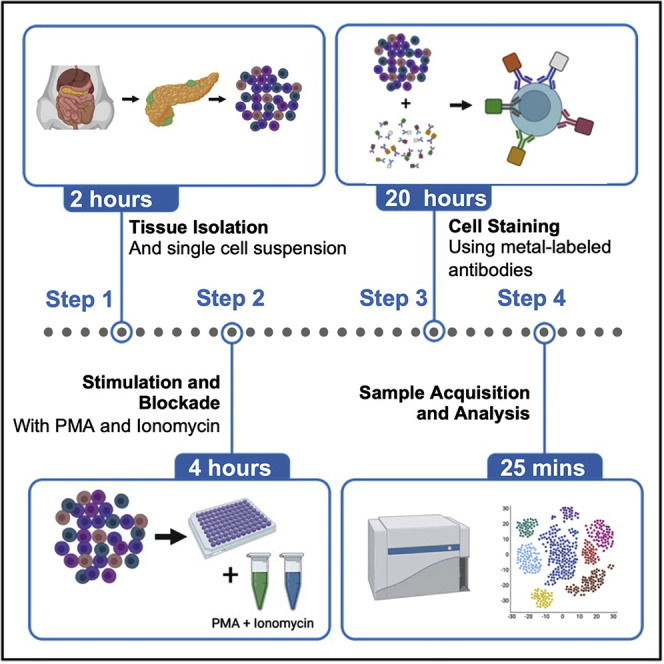
Summary
Investigating the immune attack on β cells is critical to understanding autoimmune diabetes. Here, we present a protocol to isolate immune cells from mouse pancreatic lymph nodes and whole pancreas, followed by mass cytometric analyses. This protocol can be used to analyze subsets of innate and adaptive immune cells that play critical roles in autoimmune diabetes, with as few as 5 × 105 cells. This protocol can also be adapted to study resident immune cells from other tissues.
For complete details on the use and execution of this protocol, please refer to Piñeros et al. (2022).1
Subject areas: Cell Biology, Cell isolation, Cell-based Assays, Flow Cytometry/Mass Cytometry, Immunology
Graphical abstract
Highlights
-
•
Protocol for isolating single cells from mouse pancreatic lymph nodes and whole pancreas
-
•
Steps to stimulate and stain the isolated immune cells for mass cytometric analysis
-
•
Applicable to analyze immune cells in autoimmune diabetes
-
•
Adaptable to immune cells derived from other autoimmune-disease-relevant tissues
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Investigating the immune attack on β cells is critical to understanding autoimmune diabetes. Here, we present a protocol to isolate immune cells from mouse pancreatic lymph nodes and whole pancreas, followed by mass cytometric analyses. This protocol can be used to analyze subsets of innate and adaptive immune cells that play critical roles in autoimmune diabetes, with as few as 5 × 105 cells. This protocol can also be adapted to study resident immune cells from other tissues.
Before you begin
An important driver of autoimmune diabetes is loss of immune cell tolerance that leads to the destruction of insulin-producing β cells in pancreatic islets. The long-standing perspective of the pathogenesis of autoimmune diabetes postulates that β cell antigens are presented by antigen presenting cells to naïve T cells within pancreatic lymph nodes. These activated, pathogenic T cells then migrate to the islet and initiate destruction of β cells. Recently, the concept that β cells are active participants in the disease process and promote activation of the immune system has been gaining traction in the field.2 Although the molecular mechanism(s) within the β cell that relay signals to immune cells is incompletely understood, we recently demonstrated that inflammatory signaling mediated by 12-lipoxygenase in β cells during early disease pathogenesis promotes a pathogenic immune cell profile. Elimination of this enzyme reduces the influx of pathogenic immune cells into both the pancreatic lymph nodes and islets and increases the entry of more tolerogenic immune cells into the islets.1 Given that the draining pancreatic lymph node and pancreas are key sites in the initial priming and activity of autoreactive immune cells, respectively, the precise identification and quantitation of immune cells in these two sites are crucial to understanding how the disease process is proceeding and how intervention might alter the process. In this protocol, we focus on the single cell isolation of immune cells from whole pancreas and pancreatic lymph nodes for mass cytometry, or cytometry by time of flight (CyTOF). However, with minor adjustments this protocol could also be used to isolate and analyze tissue resident immune cells from other tissues, such as liver and adipose, in the context of other autoimmune and inflammatory diseases. We then performed CyTOF mass cytometry using an approach as described previously3 for broad scale immune profiling. One major strength of this protocol is the ability to perform high dimensional and exploratory immune cell analysis with as few as 5 × 105 cells.
Institutional permissions
This protocol uses single cells isolated from whole pancreas or pancreatic lymph node. The mice used in this protocol were maintained at the University of Chicago. All experiments were approved by the Institutional Animal Care and Use Committee. Please note that an institution ethical approval of animal use is required prior to starting this protocol.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse IL-6, Clone MP5-20F3, 1:50 dilution | Fluidigm | Cat#3167003C, Label 167Er |
| Anti-mouse IL-4, Clone 11B11, 1:50 dilution | Fluidigm | Cat#3166003C, Label 166Er |
| Anti-mouse IL-10, Clone JES5-16E3, 1:50 dilution | Fluidigm | Cat#3158002C, Label 158Gd |
| Anti-mouse TNFα, Clone MP6-XT22, 1:50 dilution | Fluidigm | Cat#3162002C, Label 162Dy |
| Anti-mouse IFNγ, Clone XMG1.2, 1:50 dilution | Fluidigm | Cat#3165003C, Label 165Ho |
| Anti-mouse IL-17A, Clone TC11-18H10.1, 1:50 dilution | Fluidigm | Cat#3174002C, Label 174Yb |
| Anti-mouse CD4, Clone RM4-5, 1:50 dilution | Fluidigm | Cat#3145002C, Label 145Nd |
| Anti-mouse CD8a, Clone 53-6.7, 1:50 dilution | Fluidigm | Cat#3168003C, Label 153Eu |
| Anti-mouse CD11c, Clone N418, 1:50 dilution | Fluidigm | Cat#3142003C, Label 142Nd |
| Anti-mouse CD206, Clone C068C2, 1:50 dilution | Fluidigm | Cat#3169021C, Label 169Tm |
| Anti-mouse I-A/I-E, Clone M5/114.15.2, 1:50 dilution | Fluidigm | Cat#3209006C, Label 209Bi |
| Anti-mouse CD86, Clone GL1, 1:50 dilution | Fluidigm | Cat#3172016C, Label 172Yb |
| Anti-mouse CD25 (IL-2R), Clone 3C7, 1:50 dilution | Fluidigm | Cat#3151007C, Label 151Eu |
| Anti-mouse Ly-6C, Clone HK1.4, 1:50 dilution | Fluidigm | Cat#3150010C, Label 150Nd |
| Anti-mouse CD19, Clone 6D5, 1:50 dilution | Fluidigm | Cat#3149002C, Label 149Sm |
| Anti-mouse CD3e, Clone 145-2C11, 1:50 dilution | Fluidigm | Cat#3152004C, Label 152Sm |
| Anti-mouse CD45, Clone 30-F11, 1:50 dilution | Fluidigm | Cat#3089005C, Label 89Y |
| Anti-mouse F4/80, Clone BM8, 1:50 dilution | Fluidigm | Cat#3146008C, Label 146Nd |
| Anti-mouse PDL1, Clone 10F.9G2, 1:50 dilution | Fluidigm | Cat#3153016C, Label 153Eu |
| Anti-mouse PD1, Clone 29F.1A12, 1:50 dilution | Fluidigm | Cat#3159024C, Label 159Tb |
| Anti-mouse MHC-I, Clone 28-14-8, 1:50 dilution | Fluidigm | Cat#3143016C, Label 144Nd |
| Anti-mouse CTLA4, Clone UC10-4B9, 1:50 dilution | Fluidigm | Cat#3154008C, Label 154Sm |
| Anti-mouse iNOS, Clone CXNFT, 1:50 dilution | Fluidigm | Cat#3161011C, Label 161Dy |
| Anti-mouse CD11b, Clone M1/70, 1:50 dilution | Fluidigm | Cat#3143015C, Label 143Nd |
| Anti-mouse CD80, Clone 16-10A1, 1:50 dilution | Fluidigm | Cat#3171008C, Label 171Yb |
| Chemicals, peptides, and recombinant proteins | ||
| Cell-ID Cisplatin | Fluidigm | Cat#201064 |
| Cell-ID Intercalator-Ir 125 mM | Fluidigm | Cat#201192A |
| Maxpar Cell Staining Buffer | Fluidigm | Cat#201068 |
| Maxpar Fix and Perm Buffer | Fluidigm | Cat#201067 |
| Maxpar Fix I Buffer (5×) | Fluidigm | Cat#201065 |
| Maxpar Perm-S Buffer | Fluidigm | Cat#201066 |
| Maxpar PBS Fluidigm | Fluidigm | Cat#201058 |
| Maxpar Cell Acquisition Solution | Fluidigm | Cat#201237 |
| Maxpar Water | Fluidigm | Cat#201069 |
| RPMI Medium 1640 | Gibco | Cat#11875-093 |
| PBS | Gibco | Cat#14190-144 |
| HBSS | Gibco | Cat#14025092 |
| HEPES buffer | Corning | Cat#25-060-CI |
| Penicillin-streptomycin | Thermo Fisher Scientific | Cat#15140-122 |
| Heat-inactivated FBS | Gibco | Cat#10082-147 |
| eBioscience 1× RBC Lysis Buffer | Invitrogen | Cat#00-4333-57 |
| Ionomycin | Sigma-Aldrich | Cat#I0634 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#P8139 |
| L-Glutamine | Thermo Fisher Scientific | Cat#A2916801 |
| Trypan Blue | Gibco | Cat#1525 |
| Golgi stop | BD Biosciences | Cat#555029 |
| Histopaque-1077 | Sigma | Cat#10771 |
| Collagenase P | Sigma | Cat#C7657 |
| Experimental models: Organisms/strains | ||
| Mouse: NOD, 8–10 weeks old, female | Jackson Laboratories | Cat#1976 |
| Software and algorithms | ||
| Cytobank Premium | Beckman Coulter | https://premium.cytobank.org/cytobank/login## |
| Other | ||
| Flow tube with cell strainer snap cap | Falcon | Cat#352235 |
| Cell strainer 100 μm | Fisher Scientific | Cat#11517532 |
| Cell strainer 70 μm | Fisher Scientific | Cat#087712 |
| 96-well U bottom plate | Thermo Fisher Scientific | Cat#163320 |
| Helios mass cytometer | Fluidigm | N/A |
| CyTOF wash solution | Fluidigm | Cat#201070 |
| CyTOF tuning solution | Fluidigm | Cat#201072 |
Materials and equipment
CRITICAL: It is important that all reagents are metal-free and care is taken to avoid metal contamination (see troubleshooting).
Collagenase P digestion media
| Reagent | Final concentration | Amount |
|---|---|---|
| HBSS | N/A | 10 mL |
| Collagenase P | 1 mg/mL | 10 mg |
| Total | N/A | 10 mL |
Note: Prepare freshly on the day of experiment and store at 4°C. Warm up stock of Collagenase P before opening to avoid condensation.
Immune cell culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | N/A | 440 mL |
| HI-FBS | 10% | 50 mL |
| Penicillin-streptomycin (100×) | 1× | 5 mL |
| HEPES (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Note: Can be prepared in advance and stored at 4°C for up to 1 month.
PMA, Ionomycin, and Golgi Stop Media
| Reagent | Final concentration | Amount |
|---|---|---|
| Immune cell culture media | 4,995 μL | |
| PMA | 100 ng/mL | 500 ng |
| Ionomycin | 500 ng/mL | 5 μg |
| Golgi Stop | 5 μL | |
| Total | N/A | 5 mL |
Note: Prepare freshly on the day of experiment and store at 4°C.
Cisplatin Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Cisplatin (5 mM) | 5 μM | 5 μL |
| PBS | N/A | 4,995 μL |
| Total | N/A | 5 mL |
Note: Prepare freshly on the day of experiment and store at 4°C.
PBS + 2% FBS Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 98 mL |
| FBS | 2% | 2 mL |
| Total | N/A | 100 mL |
Note: Prepare freshly on the day of experiment and store at 4°C.
Surface marker antibody cocktail
| Reagent | Final concentration | Amount |
|---|---|---|
| Maxpar Cell Staining buffer | N/A | 960 μL |
| Surface antibody 1 | 1:50 | 20 μL |
| Surface antibody 2 | 1:50 | 20 μL |
| Total | N/A | 1,000 μL |
Note: Prepare freshly on the day of experiment and store at 4°C in the dark. Decrease Maxpar Cell Staining buffer based on number of surface antibodies added.
Cytoplasmic/secreted antibody cocktail
| Reagent | Final concentration | Amount |
|---|---|---|
| Maxpar Cell Staining buffer | N/A | 960 μL |
| Cytoplasmic/secreted antibody 1 | 1:50 | 20 μL |
| Cytoplasmic/secreted antibody 2 | 1:50 | 20 μL |
| Total | N/A | 1,000 μL |
Note: Prepare freshly on the day of experiment and store at 4°C in the dark. Decrease Maxpar Cell Staining buffer based on number of surface antibodies added.
Maxpar Fix/Perm with intercalator
| Reagent | Final concentration | Amount |
|---|---|---|
| Maxpar Fix/Perm buffer | N/A | 4,995 μL |
| Intercalator (125 mM) | 1:1000 | 5 μL |
| Total | N/A | 5,000 μL |
Note: Prepare freshly on the day of experiment and store at 4°C.
Step-by-step method details
Obtain immune cells from mouse pancreas and pancreatic lymph nodes
Timing: 6 h
These steps include how to separate immune cells from whole pancreas and pancreatic lymph nodes and stimulate them with PMA and Ionomycin.
To isolate immune cell from whole pancreas
-
1.
Euthanize mouse and open abdominal cavity.
-
2.
Identify the pancreas (Figure 1A, outline) and harvest it by careful dissection into a 60 mm culture dish containing 3 mL HBSS buffer.
Note: Fat and spleen tissue must be removed. These tissues also contain lymphocytes and will impact the results.
-
3.
Transfer pancreas into a 15 mL tube and add 3 mL collagenase P digestion media. Incubate at 37°C for 30 min on an orbital shaker at 240 rpm.
CRITICAL: Tissue should not be over-digested to ensure high cell viability (see Figure 2 and troubleshooting).
-
4.
Prepare a single cell suspension (Figure 2) by drawing up digested tissue into a 5 or 10 mL syringe and gently passing through an 18G needle into a new 15 mL tube.
-
5.
Top off with 5 mL HBSS containing 10% FBS to inactivate collagenase P.
-
6.
Pellet cells by centrifugation at 450 × g for 5 min at room temperature (15°C–25°C) and discard the supernatant.
-
7.
Resuspend cell pellet in 3 mL PBS + 2% FBS, and filter suspended cells through a 70 μm cell strainer.
-
8.
Pipette 3 mL Histopaque 1077 into a new 15 mL conical tube.
-
9.
To isolate lymphocytes from other cells of the pancreas, slowly and gently layer the suspended cells from the cell strainer of step 7 on top of Histopaque 1077.
CRITICAL: Do not allow Histopaque 1077 and pancreatic cell layers to mix, as this will adversely affect the yield and purity of isolated lymphocytes.
-
10.
Centrifuge tube at 900 × g for 20 min at room temperature in a swinging bucket rotor with the centrifuge brake turned off.
-
11.
Carefully collect the middle buffy layer (containing lymphocytes) and transfer to a new 15 mL tube with 3 mL RPMI containing 10% FBS.
-
12.
Wash the cells by adding 5 mL RPMI containing 10% FBS and pellet cells by centrifugation at 450 × g for 5 min at room temperature. Discard the supernatant.
-
13.
Repeat step 12.
-
14.
Resuspend the cells in 500 μL of Immune cell culture media.
-
15.Count cells using a hemocytometer.
-
a.Transfer 10 μL cell suspension into a 1.5 mL microcentrifuge tube.
-
b.Combine with 90 μL trypan blue, mixing well with a pipette.
-
c.Load 10 μL of this mixture into a hemocytometer.
-
d.Count the cells and calculate the total number of cells per sample.
-
a.
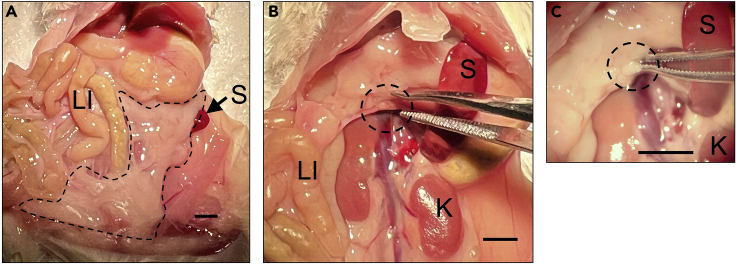
Figure 1.
Identification of pancreas and location of the pancreatic lymph node in the abdominal cavity of a 10-week-old female non-obese diabetic (NOD) mouse
(A) Image showing general location and appearance of pancreas (dotted black outline).
(B) Image showing general location and appearance of pancreatic lymph node (dotted black circle).
(C) Higher magnification image showing location and appearance of the pancreatic lymph node (dotted black circle). LI: large intestine, S: spleen, K: kidney.
Scale bar = 2 mm.
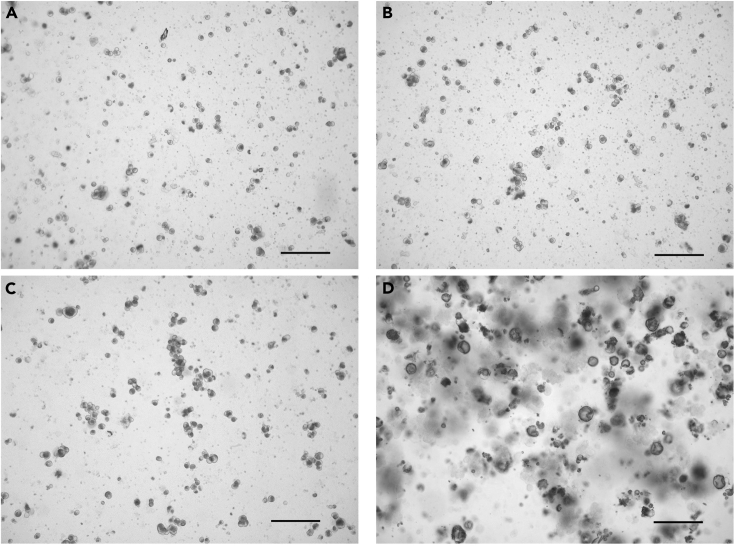
Figure 2.
Appearance of single cells from the digested pancreas of a 10-week-old female NOD mouse
(A and B) Two fields of view showing examples of single cells following collagenase P digestion of the pancreas.
(C) Example of underdigested cells.
(D) Example of over digested cells.
Scale bar = 200 μm.
To isolate immune cells from pancreatic lymph node
-
16.
Remove the pancreatic lymph nodes (see Figures 1B and 1C) and place in a 35 mm culture dish containing 2 mL cold RPMI media (no FBS).
-
17.
Press the pancreatic lymph nodes through a 100 μm cell strainer into a 50 mL conical tube using the plunger from a syringe.
-
18.
Rinse the cell strainer with 5 mL RPMI media (no FBS) and centrifuge the cell suspension at 450 × g for 5 min at 4°C.
-
19.
Discard the supernatant, resuspend the cell pellet in 1 mL red blood cell (RBC) lysis buffer, and incubate for 1 min at room temperature.
-
20.
Add 10 mL PBS containing 2% FBS and centrifuge at 450 × g for 5 min at 4°C.
-
21.
Discard the supernatant and resuspend the cells in 5 mL Immune cell culture media.
-
22.
Perform cell count using a hemocytometer and assess viability with trypan blue.
Stimulate lymphocytes with PMA and ionomycin
-
23.
Transfer an appropriate number of cells (1 × 106–3 × 106) into each well of a 96 well U-bottom plate.
-
24.
Add 100 μL PMA, Ionomycin, and Golgi Stop media.
-
25.
Incubate the plate for 4 h at 37°C under 5% CO2.
Note: Stimulation can be skipped if not staining for cytokines (steps 23–25).
Cell staining
Timing: 20 h
These steps include the procedure for staining immune cells from whole pancreas and pancreatic lymph node with metal-labeled antibodies. The protocol has been adapted from the Fluidigm MaxPar Nuclear Antigen Staining Protocol with Fresh Fix.
-
26.
Scrape the immune cells and transfer to a FACS tube.
-
27.
Add 2 mL PBS into each tube and centrifuge at 450 × g for 5 min at 4°C.
-
28.
Aspirate the supernatant and add 300 μL of the 5 μM Cisplatin Solution to each tube to discriminate viable cells from dead cells.
-
29.
Pipette to mix and incubate for 5 min at room temperature.
CRITICAL: Cell staining with cisplatin for longer than 5 min causes significant cell death.
-
30.
Add 2 mL Maxpar Cell Staining Buffer to each tube to minimize nonspecific antibody binding.
Note: Maxpar Cell Staining Buffer contains blocking protein.
-
31.
Centrifuge the cells at 300 × g for 5 min at room temperature.
-
32.
Aspirate the supernatant and flick the tube to disrupt the pellet.
-
33.
Repeat steps 30–31.
-
34.
Discard the supernatant leaving behind 50 μL Maxpar Cell Staining buffer.
-
35.
Pipette to mix and incubate for 10 min at room temperature.
-
36.
Add 50 μL of diluted Surface marker antibody cocktail to each tube.
-
37.
Pipette to mix and incubate for 30 min at room temperature.
-
38.
Add 2 mL Maxpar Cell Staining buffer and centrifuge the cells at 300 × g for 5 min at room temperature.
-
39.
Aspirate the supernatant and gently vortex to resuspend cells in residual volume.
-
40.
Fix the cells by adding 1 mL Maxpar Fix I buffer. Gently vortex and incubate for 10–30 min at room temperature.
-
41.
Wash the cells by adding 2 mL Maxpar Perm-S buffer and centrifuge at 800 × g for 5 min at room temperature.
-
42.
Aspirate the supernatant and repeat step 41.
Note: After fixation, cells are centrifuged at higher speeds to increase cell recovery and reduce cell loss.
Alternatives: Instead of using Maxpar Fix Buffer, 1.6% PFA in PBS can be used to fix the cells.
-
43.
Aspirate the supernatant leaving a residual volume of 50 μL. Gently vortex to resuspend cells.
-
44.
Add 50 μL of cytoplasmic/secreted antibody cocktail to each tube for a final volume of 100 μL.
-
45.
Gently vortex and incubate for 30 min at room temperature.
-
46.
Wash cells by adding 2 mL Maxpar Cell Staining Buffer and centrifuge at 800 × g for 5 min at room temperature. Aspirate the supernatant.
-
47.
Repeat step 46.
-
48.
Add 300 μL Maxpar Fix/Perm with DNA intercalator.
-
49.
Vortex to mix and incubate at 4°C overnight (16–20 h).
-
50.
Centrifuge the cells at 800 × g for 5 min at room temperature.
-
51.
Aspirate the supernatant and gently vortex to resuspend cells in residual volume.
-
52.
Add 2 mL Maxpar Cell Staining buffer and centrifuge at 300 × g for 5 min at room temperature.
-
53.
Aspirate the supernatant and gently vortex to resuspend cells in residual volume.
-
54.
Repeat steps 52 and 53 two more times.
-
55.
Wash the cells with 2 mL Maxpar Water and centrifuge at 800 × g for 5 min at room temperature.
-
56.
Aspirate the supernatant and gently vortex to resuspend cells in residual volume.
-
57.
Resuspend the cells with 1 mL Maxpar Water.
-
58.
Count the cells using a hemocytometer.
-
59.
Dilute 5 × 105 cells into 1 mL Maxpar Water and centrifuge the cells at 800 × g for 5 min at room temperature.
-
60.
Discard supernatant and keep cell pellet on ice until running on a mass cytometer.
Sample acquisition
Timing: 25 min
This step may differ based on the type of mass cytometer. Prior to sample acquisition, the instrument must be properly warmed up and tuned. Users should contact their service provider for detailed protocols for their specific instrument. Details below are based on using a Helios mass cytometer (Fluidigm) that had been tuned and passed quality control.
-
61.
Resuspend 5 × 105 (1 × 106 maximum) cells in 1 mL Maxpar Water containing 0.1× EQ4 beads within 10 min of running the sample.
-
62.
Filter cells through a 35 μm cell strainer cap into a 5 mL polypropylene tube.
-
63.
Place sample into the Sample Loader. Click record to start acquisition.
Expected outcomes
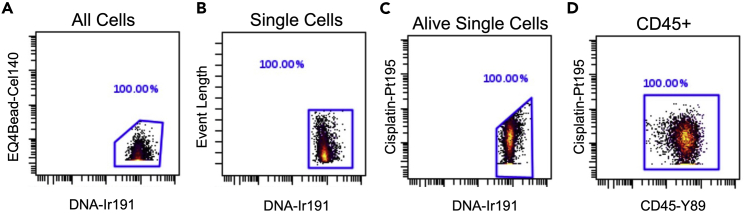
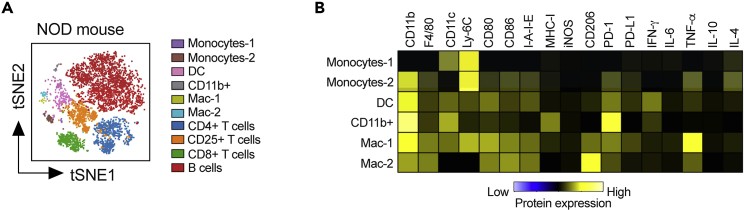
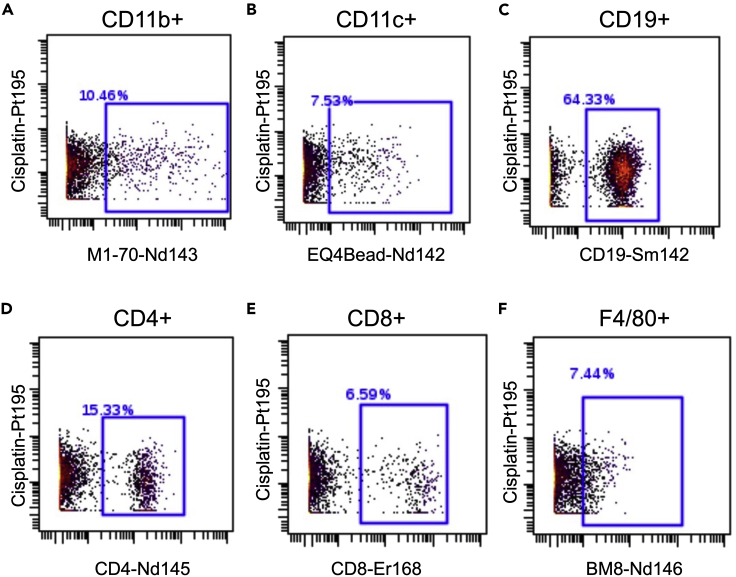
This protocol outlines the isolation of single cells from whole pancreas and pancreatic lymph nodes. Using this protocol, we expected to obtain maximum single viable immune cells and can define monocytes, dendritic cells-1, dendritic cells-2, macrophages, CD4+ T cells, CD8+ T cells, and B cells. To define the cell populations, cells are first defined by having a nucleus and being positive for DNA intercalator (Figure 3A), as well as containing a single nucleus (Figure 3B). Next, the single cells are further defined as alive when negative for cisplatin (Figure 3C). Immune cells are then characterized from the live cells when positive for CD45 (Figure 3D). After gating for live, single, CD45+ cells, the data are processed through the FlowSOM algorithm, which is provided as part of the Cytobank software platform. The FlowSOM algorithm identifies each cell cluster population by predefined markers. An example of cell clustering is shown in Figure 4A and an example of a heatmap generated for defining the cell clusters is shown in Figure 4B. Alternatively, cells can be gated manually using standard markers such as: monocytes (CD11b+ of CD45+ cells; Figure 5A), dendritic cells (CD11c+ of CD45+ cells; Figure 5B), B cells (CD19+ of CD45+ cells; Figure 5C), CD4+ T cells (CD4+ of CD45+ cells; Figure 5D), CD8+ T cells (CD8+ of CD45+ cells; Figure 5E), and macrophages (F4/80+ of CD45+ cells; Figure 5F). Further, we characterized changes in specific pancreatic immune cells under different conditions. For example, deletion of the gene Alox15 in β cells of non-obese diabetic (NOD) mice reduced the number of proinflammatory myeloid cells producing IFN-γ and increased the number of Treg cells (CD4+CD25+). These observations revealed a high degree of heterogeneity in immune cell populations during pathogenesis of autoimmune diabetes.
Figure 3.
Example of primary gating for immune cells from the pancreatic lymph node of 8-week-old female NOD mice
(A) Example of first gate for all cells using DNA-Ir191 to eliminate cellular debris.
(B) Example of second gate for single cells.
(C) Example of third gate for live cells within the single cell gate.
(D) Example of CD45+ leukocytes gating.
Figure 4.
Example of gating using FlowSOM-based cluster-assignment for immune cells from the pancreatic lymph node of 8-week-old female NOD mice
(A) Data were pre-gated on single, live, CD45+ cells as shown in Figure 3. To create a tSNE overview, data from all samples were randomly subsampled with an equal contribution of all samples. Cells are colored by their FlowSOM-based cluster-assignment.
(B) Heatmap of expression levels across all populations identified by FlowSOM.
Figure 5.
Examples of manual gating for different immune cells within the CD45+ population from the pancreatic lymph node of 8-week-old female NOD mice
(A) Example of gating for monocytes (CD11b+).
(B) Example of gating for dendritic cells (CD11c+).
(C) Example of gating for B cells (CD19+).
(D) Example of gating for CD4+ T cells (CD4+).
(E) Example of gating for CD8+ T cells (CD8+).
(F) Example of gating for macrophages (F4/80+).
Limitations
CyTOF is limited in scope to single cell suspensions and does not provide spatial information. Imaging mass cytometry and other spatial imaging systems are emerging technologies that provide additional spatial information. This protocol is restricted in use to individuals that have access to a Fluidigm mass cytometry instrument, either within their individual laboratory or within a Core facility. Additionally, the protocol is limited to using metal-labelled antibodies from Fluidigm, which are predominantly immune cell markers. Currently, there are more antibodies designed against human immune cell markers than mouse or other species.
Troubleshooting
Problem 1
Difficulty locating pancreatic lymph nodes during dissection (major step 16).
Potential solution
One or two pancreatic lymph nodes provide enough cells for mass cytometry staining. However, pancreatic lymph nodes are very small and may not be visible when dissecting the mouse. To ensure that the lymph nodes are visible, open the mouse carefully and avoid puncturing any blood vessels near the pancreas. Then, use a dissecting microscope to locate the lymph nodes. The pancreatic lymph nodes surround the entire pancreas; however, they can be easier to detect in the tail region of the pancreas in close proximity to the spleen.
Problem 2
Decrease in cell viability in pancreatic lymph node or pancreas cell preparations (major steps 15 and 22).
Potential solution
To obtain reliable CyTOF data, the single cell suspension must have at least 90% cell viability. Several conditions can affect cell viability, including the length and type of enzymatic digestion. Here are some suggestions for improving cell viability:
-
•
Pancreatic lymph nodes: After lymph node isolation and before processing, remove any residual tissues that are still attached to the lymph nodes. Pancreatic tissue especially must be cleared away to prevent pancreatic enzymes from coming in contact with the cell preparation.
-
•
Pancreas: It is critical to maintain consistency during the collagenase digestion step. Decreasing the shaker speed or reducing the length of the digestion can help to minimize cell death.
Problem 3
Formation of clumps/debris in cell suspension (major steps 15 and 22).
Potential solution
-
•
Switch to using a cell strainer with a smaller pore size to filter the samples.
-
•
When resuspending cell pellets, it is better to gently pipette up and down rather than vortexing or flicking the tube.
-
•
Viscous samples and samples containing a lot of debris/aggregates will require more dilution or additional optimization before running on the CyTOF.
Problem 4
Contamination of cell suspension with heavy metals (all steps).
Potential solution
As CyTOF relies on heavy metals to detect antibodies bound to immune cell proteins, it is essential to avoid contamination with heavy metals, which could result in high levels of background detected by the mass cytometer. Many common laboratory detergents contain heavy metals as well as potential lead from pipes.
-
•
All buffers should be made and stored in new clean Pyrex or polypropylene bottles.
-
•
Buffers should be calcium and magnesium free. Metal-free buffers are commercially available.
-
•
Avoid commercial laboratory detergents.
-
•
Carefully follow all wash steps.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Raghavendra Mirmira (mirmira@uchicago.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This research was supported by the NIH grants R01 DK060581 and R01 DK105588 (both to R.G.M.). Research core services were provided by the NIH grant P30 DK020595 (to the University of Chicago).
Author contributions
T.N., A.R.P., S.A.T., and R.G.M. conceived the study, developed the methodology, and analyzed the data; T.N., A.R.P., and S.C.M. wrote the original draft; and all authors edited and approved the final draft.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sarah A. Tersey, Email: stersey@uchicago.edu.
Raghavendra G. Mirmira, Email: mirmira@uchicago.edu.
Data and code availability
The datasets supporting the current study are available from the corresponding author on request.
References
- 1.Piñeros A.R., Kulkarni A., Gao H., Orr K.S., Glenn L., Huang F., Liu Y., Gannon M., Syed F., Wu W., et al. Proinflammatory signaling in islet β cells propagates invasion of pathogenic immune cells in autoimmune diabetes. Cell Rep. 2022;39:111011. doi: 10.1016/j.celrep.2022.111011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roep B.O., Thomaidou S., van Tienhoven R., Zaldumbide A. Type 1 diabetes mellitus as a disease of the β-cell (do not blame the immune system?) Nat. Rev. Endocrinol. 2021;17:150–161. doi: 10.1038/s41574-020-00443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannidis L.J., Mitchell A.J., Zheng T., Hansen D.S. CyTOF mass cytometry analysis of human memory CD4+ T cells and memory B cells. STAR Protoc. 2022;3:101269. doi: 10.1016/j.xpro.2022.101269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the current study are available from the corresponding author on request.