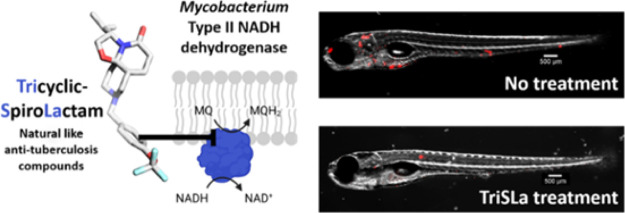
Abstract
It is critical that novel classes of antituberculosis drugs are developed to combat the increasing burden of infections by multidrug-resistant strains. To identify such a novel class of antibiotics, a chemical library of unique 3-D bioinspired molecules was explored revealing a promising, mycobacterium specific Tricyclic SpiroLactam (TriSLa) hit. Chemical optimization of the TriSLa scaffold delivered potent analogues with nanomolar activity against replicating and nonreplicating Mycobacterium tuberculosis. Characterization of isolated TriSLa-resistant mutants, and biochemical studies, found TriSLas to act as allosteric inhibitors of type II NADH dehydrogenases (Ndh-2 of the electron transport chain), resulting in an increase in bacterial NADH/NAD+ ratios and decreased ATP levels. TriSLas are chemically distinct from other inhibitors of Ndh-2 but share a dependence for fatty acids for activity. Finally, in vivo proof-of-concept studies showed TriSLas to protect zebrafish larvae from Mycobacterium marinum infection, suggesting a vulnerability of Ndh-2 inhibition in mycobacterial infections.
Introduction
While global efforts to eradicate tuberculosis (TB) by improving drug access and treatment compliance have decreased deaths by 29% over the last two decades, TB remains the leading cause of death by an infectious disease worldwide.1 With a minimum of 6 months of multidrug therapy, current TB treatment is notoriously lengthy, a feature largely attributed to the difficulty in eliminating phenotypically drug-tolerant subpopulations of the causative bacteria Mycobacterium tuberculosis (Mtb).2 However, escalating infections by multidrug-resistant TB infections (483,000 cases of rifampicin-resistant TB reported in 20201) as well as extensively drug resistant (XDR) TB (12,350 cases reported in 20193) require even longer therapy with less efficient and tolerated second-line drugs. In recognition of this global health problem, the World Health Organization has placed TB at the highest critical global priority of antibiotic-resistant bacteria for the development of new antibiotics.4
Concerted efforts to propose alternative and better antibiotics against drug-sensitive and -resistant TB have led to the approval of two novel classes of anti-TB drugs, the ATP synthase inhibitor bedaquiline,5 and the two nitroimidazole prodrugs, delamanid6 and pretomanid.7 Additional anti-TB molecules are at various levels of clinical and preclinical drug development that may feed our treatment options in the future.8−10 Despite these increased efforts, it is clear that the TB drug development pipeline requires further supplementation with additional candidates, ideally acting on novel targets (to minimize cross-resistance) and impacting on drug-tolerant bacilli to shorten the duration of the treatment.
For the discovery of novel anti-TB drugs, phenotypic screening of chemical libraries on whole bacteria has proven to be the most successful approach.11 The vast majority of synthetic molecules screened against Mtb, and hence emerged as leads, are largely flat (two-dimensional) in structure and comprise many sp2-rich aromatic cores.12 Such flat molecules are in stark contrast to the often complex three-dimensional (3D) structures of natural products, historically representing an important source of antibiotics. The molecular shape of such biomolecules plays a key role in their interaction with protein targets.13 Interestingly, the Mtb ATP synthase inhibitor bedaquiline buckles this trend as an antibiotic of synthetic origin, being highly 3D in shape, thanks to the two central adjacent highly substituted sp3-hybridized carbon atoms. With the rational that molecules with increased 3D structures will allow for the probing of previously unexplored chemical and biological space, a “natural-like” chemical library with sp3-rich synthetic molecules was assembled14−19 and evaluated for anti-TB activity.20,21
Herein, we show that the screening of chemical libraries with increased 3D diversity can result in the identification of novel chemical- and biological-space to combat Mtb. This work describes the identification of the Tricyclic SpiroLactam (TriSLa) chemical family and its chemical optimization to potent nanomolar antimycobacterial compounds with particular activity against Mtb. Target identification and validation studies revealed that TriSLas act through the inhibition of the mycobacterial type II NADH dehydrogenase. TriSLas exhibited a time-dependent bactericidal activity on Mtb and was also active on nonreplicating and intracellular bacilli. Subsequent in vivo efficacy studies of TriSLas on Mycobacterium marinum-infected zebrafish larvae confirmed efficacy and validated type-2 NADH dehydrogenase as a vulnerable and druggable target in mycobacteria.
Results
Discovery of TriSLa Antituberculosis Lead Compounds
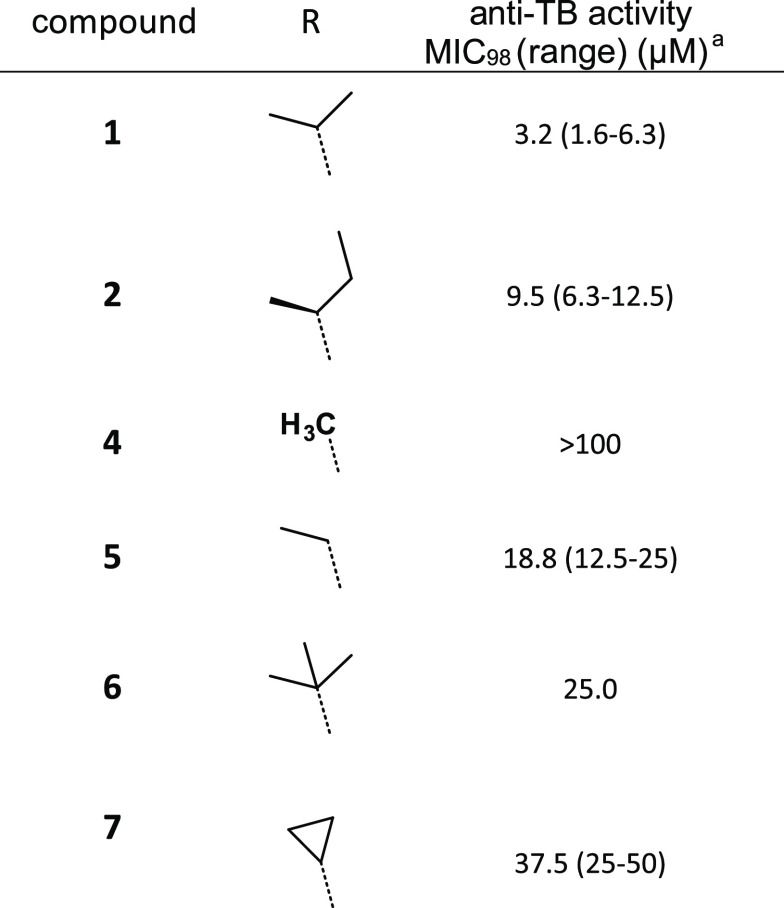
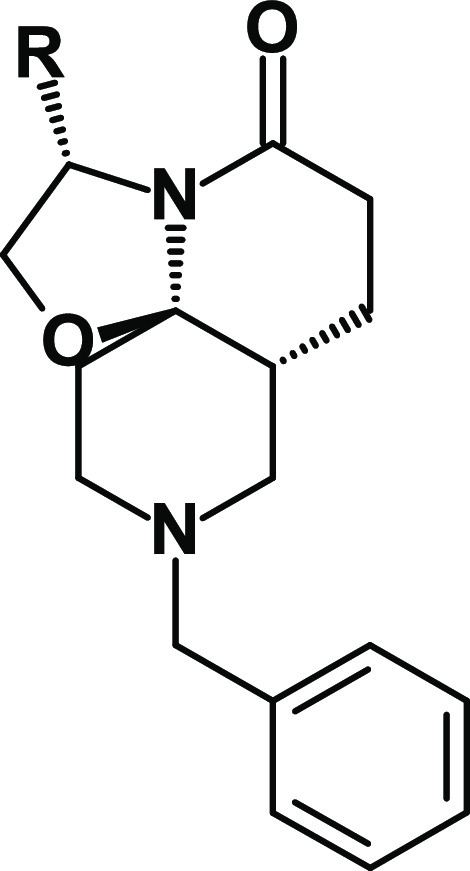
With the aim of investigating the anti-TB potential of new chemical spaces, we assembled a focused chemical library of 958 in-house synthetic “natural-like” compounds. These molecules display enhanced 3D properties, a high number of sp3-hybridized carbon atoms (≥12), a calculated molecular complexity22 greater than 0.76, and a molecular shape index lower than 0.56, reflecting a greater sphericity (as measured using DataWarrior23) (Figure S1a). Screening of this chemical library at 10 μM on replicating Mtb strain H37Rv led to the identification of compounds 1 and 2 (Table 1), two tricyclic spirolactam analogues that inhibited bacterial growth. Following de novo re-synthesis and purification of these hits, 1 and 2 were confirmed to have a minimal inhibitory concentration (MIC) against Mtb of 3.2 and 9.5 μM respectively (as measured using the resazurin reduction assay). The more potent analogue 1 is a small lead-like molecule (low molecular weight of 328 g mol–1, Lipinski and Veber rules compliant) with a high aqueous solubility (178 μM at pH 7.4) because of the basic nitrogen of the piperidine ring and favorable logD (2.7). The configuration of the stereogenic centers of 1 was confirmed by crystallization and X-ray diffraction of its hydrochloride salt (Figure S1c). To confirm the importance of the TriSLa configuration, compound 1’s enantiomer (compound 3, Figure S1b) was synthesized and found to be inactive against Mtb (MIC of 100 μM), supporting the hypothesis that the antibiotic activity is mediated by an interaction with a specific mycobacterial protein.
Table 1. Biological Activities of Compounds 1–7.
Anti-TB activity was determined on H37Rv grown in Middlebrook 7H9 media supplemented with 0.2% glycerol, 0.05% tween 80, and 10% OADC. MIC98 represents the lowest concentration of compounds that prevented 98% of resazurin turnover by H37Rv compared to the untreated bacteria.
Hit-to-Lead Optimization: Medicinal Chemistry
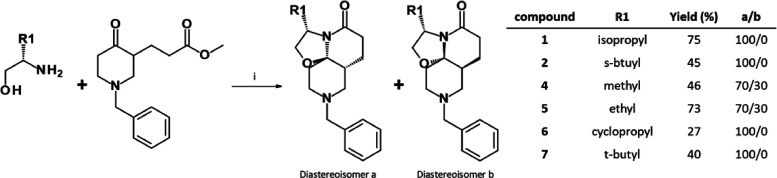
Compound 1 represented an attractive starting point for the development of a new anti-TB chemotype, though its potency required improvement. Because replacement of the isopropyl moiety by a sec-butyl (compound 2) was found to affect potency, we decided to explore other modifications in this position (Table 1). Replacement of the isopropyl with smaller alkyl substituents (a methyl for compound 4 and an ethyl for compound 5) led to a loss of activity (MIC > 100 μM and MIC = 18.8 μM, respectively). A similar result was obtained with a larger tert-butyl moiety (compound 6, MIC = 25 μM). Introduction of a cyclopropyl (compound 7, MIC = 37.5 μM) also led to a 1-log decrease in potency.
Overall, these structure–activity relationships (SARs) suggested that the isopropyl moiety of compound 1 cannot be advantageously replaced. In parallel, metabolic stability studies of compound 1 showed high clearance (Table S1) largely because of hydroxylation of the benzyl moiety (Figure S2). For these reasons, modifications of the benzyl group were next prioritized in an attempt to both metabolically stabilize compound 1 and increase its antimicrobial potency.
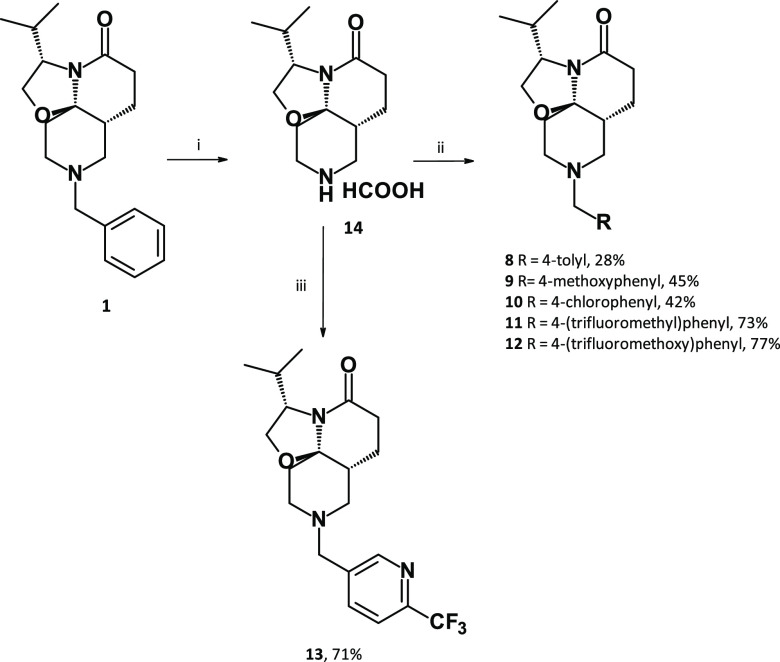
The introduction of electron-donating groups such as methyl (compound 8, MIC = 1.7 μM) or methoxy (compound 9, MIC = 4.7 μM) in position 4 of the phenyl ring did not impact antibacterial potency (Table 2). On the other hand, introduction of an electron-withdrawing chlorine atom (compound 10, MIC = 0.25 μM) led to a marked (>1-log) improvement in antibacterial potency. This effect was even more pronounced using a trifluoromethyl (compound 11, MIC = 0.10 μM) or trifluoromethoxy (compound 12, MIC = 0.13 μM) moiety. While the potency was improved, the hydrophobicity of 11 (logD7.4 = 3.9) and 12 (logD7.4 = 3.4) was increased, which had a slight impact on their solubility (148 and 133 μM, respectively). To address this, the phenyl group of 11 was replaced by a more polar pyridine group (compound 13, logD7.4 = 2.7), which indeed led to an improved solubility (>200 μM), but at the expense of anti-TB potency (MIC = 0.59 μM) (Table 2). As hypothesized, substitution of the phenyl ring prevented metabolism on this part of the molecule (Figure S2). However, microsomal stability remained only partially improved compared to compound 1. Overall, study of these first SARs for compound 1 allowed for marked improvements to the TriSLa anti-TB potency.
Table 2. Summary of TriSLa Antituberculosis Activity, Biochemical Ndh-2 Inhibition, and Physicochemical Properties.
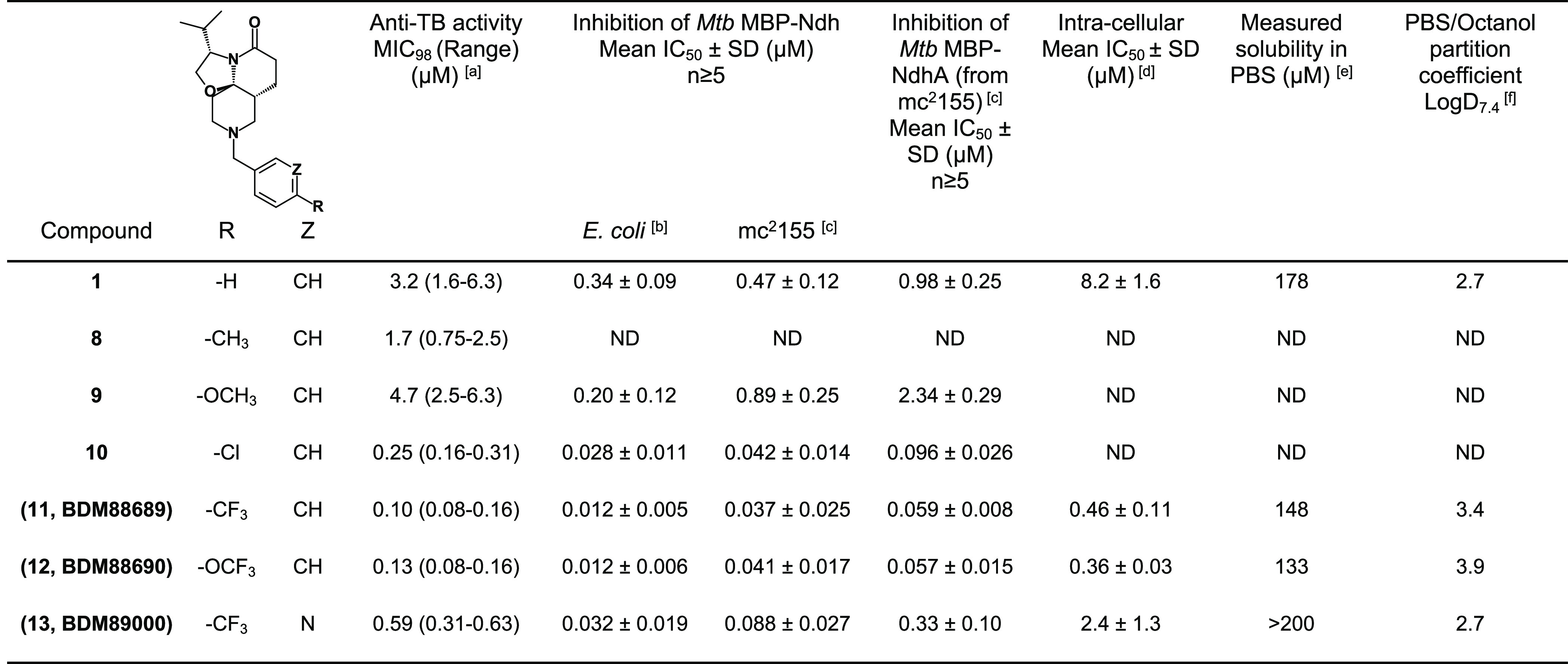
Anti-TB activity was determined on H37Rv grown in Middlebrook 7H9 media supplemented with 0.2% glycerol, 0.05% tween 80, and 10% OADC. MIC98 represents the lowest concentration of compounds that prevented 98% of resazurin turnover by H37Rv compared to the untreated bacteria.
Biochemical assays performed on recombinant MBP-Ndh purified from E. coli.
Biochemical assays performed on recombinant MBP-Ndh purified from M. smegmatis mc2155.
Intracellular activity of compounds was investigated by determining the concentration that protected 50% of the macrophages (IC50) from a lethal H37Rv infection (MOI 10), with macrophage viability determined using the resazurin reduction assay.
Solubility measured in PBS pH 7.4 starting from a 10 mM solution in DMSO of the compound.
logD was measured at pH 7.4 between PBS and octanol. All compounds were tested at the same concentration (1 μM). n.d. not determined. MBP-Ndh stands for mannose binding protein-tagged Ndh.
In Vitro Profiling of TriSLa Activity
Spectrum of TriSLa Antibiotic Activity
The spectrum of TriSLa antibiotic activity was next determined on a panel of mycobacteria as well as Gram-positive and -negative bacteria. Compounds 1, 11, and 12 showed mycobacterium-specific activity with Mtb, M. marinum, and M. avium being particularly susceptible (Table 3). Gram-positive and -negative bacteria were found to be resistant to TriSLa activity, including the efflux-deficient E. coli ΔtolC (Table 3). These three TriSLa compounds showed no apparent cytotoxicity on BALB/3T3 cells at concentrations up to 100 μM.
Table 3. Spectrum of Antibiotic Activity of TriSLa Inhibitorsa.
| bacterial strain | MIC (μM) |
||
|---|---|---|---|
| compound 1 | compound 11 | compound 12 | |
| M. tuberculosis (H37Rv)b | 3.2 (1.6–6.3) | 0.10 (0.08–0.16) | 0.13 (0.08–0.16) |
| M. marinum (M strain)b | 6.3 | 0.19 (0.16–0.31) | 0.16 |
| M. avium (TMC 724, ATCC-25291)b | 13.1 (3.1–25) | 0.55 (0.16–1.25) | 0.38 (0.16–0.63) |
| M. smegmatis (mc2155)b | 29.2 (25–50) | 1.3 (1.25–2.5) | 1.3 (1.25–2.5) |
| M. abscessus (CIP104536, Smooth variant)b | ≥100 | 5 | 8.1 (5–10) |
| M. abscessus (CIP104536, Rough variant)b | ≥100 | 6.3 (5–10) | 8.1 (5–10) |
| Bacillus subtilis (ATCC6633)c | >100 | >100 | >100 |
| Staphylococcus aureus (SH1000)c | >100 | >100 | >100 |
| Escherichia coli (BW25113)c | >100 | >100 | >100 |
| Escherichia coliΔtolC (BW25113 ΔtolC)c | >100 | 100 | 100 |
| Pseudomonas aeruginosa (POA1)c | >100 | >100 | >100 |
| Klebsiella pneumoniae (LMG 2095)c | >100 | >100 | >100 |
| Acinetobacter baumannii (LMG-17978)c | >100 | >100 | >100 |
MIC values (in μM) were determined using the resazurin reduction assay, except for M. marinum and P. aeruginosa where viability was determined visually. Data are presented as an average MIC of at least three independent biological replicates with the range of MICs (where there was a range) indicated in brackets.
Tested in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.05% tween 80, and 10% OADC.
Tested in cation-adjusted Mueller Hinton II Broth (CAMHB).
Time-Dependent Bactericidal Activity of TriSLas on Replicating and Nonreplicating M. tuberculosis
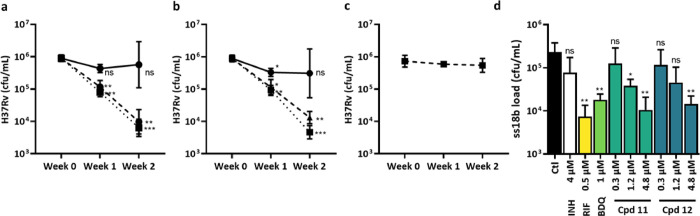
To evaluate the bactericidal activity of TriSLas in vitro, the bacterial load (measured by colony forming units, cfu) was determined after 1 and 2 weeks of compound exposure at 1×, 2×, and 4× MIC. Data revealed that 1× MIC values of both 11 and 12 (150 nM) were bacteriostatic, while exposure to 2× and 4× MIC (300 and 600 nM) led to around a 0.8-log and 2.0 log drop in cfu after 1 and 2 weeks, respectively (Figure 1a,b). In line with previous reports,24 bedaquiline at 1× and 2× MIC (250 and 500 nM, respectively) was largely bacteriostatic over the 2 week exposure period (Figure 1c).
Figure 1.
In vitro profile of TriSLas against replicating H37Rv (a–c) and nonreplicating ss18b (d). The bactericidal activity of (a) 11, (b) 12, and (c) bedaquiline was measured by cfu counting following 1 or 2 weeks of incubation with 1× MIC (filled circles, solid line), 2× MIC (filled triangles, dashed line), or 4× MIC (filled squares, dotted line). Data are mean and SEM of at least three independent biological replicates, and statistical analysis was performed with a paired t-test. (d) Bactericidal activity following 1-week incubation with TriSLAs and control antibiotics (isoniazid (INH) bedaquiline (BDQ), rifampicin (RIF)) against nonreplication ss18b-lux as measured by cfu counting. Data are mean and SEM of at least three independent biological replicates, and the statistical test to compare compound exposure to no compound control was performed using a two-tailed unpaired t test with Welch’s correction. The time-dependent decrease in ss18b-lux luminescence is available in Figure S3.
The activity of TriSLa compounds was then evaluated on nonreplicating bacteria using a luciferase-expressing streptomycin starved 18b model of nonreplicating Mtb (ss18b-lux25). By monitoring the ss18b-lux luminescence signal from the integrated luxCDABE reporter, a time-dependent decrease in luminescence was observed with rifampicin, bedaquiline, and the TriSLas 11 and 12, while isoniazid caused an increase in luminescence, as reported previously for cell wall inhibitors (Figure S3). This bactericidal activity was confirmed by plating of the remaining ss18b-lux bacteria following 7 days of antibiotic exposure on streptomycin-containing plates and cfu counting. Data clearly show that exposure to high concentrations of isoniazid had no significant bactericidal activity against nonreplicating ss18b-lux, while rifampicin and bedaquiline were bactericidal, as previously reported.25,26 In this assay, both 11 and 12 showed a concentration-dependent killing of ss18b-lux, with concentrations of 4.8 μM leading to a 1–2 log drop in bacterial counts, similar to that seen for 1 μM bedaquiline and 0.6 μM rifampicin (Figure 1d), while the potency of 11 and 12 are lower than that observed on replicating bacteria.
TriSLa Activity on Intracellular M. tuberculosis
To define the activity of the TriSLa compounds against intracellular Mtb, their ability to protect differentiated THP-1 macrophages from a lethal infection dose of H37Rv was evaluated as a proxy to the number of intracellular bacteria. Data showed that control compounds isoniazid (IC50 = 204 ± 20 nM), rifampicin (IC50 = 91 ± 32 nM), and bedaquiline (IC50 = 34 ± 3 nM) were able to fully protect the macrophages from infection, in line with previously described data (Figure S4). The TriSLa compounds tested were found to also protect macrophages from death by the intracellular infection in a similar hierarchy to that observed on extracellular H37Rv (Table 2), but protection was not absolute (did not achieve 100% protection) and required higher concentrations (Figure S4), suggesting that TriSLas are less effective on intracellular bacteria than isoniazid, rifampicin, or bedaquiline.
Target Identification of TriSLas
To gain insight into the mechanism of action of the TriSLa series, resistant Mtb mutants were isolated and characterized. After exposure to compound 1 at 14 and 28 μM, resistant H37Rv isolates were selected at a frequency of around 1 × 10–8 (six and four colonies, respectively, following the plating of 50 μL of OD600 = 50), with confirmation of resistance to 1 by REMA (MIC > 100 μM). Whole genome sequencing and variant analysis of two resistant isolates found one (RC14.2) to carry a 97 bp base-pair deletion in the ndhA (Rv0392c) promoter (including a 17 bp truncation of the 5′ end of neighboring, nonessential Rv0393 (encoding a 13E12 repeat family protein)) (Figure S5), and the other (RC28.2) to carry a nonsynonymous single nucleotide polymorphism in ndh (Rv1854c) leading to Tyr403Cys (Table S2) (sequences available at NCBI project number, PRJNA808942). Similarly, validated 11-resistant clones of H37Rv were isolated on 1 μM 11 at a frequency of resistance at around 3 × 10–7, and Sanger sequencing of ndh and ndhA (including promoter) found them to carry an alternative nonsynonymous mutation in the same ndh codon leading to Tyr403His and Tyr403Asn (Table S2). Finally, M. marinum isolates resistant to 11 were selected through sequential selection in liquid culture containing 4 μM TriSLa, followed by isolation of single clones on solid culture (direct selection on solid media led to high background). Sanger sequencing revealed these clones to similarly carry nonsynonymous mutations in ndh (MMAR_2728), but this time in conserved Gln334 codon, leading to Gln334Pro or Gln334Arg substitutions (Table S2).
Both ndh and ndhA encode for the two Mtb type II NADH dehydrogenases (Ndh-2: Ndh, and NdhA) of the bacterial electron transport chain that mediate the transfer of electrons from NADH to menaquinone. Ndh and NdhA share 67% protein identity, with both Tyr403/Gln334 and surrounding amino acids conserved (Figure S6). In the absence of a protein structure of Ndh (or NdhA), structural models were generated by AlphaFold27,28 and by homology (using SwissModel). Tyr403 and Gln334 were found in close proximity to each other (with a hydrogen bond interaction between their side chains according to AlphaFold) near the access pocket of menaquinone in the membrane-associated region of the protein (Figure S7). To date, mutations in this region of Ndh have not been associated with resistance to any antibiotic molecule. On the other hand, a deletion in the ndhA promoter (different to the one isolated here) that leads to an overexpression of ndhA has previously been isolated following selection to the 2-mercapto-quinazolinone family of Ndh-2 inhibitors.29 In line with this, RT-qPCR found that the 97-base pair deletion found in the ndhA promoter of RC 14.2 had a 23-fold increased expression of ndhA relative to the parental strain, while expression of ndh remained unchanged (Table S3).
Biochemical Inhibition of Ndh-2 by TriSLas
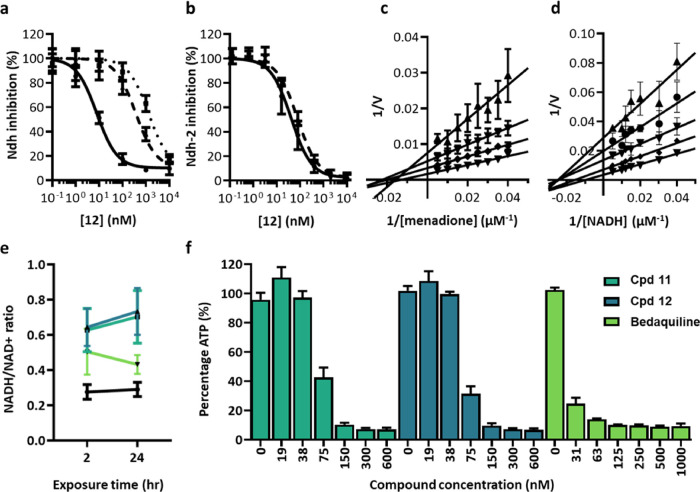
To confirm that TriSLas are Ndh-2 inhibitors, initial validation experiments focused Ndh, the primary NADH dehydrogenase in Mtb.30 Using published methodology,29 recombinant maltose-binding protein (MBP)-tagged Ndh (MBP-Ndh) was produced and purified from E. coli Rosetta cells and confirmed to be active and reduce the surrogate electron acceptor menadione in the presence of NADH. Inhibition studies found that TriSLas were a nanomolar inhibitor of NADH oxidation by MBP-Ndh (Table 1) and that this biochemical inhibition was in direct correlation with TriSLa MIC on Mtb (Figure S8). Biochemical evaluation of MBP-Ndh (Tyr403Cys) and MBP-Ndh (Gln334Pro) found these mutant proteins to be active, but 30–60 fold and 70–200 fold more resistant than MBP-Ndh to TriSLa-mediated inhibition, respectively (Figure 2a, Table S4). To determine if TriSLas also inhibit NdhA, a codon-optimized MBP-NdhA was overexpressed and purified from M. smegmatis (recombinant MBP-NdhA purified from E. coli Rosetta was found to have poor catalytic activity), and inhibition studies found TriSLas to inhibit MBP-NdhA in a similar manner to MBP-Ndh (Figure 2b). Together, these data validate TriSLas as pan-Ndh-2 inhibitors and confirm the importance of Tyr403 and Gln334 for TriSLa-mediated inhibition of Ndh.
Figure 2.
Biochemical target validation of TriSLas and impact on the Mtb electron transport chain. (a) Compound 12 biochemical inhibition of NADH oxidation by recombinant Mtb MBP-Ndh (filled circles, solid line), MBP-Ndh(Y403C) (filled triangles, dashed line), and MBP-Ndh(Q334P) (filled squares, dotted line) overexpressed and purified using E. coli. Data are presented as a mean ± SD of at least three independent biological replicates. (b) Compound 12 biochemical inhibition of NADH oxidation by recombinant Mtb MBP-Ndh (filled circles, solid line) and MBP-NdhA (filled triangles, dashed line) overexpressed and purified using M. smegmatis. (c) Lineweaver–Burk plot showing noncompetitive inhibition of Ndh by 12 relative to menadione (n ≥ 3), (d) Lineweaver–Burk plot showing noncompetitive inhibition of Ndh by 12 relative to NADH (n ≥ 3), (e) Mtb NADH/NAD+ ratios measured following 2 and 24 h exposure to 11 (green-blue, 600 nM, 4× MIC), 12 (dark blue, 600 nM, 4× MIC), and bedaquiline (green, 1 μM, 4× MIC), compared to unexposed DMSO controls (black) (n ≥ 3), and (f) Mtb ATP concentrations measured following 24 h exposure to either 11 (green-blue), 12 (dark blue), and bedaquiline (green) (n ≥ 3).
TriSLas Are Noncompetitive Inhibitors of Ndh
As mentioned above, TriSLa resistance conferring mutations in Ndh (Tyr403 and Gln334) are located near the menaquinone-binding pocket of Ndh, suggesting that TriSLas could act as competitive inhibitors for menaquinone binding. Biochemical competition assays with varying concentrations of menadione (model electron acceptor) or NADH however suggest TriSLas to not compete with either cofactor and instead inhibit Ndh through a noncompetitive or allosteric mechanism (Figure 2c,d).
Impact of TriSLas on the Bacterial Electron Transport Chain
As inhibitors of Ndh-2, TriSLas were expected to impact the bacterial NADH/NAD+ ratio and the functioning of the electron transport chain. To confirm this, NADH and NAD+ concentrations were measured in Mtb following 2 and 24 h of exposure to 4× MICs of 11 or 12 (600 nM). Relative to untreated bacteria, TriSLa-exposed bacteria showed an increase in the NADH/NAD+ ratio, driven by an increase in NADH and decrease in NAD+ (Figure 2e). As previously published,24 exposure to bedaquiline also caused an increase in the NADH/NAD+ ratio as a result from “back-pressure” on the electron transport chain, while rifampicin induced a decrease in NADH/NAD+. TriSLas were also found to impact the electron transport chain as a whole, as exposure to 12 and 11 (Figure 2f) caused a concentration-dependent shutdown in bacterial ATP concentrations, similar to that observed following exposure to bedaquiline (Figure 2f). Together, these data support that inhibition of Ndh-2 in Mtb has a major impact on the bacterial electron transport chain leading to a decrease in ATP production.
Impact of Media Composition on TriSLa Antibiotic Activity
Recent work has shown Ndh-2 to not be essential for Mtb growth in the absence of fatty acids and that the removal of all fatty acids form the culture medium rendered Ndh-2 inhibitors such as 2-mercapto-quinazolinones inactive.31 To verify this impact, the activity of TriSLas was re-evaluated in media lacking fatty acids (using fatty acid-free BSA), and data confirmed that the MIC of the compounds 1, 11, and 12 and a control 2-mercapto-quinazolinones/thioquinazoline (CBR-5992) all increased significantly, while no impact was observed for rifampicin or bedaquiline (Table S5). To confirm that this impact is not Mtb specific, the role of fatty acids in the culture medium was also evaluated on M. marinum, confirming the same dependence of fatty acids for TriSLa activity (Table S5). Other published work showed that mutations in ndh of M. smegmatis and M. bovis BCG rendered these bacteria auxotrophic for serine32 and that l-serine supplementation to the media rendered the Ndh-2 inhibitors (CBR-1825) less potent against Mtb in vitro.33 In accordance with these results, supplementation of l-serine to the growth medium also caused a loss of antibiotic activity of the TriSLa compounds against H37Rv (Table S6). Finally, the detergents (Tween 80 or tyloxapol or neither) used in the culture media were not found to have impact on TriSLa activity (Table S6).
In Vivo Efficacy on M. marinum-Infected Zebrafish
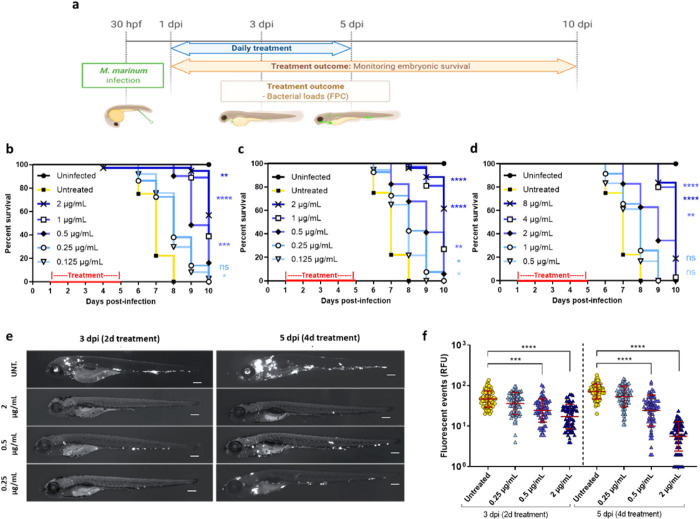
Considering the influence of media components on TriSLa antibiotic activity, it was considered a priority to evaluate the in vivo efficacy of TriSLa compounds. As the in vitro metabolic stability of the TriSLa compounds (Table S1) was too low for progression into murine efficacy studies of Mtb infection, it was decided to evaluate the in vivo efficacy of TriSLas on M. marinum infected zebrafish larvae, where compound exposure in the water is more stable. In such studies on acutely M. marinum-infected zebrafish larvae, the mycobacteria are phagocytosed by circulating macrophages and aggregate into foci that start to resemble granuloma.34−37 Initial toxicity experiments confirmed that compounds 11, 12, and 13 did not interfere with larval development or show any toxicity in noninfected zebrafish larvae (data not shown). Next, efficacy studies were performed as previously,34 where zebrafish larvae infected with M. marinum (expressing Wasabi) were exposed to different doses of TriSLas (refreshed daily) for a 4-day period (from 1 to 5 days post-infection (dpi)), followed by a 5-day period in the absence of antibiotics where survival was monitored (Figure 3a). Data confirmed that 4-day exposure compounds 11, 12, and 13 significantly prolonged the survival of the infected zebrafish in a dose-dependent manner, starting from doses of 0.3, 0.3, and 1.2 μM (respectively), with higher doses of 4.8, 4.8, and 9.6 μM (respectively) prolonging survival by 3 days (Figure 3b–d). In complement, the bacterial load of the infected zebrafish was determined by fluorescence microscopy during the treatment phase of the study (at 2 and 4 days of exposure).
Figure 3.
In vivo efficacy of TriSLas against M. marinum-infected embryonic zebrafish. (a) Schematic of the in vivo efficacy protocol showing the timeline of zebrafish embryo infection (200–250 colony-forming units (cfu) M. marinum expressing Wasabi), the exposure period to different concentrations of TriSLas (daily changes), and compound washout. hpf: hours post-fertilization; dpi: days post-infection; FPC: Fluorescence pixel count. (b–d) Survival curve ofM. marinum-infected zebrafish embryos monitored over the 10-day period post-infection, with and without 4-day treatment of (b) compound 11, (c) compound 12, and (d) compound 13. Survival curves are the cumulative results of three experiments covering more than 25 infected zebrafish per group. Statistics to compare treatment groups were performed using the log rank (Mantel–Cox) statistical test (*P < 0.05, **P < 0.01, ****P < 0.0001). (e) Representative sequential fluorescence images of M. marinum (Wasabi) infected zebrafish embryos following 2 or 4 days of treatment (3 and 5 dpi) with compound 12 at 0.25, 0.5, and 2 μg/mL, relative to untreated zebrafish (UNT). Scale bar, 210 μm. (f) M. marinum infection burden in zebrafish embryo as quantified following 2 (3 dpi) and 4 (5 dpi) days of compound 12 exposure (0.25, 0.5, and 2 μg/mL) by pixel count (fluorescent events) using the ImageJ software. The data presented are the cumulative data of three independent experiment (each containing 20 to 25 embryos per group), with each data point representing one infected zebrafish larva. The error bar represents the mean and standard deviations of the cumulative dataset. Statistical comparison of the different groups was performed using a Mann–Whitney’s t test, (***P < 0.001, ****P < 0.0001).
Images of untreated infected larvae showed an intense dissemination of the M. marinum infection, with multiple large bacterial foci that increased over time (3 and 5 dpi) (Figure 3e), while 2 or 4-day treatment with 0.25 to 2 μg/mL (0.6–4.8 μM) of 12 clearly resulting in fewer and less intense infection foci (Figure 3e). Quantification of the bacterial load on a larger number of infected zebrafish (using fluorescence pixel count determination) showed that exposure to 0.5 μg/mL (1.2 μM) of 12 was bacteriostatic, preventing an increase in bacterial load from 2–4 days of exposure, while higher doses of 2 μg/mL (4.8 μM) lead to a significant decrease in the bacterial burden between 2 and 4 days of over the same period (Figure 3f). Together, these data confirm that inhibition of Ndh-2 has a significant impact on M. marinum infection in the zebrafish model of infection.
Chemistry
The unique rigid 3D TriSLa core of the previously described molecules was obtained through stereoselective Meyers’ lactamization.17,38 This reaction uses readily accessible starting materials such as keto-esters (or keto-acids) as bi-electrophile and amino alcohols as bi-nucleophiles to generate complex-fused ring systems. Lactams 1–7 were obtained by reacting methyl 3-(1-benzyl-4-oxo-3-piperidyl)propanoate (obtained by esterification of its acid analogue, previously synthesized according to the literature)17 with the appropriate β-amino alcohol (Scheme 1) in yields ranging from 27 to 75%. Only partial stereoselectivity was observed with mildly hindered β-amino alcohols (compounds 4 and 5), while only one stereoisomer was obtained with highly hindered β-amino alcohols (compounds 1, 2, 6, and 7).
Scheme 1. Synthesis of Analogues 1–7.
Reagents and conditions: (i) methyl 3-(1-benzyl-4-oxo-3-piperidyl)propanoate (1 equiv), amino-alcohol (1.2–3 equiv), pivalic acid (1.2–3 equiv), toluene, 150 °C (thermic for 20 h or microwave irradiations for 1–4 h).
Analogues 8–13 were synthesized from compound 1 (Scheme 2), through N-debenzylation using Pd/C and ammonium formate followed by N-alkylation using nucleophilic substitutions (compounds 8–12) or reductive amination (compound 13).
Scheme 2. Synthesis of Analogues 8–13.
Reagents and conditions: (i) Pd/C 10% (10 mol %), HCO2H (5 equiv), reflux, 30 min; (ii) 14 (1 equiv), K2CO3 (3 equiv), RCH2Br (1–1.5 equiv), NaI (0–1 equiv), MeCN, RT, 1–20 h; (iii) 14 (1 equiv), 6-(trifluoromethyl)pyridine-3-carbaldehyde (3.6 equiv), Ti(OiPr)4 (1.8 equiv), NaBH(OAc)3 (3.6 equiv), dichloroethane, RT, overnight.
Discussion
Natural products and their hemisynthetic derivatives are a rich source of bioactive molecules and make up the bulk of the antibiotic classes currently used. Natural products often boast an increased chemical complexity,39,40 particularly emphasized by an abundance of chiral tetrahedral carbons. This increased 3D aspect allows natural products to occupy a different biological space compared to flatter molecules found in many drug-like screening collections. In this work, we demonstrate that screening synthetic focused chemical libraries enriched with nature-inspired scaffolds is a promising approach to identify novel antituberculosis chemical scaffolds with potent activity.
In recent years, a number of anti-TB molecules targeting Ndh-2 have been described, including 2-mercapto-quinazolinones,29,33 quinolones,41,42 and quinolinyl-pyrimidines,43,44 though no in vivo efficacy studies have been conducted thus far for these inhibitors. Overall, these inhibitors share similar structural features, being largely planar (large number of sp2-hybridized atoms) and carrying a central bicyclic aromatic ring, in sharp contrast to the highly 3D (large number of sp3-hybridized atoms) structure of the TriSLa compounds described here. Despite these structural differences, TriSLas do share common attributes with the most potent of these compounds, (2-mercapto-quinazolinones29,33). This includes comparable potency, being pan Ndh-2 inhibitors, being noncompetitive inhibitors of Ndh, displaying activity modulated by promoter mutation in ndhA, by l-serine and by fatty acid content in the media. Having structurally diverse chemical scaffolds targeting a protein is particularly worthwhile in drug development as this allows for different ADME/Tox profile and provides an alternative in case of drug development flags such as toxicity, pharmacokinetics (including tissue distribution), and off-target effects that are more often associated with the chemical scaffold. Together, this will help increase the chance of Ndh-2 inhibitors to enter clinical development. In this study, we report point mutations in Ndh that confer resistance to TriSLas, suggesting that these inhibitors act specifically at the Ndh-2 protein-membrane interface. Multiple protein alignment of Ndh-2 orthologues from a panel of bacteria shows Gln344 to be highly conserved between bacteria, while Tyr403 is conserved amongst mycobacteria, which likely explains their mycobacteria-specific antibacterial spectrum. Ndh structure models computed using Alphafold27,28 interestingly place the side chains of Tyr403 and Gln334 in close proximity to each other with possible hydrogen bonding interactions between their side chains. These residues are likely flexible, but in the model these interacting residues appear to greatly limit the size of the menaquinone access pocket (Figure S8). A hypothesis on the mechanism of Ndh-2 inhibition by TriSLas is that they bind and stabilize this closed confirmation and hence prevent the access of menaquinone, thus preventing its subsequent reduction. However, future structural biology efforts are needed to further elucidate how these two potent inhibitors are acting on Ndh-2.
Recent work demonstrated that Mtb could survive in vitro without either of its ndh-2 genes when grown in the absence of exogenous fatty acids31 and that this double ndh-2 deleted strain could still infect mice, though it was partially attenuated with a 26-fold lower maximal titre.31 As demonstrated for 2-mercapto-quinazolinones Ndh-2 inhibitors,31 the removal of all fatty acids from the media, or additional serine supplementation (greater than natural available concentration) also prevent the activity of TriSLas in vitro, confirming the vulnerability of Ndh-2 in the presence of fatty acids. Together, this work called into question the potential of Ndh-2 inhibitors in the fight against tuberculosis infections, and reasons that efficacy will depend on the fatty acid content of the compartments in which the mycobacteria reside. Data presented here highlight that despite TriSLa activity on M. marinum also being dependent on fatty acids, TriSLa treatment of M. marinum-infected zebrafish did result in concentration-dependent efficacy, both prolonging zebrafish survival and decreasing bacterial loads. This therefore evokes that, at least in this infection model, a significant proportion of M. marinum is likely to reside in a fatty acid-containing compartment, such as granuloma type structures, which are considered fatty acid-rich. Indeed, recent work has shown that the zebrafish/M. marinum granuloma contains the lipid-rich foam cells and that this differentiation of macrophages into foam cells is stimulated by the mycobacterial ESX-1 type VII secretion locus.45
This observed efficacy is, however, in contrast to the aforementioned finding that despite partial attenuation, Ndh-2 double knockout Mtb strains could still grow in C57BL/6 mice.31 Apart from possible differences in Ndh-2 vulnerability in Mtb and M. marinum, differences in the bacterial compartmental distribution (such as granulomas, intracellular, extracellular) and its associated fatty acid content may also be a factor to consider. In Mtb-infected C57BL/6 mice, the infection is largely believed to reside within macrophages, where the primary carbon source is expected to be cholesterol (depending on the macrophage) and where fatty acid availability is likely lower than in granulomas. TriSLas (and 2-mercaptoquinoxalines) are poorly active against these intracellular infections and may hence contribute to this difference. As granuloma are believed to play a major role in human Mtb infections, more work is needed to define the exact vulnerability of Ndh-2, and the potential of Ndh-2 inhibitors as anti-TB agents alone and in combination in animal models where granuloma formation is observed. For that purpose, further efforts will all focus on generating TriSLa compounds with an optimal pharmacokinetic profile.
Together, this work presents a new antimycobacterial chemical scaffold that acts through allosteric inhibition of Ndh-2. In vitro profiling of the TriSLas showed them to have a time-dependent bactericidal activity on replicating and nonreplicating bacteria, and in vivo efficacy studies confirm the vulnerability of this protein in M. marinum infected zebrafish. Future medicinal chemistry will aim to confirm Ndh-2 vulnerability for Mtb in various murine models of infection, and define TriSLa based treatment regimens.
Experimental Section
Chemical Synthesis
General Information
All reagent-grade chemicals and anhydrous solvents for synthesis, analysis, and purification were obtained from commercial suppliers and were used as received without further purification.
Flash chromatography was performed using a Puriflash PF-430 with silica gel cartridges (Buchi silica 40 μm). ELSD and UV detection (254 nm) were used to collect the desired product. Reverse flash chromatography was performed using a CombiFlash Rf200 with C18 cartridges (Buchi C18 40 μm). UV detection (215 and 254 nm) was used to collect the desired product.
1H NMR and 13C NMR spectra were recorded on a Bruker DRX-300 spectrometer. Chemical shifts (δ) are in parts per million (ppm). The 1H spectra were calibrated to signals from CD2Cl2 (δ 5.36 ppm) or CDCl3 (δ 7.26 ppm), and 13C spectra from CD2Cl2 (δ 53.5 ppm) or CDCl3 (δ 77.16 ppm). 1H NMR spectra are reported as follows: chemical shift (ppm), multiplicity (s: singlet; brs: broad singlet; d: doublet; dd: doublet of doublet; t: triplet; td: triplet of doublet; m: multiplet), coupling constants in Hertz (Hz) and integration. Proton and carbon signal assignments were established using COSY, HSQC-DEPT, and HMBC spectra.
Liquid chromatography–mass spectrometry (LC–MS) Waters system was equipped with a 2747 sample manager, a 2695 separation module, a 2996 photodiode array detector (200–400 nm), and a Micromass ZQ2000 detector (scan 100–800). An XBridge C18 column (50 mm × 4.6 mm, 3.5 μm, Waters) was used. The injection volume was 20 μL. A mixture of water and acetonitrile was used as the mobile phase in gradient-elution. The pH of the mobile phase was adjusted with HCOOH and NH4OH to form a buffer solution at pH 3.8. The analysis time was 5 min (at a flow rate of 2 mL/min), 10 min (at a flow rate of 1 mL/min), or 30 min (at a flow rate of 1 mL/min). Purity (%) was determined by reversed-phase high-performance liquid chromatography (HPLC), using UV detection (215 nm). All final compounds showed purity greater than 95%.
High-resolution mass spectra (HRMS) analysis was performed on a LC–MS system equipped with a LCT Premier XE mass spectrometer (Waters), using a XBridge C18 column (50 mm × 4.6 mm, 3.5 μm, Waters). A gradient starting from 98% H2O 5 mM ammonium formate pH 3.8 and reaching 100% MeCN 5 mM ammonium formate pH 3.8 within 3 min at a flow rate of 1 mL was used.
Methyl-3-(1-benzyl-4-oxo-3-piperidyl)propanoate
To a solution of 3-(1-benzyl-4-oxo-3-piperidyl)propanoic acid (19.3 g, 73.9 mmol) in methanol (200 mL) was added SOCl2 (5.9 mL, 81.3 mmol) dropwise at room temperature. The mixture was then stirred at 55 °C for 1 h. The solvent was removed under vacuum, and the mixture was dissolved in 0.1 N HCl (100 mL) and stirred at room temperature for 1 h. A saturated aqueous solution of Na2CO3 was added until pH 10. The solution was extracted with ethyl acetate. The organic layer was dried over MgSO4, and the solvent was removed under reduced pressure to give the crude product, which was purified by silica gel chromatography (cyclohexane/ethyl acetate: 70/30 to 0/100) to afford methyl 3-(1-benzyl-4-oxo-3-piperidyl)propanoate (6.73 g, 33%), as a colorless oil. 1H NMR (300 MHz, CD2Cl2): δ 7.36–7.25 (m, 5H), 3.64 (d, J = 13.1 Hz, 1H), 3.61 (s, 3H), 3.57 (d, J = 13.1 Hz, 1H), 3.08–2.97 (m, 2H), 2.62–2.11 (m, 7H), 2.08–1.95 (m, 1H), 1.53–1.42 (m, 1H) ppm. [ES+ MS] m/z 276 (MH+). 1H NMR data matched those reported previously (Idzik, T. J.; Myk, Z. M.; Peruzynska, M.; Maciejewska, G.; Drozdzik, M.; Sosnicki, J. G. Arylation of enelactams using TIPSOTf: reaction scope and mechanistic insight. Org. Chem. Front.2021, 8, 708–720).
General Protocol 1: Diastereoselective Meyers’ Lactamization
A solution of pivalic acid (1.2–3 equiv) in toluene (0.2 N) was added to methyl 3-(1-benzyl-4-oxo-3-piperidyl) propanoate (1 equiv). The appropriate amino-alcohol (1.2–3 equiv) was added (when the amine is used as a chlorohydrate, DIEA (1.2–3 equiv) was added). The mixture was refluxed at 150 °C (thermic for 20 h or microwave irradiations for 1–4 h). When the conversion of the keto-ester was judged completely by LC/MS, the solution was dissolved in H2O and extracted with ethyl acetate or dichloromethane. The layers were separated. The organic layer was dried over MgSO4, filtered, and concentrated under vacuum to give the crude product, which was purified, to afford the corresponding desired product.
(3S,7aR,11aR)-9-Benzyl-3-isopropyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (1)
The product 1 was obtained from l-valinol using general protocol 1. The crude product was purified by flash chromatography column over silica (cyclohexane/ethyl acetate: 1/0 to 6/4) to afford the desired product 1 (yield = 75%, white powder) as a single diastereoisomer. 1H NMR (300 MHz, CDCl3): δ 7.33–7.20 (m, 5H, ArCH), 4.13–4.03 (m, 1H, 3CH), 3.98 (dd, J = 8.6, 7.6 Hz, 1H, 2CH2), 3.77 (dd, J = 8.6, 6.2 Hz, 1H, 2CH2), 3.54 (d of AB system, J = 13.3 Hz, 1H, 12CH2), 3.41 (d of AB system, J = 13.1 Hz, 1H, 12CH2), 2.77–2.64 (m, 2H, 10CH2 and 8CH2), 2.62–2.51 (m, 1H, 6CH2), 2.49–2.29 (m, 3H, 8CH2, 7CH2 and 6CH2), 2.25–2.16 (m, 1H, 10CH2), 2.09–1.89 (m, 2H, 13CH and 11CH2), 1.83–1.73 (m, 1H, 11CH2), 1.72–1.55 (m, 2H, 7aCH, and 7CH2), 0.94 (d, J = 6.8 Hz, 3H, 14 or 15CH3), 0.90 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CDCl3): δ 170.2 (5CO), 138.9 (ArCq), 128.7, 128.3, 127.0 (ArCH), 92.7 (11aCq), 66.2 (2CH2), 62.5 (12CH2), 61.2 (3CH), 54.8 (8CH2), 50.7 (10CH2), 40.4 (7aCH), 32.4 (13CH), 32.4 (11CH2), 30.8 (6CH2), 21.9 (7CH2), 19.9 (14 or 15CH3), 18.7 (14 or 15CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C20H29N2O2, 329.2229; found 329.2227.
(3S,7aR,11aR)-9-Benzyl-3-[(1S)-1-methylpropyl]-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (2)
The product 2 was obtained from (2S,3S)-2-amino-3-methyl-pentan-1-ol using general protocol 1. The crude product was purified by preparative HPLC using MeCN + 0.1% HCOOH and H2O + 0.1% HCOOH gradient (9/1 to 0/10) to afford the product 2 (yield = 45%, yellow oil) as a single diastereoisomer. 1H NMR (500 MHz, CDCl3): δ 7.39–7.21 (m, 5H, ArCH), 4.14 (dd, J = 7.5, 7.3 Hz, 1H, 3CH), 3.97 (dd, J = 8.8, 7.5 Hz, 1H, 2CH2), 3.75 (dd, 1H, J = 8.8, 6.9 Hz, 1H, 2CH2), 3.65–3.38 (m, 2H, 12CH2), 2.76–2.20 (m, 7H, 10CH2, 8CH2, 7CH2, and 6CH2), 1.89–1.54 (m, 5H, 13CH, 11CH2, 7CH2, and 7aCH), 1.49–1.38 (m, 1H, 15CH2), 1.19–1.05 (m, 1H, 15CH2), 0.89 (t, J = 7.4 Hz, 3H, 16CH3), 0.85 (d, J = 6.8 Hz, 3H, 14CH3) ppm. 13C NMR (125 MHz, CDCl3): δ 170.3 (5CO), 138.7 (ArCq), 128.9, 128.5, 127.1 (ArCH), 92.2 (11aCq), 65.8 (2CH2), 62.5 (12CH2), 60.8 (3CH), 54.7 (10CH2), 50.7 (8CH2), 40.4 (7aCH), 38.2 (13CH), 32.0 (11CH2), 30.8 (6CH2), 27.2 (15CH2), 21.9 (7CH2), 14.9 (14CH3), 12.0 (16CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C21H31N2O2, 343.2386; found: 343.2382.
(3R,7aS,11aS)-9-Benzyl-3-isopropyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (3)
The product 3 was obtained from (2R)-2-amino-3-methyl-butan-1-ol using general protocol 1. The crude product was purified by flash chromatography column over silica (cyclohexane/ethyl acetate: 1/0 to 6/4) to afford the desired product 3 (yield = 61%, colorless oil) as a single diastereoisomer. The data obtained (NMR and HRMS) were the same as the data described for this enantiomer compound 1.
(3S,7aR,11aR)-9-Benzyl-3-methyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (4a) and (3S,7aS,11aS)-9-Benzyl-3-methyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (4b)
The products 4a and 4b were obtained from (2S)-2-aminopropan-1-ol using general protocol 1. The crude product was purified by preparative HPLC using MeCN + 0.1% HCOOH and H2O + 0.1% HCOOH gradient (9/1 to 0/10). A basification and an extraction at 10 pH with a saturated solution of Na2CO3 were carried out on the recovered fraction, and the combined organic layers were dried over MgSO4. The solvent was removed under reduced pressure the desired to afford products 4a and 4b (yield = 46%, yellow oil), as a mixture of two diastereoisomers (d.r.(4a/4b) = 70/30). Data for the major diastereoisomer 4a: 1H NMR (300 MHz, CD2Cl2): δ 7.38–7.20 (m, 5H, ArCH), 4.28–4.06 (m, 2H, 3CH and 2CH2), 3.62 (dd, J = 13.0 Hz, J = 1.3 Hz, 1H, 2CH2), 3.53 (d, J = 13.4 Hz, 1H, 12CH2), 3.42 (d, J = 13.4 Hz, 1H, 12CH2), 2.78–2.63 (m, 2H, 10CH2 and 8CH2), 2.58–2.15 (m, 5H, 10CH2, 8CH2, 7CH2 and 6CH2), 1.99–1.60 (m, 5H, 11CH2,7CH2 and 7aCH), 1.29 (d, J = 6.1 Hz, 3H, 13CH3) ppm. 13C NMR (75 MHz, CD2Cl2): δ 168.9 (5CO), 139.4 (ArCq), 129.0, 128.5, 127.3 (ArCH), 92.6 (11aCq), 69.7 (2CH2), 62.7 (12CH2), 55.2 (8CH2), 51.7 (3CH), 50.9 (10CH2), 40.7 (7aCH), 31.6 (11CH2), 31.1 (6CH2), 22.9 (7CH2), 20.2 (13CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C18H25N2O2, 301.1916; found: 301.1914.
(3S,7aR,11aR)-9-Benzyl-3-ethyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (5a) and (3S,7aR,11aR)-9-Benzyl-3-ethyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (5b)
The products 5a and 5b were obtained from (2S)-2-aminobutan-1-ol using general protocol 1. The crude product was purified by preparative HPLC using MeCN + 0.1% HCOOH and H2O + 0.1% HCOOH gradient (9/1 to 0/10) to afford products 5a and 5b (yield = 73%, yellow oil), as a mixture of two diastereoisomers (d.r.(5a/5b) = 70/30). Data for the major diastereoisomer 5a: 1H NMR (300 MHz, CD2Cl2): δ 7.42–7.28 (m, 5H, ArCH), 4.23–4.04 (m, 2H, 3CH and 2CH2), 3.76–3.66 (m, 1H, 2CH2), 3.73 (d, J = 13.3 Hz, 1H, 12CH2), 3.59 (d, J = 13.3 Hz, 1H, 12CH2), 2.97–2.75 (m, 2H, 10CH2 and 8CH2), 2.58–2.15 (m, 5H, 10CH2, 8CH2, 7CH2 and 6CH2), 2.09–1.67 (m, 5H, 13CH2,11CH2,7CH2 and 7aCH), 1.50–1.34 (m, 1H, 13CH2), 0.92 (t, J = 7.5 Hz, 3H, 14CH3) ppm. 13C NMR (75 MHz, CD2Cl2): δ 169.0 (5CO), 136.7 (ArCq), 129.3, 128.3, 127.5 (ArCH), 91.6 (11aCq), 70.0 (2CH2), 61.9 (12CH2), 57.0 (3CH), 54.1 (8CH2), 50.0 (10CH2), 39.9 (7aCH), 30.6 (11CH2), 30.6 (6CH2), 27.8 (13CH2), 22.1 (7CH2), 10.2 (14CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C19H27N2O2, 351.2017; found: 351.2093.
(3S,7aR,11aR)-9-Benzyl-3-cyclopropyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (6)
The product 6 was obtained from (2S)-2-amino-2-cyclopropyl-ethanol;hydrochloride using general protocol 1. The crude product was purified by preparative HPLC using H2O + 0.1% HCOOH/MeCN + 0.1% HCOOH (90/10 to 0/100) to afford the desired product 6 (yield = 27%, brown oil), as a single diastereoisomer. 1H NMR (300 MHz, CDCl3): δ 7.36–7.23 (m, 5H, ArCH), 4.15 (dd, J = 8.5 Hz, J = 8.4 Hz, 1H, 2CH2), 3.86 (dd, J = 8.9 = 5 Hz, J = 6.8 Hz, 1H, 2CH2), 3.75–3.62 (m, 1H, 3CH), 3.58 (d of AB system, J = 13.4 Hz, 1H, 12CH2), 3.44 (d of AB system, J = 13.4 Hz, 1H, 12CH2), 2.82–2.18 (m, 6H, 10CH2, 8CH2, 7CH2 and 6CH2), 2.11–1.89 (m, 2H, 11CH2), 1.74–1.54 (m, 2H, 7aCH and 7CH2), 0.93–0.72 (m, 2H, 14 or 15CH2 and 13CH), 0.66–0.41 (m, 2H, 14 or 15CH2), 0.30–0.17 (m, 1H, 14 or 15CH2) ppm. 13C NMR (75 MHz, CDCl3): δ 169.4 (5CO), 138.9 (ArCq), 128.8, 128.4, 127.1 (ArCH), 92.6 (11aCq), 68.0 (2CH2), 62.6 (12CH2), 59.9 (3CH), 54.9 (8CH2), 50.8 (10CH2), 40.4 (7aCH), 31.8 (11CH2), 31.1 (6CH2), 22.5 (7CH2), 15.8 (13CH), 4.6 (14 or 15CH2), 2.7 (14 or 15CH2) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C20H27N2O2, 327.2071; found: 327.2073.
(3S,7aR,11aR)-9-Benzyl-3-tert-butyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (7)
The product 7 was obtained from (2S)-2-amino-3,3-dimethyl-butan-1-ol using general protocol 1. The crude product was purified by preparative HPLC using H2O + 0.1% HCOOH/MeCN +0.1% HCOOH (90/10 to 0/100) to afford the desired product 7 (yield = 40%, yellow oil), as a single diastereoisomer. 1H NMR (300 MHz, CD2Cl2): δ 7.36–7.19 (m, 5H, ArCH), 4.11 (t, J = 6.9 Hz, 1H, 3CH), 3.85 (d, J = 6.5 Hz, 2H, 2CH2), 3.53 (d, J = 13.3 Hz, 1H, 12CH2), 3.41 (d, J = 13.3 Hz, 1H, 12CH2), 2.77–2.69 (m, 1H, 10CH2), 2.61–2.57 (m, 1H, 8CH2), 2.58–2.49 (m, 1H, 6CH2), 2.45–2.27 (m, 3H, 8CH2,7CH2, and 6CH2), 2.20 (dt, J = 12.0 Hz, J = 3.4 Hz, 1H, 10CH2), 1.98 (dt, J = 13.3 Hz, J = 4.7 Hz, 1H, 11CH2), 1.90–1.82 (m, 1H, 11CH2), 1.73–1.52 (m, 2H, 7CH2 and 7aCH), 0.92 (s, 9H, 14CH3,15CH3 and 16CH3) ppm. 13C NMR (75 MHz, CD2Cl2): δ 172.7 (5CO), 139.5 (ArCq), 129.1, 128.5, 127.2 (ArCH), 94.3 (11aCq), 64.8 (3CH), 64.5 (2CH2), 62.7 (12CH2), 54.9 (8CH2), 50.9 (10CH2), 40.2 (7aCH), 34.8 (13Cq), 32.7 (11CH2), 31.0 (6CH2), 27.8 (14CH3,15CH3 and 16CH3), 20.9 (7CH2) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C21H31N2O2, 343.2386; found: 343.2394.
(3S,7aR,11aR)-3-Isopropyl-3,6,7,7a,8,9,10,11-octahydro-2H-oxazolo[2,3-j][1,6]naphthyridin-5-one; Formic Acid (14)
Compound 1 (655 mg, 1.99 mmol, 1 equiv) was dissolved in methanol (20 mL), and then Pd/C 10% (127 mg, 1.20 mmol, 0.12 mmol, 10 mol %) and ammonium formate (629 mg, 9.97 mmol, 5 equiv) were added. The mixture was refluxed for 30 min. The solution was filtered over celite, and then the filtrate was concentrated under reduced pressure to afford the desired product 14 with a quantitative yield as a white powder. 1H NMR (300 MHz, CD2Cl2): δ 8.44 (brs, 1H), 4.13–3.99 (m, 2H), 3.77 (dd, J = 8.3, 5.6 Hz, 1H), 3.35–3.11 (m, 3H), 2.99 (td, J = 13.1, 3.2 Hz, 1H), 2.68–2.55 (m, 1H), 2.48–2.20 (m, 2H), 2.10 (td, J = 14.4, 4.6 Hz, 1H), 2.01–1.70 (m, 4H), 0.92 (d, J = 6.4 Hz, 3H), 0.90 (d, J = 6.4 Hz, 3H) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C13H22N2O2, 239.1760; found 239.1759.
General Protocol 2: Alkylation of Compound 14
Compound 14 (1 equiv) was dissolved in MeCN (0.1 N), and then K2CO3(3 equiv) and the appropriate bromide (1–1.5 equiv) and NaI (0–1 equiv) were added. The solution was stirred at room temperature. When the conversion of 14 into the desired product was judged completely by LC/MS or TLC, the solvent was removed under reduced pressure. The residue obtained was dissolved in H2O and extracted with dichloromethane. The organic layer was dried over MgSO4, filtered, and concentrated under vacuum to give the crude product, which was purified by flash chromatography column over silica gel using cyclohexane/ethyl acetate (1/0 to 0/1) to afford the corresponding desired lactams.
(3S,7aR,11aR)-3-Isopropyl-9-(p-tolylmethyl)-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (9)
The product 9 was obtained from 1-(bromomethyl)-4-methyl-benzene using general protocol 2 (yield = 28%), as a white powder. 1H NMR (300 MHz, CDCl3): δ 7.19 (d of AB system, J = 8.0 Hz, 2H, ArCH), 7.11 (d of AB system, J = 8.0 Hz, 2H, ArCH), 4.12–4.04 (m, 1H, 3CH), 3.97 (dd, J = 8.6, 7.6 Hz, 1H, 2CH2), 3.76 (dd, J = 8.6, 6.2 Hz, 1H, 2CH2), 3.50 (d of AB system, J = 13.1 Hz, 1H, 12CH2), 3.36 (d of AB system, J = 13.1 Hz, 1H, 12CH2), 2.77–2.62 (m, 2H, 10CH2, and 8CH2), 2.62–2.50 (m, 1H, 6CH2), 2.46–2.34 (m,3H, 8CH2, 7CH2, and 6CH2), 2.30 (s, 3H, 16CH3), 2.24–2.13 (m, 1H, 10CH2), 2.07–1.88 (m, 2H, 13CH, and 11CH2), 1.84–1.71 (m, 1H, 11CH2), 1.70–1.54 (m, 2H, 7aCH, and 7CH2), 0.94 (d, J = 6.8 Hz, 3H, 14 or 15CH3), 0.89 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CDCl3): δ 170.3 (5CO), 136.7, 135.9 (ArCq), 129.0, 128.7 (ArCH), 92.8 (11aCq), 66.2 (2CH2), 62.3 (12CH2), 61.3 (3CH), 54.8 (8CH2), 50.7 (10CH2), 40.5 (7aCH), 32.5 (13CH), 32.4 (11CH2), 30.9 (6CH2), 22.0 (7CH2), 21.2 (16CH3), 19.9 (14 or 15CH3), 18.7 (14 or 15CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C21H30N2O2, 343.2386; found 343.2398.
(3S,7aR,11aR)-3-Isopropyl-9-[(4-methoxyphenyl)methyl]-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (10)
The product 10 was obtained from 1-(bromomethyl)-4-methoxy-benzene using general protocol 2 (yield = 45%), as a colorless oil. 1H NMR (300 MHz, CDCl3): δ 7.21 (d of AB system, J = 8.6 Hz, 2H, ArCH), 6.85 (d of AB system, J = 8.6 Hz, 2H, ArCH), 4.14–4.03 (m, 1H, 3CH), 3.98 (dd, J = 8.6, 7.6 Hz, 1H, 2CH2), 3.80 (s, 3H, 16CH3), 3.79–3.73 (m, 1H, 2CH2), 3.48 (d of AB system, J = 13.0 Hz, 1H, 12CH2), 3.34 (d of AB system, J = 13.0 Hz, 1H, 12CH2), 2.76–2.63 (m, 2H, 10CH2, and 8CH2), 2.61–2.51 (m, 1H, 6CH2), 2.45–2.28 (m, 3H, 8CH2, 7CH2 and 6CH2), 2.24–2.13 (m, 1H, 10CH2), 2.07–1.89 (m, 2H, 13CH and 11CH2), 1.82–1.71 (m, 1H, 11CH2), 1.70–1.54 (m, 2H, 7aCH and 7CH2), 0.94 (d, J = 6.9 Hz, 3H, 14 or 15CH3), 0.90 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CDCl3): δ 170.3 (5CO), 158.8, 130.9 (ArCq), 129.9, 113.7 (ArCH), 92.8 (11aCq), 66.2 (2CH2), 61.9 (12CH2), 61.3 (3CH), 55.4 (16CH3), 54.7 (8CH2), 50.6 (10CH2), 40.4 (7aCH), 32.5 (13CH), 32.4 (11CH2), 30.9 (6CH2), 22.0 (7CH2), 19.9 (14 or 15CH3), 18.7 (14 or 15CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C21H30N2O3, 359.2335; found 359.2314.
(3S,7aR,11aR)-9-[(4-Chlorophenyl)methyl]-3-isopropyl-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (11)
The compound 11 was obtained from 1-chloro-4-(chloromethyl)benzene using general protocol 2 (yield = 42%), as a colorless oil. 1H NMR (300 MHz, CDCl3): δ 7.30–7.21 (m, 4H, ArCH), 4.14–4.05 (m, 1H, 3CH), 3.98 (dd, J = 8.6, 7.6 Hz, 1H, 2CH2), 3.76 (dd, J = 8.6, 6.2 Hz, 1H, 2CH2), 3.49 (d of AB system, J = 13.5 Hz, 1H, 12CH2), 3.37 (d of AB system, J = 13.5 Hz, 1H, 12CH2), 2.74–2.61 (m, 2H; 10CH2 and 8CH2), 2.60–2.52 (m, 1H, 6CH2), 2.50–2.29 (m, 3H, 8CH2, 7CH2 and 6CH2), 2.26–2.15 (m, 1H, 10CH2), 2.08–1.89 (m, 2H, 13CH and 11CH2), 1.84–1.76 (m, 1H, 11CH2), 1.72–1.56 (m, 2H, 7aCH and 7CH2), 0.94 (d, J = 6.8 Hz, 3H, 14 or 15CH3), 0.89 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CDCl3): δ 170.2 (5CO), 137.5, 132.7 (ArCq), 130.0, 128.5 (ArCH), 92.6 (11aCq), 66.2 (2CH2), 61.8 (12CH2), 61.3 (3CH), 54.8 (8CH2), 50.7 (10CH2), 40.4 (7aCH), 32.4 (13CH), 32.4 (11CH2), 30.8 (6CH2), 22.0 (7CH2), 19.9 (14 or 15CH3), 18.7 (14 or 15CH3) ppm. HRMS (ESI, m/z): [M + H]+ calcd for C20H28N2O2Cl, 363.1839; found 363.1872.
(3S,7aR,11aR)-3-Isopropyl-9-[[4-(trifluoromethyl)phenyl]methyl]-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (12)
The product 12 was obtained from 1-(bromomethyl)-4-(trifluoromethyl)benzene using general protocol 2 (yield = 73%), as a white powder. 1H NMR (300 MHz, CD2Cl2): δ 7.57 (d of AB system, J = 8.1 Hz, 2H, ArCH), 7.47 (d of AB system, J = 8.1 Hz, 2H, ArCH), 4.07–3.94 (m, 2H, 3CH and 2CH2), 3.75 (dd, J = 8.0, 5.7 Hz, 1H, 2CH2), 3.58 (d of AB system, J = 13.9 Hz, 1H, 12CH2), 3.48 (d of AB system, J = 13.9 Hz, 1H, 12CH2), 2.73–2.61 (m, 2H, 10CH2 and 8CH2), 2.57–2.45 (m, 2H, 8CH2 and 6CH2), 2.41–2.28 (m, 2H, 7CH2 and 6CH2), 2.27–2.18 (m, 1H, 10CH2), 2.03–1.90 (m, 2H, 13CH and 11CH2), 1.82–1.74 (m, 1H, 11CH2), 1.69–1.56 (m, 2H, 7aCH and 7CH2), 0.91 (d, J = 6.8 Hz, 3H, 14 or 15CH3), 0.88 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CD2Cl2): δ 170.0 (5CO), 144.0 (ArCq), 129.2 (q, J = 32.2 Hz, ArCq), 129.2 (ArCH), 125.4 (q, J = 3.7 Hz, ArCH), 124.8 (q, J = 272.1 Hz, CF3Cq), 92.7 (11aCq), 66.4(2CH2), 62.2 (12CH2), 61.5 (3CH), 55.3 (8CH2), 51.0 (10CH2), 40.8 (7aCH), 32.8 (13CH), 32.6 (11CH2), 31.2 (6CH2), 32.3 (7CH2), 19.9 (14 or 15CH3), 18.7 (14 or 15CH3) ppm. LCMS (ESI, m/z): tr = 2.83 min, [M + H]+ = 397. HRMS (ESI, m/z): [M + H]+ calcd for C21H27N2O2F3, 397.2103; found 396.2101.
(3S,7aR,11aR)-3-Isopropyl-9-[[4-(trifluoromethoxy)phenyl]methyl]-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (13)
The product 13 was obtained from 1-(bromomethyl)-4-(trifluoromethoxy)benzene using general protocol 2 (yield = 77%), as a colorless oil. 1H NMR (300 MHz, CD2Cl2): δ 7.38 (d of AB system, J = 8.6 Hz, 2H, ArCH), 7.17 (d of AB system, J = 8.6 Hz, 2H, ArCH), 4.07–3.94 (m, 2H, 3CH and 2CH2), 3.75 (dd, J = 8.0, 5.7 Hz, 1H, 2CH2), 3.52 (d of AB system, J = 13.6 Hz, 1H, 12CH2), 3.42 (d of AB system, J = 13.6 Hz, 1H, 12CH2), 2.74–2.60 (m, 2H, 10CH2 and 8CH2), 2.58–2.42 (m, 2H, 8CH2 and 6CH2), 2.40–2.26 (m, 2H, 7CH2 and 6CH2), 2.26–2.14 (m, 1H, 10CH2), 2.04–1.87 (m, 2H, 13CH and 11CH2), 1.83–1.74 (m, 1H, 11CH2), 1.72–1.54 (m, 2H, 7aCH, and 7CH2), 0.91 (d, J = 6.7 Hz, 3H, 14 or 15CH3), 0.88 (d, J = 6.7 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CD2Cl2): δ 170.0 (5CO), 148.5, 138.6 (ArCq), 130.3, 121.1 (ArCH), 120.7 (q, J = 250.1 Hz, CF3Cq), 92.8 (11aCq), 66.4 (2CH2), 61.9 (12CH2), 61.6 (3CH), 55.3 (8CH2), 50.9 (10CH2), 40.8 (7aCH), 32.8 (13CH), 32.7 (11CH2), 31.2 (6CH2), 32.3 (7CH2), 20.0 (14 or 15CH3), 18.8 (14 or 15CH3) ppm. LCMS (ESI, m/z): tr = 2.82 min, [M + H]+ = 413. HRMS (ESI, m/z): [M + H]+ calcd for C21H28N2O3F3, 413.2052; found 413.2056.
(3S,7aR,11aR)-3-Isopropyl-9-[[6-(trifluoromethyl)-3-pyridyl]methyl]-2,3,6,7,7a,8,10,11-octahydrooxazolo[2,3-j][1,6]naphthyridin-5-one (13)
To a solution of compound 14 (500 mg, 1.76 mmol, 1 equiv) in dichloroethane (4 mL) were added 6-(trifluoromethyl)pyridine-3-carbaldehyde (1.1 g, 6.29 mmol, 3.6 equiv), titanium isopropoxide (937 μL, 3.15 mmol, 1.8 equiv), and NaBH(OAc)3 (1.33 g, 6.29 mmol, 3.6 equiv). The mixture was stirred at room temperature overnight. The solvent was removed under vacuum, and the mixture was then taken up in CH2Cl2 and washed with a saturated aqueous solution of Na2CO3. The organic layer was dried over magnesium sulfate, filtered, and concentrated under vacuum. The product was purified by flash chromatography column over silica gel using cyclohexane/ethyl acetate (1/0 to 0/1) to afford the desired product 13 (493 mg, 71%) as a white powder. 1H NMR (300 MHz, CDCl3): δ 8.66 (brs, 1H, ArCH), 7.85 (dd, J = 8.0, 1.3 Hz, 1H, ArCH), 7.64 (d, J = 8.0 Hz, 1H, ArCH), 4.14–4.06 (m, 1H, 3CH), 3.99 (dd, J = 8.5, 8.0 Hz, 1H, 2CH2), 3.78 (dd, J = 8.5, 6.2 Hz, 1H, 2CH2), 3.62 (d of AB system, J = 14.1 Hz, 1H, 12CH2), 3.52 (d of AB system, J = 14.1 Hz, 1H, 12CH2), 2.75–2.52 (m, 4H, 10CH2, 8CH2 and 6CH2), 2.48–2.25 (m, 3H, 10CH2, 7CH2 and 6CH2), 2.07–1.91 (m, 2H, 13CH and 11CH2), 1.85–1.77 (m, 1H, 11CH2), 1.75–1.59 (m, 2H, 7aCH and 7CH2), 0.95 (d, J = 6.8 Hz, 3H, 14 or 15CH3), 0.91 (d, J = 6.8 Hz, 3H, 14 or 15CH3) ppm. 13C NMR (75 MHz, CDCl3): δ 170.1 (5CO), 129.2 (ArCH), 147.2 (q, J = 33.5 Hz, ArCq), 137.9 (ArCq), 137.5 (ArCH), 121.5 (q, J = 272.0 Hz, CF3Cq), 120.3 (q, J = 3.7 Hz, ArCH), 92.3 (11aCq), 66.3 (2CH2), 61.3 (12CH2), 59.4 (3CH), 55.0 (8CH2), 50.8 (10CH2), 40.3 (7aCH), 32.4 (13CH), 32.3 (11CH2), 30.7 (6CH2), 21.9 (7CH2), 19.9, 18.7 (14 and 15CH3) ppm. LCMS (ESI, m/z): tr = 2.77 min, [M + H]+ = 398. HRMS (ESI, m/z): [M + H]+ calcd for C20H27N3O2F3, 398.2055; found 398.2044.
Acknowledgments
We thank F. Capet and P. Roussel from the X-ray platform of Chevreul Institute (FR CNRS 2638), for supporting data collection and solving the x-ray crystal structure. We thank V. Landry (U1177- Drugs and Molecules for Living Systems) for cytotoxicity experiments. We thank BLS-3 Facility staff (N. Vandenabeele, R. Prath, and S. Marin) at the Institut Pasteur de Lille for technical support. We thank E. Anoz Carbonell for unbiased MIC analysis. We are grateful to P. Richard and M. Plays for zebrafish husbandry. This research was financially cofunded by NL4Tb, Atip-Avenir, SMARt-Lab, and CPER grants. The NL4Tb grant was funded by the French National Research Agency (ANR-19-CE18-0034-01). The SMARt-Lab grant was funded by European Union under the European Regional Development Fund (ERDF), by the Hauts De France Regional Council (Contract n°NP0020070), and by I-Site ULNE (ANR-16-IDEX-0004 ULNE). The CPER grants were funded by European Union under the European Regional Development Fund (ERDF) and by the Hauts de France regional Council (contract n°20002842 and contract n° 18006176), the MEL (contract n°2017_ESR_14 and contract_2020_ESR_06), and the French State (contract n° 2018-R3-CTRL-Phase2 and contract n°2020-R3-CTRL_IPL_Phase4). The compound management and physicochemical/ADME property measurement were supported by ChemBioFrance through the ARIADNE-ADME platform (Lille, France). We thank the NMR platform of the faculty of pharmacy, Lille University for the use of the 300 MHz NMR facilities, funded by the Région Hauts-De-France, the ministère de la jeunesse de l’éducation Nationale et de la Recherche (MJENR) and the fonds Européens de développement Régional (FEDER).
Glossary
Abbreviations
- BDQ
bedaquiline
- cfu
colony forming unit
- dpi
days post-infection
- FPC
fluorescent pixel count
- hpf
hours post-fertilization
- INH
isoniazid
- MBP
maltose-binding protein
- MDR
multidrug resistant
- Mtb
Mycobacterium tuberculosis
- REMA
resazurin microtiter assay
- RIF
rifampicin
- XDR
extensively drug resistant
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c01493.
Author Contributions
S.D., S.T., C.H., T.A., L.F., L.K., N.W., B.V., and R.C.H. designed experiments. N.W. and B.D. designed, generated, and compiled the screened chemical library. S.T., T.A., L.F. M.F., B.D., N.W., and B.V. designed, synthesized, and characterized TriSLa analogues. A.H., F.L., N.W., and R.C.H. performed the chemical library screening. Selection of TriSLa-resistant isolates was performed by SD and RCH. Metabolic stability was determined by C.P., M.E., and B.V. Phys-chem properties were determined by C.P., M.E., and B.V. Antibacterial In vitro profiling of TriSLa compounds was performed by S.D. and R.C.H. Biochemical validation was performed by S.D., P.C., and R.C.H. In vivo efficacy studies on M. marinum-infected zebrafish and analysis were performed by C.H. and L.K. S.D., S.T., C.H., L.F., M.F., B.D., L.K., N.W., B.V., and R.C.H. wrote the paper. N.W., B.V., and R.C.H. obtained funding for this work. S.D., S.T., and C.H. contributed equally.
The authors declare the following competing financial interest(s): SD, ST, TA, LF, MF, BD, NW, BV and RCH, are inventors on patent application covering the TriSLa molecules described in this manuscript. The remaining authors declare no competing interests.
Notes
Atomic coordinates and structure factors reported in this paper (crystal structure of BDM44410, 1) have been deposited in the Cambridge Structural Database under accession number CCDC 2143694. Whole genome sequencing data (fastq files) for parental and BDM44410 (1) resistant H37Rv isolates (RC14.2, RC28.1) have been deposited at NCBI (BioProject ID: PRJNA808942).
Supplementary Material
References
- World Health Organization . Global Tuberculosis Report 2021: Geneva, 2021. [Google Scholar]
- Connolly L. E.; Edelstein P. H.; Ramakrishnan L. Why Is Long-Term Therapy Required to Cure Tuberculosis?. PLoS Med. 2007, 4, e120 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Global Tuberculosis Report 2020: Geneva, 2020. [Google Scholar]
- World Health Organization . Global Priority List Of Antibiotic-Resistant Bacteria To Guide Research, Discovery And Development Of New Antibiotics, 2017.
- Palomino J. C.; Martin A. TMC207 Becomes Bedaquiline, a New Anti-TB Drug. Future Microbiol. 2013, 8, 1071–1080. 10.2217/fmb.13.85. [DOI] [PubMed] [Google Scholar]
- Ryan N. J.; Lo J. H. Delamanid: First Global Approval. Drugs 2014, 74, 1041–1045. 10.1007/s40265-014-0241-5. [DOI] [PubMed] [Google Scholar]
- Keam S. J. Pretomanid: First Approval. Drugs 2019, 79, 1797–1803. 10.1007/s40265-019-01207-9. [DOI] [PubMed] [Google Scholar]
- Tiberi S.; du Plessis N.; Walzl G.; Vjecha M. J.; Rao M.; Ntoumi F.; Mfinanga S.; Kapata N.; Mwaba P.; McHugh T. D.; Ippolito G.; Migliori G. B.; Maeurer M. J.; Zumla A. Tuberculosis: Progress and Advances in Development of New Drugs, Treatment Regimens, and Host-Directed Therapies. Lancet Infect. Dis. 2018, 18, e183–e198. 10.1016/S1473-3099(18)30110-5. [DOI] [PubMed] [Google Scholar]
- Tornheim J. A.; Dooley K. E. The Global Landscape of Tuberculosis Therapeutics. Annu. Rev. Med. 2019, 70, 105–120. 10.1146/annurev-med-040717-051150. [DOI] [PubMed] [Google Scholar]
- Libardo M. D. J.; Boshoff H. I.; Barry C. E. The Present State of the Tuberculosis Drug Development Pipeline. Curr. Opin. Pharmacol. 2018, 42, 81–94. 10.1016/j.coph.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikušová K.; Ekins S. Learning from the Past for TB Drug Discovery in the Future. Drug Discovery Today 2017, 22, 534–545. 10.1016/j.drudis.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballell L.; Bates R. H.; Young R. J.; Alvarez-Gomez D.; Alvarez-Ruiz E.; Barroso V.; Blanco D.; Crespo B.; Escribano J.; González R.; Lozano S.; Huss S.; Santos-Villarejo A.; Martín-Plaza J. J.; Mendoza A.; Rebollo-Lopez M. J.; Remuiñan-Blanco M.; Lavandera J. L.; Pérez-Herran E.; Gamo-Benito F. J.; García-Bustos J. F.; Barros D.; Castro J. P.; Cammack N. Fueling Open-Source Drug Discovery: 177 Small-Molecule Leads against Tuberculosis. ChemMedChem 2013, 8, 313–321. 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Zhang K. Y. J. Advances in the Development of Shape Similarity Methods and Their Application in Drug Discovery. Front. Chem. 2018, 6, 1–21. 10.3389/fchem.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez-Poulain R.; Willand N.; Boutillon C.; Nowogrocki G.; Azaroual N.; Deprez B. A Simple Reaction to Produce Small Structurally Complex and Diverse Molecules. Tetrahedron Lett. 2004, 45, 5287–5290. 10.1016/j.tetlet.2004.05.008. [DOI] [Google Scholar]
- Willand N.; Beghyn T.; Nowogrocki G.; Gesquiere J.-C.; Deprez B. Synthesis and Structural Studies of a Novel Scaffold for Drug Discovery: A 4,5-Dihydro-3H-Spiro[1,5-Benzoxazepine-2,4′-Piperidine]. Tetrahedron Lett. 2004, 45, 1051–1054. 10.1016/j.tetlet.2003.11.079. [DOI] [Google Scholar]
- Willand N.; Folléas B.; Boutillon C.; Verbraeken L.; Gesquière J.-C.; Tartar A.; Deprez B. Efficient, Two-Step Synthesis of N-Substituted Nortropinone Derivatives. Tetrahedron Lett. 2007, 48, 5007–5011. 10.1016/j.tetlet.2007.05.110. [DOI] [Google Scholar]
- Malaquin S.; Jida M.; Courtin J.; Laconde G.; Willand N.; Deprez B.; Deprez-Poulain R. Water-Based Conditions for the Microscale Parallel Synthesis of Bicyclic Lactams. Tetrahedron Lett. 2013, 54, 562–567. 10.1016/j.tetlet.2012.11.082. [DOI] [Google Scholar]
- Tran N. C.; Dhondt H.; Flipo M.; Deprez B.; Willand N. Synthesis of Functionalized 2-Isoxazolines as Three-Dimensional Fragments for Fragment-Based Drug Discovery. Tetrahedron Lett. 2015, 56, 4119–4123. 10.1016/j.tetlet.2015.05.035. [DOI] [Google Scholar]
- Prevet H.; Flipo M.; Roussel P.; Deprez B.; Willand N. Microwave-Assisted Synthesis of Functionalized Spirohydantoins as 3-D Privileged Fragments for Scouting the Chemical Space. Tetrahedron Lett. 2016, 57, 2888–2894. 10.1016/j.tetlet.2016.05.065. [DOI] [Google Scholar]
- Blondiaux N.; Moune M.; Desroses M.; Frita R.; Flipo M.; Mathys V.; Soetaert K.; Kiass M.; Delorme V.; Djaout K.; Trebosc V.; Kemmer C.; Wintjens R.; Wohlkönig A.; Antoine R.; Huot L.; Hot D.; Coscolla M.; Feldmann J.; Gagneux S.; Locht C.; Brodin P.; Gitzinger M.; Déprez B.; Willand N.; Baulard A. R. Reversion of Antibiotic Resistance in Mycobacterium Tuberculosis by Spiroisoxazoline SMARt-420. Science 2017, 355, 1206–1211. 10.1126/science.aag1006. [DOI] [PubMed] [Google Scholar]
- Prevet H.; Moune M.; Tanina A.; Kemmer C.; Herledan A.; Frita R.; Wohlkönig A.; Bourotte M.; Villemagne B.; Leroux F.; Gitzinger M.; Baulard A. R.; Déprez B.; Wintjens R.; Willand N.; Flipo M. A Fragment-Based Approach towards the Discovery of N-Substituted Tropinones as Inhibitors of Mycobacterium Tuberculosis Transcriptional Regulator EthR2. Eur. J. Med. Chem. 2019, 167, 426–438. 10.1016/j.ejmech.2019.02.023. [DOI] [PubMed] [Google Scholar]
- von Korff M.; Sander T.. About Complexity and Self-Similarity of Chemical Structures in Drug Discovery. In Chaos and Complex Systems; Springer, Berlin, Heidelberg: Berlin, Heidelberg, 2013; pp 301–306. [Google Scholar]
- Sander T.; Freyss J.; von Korff M.; Rufener C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- Koul A.; Vranckx L.; Dhar N.; Göhlmann H. W. H.; Özdemir E.; Neefs J.-M.; Schulz M.; Lu P.; Mørtz E.; McKinney J. D.; Andries K.; Bald D. Delayed Bactericidal Response of Mycobacterium Tuberculosis to Bedaquiline Involves Remodelling of Bacterial Metabolism. Nat. Commun. 2014, 5, 3369. 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocat A.; Hartkoorn R. C.; Lechartier B.; Zhang M.; Dhar N.; Cole S. T.; Sala C. Bioluminescence for Assessing Drug Potency against Nonreplicating Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 4012–4019. 10.1128/AAC.00528-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Sala C.; Hartkoorn R. C.; Dhar N.; Mendoza-Losana A.; Cole S. T. Streptomycin-Starved Mycobacterium Tuberculosis 18b, a Drug Discovery Tool for Latent Tuberculosis. Antimicrob. Agents Chemother. 2012, 56, 5782–5789. 10.1128/AAC.01125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M.; Anyango S.; Deshpande M.; Nair S.; Natassia C.; Yordanova G.; Yuan D.; Stroe O.; Wood G.; Laydon A.; Žídek A.; Green T.; Tunyasuvunakool K.; Petersen S.; Jumper J.; Clancy E.; Green R.; Vora A.; Lutfi M.; Figurnov M.; Cowie A.; Hobbs N.; Kohli P.; Kleywegt G.; Birney E.; Hassabis D.; Velankar S. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan D.; Ray P. C.; Bayliss T.; Prosser G. A.; Harrison J. R.; Green K.; Soares de Melo C.; Feng T.-S.; Street L. J.; Chibale K.; Warner D. F.; Mizrahi V.; Epemolu O.; Scullion P.; Ellis L.; Riley J.; Shishikura Y.; Ferguson L.; Osuna-Cabello M.; Read K. D.; Green S. R.; Lamprecht D. A.; Finin P. M.; Steyn A. J. C.; Ioerger T. R.; Sacchettini J.; Rhee K. Y.; Arora K.; Barry C. E.; Wyatt P. G.; Boshoff H. I. M. 2-Mercapto-Quinazolinones as Inhibitors of Type II NADH Dehydrogenase and Mycobacterium Tuberculosis: Structure–Activity Relationships, Mechanism of Action and Absorption, Distribution, Metabolism, and Excretion Characterization. ACS Infect. Dis. 2018, 4, 954–969. 10.1021/acsinfecdis.7b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C.; Weinrick B.; Leung L. W.; Jacobs W. R. Plasticity of Mycobacterium Tuberculosis NADH Dehydrogenases and Their Role in Virulence. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 1599–1604. 10.1073/pnas.1721545115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites T.; O’Brien K.; Tiwari D.; Engelhart C. A.; Walters S.; Andrews J.; Yang H.-J.; Sutphen M. L.; Weiner D. M.; Dayao E. K.; Zimmerman M.; Prideaux B.; Desai P. V.; Masquelin T.; Via L. E.; Dartois V.; Boshoff H. I.; Barry C. E.; Ehrt S.; Schnappinger D. Plasticity of the Mycobacterium Tuberculosis Respiratory Chain and Its Impact on Tuberculosis Drug Development. Nat. Commun. 2019, 10, 4970. 10.1038/s41467-019-12956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel L.; Weisbrod T. R.; Marcinkeviciene J. A.; Bittman R.; Jacobs W. R. NADH Dehydrogenase Defects Confer Isoniazid Resistance and Conditional Lethality in Mycobacterium Smegmatis. J. Bacteriol. 1998, 180, 2459–2467. 10.1128/JB.180.9.2459-2467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbut M. B.; Yang B.; Liu R.; Yano T.; Vilchèze C.; Cheng B.; Lockner J.; Guo H.; Yu C.; Franzblau S. G.; Petrassi H. M.; Jacobs W. R.; Rubin H.; Chatterjee A. K.; Wang F. Small Molecules Targeting Mycobacterium Tuberculosis Type II NADH Dehydrogenase Exhibit Antimycobacterial Activity. Angew. Chem., Int. Ed. 2018, 57, 3478–3482. 10.1002/anie.201800260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki K.; Cosma C. L.; Troll M. A.; Ramakrishnan L. An In Vivo Platform for Rapid High-Throughput Antitubercular Drug Discovery. Cell Rep. 2012, 2, 175–184. 10.1016/j.celrep.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H.; Volkman H. E.; Ramakrishnan L. Tumor Necrosis Factor Signaling Mediates Resistance to Mycobacteria by Inhibiting Bacterial Growth and Macrophage Death. Immunity 2008, 29, 283–294. 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman H. E.; Clay H.; Beery D.; Chang J. C. W.; Sherman D. R.; Ramakrishnan L. Tuberculous Granuloma Formation Is Enhanced by a Mycobacterium Virulence Determinant. PLoS Biol. 2004, 2, e367 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibaud L.; Rombouts Y.; Trivelli X.; Burguière A.; Cirillo S. L. G.; Cirillo J. D.; Dubremetz J.-F.; Guérardel Y.; Lutfalla G.; Kremer L. A Mycobacterium Marinum TesA Mutant Defective for Major Cell Wall-Associated Lipids Is Highly Attenuated in Dictyostelium Discoideum and Zebrafish Embryos. Mol. Microbiol. 2011, 80, 919–934. 10.1111/j.1365-2958.2011.07618.x. [DOI] [PubMed] [Google Scholar]
- Beghyn T.; Deprez-Poulain R.; Willand N.; Folleas B.; Deprez B. Natural Compounds: Leads or Ideas? Bioinspired Molecules for Drug Discovery. Chem. Biol. Drug Des. 2008, 72, 3–15. 10.1111/j.1747-0285.2008.00673.x. [DOI] [PubMed] [Google Scholar]
- Lee M.-L.; Schneider G. Scaffold Architecture and Pharmacophoric Properties of Natural Products and Trade Drugs: Application in the Design of Natural Product-Based Combinatorial Libraries. J. Comb. Chem. 2001, 3, 284–289. 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- Henkel T.; Brunne R. M.; Müller H.; Reichel F. Statistical Investigation into the Structural Complementarity of Natural Products and Synthetic Compounds. Angew. Chem., Int. Ed. 1999, 38, 643–647. . [DOI] [PubMed] [Google Scholar]
- Hong W. D.; Gibbons P. D.; Leung S. C.; Amewu R.; Stocks P. A.; Stachulski A.; Horta P.; Cristiano M. L. S.; Shone A. E.; Moss D.; Ardrey A.; Sharma R.; Warman A. J.; Bedingfield P. T. P.; Fisher N. E.; Aljayyoussi G.; Mead S.; Caws M.; Berry N. G.; Ward S. A.; Biagini G. A.; O’Neill P. M.; Nixon G. L. Rational Design, Synthesis, and Biological Evaluation of Heterocyclic Quinolones Targeting the Respiratory Chain of Mycobacterium Tuberculosis. J. Med. Chem. 2017, 60, 3703–3726. 10.1021/acs.jmedchem.6b01718. [DOI] [PubMed] [Google Scholar]
- Heikal A.; Nakatani Y.; Jiao W.; Wilson C.; Rennison D.; Weimar M. R.; Parker E. J.; Brimble M. A.; Cook G. M. ‘Tethering’ Fragment-Based Drug Discovery to Identify Inhibitors of the Essential Respiratory Membrane Protein Type II NADH Dehydrogenase. Bioorg. Med. Chem. Lett. 2018, 28, 2239–2243. 10.1016/j.bmcl.2018.05.048. [DOI] [PubMed] [Google Scholar]
- Shirude P. S.; Paul B.; Roy Choudhury N.; Kedari C.; Bandodkar B.; Ugarkar B. G. Quinolinyl Pyrimidines: Potent Inhibitors of NDH-2 as a Novel Class of Anti-TB Agents. ACS Med. Chem. Lett. 2012, 3, 736–740. 10.1021/ml300134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.; Åkerbladh L.; Ahmad S.; Konda V.; Cao S.; Vocat A.; Maes L.; Cole S. T.; Hughes D.; Larhed M.; Brandt P.; Karlén A.; Mowbray S. L. Synthesis and In Vitro Biological Evaluation of Quinolinyl Pyrimidines Targeting Type II NADH-Dehydrogenase (NDH-2). ACS Infect. Dis. 2022, 8, 482. 10.1021/acsinfecdis.1c00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M. D.; Kasparian J. A.; Hortle E.; Britton W. J.; Purdie A. C.; Oehlers S. H. Mycobacterium Marinum Infection Drives Foam Cell Differentiation in Zebrafish Infection Models. Dev. Comp. Immunol. 2018, 88, 169–172. 10.1016/j.dci.2018.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.