Abstract
Over the course of the Corona Virus Disease-19 (COVID-19) pandemic in 2020–2022, monitoring of the severe acute respiratory syndrome coronavirus 2 ribonucleic acid (SARS-CoV-2 RNA) in wastewater has rapidly evolved into a supplementary surveillance instrument for public health. Short term trends (2 weeks) are used as a basis for policy and decision making on measures for dealing with the pandemic. Normalisation is required to account for the dilution rate of the domestic wastewater that can strongly vary due to time- and location-dependent sewer inflow of runoff, industrial discharges and extraneous waters. The standard approach in sewage surveillance is normalisation using flow measurements, although flow based normalisation is not effective in case the wastewater volume sampled does not match the wastewater volume produced. In this paper, two alternative normalisation methods, using electrical conductivity and crAssphage have been studied and compared with the standard approach using flow measurements. For this, a total of 1116 24-h flow-proportional samples have been collected between September 2020 and August 2021 at nine monitoring locations. In addition, 221 stool samples have been analysed to determine the daily crAssphage load per person. Results show that, although crAssphage shedding rates per person vary greatly, on a population-level crAssphage loads per person per day were constant over time and similar for all catchments. Consequently, crAssphage can be used as a quantitative biomarker for populations above 5595 persons. Electrical conductivity is particularly suitable to determine dilution rates relative to dry weather flow concentrations. The overall conclusion is that flow normalisation is necessary to reliably determine short-term trends in virus circulation, and can be enhanced using crAssphage and/or electrical conductivity measurement as a quality check.
Keywords: Sewage surveillance, COVID-19, Public health, Normalisation
Graphical abstract
1. Introduction
Over the course of the Corona Virus Disease-19 (COVID-19) pandemic in 2020–2022, monitoring of the severe acute respiratory syndrome coronavirus 2 ribonucleic acid (SARS-CoV-2 RNA) in wastewater has rapidly evolved into a supplementary surveillance instrument for public health (Lodder and de Roda Husman, 2020; Medema et al., 2020b; Kirby et al., 2021). It is currently used in many countries (COVIDPoops19 Dashboard | covid19wbec.org) at different scales, from national surveillance at all (Virus particles in wastewater | Coronavirus Dashboard | Government.nl) or selected wastewater treatment plants (WWTPs) (Szennyvizekben mért SARS-CoV-2 vírus koncentrációja (gov.hu); Koronaviruksen jätevesiseurannan viikkoraportti (thl.fi); Données ouvertes - Réseau OBEPINE (reseau-obepine.fr); Coronastep | Luxembourg Institute of Science and Technology (list.lu); SARS-CoV-2 in Wastewater (sensors-eawag.ch)) to regional (Wolfe et al., 2021; Ai et al., 2021; Sarsaigua (icra.cat)), (sub) city level (Yaniv et al., 2021; Rodríguez Rasero et al., 2022) and building level (Davo et al., 2021; Sweetapple et al., 2021; Betancourt et al., 2021). The concentration of SARS-CoV-2 RNA in wastewater has shown to reflect and even precede the trends of the newly reported cases or COVID-19 hospitalizations (Medema et al., 2020a; Prado et al., 2021; Ho et al., 2021). In situations with low COVID-19 prevalence (and a limited willingness and/or capacity for SARS-CoV-2 testing), wastewater surveillance is being used as an early warning system (Medema et al., 2020b; Betancourt et al., 2021; Ahmed et al., 2020) enabling rapid and targeted measures to limit the SARS-CoV-2 transmission. Wastewater surveillance is also an efficient tool to monitor the emergence of (signature mutations of) new variants-of-concern in communities, using targeted sequencing (Rios et al., 2021; Rothman et al., 2021; Jahn et al., 2022) or targeted PCR methods (Graber et al., 2021; Heijnen et al., 2021). A key advantage of wastewater surveillance is the possibility to obtain objective information about virus circulation in a community. Surveillance of reported cases is subject to bias by (changes in) testing strategies, access to testing, and compliance of communities, and reflect only those that get tested. SARS-CoV-2 concentrations in community wastewater are independent of testing behaviour, thereby providing a more complete and objective image of the virus circulation in the community provided there is unrestricted access to sanitation. Public health agencies use the wastewater signal to spot changes in testing behaviour over time or between communities (De Graaf et al., 2022) and to verify the trends in reported cases in communities. Decision-making on (partial) lockdowns and travel restrictions requires up-to-date and reliable information on trends in virus circulation, preferably at a high spatial resolution. The relevant time window of many of these decisions is one to two antecedent weeks; examples are the weekly updates of the epidemiological situation in e.g. the Netherlands (https://www.rivm.nl/en/coronavirus-covid-19/weekly-figures) and the maps in support of the Council Recommendation on a coordinated approach to travel measures in the EU (https://www.ecdc.europa.eu/en/covid-19/situation-updates/weekly-maps-coordinated-restriction-free-movement) that are based on the testing, positivity and notification rate over a time frame of two weeks.
To monitor trends in SARS-CoV-2 circulation via wastewater in a similar time frame, frequent and representative sampling of a community is necessary. Sample collection typically takes place as 24 h composite samples to account for the typical diurnal pattern associated with wastewater production and toilet use (Ort et al., 2010; Medema et al., 2020b). The concentration of SARS-CoV-2 RNA in wastewater can be affected by the dilution of domestic wastewater with water from other sources that normally do not contain SARS-CoV-2 RNA such as stormwater runoff, infiltrating groundwater and/or industrial wastewater. Dilution rates may vary between catchment areas (e.g. different contributions of industrial wastewater) as well as in time (wet weather flows can be many times the normal dry weather flow of sewers). To account for this variable dilution, normalisation of the measured concentrations is a common procedure in sewage surveillance and Wastewater Based Epidemiology (WBE), building on methods and procedures developed over the last decade mainly for drug use monitoring (Castiglioni et al., 2014).
Standard SARS-CoV-2 RNA normalisation follows the approach as presented in eq. (1) using the total volume of wastewater over the 24 h sampling period (V 24h) and the number of inhabitants in the catchment area (# inh) as normalisers to get from the SARS-CoV-2 concentration in the sample (c sample) to a viral load per 24 h and per capita (L 24h,cap) that can be compared in both time and space to other values:
| (1) |
with L 24h,cap = viral load per 24 h and per capita;
csample = concentration SARS-CoV-2 RNA in 24 h composite sample (unnormalised, ‘raw’ value);
V24h = measured wastewater volume in same 24 h sampling period;
#inh = number of inhabitants in the catchment area.
At some locations with SARS-CoV-2 monitoring, however, flow measurements are unavailable. Typically, flow sensors are only installed at wastewater treatment plants (WWTP) and large sewer pumping stations (ps). At smaller, more upstream locations such as residence areas, university campuses, schools, hospitals, nursing homes, etc. flow monitoring in sewer systems is very uncommon (and often technically challenging). For these locations, an alternative normalisation approach is needed using a parameter other than flow. Ideally, such an alternative normalisation approach would yield normalised values that can be directly compared to normalised values using the flow-based approach.
Also in situations where flow measurement are available, flow normalisation may not always be representative. For instance, as a result of commuters or tourists, the registered number of inhabitants in an area can differ significantly from the actual number of shedders to the sewer system. Also, sewer system dynamics can result in situations where the wastewater flow at the monitoring location, for instance the inlet of the WWTP, is not representative for wastewater that is produced in the catchment area on that day. Examples of such dynamics are (1) wastewater spills from sanitary sewer overflows (SSOs) due to blockages in the sewer system, (2) wastewater spills from combined sewer overflows (CSOs) during heavy rainfall, (3) transport delays in pressure mains that can strongly vary between dry weather and wet weather conditions and (4) delays in the transport of wastewater to the WWTP as a result of pump failure, maintenance work or other operational issues. In these cases, wastewater (and the associated SARS-CoV-2 RNA load) from the catchment area arrives earlier, later or not at all at the WWTP, and is hence not well represented in the 24 h composite samples. Flagging these situations using an additional, independently measured parameter could serve as a quality control for the standard flow normalisation.
Many parameters that may be used to normalise wastewater samples have already been studied. Launay et al. (2016) propose the use of electrical conductivity (EC) as a proxy of the dilution of wastewater with stormwater based on the fact that typical EC values for urban stormwater runoff (∼200–250 μS/cm) differ significantly from those of wastewater (∼1000–1500 μS/cm). They showed in their study that the dilution rate based on EC is similar to the rate based on inert human wastewater tracers such as ibuprofen, naproxen and diclofenac. In our study, EC was selected as a potential alternative parameter for SARS-CoV-2 normalisation, given its ability to monitor dilution of sewage due to storm events and the broad experience in monitoring sewage with EC sensors.
Choi et al. (2018), Tandukar et al. (2020), Bivins et al. (2020) and Medema et al. (2020b) assessed the applicability of several chemical and biological markers for human input in wastewater. Like others (Crank et al., 2020; Hillary et al., 2021; Wilder et al., 2021; Heijnen et al., 2021), we also selected crAssphage for the current study, given it is a highly abundant virus (almost) exclusively found in human faeces and is present in humans world-wide (Edwards et al., 2019). High crAssphage concentrations are reported in domestic wastewater globally, and loads are reported to not show significant seasonal variation (Ballesté et al., 2019), making them a potentially useful index for the human faecal fraction of wastewater. Many other possible tracers of human (faeces) origin are less well-suited due to e.g. a higher background variation or degradation in the sewer (Gao et al., 2017).
The aim of this study was to assess alternative normalisation methods for wastewater samples using EC and crAssphage and to compare the results to the standard normalisation approach using flow measurements and number of inhabitants. We considered these alternative approaches for two different purposes: (1) a quality check for the standard flow normalisation of SARS-CoV-2 RNA, and (2) normalisation of SARS-CoV-2 RNA concentration values at locations without flow monitoring.
2. Material and methods
2.1. Sampling locations Rotterdam Rijnmond
In the Rotterdam Rijnmond region (the Netherlands) nine catchment areas were selected for wastewater sampling (Fig. 1 ). Population sizes have been derived matching the sewer catchment areas with the (highly detailed) Municipal Administration (CBS, 2020). All four catchment areas of the WWTP Dokhaven (INF2, INF3, INF4 and INF5–6) were sampled from the influent pressure mains arriving at the WWTP. These four areas make up about half of the urban area of the city of Rotterdam with residential populations of 27,044, 121,118, 36,011 and 138,280 inhabitants, respectively. Within the INF3 catchment area, subcatchment Pretorialaan (71,325 inhabitants) was selected for surveillance at the wastewater pumping station (ps). Within the Pretorialaan catchment the wastewater from an even smaller sub-subcatchment (Katendrecht, 5595 inhabitants) was also sampled at a pumping station.
Fig. 1.
Catchment areas in the Rotterdam Rijnmond area used for wastewater surveillance with locations of wastewater treatment plants (WWTP) and sewer pumping stations (ps).
Outside the WWTP Dokhaven area, three predominantly residential areas were selected for surveillance: Ommoord (28,434 inhabitants), Bergschenhoek (18,750 inhabitants) and Rozenburg (12,374 inhabitants). Ommoord and Bergschenhoek were sampled at their respective sewer pumping stations, Rozenburg at the local WWTP that serves only the Rozenburg area.
The sewer network in Rotterdam is predominantly combined, with separate sewers only in more recently developed and renovated areas. All monitoring locations (WWTPs as well as pumping stations) are equipped with flow sensors that give continuous or high-frequent flow measurements.
2.2. Wastewater sampling
At all sampling sites automated composite samplers (Endress+Hauser Liquistation CSF48 and ASP2000 stations) were used to collect flow-proportional 24 h composite wastewater samples. The autosamplers were programmed to collect 50 ml aliquots per fixed volume of wastewater (as measured by the flow sensors at the WWTP/pumping station). These fixed volumes varied between 12 m3 (Katendrecht) and 170 m3 (Dokhaven INF3) and were selected to ensure a minimum of 100 aliquots (5 L of sample) for each 24 h sample throughout the year (NEN, 2019). All locations followed the same sampling schedule: three 24 h samples per week (Sun 08 h00 - Mon 08 h00, Tue 08 h00 - Wed 08 h00 and Thu 08 h00 - Fri 08 h00) from September 2020 (Bergschenhoek: January 2021) up to and including August 2021. Samples were stored inside the autosampler at a temperature between 1 °C and 5 °C until sample collection (Mon, Wed and Fri between 08 h00 and 16 h00). At the time of sample collection, the collected composite sample was manually stirred and an aliquot of 250 ml was collected from the container in the autosampler, stored in a sterile flask and transported at 4 °C to the laboratory.
Upon sample collection the quality of each sample was assessed comparing the theoretical sample volume (number of aliquots in 24 h, multiplied by aliquot volume) with the actual sample volume. In case of sampling ‘failure’ (> 7.5 % difference according to NEN, 2019), details of the sampling process were further studied to determine whether the sample could still be included in the data set. Overall, 6–10 % (depending on the location) of all sampling failed completely (not a single aliquot in the container in a 24 h period) due to a power cut, maintenance activities, complete clogging of the autosampler, public holidays, a prolonged cold period with frozen suction hoses, etc. For another 4–19 % of samples a deviation of >7.5 % was observed. These were caused mainly by (partial or temporal) clogging of the equipment (relatively often coinciding with storm events) and logistical errors in the field. In total, 1116 samples were available for further processing. An overview of sampling statistics is given in Supplementary Materials S1.
2.3. Wastewater sample processing
2.3.1. Virus concentration and nucleic acid extraction
Samples were processed within one week after sampling using the procedure as previously described (Medema et al., 2020a). In short, centrifugation was used as pre-treatment to remove larger particles. Virus particles were concentrated from 50 ml supernatant by ultrafiltration through Centricon® Plus-70 centrifugal ultrafilters with a cut-off of 30 kDa (Millipore, Amsterdam, Netherlands). Mouse Hepatitis Virus (MHV)-A59 (Department Medical Microbiology, Leiden University Medical Center, Leiden, Netherlands) was spiked to each concentrate as quality control. Nucleic acid was extracted from the concentrate with the Biomerieux Nuclisens kit (Biomerieux, Amersfoort, Netherlands) in combination with the semi-automated KingFisher ml (Thermo Scientific, Bleiswijk, Netherlands) as previously described. Extracted nucleic acid was eluted in a volume of 100 μl.
2.3.2. RT-qPCR for SARS-CoV-2 RNA-quantification in wastewater
The N2 and E_Sarbecco gene fragments of SARS-CoV-2 were used as qRT-PCR targets. N2 was used for quantification, E_Sarbecco for confirmation. Reagents and reaction conditions were as previously described (Medema et al., 2020a; Heijnen et al., 2021). All RT-PCR's were run as technical duplicates on 5 μl extracted nucleic acid. Reactions were considered positive if the cycle threshold was below 40 cycles. Spiked MHV-A59 RNA was detected by performing an MHV-A59 specific RT-qPCR targeting the N-gene using the primers and reaction profile described by Raaben et al. (2007).
2.3.3. PCR for crAssphage quantification in wastewater
A crAssphage CPQ_064 specific PCR (Stachler et al., 2017) was used to quantify this DNA-virus. Assays were performed in duplicate on 5 μl 1:10 diluted extracted nucleic acid as described previously (Heijnen et al., 2021). The concentration of the gBlock used for quantification was checked with digital droplet qPCR.
2.4. PCR for crAssphage quantification in stool samples
Faeces swabs from 221 patients from the Rotterdam Rijnmond region that were tested positive for SARS-CoV-2 with nose-swabs were collected in 3 ml of viral transport medium. Samples were spun down for 5 min at 17,000 g, nucleic acids were isolated from 200 μl supernatant using the high pure RNA isolation kit (Roche) while omitting the DNAse I step. The same RT-PCR as used for wastewater samples was used to quantify levels of crAssphage. This study was approved by the Medical Ethical Committee of Erasmus MC under MEC-2020-0617.
2.5. Multiplication factors for comparison of normalisation parameters
Flow-based normalisation.
The standard flow-based normalisation for SARS-CoV-2 values follows the approach as presented in eq. (1). It uses the ratio of the total volume of wastewater over the 24 h sampling period (V 24h) to the number of inhabitants in the catchment area (# inh) as normaliser to get from the SARS-CoV-2 RNA concentration in the sample (c sample) to a viral load per 24 h per capita (L 24h,cap) For comparison with other normalisation approaches, instead of using the number of inhabitants (# inh) only, the average amount of domestic dry weather flow these inhabitants was used (V ddwf). For this, the number of inhabitants was multiplied with their average daily domestic wastewater production (in the Netherlands a stable 0.12 m3 per capita per day (RIONED, 2013), but values can vary per country or region). With this adaptation, the normaliser in eq. (1) changes to a dimensionless multiplication factor that expresses the number of times the measured 24 h volume is larger than the ‘expected’ volume of domestic wastewater:
| (2) |
with V 24h = measured volume of wastewater in 24 h sampling period;
Vddwf = estimated domestic dry weather flow (# inhabitants * 0.12 m3 per capita per day).
This factor is an estimate of how much higher the concentration SARS-CoV-2 RNA was in the original domestic wastewater before it was mixed with other sources of (waste)water that do not contain SARS-CoV-2 RNA, such as groundwater, industrial wastewater and stormwater run-off. The reciprocal of this factor expresses the share of domestic wastewater in each of the collected 24 h composite samples. Note that the multiplication factor equals the original normaliser in eq. (1) except for a (fixed) offset of 1/0.12 ≈ 8.33. The multiplication factor (or the share of domestic wastewater in the 24 h sample) forms the basis for comparison with the other normalisation approaches.
Rewriting eq. (2) into eq. (3) allows differentiation between dry weather and wet weather dilution processes of the domestic wastewater:
| (3) |
with V 24h = measured volume of wastewater in 24 h sampling period;
Vddwf = estimated domestic dry weather flow (# inhabitants * 0.12 m3 per capita per day);
VDWF,ref = average wastewater volume during dry weather flow (details below).
The first term in the equation (V DWF,ref / V ddwf) indicates how much larger the ‘average’ dry weather flow is compared to the expected domestic wastewater volume. Hence, it gives a (constant and site-specific) multiplication factor to correct for mixing of domestic wastewater in dry weather conditions with industrial wastewater, groundwater infiltration, etc. The second term (V 24h / V DWF,ref) indicates how the actual (measured) 24 h wastewater volume compares to the ‘average’ wastewater volume during dry weather. This terms gives the (varying) multiplication factor to account for any additional mixing of the dry weather flow with stormwater run-off. The reference dry weather values (V DWF,ref) was defined as the 40th percentile from a 1-year time series of flow data at each sampling site (Mulder et al., 2020). The choice for the 40th percentile is arbitrary, but is generally considered the ‘representative’ value for dry weather flow. Also, for most monitoring locations the difference between the 30th and 50th percentile value is relatively small (see the S-curves for all monitoring locations in the Supplementary Materials S2a and an overview of values in S2b). This assessment is straightforward using the methods as described in Weiß et al. (2002). For catchments with a strong seasonal variation in the amount of extraneous waters (such as ps Ommoord, see Supplementary Materials S8), it may be necessary to derive different reference values per season.
2.5.1. EC- and crAssphage-based normalisation
The multiplication factors based on EC and crAssphage were derived using the same approach as for the flow-based normalisation:
| (4) |
| (5) |
with V DWF,ref = average wastewater volume during dry weather flow;
Vddwf = estimated domestic dry weather flow (# inhabitants * 0,12 m3 per capita per day);
ECDWF,ref or crAss DWF,ref = average EC or crAssphage concentration during dry weather flow;
ECsample or crAss sample = measured EC or crAssphage concentration in 24 h composite sample.
Eqs. (4), (5) also estimate the dilution of the original volume of domestic wastewater with other, non-SARS-CoV-2 or crAssphage containing water sources. The first term in both equations (V DWF,ref / V ddwf) is equal to the first term in eq. (3) and again gives the (constant) multiplication factor to correct for dilution during dry weather flow (with industrial wastewater, groundwater, etc.). The second terms (EC DWF,ref/EC sample and crAss DWF,ref (conc.)/crAss sample (conc.)) again give the additional and varying multiplication factors to account for the mixing of the dry weather flow with stormwater run-off, now based on EC and crAssphage measurements.
The reference dry weather values (EC DWF,ref and crAss DWF,ref) were defined as the median of EC and crAssphage concentrations during dry weather flow. For this, all days were selected with a daily flow volume ranging between the 10th and 50th percentile in a 1-year time series. The rationale behind these ‘safe’ threshold values is that up to the lowest 10 % of values could be affected by operational irregularities such as pump failures or maintenance activities, while flows larger than the 50th percentile could already be affected by (small) storm events. The Supplementary Materials (S2a) give the S-curves per catchment area for all EC and crAssphage values as well as those values used to derive the dry weather flow references. An overview of values of EC DWF,ref and crAss DWF,ref are given in S2b.
Comparison of normalisation approaches.
For all three normalisation approaches the actual normalisation of the SARS-CoV-2 RNA concentration in wastewater was done by multiplying the ‘raw’ concentration values with the multiplication factors in eqs. (3), (4), (5). These values are referred to as “Q-normalised”, “EC-normalised” and “crAss-normalised”, respectively, as opposed to the standard flow -normalised values following eq. (1).
3. Results and discussion
3.1. Variation of Q-based multiplication factors across catchment areas
The Q-based multiplication factor values range between a minimum of approximately 2 up to a maximum of 5–15, depending on the catchment, see Fig. 2 . These large ranges are explained by the fact that all catchments are served by combined sewer systems with increasing flows, and hence increasingly high dilutions rates, during storm events. The variation in maximum values across catchment areas is in line with the variation in ratio between domestic wastewater production and typical design based maximum daily wastewater volumes. For instance, the pumping capacity of catchment WWTP Dokhaven INF2 (2500 m3/h) allows a daily maximum of roughly 60.000 m3 of wastewater, which results (with a domestic wastewater production of 3245 m3) in a maximum multiplication factor of 18.5. For catchment INF3 this maximum factor lies much lower at 9.9 (domestic wastewater production of 14,534 m3 on a maximum daily volume of 144,000 m3). The observed maximum values for both catchments, 14.6 and 8.5, respectively, are both around 80 % of their theoretical maximum values.
Fig. 2.
Boxplot with Q-based multiplication factors across catchment areas. Each boxplot gives the minimum, 25th percentile, the median, the 75th percentile and the maximum value by the whiskers.
The minimum factors vary between 1.5 and 2.5, and median values between 2.0 and 3.5. This indicates that also during dry weather a significant proportion of the sampled wastewater consists of non-domestic sources, such as extraneous waters and industrial wastewater. Earlier studies showed that at WWTP Dokhaven the average amount of extraneous waters during dry weather flow is 43 % of the total influent (Vosse, 2013). With the majority of multiplication factors associated with dry weather conditions, all catchments show a skewed distribution with much smaller interquartile lengths between minimum and median than between median and maximum.
The large variations in multiplication factors confirm that direct comparison of measured SARS-CoV-2 concentrations without normalisation would result in an incorrect assessment of the relative concentration levels, both in time (trends per catchment) and in space (between catchments).
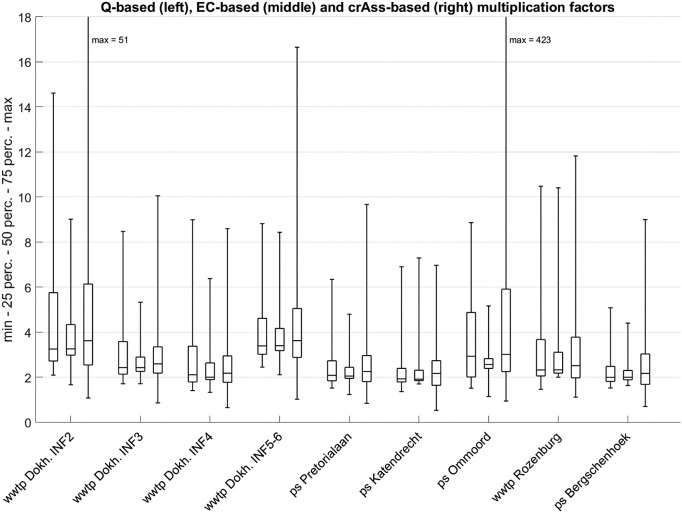
3.2. Comparison of Q-based, EC-based and crAssphage-based multiplication factors
Fig. 3 presents the same Q-based multiplication factors as in Fig. 2 for all catchments, but now compared to the EC- and crAssphage-based multiplication factors. The median values for the three parameters are comparable per catchment (e.g. for WWTP Dokhaven INF2 Q-based: 3.3, EC-based: 3.3 and crAss-based: 3.6), which is in line with all three approaches using a measured value relative to a reference value for dry weather flow.
Fig. 3.
Boxplot with Q-based (left), EC-based (middle) and crAssphage-based (right) multiplication factors per catchment area. Each boxplot gives the minimum, 25th percentile, the median, the 75th percentile and the maximum value by the whiskers.
Relative to the Q-based factors, the EC-based factors show a similar or smaller range of values and the range shown for crAssphage is similar or (much) larger. This is due to the higher variability in crAssphage concentrations compared with the EC measurements, both for dry and wet weather samples. This can be observed by comparing the S-curves for EC and crAssphage in Supplemental Fig. S2: which shows that the range of values associated with dry weather flow volumes (in red) for crAssphage is much larger than for EC.
The largest EC-based multiplication factors are the result of the lowest measured EC-values in the wastewater samples. EC values have a (theoretical) lower limit of 200–250 μS/cm (typical values for urban stormwater runoff) with associated (theoretical) maximum wet weather multiplication factors of approximately 4–5. Measurements show the lowest EC values in the range of 315 μS/cm (ps Katendrecht) to 600 μS/cm (ps Bergschenhoek), resulting in lower maximum wet weather factors.
Fig. 4 shows catchment WWTP Dokhaven INF4 as an example of the comparison between the sets of multiplication factors. The 100 % fit between flow-based and Q-based values is a logical results of the fixed offset between the two values. The linear fit between flow-based and EC-based values gives an R2 of 0.84. These value ranges roughly between an R2 of 0.70 and 0.90 for the other catchment areas except for the smallest catchment ps Katendrecht (R2 = 0.37) and ps Ommoord (see S8). The linear fit for crAssphage-based values gives a much weaker R2 of 0.53, also for the other areas the R2 for crAssphage is much lower with values ranging between 0.30 and 0.60. Again, areas ps Katendrecht (R2 = 0.19) and ps Ommoord give the lowest coefficients of determination. The other catchment areas give similar results (Supplementary Materials S4).
Fig. 4.

Relations between multiplication factors derived from Q-, EC- and crAssphage-based normalisation for catchment WWTP Dokhaven INF4.
The better fit for the EC-based multiplication factors means that EC-normalised SARS-CoV-2 RNA values are generally in better agreement with the ‘targeted’ Q-normalised values than the crAssphage-normalised values. Fig. 5 presents an example for catchment area WWTP Dokhaven INF3 for a few months in 2020, and shows the general need for normalisation of values. The apparently constant virus circulation in raw (unnormalized) values in, for instance, mid-November and the last half of December 2020 changes into a rising trend after normalisation, because rainfall diluted the domestic wastewater. Considering the Q-normalised values as the reference, the EC-normalised values show a better fit in terms of absolute values and trends compared to crAssphage-normalised values for most sampling events. The trend in the SARS-CoV-2 RNA signal, however, dominated the differences between the normalisation approaches.
Fig. 5.

SARS-CoV-2 RNA concentrations in wastewater (raw values and Q-, EC- and crAssphage- normalised values) and wastewater volumes for catchment WWTP Dokhaven INF3.
The other catchments show similar results (Supplementary Materials S5).
3.3. Quality control: Comparing multiplication factors per sampling event
A comparison between multiplication factors per sampling event allowed flagging events for which the multiplication factors differed significantly from each other. Given the correlations between Q-based, EC-based and/or crAssphage-based factors presented above, a relatively large difference between factors suggests that at least one of the three parameters might show an ‘abnormal’ result. Such an ‘abnormality’ can be caused by (1) an erroneous measurement or laboratory analysis or (2) a process in or around the sewer system. As some of these causes influence the representativeness of the sample and normal WBE-normalisation, flagged events require further investigation.
As an example, Fig. 6 shows the difference in the share of domestic wastewater in each sample (the reciprocal of the multiplication factors) for catchment Katendrecht as calculated using the Q-based and EC-based approaches. Similar results were obtained in the other catchment areas (Supplementary Materials S4b).
Fig. 6.

Differences in percentage domestic wastewater (reciprocal of multiplication factors) between Q-based and EC-based normalisation for catchment area Katendrecht.
For EC-based factors, the majority of values are within a 10 % difference compared to Q-based values, and are hence in good accordance. On a number of occasions, however, differences larger than 10 % and/or sudden changes in differences were observed. Most striking was the difference on January 20th, 2021 (−41 %). On this day the EC-value (315 μS/cm) was typical for wet weather flow, but the measured 24 h wastewater volume (1215 m3) was more typical for a dry weather day. Further assessment revealed that it was indeed a wet weather day, but that halfway the sampling event a pump failure caused the pumping station to cease operation for about 48 h (for details, see Fig. S4c.2 in the Supplementary Materials). As a result, the flow proportional sampling only collected samples until the pumps stopped and the sample hence only represents a fraction of the full wastewater production of the area on that day (the other fraction being stored in the sewer system during the pump failure). Interestingly, during the following sampling event (January 22nd) the opposite occurred: after the re-start, the pumping station processed all wastewater of the preceding 48 h that had been stored in the sewer system, leading to a 24 h sample and measured wastewater volume that were again not representative of the wastewater production in the area in the targeted 24 h.
In the Supplementary Materials (S4a) an overview is given of other flagged events (based on EC) with their possible causes and effects on representativeness and WBE-normalisation. In total, 7 events have been flagged for which the sample and measured 24 h volume were not representative for the targeted 24 h period as a result of maintenance or failure of a pumping station. Hence, these results were excluded from trend analysis. Other flagged events were related to high EC values (after road de-icing) or very low flow values (in summer periods without rain). In these cases there was no need to exclude results as the representativeness of the sample and the associated flow volume were not affected.
Flagging based on comparison of Q-based and crAssphage-based values proved valuable in identifying samples with unexpected low recovery of viruses from wastewater (see results in Supplementary Materials S4, particularly in Ommoord), poorly performing lab filter batches (data not included) and deviations (mainly in the period 8–15 March 2021 and end of August 2021) associated with erroneous laboratory results of crAssphage. Most anomalies observed with EC were not seen with crAssphage, as they were masked by the inherent large(r) variation of measured crAssphage concentrations.
3.4. Quality control: crAssphage loads to verify number of shedders
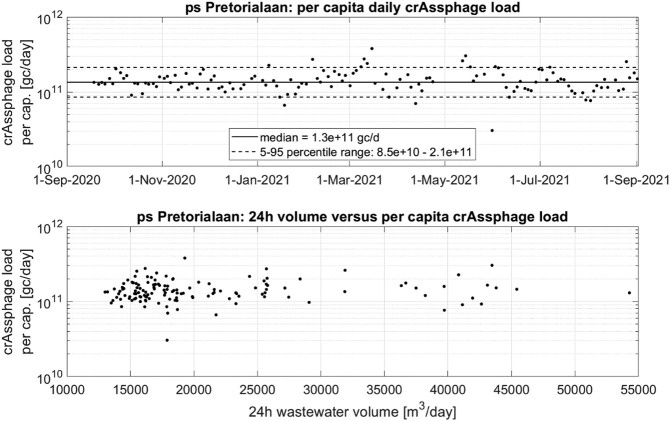
For each catchment area the crAssphage load per day was calculated by multiplying the (measured) crAssphage concentration of the sample with the associated (measured) 24 h wastewater volume. Dividing this daily load over the (administrative) number of inhabitants yielded a time-series of crAssphage loads per capita for each catchment area. Fig. 7 presents an example for catchment area ps Pretorialaan (other catchment areas in Supplementary Materials S6).
Fig. 7.
CrAssphage daily load per capita for catchment ps Pretorialaan: chronologically (upper graph) and comparison with 24 h wastewater volumes (lower graph).
The per capita crAssphage load showed little variation and no long term trend in time (Fig. 7, upper graph) with a median value of 1.3 * 1011 gc/(cap*day) and a 5–95 percentile range between 8.5*1010–2.1 * 1011 gc/(cap*day). Other catchment areas (except ps Ommoord) showed similar results with median values ranging between 1.2 and 1.6 * 1011 gc/(cap*day). The lack of correlation between daily flow and crAssphage load per capita (lower graph) suggests that there is no significant build-up of crAssphage in sediments or biofilm in the sewer network during dry weather that is flushed out at higher rates during wet weather events (Ballesté et al., 2019). This is unlike other wastewater parameters such as Chemical Oxygen Demand (COD) and Total Suspended Solids (TSS) (Schilperoort et al., 2012), or pollutants that show a strong adsorption to TSS (Gasperi et al., 2014).
Given the small range of median values for the per capita crAssphage loads across the studied catchment areas, it was possible to flag any (sufficiently large) deviations from this median. As an illustration: during data processing, the median value for catchment area ps Katendrecht was initially calculated as 2.0 * 1011 gc/(cap*day), which was relatively far outside the range for the other catchment areas (1.2–1.6 * 1011 gc/(cap*day)). Further assessment showed that the number of inhabitants in the Katendrecht catchment area had increased from the 4884 based on census data of 2019 to 5595 based on census data in 2020, i.e. by approximately 20 %, due to urban development not long before the monitoring started. The administrative data that were used initially had not been updated. The anomalous per capita crAssphage load in this case flagged the use of outdated administrative information.
Stikkers (2022) demonstrated for a number of WWTPs in the Netherlands that traditional biomarkers N (nitrogen) and P (phosphate) can be used to determine temporal variations due to tourism in the number of shedders. N and P are routinely monitored at WWTP Dokhaven, while ammonium, the main fraction of N in WWTP influent, can be considered to be a very good biomarker (Choi et al. (2018), and has been used as a population normaliser in recent studies (e.g. Aberi et al., 2021). For the main summer holiday period (15 July 2021–15 August 2021) we calculated a decrease of 14 % in the median daily crAssphage load relative to the annual median daily crAssphage load on DWF days, while the decrease in P load was 11 % and the decrease in N load was 8 %. As crAssphage is only found in domestic wastewater, while N and P may also originate from industrial sources (Zessner and Lindtner, 2005), a higher reduction in the crAssphage load compared to the nutrient load was to be expected. This illustrates the potential of crAssphage as a biomarker for temporal variations in the number of shedders.
3.5. CrAssphage shedding: Individual versus population shedding
In addition to the population shedding from catchment areas, crAssphage shedding of individuals was studied. Stool samples were collected from SARS-CoV-2 positive adults (n = 221) that notified their general practitioner of a positive SARS-CoV-2 test. The crAssphage concentrations in these stool samples showed a very large variation, spanning >10 log10 units, see Fig. 8 . A small percentage (n = 10 or 4.5 %) had no detectable (<1090 gc/ml faeces) crAssphage in their stool sample. In the positive stool samples, a bimodal distribution was observed, with peaks around 104.5 and 109.3 gc/ml faeces.
Fig. 8.
Distribution of crAssphage concentration in human stool samples (n = 221) from the Rotterdam Rijnmond area. Data below the detection limit (n = 103 gc/ml) is not shown in the graph. Normal distributions are fitted through the log10-transformed concentrations in stool samples of low shedder (<107.4 gc/ml; mean: 104.5; standard deviation 100.82; in black) and high shedder (≥107.4 gc/ml; mean: 109.3; standard deviation 100.96; in red).
The arithmetic average concentration of crAssphage in stool samples is 2.64 × 1010 GC/ml. This average was largely determined by a single stool sample that contained an extremely high concentration of 1013gc/ml). The high concentration was confirmed by re-analysis of the sample. To be less dependent on data from a single stool sample, the shedding data were fitted to two separate Normal distribution after log-transformation, one for the low shedding part of the population (66.1 %, including the non-detects), with an average of 104.5 GC/ml and a standard deviation of 100.82 GC/ml, and one for the high shedding part (33.9 %, with an average of 109.3 GC/ml and a standard deviation of 100.96 GC/ml. In order to compare the crAssphage load in stool samples and in sewage, the daily load in stool has been calculated by multiplying the measured concentration in the stool samples with an average daily stool production of 128 g wet weight (≈ml) per person per day (pppd) (Rose et al., 2015). The resulting calculated average crAssphage load of the stool sample population would be 8.7 × 1010 GC pppd. This is in line with the median per capita crAssphage load derived from the measurements in the Rotterdam wastewater (1.2–1.6 * 1011 gc/(cap*day)). The two values being of the same order of magnitude confirms there is no significant decay of crAssphage in the sewer environment (Ballesté et al., 2019).
Compared to the variability of crAssphage shedding per person, the observed crAssphage concentration in wastewater was very comparable between different city areas and over time. This indicates that, even though the variation in crAssphage shedding between individuals is very high, the ‘population shedding’ is fairly constant in the population sizes of this project from 5.595 and above. Although crAssphage has been shown to reside in the human gut for months (Siranosian et al., 2020; Honap et al., 2020), little is known about the crAssphage shedding dynamics per person over time. A longitudinal study in three infants showed considerable variation over the first year of life (Taboada et al., 2021), but a study among ten healthy adults showed that the human gut virome composition is personalized and relatively stable over time, including for crAssphage (Shkoporov et al., 2019). The high variability in crAssphage concentration in stool samples observed in this study, would imply that crAssphage would become less suitable as normaliser in small populations. More research on smaller populations is required to answer this question.
3.6. Normalisation without flow
For locations where flow measurements are not available, an alternative normalisation approach is developed using EC and crAssphage concentration measurements only. The developed “no-Q”-normalisation approach is given in eq. (6):
| (6) |
with V DWF,ref,noQ = estimated mean wastewater volume during dry weather flow, using crAssphage;
Vddwf = estimated domestic dry weather flow (# inhabitants * 0,12 m3 per capita per day);
ECDWF,ref,noQ = estimated mean EC concentration during dry weather flow;
ECsample = measured EC in 24 h composite sample.
Similar to eqs. (3), (4), (5) the first term of the equation (V DWF,ref,noQ / V ddwf) describes the dilution of domestic wastewater during dry weather, and the second term (EC DWF,ref,noQ / EC sample) the dilution during wet weather. The difference, however, lies in the way the dry weather reference values (V DWF,ref,noQ and EC DWF,ref,noQ) are calculated. To describe the wet weather dilution EC measurements were selected as these proved to be a better descriptor of this dilution process than crAssphage. To derive a dry weather reference value without flow information (EC DWF,ref,noQ), we used the 40th percentile value of all EC measurements (in contrast to the earlier method of using only values associated with dry weather flow volumes). For catchment WWTP Dokhaven INF2 this gave a reference value of 1.115 μS/cm (Fig. 9 ). This value differed slightly (−3 %) from the reference value determined with the EC-based approach (1.155 μS/cm) using only EC values associated with dry weather flows.
Fig. 9.
EC values of wastewater samples from catchment area WWTP Dokhaven INF2 and dry weather reference values with (ECDWF,ref) and without (ECDWF,ref,noQ) flow information.
To describe the dry weather dilution, measured crAssphage concentrations were combined with the number of inhabitants in the catchment areas (CBS, 2020) (# inh) and a constant value for shedded crAssphage load per capita per day (1.85 * 1010). The ‘expected’ daily crAssphage load from a catchment area was calculated (eq. 8a). Then, given the measured crAssphage concentration, this expected load was used to estimate the associated volume of wastewater in 24 h (V 24h,est) for each wastewater sample following eq. (8b). Finally, the time-series of estimated 24 h wastewater volumes was used to derive a reference dry weather value (V DWF,ref,noQ) using the 40th-percentile value of that time-series.
| (7a) |
| (7b) |
with crAss (load) = estimated daily crAssphage load from a catchment area;
# inh = number of inhabitants/shedders in an area, based on Municipal Administration;
crAss (daily load per capita) = constant value (1.85 * 1010), see results;
crAsssample(conc.) = measured crAssphage concentration in 24 h composite sample;
V24h,est = estimated 24 wastewater volume.
The expected daily crAssphage load from the WWTP Dokhaven INF2 catchment area was calculated to be 3.8 * 1015 gc/day (27.044 inhabitants × 1.4 * 1011 daily load per capita). Using this (fixed) load and the (varying) measured crAssphage concentration per wastewater sample, a 24 h wastewater volume was estimated for each sample (V 24h,est) using eq. 7b. Comparison of estimated wastewater volumes with the actual measured volumes (see Fig. 10 ) showed a reasonable fit for most samples, but also occasional large differences between estimated and measured values. More specifically, of all 1138 samples from the nine catchment areas 20 %–35 % showed a difference smaller than 10 %, and 80 %–90 % of all samples showed a difference smaller than 50 % between estimated and measured wastewater volumes (for details, see S7b).
Fig. 10.
Measured and estimated 24 h wastewater volumes of catchment WWTP Dokhaven INF2 (upper graph) and their relative differences (lower graph).
Finally, using the time-series of estimated 24 h wastewater volumes (V 24h,est) the reference dry weather volume was derived using the 40th-percentile of this time-series. For catchment WWTP Dokhaven INF2 this gave a reference value (V DWF,ref,noQ) of 8442 m3/d, about 13 % less than the reference value for the Q-based approach (V DWF,ref = 9754 m3/d). For other catchment areas reference values differed between 1 % and 15 %.
Finally, we calculated the “no-Q" multiplications factors using eq. 6. Fig. 11 shows the distribution of factors per catchment area as well as a comparison with the Q-based and EC-based normalisation factors (as presented in Fig. 2, Fig. 3). The width of the range of no-Q multiplication factors was very similar to the EC-based ranges. This is in line with the small differences (1–3 %) between the dry weather reference values for EC as determined wíth (EC DWF,ref) and without (EC DWF,ref,noQ) flow information. As a result, the second terms in eqs. (4), (6) (representing the wet weather dilution) did not differ much.
Fig. 11.
Boxplot with Q-based (left), EC-based (middle) and noQ-based (right) multiplication factors per catchment area. Each boxplot gives the minimum, 25th percentile, the median, the 75th percentile and the maximum value.
The dry weather dilution, on the other hand, did show larger differences for some catchment areas (WWTP Dokhaven INF2, INF4 and ps Katendrecht). For catchment WWTP Dokhaven INF2, for instance, the dry weather reference values estimated based on crAssphage (V DWF,ref,noQ) was 13 % smaller than the reference based on measured wastewater volumes (V DWF,ref) resulting in ditto smaller “no-Q” multiplication factors. For WWTP Dokhaven INF4 the difference is +15 %, resulting in overall higher “no-Q” multiplication factors.
The normalisation method that can be applied in absence of flow data (a frequent situation for sewer surveillance more upstream in a catchment), named ‘no-Q normalisation’, provided results comparable to the traditional flow based normalisation. The presented no-Q normalisation combines the well know potential of EC to describe dilution during wet weather flow (WWF) and the potential of crAssphage to describe the dilution during DWF due to e.g. industrial discharges and extraneous waters. The largest uncertainty in the no-Q approach is related to the estimation of the average daily crAssphage load, which is based on the number of inhabitants and a fixed daily load per capita. The latter may be uncertain for smaller populations than the smallest catchment (5595 inhabitants) of this study.
4. Conclusions
Wastewater surveillance has evolved rapidly during the COVID-19 pandemic as an important source of information on the SARS-CoV-2 circulation. The long term SARS-CoV-2 monitoring project in the Rotterdam Rijnmond region compared three normalisation methods using flow, electrical conductivity and crAssphage. Based on the findings in this project, we conclude the following:
Unnormalised SARS-CoV-2 sewage data may misrepresent the actual short-term trends of SARS-CoV-2 circulation in the population due to the impact of rain and snowmelt in a combined sewer network. Unnormalised data do show similar long-term trends as normalised data, but it is the short-term (1–2 weeks) trends are the most relevant to support public health actions to limit the transmission of the virus and where discrepancies were considerable.
Flow normalisation, being the standard in sewage surveillance, is an appropriate normaliser as long as the basic assumption that the sample taken is representative of 24 h of shedding of the population sampled, is not violated. Pump failures have been demonstrated to violate this assumption. Moreover, flow normalisation is not suitable to back calculate the number of shedders.
EC normalisation yielded comparable results as flow normalisation in terms of being able to calculate the dilution due to rainfall. EC was not an appropriate normaliser in case of time varying disturbing factors, such as road de-icing and intrusion of brackish groundwater.
CrAssphage could be used as a quantitative biomarker to normalise the population of shedders. Our results showed that despite inherent variation, errors in the number of inhabitants of 20 % s and a time varying number of shedders during summer holidays of 15 % could be detected. Significant deviations in recovery efficiency of the laboratory methods could also be detected. The high variation in crAssphage shedding per person observed in stool samples (n = 221) suggests that the potential of crAssphage as a quantitative biomarker for normalisation of wastewater samples for populations smaller than 5595 needs to be subject of further research.
The parallel use of the three normalisation methods, as applied in this study, provides a very strong quality check of the wastewater surveillance data. Differences between the three methods helped to identify issues with sampling, lab analysis or specific situations such as road de-icing.
Moreover, in absence of flow data, crAssphage and EC offer an appropriate alternative to normalise samples taken as part of wastewater surveillance, where the strength of EC to capture dilution due to rainfall and the strength of crAssphage to capture differences in the number of shedders are combined.
CRediT authorship contribution statement
Jeroen Langeveld: Writing – original draft, Conceptualization, Methodology. Remy Schilperoort: Writing – original draft, Formal analysis, Visualization. Leo Heijnen: Investigation. Goffe Elsinga: Investigation. Claudia E.M. Schapendonk: Resources. Ewout Fanoy: Resources, Funding acquisition. Evelien I.T. de Schepper: Resources. Marion P.G. Koopmans: Supervision. Miranda de Graaf: Writing – review & editing. Gertjan Medema: Writing – review & editing, Conceptualization, Methodology, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are very grateful for the assistance of the Water Authorities and WWTP and pumping station operators of Waterschap Hollandse Delta and municipality of Rotterdam, staff of IMD for installing and maintaining the autosamplers and sampling specialists of AQUON, Toke Mulder – van Kempen and Eline Hoogteijling for assisting with the inclusion of the patients for the faecal sampling study. This study was financed by STOWA, TKI Watertechnology in collaboration with Erasmus Foundation, Adessium Foundation, European Union's Horizon H2020 grant VEO (grant no. 874735), Ministry of Health, Welfare and Sport, H2020 and Waterboards Waterschap Hollandse Delta, Hoogheemraadschap van Delfland and Hoogheemraadschap Schieland en Krimpenerwaard. The authors would like to thank all the GP-practices and patients that contributed to the faecal sampling study.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.161196.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Aberi P., Arabzadeh R., Insam H., Markt R., Mayr M., Kreuzinger N., Rauch W. Quest for optimal regression models in SARS-CoV-2 wastewater based epidemiology. Int. J. Environ. Res. Public Health. 2021;18(20):10778. doi: 10.3390/ijerph182010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O'Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2020;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Ru P., Pan X., Bohrerova Z., Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesté E., Pascual-Benito M., Martín-Díaz J., Blanch A.R., Lucena F., Muniesa M., Jofre J., García-Aljaro C. Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 2019;155:233–244. doi: 10.1016/j.watres.2019.02.042. [DOI] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Crank K., Greaves J., North D., Wu Z., Bibby K. Cross-assembly phage and pepper mild mottle virus as viral water quality monitoring tools—potential, research gaps, and way forward. Curr. Opin. Environ. Sci. Health. 2020;16:54–61. doi: 10.1016/j.coesh.2020.02.001. [DOI] [Google Scholar]
- Castiglioni S., Thomas K.V., Kasprzyk-Hordern B., Vandam L., Griffiths P. Testing wastewater to detect illicit drugs: state of the art, potential and research needs. Sci. Total Environ. 2014;487:613–620. doi: 10.1016/j.scitotenv.2013.10.034. [DOI] [PubMed] [Google Scholar]
- CBS Central Bureau of Statistics in the Netherlands. 2020. https://cbsinuwbuurt.nl/#sub-buurten2020_aantal_inwoners (accessed March 2022)
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 2018;2018(105):453–469. [Google Scholar]
- Crank K., Xiang L., North D., Bonanno Ferraro G., Iaconelli M., Mancini P., La Rosa G., Bibby K. CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116161. [DOI] [PubMed] [Google Scholar]
- Davo L., Seguí R., Botija P., Beltr M.J., Albert E., Torres I., Lopez-Fernandez P.A., Ortí R., Maestre J.F., Sanchez G., Navarro D. Early detection of SARS-CoV-2 infection cases or outbreaks at nursing homes by targeted wastewater tracking. Clin. Microbiol. Infect. 2021;27(2021):1061–1063. doi: 10.1016/j.cmi.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graaf M., Langeveld J., Post J., Balmont C.C., Franz E., Izquirdo-Lara R.W., Elsinga G., Heijnen L., Been F., Beek J.van, Schilperoort R., Vriend R., Fanoy E., de Schepper E.I.T., Koopmans M.P.G., Medema G.J. medRxiv; 2022. Capturing the SARS-CoV-2 Infection Pyramid Within the Municipality of Rotterdam. 2022.06.27.22276938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.A., Vega A.A., Norman H.M., Ohaeri M., Levi K., Dinsdale E.A., Dutilh B.E.… Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019;4(10):1727–1736. doi: 10.1038/s41564-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Banks A., Li J., Jiang G., Lai F.Y., Mueller J.F., Thai P.K. Evaluation of in-sewer transformation of selected illicit drugs and pharmaceutical biomarkers. Sci. Total Environ. 2017;609:1172–1181. doi: 10.1016/j.scitotenv.2017.07.231. [DOI] [PubMed] [Google Scholar]
- Gasperi J., Sebastian C., Ruban V., Delamain M., Percot S., Wiest L., Mirande C., Caupos E., Demare D., Diallo Kessoo M., Saad M., Schwartz J.J., Dubois P., Fratta C., Wolff H., Moilleron R., Chebbo G., Cren C., Millet M., Barraud S., Gromaire M.C. Micropollutants in urban stormwater: occurrence, concentrations, and atmospheric contributions for a wide range of contaminants in three french catchments. Environ. Sci. Pollut. Res. 2014;21:5267–5281. doi: 10.1007/s11356-013-2396-0. [DOI] [PubMed] [Google Scholar]
- Graber T.E., Mercier E., Bhatnagar K., Fuzzen M., D'Aoust P.M., Hoang H.D., Tian X., Towhid S.T., Plaza-Diaz J., Eid W., Alain T., Butler A., Goodridge L., Servos M., Delatolla R. Near real-time determination of B.1.1.7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Elsinga G.S., de Graaf M., Molenkamp R., Koopmans M.P.G., Medema G.J. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., McDonald J.E., Malham S.K., Jones D.L. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Stange C., Suhrborg R., Wurzbacher C., Drewes J.E., Tiehm A. SARS-CoV-2 wastewater surveillance in Germany: long-term PCR monitoring, suitability of primer/probe combinations and biomarker stability. Water Res. 2021;210 doi: 10.1016/j.watres.2021.117977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honap T.P., Sankaranarayanan K., Schnorr S.L., Ozga A.T., Warinner C., et al. Biogeographic study of human gut-associated crAssphage suggests impacts from industrialization and recent expansion. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0226930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., Bänziger C., Devaux A.J., Stachler E., Caduff L., Cariti F., Corzón A.T., Fuhrmann L., Chen C., Jablonski K.P., Nadeau S., Feldkamp M., Beisel C., Aquino C., Stadler T., Ort C., Kohn T., Julian T.R., Beerenwinkel N. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat. Microbiol. 2022;7(8):1151–1160. doi: 10.1038/s41564-022-01185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A.E., Walters M.S., Jennings W.C., Fugitt R., LaCross N., Mattioli M., Marsh Z.A., Roberts V.A., Mercante J.W., Yoder J., Hill V.R. Using wastewater surveillance data to support the COVID-19 response - United States, 2020–2021. Morb. Mortal. Weekly Rep. 2021;70(36):1242–1244. doi: 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay M.A., Dittmer U., Steinmetz H. Organic micropollutants discharged by combined sewer overflows - characterisation of pollutant sources and stormwater-related processes. Water Res. 2016;104:82–92. doi: 10.1016/j.watres.2016.07.068. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G.J., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G.J., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mulder M., van Voorthuizen E., Langeveld J. STOWA; Amersfoort, the Netherlands: 2020. Verwijdering van organische microverontreinigingen (in Dutch: Removal of organic micropollutants). STOWA Rapport 2020-06. [Google Scholar]
- NEN . 2019. Nederlandse Norm 6600-1:2019. Water - Monsterneming - Deel 1: Afvalwater (in Dutch: Dutch Standard 6600-1:2019. Water - Sampling - Part 1: Wastewater) [Google Scholar]
- Ort C., Lawrence M.G., Reungoat J., Mueller J.F. Sampling for PPCPs in wastewater systems: comparison of different sampling modes and optimization strategies. Environ. Sci. Technol. 2010;44(2010):6289–6296. doi: 10.1021/es100778d. [DOI] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Chagas do Vale V.H., Braz R.M.S., Maranhão A.G., Miagostovich M.P., de Andrade J.d.S.R. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Research. 2021;191:116810. doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Einerhand A.W.C., Taminiau L.J.A., van Houdt M., Bouma J., Raatgeep R.H., Büller H.A., de Haan C.A.M., Rossen J.W.A. Cyclooxygenase activity is important for efficient replication of mouse hepatitis virus at an early stage of infection. Virol. J. 2007;4:55. doi: 10.1186/1743-422X-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIONED . Stichting RIONED; Ede, the Netherlands: 2013. Riolering in Beeld. Benchmark Riolering 2013 (in Dutch: Basic Sewer Information in the Netherlands ) [Google Scholar]
- Rios G., Lacoux C., Leclercq V., Diamant A., Lebrigand K., Lazuka A., Soyeux E., Lacroix S., Fassy J., Couesnon A., Thiery R., Mari B., Pradier C., Waldmann R., Barbry P. Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing. Lancet Reg. Health Eur. 2021;10 doi: 10.1016/j.lanepe.2021.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Rasero F.J., Moya Ruano L.A., Rasero Del Real P., Cuberos Gómez L., Lorusso N. Associations between SARS-CoV-2 RNA concentrations in wastewater and COVID-19 rates in days after sampling in small urban areas of Seville: a time series study. Sci. Total Environ. 2022;806(Pt 1) doi: 10.1016/j.scitotenv.2021.150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.A., Loveless T.B., Kapcia J., III, Adams E.D., III, Steele J.A., III, Zimmer-Faust A.G., III, Langlois K., III, Wanless D., III, Griffith M., III, Mao L., III, Chokry J., III, Griffith J.F., III, Whiteson K.L., III RNA viromics of southern california wastewater and detection of SARS-CoV-2 single-nucleotide variants. Appl. Environ. Microbiol. 2021;87(23) doi: 10.1128/AEM.01448-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilperoort R.P.S., Dirksen J., Langeveld J.G., Clemens F.H.L.R. Assessing characteristic time and space scales of in-sewer processes by analysis of one year of continuous in-sewer monitoring data. Water Sci. Technol. 2012;66(8):1614–1620. doi: 10.2166/wst.2012.115. [DOI] [PubMed] [Google Scholar]
- Shkoporov A.N., Clooney A.G., Sutton T.D.S., Ryan F.J., Daly K.M., Nolan J.A., McDonnell S.A., Khokhlova E.V., Draper L.A., Forde A., Guerin E., Velayudhan V., Ross R.P., Hill C. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–541.e5. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- Siranosian B.A., Tamburini F.B., Sherlock G., Bhatt A.S. Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat. Commun. 2020;11(1):280. doi: 10.1038/s41467-019-14103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative crAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51(16):9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stikkers A.B. Delft University of Technology; 2022. Monitoring of SARS-CoV-2 From Wastewater Factors Affecting the Representativeness of Wastewater-based Epidemiology for the Monitoring of the SARS-CoV-2 Virus. MSc thesis. [Google Scholar]
- Sweetapple C., Melville-Shreeve P., Chen A.S., Grimsley J.M.S., Bunce J.T., Gaze W., Fielding S., Wade M.J. Building knowledge of university campus population dynamics to enhance near-to-source sewage surveillance for SARS-CoV-2 detection. Sci. Total Environ. 2021;806(Pt 1) doi: 10.1016/j.scitotenv.2021.150406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada B., Morán P., Serrano-Vázquez A., Iša P., Rojas-Velázquez L., Pérez-Juárez H., López S., Torres J., Ximenez C., Arias C.F. The gut virome of healthy children during the first year of life is diverse and dynamic. PLoS One. 2021;16(4) doi: 10.1371/journal.pone.0240958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S., Sherchan S.P., Haramoto E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 2020;10:3616. doi: 10.1038/s41598-020-60547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosse M. 2013. Rioolvreemd water rioolwaterzuiveringsinrichting Dokhaven (in Dutch: Extraneous Waters at Wastewater Treatment Plant Dokhaven). Technical Report of RoyalHaskoningDHV. [Google Scholar]
- Weiß G., Brombach H., Haller B. Infiltration and inflow in combined sewer systems: long-term analysis. Water Sci. Technol. 2002;45(7):11–19. doi: 10.2166/wst.2002.0112. [DOI] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Topol A., Knudson A., Simpson A., White B., Vugia D.J., Yu A.T., Li L., Balliet M., Stoddard P., Han G.S., Wigginton K.R., Boehm A.B. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in northern California shows strong association with COVID-19 incidence. mSystems. 2021;6(5) doi: 10.1128/mSystems.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Shagan M., Lewis Y.E., Kramarsky-Winter E., Weil M., Indenbaum V., Elul M., Erster O., Brown A.S., Mendelson E., Mannasse B., Shirazi R., Lakkakula S., Miron O., Rinott E., Baibich R.G., Bigler I., Malul M., Rishti R., Brenner A., Friedler E., Gilboa Y., Sabach S., Alfiya Y., Cheruti U., Moran-Gilad J., Berchenko Y., Bar-Or I., Kushmaro A. City-level SARS-CoV-2 sewage surveillance. Chemosphere. 2021;283 doi: 10.1016/j.chemosphere.2021.131194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zessner M., Lindtner S. Estimations of municipal point source pollution in the context of river basin management. Water Sci. Technol. 2005;52(9):175–182. doi: 10.2166/wst.2005.0313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.