed a greater than 98% identity to the eight isolates. That notwithstanding, all eight isolates sub clustered together with isolate from Nyankpala clustering differently from the other seven. Among all eight isolates, sequences of the isolates from Manga and Nafkulga appears to be the most identical/similar.
4.1. Resistance of cowpea lines to MRRD
Significant differences (P ≤ 0.05) in MRRD were observed amongst the 49 cowpea lines screened for resistance (Table 4). However, the most resistant lines based on the 1–9 reaction scale were Suvita 2, Apagbaala, 1793K-503-1 and IT99K573-1-1, with disease severity scores of 1.0, 1.8, 2.2 and 2.6, respectively. Cowpea lines Songotra, Vita 7, CB27, Padituya, KVX295-2-124-99 and V2 3274 were the most susceptible, with disease severity scores of nine (9). Similarly, the resistant lines Suvita 2, Apagbaala, 1793K-503-1, and IT99K573-1-1 recorded lower AUDPC values of 28.0, 40.6, 51.8 and 44.8. The most susceptible lines (Songotra, Vita 7, CB27, Padituya, KVX295-2-124-99 and V2 3274) recorded higher AUDPC that ranged from 203.0 to 232.4. The values for the AUDPC also revealed that most of the cowpea lines were susceptible to the MRRD. For cowpea lines BC1F1 (499 × Sanzi) 499 (2) and F4 (Sanzi × 499) (4), however, although they recorded a mean disease severity score of 7.0, which implied they were susceptible, they recorded an AUDPC of 89.6 and 81.2 which suggested they were resistant to the Macrophomina root rot pathogen.
Table 4.
Mean disease severity scores for MRRD and AUDPC of 49 cowpea lines inoculated with M. phaseolina.
| Cowpea line | Mean disease severity | AUDPC | Host reaction | Cowpea line | Mean disease severity | AUDPC | Host reaction |
|---|---|---|---|---|---|---|---|
| Suvita 2 | 1.0 | 28.0 | HR | F4(IT97K-499–35 × Sanzi) (5) | 7.0 | 130.2 | S |
| Apagbaala | 1.8 | 40.6 | HR | F4(Sanzi × 499) (2) | 7.0 | 166.6 | S |
| 1T93K-503-1 | 2.2 | 51.8 | HR | SARC1-91-1 | 7.0 | 177.8 | S |
| IT99K 573-1-1 | 2.6 | 44.8 | HR | Videza | 7.0 | 162.4 | S |
| AV2 3224 | 3.4 | 39.2 | R | 374 × Padituya | 7.4 | 183.4 | S |
| T2T4 | 3.4 | 44.8 | R | AV2 3181 | 7.4 | 141.4 | S |
| Asetenapa | 3.8 | 60.2 | R | Av2 3227 | 7.4 | 172.2 | S |
| Hewale | 4.2 | 70.0 | R | BC1F4 (Sanzi × SARC) Sanzi | 7.4 | 98.0 | S |
| Marfotuya | 4.2 | 89.6 | R | BC1 (Sanzi x 499) Sanzi | 7.8 | 119.0 | S |
| Nhyria | 4.2 | 92.4 | R | F4 (499 × Sanzi) (1) | 7.8 | 197.4 | S |
| 503 × Omon | 4.6 | 84.0 | R | 499 x Apagbaala | 8.2 | 218.4 | S |
| 499 × Omon | 5.0 | 98.0 | T | IT89D-374-57 | 8.2 | 196.0 | S |
| 503 × Apagbaala | 5.4 | 134.4 | T | IT99K-566-6 | 8.2 | 166.6 | S |
| 568 × Padituya | 5.4 | 149.8 | T | Zaayura | 8.2 | 197.4 | S |
| IT99K 573-3-2-1 | 5.4 | 137.2 | T | AV1 3144 | 8.6 | 170.8 | S |
| Yacine | 5.4 | 141.4 | T | Bautawuta | 8.6 | 210.0 | S |
| 374 × Apagbaala | 5.8 | 140.0 | T | Brown eye | 8.6 | 210.0 | S |
| Asomdwie | 5.8 | 152.6 | T | Sanzi | 8.6 | 176.4 | S |
| Av1 × 3161 | 5.8 | 130.2 | T | CB 27 | 9.0 | 232.4 | HS |
| IT97K-499-35 | 6.6 | 119.0 | T | KVX295-2-124-99 | 9.0 | 231.0 | HS |
| SARC1 - 57-2 | 6.6 | 127.4 | T | Padituya | 9.0 | 203.0 | HS |
| T2T1 | 6.6 | 141.4 | T | Songotra | 9.0 | 225.4 | HS |
| AV2 3223 | 7.0 | 159.6 | S | V2 3274 | 9.0 | 221.2 | HS |
| BC1F1 (499 × Sanzi) 499 (2) | 7.0 | 89.6 | S | Vita 7 | 9.0 | 228.2 | HS |
| F4 (Sanzi × 499) (4) | 7.0 | 81.2 | S | ||||
| Mean | 6.4 | 140.5 | |||||
| L.S. D (P = 0.05) | 2.3 | 71.1 | |||||
| CV% | 29.3 | 40.6 |
Mean disease severity scored at flowering based on 1–9 disease scale.
4.2. Evaluation of nine (9) cowpea lines against ten isolates of M. Phaseolina collected from Northern Ghana
From the host reaction results (Table 4), most of the cowpea lines evaluated against the Manga isolate of M. phaseolina pathogen were susceptible or highly susceptible to the MRRD. Only 10 of the cowpea lines were found to be resistant based on the 1–9 disease severity scale.
There was no interaction between the root rot fungal isolates and the cowpea lines (P ≥ 0.05) (Table 5). There were, however, significant (P ≤ 0.05) differences amongst the isolates and the cowpea lines. The isolate from Manga was the most aggressive based on mean severity (3.76), whilst the Botanga isolate expressed the least aggression (2.11). However, the Manga isolate did not vary significantly (P ≥ 0.05) from the Feo, Silbelle, Asumsapeliga, Chinchan and Arigu, which recorded mean reactions of 3.58, 3.42, 3.40, 3.22 and 3.18, respectively.
Table 5.
Mean severity of MRRD on nine cowpea lines inoculated with isolates of the pathogen from ten locations in Northern Ghana.
| M. phaseolina Isolates | Cowpea lines |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Apagbaala | AV22224 | Hewale | IT93K503-1 | IT99K573-1-1 | Nhyira | Songotra (susceptible check) | Suvita2 | T2T4 | Mean virulence | |
| Arigu | 1.40 | 3.40 | 3.00 | 2.60 | 3.00 | 3.40 | 7.00 | 1.80 | 3.00 | 3.18 |
| Asumsapeliga | 2.20 | 3.00 | 3.00 | 3.00 | 2.60 | 3.40 | 8.20 | 1.80 | 3.40 | 3.40 |
| Botanga | 1.80 | 1.80 | 1.40 | 1.80 | 1.00 | 3.00 | 5.80 | 1.40 | 1.00 | 2.11 |
| Chinchan | 2.20 | 3.40 | 3.40 | 2.60 | 2.60 | 3.40 | 7.00 | 1.40 | 3.00 | 3.22 |
| Feo | 2.20 | 3.40 | 3.40 | 3.00 | 2.60 | 3.00 | 8.60 | 2.60 | 3.40 | 3.58 |
| Manga | 3.00 | 3.40 | 3.40 | 3.00 | 3.00 | 3.40 | 9.00 | 2.20 | 3.40 | 3.76 |
| Nafkuluga | 2.20 | 3.80 | 2.60 | 2.60 | 2.20 | 2.20 | 7.00 | 1.80 | 3.40 | 3.09 |
| Nyankpala | 1.00 | 1.80 | 1.80 | 1.80 | 1.80 | 2.60 | 6.60 | 1.80 | 1.40 | 2.29 |
| Sakpari | 1.80 | 2.20 | 3.00 | 2.60 | 2.20 | 3.40 | 6.20 | 1.40 | 3.00 | 2.87 |
| Silbelle | 2.60 | 3.40 | 3.40 | 2.60 | 2.20 | 3.80 | 7.80 | 1.80 | 3.20 | 3.42 |
| Mean disease severity | 2.04 | 2.96 | 2.84 | 2.56 | 2.32 | 3.16 | 7.32 | 1.80 | 2.82 | 3.09 |
L.S.D (P = 0.05) genotype = 0.63; isolate = 0.66; genotype × isolate = NS. CV (%) = 22.2.
(NS) = not significant.
All the lines, except for Songotra (the susceptible check), showed resistance stability (1–3 disease score) to the root rot pathogen (Table 5). Suvita 2 was, however the most resistant (1.8) and was significantly lower (P ≤ 0.05) to T2T4 (2.82), Hewale (2.84), AV2 3224 (2.96), Nhyira (3.16) and Songotra (7.32). It however did not vary significantly (P ≥ 0.05) from Apagbaala (2.04), IT99K573-1-1 (2.32) and IT93K503-1 (2.56).
5. Discussion
The approximately 340 bp observed in the PCR analysis agreed with the findings of Babu et al. (2007), which confirmed the root rot pathogens from the various locations to be M. phaseolina. The phylogenetic analysis of the Macrophomina root rot isolates from different locations also showed high identity (98%) with sequences of M. phaseolina deposited at the NCBI database. They further confirmed the isolates to be M. phaseolina and the actual cause of the root rot disease of cowpea observed on farmers' fields in 2016 and 2017 (Lamini et al., 2020). It also revealed the pathogen to be the same without any species, thus confirming the pathogen as a monotypic fungus as earlier reported (Wyllie, 1993; Babu et al., 2007; Kaur et al., 2012; Phillips et al., 2013).
The current effects of climate change are expected to negatively affect crop production due to the increasing occurrence of high temperature and drought conditions (Medhaug et al., 2017). This phenomenon culminates in biotic constraints, including pests and diseases in locations where they were previously absent (Zayan, 2018). The recent emergence of M. phaseolina in several locations globally is an example of a pathogen associated with climate change, and its associated disease incidence and severity may worsen (Medhaug et al., 2017). The pathogen is associated with an extensive yield loss of many crops, including grain legumes (Mengistu et al., 2015; Bodah, 2017). Its control is challenging due to its versatile nature, thus making current control methods ineffective (García et al., 2019; Lodha and Mawar, 2020). The recent occurrence of the disease in farmers' fields in Northern Ghana (Lamini et al., 2020) requires the development of disease mitigation strategies based on an integrated approach to safeguard the yields of resource-poor farmers. Host plant resistance offers the most practical and low-cost option for its control; however, sources of resistance are inadequate and, in most cases, lacking (Ndiaye et al., 2010). Identifying sources of resistance requires a robust system of screening genotypes against root rot pathogens such as M. phaseolina (Gbaguidi et al., 2013).
Screening the 49 cowpea lines for resistance to MRRD revealed variations in levels of disease severity against the Macrophomina root rot pathogen. The observed variation in disease severity corroborates with findings from Muchero et al. (2011) on cowpea and for other grain legumes evaluated for the disease, including bean (Mayek-Pérez et al., 2001), and soybean (Mengistu et al., 2013). The fact that within the variation observed in disease severity, some genotypes were resistant is encouraging because it offers the opportunity to select good sources of cowpea genotypes with resistance to MRRD. Generally, however, relatively few genotypes were resistant to the disease, with only 10 out of 49 cowpea lines showing resistance, whilst the rest were tolerant or susceptible to the disease. This relatively low number of cowpea lines expressing resistance to the root rot disease demonstrates the difficulty in sourcing cowpea with high resistance levels to M. phaseolina (Ndiaye, 2007; Ndiaye et al., 2010; Muchero et al., 2011; Ouédraogo et al., 2021). The challenge faced in finding sources of resistance to M. phaseolina is, however, not limited to cowpea since similar challenges have been reported for other grain legumes such as soybean (Smith and Carvil, 1997; Pawlowski et al., 2015; Coser et al., 2017; Amrate et al., 2019), bean (Mayek-Pérez et al., 2001a; Hernandez-Delgado et al., 2009), mungbean (Pandey et al., 2021) and black gram (Iqbal et al., 2010; Elmerich et al., 2022). The relatively few genotypes with resistance to the root rot disease also further emphasise the challenges faced in getting sources of resistance that may be used to improve field resistance of farmer-preferred but susceptible cultivars to the disease.
Generally, all the 10 isolates of M. phaseolina collected from the different locations in Northern Ghana caused disease on the nine cowpea lines they were screened against. The level of disease severity, however, varied amongst the isolates on the genotypes. However, isolates from Manga, Feo, Silbelle, Chinchan, and Asumsapeliga were the most aggressive. The variability in pathogenicity amongst the isolates strongly suggests that cowpea farmers are dealing with Macrophomina root rot pathogens with different levels of aggressiveness, and more isolates should be collected from other locations to determine the range of variability. Efforts at selecting genotypes for resistance to MRRD should therefore consider this variability in pathogenicity.
In assessing resistant stability (a near-constant disease resistance irrespective of the variant of the pathogen isolates) of the eight cowpea lines previously identified to be resistant, all the cowpea lines except for Songotra (the susceptible check) were resistant to all the 10 root rot isolates. Hence, confirming the lines stability in resistance to MRRD. The lines Suvita 2, Apagbaala, Hewale, AV2 3224, Nhyira, IT99K573-1-1, IT93K303-1 and T2T4 were previously resistant to Macrophomina root rot in an earlier screening was also found to be resistant to 10 isolates of M. phaseolina collected from different locations in Northern Ghana (Lamini et al., 2020. Unpublished doctoral thesis). In a previous experiment, Muchero et al. (2011) also reported cowpea genotypes Suvita 2 and IT93K-503-1 (included in this study) to be resistant to M. phaseolina in screening studies at the University of California, Riverside. The general resistance of the cowpea lines to the different Macrophomina isolates in this study offers the hope of improving cowpea cultivars in Northern Ghana for field resistance to the root rot disease.
6. Conclusion
This study confirmed the causal pathogen of MRRD M. phaseolina which was also monotypic based on the molecular confirmation and characterisation. The identified sources resistance provides a valuable resource to plant breeders to improve field resistance of farmer preferred cowpea cultivars to the MRRD. Eight cowpea genotypes which were further screened against 10 isolates of M. phaseolina for resistant stability were found near constant in their disease resistance (stable) regardless of the variant of the pathogen isolates.
Declarations
Author contribution statement
Salim Lamini, PhD: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Francis Kusi; Eric William Cornelius, Ph. D; Agyemang Danquah, Ph. D: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Patrick Attamah; Justice Frederick Awuku: Performed the experiments; Analyzed and interpreted the data.
Zakaria Mukhtaru; Emmanuel Yaw Owusu; Mavis Agyeiwaa Acheampong; Gloria Mensah: Performed the experiments.
Funding statement
Mr Salim Lamini was supported by Kirkhouse Trust [SC 047432].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to acknowledge the Kirkhouse Trust, United Kingdom for their support and funding for this study.
References
- Abawi G.S., Pastor-Corrales M.A. Root rot of beans in Latin American and Africa: diagnosis, research, methodologies, and management strategies. CIAT, Cali, Colombia. Resúmenes Analíticos sobre Frijol. 1990;3(68):114. [Google Scholar]
- Adejuwon J., Barros V., Burton I., Kulkarni J., Lasco R., Leary N. Routledge; 2008. Climate Change and Adaptation. [Google Scholar]
- Alemu M., Asfaw Z., Woldu Z., Fenta B.A., Medvecky B. Cowpea (Vigna unguiculata (L.) Walp.)(Fabaceae) landrace diversity in northern Ethiopia. Int. J. Biodivers. Conserv. 2016;8(11):297‒309. [Google Scholar]
- Amrate P.K., Shrivastava M.K., Bhale M.S. Resistance in soybean varieties against charcoal rot disease caused by Macrophomina phaseolina. Plant Dis. Res. 2019;34(2):124–128. [Google Scholar]
- Ayanwuyi E., Akintonde J.O. Effect of climate change on cowpea production in Kuje Area council, Abuja, Nigeria. Int. J. Adv. Res. Manag. Soc. Sci. 2012;1(2):273–283. [Google Scholar]
- Babu B.K., Saxena A.K., Srivastava A.K., Arora D.K. Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia. 2007;99(6):797–803. doi: 10.3852/mycologia.99.6.797. [DOI] [PubMed] [Google Scholar]
- Bedawy I., Moharm M.H. Reaction and performance of some sesame genotypes for resistance to Macrophomina phaseolina, the incitant of charcoal rot disease. Alexandria Sci. Exchange J. 2019;40:12‒18. [Google Scholar]
- Bodah E.T. Root rot diseases in plants: a review of common causal agents and management strategies. Agric. Res. Technol. 2017;5(3):555‒661. [Google Scholar]
- Boukar O., Bhattacharjee R., Fatokun C., Kumar P.L., Gueye B. Elsevier Incorporation; 2013. Genetic and Genomic Resources of Grain Legume Improvement. [Google Scholar]
- Boukar O., Belko N., Chamarthi S., Togola A., Batieno J., Owusu E., Haruna M., Diallo S., Umar M.L., Olufajo O., Fatokun C. Cowpea (Vigna unguiculata): genetics, genomics and breeding. Plant Breed. 2019;138(4):415–424. [Google Scholar]
- Campbell C.L., Madden L.V. John Wiley and Sons; 1990. Introduction to Plant Disease Epidemiology. [Google Scholar]
- Coser S.M., Chowda Reddy R.V., Zhang J., Mueller D.S., Mengistu A., Wise K.A., Allen T.W., Singh A., Singh A.K. Genetic architecture of charcoal rot (Macrophomina phaseolina) resistance in soybean revealed using a diverse panel. Front. Plant Sci. 2017;8:1626. doi: 10.3389/fpls.2017.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmerich C., Pandey A.K., Vemula A., Rathore A., Nair R.M. Blackgram-macrophomina phaseolina interactions and identification of novel sources of resistance. Plant Dis. 2022;106(11):2911–2919. doi: 10.1094/PDIS-11-21-2588-RE. [DOI] [PubMed] [Google Scholar]
- EPA-Ghana . 2021. Environmental Protection Agency: Pesticide Registration Manual. Accra, Ghana. [Google Scholar]
- Farr D.F., Rossman A.Y. USDA; 2018. Fungal Databases, US National Fungus Collections, ARS.https://nt.ars-grin.gov/fungaldatabases/ Retrieved December 20, 2020, from. [Google Scholar]
- García M.E.M., Sousa E.S., Silva J.D.D.C., Melo M.P.D., Mota J.M.D., Almeida Neto A.D.D., Gomes R.L.F., Beserra J.E.A., Jr. Reaction of lima bean genotypes to Macrophomina phaseolina. Summa Phytopathol. 2019;45(1):11‒17. [Google Scholar]
- Gbaguidi A.A., Dansi A., Loko L.Y., Dansi M., Sanni A. Diversity and agronomic performances of the cowpea (Vigna unguiculata Walp.) landraces in southern Benin. Int. Res.J. Agric. Sci. Soil Sci. 2013;3(4):121–133. [Google Scholar]
- Hall B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013;30(5):1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- Hernandez-Delgado S., Reyes-Valdés M.H., Rosales-Serna R., Mayek-Perez N. Molecular markers associated with resistance to Macrophomina phaseolina (Tassi) Goid. in common bean. J. Plant Pathol. 2009:163‒170. [Google Scholar]
- Horn L.N., Shimelis H. Production constraints and breeding approaches for cowpea improvement for drought prone agro-ecologies in sub Saharan Africa. Ann. Agric. Sci. 2020;65(1):83‒91. [Google Scholar]
- Iqbal U., Mukhtar T., Iqbal S.M., Ul-Haque I., Malik S.R. Host plant resistance in blackgram against charcoal rot (Macrophomina phaseolina (Tassi) Goid) Pakistan J. Phytopathol. 2010;22(1):126‒129. [Google Scholar]
- Islam M.S., Haque M.S., Islam M.M., Emdad E.M., Halim A., Hossen Q.M., Hossain M.Z., Ahmed B., Rahim S., Rahman M.S., Alam M.M. Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genom. 2012;13(1):493. doi: 10.1186/1471-2164-13-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamara A.Y., Omoigui L.O., Kamai N., Ewansiha S.U., Ajeigbe H.A. Burleigh Dodds Science Publishing; 2018. Improving cultivation of cowpea in West Africa; pp. 1–18. (Achieving Sustainable Cultivation of Grain Legumes Volume 2: Improving Cultivation of Particular Grain legumes. Burleigh Dodds Series in Agricultural Science). [Google Scholar]
- Kaur S., Dhillon G.S., Brar S.K., Vallad G.E., Chand R., Chauhan V.B. Emerging phytopathogen Macrophomina phaseolina: biology, economic importance and current diagnostic trends. Crit. Rev. Microbiol. 2012;38(2):136‒151. doi: 10.3109/1040841X.2011.640977. [DOI] [PubMed] [Google Scholar]
- Kebede E., Bekeko Z. Expounding the production and importance of cowpea (Vigna unguiculata (L.) Walp.) in Ethiopia. Cogent Food Agric. 2020;6(1) [Google Scholar]
- Khan K.A., Shoaib A., Akhtar S. Response of Vigna radiata (L.) Wilczek genotypes to charcoal rot disease. Mycopathology. 2016;14(1&2):1–7. [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinf. 2008;9(4):299‒306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547‒1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamini S., Cornelius E.W., Kusi F., Danquah A., Attamah P., Mukhtaru Z., Awuku J.F., Mensah G. Prevalence, incidence and severity of a new root rot disease of cowpea caused by Macrophomina phaseolina (Tassi) Goid in Northern Ghana. West Afr. J. Appl. Ecol. 2020;28(2):140–154. [Google Scholar]
- Lodha S., Mawar R. Population Dynamics of Macrophomina phaseolina in Relation to disease management: A review. J. Phytopathol. 2020;168(1):1–17. [Google Scholar]
- Mayek-Pérez N., López-Castañeda C., López-Salinas E., Cumpián-Gutiérrez J., Acosta-Gallegos J.A. Macrophomina phaseolina resistance in common bean under field conditions in Mexico. Agrociencia. 2001;35:649–661. [Google Scholar]
- Mbong G.A., Fokunang C.N., Emechebe A.M., Alabi O., Alegbejo M.D., Fontem D.A. The effect of Sphaceloma sp causal agent of scab infection on grain yield of cowpea (Vigna unguiculata) in northern Nigeria. Int. Res. J. Biochem. Bioinf. 2012;2(5):98–104. [Google Scholar]
- Medhaug I., Stolpe M.B., Fischer E.M., Knutti R. Reconciling controversies about the ‘global warming hiatus’. Nature. 2017;545(7652):41–47. doi: 10.1038/nature22315. [DOI] [PubMed] [Google Scholar]
- Mengistu A., Arelli P., Bond J., Nelson R., Rupe J., Shannon G., Wrather A. Identification of soybean accessions resistant to Macrophomina phaseolina by field screening and laboratory validation. Plant Health Prog. 2013;14(1):25. [Google Scholar]
- Mengistu A., Wrather A., Rupe J.C. In: Compendium of Soybean Diseases and Pests. fifth ed. Hartman G.L., Rupe J.C., Sikora E.J., Domier L.L., Davis J.A., Steffey K.L., editors. APS Press; St. Paul (MN): 2015. Charcoal rot; pp. 67–69. [Google Scholar]
- MOFA . 2019. Ministry of Food and Agriculture, Agriculture in Ghana: Facts and Figures. Statistics, Research and Information Directorate (SRID) [Google Scholar]
- Muchero W., Ehlers J.D., Close T.J., Roberts P.A. Genic SNP markers and legume synteny reveal candidate genes underlying QTL for Macrophomina phaseolina resistance and maturity in cowpea [Vigna unguiculata (L) Walp.] BMC Genom. 2011;12(1):8. doi: 10.1186/1471-2164-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseri B. Charcoal rot of bean in diverse cropping systems and soil environments. J. Plant Dis. Prot. 2014;121(1):20–25. [Google Scholar]
- Ndiaye M. Wageningen University and Research Centre; Wageningen, The Netherlands: 2007. Ecology and Management of Charcoal Rot (Macrophomina phaseolina) on Cowpea in the Sahel. Ph.D. thesis. [Google Scholar]
- Ndiaye M., Termorshuizen A.J., Van Bruggen A.H.C. Effects of compost amendment and the biocontrol agent Clonostachys rosea on the development of charcoal rot (Macrophomina phaseolina) on cowpea. J. Plant Pathol. 2010:173–180. [Google Scholar]
- Negreiros A.M.P., Sales Júnior R., León M., de Assis Melo N.J., Michereff S.J., de Queiroz Ambrósio M.M., de Sousa Medeiros H.L., Armengol J. Identification and pathogenicity of Macrophomina species collected from weeds in melon fields in northeastern Brazil. J. Phytopathol. 2019;167(6):326‒337. [Google Scholar]
- Oladimeji A., Balogun O.S., Busayo T.S. Screening of cowpea genotypes for resistance to Macrophomina phaseolina infection using two methods of inoculation. Pathology. 2012;6(1):13–18. [Google Scholar]
- Ouédraogo N., Zida E.P., Ouédraogo H.M., Sawadogo N., Soalla R.W., Batieno J.B.T., Ouoba A., Ouédraogo M., Sawadogo M. Identification of resistant cowpea (Vigna unguiculata (L.) Walp.) genotypes against charcoal under artificial inoculation. Asian J. Plant Sci. 2021;20:271–280. [Google Scholar]
- Owusu E.Y., Akromah R., Denwar N.N., Adjebeng-Danquah J., Kusi F., Haruna M. Inheritance of early maturity in some cowpea (Vigna unguiculata (L.) Walp.) genotypes under rain fed conditions in northern Ghana. Adv. Agric. 2018;2018:1–10. [Google Scholar]
- Pandey A.K., Burlakoti R.R., Rathore A., Nair R.M. Morphological and molecular characterization of Macrophomina phaseolina isolated from three legume crops and evaluation of mungbean genotypes for resistance to dry root rot. Crop Protect. 2020;127 [Google Scholar]
- Pandey A.K., Yee M., Win M.M., Lwin H.M.M., Adapala G., Rathore A., Sheu Z.M., Nair R.M. Identification of new sources of resistance to dry root rot caused by Macrophomina phaseolina isolates from India and Myanmar in a mungbean mini-core collection. Crop Protect. 2021;143 [Google Scholar]
- Pawlowski M.L., Hill C.B., Hartman G.L. Resistance to charcoal rot identified in ancestral soybean germplasm. Crop Sci. 2015;55(3):1230‒1235. [Google Scholar]
- Phillips A.J., Alves A., Abdollahzadeh J., Slippers B., Wingfield M.J., Groenewald J.Z., Crous P.W. The Botryosphaeriaceae: genera and species known from culture. Stud. Mycol. 2013;76(1):51–167. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.K., Chittpurna A., Sharma V., Patil P.B., Korpole S. Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.S., Carvil O.N. Field screening of commercial and experimental soybean cultivars for their reaction to Macrophomina phaseolina. Plant Dis. 1997;81(4):363–368. doi: 10.1094/PDIS.1997.81.4.363. [DOI] [PubMed] [Google Scholar]
- Umesha S., Manukumar H.M., Raghava S. A rapid method for isolation of genomic DNA from food-borne fungal pathogens. 3 Biotech. 2016;6(2):123. doi: 10.1007/s13205-016-0436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagan K.H., Khaskheli M.I., Hajano J.D., Lanjar A.G. Population density and aggressiveness of Macrophomina phaseolina isolates from Sindh, Pakistan. Sarhad J. Agric. 2019;35(2):400–407. [Google Scholar]
- Wiggins S., Sharda K. Overseas Development Institute (ODI); 2013. Looking back, peering forward: what has been learned from the food-price spike, 2007–2008. (ODI Briefing 81). [Google Scholar]
- Wyllie T.D. In: Compendium of Soybean Diseases. third ed. Sinclair J.B., Backman P.A., editors. APS Press; St. Paul, MN: 1993. Charcoal rot. [Google Scholar]
- Zayan S.A. Impact of climate change on plant diseases and IPM strategies. Arab J. Plant Protect. 2018;36(1):75–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.