Abstract
Site-specific recombinase Int mediates integration of the bacteriophage λ genome into the Escherichia coli chromosome. Integration occurs once the Int tetramer, assisted by the integration host factor IHF, forms the intasome, a higher order structure, within which Int, a heterobivalent protein, interacts with two nonhomologous DNA sequences: the core recombination sites and the accessory arm sites. The binding to these sites is mediated by the catalytic C-terminal domain (CTD) and the regulatory N-terminal domain (NTD) of Int, respectively. Within Int, the NTD can activate or inhibit the recombination activity of the CTD depending on whether the NTD is bound to the arm sites. The CTD alone cannot mediate recombination, and even when the NTD and the CTD are mixed together as individual polypeptides, the NTD cannot trigger recombination in the CTD. In this work, we set to determine what modifications can unlock the recombination activity in the CTD alone and how the CTD can be modified to respond to recombination-triggering signals from the NTD. For this, we performed a series of genetic analyses, which showed that a single mutation that stabilizes the CTD on DNA, E174K, allows the CTD to recombine the core DNA sequences. When the NTD is paired with the CTD (E174K) that also bears a short polypeptide from the C terminus of the NTD, the resulting binary Int can recombine arm-bearing substrates. Our results provide insights into the molecular basis of the regulation of the Int activity and suggest how binary recombinases of the integrase type can be engineered.
Keywords: DNA recombination, protein-DNA interaction, protein-protein interaction, protein engineering, structure-function, synthetic biology
Abbreviations: CTD, C-terminal domain; NTD, N-terminal domain
Bacteriophage λ integrase, Int, is a founding member of the tyrosine family of site-specific DNA recombinases that houses such popular DNA recombinases as Cre and Flp as well as about 1000 other recombinases that share the chemistry of the DNA exchange reaction and the overall three-dimensional organization (1). The function of λ Int is to integrate the bacteriophage λ genome into and to excise it from the Escherichia coli chromosome (for recent review on λ Int see Landy, 2015 (2)). In contrast to simple members of the tyrosine family, such as Cre and Flp, which do not require any accessory proteins to perform recombination, both the integration and excision activities of Int depend on the host protein IHF (integration host factor), while the excision activity also depends on the phage encoded protein Xis. IHF and Xis are the DNA-bending proteins that bind within the phage genome region called attP (the phage attachment site) and bring closer two groups of sequences that have different functional role: the core recombination sites and the accessory arm sites (Fig. 1A). Two sets of the arm sites (P and P′) are located upstream and downstream from the core sites and are separated from them by ∼100 bp and ∼50 bp, respectively. Once DNA within attP is properly folded, a tetramer of Int, a bivalent protein, can bind both the arm sites and the core sites forming a higher order, recombination-competent structure called the intasome (Fig. 1B). Functional analysis of the polypeptides that were obtained by limited proteolytic digestion of Int demonstrated that the N-terminal domain of Int (NTD, residues 1–64, Fig. 1C) mediates binding to the arm sites of attP while the C-terminal domain of Int (CTD, residues 65–356, Fig. 1C) mediates binding to the core sites of attP (3).
Figure 1.
Bacteriophage lambda pathway, structure of Int tetramer bound to cognate DNA sequences, and structure of Int monomer.A, schematics of integration/excision of the bacteriophage λ chromosome into/from the E. coli chromosome. The relative location of the core recombination sites (COC’ and BOB′) and the accessory arm sites (P and P′) are indicated. B, 3D structure of the Int tetramer bound to the Holliday structure intermediate; PDB code 1z1g (28). C, detailed view of the Int monomer. The location of Int domains, linkers, and several critical residues identified in this work is pointed by arrows. PDB, Protein Data Bank.
The CTD is responsible for the recombination activity of Int but this domain can recombine the core sites only when it is the integral part of Int, that is, covalently connected to the NTD. When expressed as an individual protein, the CTD can bind to the core sites, cleave them, and resolve premade Holliday junction recombination intermediates that contain the core sites but cannot perform the actual recombination reaction (4, 5).
Within Int, the NTD inhibits all activities of the CTD unless the NTD is bound to the arm sites. The binding to the arm sites changes the regulatory mode of the NTD so it can then stimulate the activity of the CTD. Remarkably, when the NTD and the CTD are combined as individual proteins in vitro, the NTD is able to stimulate the DNA binding and cleavage activities of the CTD even if the NTD is not bound to the arm sites (4).
Cumulative data on the activities of the NTDs and CTDs of Int and their functional interactions suggest that the ability of the CTD to recombine the core sites are weakened to allow the NTD to regulate this activity of CTD once the NTD binds to the arm sites (2). Such dependency on the NTD binding restricts the recombination activity of Int to the settings that support the formation of the intasome. Understanding the molecular mechanism of the regulatory interactions between the NTDs and CTDs is important not only for understanding the molecular mechanism of the functional regulation of Int but also for designing recombination enzymes, activity of which can be similarly controlled.
Deciphering the molecular basis of the interactions between the NTD and CTD depends on how the functional boundaries of these domains are defined and whether they overlap. The original definition of the domains—NTD(1–64) and CTD(65–356)—was later adjusted since it was shown that for the NTD to be fully functional (that is, to be able to bind to the arm sites cooperatively), the NTD(1–64) needs six additional residues to become NTD(1–70) (5). It was also shown that the length of the CTD(65–356) can be reduced by 10 amino acid residues and the resulting CTD(75–356) polypeptide essentially as active in the binding and cleavage activities as the CTD(65–356) (5). Moreover, the CTD(75–356) was capable of cocrystallizing with the cleaved DNA substrate (6).
The crystal structures of Int complexed with both the core and the arm sites showed that the NTD and the CTD do not apparently interact directly if the C-terminal of the NTD is considered to end at the residue 70 and the N terminus of the CTD to start at the residue 75 (7). On the other hand, it was suggested that different domain interfaces might form at different reaction steps (8). It is therefore possible that the interaction snapshots trapped in the crystal structures may not represent all functional interactions especially because the Int/DNA cocrystal was formed by the arm sites with unexpected dyad symmetry of the arm sites. It is also possible that not all identified interactions are functional since the NTD(1–62) is able to interact with the CTD(75–356) (8) but the molecular basis for these interactions cannot be easily inferred from the Int/DNA cocrystal (7).
To get insight into the molecular basis of the functional interactions between the NTD and CTD, we wanted to analyze what modifications can trigger the recombination reaction when the two domains communicate. These interactions can be probed by modifying the domains and testing how these modifications affect the efficiency of the recombination reaction. In principle, interactions between the NTDs and CTDs can be tested either when these domains are covalently connected (that is, when they are the integral part of the Int molecule) or when they are combined as separately expressed polypeptides. The latter approach has an obvious advantage since the CTD is recombinationally inactive and therefore if the modification of the domains can trigger even a subtle recombination activity, it can be detected, especially using the in vivo approaches.
In this work we asked two main questions (1): what modifications can unlock the recombination activity in the CTD in the absence of the NTD and (2) how the CTD can be modified to efficiently respond to recombination triggering signals from the NTD. To answer these questions, we performed a series of genetic analyses that utilized the reporter vectors with different recombination substrate pairs and the vectors that expressed variants of the NTD and the CTD. We found that a single modification that stabilizes the CTD on DNA is sufficient to allow this domain to recombine the core DNA sequences: the int-h mutation, E174K (9, 10, 11). We also found that when the NTD is paired with the CTD that has the int-h mutation and a short polypeptide from the C terminus of the NTD, the resulting binary Int can recombine several arm site–bearing substrates. The functional performance of binary Int provides insights into the molecular basis of the regulation of the λ integrase activity and suggests how binary recombinases of the integrase type can be engineered.
Results
CTD of Int with int-h mutation can recombine core DNA sites
Since the CTD of Int is not capable of performing recombination (4), the experiments to determine what modifications are needed to trigger its recombination activity are likely to yield the CTD variants with only marginal activity. Consequently, these variants will be difficult to detect using the in vitro assays and thus the in vivo approaches, which can identify variants with quite low recombination activity, are preferred. To this end, we utilized the bacterial ‘positive’ recombination screening system, which is capable of detecting recombinase variants even if their activity is less than 0.01% of that of the WT recombinase (12). The positive screening system is composed of the reporter and expression vectors that can coexist in E. coli. The positive reporter vector contains the lacZα expression cassette, which is interrupted by the transcription STOP cassette that is flanked by a recombination substrate pair in the direct orientation (Fig. 2A). Excision of the STOP cassette by the recombination capable variants leads to the expression of the lacZα gene, which is signaled by the appearance of the blue colonies when bacteria are spread on the plates containing chromogenic substrate X-gal (12).
Figure 2.
Analysis of the activity of the Int and CTD variants on the attP/attB and COC’/COC’ substrate pairs using the positive reporter.A, schematics of the deletion assay. In the positive reporter vector, the lacZα expression module is interrupted by the rrnB STOP cassette, which is flanked by the recombination targets (RT1 and RT2) in the head-to-tail orientation. Recombination competent Int variants can excise the STOP cassette leading to the expression of the lacZα gene. B, analysis of the activity of Int, Int(E174K), and the CTD variants C65 and C75 on the attP/attB substrate pair. C, PCR analysis of the plasmid DNA isolated from the white (W) or blue (B) colonies from the respective plates shown in (B). D, analysis of the activity of Int, Int(E174K), Int(E174K, E218K), C65(E174K), C65(E174K, E218K), and C75(E174K) on the COC’/COC’ substrate pair. The NTD domain N64 was added to C65(E174K) and C75(E174K) were indicated. E, PCR analysis of the plasmid DNA isolated from the white or blue colonies from the respective plates shown in (D). In the experiments with Int and Int(E174K), two different types of the PCR results were observed, which are marked as B1 and B2. B1-type PCR product was seen in more than 80% of the blue colonies in the experiments with Int but only in about 20% of the blue colonies in the experiments with Int(E174K). rep, reporter vector; empty_rep, ‘empty’ reporter vector that lacks the deletion cassette and the recombination sites; M, DNA ladder. The scale bar on the plate images is 1 cm. CTD, C-terminal domain; NTD, N-terminal domain.
The functionality of the positive reporter system was tested using the attP/attB substrate pair and the following recombinases and domains: Int and Int(E174K) and the CTD variants, CTD(65–356) and CTD(75–356) or C65 and C75 for short. As expected, in the experiments with Int and Int(E174K), which is known to be more active than Int (9), essentially all bacterial colonies were blue, while in the experiments with both C65 and C75, no blue colonies were observed (Fig. 2B and Table 1). The deletion of the STOP cassette from the reporter in the blue colonies and the absence of this deletion in the white colonies was confirmed by the PCR analysis (Fig. 2C) and sequencing of the respective diagnostic PCR bands.
Table 1.
Recombination activity of Int and some of its variants
| Recombinase | Substrate (positive reporter) |
|||
|---|---|---|---|---|
| attP/attB | COC’/COC’ | P-COC’/COC’ | COC’-P’/COC’ | |
| Int | <99% | ∼20% | ∼25% | <99% |
| Int(E174K) | <99% | <95% | <95% | <99% |
| Int(E174K, E218K) | <99% | <95% | ∼100% | ∼100% |
| C65 | 0% | 0% | nt | nt |
| C65(E174K) | 0% | ∼0.2% | nt | nt |
| C65(E174K, E218K) | 0% | ∼3% | nt | nt |
| C57 | 0% | 0% | nt | nt |
| C57(E174K) | 0% | ∼0.1% | nt | nt |
| C57(E174K, E218K) | 0% | ∼0.1% | nt | nt |
| [C65 + N64] | 0% | 0% | nt | nt |
| [C65(E174K) + N64] | 0% | ∼0.2% | nt | nt |
| [C65(E174K, E218K) + N64] | 0% | ∼5% | nt | nt |
| [C57 + N64] | 0% | 0% | nt | nt |
| [C57(E174K) + N64] | ∼2% | ∼0.1% | ∼0.1% | ∼10% |
| [C57(E174K, E218K) + N64] |
∼8% |
∼0.1% |
∼0.2% |
∼25% |
| Substrate (negative reporter) |
||||
|---|---|---|---|---|
| attP/attB | COC’/COC’ | P-COC’/COC’ | COC’-P’/COC’ | |
| Int | >99% | 0% | ∼0.1% | ∼99% |
| Int(E174K) | ∼97% | ∼50% | ∼50% | ∼95% |
| Int(E174K, E218K) | ∼95% | ∼90% | ∼70% | ∼95% |
| [C57(E174K) + N64] | ∼40% | 0% | 0% | ∼3% |
| [C57(E174K, E218K) + N64] | ∼70% | 0% | 0% | ∼10% |
nt, not tested.
Next, we tested whether a mutation that can potentially stabilize the CTD on DNA could help trigger the recombination activity of the CTD. For this, we first tried the int-h mutation, E174K, which was shown to relax the requirement for the integration host factor (IHF) during recombination and to increase the binding affinity of Int (9, 10, 11). We thus added the int-h mutation to C65 and C75 and tested these modified CTD variants, C65(E174K) and C75(E174K), respectively, using the positive reporter vector that contained the attP/attB substrate pair. We found that the int-h mutation was not sufficient to allow C65 and C75 to recombine this vector (data not shown and Table 1). We also noted that the addition of the NTD of Int, N64 (4), did not apparently stimulate the recombination activity of either the original or the modified CTD variants on the attP/attB substrate pair (data not shown and Table 1).
To increase the likelihood of the recombination reaction by C65(E174K) and C75(E174K), we tested them on the armless COC’/COC’ substrate pair since COC’ is known to be a better substrate for Int than attB and since the CTD variants cannot bind the arm sites of attP so the arm-bearing substrates offer no advantage over the armless ones. We therefore tested C65 and C75, C65(E174K) and C75(E174K), as well as the control recombinases Int and Int(E174K) using the positive reporter vector, in which the STOP cassette is flanked by the COC’/COC’ substrate pair. In these experiments, the ‘control’ recombinases behaved as expected, Figure 2D and Table 1, the activity of Int on the armless COC’/COC’ substrate pair was significantly lower than that on the attP/attB substrate pair while the activity of Int(E174K) was dramatically higher than that of Int (9, 10). In contrast, both C65 and C75 remained recombinationally inactive (data not shown and Table 1).
To our delight, C65(E174K) was able to recombine COC’/COC’ with low but detectable activity: about 0.2% of the colonies were blue (Fig. 2D and Table 1). On the other hand, under the same conditions C75(E174K) remained recombinationally inactive. We also found that the expression of N64 in addition to C65(E174K) and C75(E174K) did not noticeably affect their activity (Fig. 2D).
The results of the PCR analysis of the plasmid DNA isolated from the blue colonies, which were obtained in the COC’/COC’ deletion experiments, were unexpected (Fig. 2E). In contrast to just one band that corresponds to the deletion product that was obtained in the experiments with Int and the attP/attB substrate pair (Fig. 2C), we observed several PCR bands when we analyzed the majority of the blue colonies in the experiments with the COC’/COC’ substrate pair: the bands that correspond to the deletion product and the original vector but also several other bands (Fig. 2E). The only exception was noted in the experiments with Int(E174K): the multiband PCR product was seen in about 20% of the blue colonies that we analyzed (Fig. 2E). We were not able to determine the identity of these extra PCR bands because the high quality sequencing data could not be obtained from these bands and re-PCR of the individual bands did not result in a single band (data not shown).
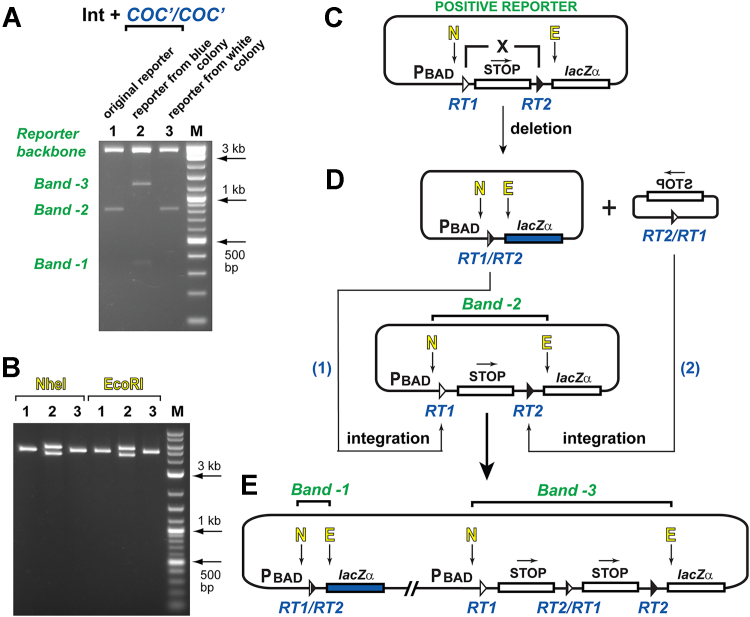
To determine the nature of the recombination product(s) in the experiments with the COC’/COC’ substrate pair, we performed diagnostic digestion of the reporter vectors that were isolated from the blue colonies, which were obtained in the experiments with Int and which generated the unexpected PCR products (Fig. 3). The analysis of the digestion patterns suggests that the main recombination product in these experiments is a dimer of two reporter molecules: one molecule that lacks the reporter cassette (that is, the plasmid with the expected deletion of the STOP cassette) and the other molecule that contains two copies of the STOP cassette. One of the simplest explanations of how two copies of the STOP cassette have appeared in the second reporter molecule is to assume that the deleted STOP cassette integrated into one of the COC’ sites, which flank the original reporter cassette (Fig. 3, D and E). Because we observed unexpected recombination products in the experiments with the positive reporter and the armless substrate, in the subsequent experiments, we analyzed how the vector design and the recombinase activity can influence the potential of fusion between the reporter vector molecules and the reintegration of the deleted reporter cassette.
Figure 3.
Restriction digestion analysis of the reporter plasmids isolated from the recombination-positive blue colony and recombination-negative white colony obtained in the experiments with Int, positive reporter, and the COC’/COC’ substrate pair.A, analysis of the reporter vectors obtained in the recombination experiments with Int and the COC’/COC’ substrate pair. In this analysis, the blue colony that generates the B1-type PCR outcome was used, see Figure 2E. Digestion of the representative reporters isolated from the blue and white colonies were performed with both NheI and EcoRI. ‘Band-1’ marks the DNA fragment that corresponds to the deleted STOP cassette (normal, expected outcome of the recombination reaction). ‘Band-2’ marks the DNA fragment that corresponds to the original STOP cassette, which was not deleted from the reporter vector. ‘Band-3’ marks the DNA fragment that corresponds to the duplicated STOP cassette. See also panels (C), (D), and (E). B, digestion of the same reporter plasmids as in (A) but either with NheI or EcoRI (lane numbering is same as in A). About equal intensity of the bands in lanes ‘2’ indicates that the core unit of the recombined reporter is a dimer. The locations of the NheI and EcoRI recognition sequences are indicated as ‘N’ and ‘E’, respectively, and are shown in the original positive reporter vector (C), in the deletion products (D), and in one of the simplest versions of the fusion product that is formed between the original reporter vector, the reporter vector, in which the STOP cassette is excised, and the excised STOP cassette (E).
In addition to the E174K mutation, we tested if the E218K mutation, which was suggested to increase the activity of Int and possibly its affinity for the core sites (13), can stimulate the recombination activity in C65 or C75. We found that in contrast to E174K, E218K was not able to stimulate the recombination activity of either of the CTD variants on the COC’/COC’ substrate pair (data not shown). On the other hand, we found that the doubly mutated variant C65(E174K, E218K) was noticeably more active than C65(E174K): about 3% of the colonies were blue, Figure 2D. Still, C75(E174K, E218K) remained inactive (data not shown). Overall, these results and the results shown later are more consistent with the role of the E218K mutation as a general enhancer of the recombination reaction rather than a mutation that can enhance affinity of Int for the core sites.
In conclusion, the aforementioned experiments demonstrated that the recombination activity of the CTD of Int can be indeed activated. These experiments also demonstrated that although C75 is a minimal CTD that can bind and cleave DNA and resolve premade Holliday structure intermediates (5), its recombination activity cannot be activated as easily as in C65, which has longer N terminus.
NTD can stimulate recombination activity of CTD if C-terminal residues from NTD are added to CTD
Since the NTD of Int was not able to enhance the recombination activity of C65(E174K) on the attP/attB substrate pair, we tested if the variants of the CTD that are extended toward the NTD could respond to the stimulation by the NTD. The rationale for these experiments is based on the analysis of the Int/DNA cocrystal (7) which shows that the polypeptide at the C terminus of the NTD that is located between residues 57 and 64 (Fig. 1C) makes intramonomer and intermonomer contacts with various polypeptides of the NTD, including itself. These interactions seem strategic as their profile differs depending on whether the Int monomers that form a recombination competent tetramer are in the recombination active or recombination inactive configuration. In principle, if the polypeptide 57 to 64 is added to C65, this extended CTD variant might be able to interact with at least some of the NTD polypeptides that are normally contacted by the polypeptide 57 to 64 during Int tetramer assembly. These interactions could help the extended CTD variant to functionally communicate with the NTD and stabilize the tertiary structure of the two-domain complex which, in turn, might trigger the recombination activity of the CTD on the substrates that contain the arm sites.
To test this hypothesis, we constructed two sets of the CTD variants, with full or partial 57 to 64 polypeptide and with or without the int-h mutation: C57, C58, and C60 and C57(E174K), C58(E174K), and C60(E174K). C57 (as well as C58 and C60, data not shown) was not able to recombine the attP/attB substrate pair and its recombination activity was not stimulated by the addition of the NTD (Fig. 4A). C57(E174K), as well as C58(E174K) and C60(E174K), data not shown, was also incapable of recombining attP/attB, Figure 4A. However, the recombination was indeed triggered when these CTD variants were supplemented with the NTD: the ratio of the recombination positive blue colonies was about the same for C60(E174K), C58(E174K), and C57(E174K), ∼ 1% to 3% (Fig. 4A and data not shown). When paired with N64, C57(E174K, E218K) was about four times more active than C57(E174K): 7% to 9%, Figure 4A. The deletion recombination in the blue colonies was confirmed by PCR and sequencing of the diagnostic PCR bands, Figure 4B.
Figure 4.
Complementation between the CTD and NTD variants of different lengths.A, the recombination activity of the CTD variants C57(E174K) and C57(E174K, E218K) is stimulated by the addition of the NTD variant N64 in the experiments with the positive reporter and the attP/attB substrate pair. B, PCR analysis of the plasmid DNA isolated from the white (W) or blue (B) colonies from the respective plates shown in (A). C, CTD variants C57(E174K) and C56(E174K) have about the same activity when the attP/attB substrate pairs are used while longer CTD variants C55(E174K) and C53(E174K) gradually loose the ability to trigger recombination in the presence of N64. D, CTD variant C57(E174K) can be activated only by the NTD variants N64 and N60 when the attP/attB substrate pairs are used. rep, reporter vector; empty_rep, ‘empty’ reporter vector that lacks the deletion cassette and the recombination sites; C, positive control PCR product similar to that shown in Figure 2C, lane [Int (B)]; M, DNA ladder. The scale bar on the plate images is 1 cm. CTD, C-terminal domain; NTD, N-terminal domain.
We next tested whether the recombination activity of the CTD can be further stimulated if, in addition to the 57 to 64 polypeptide, a partial or full alpha helix of the NTD (residues 56 through 41, Fig. 1C) is added to the CTD. For this, we created several CTD(E174K) variants that start at positions 56, 55, 53, 50, 47, 45, and 41, complemented them with N64, and tested the resulting modified Int domain combinations on the attP/attB substrate pair (Fig. 4C). The results showed that C56(E174K) was about as active as C57(E174K) and C58(E174K) while the other variants quickly lost the ability to get activated by the N64, whereas C50(E174K), C47(E174K), C45(E174K), and C41(E174K) appeared recombinationally inactive (Fig. 4C and data not shown).
N64 with shorter or longer C terminus lose the ability to stimulate recombination activity of CTD
We then analyzed whether the length of the C terminus of N64 affects its ability to stimulate the recombination activity of the extended CTD variant. To this end, we generated several shorter and longer variants of N64 that either lacked the 57 to 64 polypeptide completely (N56) or retained partial polypeptide 57 to 64 (N57, N58, and N60) or in which the 57 to 64 polypeptide was extended toward the CTD (N70 and N74).
These NTD variants were paired with C57(E174K) and tested for their ability to recombine the attP/attB substrate pair (Fig. 4D). These experiments showed that N56, which completely lacks the 57 to 64 polypeptide, and the variants with the truncated 57 to 64 polypeptide (N57 and N58) were not able to stimulate the recombination activity of C57(E174K). The NTD variant N60, which also has the truncated 57 to 64 polypeptide, was the only NTD variant that was able to trigger recombination in C57(E174K) to some extent: ∼1%, Figure 4D. On the other hand, the variants with the longer C-terminal polypeptide of the NTD, N70 and N74, lost the ability to stimulate the recombination activity of C57(E174K), data not shown.
Recombination activity of CTD is stimulated by NTD when substrates contain P′ arm but not P arm
Our results indicate that the recombination activity of the CTD variant C57(E174K), as well as several others, can be stimulated by the NTD variant N64 when the attP/attB substrate pair, which contains the arm sites, is used, but not the armless COC’/COC’ substrate pair. Since both P and P′ arms of attP contain the N64-binding sequences, both arms, in principle, can partake in stimulating the recombination activity of the CTD when it is paired with the NTD, even though P and P′ arms are not functionally equal (2). We therefore wanted to assess the relative contribution of each arm into the recombination stimulation process. To this end, we constructed two reporter vectors, with the P-COC’/COC’ and with the COC’-P’/COC’ substrate pairs; in these vectors, we used COC’ instead of attB as a partner substrate to increase the efficiency of recombination. The recombination experiments were performed with two CTD variants, C57(E174K) and C57(E174K, E218K), which were paired with the NTD variant N64. As controls, we used Int, Int(E174K), and Int(E174K, E218K).
We found that the profile of the recombination activity of the control and experimental recombinases differ for the P-COC’/COC’ and COC’-P’/COC’ substrate pairs: the behavior of the P-COC’/COC’ substrate pair resembled that of COC’/COC’, while the behavior of the COC’-P’/COC’ substrate pair was similar to that of attP/attB (Figs. 2 and 5).
Figure 5.
Activity of Int and CTD variants on P-COC’/COC’ and COC’-P’/COC’ substrate pairs located on the positive reporter.A, on the P-COC’/COC’ substrate pair, the addition of the int-h (E174K) mutation to Int increases its activity. The addition of the E218K mutation to the E174K mutation does not noticeably modify the activity of Int. The activity of the CTD variants C57(E174K) and C57(E174K, E218K) on the P-COC’/COC’ substrate pair was found to be about the same. B, PCR analysis of the plasmid DNA isolated from the blue colonies obtained from the respective plates shown in (A). C, the activity of Int, Int(E174K), and Int (E174K, E218K) on the COC’/COC’-P′ substrate pair is about the same. The CTD variant C57(E174K, E218K) is about twice as more active as C57(E174K). D, PCR analysis of the plasmid DNA isolated from the blue colonies obtained from the respective plates shown in (C). rep, reporter vector; empty_rep, ‘empty’ reporter vector that lacks the deletion cassette and the recombination sites; M, DNA ladder. The scale bar on the plate images is 1 cm. CTD, C-terminal domain; NTD, N-terminal domain.
The recombination activity of both C57(E174K) and C57(E174K, E218K) on P-COC’/COC’ was quite low (∼0.1%) and was about the same; although in some experimental series, we observed higher activity of C57(E174K, E218K). In contrast, the activity of C57(E174K) on the COC’-P’/COC’ substrate pair was substantially higher (∼10%) and its recombination activity increased 2- to 3-fold when the E218K mutation was added, Figure 5C.
These experiments did not address cooperativity between the NTD monomers bound to P and P′ arms within the intasome, which can enhance the activity of the binary Int. Nevertheless, the results of these experiments demonstrate that the higher recombination activity of the substrate pairs that contained the P′ arm compared to those with the P arm correlates well with the number of the sites that the NTD can bind in the P′ and P arms during the integration reaction: 3 versus 1, respectively (2).
Recombination outcome depends on vector design and activity of recombinases
Since we encountered an unexpected type of the reporter vector dimerization in the experiments in which the ‘positive’ reporter with the COC’/COC’ substrate pair was used (Figs. 2E and 3) and since this phenomenon can impact the way our results are interpreted, we wanted to test whether the observed reporter vector dimerization reflects the functional properties of Int and its variants and/or a particular reporter vector design. For this, we compared Int-mediated deletion using different substrate pairs using two different reporter formats: ‘positive’ and ‘negative’ (14). The key design difference between the two reporters is the relative location of the strong constitutive Plac promoter: downstream from the 5′ recombination target (that is, inside the excision lacZα cassette) in the negative reporter (Fig. 6A) or upstream of this recombination target (that is, upstream the excision STOP cassette) in the positive reporter (Figs. 2A and 6B). In both reporters, the strong pBAD promoter, which is located upstream of the recombination sites, is fully active only for ∼2.5 h during transformation of the vectors into bacterial cells when arabinose, its inducer, is added to the outgrowth medium.
Figure 6.
Comparison of recombination activity of Int on attP/attB substrate pair located either on the negative or the positive reporter. Schematics of the negative (A) and positive (B) reporters. The location of the Plac promoter in the reporters is indicated by vertical green arrow. C, electrophoregram of the undigested plasmid DNA isolated from the recombination-positive colonies obtained in the [Int + attP/attB] experiments. Green and white arrows point to the monomeric reporter and Int expression vectors, respectively. C1 and C2, original reporter and Int expression vectors, respectively. RT1 and RT2, recombination targets that flank the deletion cassette.
We found that in the experiments with the attP/attB substrate pair and Int, the recombination products of the negative reporter exist primarily as monomers, while that of the positive reporter, mostly as oligomers (Fig. 6C). On the other hand, when Int(E174K) and Int(E174K, E218K) were tested using the negative reporter, more oligomerized reporter were noted (Fig. 7D). Regardless, in the experiments with Int, Int(E174K), and Int(E174K, E218K) and the attP/attB substrate pair located on either of the reporters, no integration of the deleted cassette was noted (Figs. 2C, 6C and 7D).
Figure 7.
Recombination activity of Int, Int(174), and Int(174,218) on attP/attB and COC’/COC’ substrate pairs using negative reporter.A, schematics of the negative reporter. The location of the EcoRV and HindIII recognition sequences is marked as ‘R’ and ‘H’, respectively. B, activity of Int and Int variants on the attP/attB substrate pair. In these experiments, essentially all colonies were white. C, activity of Int and Int variants on the COC’/COC’ substrate pair. While no white colonies were observed in the [Int + COC’/COC’] experiments, about 50% and 80% of the colonies in the [Int(174) + COC’/COC’] and [Int(174, 218) + COC’/COC’] experiments, respectively, were white. D and E, show the electrophoregram of the plasmid DNA isolated from the respective recombination positive colonies. In these experiments, the reporter and the expression vectors were not separated. Note that no recombination-positive colonies were present in the [Int + COC’/COC’] experiments (E). Top panels display undigested plasmid DNA while bottom panels show plasmid DNA digested with EcoRV and HindIII. Green and white arrows in the top panels point to the monomers of the reporter and recombinase expression vectors, respectively, while in the bottom panels—to the DNA bands that resulted after digesting the reporter and expression vectors with EcoRV and HindIII. Note that in these experiments no abnormal recombination products in the negative reporters were observed. C1 and C2, negative reporter and Int expression vectors, respectively. RT1 and RT2, recombination targets that flank the deletion cassette. M, DNA ladder. The scale bar on the plate images is 1 cm.
In the experiments using the positive reporter, Int, and the COC’/COC’ substrate pair, the recombination products were oligomers and reinsertion of the deleted cassette was observed in more than 75% of the blue colonies, while in the same experimental settings with Int(E174K), the reinsertion product was hardly detectable (Fig. 8B). In the similar experiments using the negative reporter, Int appeared inactive and no apparent reintegration activity was noted (Fig. 7E). On the other hand, both Int(E174K) and Int(E174K, E218K) were able to excise the reporter cassette in the negative reporter generating white colonies (Fig. 7E). We also noted that although the experiments with the positive reporter (either with the attP/attB substrate pair, data not shown, or with the COC’/COC’ substrate pair, Fig. 2D) did not indicate that Int(E174K, E218K) is more active than Int(E174K), the experiments with the negative reporter and the COC’/COC’ substrate pair showed that Int(E174K, E218K) is about twice as more active as Int(E174K), (Fig. 7, C and E and Table 1).
Figure 8.
Analysis of recombination activity of Int, binary Int, and their variants that contain activity enhancing mutations on various substrate pairs using positive reporter.A, schematics of the positive reporter. The location of the NheI and EcoRI recognition sequences is marked as ‘N’ and ‘E’, respectively. NheI and EcoRI were used to digest the reporter plasmids that were separated from the expression plasmids. B–D, restriction analysis of the reporter plasmids obtained in the [Int + COC’/COC’] and [Int(E174K) + COC’/COC’] experiments; in the [Int + COC’-P’/COC’] and [Int + P-COC’/COC’] experiments; and in the [Int(E174K) + P-COC’/COC’] and [Int(E174K, E218K) + P-COC’/COC’] experiments, respectively. See Figure 3 for the definitions of the ‘Bands’. E, restriction analysis of the reporter plasmids obtained in the experiments with binary Int [57(E174K) + N64] and the following substrate pairs: COC’/COC’, P-COC’/COC’, attP/attB, and COC’-P’/COC’. In the electrophoregram, white and green arrows point to the fusion recombination products that contain ‘Band-3’ and ‘Band-2’, respectively. C4, C5, and C6 are control reporter vectors: ‘empty’ reporter vector, positive reporter vector, in which the STOP cassette is flanked by the COC’-P’/COC' substrate pair, and the plasmid DNA isolated from the recombination positive colony in the [Int + COC’/COC’] experiments (see Fig. 3), respectively. F and G, restriction analysis of the reporter plasmids obtained in the experiments with binary Int variants [57(E174K) + N64] and [57(E174K, E218K) + N64] that were tested on the attP/attB and COC’-P’/COC' substrate pairs, respectively. M, DNA ladder, RT1 and RT2, recombination targets that flank the deletion cassette.
The results of the experiments with Int, the positive reporter, and the P-COC’/COC’ and COC’-P’/COC’ substrate pairs showed that the latter substrate pair does not lead to any apparent reintegration products while the former substrate pair usually does (Fig. 8C). These results correlated well with the results shown in Figure 5, B and D.
Finally, we performed the simultaneous analysis of the activity of Int and the binary Int variant [57(E174K)+N64] using the negative reporter and all four substrate pairs: attP/attB, P-COC’/COC’, COC’-P’/COC’, and COC’/COC’. In the experiments with Int, we were able to get white colonies for all substrate pairs except for COC’/COC’, while in that with the binary Int, white colonies were observed only in the experiments with the attP/attB and COC’-P’/COC’ substrates pairs (Fig. 9 and Table 1). The analysis of the reporters isolated from the respective recombination-positive white colonies showed just the deletion product and no apparent reintegration product(s), Figure 9.
Figure 9.
Recombination activity of Int and binary Int that were tested using negative reporter.A, schematics of the negative reporter. The location of the EcoRV and HindIII recognition sequences is marked as ‘R’ and ‘H’, respectively. B, activity of Int on the P-COC’/COC’ and COC’-P’/COC’ substrate pairs. C, activity of binary Int [57(174) + N64] on the attP/attB and COC’-P’/COC’ substrate pairs. D, restriction digestion of the reporters that were isolated from the recombination positive white colonies and then separated from the expression vectors and digested with EcoRV and HindIII. The respective substrate pairs, attP/attB, P-COC’/COC’, and COC’-P’/COC’ for Int and attP/attB and COC’-P’/COC’ for binary Int [57(174) + N64], are indicated above the corresponding lanes. The electrophoregram shows that the reporters do not contain any apparent reintegration products. C1, C2, and C3 are original reporter vectors in which the lacZα cassette is flanked by the attP/attB, P-COC’/COC’, and COC’-P’/COC’ substrate pairs, respectively. The scale bar on the plate images is 1 cm.
In contrast, the analysis of the reporters isolated from the recombination-positive blue colonies in the experiments with the positive reporter and the binary Int, all recombinase pairs, except for COC’-P’/COC’, had the reintegrated deleted reporter cassette (Fig. 8E). It is interesting that the recombination product generated in the experiments with the latter substrate pair was just a dimer of the original reporter vector and the reporter vector with the deleted reporter cassette (Fig. 8E). It is also interesting that the addition of the E218K mutation to the binary Int was sufficient to prevent generation of this vector dimer (Fig. 8G) but was insufficient to prevent generation of the reintegration product in all other substrate pairs (Fig. 8F and data not shown).
Taken together, the results obtained with different Int variants and the negative and positive reporters suggest that both the vector design and the activity of the recombinase influence the recombination outcome.
Discussion
λ Int is a bidomainal DNA rearranging enzyme in which the recombination activity of its catalytic CTD is controlled by its regulatory NTD (2, 4). Counterintuitive, the NTD is capable of regulating the activity of not just the CTD but also of Cre, a simple recombinase, which is not normally regulated by any extra domain or protein (15). This apparently relaxed specificity of the NTD implies that at least some functions of this Int domain are exerted mainly via interactions within the NTD itself and not via interactions with the CTD. In the present work, we analyzed some of these interactions and uncovered the critical role of the polypeptide 57 to 64 that is located at the C terminus of the NTD in stimulating the recombination activity of the CTD. By adding the int-h mutation to the CTD(65–356) and thus stabilizing it on DNA (and, possibly, facilitating the formation of the CTD tetramers and/or enhancing the overall catalytic activity of the CTD) and transferring the 57 to 64 polypeptide from the NTD to the CTD and combining this modified CTD(57–356) with the NTD(1–64), we created a binary version of Int (Fig. 10) that functions in all in vivo reaction tested albeit with somewhat lower efficiency than its WT counterpart. Our results provide insights into the molecular basis of the regulation of the λ integrase activity and suggest how binary recombinases of the integrase type can be engineered. After further research and development, these binary integrases could be potentially used to tightly control the recombination activity in the genome editing and genome engineering applications by, for example, designing their photoactivatable versions that will be functionally similar to the split variants of Cre and Flp (16, 17, 18, 19, 20, 21, 22). If successful, the activity of these modified binary integrases could be improved and controlled by illuminating cells with blue light to help predictably associate the two integrase domains.
Figure 10.
Binary Int recombinase. This hybrid recombinase is composed of the N-terminal domain (NTD) of Int NTD(1–64) and the modified C-terminal domains (CTDs) of Int CTD(57–356). These two domains interact via polypeptide 57 to 64, which is schematically indicated by black dotted lines between the domains. CTD(57–356) also contains the int-h mutation E174K, which can stabilize the CTD on its cognate core sites, improve its monomer-monomer interactions, and/or enhance its overall recombination activity. These potential consequences of the E174K mutation are schematically indicated as red chain links and the ‘energy’ symbol.
In this work, we found that the int-h mutation (E174K) allows the recombinationally inactive CTD variant C65 (but not C75) to recombine the armless COC’/COC’ substrate pair (Figs. 1 and 2). Int with the int-h mutation was shown to perform recombination without the accessory protein IHF, which bends DNA within attP, thus helping Int to assemble the intasome and to have increased binding affinity for the recombination targets (10, 11). The other mutation that we tested, E218K (13), which was suggested to increase the overall activity of Int and possibly its binding affinity, was not able to stimulate the recombination activity of C65 if added alone but was able to enhance the activity of C65(E174K), Figure 2D. Off note, it is interesting that E174K and E218K allow Int to perform recombination without the bacterial accessory proteins not only in bacterial cells but also in mammalian cells (23).
The int-h mutation (E174K) modifies the amino acid residue that is located on the CB-CD linker that connects the core binding domain (CB) and the catalytic domain (CD) of the CTD (Fig. 1C). This long interdomain linker is located between residues 157 and 181; for the sake of the analysis, we consider that the CB-CD linker spans the entire polypeptide and does not have a well-defined secondary structure (6). The CB-CD linker occupies a ‘strategic’ position as it makes multiple direct contacts with the CB and the CD domains and DNA (7). In the CD domain, the linker is positioned between the loop that connects alpha helixes J and K, helix K, which bears catalytic residues H308 and R311, and the disordered structure between β strands 6 and 7 that harbors the catalytic residue Y342 (6, 7). Moreover, the residues in the CB-CD linker participate in the monomer-monomer interactions (6, 7). As such, the functional consequence of the E174K mutation may be multilayered and include the facilitation of the assembly of the functional tetramer complex and the transduction of the tetramer ‘assembly signal’ to the catalytic residues thus enhancing the catalytic activity of the CTD.
The E218K mutation also occupies a ‘strategic’ position as it is located next to beta sheet that is formed by β1, β2, and β3 strands and which contacts both DNA and the C-terminal tail of the neighboring monomer (6, 7). Moreover, E218K is located near the catalytic residue R212. Consequently, the E218K mutation can, in principle, affect the activity of Int, which is supported by our results.
We show that the recombination activity of the CTD variant C57(E174K), as well as of some other CTD variants that bear the 57 to 64 polypeptide, depends on the presence of the NTD variant N64 and the recombination substrate pair used. On the armless COC’/COC’ substrate, with or without N64, the activity of C57(E174K) is about the same as that of C65(E174K), which lacks the 57 to 64 polypeptide (Fig. 2D and data not shown). On the arm site bearing attP/attB substrate pair, in the absence of N64, both these CTD variants do not show detectable recombination activity but when N64 is added, the recombination activity is triggered in C57(E174K) but not in C65(E174K), Figures 2 and 4.
We also show that P and P′ arms of attP are not equivalent in stimulating the recombination activity of C57(E174K) and that this activity correlated with the number of the arm type–binding sites for the N64 domain, Figure 5. The P′ arm (which has three ‘integration-positive’ N64-binding sites) strongly activated the activity of C57(E174), while the P arm (which has one ‘integration-positive’ N64 binding site) was essentially as weak as the armless COC’/COC’ substrate. Taken together, these results suggest that the mere presence of the 57 to 64 polypeptide in a CTD variant does not stimulate its recombination activity even when the NTD is present but not bound to the arm sites. However, the 57 to 64 polypeptide is essential for stimulating recombination when the NTD can bind to the arm sites and the number (and, possibly, the functional properties) of the NTD-binding sites determines the level of the recombination activity of the CTD (Figs. 4A and 5).
We found that shortening or lengthening N64 had negative effect on the stimulation of the recombination activity of C57(E174K). It is interesting that among different longer NTD variants, only N60 was able to stimulate the recombination activity of C57(E174K), Figure 4D. These results suggest that the amino acid residues that are located downstream from the residue 64 participate in the intermonomer or intramonomer interactions in the NTD, which can block the access to the 57 to 64 polypeptide that is located in C57(E174K). These potential interactions, however, are not apparent in the Int/DNA cocrystal (7).
Our results indicate that both the CTD and the NTD should have the 57 to 64 polypeptide to trigger the recombination activity in the CTD, which points to the critical intermonomer interactions between the domains in this region (7). In principle, our data can be interpreted that these intermonomer interactions affect the recombination activity through transducing the arm site–binding signal from the NTD to the catalytic residues in the CTD. Another, nonexclusive possibility is that the 57 to 64 polypeptide stabilizes the Int tetramer, which, in turn, can affect the recombination activity of the CTD.
In this work, we encountered a previously unnoticed phenomenon; the outcome of the recombination reaction mediated by WT Int as well as by binary Int depends on the following parameters (1): the location of the strong constitutive promoter, upstream of the reporter cassette as in the negative reporter or inside the reporter cassette as in the positive reporter (2), the presence (and the type) of the arm-type sequences in the recombination substrates that flank the reporter cassette, and (3) the presence of the activity enhancing mutations E174K and E218K in the recombinases, (Figs. 2, 3, 7 and 8).
We noted that the vector type apparently leads only (or almost exclusively) to the expected deletion end product of the recombination reaction in the experiments with Int and the attP/attB or the right arm containing COC’-P’/COC’ substrate pairs (Figs. 2B, 7C, 8C, and 9B).
The unusual recombination product is present when Int is tested using the positive reporter in which the excision cassette was flanked by the COC’/COC’ and the P-COC’/COC’ substrate pairs, Figures 3 and 8, B and C. The analysis showed that this recombination product is composed of three DNA fragments: the reporter molecule from which the reporter cassette was deleted, the original reporter molecule, and the deleted cassette that integrated into one of the recombination sites of the original reporter molecule (Fig. 3). Binary Int also generated similar recombination product when tested using the positive reporter and all substrate pairs except for COC’-P’/COC’ (Fig. 8E). In the latter case, the recombination product contained only two DNA fragments: the reporter molecule from which the reporter cassette was deleted and the original reporter molecule (Fig. 8E).
The abnormal recombination products that are formed in the experiments with Int and the COC’/COC’ and the P-COC’/COC’ substrate pairs are usually absent if the experiments are performed with Int that contains the activity-enhancing mutations E174K and E218K (Fig. 8, B and D). We noted, however, a puzzling difference between the two substrate pairs: P-COC’/COC’ is more prone to form abnormal recombination products than COC’/COC’ when tested not only with Int but also with Int(E174K) and even with Int(E174K, E218K). Nevertheless, Int(E174K, E218K) is eager to form just the desired deletion product than Int(E174K), Figure 8D. These results suggest the P arm might negatively affect the formation of the correct recombination competent complex, which in turn reduces the recombination activity with the P-COC’/COC’ substrate pair.
The addition of E218K to binary Int [C57(174) + N64] was helpful only when the experiments were performed with the COC’-P’/COC’ substrate pair (Fig. 8, F and G and data not shown). This result is surprising since in these experiments, attP/attB offered no advantage over COC’-P’/COC’, although attP is considered the best recombination target. One potential explanation of this result is that the activity of the substrate pairs that contain attB is much lower than those that contain COC’.
Our final set of experiments established an important result: no abnormal recombination products are present, regardless of the substrate pairs used, when Int and binary Int are tested using the negative reporter (Fig. 9).
Several general conclusions can be made by comparing the results obtained using the positive and negative reporter configurations (1): in the positive reporter configuration, the strong constitutive promoter located upstream of the 5′ recombination site apparently influences the reaction outcome leading to abnormal fusions between the reporters and/or the deleted reporter cassette; (1a), in this type of reporter, the higher activity of the recombinases is usually needed to prevent the formation of the reporter vector fusions (2); in the negative reporter configuration, in which the strong constitutive promoter located downstream from the 5′ recombination site, fusions between the reporter vectors are not formed, even in the experiments with binary Int, which has lower activity than Int. These conclusions suggest that although the design of the positive reporter allows it to identify recombinase variants with quite low activity, the positive reporter configuration itself is paradoxically less recombinationally active than that of the negative reporter configuration.
Since the main difference between the two vector designs is the relative location of the strong constitutive promoter, it is reasonable to assume that it is the ‘upstream’ location of this promoter that makes the positive reporter configuration less recombinationally active. The promoter location differently affects the topology of DNA upstream and downstream of a transcription bubble: DNA is overwound downstream of the bubble while it is underwound upstream of it (24). Since in the experiments with the negative reporter (in which the 5′ recombination target likely experiences underwinding) Int was able to generate the correct recombination outcome with all recombination pairs tested, we hypothesize that it is the recombination target underwinding that positively affects the recombination efficiency. One possibility is that underwinding of the recombination target simply allows for easier assembly of the recombination competent complex and/or for facilitating the first catalytic single strand exchange reactions.
Alternatively, regardless of the DNA topology state, the mere activity of the ‘upstream’ strong constitutive promoter in the positive reporter (that is, binding of the RNA polymerase and subsequent mRNA elongation) might reduce the probability of binding of WT and binary Int to their targets, which will, in turn, lower their recombination activity. The lower recombination activity might result in shifting the recombination outcome from deletion to integration, thus leading to fusions between the reporter vectors; it is well known from the integration experiments with simple recombinases (Flp and Cre, for example) that the efficiency of integration can be increased by lowering the concentration of these recombinases.
We also want to note here that we observed fusions between the reporter vectors in the experiments with the Flp variants and the positive reporter in which the deletion cassette was flanked by the FRT-like sequences (12). This suggests that the reporter fusion is a general property of the positive reporter configuration, which is especially pronounced when the low activity recombinases are used.
Experimental procedures
Bacterial strain
E. coli strain NEB 10-beta (araD139 Δ(ara-leu)7697 fhuA lacX74 galK (φ80 Δ(lacZ)M15) mcrA galU recA1 endA1 nupG rpsL (StrR) Δ(mrr-hsdRMS-mcrBC)), purchased from New England Biolabs, was used in all experiments. NEB 10-beta cells are not known to have mutations in the IHF subunits, so we assume that this strain has WT IHF.
Reporter and expression vectors
Two types of the reporter vectors were used to analyze the functional performance of the CTD variants and the [CTD + NTD] variant combinations (Figs. 2A and 6, A and B): ‘positive’ and ‘negative’. The positive reporters are based on the vector for positive screening, which was described in Bolusani et al., 2006 (12) and is a derivative of pBAD24 (AmpR, ColE1 origin of replication (25)). The reporter vectors of this type contain the lacZα expression unit that is interrupted by the rrnB STOP cassette, which is flanked by different sets of the recombination sites. To construct these reporter vectors, the respective lacZα-rrnB cassettes were assembled by PCR and cloned into the core vector (12).
The negative reporter vectors also derive from pBAD24 and utilize the negative screening configuration, which was described in Voziyanov et al., 2002 (14). The reporter vectors of this type contain the lacZα expression unit that is flanked by different sets of the recombination sites. To construct these reporter vectors, the lacZα expression unit was PCR-amplified using primers that contained the recombination sequences and cloned into the MSC region of pBAD24.
Two types of expression vectors were used in this study. The vector that was used to express Int, Int variants, and the CTD variants is a derivative of pBAD33 (CmR, p15a origin of replication (25, 26)). The vector that was used to express the NTD variants is a derivative of pBBR1MCS-2 (KmR, R1 origin of replication (14, 27)). In both vectors, the recombinase genes were expressed from the pBAD promoter.
Recombination experiments in E. coli
The recombination experiments in bacterial cells were performed essentially as described in Voziyanov et al., 2002 (14). In brief, competent bacterial cells were transformed with two sets of vectors (1): with the reporter vector and with the vector that expresses either Int (or its variants) or the CTD variant or (2) with the reporter vector and with two vectors that express a CTD variant and an NTD variant (or an empty expression vector as a control). In the experiments with a reporter vector and only one expression vector, the input of each vector during transformation was 1 ng. In the experiments with a reporter vector and two expression vectors, the input of each vector was 10 ng. Transformed bacterial cells were incubated with the inducer L-arabinose at the final concentration 0.02% for 2.5 h and then cells were plated onto LB/agar plates that contained X-gal to visualize the colonies in which, depending on the type of the reporter vector used, either the STOP cassette or the lacZα cassette was deleted.
Separation of plasmids from plasmid mixture by dilution and retransformation
To determine the nature of the individual reporter plasmid in the experiments described in Figures 3, 8 and 9, the plasmid mixture that contained the expression and reporter vectors was isolated from the original blue colonies and then the isolated plasmids were separated by diluting them 2000-fold, transforming them into competent E. coli cells, and selecting the reporter containing colonies on ampicillin (since the reporter vectors are AmpR). This ‘vector separation’ protocol dramatically reduces the probability that more than one vector molecule enters a bacterial cell. Consequently, each resultant AmpR colony likely contains copies of only one reporter plasmid (14). In the ‘reporter separation’ experiments, we obtained both blue and white colonies and, depending on the goal, isolated and analyzed plasmids from each colony type.
In the positive reporter separation experiments, the ratio of the blue to white colonies correlated with the color intensity of the original blue colony and was somewhere between ∼30% and ∼95%.
In the negative reporter separation experiments, the ratio of the white to blue colonies was usually about 100%.
Other methods
Plasmid DNA was isolated using E.Z.N.A. Plasmid Mini Kit (Omega Bio-Tek). Amplification of the DNA fragments that were used for cloning was performed using PfuUltra polymerase (Agilent) and Q5 polymerase (New England Biolabs). Site-directed mutagenesis of Int was performed using PfuUltra polymerase. PCR analysis of the reporter vectors was performed using GoTaq Green Master Mix (Promega). General genetic engineering experiments were performed as described in Molecular Cloning Manual (28) and in the manufacturer’s protocols. 3D structure of Int was analyzed using Swiss-Pdb-Viewer (29) and visualized using UCSF Chimera (30) to prepare Figure 1, B and C.
Data availability
All data are contained within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by Louisiana Tech University through Marvin T. Green, Sr. Endowed Professorship in Premedicine which was made available through the State of Louisiana Board of Regents Support Funds.
Author contributions
Y. V. conceptualization; Y. V. methodology; Y. V. validation; Y. V. formal analysis; J. D. W., E. V., and Y. V. investigation; Y. V. writing–original draft; Y. V. writing–review & editing; Y. V. visualization; Y. V. supervision; Y. V. project administration; Y. V. funding acquisition.
Edited by Craig Cameron
References
- 1.Grindley N.D., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Landy A. The lambda integrase site-specific recombination pathway. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0051-2014. MDNA3-0051-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moitoso de Vargas L., Pargellis C.A., Hasan N.M., Bushman E.W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 4.Sarkar D., Radman-Livaja M., Landy A. The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions. EMBO J. 2001;20:1203–1212. doi: 10.1093/emboj/20.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkar D., Azaro M.A., Aihara H., Papagiannis C.V., Tirumalai R., Nunes-Duby S.E., et al. Differential affinity and cooperativity functions of the amino-terminal 70 residues of lambda integrase. J. Mol. Biol. 2002;324:775–789. doi: 10.1016/s0022-2836(02)01199-3. [DOI] [PubMed] [Google Scholar]
- 6.Aihara H., Kwon H.J., Nunes-Duby S.E., Landy A., Ellenberger T. A conformational switch controls the DNA cleavage activity of lambda integrase. Mol. Cell. 2003;12:187–198. doi: 10.1016/s1097-2765(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 7.Biswas T., Aihara H., Radman-Livaja M., Filman D., Landy A., Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren D., Lee S.Y., Landy A. Mutations in the amino-terminal domain of lambda-integrase have differential effects on integrative and excisive recombination. Mol. Microbiol. 2005;55:1104–1112. doi: 10.1111/j.1365-2958.2004.04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller H.I., Mozola M.A., Friedman D.I. int-h: an int mutation of phage lambda that enhances site-specific recombination. Cell. 1980;20:721–729. doi: 10.1016/0092-8674(80)90318-9. [DOI] [PubMed] [Google Scholar]
- 10.Patsey R.L., Bruist M.F. Characterization of the interaction between the lambda intasome and attB. J. Mol. Biol. 1995;252:47–58. doi: 10.1006/jmbi.1995.0474. [DOI] [PubMed] [Google Scholar]
- 11.Lange-Gustafson B.J., Nash H.A. Purification and properties of int-h, a variant protein involved in site-specific recombination of bacteriophage lambda. J. Biol. Chem. 1984;259:12724–12732. [PubMed] [Google Scholar]
- 12.Bolusani S., Ma C.H., Paek A., Konieczka J.H., Jayaram M., Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Gumport R.I., Gardner J.F. Genetic analysis of second-site revertants of bacteriophage lambda integrase mutants. J. Bacteriol. 1997;179:4030–4038. doi: 10.1128/jb.179.12.4030-4038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voziyanov Y., Stewart A.F., Jayaram M. A dual reporter screening system identifies the amino acid at position 82 in Flp site-specific recombinase as a determinant for target specificity. Nucleic Acids Res. 2002;30:1656–1663. doi: 10.1093/nar/30.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren D., Laxmikanthan G., Landy A. A chimeric Cre recombinase with regulated directionality. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18278–18283. doi: 10.1073/pnas.0809949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung H., Kim S.W., Kim M., Hong J., Yu D., Kim J.H., et al. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 2019;10:314. doi: 10.1038/s41467-018-08282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian X., Zhou B. Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J. Biol. Chem. 2021;296:100509. doi: 10.1016/j.jbc.2021.100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano F., Okazaki R., Yazawa M., Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 2016;12:1059–1064. doi: 10.1038/nchembio.2205. [DOI] [PubMed] [Google Scholar]
- 19.Meador K., Wysoczynski C.L., Norris A.J., Aoto J., Bruchas M.R., Tucker C.L. Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res. 2019;47:e97. doi: 10.1093/nar/gkz585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa K., Furuhashi K., de Sena-Tomas C., Garcia-Garcia A.L., Bekdash R., Klein A.D., et al. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 2020;11:2141. doi: 10.1038/s41467-020-16030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolot M., Malchin N., Elias A., Gritsenko N., Yagil E. Site promiscuity of coliphage HK022 integrase as a tool for gene therapy. Gene Ther. 2015;22:521–527. doi: 10.1038/gt.2015.9. [DOI] [PubMed] [Google Scholar]
- 22.Elias A., Kassis H., Elkader S.A., Gritsenko N., Nahmad A., Shir H., et al. HK022 bacteriophage Integrase mediated RMCE as a potential tool for human gene therapy. Nucleic Acids Res. 2020;48:12804–12816. doi: 10.1093/nar/gkaa1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorbach E., Christ N., Schwikardi M., Droge P. Site-specific recombination in human cells catalyzed by phage lambda integrase mutants. J. Mol. Biol. 2000;296:1175–1181. doi: 10.1006/jmbi.2000.3532. [DOI] [PubMed] [Google Scholar]
- 24.Lansing F., Mukhametzyanova L., Rojo-Romanos T., Iwasawa K., Kimura M., Paszkowski-Rogacz M., et al. Correction of a Factor VIII genomic inversion with designer-recombinases. Nat. Commun. 2022;13:422. doi: 10.1038/s41467-022-28080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konieczka J.H., Paek A., Jayaram M., Voziyanov Y. Recombination of hybrid target sites by binary combinations of Flp variants: mutations that foster interprotomer collaboration and enlarge substrate tolerance. J. Mol. Biol. 2004;339:365–378. doi: 10.1016/j.jmb.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 27.Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., 2nd, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D.W. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 29.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.