Abstract
The vacA gene of Helicobacter pylori strain 60190 encodes a 1,287-amino-acid protoxin, which undergoes cleavage of a 33-amino-acid amino-terminal signal sequence and carboxy-terminal proteolytic processing to yield a mature secreted toxin. Several features of VacA suggest that it belongs to the autotransporter family of gram-negative bacterial secreted proteins. Based on matrix-assisted laser desorption ionization–time of flight mass spectrometric analysis, we calculate that the mature toxin has a mass of 88.2 ± 0.2 kDa and consists of approximately 821 amino acids.
Chronic colonization of the human gastric mucosa by Helicobacter pylori is associated with gastritis and an increased risk for development of peptic ulcer disease and gastric malignancies (5). Most H. pylori strains secrete a toxin (VacA) that induces multiple structural and functional alterations in eukaryotic cells (see references 1 and 10 for reviews). H. pylori vacA encodes an ∼139-kDa protoxin, which undergoes cleavage of a 33-amino-acid amino-terminal signal sequence and carboxy-terminal proteolytic processing to yield a mature secreted toxin (2, 4, 14, 15) (see GenPept accession number B53739). In various studies, the reported masses of mature secreted VacA toxins have varied from 87 to 95 kDa, based on analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2, 4, 14, 15). Thus, the site (or sites) at which the VacA protoxin undergoes carboxy-terminal proteolytic processing is not known. In this study, we used matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry to determine the precise molecular mass of the secreted VacA toxin.
H. pylori strain 60190 (ATCC 49503) was cultured for 48 h at 37°C in sulfite-free brucella broth containing 0.5% charcoal. VacA was purified in an oligomeric form from the culture supernatant, as described previously (3). Purified VacA preparations were analyzed by MALDI-TOF mass spectrometry, using a Voyager Elite (PerSeptive Biosystems, Framingham, Mass.) instrument equipped with a pulsed nitrogen laser source operating at 337 nm. Mass spectra were obtained in the delayed-extraction positive ion mode with an accelerating voltage of 25 kV. Sinapinic acid (10 mg/ml in 70:30 [vol/vol] acetonitrile–0.1% trifluoroacetic acid) was used as a matrix additive. The instrument was calibrated externally with bovine serum albumin (MH+ = 66,431) for analysis of intact VacA and was calibrated with bovine insulin (MH+ = 5,734.6) for analysis of VacA peptides. The molecular masses reported in this paper are those of single-protonated species (MH+).
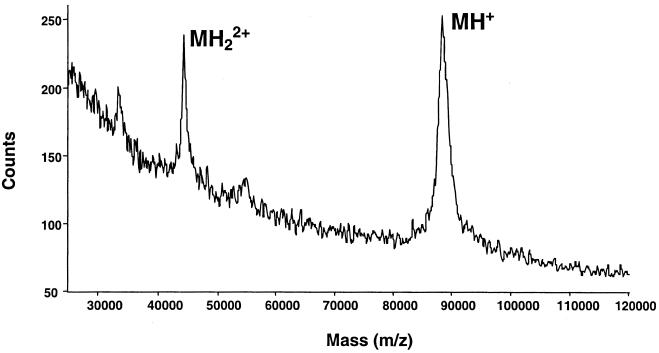
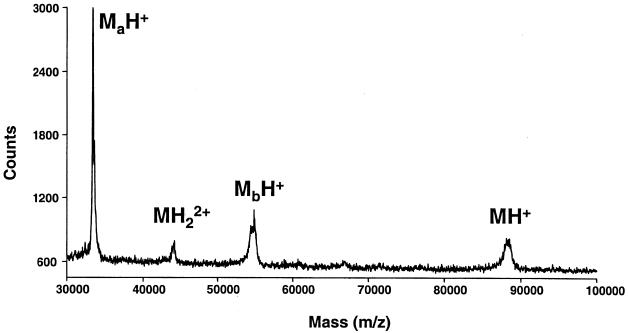
Based on MALDI-TOF analysis of purified VacA, we calculated the molecular mass of VacA monomers to be 88.2 ± 0.2 kDa (value is mean ± standard deviation, as are values given below) (Fig. 1). Purified VacA degrades during prolonged storage into two fragments (of about 34 and 58 kDa, respectively) which are derived from the amino terminus and the carboxy terminus of the toxin, respectively (3, 15). Mass spectrometric analysis of partially proteolysed VacA preparations demonstrated two major peaks representing proteins with average molecular masses of 33.4 ± 0.08 and 54.8 ± 0.1 kDa (Fig. 2). Thus, the experimentally determined mass of intact VacA (88.2 kDa) corresponded closely to the sum of the masses of two proteolytic fragments. The mass of the amino-terminal fragment is consistent with proteolytic cleavage between amino acid 344 (alanine) and amino acid 345 (lysine) of the VacA protoxin (4, 15). The 54.8-kDa fragment sometimes could be resolved into a series of clustered peaks with very similar molecular masses (Fig. 2), which suggests the occurrence of proteolytic cleavage at multiple sites. Clustered peaks were not readily detectable for the intact 88.2-kDa VacA protein (Fig. 1), but this may simply reflect limited resolution with spectrometric analysis of this relatively large protein.
FIG. 1.
MALDI-TOF mass spectrum of secreted, intact VacA from H. pylori 60190 (MH+ = 88.2 ± 0.2 kDa). MH22+ denotes a peak corresponding to doubly protonated VacA.
FIG. 2.
MALDI-TOF mass spectrum of spontaneously arising proteolytic degradation products of VacA (MaH+ = 33.4 ± 0.08 kDa and MbH+ = 54.8 ± 0.1 kDa).
The spectrometric analyses shown in Fig. 1 and 2 were not sufficiently precise to permit identification of the exact site of carboxy-terminal VacA processing. Therefore, purified VacA was treated with cyanogen bromide (CNBr) in 70% trifluoroacetic acid in water overnight at room temperature, and the resulting peptides were dried and dissolved in a 0.1% trifluoroacetic acid aqueous solution prior to mass spectrometric analysis. The molecular masses of six prominent peaks (1,692, 2,525, 4,331, 8,041, 10,618, and 11,220 Da) corresponded to predicted cyanogen bromide products (amino acids 769 to 784, 137 to 160, 655 to 695, 697 to 768, 161 to 259, and 34 to 136 of the VacA protoxin, respectively) (Fig. 3 and data not shown). Peaks corresponding to three other predicted CNBr fragments (672, 1,274, and 40,783 Da) were not successfully detected. In addition, seven peaks of similar size (ranging from 5,922 to 6,803 Da), all of which can be derived only from a CNBr product that contains valine-792 at its amino terminus (Fig. 3), were visualized. These peptides are predicted to result from proteolytic cleavage after amino acid residues 846, 847, 848, 849, 850, 851, and 854 (4). Similar ragged ends were not detected for any of the other CNBr-generated peptides (Fig. 3). Notably, there are no methionine residues located in the region between amino acids 846 and 855 (4), which indicates that these peptides did not result solely from CNBr-mediated proteolytic cleavage. Therefore, we postulate that the family of seven similar-size peptides are derived from the carboxy terminus of the secreted VacA toxin. The predicted molecular mass of a VacA protein containing amino acids 34 to 854 of the protoxin is 88.3 kDa, a result that compares favorably with the experimentally determined molecular mass of intact VacA, as described above. The mature VacA toxin probably forms following a proteolytic cleavage event between amino acids 854 and 855 (alanine and leucine), and several individual amino acids are thereafter susceptible to further nonspecific proteolysis. In agreement with this interpretation, some preparations of CNBr-digested VacA exhibited a single peak corresponding to amino acids 792 to 854, without associated ragged ends (data not shown). Alternatively, the ragged ends may result from nonspecific endoproteolysis at an exposed loop in the protoxin.
FIG. 3.
MALDI-TOF mass spectrum of CNBr-digested VacA. The peaks in the inset represent peptides derived from the C terminus of VacA. The numbers indicate the location of peptides in the amino acid sequence of the VacA protoxin (4).
Proteins that are secreted into the extracellular space by gram-negative bacteria must cross two different lipid bilayers of the cell envelope. Four classes of secretory pathways (termed types I, II, III, and IV, respectively) utilize accessory proteins in the export process. In contrast, certain autotransporter proteins are secreted via a pathway that does not require any accessory proteins (6, 7, 9). The prototype of the autotransporter protein family is immunoglobulin A1 (IgA1) protease from Neisseria gonorrhoeae (8, 11). Autotransporter proteins are typically comprised of three functional domains, as follows: (i) an amino-terminal signal sequence, (ii) a passenger domain (corresponding to the mature secreted protein), and (iii) a carboxy-terminal domain that is rich in amphipathic β-sheets (6, 7, 9). It is presumed that the carboxy-terminal domain forms a β-barrel structure consisting of antiparallel amphipathic β-sheets in the outer membrane and that this structure mediates translocation of the secreted passenger domain from the periplasm to the bacterial cell surface (6–9).
Based on various features of the vacA gene structure, H. pylori VacA has been classified in the autotransporter family of secreted proteins (4, 6, 7, 9, 14). Secondary structure predictions suggest that a 35-kDa portion of the VacA carboxy-terminal domain is rich in amphipathic β-sheets, and this region exhibits low-level homology to members of the family of autotransporter proteins (6, 9, 17). In addition, at the carboxy terminus of VacA there is a phenylalanine-containing motif that is commonly found in autotransporter proteins, as well as in numerous gram-negative bacterial outer membrane proteins (4, 7). Isogenic H. pylori mutant strains in which the carboxy-terminal VacA domain is disrupted fail to express or secrete any detectable VacA, which is probably attributable to the degradation of export-incompetent toxin precursors within the periplasm (14).
The molecular mass of the carboxy-terminal VacA domain that remains associated with bacterial cells is reported to be 33 kDa (15). Interestingly, the combined masses of known VacA domains (i.e., the N-terminal signal sequence [3 kDa], the secreted toxin [88.2 kDa], and the carboxy terminal domain [33 kDa]) is only 124 kDa, which is considerably less than the predicted mass of the VacA protoxin (139 kDa). We speculate that the VacA protoxin may undergo proteolytic cleavage at multiple sites downstream from amino acid 854 of the protoxin, which would yield the previously identified 33-kDa cell-associated domain (15), as well as a fragment of ∼15 kDa. Similar carboxy-terminal proteolytic processing at multiple sites has been described for N. gonorrhoeae IgA1 protease (8, 11).
Following translocation of the passenger domain through the outer membrane, several autotransporter proteins, including N. gonorrhoeae IgA1 protease, undergo autoproteolytic cleavage, which results in release of the secreted protein from the cell-associated carboxy-terminal β-barrel domain (6). Thus, there has been speculation that VacA might possess similar autoproteolytic activity. In accordance with this hypothesis, the cytotoxic activity of VacA can be blocked by treatment of VacA with 3,4-dichloro-isocoumarin (a serine protease inhibitor), and some features of the amino-terminal portion of VacA are related to serine proteases (13). However, at present there is no direct experimental evidence that VacA possesses proteolytic activity (10, 13). Moreover, H. pylori mutant strains, constructed with in-frame deletion mutations in the portion of vacA that encodes the amino-terminal portion of the toxin, express and secrete truncated vacA products that undergo carboxy-terminal proteolytic processing (12, 16). This indicates that an intact amino-terminal portion of VacA is not required for proteolytic processing of the protoxin. At present, we favor the hypothesis that carboxy-terminal proteolytic processing of VacA is mediated by an independent membrane-associated protease, rather than occurring as an autoproteolytic event. Potentially, the identification of the site of carboxy-terminal VacA processing will permit future studies in which the relevant H. pylori protease can be identified.
Acknowledgments
This work was supported in part by the National Institutes of Health (R01 AI-39657 and DK-53623) and the Department of Veterans Affairs.
REFERENCES
- 1.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 2.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 3.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 5.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 7.Jose J, Jahnig F, Meyer T F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol. 1995;18:378–380. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 8.Klauser T, Pohlner J, Meyer T F. The secretion pathway of IgA protease-type proteins in Gram-negative bacteria. Bioessays. 1993;15:799–805. doi: 10.1002/bies.950151205. [DOI] [PubMed] [Google Scholar]
- 9.Loveless B J, Saier M H. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membrane Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 10.Montecucco C, Papini E, de Bernard M, Telford J L, Rappuoli R. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors. In: Alouf J E, Freer J H, editors. The comprehensive sourcebook of bacterial protein toxins. San Diego, Calif: Academic Press; 1999. pp. 264–283. [Google Scholar]
- 11.Pohlner J, Halter R, Beyreuther K, Meyer T F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 12.Reyrat J-M, Lanzavecchia S, Lupetti P, de Bernard M, Pagliaccia C, Pelicic V, Charrel M, Ulivieri C, Norais N, Ji X, Cabiaux V, Papini E, Rappuoli R, Telford J L. 3D imaging of the 58 kDa cell binding subunit of the Helicobacter pylori cytotoxin. J Mol Biol. 1999;290:459–470. doi: 10.1006/jmbi.1999.2877. [DOI] [PubMed] [Google Scholar]
- 13.Rossetto O, de Bernard M, Pellizzari R, Vitale G, Caccin P, Schiavo G, Montecucco C. Bacterial toxins with intracellular protease activity. Clin Chim Acta. 2000;291:189–199. doi: 10.1016/s0009-8981(99)00228-4. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 15.Telford J L, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinion A D, McCLain M S, Czajkowsky D M, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke S R, Shao Z, Cover T L. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J Biol Chem. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 17.Wang H-J, Chang P C L, Kuo C-H, Tzeng C-S, Wang W-C. Characterization of the C-terminal domain of Helicobacter pylori vacuolating toxin and its relationship with extracellular toxin production. Biochem Biophys Res Commun. 1998;250:397–402. doi: 10.1006/bbrc.1998.9228. [DOI] [PubMed] [Google Scholar]