Abstract
Breast cancer is the most common malignancy and the second leading cause of cancer-related death in women. Recent studies have indicated that aberrant activation of Rho GTPases relates to the malignant properties of breast cancer cells. As the guanine nucleotide exchange factor of Rho GTPases, the role of ECT2 (epithelial cell transforming 2) in breast cancer is still unclear. Tissue microarrays and multiple public databases were utilized to investigate the relationship between ECT2 level and clinical-pathological features of breast cancer patients. Kaplan Meier-plotter online tool and tissue microarray with survival information were used to investigate the predictive value for breast cancer. Here, we found increased ECT2 level was highly associated with advanced TNM stage, poor differentiation, and loss of hormone receptors of breast cancer. Gene expression profile showed that ECT2 level was closely correlated to cell-proliferation-associated pathways. Integration analysis using public databases and tissue microarray indicated that high ECT2 was an adverse prognostic factor for breast cancer patients. We believe the ECT2 level might be a valuable complement for commercially available predictors such as the 21 genes test. Furthermore, ECT2 would be a novel target for drug development for breast cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40164-022-00361-3.
Keywords: ECT2, Breast cancer, Predictive biomarker, Rho GTPases, Prognostic factor
Introduction
Breast cancer is the most common malignancy and the second leading cause of cancer-related death in women [1–4]. According to the level of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor 2 (Her2), breast cancers are classified into multiple subtypes: Luminal, Her2 overexpression, Basal-like, and Normal-like [5]. Propelled by the fundamental investigation of tumor biology, breast cancer therapeutics have progressed substantially in the past decades [6]. Locoregional tumor load, together with molecular subtype, determines the therapeutic regimen of breast cancer [7–10]. Further exploration of molecular mechanisms contributing to tumor progression is meaningful to the development of novel therapies [11–15]. Notably, immunotherapy, especially immune checkpoint inhibitors, brings new hope to the treatment of breast cancer [16–20]. Recent studies have indicated that aberrant activation of Rho GTPases relates to the malignant biological behavior of breast cancer cells [21–23].
ECT2 (epithelial cell transforming 2) is a guanine nucleotide exchange factor of Rho GTPases (e.g. RhoA, Rac1, and Cdc42) [24]. ECT2 converts the status of Rho GTPases from inactive state to active state via replacing bound GDP with GTP, further promoting actin remodeling [25]. It has been verified that ECT2 mainly plays a vital role in cytokinesis. In M phase, ECT2 is recruited to central spindle and activates Rho signaling pathway, subsequently inducing contractile ring formation and contraction [26]. Loss of ECT2 interferes with cytokinesis without remarkable effects on mitosis, which ultimately promotes the generation of binucleate cells [27].
Besides normal cellular activity, ECT2 also participates in malignant transformation, tumor initiation, and metastasis [28]. The tumorigenic role of ECT2 changes along with its subcellular location. Full-length ECT2 protein contains a nuclear localization signal (N-terminal) and dominantly distributes in the nucleus [29]. Independent of cytokinesis regulation, nuclear ECT2 promotes malignant transformation in other manners [30]. In lung adenocarcinoma cells, nuclear ECT2 recruits and activates Rac1, which increases ribosome biogenesis and supports transformed growth (anchorage-independent growth) [31, 32]. Besides, in ovarian cancer cells, ECT2 is retained in cytoplasm by binding to protein kinase Cι-Par6α complex [30]. Cytoplasmic ECT2-protein kinase Cι-Par6α complex subsequently activates Rac1-Pak-Mek-Erk signaling pathway, which promotes cell cycle progression and proliferation [30]. Moreover, explorations in human hepatocellular carcinoma cell confirmed ECT2 as a multiple-functional oncogenic protein that upregulates Rho-Erk signal, promotes cell proliferation, suppresses apoptosis, and induces distant metastasis [33]. It has been detected that ECT2 was overexpressed in various cancers, including non-small lung cell cancer, glioblastomas, and prostate cancer [31, 34–36]. Generally, high ECT2 level is an unfavorable factor for patient prognosis [37, 38]. However, there are rare studies investigating the role of ECT2 in breast cancer.
In this study, by combining samples from tissue microarrays and gene expression data from public databases, we retrospectively evaluated the expression of ECT2 in breast cancer and non-cancer tissues. Moreover, we explored the relationship between ECT2 abundance and clinic-pathologic features as well as clinical outcomes of patients with breast cancer. Collectively, we explored the involvement of ECT2 in breast cancer development which would be a potential predictive biomarker and treatment target.
Materials and methods
Breast tissue samples
Tissue microarray Br2082a was obtained from Xi’an Alena-bio Ltd which contained 32 metastatic carcinoma, 120 primary carcinoma, 8 fibroadenoma, 16 hyperplasia, 16 inflammation, and 16 adjacent normal breast tissues. However, some clinical-pathological parameters and surviving data of samples were not given in Br2082a. Therefore, another tissue microarray with detailed pathological parameters (HBreD145Su01, Shanghai Outdo Biotech Ltd.) was involved for further analysis. The clinical-pathological features of breast cancer patients in HBreD145Su01 cohorts are shown in Table 1.
Table 1.
Correlations between ECT2 expression and clinic-pathological features of 120 breast cancer patients in immunohistochemistry chip (HBreD145Su01)
| Variables | N | ECT2 expression | P value | |
|---|---|---|---|---|
| < 8 (low expression) | ≥ 8 (high expression) | |||
| Age | 0.224a | |||
| < Median(51) | 57 | 28 | 29 | |
| ≥ Median (51) | 63 | 24 | 39 | |
| Tumor location | 0.961a | |||
| Left | 58 | 25 | 33 | |
| Right | 62 | 27 | 35 | |
| Grade | 0.073a | |||
| Grade 1 | 25 | 16 | 9 | |
| Grade 2 | 90 | 44 | 56 | |
| Stage | 0.043a | |||
| Stage I–II | 75 | 37 | 38 | |
| Stage III–IV | 43 | 13 | 30 | |
| Subtype | 0.0041a | |||
| Luminal | 89 | 46 | 43 | |
| Her2 enriched | 13 | 4 | 9 | |
| Basal | 18 | 2 | 16 | |
aPearson Chi-square test
Public breast cancer datasets acquisition and process
We searched GEO database (http://www.ncbi.nlm.nih.gov/geo/) to select eligible breast cancer datasets for pooled analysis. The searching and selecting strategies followed the methods we previously described [39]. The detailed information of involved datasets in the meta-analysis was listed in Additional file 1: Table S1. Odds ratio (OR) with 95% confidence interval (95% CI) was utilized to assess the correlation between ECT2 mRNA level and clinic-pathological markers. Patient outcomes, including overall survival (OS), relapse-free survival (RFS) and metastasis-free survival (MFS) were evaluated by hazard ratio (HR) and 95% CI. The Stata software (version 12.0) was used in this meta-analysis. Transcriptional profiling of ECT2 gene and other genes was downloaded from TCGA database (https://xenabrowser.net/), processed and analyzed by SPSS software (version 24.0).
Online analysis tool
Online analysis tool Kaplan Meier-plotter could plot Kaplan–Meier survival curve with log-rank test analysis (http://kmplot.com). The background data of Kaplan Meier-plotter are obtained from GEO and TCGA databases. Adopting median ECT2 expression as cutoff value, Kaplan–Meier survival curves were generated and downloaded from this website.
Immumohistochemical staining assay
Immunohistochemical (IHC) staining was conducted following the standard protocol described previously [40]. The specific primary antibody against ECT2 (catalog no. 07-1364; Millipore Corporation, Billerica, MA, USA, 1:150) was used for the IHC assay. The stained images were captured by a light microscope with the image processing system (Sunny Optical, China). The IHC scores were evaluated by two experienced pathologists without patient information. Based on Fromowitz Standard, the multiplication of intensity and proportion of positively stained cancer cells represents the abundance of ECT2. The intensity was scored according to color shade: 0 (no staining), 1 (light yellow), 2 (yellow–brown), and 3 (brown). The proportion of stained cancer cells was scored as: 1 (0–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%).
Bioinformatics analysis
The biological significance of ECT2 for breast cancer was explored using bioinformatics analysis. Based on TCGA and GSE25066 datasets, the enriched Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in high ECT2 tumors were calculated. Pathways or Terms with adjusted P value < 0.05 and fold change > 2 were regarded as statistically significant [18, 41]. Gene Set Enrichment Analysis (GSEA) was performed to explore the enriched pathways in high ECT2 tumors [19, 42]. R software (4.1.2) and packages DESeq2, ggplot2, and ClusterProfile were used in this assay.
Statistical analysis
The Student's t-test was used to compare the difference between two groups with the significance cutoff as 0.05. Correlations were analyzed using Person χ2 test. The cumulative survival was analyzed using Kaplan–Meier curve with log-rank test. Univariate and multivariate analyses were conducted by the Cox proportional hazards regression model.
Results
Increased ECT2 expression in breast cancer
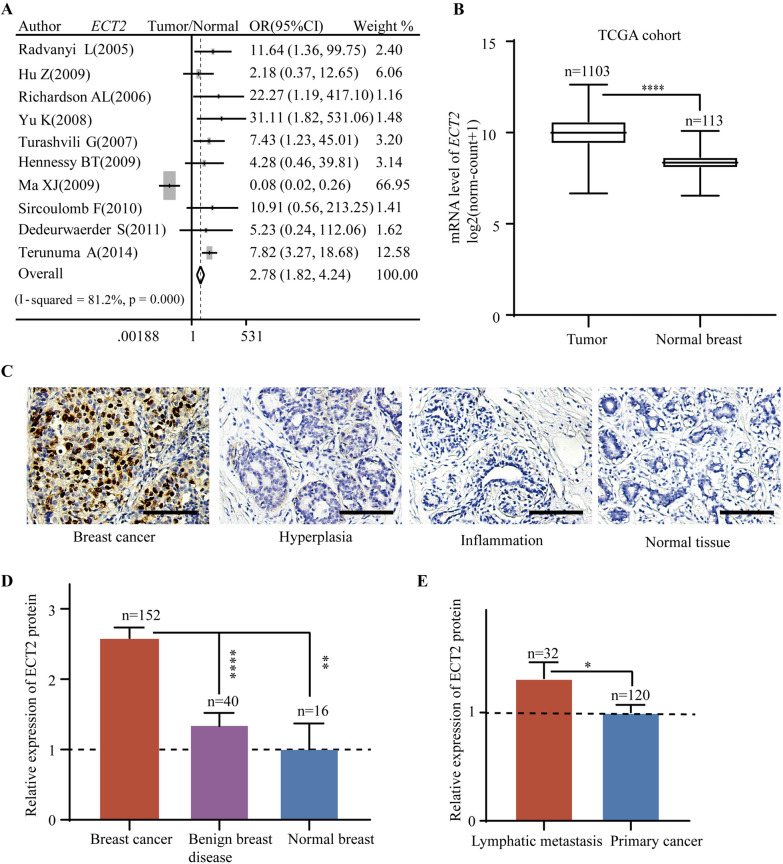
To explore the role of ECT2 in breast cancer, we first assessed the ECT2 expression in breast cancers and normal breast tissues. Pooled analysis of GEO datasets demonstrated that ECT2 mRNA level was upregulated in breast cancer tissues relative to normal tissues (OR = 2.78, 95%CI = 1.82–4.24) (Fig. 1A). ECT2 mRNA data from TCGA showed significantly elevated ECT2 mRNA in breast cancer tissues as well (P < 0.0001) (Fig. 1B). Breast samples of the tissue microarray contained breast cancer, benign disease, and normal breast tissues. IHC results indicated that ECT2 protein was increased in breast cancer tissues compared with non-cancer tissues (Fig. 1C, D). Among 152 breast cancer tissues, samples from lymphatic metastasis exhibited higher ECT2 expression relative to primary breast cancer (Fig. 1E).
Fig. 1.
The expression level of ECT2 in breast cancer, benign disease, and normal breast tissues. A Pooled analysis of GEO datasets showing ECT2 mRNA level in breast cancer and normal tissues. B TCGA breast cancer dataset showing the level of ECT2 mRNA level in breast cancer and normal tissues. C The representative immunohistochemical staining images showing ECT2 protein expression in breast cancer and normal tissues. D Histogram showing ECT2 protein expression in breast cancer, benign disease, and normal breast tissues. E Histogram showing ECT2 protein expression in lymphatic metastasis and primary breast cancer tissues. Scale bar: 100 μm
Correlations between ECT2 expression and clinic-pathologic features of breast cancer patients
In order to further verify the clinical significance of increased ECT2 for breast cancer patients, we interrogated the relationship between ECT2 expression and clinic-pathologic parameters, including TNM stage (based on tumor size, lymph node involvement and distant metastasis), and molecular subtype.
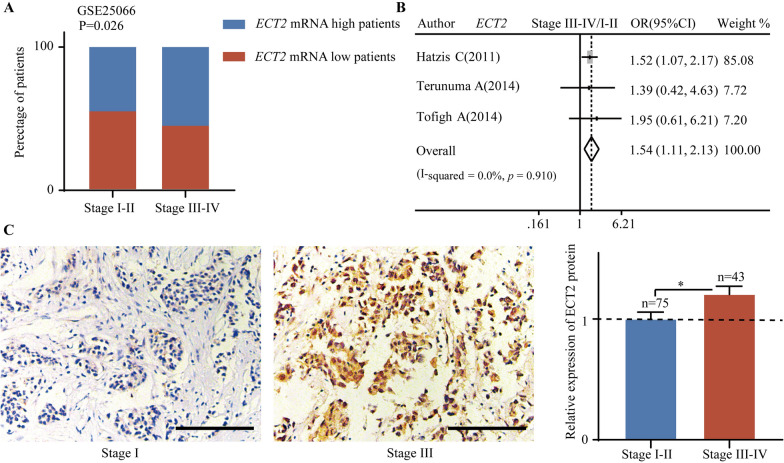
ECT2 expression was related to TNM stage. Gene expression profile from GSE25066 showed that the percentage of patients with upregulated ECT2 mRNA was higher in the stage III–IV group (Fig. 2A). Pooled analysis showed that increased ECT2 mRNA significantly correlated with advanced TNM stage (Pooled OR = 1.54, 95%CI 1.11–2.13) (Fig. 2B). ECT2 protein abundance data from tissue microarray demonstrated that upregulated ECT2 protein correlated with advanced TNM stage (P < 0.05) (Fig. 2C).
Fig. 2.
The correlation between ECT2 expression and TNM stage. A Gene expression profile from GSE25066 showing that the percentage of patients with high ECT2 mRNA and low high ECT2 mRNA in different TNM stage groups. B Pooled analysis showing the correlation between ECT2 mRNA and TNM stage. C Data from tissue microarray showing the relationship between ECT2 protein level and TNM stage. Scale bar: 100 μm
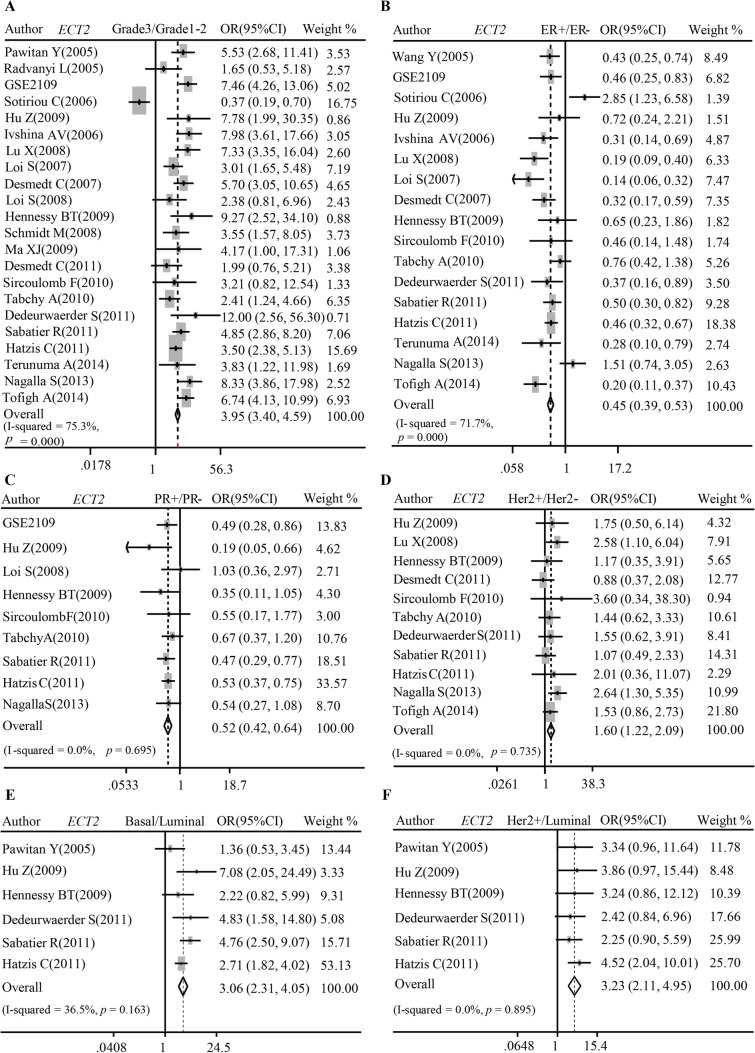
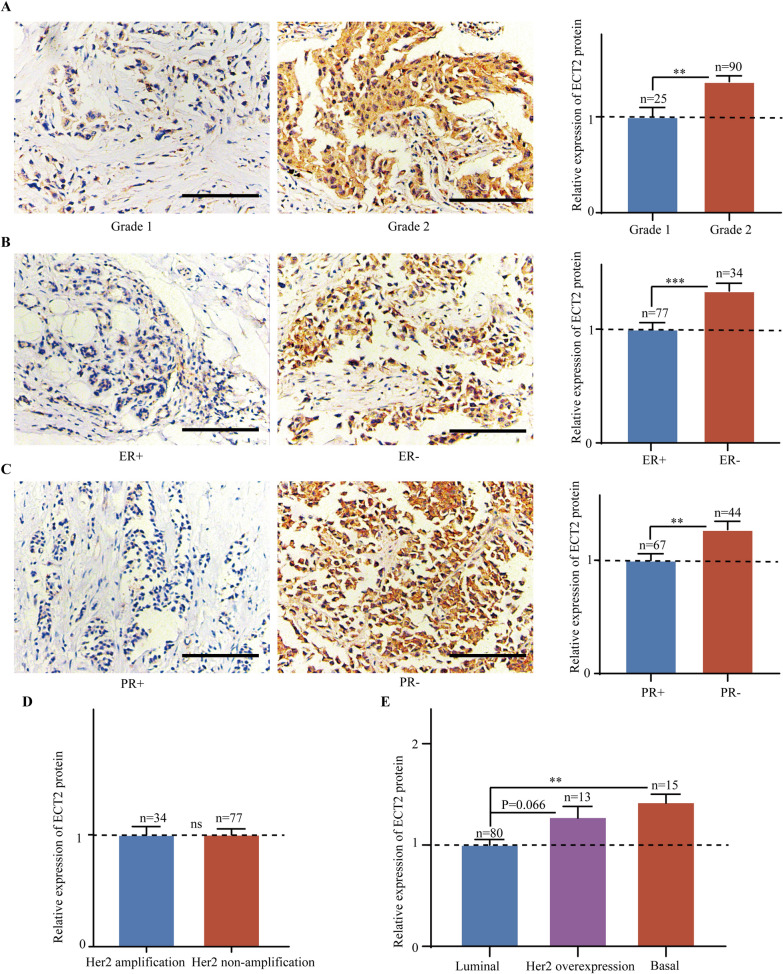
Elevated ECT2 expression was correlated to molecular biomarkers and subtypes of breast cancer. ECT2 manifested remarkably high transcriptional level in poorly differentiated tumor (Pooled OR = 3.95, 95%CI 3.40–4.59) (Fig. 3A) but low transcriptional level in ER + (Pooled OR = 0.45, 95%CI 0.39–0.53) (Fig. 3B) and PR + (Pooled OR = 0.52, 95%CI 0.42–0.64) patients (Fig. 3C). On the contrary, ECT2 mRNA was upregulated in Her2-overexpressed tissue (Pooled OR = 1.60, 95%CI 1.22–2.09) (Fig. 3D), Basal-like subtype (Pooled OR = 3.06, 95%CI 2.31–4.05) (Fig. 3E), as well as Her2 overexpression subtype (Pooled OR = 3.23, 95%CI 2.11–4.95) (Fig. 3F). IHC scoring results showed that the abundance of ECT2 protein was markedly upregulated in poorly differentiated (Fig. 4A), ER- (Fig. 4B), PR- (Fig. 4C) breast cancers. However, there was no significant difference between Her2-amplificated and non-amplificated cancers (Fig. 4D). Additionally, ECT2 protein level was highest in Basal-like subtype breast cancers (Fig. 4E).
Fig. 3.
Pooled analysis showing the correlations between ECT2 mRNA and molecular biomarkers of breast cancer. A Pooled analysis showing the correlations between ECT2 mRNA level and differentiation grade. B Pooled analysis showing the correlations between ECT2 mRNA level and ER status. C Pooled analysis showing the correlations between ECT2 mRNA level and PR status. D Pooled analysis showing the correlations between ECT2 mRNA level and HER2 status. E, F Pooled analysis showing the correlations between ECT2 mRNA level and subtypes of breast cancers
Fig. 4.
Data from tissue microarray showing the relationship between ECT2 protein level and molecular biomarkers of breast cancer. A The representative immunohistochemical staining images showing ECT2 protein expression in Grade 1 and Grade 2 breast cancers. B The representative immunohistochemical staining images showing ECT2 protein expression in ER+ and ER− breast cancers. C The representative immunohistochemical staining images showing ECT2 protein expression in PR+ and PR− breast cancers. D Histogram showing ECT2 protein expression in Her2-amplificated and non-amplificated breast cancers. E Histogram showing ECT2 protein expression in different subtypes of breast cancers. Scale bar: 100 μm
The biological significance of ECT2 for breast cancer
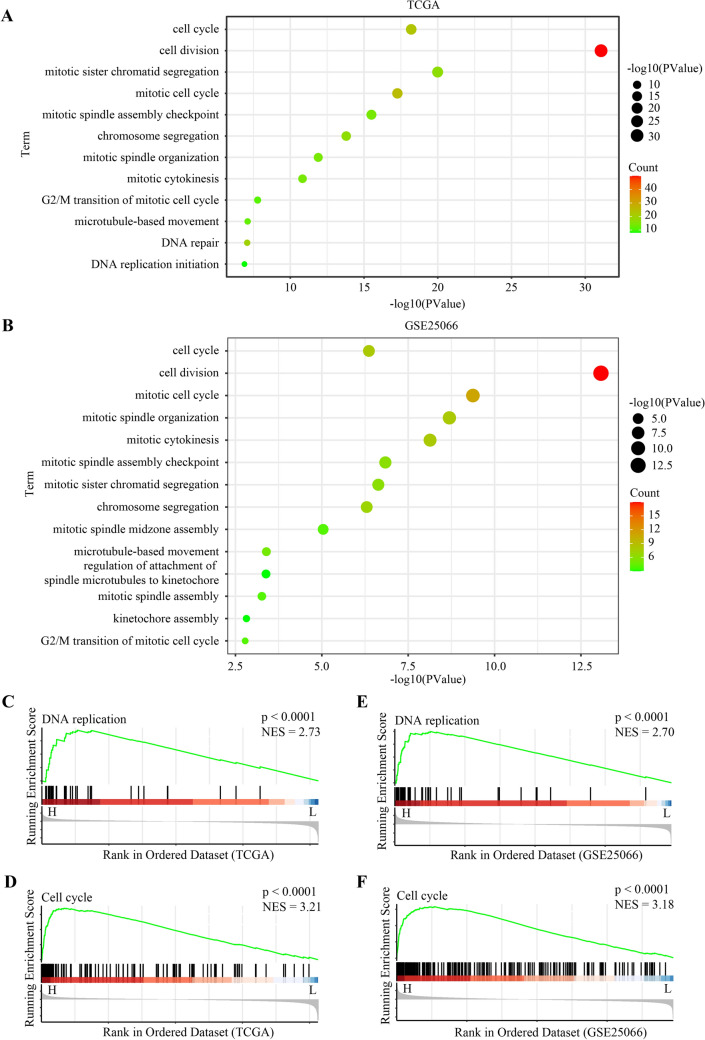
To investigate the biological significance of ECT2 for breast cancer, we performed KEGG and GO enrichment analysis using TCGA and GEO databases. The results showed cell proliferation-associated pathways were significantly enriched in high ECT2 tumors, including cell cycle, cell division, mitotic sister chromatid segregation, mitotic spindle assembly checkpoint, chromosome segregation, mitotic spindle organization, G2/M transition of mitotic cell cycle, DNA replication initiation, regulation of attachment of spindle microtubules to kinetochore, and kinetochore assembly (Fig. 5A, B). The results of GSEA also demonstrated that DNA proliferation and cell cycle were significantly enriched in high ECT2 tumors (Fig. 5C–F).
Fig. 5.
KEGG and GO enrichment analysis using TCGA and GEO databases. A Data from TCGA showing enriched pathways or termed in high ECT2 breast cancers. B Data from GSE25066 showing enriched pathways or termed in high ECT2 breast cancers. C GSEA indicating the enrichment of DNA replication pathway in high ECT2 breast cancers (Based on TCGA). D GSEA indicating the enrichment of cell cycle pathway in high ECT2 breast cancers (Based on TCGA). E GSEA indicating the enrichment of DNA replication pathway in high ECT2 breast cancers (Based on GSE25066). F GSEA indicating the enrichment of DNA replication pathway in high ECT2 breast cancers (Based on GSE25066)
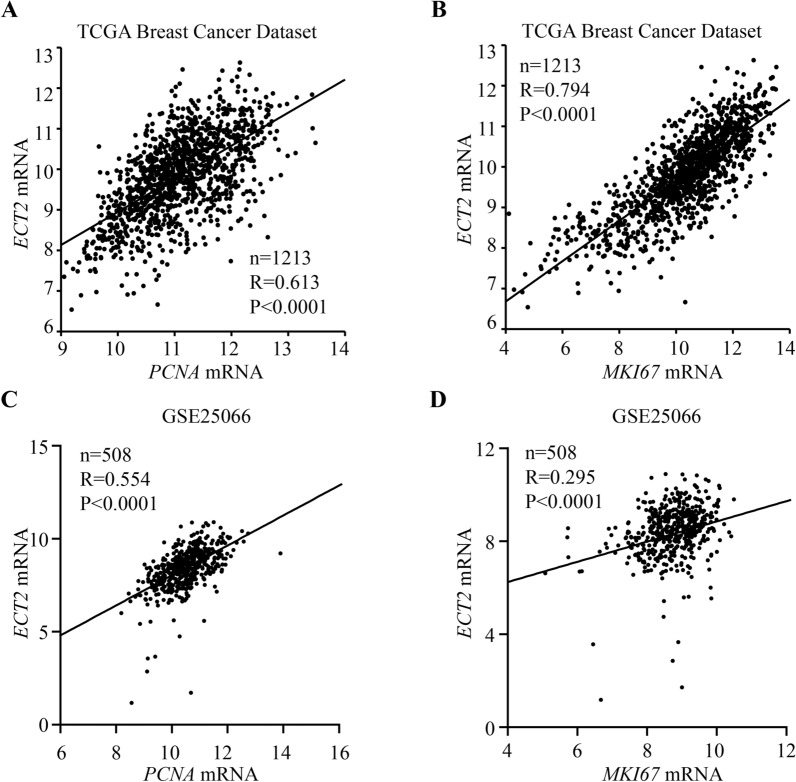
Additionally, as mentioned above, ECT2 plays a vital role in cell proliferation by regulating cytokinesis. PCNA and MKI67 are well-accepted proliferation-associated markers. Therefore, we explored the relationship between ECT2 expression and PCNA and MKI67. We found that ECT2 expression was highly correlated with cell proliferation-associated markers such as PCNA and MKI67 (Fig. 6A–D). Our results indicated ECT2 participated in cell proliferation, which might contribute to the malignant biological properties of breast cancer.
Fig. 6.
The correlation between ECT2 expression and cell proliferation-associated markers. A Data from TCGA showing correlation between ECT2 expression and PCNA level. B Data from TCGA showing correlation between ECT2 expression and MKI67 level. C Data from GSE25066 showing correlation between ECT2 expression and PCNA level. D Data from GSE25066 showing correlation between ECT2 expression and MKI67 level
Elevated ECT2 expression heralded poor prognosis of breast cancer patients
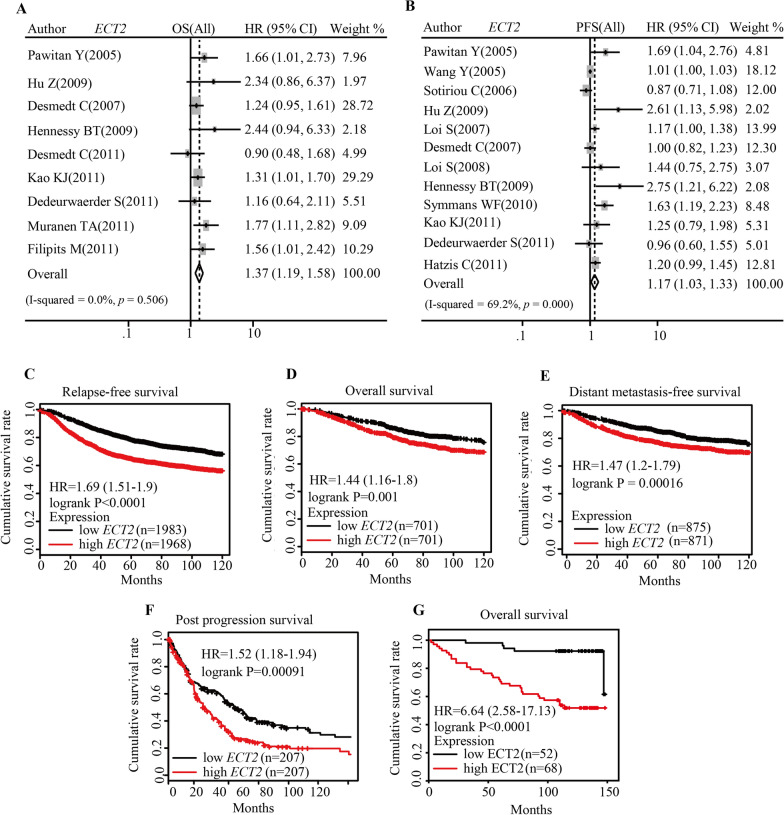
To assess whether ECT2 expression could predict the prognosis of breast cancer patients, we conducted pooled analysis and found that higher ECT2 mRNA level related to shorter OS (Pooled HR = 1.37, 95%CI 1.19–1.58) (Fig. 7A) and RFS (Pooled HR = 1.17, 95%CI 1.03–1.33) (Fig. 7B). Survival analysis by online tool Kaplan Meier-plotter showed elevated ECT2 mRNA level indicated poor prognosis of breast cancer patients (PFS: HR = 1.69, log-rank P < 0.0001; OS: HR = 1.44, log-rank P = 0.001; Distant metastasis-free survival: HR = 1.47, log-rank P = 0.00016; Post progression survival: HR = 1.52, log-rank P = 0.00091) (Fig. 7C–F). Based on patient survival data and ECT2 protein level, survival analysis demonstrated that patients with high ECT2 had shorter OS (HR = 6.64, log-rank P < 0.0001). Cox regression analysis showed that ECT2 expression is an independent prognostic factor for breast cancer patients (Table 2).
Fig. 7.
The predictive value of ECT2 for the prognosis of breast cancer patients. A Pooled analysis showing the relationship between ECT2 mRNA level and overall survival. B Pooled analysis showing the relationship between ECT2 mRNA level and progression-free survival. C Survival curves showing the relationship between ECT2 mRNA level and relapse-free survival (data from Kaplan–Meier plotter). D Survival curves showing the relationship between ECT2 mRNA level and overall survival (data from Kaplan–Meier plotter). E Survival curves showing the relationship between ECT2 mRNA level and distant metastasis-free survival (data from Kaplan–Meier plotter). F Survival curves showing the relationship between ECT2 mRNA level and post-progression survival (data from Kaplan–Meier plotter). G Survival curves showing the relationship between ECT2 protein level and overall survival (data from tissue microarray)
Table 2.
ECT2 expression is an independent prognostic factor for breast cancer patients (HBreD145Su01)
| Variables | Univariate analysis | Variable selection | ||
|---|---|---|---|---|
| HR (95%CI) | P Value | HR (95%CI) | P value | |
| Age | 0.998 (0.970–1.027) | 0.895 | ||
| Tumor location (left vs. right) | 2.077 (0.930–4.638) | 0.074 | ||
| Grade (grade 2 vs. 1) | 1.575 (0.620–3.999) | 0.339 | ||
| TNM stage (III–IV vs. I–II) | 1.979 (0.988–3.966) | 0.054 | ||
| ER (ER+ vs. ER−) | 0.348 (0.157–0.768) | 0.009 | 0.414 (0.185–0.927) | 0.032 |
| PR (PR+ vs. PR−) | 0.385 (0.173–0.858) | 0.020 | ||
| Her2 (Her2+ vs. Her2−) | 0.906 (0.338–2.428) | 0.845 | ||
| ECT2 level (high vs. low) | 6.198 (2.129–18.043) | 0.0001 | 4.899 (1.668–14.385) | 0.004 |
Discussion
In this study, by comprehensive analysis of GEO database, TCGA breast cancer dataset, and samples of tissue microarrays, we found that ECT2 expression (at transcriptional and translation levels) was significantly increased in breast cancers compared with non-cancer tissues. Moreover, increased ECT2 expression was related to clinic-pathologic parameters especially advanced TNM stage. We also found ECT2 was remarkably higher in breast cancers belonging to Her2 overexpression and Basal-like subtypes. In general, elevated ECT2 level was a potential biomarker predicting poor prognosis of breast cancer patients. The oncogenic role of ECT2 could be partly fulfilled by enhancing malignant transformation, invasion, and migration activities.
ECT2 is a vital regulator of cell division via modifying the activation of small GTPases of Rho family (e.g. RhoA, Rac1, and Cdc42) [43]. It has been well established that ECT2 activates Rho-Citron kinase pathway which subsequently phosphorylates the myosin heavy chain kinase, induces the formation of contractile ring, and promotes cytokinesis [44]. Apart from cytokinesis, ECT2 also participates in mitosis by impairing the attachment between mitotic spindles and kinetochores [45]. In situ hybridization confirmed that ECT2 level was a biomarker reflecting the proportion of cells undergoing mitosis [46]. In line with the role of ECT2 in cell proliferation, we also found a positive correlation between ECT2 abundance and proliferation-associated biomarkers. Besides, it has been verified that increased ECT2 abundance directly activates the downstream MAPK signaling pathway, further accelerating cell cycle progression and proliferation [33, 47]. Cell proliferation could be effectively inhibited by interfering ECT2 expression [48–50]. Accumulated ECT2 in multiple cancers indicates hyperactive cell division and proliferation [37, 51]. Intervention targeting factors driving the uncontrolled proliferation and disordered cell cycle have always been a hot topic for developing anti-tumor agents [52–55]. Blocking ECT2 and its downstream signaling pathway would be meaningful to transform tumor cells from hyperactive proliferation towards non-division state.
Mis-localized ECT2 relates to malignant transformation, aberrant cell proliferation, and distant metastasis [56]. In cancer tissues, we found abnormal ECT2 staining in cytoplasm. However, in non-cancer tissue, ECT2 staining was only detected in nuclear. It was reported that the splicing variant of ECT2 lacked nuclear localization signal [57]. Even though the oncogenic role of splicing variant of ECT2 without nuclear localization signal was verified in mice model, human cancer cells express full-length ECT2. It is generally believed that transforming ECT2 variant is not related to human cancers. Other factors influencing the subcellular location of ECT2 such as Protein kinase Cι-Par6α complex, are meaningful to counteract malignant behaviors of breast cancers [31, 58].
Besides promoting cell proliferation, the oncogenic role of ECT2 could be attributed to ECT2-Rho pathway-mediated cellular transformation and metastasis [30]. In mice fibroblast models, cytoplasmic ECT2 showed constitutive guanine nucleotide exchange factor activity and effectively induced transformation, while nuclear ECT2 exhibited no malignant transformation activity [28]. In the interphase, due to containing nuclear localization signal, ECT2 is generally separated in nucleus to avoid the activation of downstream Rho GTPases [59]. However, accumulated ECT2 in the cytoplasm of cancer cell leads to hyperactive Rho GTPases, which promotes epithelial-to-mesenchymal transition, loss of cell polarization, formation of invadopodia/ lamellipodia/ filopodia, and tail retraction [24, 30]. As a result, cancer cells undergo malignant transformation with enhanced capabilities of migration, invasion, and metastasis. We found ECT2 expression was markedly elevated in metastatic breast cancer tissues relative to non-metastatic tissues. It has been documented inhibiting the activity of ECT2-Rho pathway could effectively inhibit breast cancer metastasis via modifying actin cytoskeleton remodeling [60]. Given that distant metastasis, together with recurrence, are major causes of breast cancer-related deaths, upregulated ECT2 signaling is a potential target to inhibit the generation of metastatic lesions.
Collectively, increased ECT2 level is highly associated with advanced TNM stage, poor differentiation, and loss of hormone receptors of breast cancer. Integration analysis using GEO public database and tissue microarray indicates that high ECT2 is an adverse prognostic factor for breast cancer patients. We believe the ECT2 level might be a valuable complement for commercially available predictors such as the 21 genes test. Besides, ECT2 would be a novel target for drug development for breast cancer.
Supplementary Information
Additional file 1: Table S1: Characteristics of studies involved in meta-analysis.
Acknowledgements
Not applicable.
Abbreviations
- CI
Confidence interval
- ECT2
Epithelial cell transforming 2
- ER
Estrogen receptor
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- Her2
Human epidermal growth factor 2
- HR
Hazard ratio
- IHC
Immunohistochemical
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OR
Odds ratio
- OS
Overall survival
- MFS
Metastasis-free survival
- RFS
Relapse-free survival
- PR
Progesterone receptor
Author contributions
MY and DZ conducted the experiments and drafted the manuscript. BS, BZ, MN, and YW participated in the analysis and interpretation of data. KW and ZD designed the work. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81874120 and 82272794).
Availability of data and materials
The TCGA datasets for this study can be found in the https://xenabrowser.net/datapages/. The GSE25066 datasets for this study can be found in the https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse25066.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of the Tongji Hospital of Huazhong University of Science and Technology.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. As the editorial board member of Experimental Hematology & Oncology, KW decleare that he has no competing interests and is excluded from the peer review process. This submission is not given any priority over other manuscripts, and editorial board member status has no bearing on editorial consideration.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ming Yi and Di Zhang are contributed equally to this work
Contributor Information
Ming Yi, Email: 1978135000@qq.com.
Di Zhang, Email: zhangdi1001@zju.edu.cn.
Bin Song, Email: songbintara@163.com.
Bin Zhao, Email: zhaobinxhb@163.com.
Mengke Niu, Email: niumengke9505@163.com.
Yuze Wu, Email: wuyz0304@163.com.
Zhijun Dai, Email: dzj0911@126.com.
Kongming Wu, Email: kmwu@tjh.tjmu.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Yi M, Li T, Niu M, Luo S, Chu Q, Wu K. Epidemiological trends of women's cancers from 1990 to 2019 at the global, regional, and national levels: a population-based study. Biomark Res. 2021;9:55. doi: 10.1186/s40364-021-00310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li N, Deng Y, Zhou L, Tian T, Yang S, Wu Y, et al. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the Global Burden of Disease Study 2017. J Hematol Oncol. 2019;12:140. doi: 10.1186/s13045-019-0828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Li Z, Yan L, Liu Y, Yang H, Li H. Global, regional, and national cancer incidence and death for 29 cancer groups in 2019 and trends analysis of the global cancer burden, 1990–2019. J Hematol Oncol. 2021;14:197. doi: 10.1186/s13045-021-01213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Tian Y, Yuan X, Liu Y, Wu H, Liu Q, et al. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco Targets Ther. 2016;9:431–444. doi: 10.2147/OTT.S97192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 7.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 8.Turner NC, Neven P, Loibl S, Andre F. Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet. 2017;389:2403–2414. doi: 10.1016/S0140-6736(16)32419-9. [DOI] [PubMed] [Google Scholar]
- 9.Pernas S, Tolaney SM. HER2-positive breast cancer: new therapeutic frontiers and overcoming resistance. Ther Adv Med Oncol. 2019;11:1758835919833519. doi: 10.1177/1758835919833519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng F, Wei T, Wang X, Du F, Yue J, Yuan P. Disease-free survival landmark analysis: a potential critical endpoint in triple-negative breast cancer. Exp Hematol Oncol. 2022;11:65. doi: 10.1186/s40164-022-00308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu S, Zhang J, Yan Y, Yao X, Fang L, Xiong H, et al. A novel asymmetrical anti-HER2/CD3 bispecific antibody exhibits potent cytotoxicity for HER2-positive tumor cells. J Exp Clin Cancer Res. 2019;38:355. doi: 10.1186/s13046-019-1354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu T, Yu S, Zhang J, Wu S. Dysregulated tumor-associated macrophages in carcinogenesis, progression and targeted therapy of gynecological and breast cancers. J Hematol Oncol. 2021;14:181. doi: 10.1186/s13045-021-01198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Liu J, Chen J, Zhou Q. The developing landscape of combinatorial therapies of immune checkpoint blockade with DNA damage repair inhibitors for the treatment of breast and ovarian cancers. J Hematol Oncol. 2021;14:206. doi: 10.1186/s13045-021-01218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Criscitiello C, Morganti S, Curigliano G. Antibody-drug conjugates in solid tumors: a look into novel targets. J Hematol Oncol. 2021;14:20. doi: 10.1186/s13045-021-01035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong B, Yi M, Luo S, Li A, Wu K. RDGN-based predictive model for the prognosis of breast cancer. Exp Hematol Oncol. 2020;9:13. doi: 10.1186/s40164-020-00169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17:129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi M, Zhang J, Li A, Niu M, Yan Y, Jiao Y, et al. The construction, expression, and enhanced anti-tumor activity of YM101: a bispecific antibody simultaneously targeting TGF-β and PD-L1. J Hematol Oncol. 2021;14:27. doi: 10.1186/s13045-021-01045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi M, Niu M, Zhang J, Li S, Zhu S, Yan Y, et al. Combine and conquer: manganese synergizing anti-TGF-β/PD-L1 bispecific antibody YM101 to overcome immunotherapy resistance in non-inflamed cancers. J Hematol Oncol. 2021;14:146. doi: 10.1186/s13045-021-01155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi M, Zheng X, Niu M, Zhu S, Ge H, Wu K. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer. 2022;21:28. doi: 10.1186/s12943-021-01489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen S, Gosens R, Wieland T, Schmidt M. Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacol Ther. 2018;183:1–21. doi: 10.1016/j.pharmthera.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Lawson CD, Fan C, Mitin N, Baker NM, George SD, Graham DM, et al. Rho GTPase transcriptome analysis reveals oncogenic roles for Rho GTPase-activating proteins in Basal-like breast cancers. Cancer Res. 2016;76:3826–3837. doi: 10.1158/0008-5472.CAN-15-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphries B, Wang Z, Yang C. Rho GTPases: big players in breast cancer initiation, metastasis and therapeutic responses. Cells. 2020;9:2167. doi: 10.3390/cells9102167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 26.Niiya F, Tatsumoto T, Lee KS, Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 2006;25:827–837. doi: 10.1038/sj.onc.1209124. [DOI] [PubMed] [Google Scholar]
- 27.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- 29.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 30.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, et al. Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell. 2017;31:256–269. doi: 10.1016/j.ccell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huff LP, Decristo MJ, Trembath D, Kuan PF, Yim M, Liu J, et al. The role of Ect2 nuclear RhoGEF activity in ovarian cancer cell transformation. Genes Cancer. 2013;4:460–475. doi: 10.1177/1947601913514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Xia H, Zhang X, Karthik S, Pratap SV, Ooi LL, et al. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol. 2015;62:1287–1295. doi: 10.1016/j.jhep.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Bai X, Yi M, Xia X, Yu S, Zheng X, Wu K. Progression and prognostic value of ECT2 in non-small-cell lung cancer and its correlation with PCNA. Cancer Manag Res. 2018;10:4039–4050. doi: 10.2147/CMAR.S170033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortin SP, Ennis MJ, Schumacher CA, Zylstra-Diegel CR, Williams BO, Ross JT, et al. Cdc42 and the guanine nucleotide exchange factors Ect2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol Cancer Res. 2012;10:958–968. doi: 10.1158/1541-7786.MCR-11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Z, Chen X, Du T, Zhu D, Lai Y, Dong W, et al. Elevated levels of epithelial cell transforming sequence 2 predicts poor prognosis for prostate cancer. Med Oncol. 2017;34:13. doi: 10.1007/s12032-016-0872-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhou S, Wang P, Su X, Chen J, Chen H, Yang H, et al. High ECT2 expression is an independent prognostic factor for poor overall survival and recurrence-free survival in non-small cell lung adenocarcinoma. PLoS ONE. 2017;12:e0187356. doi: 10.1371/journal.pone.0187356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Liu J, Zhang Y. Role of epithelial cell transforming sequence 2 (ECT2) in predicting prognosis of osteosarcoma. Med Sci Monit. 2017;23:3861–3868. doi: 10.12659/MSM.905951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Jiao Y, Yi M, Zhao W, Wu K. EYA2 correlates with clinico-pathological features of breast cancer, promotes tumor proliferation, and predicts poor survival. Front Oncol. 2019;9:26. doi: 10.3389/fonc.2019.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Li A, Yu S, Qin S, Han N, Pestell RG, et al. DACH1 antagonizes CXCL8 to repress tumorigenesis of lung adenocarcinoma and improve prognosis. J Hematol Oncol. 2018;11:53. doi: 10.1186/s13045-018-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi M, Niu M, Wu Y, Ge H, Jiao D, Zhu S, et al. Combination of oral STING agonist MSA-2 and anti-TGF-β/PD-L1 bispecific antibody YM101: a novel immune cocktail therapy for non-inflamed tumors. J Hematol Oncol. 2022;15:142. doi: 10.1186/s13045-022-01363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JE, Billadeau DD, Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J Biol Chem. 2005;280:5733–5739. doi: 10.1074/jbc.M409298200. [DOI] [PubMed] [Google Scholar]
- 44.Kamijo K, Ohara N, Abe M, Uchimura T, Hosoya H, Lee JS, et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. doi: 10.1091/mbc.e05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oceguera-Yanez F, Kimura K, Yasuda S, Higashida C, Kitamura T, Hiraoka Y, et al. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J Cell Biol. 2005;168:221–232. doi: 10.1083/jcb.200408085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakata H, Rubin JS, Taylor WG, Miki T. A Rho-specific exchange factor Ect2 is induced from S to M phases in regenerating mouse liver. Hepatology. 2000;32:193–199. doi: 10.1053/jhep.2000.8271. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Hill KS, Fields AP. PKCiota maintains a tumor-initiating cell phenotype that is required for ovarian tumorigenesis. Mol Cancer Res. 2013;11:1624–1635. doi: 10.1158/1541-7786.MCR-13-0371-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y, Cai O, Wu P, Tan S. MiR-490-5p inhibits the stemness of hepatocellular carcinoma cells by targeting ECT2. J Cell Biochem. 2019;120:967–976. doi: 10.1002/jcb.27459. [DOI] [PubMed] [Google Scholar]
- 49.Huff LP, Kikuchi DS, Faidley E, Forrester SJ, Tsai MZ, Lassegue B, et al. Polymerase delta interacting protein 2 activates the RhoGEF epithelial cell transforming sequence 2 in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2019;316:C631. doi: 10.1152/ajpcell.00208.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Z, Chen D, Zhang W, Zhao J, Zhi L, Huang F, et al. Modulation of alternative splicing induced by paclitaxel in human lung cancer. Cell Death Dis. 2018;9:491. doi: 10.1038/s41419-018-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang HK, Liang JF, Zheng HX, Xiao H. Expression and prognostic significance of ECT2 in invasive breast cancer. J Clin Pathol. 2018;71:442–445. doi: 10.1136/jclinpath-2017-204569. [DOI] [PubMed] [Google Scholar]
- 52.Majeed U, Manochakian R, Zhao Y, Lou Y. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol. 2021;14:108. doi: 10.1186/s13045-021-01121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han J, Liu Y, Yang S, Wu X, Li H, Wang Q. MEK inhibitors for the treatment of non-small cell lung cancer. J Hematol Oncol. 2021;14:1. doi: 10.1186/s13045-020-01025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng LX, Jie GL, Li AN, Liu SY, Sun H, Zheng MM, et al. MET amplification identified by next-generation sequencing and its clinical relevance for MET inhibitors. Exp Hematol Oncol. 2021;10:52. doi: 10.1186/s40164-021-00245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmadi SE, Rahimi S, Zarandi B, Chegeni R, Safa M. MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J Hematol Oncol. 2021;14:121. doi: 10.1186/s13045-021-01111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook DR, Kang M, Martin TD, Galanko JA, Loeza GH, Trembath DG, et al. Aberrant expression and subcellular localization of ECT2 drives colorectal cancer progression and growth. Cancer Res. 2022;82:90–104. doi: 10.1158/0008-5472.CAN-20-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito S, Tatsumoto T, Lorenzi MV, Chedid M, Kapoor V, Sakata H, et al. Rho exchange factor ECT2 is induced by growth factors and regulates cytokinesis through the N-terminal cell cycle regulator-related domains. J Cell Biochem. 2003;90:819–836. doi: 10.1002/jcb.10688. [DOI] [PubMed] [Google Scholar]
- 58.Justilien V, Lewis KC, Meneses KM, Jamieson L, Murray NR, Fields AP. Protein kinase Cι promotes UBF1-ECT2 binding on ribosomal DNA to drive rRNA synthesis and transformed growth of non-small-cell lung cancer cells. J Biol Chem. 2020;295:8214–8226. doi: 10.1074/jbc.RA120.013175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kosibaty Z, Murata Y, Minami Y, Dai T, Kano J, Matsuoka R, et al. Cytoplasmic expression of epithelial cell transforming sequence 2 in lung adenocarcinoma and its implications for malignant progression. Lab Invest. 2019;99:551–567. doi: 10.1038/s41374-018-0142-4. [DOI] [PubMed] [Google Scholar]
- 60.Mansour M, Haupt S, Chan AL, Godde N, Rizzitelli A, Loi S, et al. The E3-ligase E6AP represses breast cancer metastasis via regulation of ECT2-Rho signaling. Cancer Res. 2016;76:4236–4248. doi: 10.1158/0008-5472.CAN-15-1553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Characteristics of studies involved in meta-analysis.
Data Availability Statement
The TCGA datasets for this study can be found in the https://xenabrowser.net/datapages/. The GSE25066 datasets for this study can be found in the https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse25066.