Abstract
Plant homeodomain (PHD) transcription factors (TFs) are a class of proteins with conserved Cys4-His-Cys3 domains that play important roles in plant growth and development and in response to abiotic stresses. Although characterization of PHDs has been performed in plants, little is known about their function in wheat (Triticum aestivum L.), especially under stress conditions. In the present study, 244 TaPHDs were identified in wheat using comparative genomics. We renamed them TaPHD1-244 based on their chromosomal distribution, and almost all PHD proteins were predicted to be located in the nucleus. According to the unrooted neighbor-joining phylogenetic tree, gene structure, and motif analyses, PHD genes were divided into four clades. A total of 149 TaPHD genes were assigned to arise from duplication events. Furthermore, 230 gene pairs came from wheat itself, and 119, 186, 168, 7, 2, and 6 gene pairs came from six other species (Hordeum vulgareto, Zea mays, Oryza sativa, Arabidopsis thaliana, Brassica rapa, and Gossypium raimondii, respectively). A total of 548 interacting protein branches were identified to be involved in the protein interaction network. Tissue-specific expression pattern analysis showed that TaPHDs were highly expressed in the stigma and ovary during flowering, suggesting that the TaPHD gene plays an active role in the reproductive growth of wheat. In addition, the qRT-PCR results further confirmed that these TaPHD genes are involved in the abiotic stress response of wheat. In conclusion, our study provides a theoretical basis for deciphering the molecular functions of TaPHDs, particularly in response to abiotic stress.
Keywords: PHD-finger genes, wheat, phylogenetic analysis, expression patterns, abiotic stress
Introduction
Plants encounter various unfavorable growth conditions during their life cycle, such as pests and diseases, drought, and extreme temperatures. In response to adverse external environments, plants activate in vivo defense response mechanisms by inducing stress-responsive gene expression (Fujita et al., 2006; Zhu et al., 2019). Many plant-specific transcription factor (TF) family members are involved in plant-specific developmental processes and participate in and regulate the stress response of plants to the external environment, thereby improving their adaptation to adversity (Yamasaki et al., 2013). To date, some such transcription factors have been successively isolated from many species of plants, such as AP2/ERF (Mizoi et al., 2012), bHLH (Sun et al., 2018), MYB (Li et al., 2015), and WRKY (Rushton et al., 2010). Among these, the plant homeodomain (PHD)-finger transcription factor family is tissue-specific and plays an important role in plant growth, development, and transcriptional regulation by adversity. The PHD is a conserved zinc finger structural domain in biological evolution and is commonly distributed in eukaryotes ranging from yeast to plants and animals (Ogryzko et al., 1996; Gibbons et al., 1997; Kehle et al., 1998; Papoulas et al., 1998; Martin et al., 2006). A typical PHD domain consists of 50-80 amino acid residues with a characteristic Cys4-His-Cys3 sequence, which is arranged in a manner similar to RING (Cys3-His-Cys4) and LIM (Cys2-His-Cys5) (Aasland et al., 1995; Borden and Freemont, 1996). The most important function of the PHD domain is the specific recognition of various histone modifications and DNA sequences, thus acting in transcriptional regulation and participating in various biological processes in organisms (Li et al., 2006; Hu et al., 2011; Xi et al., 2011). For example, previous studies have shown that, in model plants, proteins containing PHD domains are involved in embryonic meristem germination, root development, photoperiod, vernalization, meiosis, and post-meiotic pollen development. PHD domains play an important role in plant growth and development (Mouriz et al., 2015).
PHD domains are a class of relatively small protein domains. Their relatively conserved cysteine and histidine can stabilize the normal spatial structure by binding zinc ions, so that the three-dimensional conformation of the entire domain is basically spherical (Kwan et al., 2003). In addition to the conserved Cys4-His-Cys3 residues, PHD proteins usually contain highly diverse sequences. These diverse sequences form genes with different biological functions within the PHD-finger family. For example, the PHD domain–containing protein MMD1 is involved in essential chromatin remodeling and transcriptional events during male meiosis (Yang et al., 2003). In Arabidopsis, the ALFIN1-like (AL) protein, which contains the PHD domain, plays a key role in seed germination (Molitor et al., 2014). Furthermore, the PHD-finger protein VIL1 is involved in the photoperiod and vernalization pathways, as it regulates the expression of related floral repressors (Sung and Amasino, 2004). ATX1 and ATX2 have histone methyltransferase activities and regulate the development of roots, leaves, and floral organs, as well as the transcription of some stress genes (Saleh et al., 2008).
Since Schinder first discovered and identified PHD proteins in plants (Schindler et al., 1993), an increasing number of PHDs have been reported. To date, 59 Oryza stiva members (Sun et al., 2017), 108 Gossypium hirsutum members (Wu et al., 2021), 72 Solanum tuberosum members (Qin et al., 2019), 60 Phyllostachys edulis members (Gao et al., 2018), and 67 Zea mays members (Wang et al., 2015a) have been identified. It is known that PHD proteins not only participate in the regulation of plant growth and development but also play an important role in stress response, especially to abiotic stresses such as salt, high-temperature, low-temperature, and drought stress. In rice, overexpression of the OsPHD1 gene can significantly improve resistance to low-temperature, high-salt, and drought stress (Liu et al., 2011). Overexpression of the PHD-finger transcription factor gene OsMsr16 can enhance salt resistance in rice plants (Zhang et al., 2016). Wei et al. also found that Arabidopsis thaliana transgenic plants overexpressing soybean GmPHD2 exhibited higher salt resistance, possibly because overexpression of GmPHD2 enhanced the scavenging of oxidative substances (Wei et al., 2009). Furthermore, under abiotic stress, genes in the PHD-finger family in maize, cotton, and poplar show differential expression under salt, drought, and cold stress (Wang et al., 2015a; Wu et al., 2016; Wu et al., 2021). Thus, it can be seen that the PHD family genes play a crucial role in regulating plant resistance to stress.
Wheat is a major food crop worldwide and plays a crucial role in global food security. It is especially important to tap important resistance genes, breed new resistant wheat varieties, and improve the resistance of wheat itself (He et al., 2011). The PHD-finger gene family, which is essential for growth and development, has been identified and studied in many crops, but no systematic studies of the PHD gene family in wheat have been performed. In the present study, we identified PHD-finger family members in wheat for the first time and performed a comprehensive and systematic genome-wide analysis, including gene conserved motif analysis, phylogenetic relationships, Gene Ontology (GO) annotation analysis, covariance analysis, reciprocal relationship analysis, and subcellular localization. We also investigated the expression of PHD family proteins during growth and development, their specific expression in each organ, and their expression under multiple stresses of low temperature, high temperature, and drought. We lay the foundation for analyzing the functions of PHD proteins and regulating stress resistance and also provide theoretical references for the excavation of stress resistance genes and stress resistance breeding in wheat.
Materials and methods
Identification and classification analysis of PHD family genes in wheat
To identify PHD gene family members from wheat, whole genome data for T. aestivum (IWGSC RefSeq_v1.1) were obtained from the Ensembl plant database (http://plants.ensembl.org/info/website/ftp/index.html), and the PHD-finger domain (PF00628) was downloaded from the PFAM database (https://pfam.xfam.org/). The PHD protein sequences from A. thaliana (70) and O. sativa (59) ( Supplementary Table S1 ) (Sun et al., 2017) were used as query sequences to search against the wheat protein dataset using the BLASTP program, and the threshold was set as E-value < 1e-5. The NCBI-Batch CD-Search (Marchler-Bauer et al., 2017) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), PFAM database, and SMART database (http://smart.embl.de/) were used to further confirm the candidate PHD-finger genes of T. aestivum. There were other spliced transcripts in the candidate genes of these species, and we selected the first splice variant as a representative for subsequent analysis.
The protein sequences of TaPHDs were computed using the ExPASy server (Artimo et al., 2012) to obtain the theoretical isoelectric point (pI), molecular weight (MW), instability index (II), aliphatic index (AI), and grand average hydrophobicity (GRAVY). Plant-mPLoc (Chou and Shen, 2010) (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantmPLoc.cgi) and BUSCA (Savojardo et al., 2018) (Bologna Unified Subcellular Component Annotator, http://busca.biocomp.unibo.it) were used to predict the subcellular localization of the TaPHD proteins.
Phylogenetic analyses of TaPHD genes
The PHD-finger protein sequences of T. aestivum, A. thaliana, and O. sativa were used for phylogenetic analysis. Jalview 2.11 software (http://www.jalview.org/) with the MUSCLE method with default parameters was utilized to conduct multiple sequence alignment. Evolutionary analysis involved 342 amino acid sequences (all wheat PHD genes, and most rice and Arabidopsis PHD genes). These analyses were conducted in MEGA X (Kumar et al., 2018) using the neighbor-joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method and were expressed as the number of amino acid substitutions per site. The iTOL website (http://itol.embl.de/) was used to visualize the phylogenetic tree.
Gene duplication and Ka/Ks analysis of TaPHD genes
MCScanX software (Wang et al., 2012) was used to detect collinear regions between TaPHD genes as well as collinear blocks of TaPHDs with three monocotyledons (H. vulgareto, Z. mays, and O. sativa) and three dicotyledons (A. thaliana, B. rapa, and G. raimondii). Whole genome data for H. vulgareto, Z. mays, O. sativa, A. thaliana, B. rapa, and G. raimondii were obtained from the Ensembl plant database (http://plants.ensembl.org/info/website/ftp/index.html). All TaPHD genes were mapped to their respective loci in the wheat genome in a circular diagram using shinyCircos (Yu et al., 2018). Gene duplication events of TaPHDs and synteny relationships between the aforementioned species were visualized using TBtools (v1.082) (Chen et al., 2020). The Ka/Ks values (non-synonymous substitution rate/synonymous substitution rate) were calculated after identification of duplicated genes, using the method of Nei and Gojobori as implemented in KaKs_calculator (Zhang et al., 2006) based on the coding sequence alignments. Subsequently, the divergence time of collinear gene pairs was calculated using the duplication events formula T = Ks/(2λ × 10-6) in millions of years (Mya), with λ = 6.5 × 10-9 (Wang et al., 2015b).
GO annotation and protein-protein interaction network analysis of TaPHD genes
GO annotation of TaPHD proteins was available from the KOBAS database (http://kobas.cbi.pku.edu.cn/kobas3) (Xie et al., 2011). The full-length amino acid sequences of TaPHD proteins were uploaded to the original program, followed by drawing and annotation. GO annotations were performed for three types of analyses: biological processes, molecular functions, and cellular composition. The GO annotation results were visualized using the online tool OmicStudio (https://www.omicstudio.cn/tool) (Ye et al., 2018). All the predicted TaPHD proteins were submitted to the STRING database (https://string-db.org/cgi/input.pl). The minimum required interaction score was set to a high confidence (0.700). The maximum number of interactors was no more than 10 on the first shell.
Expression of TaPHD genes
Transcriptional data for TaPHDs were obtained from the wheat expression website (http://www.wheat-expression.com/download) (Borrill et al., 2016; Ramírez-González et al., 2018) and were used to explore the potential biological functions of TaPHD genes in growth and development, abiotic and biotic stress, and other conditions. Systematic clustering analysis was performed based on the log2 of transcripts per million (TPM) values for the 244 TaPHD genes. R was used to display the expression patterns in a heat map, and OmicStudio (https://www.omicstudio.cn/tool) was used to display the histogram, volcano plot, and Venn diagram.
Quantitative real-time PCR analyses (qRT-PCR) of TaPHD genes in response to environmental stresses
In this study, the seeds of the hexaploid common wheat variety “Zhengmai 7698” were surface-sterilized with 2% hydrogen peroxide, rinsed thoroughly with distilled water, and germinated with water saturation at 25°C for two days in Petri dishes on three layers of filter paper. The young seedlings were transformed and grown in 1/2 Hoagland’s culture solution under a 14 h light (25°C)/10 h dark (20°C) photoperiod. When the wheat grew to two leaves and one heart, the plants were subsequently treated with 16% polyethylene glycol 6000. For cold stress, wheat seedlings were exposed to 4°C for 12 h. For heat stress, wheat seedlings were exposed to 40°C for 12 h. New leaves of the three seedlings were collected as biological replicates, and each treatment had three replicates.
Total RNA was extracted using RNAiso Reagent (TaKaRa, Beijing, China) and Cdna was synthesized using the RT Master Mix Perfect RealTime kit (TaKaRa, Beijing, China). Quantitative real-time PCR was performed using the CFX Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) and the SG Fast Qpcr Master Mix (Sangon Biotech, Shanghai, China). Relative expression levels were determined using the 2(-ΔΔCt) method (Livak and Schmittgen, 2001), and β-actin was used as the internal control to normalize the expression levels of TaPHD genes. Specific primers used for qRT-PCR are listed in Supplementary Table S2 .
Determination of subcellular localization of TaPHD11, TaPHD19, and TaPHD133
Full-length open reading frames of TaPHD11, TaPHD19, and TaPHD133 were obtained from “Zhengmai 7698” Cdna ( Supplementary Table S2 ). The Coding sequence (CDS)of TaPHD11, TaPHD19, and TaPHD133 were cloned into the pJIT16318 vector at the BamHI site using specific primers ( Supplementary Table S2 ). The pJIT16318 vector contained a CaMV 35S promoter and C-terminal GFP. Transient expression assays were conducted as described by Cui et al. (2019). Approximately 4 × 104 mesophyll protoplasts were isolated from 12-day-old wheat seedlings. The transfected protoplasts were incubated at 23°C for 12 h. The GFP fluorescence in the transformed protoplasts was imaged using a confocal laser-scanning microscope (LSM 700; Zeiss).
Results
Identification and classification analysis of PHD genes in wheat
In this study, 244 T. aestivum genes were designated PHD genes with two query methods; HMM and BLASTP were used for identification, and three websites, NCBI-Batch CD-Search, PFAM database, and SMART database, were used for confirmation ( Supplementary Table S3 ). These PHD genes were renamed TaPHD1 to TaPHD244, based on the order of their chromosomal locations and physical positions.
To further determine the characteristics of TaPHD genes, the ExPASy Server online tool was used to analyze the protein characteristics ( Supplementary Table S3 ). The shortest protein contained 216 amino acids (TaPHD158, TaPHD175) and the longest protein contained 2853 amino acids (TaPHD204); the molecular weight was between 24567.82 Da (TaPHD158) and 310347.53 Da (TaPHD204). The protein instability index showed that all PHD genes were unstable proteins. The isoelectric point of TaPHD genes varied markedly from 4.42 (TaPHD36) to 9.65 (TaPHD78), and the aliphatic index varied significantly from 48.13 (TaPHD26/39/51) to 97.51 (TaPHD42). The GRAVY of TaPHD proteins in wheat varied from 0.016 (TaPHD160) to -1.285 (TaPHD23), indicating that they were all hydrophilic proteins, except for TaPHD160 ( Supplementary Table S3 ). We used two methods (Plant-mPLoc and BUSCA) to predict the subcellular localization of the TaPHD proteins. The results showed that a few TaPHDs may be localized in the chloroplast, mitochondrion, or cytoplasm, and most members were predicted to be located in the nucleus ( Supplementary Table S3 ).
Multiple sequence alignment and phylogenetic analysis of PHD genes
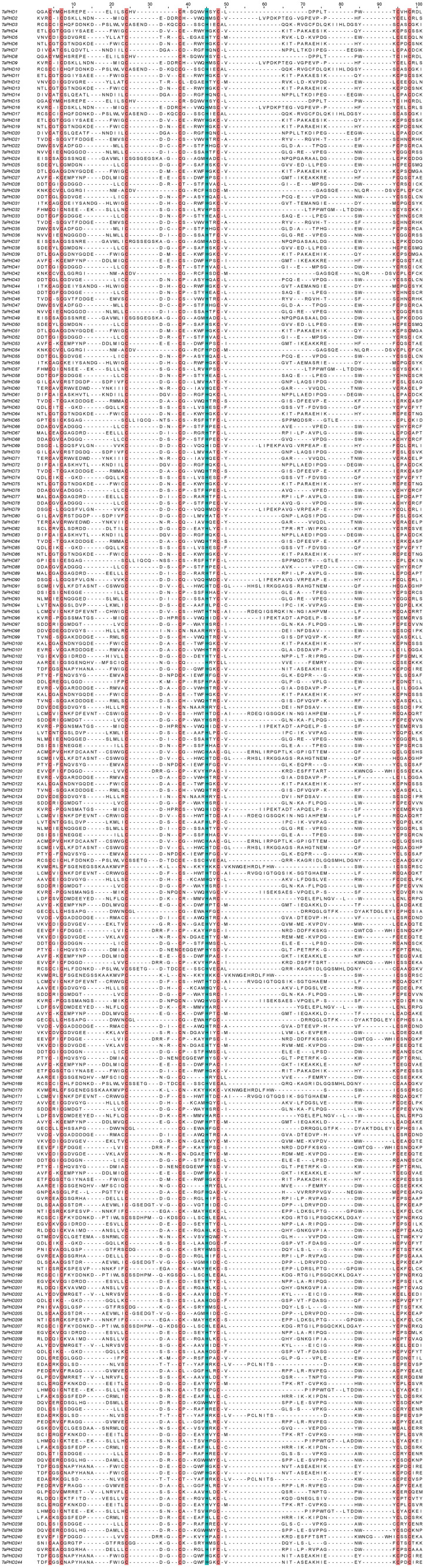
Multiple sequence alignments of PHD domains were performed ( Figure 1 ). Approximately 60 amino acids (aa) comprised a PHD domain containing basic Cys4-His-Cys3 sequence motifs in each TaPHD.
Figure 1.
Protein sequence multiple alignment of the PHD-finger domains in TaPHD family proteins. The multiple alignment was conducted with the amino acid sequences within the predicted PHD domains by using Jalview software. The conserved amino acids (Cys4-His-Cys3) within the PHD-finger domains are shaded in red and blue.
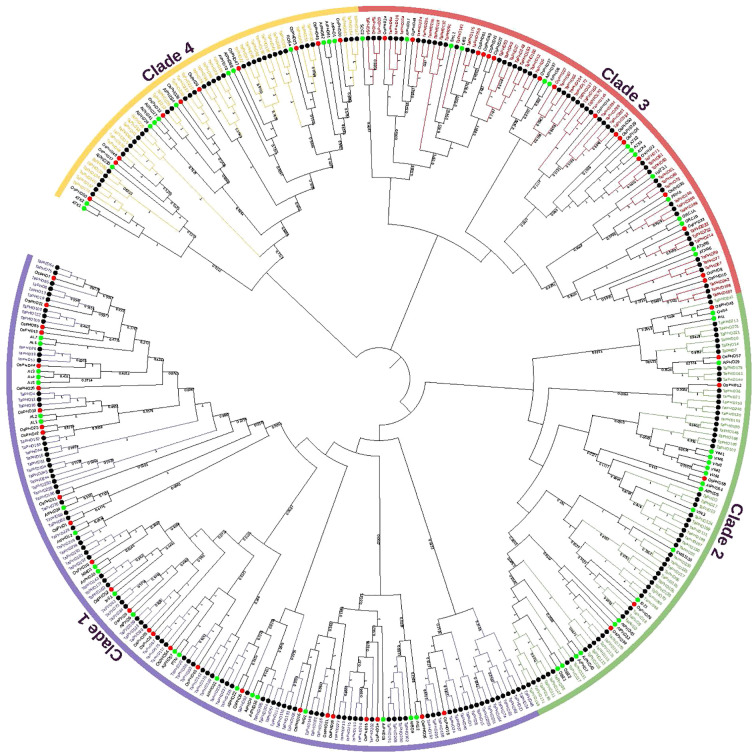
To evaluate the evolutionary relationships of PHD genes in T. aestivum, O. sativa, and A. thaliana, a neighbor-joining phylogenetic tree was constructed using full-length PHD proteins ( Figure 2 and Supplementary Table S1 ). Phylogenetic analysis showed that PHD family proteins can be divided into four clades (clades 1 to 4). TaPHD members were found in all clades. Clade 1 was the largest, with 95 TaPHD members, and clade 4 was the smallest, with only 38 members. The results showed that there were many small branches under each clade, and almost every small branch had corresponding genes of rice and Arabidopsis. This indicates that the TaPHD gene is not an evolutionary characteristic of monocotyledonous and dicotyledonous plants, and that the PHD gene family was formed before the differentiation of these two types of plants.
Figure 2.
Phylogenetic tree of PHD genes in wheat, rice, and Arabidopsis. The tree was analyzed in MEGA X by using the neighbor-joining method. The PHDs from wheat, rice, and Arabidopsis are distinguished with black, red, and green dots. The PHD proteins were grouped into four distinct clades (clades 1-4), which are indicated by colored branches.
Protein domains are often functional carriers. According to phylogenetic and domain analyses (NCBI-Batch CD-Search, PFAM, and SMART database), 30 dominant types were identified in all wheat PHD proteins ( Table 1 ). The results showed that among all wheat PHD proteins, 43 contained a typical PHD domain. The next most common, the jas-PHD and alifn-PHD domains, had 28 and 25 members, respectively; the PHD-Oberon_cc domain and the PHD-RING domains had 11 members, and the remaining domain types had less than ten members. The results showed that wheat PHD proteins contained a canonical PHD domain or double PHD domains. Owing to their different domains, differentiation in function was achieved.
Table 1.
Types, names, and numbers of wheat PHD-finger genes.
| Domain type | Wheat triad | Rice orthologs | Arabidopsis thaliana orthologs | Gene number | Chr | Genomes |
|---|---|---|---|---|---|---|
| PHD | TaPHD1/TaPHD8/TaPHD15 | 3 | 1 | ABD | ||
| TaPHD5/TaPHD12 | 2 | 1 | AB | |||
| TaPHD21/TaPHD34/TaPHD46 | 3 | 2 | ABD | |||
| TaPHD62/TaPHD73/TaPHD84 | OsPHD5 | 3 | 3 | ABD | ||
| TaPHD82 | 1 | 3 | D | |||
| TaPHD94/TaPHD114/TaPHD128 | OsPHD24 | 3 | 4 | ABD | ||
| TaPHD218/TaPHD226/TaPHD237 | OsPHD24 | 3 | 7 | ABD | ||
| TaPHD98/TaPHD110/TaPHD124 | 3 | 4 | ABD | |||
| TaPHD99/TaPHD109/TaPHD123 | OsPHD55 | AtPHD6 | 3 | 4 | ABD | |
| TaPHD101/TaPHD107/TaPHD121 | OsPHD19 | MS1,MMD1 | 3 | 4 | ABD | |
| TaPHD143/TaPHD160/TaPHD177 | OsPHD52 | MS1,MMD1 | 3 | 5 | ABD | |
| TaPHD103 | 1 | 4 | A | |||
| TaPHD137/TaPHD154/TaPHD172 | 3 | 5 | ABD | |||
| TaPHD146/TaPHD163/TaPHD180 | OsPHD58 | AtPHD54 | 3 | 5 | ABD | |
| TaPHD168/TaPHD185 | 2 | 5 | BD | |||
| TaPHD186 | OsPHD11 | 1 | 5 | D | ||
| TaPHD192/TaPHD201/TaPHD209 | OsPHD14,OsPHD37 | AtPHD8,AtPHD37 | 3 | 6 | ABD | |
| PHD-PHD | TaPHD135/TaPHD152/TaPHD170 | OsPHD59 | AtPHD32,AtPHD40 | 3 | 5 | ABD |
| TaPHD144/TaPHD161/TaPHD178 | AtPHD29 | 3 | 5 | ABD | ||
| TaPHD216/TaPHD224/TaPHD235 | OsPHD48 | 3 | 7 | ABD | ||
| Alifn-PHD | TaPHD4/TaPHD11/TaPHD18 | OsPHD30 | AL1,AL2 | 3 | 1 | ABD |
| TaPHD6/TaPHD13/TaPHD19 | OsPHD31 | AL6,AL7 | 3 | 1 | ABD | |
| TaPHD26/TaPHD39/TaPHD51 | OsPHD44 | AL3.AL4,AL5 | 3 | 2 | ABD | |
| TaPHD31/TaPHD44/TaPHD56 | AL1,AL2,AL3.AL4,AL5,AL6,AL7 | 3 | 2 | ABD | ||
| TaPHD64/TaPHD75/TaPHD86 | OsPHD7 | AL6,AL7 | 3 | 3 | ABD | |
| TaPHD100/TaPHD108/TaPHD122 | OsPHD56 | AL6,AL7 | 3 | 4 | ABD | |
| TaPHD167/TaPHD184 | OsPHD23,OsPHD42 | AL1,AL2 | 2 | 5 | BD | |
| TaPHD104/TaPHD229/TaPHD230/TaPHD243/TaPHD244 | AL1,AL2,AL3.AL4,AL5,AL6,AL7 | 5 | 4(7) | A(DD)UU | ||
| ARID-PHD | TaPHD142/TaPHD159/TaPHD176 | 3 | 5 | ABD | ||
| RING-PHD | TaPHD23/TaPHD36/TaPHD48 | OsPHD46 | AtPHD30 | 3 | 2 | ABD |
| TaPHD93/TaPHD115/TaPHD129 | OsPHD17 | AtPHD30 | 3 | 4 | ABD | |
| ING-PHD | TaPHD105/TaPHD119/TaPHD133 | OsPHD16 | ING1 | 3 | 4 | ABD |
| TaPHD148/TaPHD165/TaPHD182 | OsPHD21 | ING2 | 3 | 5 | ABD | |
| BAH-PHD | TaPHD27/TaPHD40/TaPHD53 | OsPHD41 | SHL1 | 3 | 2 | ABD |
| TaPHD141/TaPHD158/TaPHD175 | OsPHD49,OsPHD51 | EBS | 3 | 5 | ABD | |
| TaPHD149/TaPHD166/TaPHD183 | OsPHD22 | SHL1 | 3 | 5 | ABD | |
| Jas-PHD | TaPHD28/TaPHD41/TaPHD52 | OsPHD40 | AtPHD1,AtPHD61,AtPHD62 | 3 | 2 | ABD |
| TaPHD147/TaPHD164/TaPHD181 | OsPHD20 | AtPHD1,AtPHD61,AtPHD62 | 3 | 5 | ABD | |
| TaPHD30/TaPHD43/TaPHD55 | OsPHD25 | 3 | 2 | ABD | ||
| TaPHD33/TaPHD45/TaPHD58 | ROS4 | 3 | 2 | ABD | ||
| TaPHD22/TaPHD35/TaPHD47 | OsPHD47 | AtPHD68,AtPHD70 | 3 | 2 | ABD | |
| TaPHD92/TaPHD116/TaPHD130 | OsPHD47 | AtPHD68,AtPHD70 | 3 | 4 | ABD | |
| TaPHD66/TaPHD76/TaPHD88 | OsPHD9 | 3 | 3 | ABD | ||
| TaPHD68/TaPHD78 | OsPHD9 | 2 | 3 | AB | ||
| TaPHD106/TaPHD212 | OsPHD32 | AtPHD24,AtPHD26,AtPHD27,AtPHD41 | 2 | 4(7) | A(A) | |
| TaPHD220/TaPHD227/TaPHD238 | OsPHD38 | AtPHD24,AtPHD26,AtPHD27,AtPHD41 | 3 | 7 | ABD | |
| DDT-PHD | TaPHD25/TaPHD38/TaPHD50 | OsPHD45 | DDP1,DDP2 | 3 | 2 | ABD |
| TaPHD97/TaPHD112/TaPHD125 | OsPHD54 | DDP3 | 3 | 4 | ABD | |
| TaPHD138/TaPHD155/TaPHD173 | OsPHD54 | DDP3 | 3 | 5 | ABD | |
| zf-HC5HC2H-PHD | TaPHD59/TaPHD70/TaPHD80 | OsPHD1 | 3 | 3 | ABD | |
| PHD-Oberon_cc | TaPHD91/TaPHD118/TaPHD132 | 3 | 4 | ABD | ||
| TaPHD95/TaPHD111/TaPHD127 | OBE1,OBE2 | 3 | 4 | ABD | ||
| TaPHD136/TaPHD153/TaPHD171 | OBE1,OBE2 | 3 | 5 | ABD | ||
| TaPHD117/TaPHD131 | 2 | 4 | BD | |||
| PHD-FN3 | TaPHD3/TaPHD10/TaPHD17 | VIN3 | 3 | 1 | ABD | |
| TaPHD134/TaPHD151/TaPHD169 | VIN3 | 3 | 5 | ABD | ||
| TaPHD190/TaPHD199/TaPHD207 | VIN3 | 3 | 6 | ABD | ||
| PHD-SANT | TaPHD63/TaPHD74/TaPHD85 | 3 | 3 | ABD | ||
| TaPHD194/TaPHD203/TaPHD211 | 3 | 6 | ABD | |||
| PHD-WHIM1 | TaPHD102/TaPHD191/TaPHD200/TaPHD208 | MBD9 | 4 | 6(4) | (A)ABD | |
| PHD-SET | TaPHD67/TaPHD77/TaPHD89 | OsPHD8 | ATXR5,ATXR6 | 3 | 3 | ABD |
| TaPHD187/TaPHD196/TaPHD242 | OsPHD10 | ATXR5,ATXR6 | 3 | 6 | AB(U) | |
| PWWP-PHD-SET | TaPHD60/TaPHD71/TaPHD81 | OsPHD2,OsPHD4 | ATX3,ATX4,ATX5 | 3 | 3 | ABD |
| PWWP-FYRN-FYRC-PHD-SET | TaPHD140/TaPHD157/TaPHD174 | OsPHD50 | ATX1,ATX2 | 3 | 5 | ABD |
| PHD-BAH | TaPHD234 | 1 | 7 | D | ||
| PHD-BAH-AAA | TaPHD214/TaPHD222/TaPHD232 | OsPHD33 | ORC1A,ORC1B | 3 | 7 | ABD |
| PHD-homeodomain | TaPHD7/TaPHD14/TaPHD20 | PRHA | 3 | 1 | ABD | |
| TaPHD61/TaPHD72/TaPHD83 | OsPHD35 | HAT3.1 | 3 | 3 | ABD | |
| TaPHD189/TaPHD198/TaPHD206 | 3 | 6 | ABD | |||
| PHD-PLN03142 | TaPHD65/TaPHD87 | 2 | 3 | AD | ||
| TaPHD195/TaPHD204/TaPHD241 | 3 | 6 | ABD | |||
| PHD-RING | TaPHD2/TaPHD9/TaPHD16 | OsPHD29 | SIZ1 | 3 | 1 | ABD |
| TaPHD69/TaPHD79/TaPHD90 | SIZ1 | 3 | 3 | ABD | ||
| TaPHD96/TaPHD113/TaPHD126 | SIZ1 | 3 | 4 | ABD | ||
| TaPHD139/TaPHD156 | SIZ1 | 2 | 5 | AB | ||
| PHD-JmjC-PLU1 | TaPHD219/TaPHD228/TaPHD239 | 3 | 7 | ABD | ||
| AAA_34-PHD-Helicase_C_4 | TaPHD32/TaPHD57 | OsPHD27 | EMB1135 | 2 | 2 | AD |
| TaPHD217/TaPHD225/TaPHD236 | OsPHD27 | EMB1135 | 3 | 7 | ABD | |
| PHD-zf-HC5HC2H-zf-HC5HC2H | TaPHD193/TaPHD202/TaPHD210 | OsPHD15,OsPHD34 | AtPHD18 | 3 | 6 | ABD |
| TaPHD215/TaPHD223/TaPHD233 | 3 | 7 | ABD | |||
| BRCT-BRCT-PHD | TaPHD24/TaPHD37/TaPHD49 | OsPHD18 | 3 | 2 | ABD | |
| TaPHD188/TaPHD197/TaPHD205 | OsPHD18 | 3 | 6 | ABD | ||
| PHD-SWIB-GYF-Plus3 | TaPHD120/TaPHD150/TaPHD240 | 3 | 4(5) | (A)BU | ||
| PHD-SWIB-Plus3-GYF | TaPHD145/TaPHD162/TaPHD179 | 3 | 5 | ABD | ||
| PHD-Chromo-Helicase_C-DUF | TaPHD213/TaPHD221/TaPHD231 | PKL | 3 | 7 | ABD | |
| PHD-Cohesin_HEAT-Nipped-B_C | TaPHD29/TaPHD42/TaPHD54 | EMB2773 | 3 | 2 | ABD | |
To better understand why PHD-finger genes are abundant in the wheat genome, we analyzed the homoeologous groups in detail ( Table 2 ). A total of 35.8% of wheat genes were present in homoeologous groups of three, also termed triads (A:B:D = 1:1:1) (Consortium et al., 2018). In contrast, 84.8% of the PHD-finger genes identified were present in triads ( Table 2 ). Also, the percentage of PHD-finger genes with homoeolog-specific duplications was lower for PHD-finger genes than for all wheat genes (1.6% vs 5.7%; Table 2 ). Loss of one homoeolog, on the other hand, was less pronounced in PHD-finger genes (6.6% vs 13.2%; Table 2 ). Only four PHD-finger genes were orphans/singletons. Thus, the high homoeolog retention rate could partly explain the high number of wheat PHD-finger genes.
Table 2.
Groups of homoeologous PHD-finger genes in wheat.
| Homoeologous group (A: B: D) | All wheat genes1 | Wheat PHD-finger genes (all) | ||
|---|---|---|---|---|
| Number of groups | Number of genes | % of genes2 | ||
| 1: 1: 1 | 35.8% | 69 | 207 | 84.8 |
| n: 1: 1/1: n: 1/1: 1: n3 | 5.7% | 1 | 4 | 1.6 |
| 1: 1: 0/1: 0: 1/0: 1: 1 | 13.2% | 8 | 16 | 6.6 |
| Other ratios4 | 8.0% | 3 | 11 | 4.5 |
| Orphans/singletons | 37.1% | 4 | 4 | 1.6 |
| Not categorized5 | - | - | 2 | 0.8 |
| 99.8% | 244 | 100.0 | ||
Chromosomal location, gene duplication, and synteny analysis of TaPHD genes
Based on the reference GFF3 files, the physical positions of PHD genes on the corresponding chromosomes are shown in Figure 3 . The identified TaPHDs could be mapped on every chromosome and evenly across the three sub-genomes. The map shows that chromosomes 5B and 5D harbor the largest number of TaPHD genes (18), whereas chromosome 1D contains the least (6).
Figure 3.
Chromosomal localization of TaPHDs. The light blue column represents the chromosome. The depth of blue in the columns represent the density of genes on the chromosome.
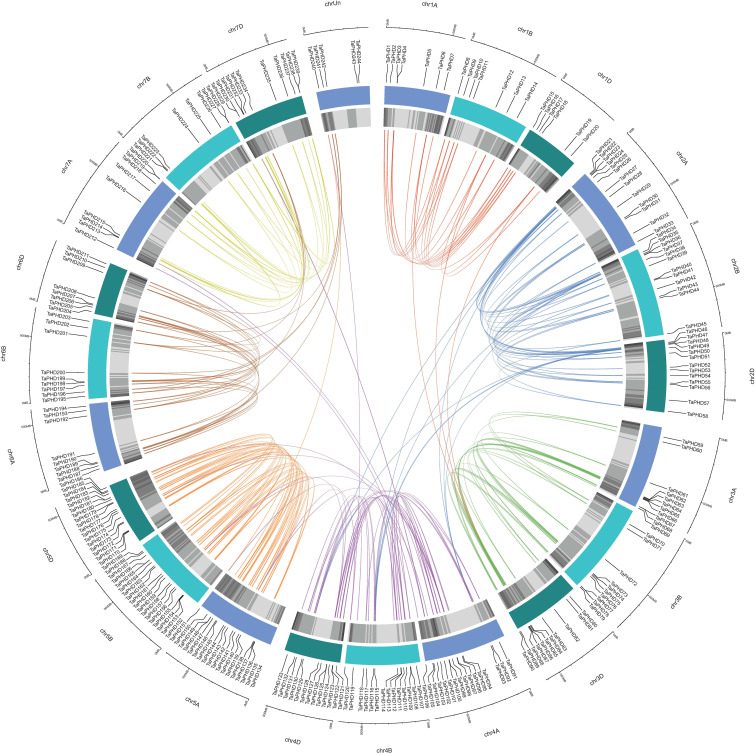
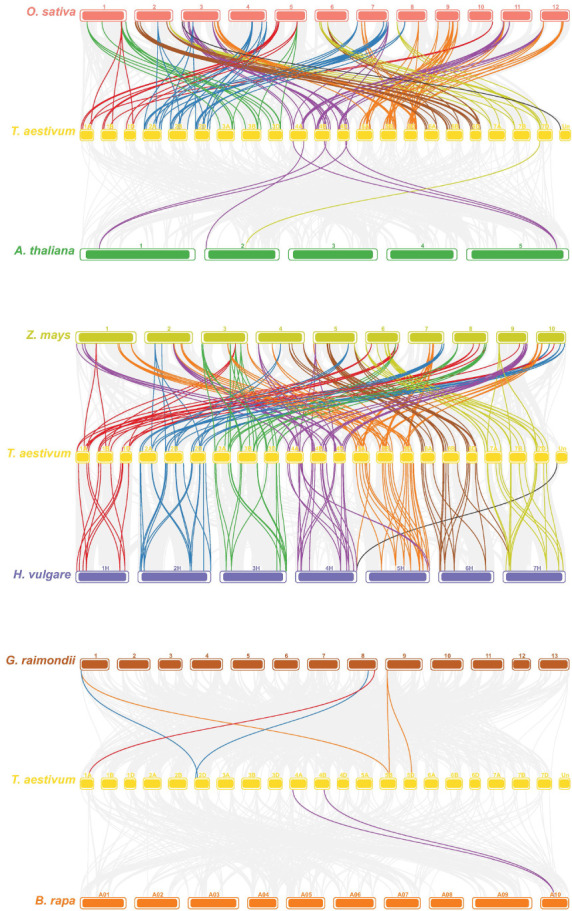
Gene duplication is an indispensable mechanism by which organisms create new genes with similar or different functions (Song et al., 2019). Therefore, we analyzed the duplication events that occurred in the TaPHD gene family. A total of 230 PHD gene pairs from wheat were identified as duplicated ( Figure 4 and Supplementary Table S4 ). These similar PHD gene pairs had the same domain type and appeared in the same branch of the phylogenetic tree. Tandem and segment duplications are critical for the evolution of gene families to adapt to different environmental conditions. Interestingly, all the TaPHD gene pairs were associated with segmental duplication events. This suggests that this was the main route for expanding PHD genes in wheat and the many homologous genes on different wheat chromosomes suggest the high conservation of the family. To further infer the evolutionary origin and homology of the wheat PHD family, we constructed a collinear chart comparing six species with wheat, including three monocotyledons (H. vulgareto, Z. mays, and O. sativa) and three dicotyledons (A. thaliana, B. rapa, and G. raimondii) ( Figure 5 and Supplementary Table S4 ). We identified pairwise homologues of the TaPHD genes and detected 119, 186, 168, 7, 2, and 6 pairs of homologous genes from H. vulgareto, Z. mays, O. sativa, A. thaliana, B. rapa, and G. raimondii, respectively ( Figure 5 and Supplementary Table S4 ). This implies that TaPHD genes share a strong evolutionary relationship with ZmPHDs, HvPHDs, and OsPHDs. Furthermore, these results indicated that the PHD gene family was differentiated between monocotyledonous and dicotyledonous plants. This also indicated that TaPHD genes had a strong evolutionary relationship with ZmPHDs, HvPHDs, and OsPHDs. The average differentiation time was: barley (12.78 Mya) < rice (22.09 Mya) < maize (60.87 Mya).
Figure 4.
Synteny analysis of PHD genes in wheat. All TaPHD genes were mapped to their respective locus in the wheat genome in a circular diagram using shinyCircos (Yu et al., 2018). Subgenomes are indicated by different shades of blue (outer track), and chromosomal segments are indicated by shades of gray (inner track). Homoeologous PHD genes were inferred by phylogeny (for details see the Materials and Methods section) and linked with chromosome-specific colors.
Figure 5.
Synteny analysis of PHD genes between wheat and six representative plants (maize, barley, rice, Arabidopsis, cotton, and Brassica rapa). Each different species is replaced with a different color. The gray line in the background indicates a collinear block in the genome of wheat and other plants, while the line highlights the isomorphic PHD gene pair. Homoeologous PHD genes were inferred by phylogeny (for details see the Materials and Methods section) and linked with chromosome-specific colors.
Ka/Ks, the non-synonymous substitution ratio, determines the selection pressure for duplicated genes. According to the results ( Supplementary Table S4 ), only a very few TaPHD gene pairs had Ka/Ks ratios >1, suggesting that the evolution of TaPHD genes was accompanied by strong purifying selection. The Ka/Ks ratios between wheat and three monocotyledonous plants were calculated based on the collinear gene pairs. Except for very few genes, the values of the other collinear gene pairs were all below 1, which confirmed that the evolution of the wheat PHD gene family underwent strong purifying selection. However, the Ka/Ks ratios of the collinear gene pairs between wheat and the three dicots could not be calculated properly. This is because most synonymous mutation sites have synonymous mutations; that is, the degree of sequence divergence and evolutionary distance is too large. Some TaPHD genes have formed at least five homologous gene pairs, such as TaPHD9, which may have played key roles in the evolution of the PHD gene family ( Figure 5 and Supplementary Table S4 ).
GO annotation analysis and protein-protein interaction network of TaPHD genes
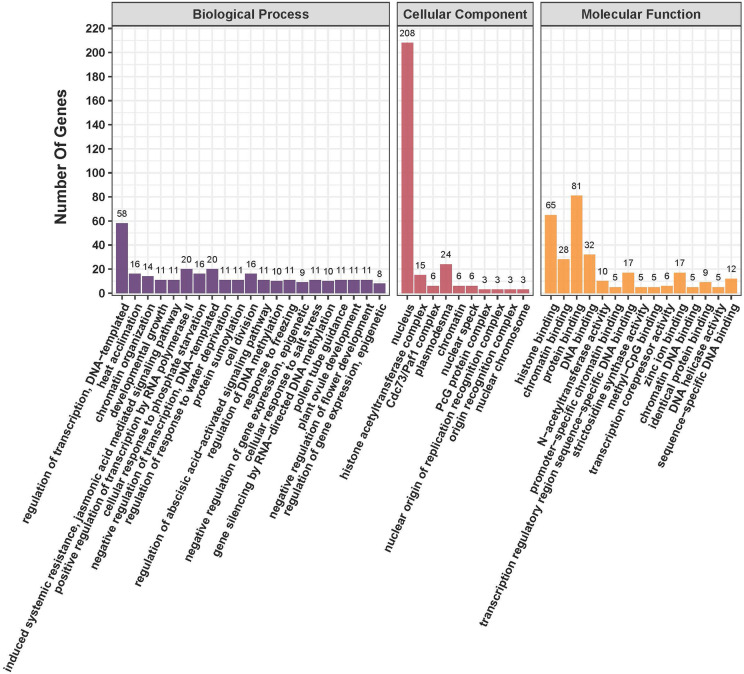
We performed GO annotation analysis of the 244 TaPHD proteins, revealing that they may participate in a range of cellular components, molecular functions, and biological processes ( Figure 6 and Supplementary Table S5 ). The 244 TaPHD proteins were assigned a total of 105 GO terms. In biological processes, the three most highly enriched categories were related to the regulation of DNA-templated transcription, heat acclimation, and chromatin organization. Developmental growth and jasmonic acid-mediated systemic resistance were also particularly enriched. In the cellular component category, the most highly enriched categories were related to the nucleus, and 85% of the TaPHDs could participate in this process, whereas less than 10% of TaPHDs were involved in plasmodesma. Regarding molecular functions, the 65 most enriched TaPHDs were involved in histone binding, 28 TaPHDs were involved in chromatin binding, and 81 TaPHDs were related to protein binding.
Figure 6.
Functional annotation analysis of TaPHD genes. Gene Ontology (GO) classification based on TaPHD gene annotation. The GO terms are grouped into three main categories: purple for Biological Processes, red for Cellular Components, and yellow for Molecular Function.
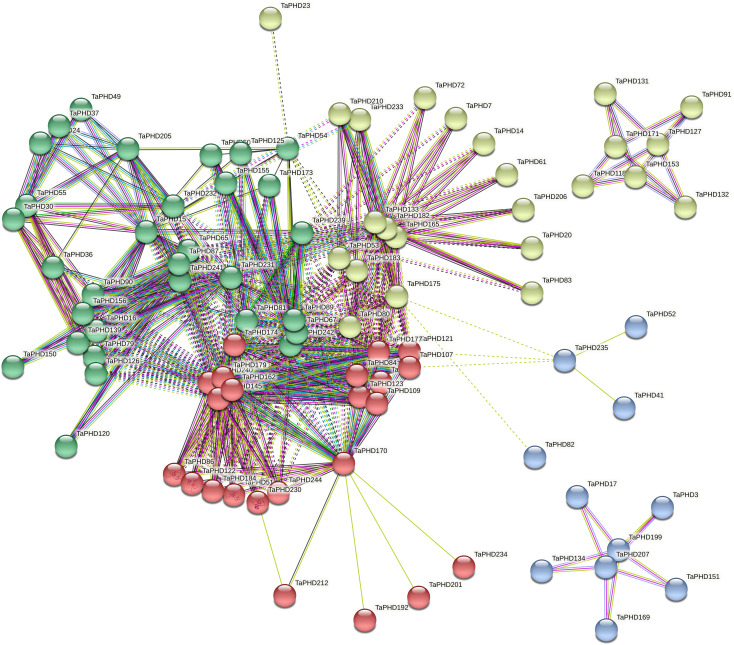
To understand protein-protein interactions between TaPHDs and other proteins in wheat, we constructed a protein-protein interaction network ( Figure 7 and Supplementary Table S6 ). A total of 89 TaPHD proteins and 548 interacting protein branches were identified. According to the strength of the interaction, we divided the 89 proteins into four interaction regions, which are represented by different colors, as shown in Figure 7 . Some TaPHDs, such as TaPHD15, TaPHD145, and TaPHD162, could interact with up to 28 proteins, suggesting that these TaPHD proteins play a significant role in the regulation of protein networks. Notably, we found that these proteins had a PHD domain or a PHD-SWIB-Plus3-GYF domain. Therefore, we believe that such domains are likely to play an important role in the PHD family.
Figure 7.
Predicted protein association networks analyses of TaPHD proteins. The four colors represent different interaction areas. The nodes represent the proteins, and the lines represent the protein-protein associations. Light blue and purple lines represent the known interactions from the curated database or experimentally determined interactions; green, red, and blue lines represent gene neighborhood, gene fusions, and gene co-occurrence, indicating that the proteins have the predicted interactions; yellow, black, and light blue lines represent textmining, co-expression, and protein homology, respectively.
Expression analysis of TaPHD genes during growth and development
RNA-sequencing is a powerful tool for exploring certain gene transcription patterns using high-throughput sequencing methods (Wang et al., 2009). Systematic clustering analysis was performed based on the log2 of TPM values for 244 TaPHD genes ( Figure 8A and Supplementary Table S7 ). The data showed that TaPHD gene expression showed great differences with the change in the growth period. In general, the expression of TaPHDs can be divided into three categories: the first group contains members that are widely expressed in many tissues under multiple developmental stage conditions; the second group contains those that are highly induced only at specific growth and development stages; and the last group includes members that do not appear to be expressed during growth and development. For example, TaPHD100, TaPHD108, and TaPHD122 had high expression during most growth and developmental processes, except in the endosperm. There were also some genes (TaPHD222 and TaPHD232) that had higher expression only in shoots and roots. Furthermore, some genes, such as TaPHD68, TaPHD78, and TaPHD86, were not expressed, which implies that these genes may have functional redundancy.
Figure 8.
Transcriptome analyses of TaPHDs in different tissues. (A) Heat map of expression profiles for 244 TaPHD genes in different tissues. Red color indicates high expression levels; blue color indicates low expression levels. The gradual change of the color indicates different levels of gene log2-transformed expression. (B) Numbers of expressed genes in different tissues. High: TPM values >10, medium: 10 ≥ TPM values > 1, low: 1 ≥ TPM values > 0, none: TPM values = 0.
To further study the expression differences of this family in different stages and organs of wheat, we counted the number of high, medium, and low expression genes in each period and organ ( Figure 8B ). The data showed that the number of highly expressed genes was the largest in the stigma and ovary, reaching as high as 60, followed by a spike in the boot period, reaching 41. The lowest number of highly expressed genes (none) was found in the flag leaf blade at night in the flag leaf stage. Our results suggest that some TaPHDs may play important roles in many biological processes during wheat growth, especially during anthesis.
Expression responses of TaPHD genes to abiotic/biotic stress
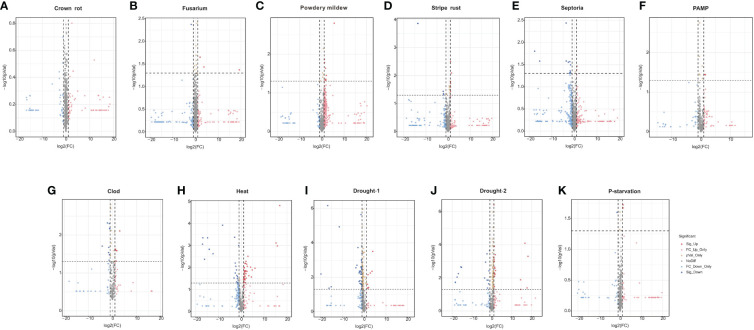
The differential expression of TaPHDs under different conditions is shown in Figures 9A–F and Supplementary Table S8 . During biological stress, we found that inoculation with Fusarium, powdery mildew, pathogen associated molecular patterns (PAMP), crown rot, Septoria, or stripe rust caused few changes in the expression of TaPHD genes. This suggests that TaPHD family members may not be associated with disease resistance.
Figure 9.
Expression of TaPHDs during different biological stress. Volcano map of expression profiles for 244 TaPHD genes under different biological/abiotic stresses, including (A) crown rot infection, (B) Fusarium infection, (C) powdery mildew infection, (D) stripe rust infection, (E) Septoria (Septoria tritici infection and Zymoseptoria tritici infection), (F) PAMP (chitin and flg22 infection), (G) cold stress, (H) drought-1 (drought stress in Giza 168), (I) drought-2 (drought stress in Gemmiza 10), (J) heat stress, and (K) P-starvation. DEGs were defined as Fold Change > 1 and FDR < 0.05.
Under abiotic stress, there are many TaPHD genes whose expression changes are more obvious under high-temperature, drought, and cold conditions ( Figures 9G–K and Supplementary Table S8 ). For example, after high-temperature treatment, the expression levels of many TaPHD genes (TaPHD26, TaPHD75, TaPHD100, TaPHD115, TaPHD117, and TaPHD167) were significantly altered compared to those in the experimental control group. In the drought starvation treatment, TaPHD11, TaPHD19, TaPHD99, TaPHD141, TaPHD153, and TaPHD171 expression levels changed significantly. However, in the phosphorus starvation treatment, there were few changes in the expression of TaPHD genes. To further understand whether there is an intersection between the differential genes of the PHD family under drought, high-temperature, and low-temperature treatments, we drew a Venn diagram of DEGs in TaPHD genes during the four different transcriptomes ( Figure 10 , Supplementary Table S9 ). The data showed that TaPHD215 and TaPHD223 were significantly altered in every treatment. TaPHD30, TaPHD96, TaPHD180, TaPHD174, and TaPHD239 gene expression varied greatly between the two drought and heat treatments. In addition, in cold and heat stress environments, the expression levels of five genes (TaPHD109, TaPHD118, TaPHD120, TaPHD167, and TaPHD178) were significantly changed.
Figure 10.
Venn diagram of DEGs in TaPHD genes during different abiotic stress. DEGs of TaPHD genes in different abiotic stress conditions, including cold stress, drought-1 (drought stress in Giza), drought-2 (drought stress in Gemmiza), and heat stress.
qRT-PCR confirmed the response capability of TaPHD genes to abiotic stress conditions
To elucidate the possible regulatory mechanisms of TaPHD genes under cold, drought, and heat conditions, we performed qRT-PCR analysis of 20 genes ( Figure 11 ). The results showed that all 20 TaPHDs responded to different stress conditions and had different manifestations. Under low temperature stress induced by 4°C, the expression of five TaPHDs was significantly upregulated at different time points, and the expression of six TaPHDs was significantly downregulated at different time points compared with the control. In contrast, under 40°C-induced high-temperature stress, the expression of 12 TaPHDs was significantly upregulated at different time points compared with the control. The expression of five TaPHDs was inhibited at different time points. This indicated that compared with low temperature stress, high temperature stress could induce more changes in the expression of TaPHDs and could upregulate the expression more. In wheat under 16% PEG stress, the expression of ten TaPHDs was significantly upregulated at different time points. The expression of seven TaPHDs was inhibited at different time points. Among them, TaPHD72 was most significantly inhibited, and it was downregulated four-fold at 6 and 12 h after treatment. The expression levels of TaPHD69 and TaPHD135 significantly increased after the three treatments. However, the expression levels of TaPHD23 and TaPHD141 significantly decreased after the three treatments. In addition, TaPHD99 was strongly upregulated or downregulated by high temperature, low temperature, and PEG, and we speculated that this might be a key regulator of abiotic induction. In conclusion, we verified the effect of PHD-finger gene expression on the effect of three abiotic stresses in wheat using qRT-PCR. These results indicate that PHD-finger genes play an important role in coping with abiotic stress in wheat.
Figure 11.
Relative expression levels of 20 genes under three different treatments. Expression of TaPHD genes in wheat were detected after 4°C, 16% PEG, and 40°C treatments for 0, 1, 6, and 12 h. Significant differences were determined by one-way ANOVA test: * p < 0.05; ** p < 0.01.
Subcellular localization of TaPHD11, TaPHD19, and TaPHD133
Previous studies have shown that most PHD finger proteins are localized in the nucleus, and only a few are localized in the membranes or other organelles (Gozani et al., 2003; Wu et al., 2016; Sun et al., 2017). For example, ZmPHD14 and ZmPHD19 are localized to the nucleus (Wang et al., 2015a). Also, GmPHD1 to GmPHD6 target the nucleus, and their nuclear localization requires the PHD domain (Wei et al., 2009). To better understand the functions of TaPHDs, we used Plant-mPLoc and BUSCA to predict their subcellular localization. The results showed that more than 90% of the TaPHD proteins were localized in the nucleus ( Table S1 ). In Arabidopsis thaliana, the PHD genes AL5 and AL6 play a very important role in improving the resistance of plants to abiotic stress. Therefore, we selected TaPHD11 and TaPHD19, which are highly homologous to AtALs, for subcellular localization of wheat protoplasts. As shown in Figure 12 , this suggests that, in wheat, the proteins TaPHD11 and TaPHD19 not only function in the nucleus but also in the membrane. In addition, research has shown that PHD finger ING2 is a phosphoinositide binding module and a nuclear PtdInsP receptor and suggests that PHD-phosphoinositide interactions directly regulate nuclear responses to DNA damage (Gozani et al., 2003). However, we studied the protein TaPHD133, which is highly homologous to ING1, and found that it is localized not only in the nucleus but also in the membrane. In summary, the subcellular localization of PHD proteins in wheat differs from that in other species.
Figure 12.
Subcellular location of TaPHD11, TaPHD19, and TaPHD133. Localization of TaPHD proteins under normal conditions. Images were observed under a confocal laser scanning microscope (LSM 700, Zeiss). Scale bars = 10 μm.
Discussion
As an important transcription factor in organisms, the PHD gene family not only plays a key role in regulating plant growth and development but also an important regulatory role when plants face biotic and abiotic stresses (Mouriz et al., 2015). In this study, we identified 244 TaPHD gene members in the wheat genome for the first time ( Supplementary Table S3 ), which we divided into four large evolutionary branches. In terms of the number of genes, compared with the 59 and 67 PHD members in the diploid gramineous crops rice and maize, the PHD gene in wheat has a more exaggerated expansion and evolution. This is not only because the origin of wheat involves two polyploidy events, resulting in the existing allohexaploid bread wheat, but also because segmental duplication contributes to the amplification of TaPHD genes. Gene duplication events are important for the rapid expansion and evolution of plant gene families (Cannon et al., 2004). Approximately 70%–80% of angiosperms experience duplication events (Blanc et al., 2003; Bowers et al., 2003), and in common wheat (Triticum aestivum L.), more than 85% of the sequences are duplicates (Walkowiak et al., 2020). Our research revealed the presence of several segmental duplication events during the evolution of TaPHD genes ( Figures 4 , 5 ). The proportion of TaPHDs with a 1:1:1 ratio of the three subgenomes A:B:D accounted for 84.8% of the total proportion ( Table 2 ), which was much higher than the 35.8% observed for the whole wheat genome, indicating that the PHD gene family is highly conserved in the three subgenomes. When the PHD genes with different chaperone structural domains were subdivided ( Table 1 ), the fold divergences were also different; for example, ING1, ING2, ROS1, EBS, and PKL were expanded 3-fold, while SHL1 was expanded 6-fold, and VIN3 and SIZ1 were expanded 9-fold and 11-fold, respectively. It is likely that the presence of many redundant genes has contributed to the stability of the genome of the hexaploid wheat species (Consortium et al., 2018). In terms of the covariance and evolutionary relationship of wheat PHD genes among species ( Figure 5 ), the PHD-finger family diverged between monocotyledonous and dicotyledonous species, with the average divergence time from the monocotyledonous species in the order of barley (12.78 Mya) < rice (22.09 Mya) < maize (60.87 Mya), indicating a more similar genetic structure to barley in terms of PHD genes.
Genes perform their functions through transcription and translation, and the expression patterns of genes reflect their function. PHD genes can regulate the growth and development of plants. Therefore, their expression in different plant tissues has also attracted much attention. Studies have shown that the expression patterns of the PHD gene family in different species are concentrated in different tissue types (Sun et al., 2017; Qin et al., 2019; Wu et al., 2021). Most of them have a high level of expression in reproductive organs, including rice (Sun et al., 2017) and potato (Qin et al., 2019). However, in cotton, GhPHDs genes have the highest expression levels in ovule and fiber tissues (Wu et al., 2021). This study showed that the TaPHD expression in various tissues of wheat showed great differences with the growth period. In particular, TaPHD expression was highest in the stigma and ovary at the flowering stage. A large number of PHD proteins regulate plant reproductive and developmental processes, which indicates that TaPHDs may play the same role in rice and potato. It also has a similar expression pattern in Arabidopsis thaliana, the model plant with the most in-depth research. Some genes have been identified as having key functions. For example, MMD1, MS1, VIM1, and SHL1 in Arabidopsis have been shown to play key roles in the reproductive growth stage (Yang et al., 2003; Woo et al., 2007; Fernández Gómez and Wilson, 2014). Moreover, TaPHD100, TaPHD108, and TaPHD122, which were highly orthologous to AtAL6 and AtAL7 are highly expressed during the whole growth period. In Arabidopsis, AtAL6 and AtAL7 are methylated by histones via the PHD domain, and the modification sites H3K4me3 and H3K4me2 bind to regulate the expression of target genes. Alifn-PHD domain proteins bind to di- or trimethylated histone H3 (H3K4me3/2) and affect plant growth and development in Arabidopsis (Winicov, 2000). It can be seen that these three genes may play an important role in the growth and development of wheat via methylated histones. Furthermore, we can also speculate the function of the PHD gene in wheat through the expression mode of a more highly homologous PHD gene. PWWP-PHD-SET domain proteins have histone methyltransferase activities and regulate the development of roots, leaves, and floral organs, as well as the transcription of some stress genes (Saleh et al., 2008). Therefore, TaPHD100, TaPHD108, and TaPHD122, which have high coincidence with the PWWP-PHD-SET domain, may play important roles in regulating the growth and development of wheat histone methylation (Lee et al., 2009). In addition, TaPHD222 and TaPHD232 are only highly expressed in shoots and roots; these two genes are highly orthologous to ORC1A/B. In contrast, in Arabidopsis, the ORC1A/B protein binds methyl groups through the PHD domain and functions as a transcriptional activator (De La Paz Sanchez and Gutierrez, 2009). Therefore, we infer that TaPHD222 and TaPHD232 are essential for root and shoot development. However, their function during development requires further verification.
The PHD family not only regulates plant growth and development but also responds to abiotic stresses. Existing research shows that the PHD genes AL5 and AL6 in Arabidopsis bind to the promoter regions of downstream target genes, thereby inhibiting various signaling pathways to improve the resistance of plants to abiotic stresses such as low temperature, drought, and high salt (Gozani et al., 2003; Wei et al., 2015). Notably, in this study, TaPHD11 and TaPHD19, which are highly homologous to ALs, were upregulated only under induction by PEG treatment. Through qRT-PCR analysis, we also found that TaPHD11 and TaPHD19, which are highly homologous to AtALs, were significantly upregulated only under drought treatment. This finding is different from the results of the previous study in Arabidopsis, indicating that ALs seem to have different responses to abiotic stress in monocotyledonous and dicotyledonous plants. Meanwhile, subcellular localization experiments also showed that TaPHD11 and TaPHD19 were localized in the nucleus and cell membrane, indicating that they function not only in the nucleus but also in the cell membrane of wheat. This suggests that there are differences in the responses of PHD genes to abiotic stresses among species.
This does not mean that the PHD gene expression pattern of monocotyledons and dicotyledons is completely different. TaPHD69, which is highly homologous to AtSIZ1, can be significantly upregulated under low-temperature, drought, and high-temperature conditions. AtSIZ1 accumulates high levels of SUMOylated proteins through an ABA-independent pathway in response to abiotic stresses such as drought, low temperature, and heat shock (Catala et al., 2007). The accumulation of TaPHD69 seems to be beneficial for plants to cope with abiotic stress, which is similar to the function of AtSIZ1 in Arabidopsis. In rice, the cis-acting elements DRE/CRT in the OsPHD13 and OsPHD52 promoters are upregulated by as much as 15-fold under low-temperature stress. Overexpression of OsPHD1 can significantly improve plant resistance to stress (drought, high salt, and low temperature) (Liu et al., 2011; Ahmar and Gruszka, 2022). In maize, the expression of subfamily IX TaPHDs responds to salt, drought, and ABA stress (Wang et al., 2015a). Among TaPHDs, 45 TaPHDs genes were significantly changed under two or three treatments, indicating that TaPHDs play an active role in plant responses to low-temperature, drought, or high-temperature stress. TaPHD117 was significantly upregulated under high-temperature and drought treatments and significantly downregulated under low-temperature treatment and had distinct expression patterns in response to different treatments. Therefore, whether TaPHDs act as key genes in the roots to cope with abiotic stress requires further verification, but our results suggest that TaPHDs have potential functions in plant responses to abiotic stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author contributions
FP, ZL, and ZW designed the study. FP and MS conducted the experiments. JN and SN analyzed the data. FP, JN, ZL, and ZW wrote the manuscript. ZL and ZW revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The present study was supported by the Scientific Startup Foundation for Doctors of Yulin Normal University (CN) (Grant No. G2020ZK13).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1016831/full#supplementary-material
References
- Aasland R., Gibson T. J., Stewart A. F. (1995). The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 56–59. doi: 10.1016/S0968-0004(00)88957-4 [DOI] [PubMed] [Google Scholar]
- Ahmar S., Gruszka D. (2022). In-silico study of brassinosteroid signaling genes in rice provides insight into mechanisms which regulate their expression. Front. Genet. 13, 953458. doi: 10.3389/fgene.2022.953458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., De Castro E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K. H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. doi: 10.1101/gr.751803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L., Freemont P. S. (1996). The RING finger domain: a recent example of a sequence–structure family. Curr. Opin. Struct. Biol. 6, 395–401. doi: 10.1016/S0959-440X(96)80060-1 [DOI] [PubMed] [Google Scholar]
- Borrill P., Ramirez-Gonzalez R., Uauy C. (2016). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170, 2172–2186. doi: 10.1104/pp.15.01667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J., Paterson A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521 [DOI] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I. A., Hu Y., Seo H., Zhang X., et al. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19, 2952–2966. doi: 10.1105/tpc.106.049981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chou K.-C., Shen H.-B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335. doi: 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium I. W. G. S., Appels R., Eversole K., Stein N., Feuillet C., Keller B., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Cui X.-Y., Gao Y., Guo J., Yu T.-F., Zheng W.-J., Liu Y.-W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1 . Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Paz Sanchez M., Gutierrez C. (2009). Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription. Proc. Natl. Acad. Sci. 106, 2065–2070. doi: 10.1073/pnas.0811093106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Gómez J., Wilson Z. A. (2014). A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777. doi: 10.1111/pbi.12181 [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Gao Y., Liu H., Wang Y., Li F., Xiang Y. (2018). Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 123, 378–391. doi: 10.1016/j.plaphy.2017.12.034 [DOI] [PubMed] [Google Scholar]
- Gibbons R. J., Bachoo S., Picketts D. J., Aftimos S., Asenbauer B., Bergoffen J., et al. (1997). Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 17, 146–148. doi: 10.1038/ng1097-146 [DOI] [PubMed] [Google Scholar]
- Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., et al. (2003). The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114, 99–111. doi: 10.1016/S0092-8674(03)00480-X [DOI] [PubMed] [Google Scholar]
- He Z., Xia X., Chen X., Zhuang Q. (2011). Progress of wheat breeding in China and the future perspective. Acta Agronom. Sin. 37, 202–215. doi: 10.3724/SP.J.1006.2011.00202 [DOI] [Google Scholar]
- Hu L., Li Z., Wang P., Lin Y., Xu Y. (2011). Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 21, 1374–1378. doi: 10.1038/cr.2011.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J. A., Bienz M., et al. (1998). dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science 282, 1897–1900. doi: 10.1126/science.282.5395.1897 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan A. H., Gell D. A., Verger A., Crossley M., Matthews J. M., Mackay J. P. (2003). Engineering a protein scaffold from a PHD finger. Structure 11, 803–813. doi: 10.1016/S0969-2126(03)00122-9 [DOI] [PubMed] [Google Scholar]
- Lee W. Y., Lee D., Chung W. I., Kwon C. S. (2009). Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58, 511–524. doi: 10.1111/j.1365-313X.2009.03795.x [DOI] [PubMed] [Google Scholar]
- Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., et al. (2006). Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95. doi: 10.1038/nature04802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ng C. K.-Y., Fan L.-M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014 [DOI] [Google Scholar]
- Liu Y., Liu C., Li Z., Xia M., Jiang H., Cheng B., et al. (2011). Overexpression of a plant homedomain (PHD)-finger transcription factor, OsPHD1, can enhance stress tolerance in rice. J. Agric. Biotechnol. 19, 462–469. doi: 10.3969/j.issn.1674-7968.2011.03.009 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. G., Baetz K., Shi X., Walter K. L., Macdonald V. E., Wlodarski M. J., et al. (2006). The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26, 7871–7879. doi: 10.1128/MCB.00573-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Molitor A. M., Bu Z., Yu Y., Shen W.-H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10, e1004091. doi: 10.1371/journal.pgen.1004091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouriz A., López-González L., Jarillo J. A., Piñeiro M. (2015). PHDs govern plant development. Plant Signaling Behav. 10, e993253. doi: 10.4161/15592324.2014.993253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959. doi: 10.1016/S0092-8674(00)82001-2 [DOI] [PubMed] [Google Scholar]
- Papoulas O., Beek S. J., Moseley S. L., Mccallum C. M., Sarte M., Shearn A., et al. (1998). The drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125, 3955–3966. doi: 10.1242/dev.125.20.3955 [DOI] [PubMed] [Google Scholar]
- Qin M., Luo W., Zheng Y., Guan H., Xie X. (2019). Genome-wide identification and expression analysis of the PHD-finger gene family in Solanum tuberosum . PloS One 14, e0226964. doi: 10.1371/journal.pone.0226964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-González R., Borrill P., Lang D., Harrington S., Brinton J., Venturini L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089. doi: 10.1126/science.aar60 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Yilmaz M., Le O., Hou G., Sadder M., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20, 568–579. doi: 10.1105/tpc.107.056614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savojardo C., Martelli P. L., Fariselli P., Profiti G., Casadio R. (2018). BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 46, W459–W466. doi: 10.1093/nar/gky320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U., Beckmann H., Cashmore A. R. (1993). HAT3. 1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150. doi: 10.1046/j.1365-313X.1993.04010137.x [DOI] [PubMed] [Google Scholar]
- Song S., Hao L., Zhao P., Xu Y., Zhong N., Zhang H., et al. (2019). Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum group phureja). Sci. Rep. 9, 1755. doi: 10.1038/s41598-018-37923-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. doi: 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- Sun M., Jia B., Yang J., Cui N., Zhu Y., Sun X. (2017). Genome-wide identification of the PHD-finger family genes and their responses to environmental stresses in oryza sativa l. Int. J. Mol. Sci. 18, 2005. doi: 10.3390/ijms18092005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang Y., Sui N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123 [DOI] [PubMed] [Google Scholar]
- Walkowiak S., Gao L., Monat C., Haberer G., Kassa M. T., Brinton J., et al. (2020). Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283. doi: 10.1038/s41586-020-2961-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu J., Wang Y., Zhao Y., Jiang H., Cheng B. (2015. a). Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. Int. J. Mol. Sci. 16, 23517–23544. doi: 10.3390/ijms161023517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Q., Zhao Y., Han G., Zhu S. (2015. b). Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566, 95–108. doi: 10.1016/j.gene.2015.04.041 [DOI] [PubMed] [Google Scholar]
- Wei W., Huang J., Hao Y.-J., Zou H.-F., Wang H.-W., Zhao J.-Y., et al. (2009). Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic arabidopsis plants. PloS One 4, e7209. doi: 10.1371/journal.pone.0007209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Zhang Y. Q., Tao J. J., Chen H. W., Li Q. T., Zhang W. K., et al. (2015). The alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in arabidopsis. Plant J. 81, 871–883. doi: 10.1111/tpj.12773 [DOI] [PubMed] [Google Scholar]
- Winicov I. (2000). Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta 210, 416–422. doi: 10.1007/PL00008150 [DOI] [PubMed] [Google Scholar]
- Woo H. R., Pontes O., Pikaard C. S., Richards E. J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21, 267–277. doi: 10.1101/gad.1512007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wu M., Dong Q., Jiang H., Cai R., Xiang Y. (2016). Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa . Gene 575, 75–89. doi: 10.1016/j.gene.2015.08.042 [DOI] [PubMed] [Google Scholar]
- Wu H., Zheng L., Qanmber G., Guo M., Wang Z., Yang Z. (2021). Response of phytohormone mediated plant homeodomain (PHD) family to abiotic stress in upland cotton (Gossypium hirsutum spp.). BMC Plant Biol. 21, 13. doi: 10.1186/s12870-020-02787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q., Wang Z., Zaromytidou A.-I., Zhang X. H.-F., Chow-Tsang L.-F., Liu J. X., et al. (2011). A poised chromatin platform for TGF-β access to master regulators. Cell 147, 1511–1524. doi: 10.1016/j.cell.2011.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., Kigawa T., Seki M., Shinozaki K., Yokoyama S. (2013). DNA-Binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Yang X., Makaroff C. A., Ma H. (2003). The arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15, 1281–1295. doi: 10.1105/tpc.010447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zhang Y., Cui H., Liu J., Wu Y., Cheng Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotation 2018 update. Nucleic Acids Res. 46, W71–W75. doi: 10.1093/nar/gky400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ouyang Y., Yao W. (2018). shinyCircos: an R/Shiny application for interactive creation of circos plot. Bioinformatics 34, 1229–1231. doi: 10.1093/bioinformatics/btx763 [DOI] [PubMed] [Google Scholar]
- Zhang X., Li M., Zhang B., Yin X., Wang M., Xia X. (2016). Possibility study on improving salt tolerance of rice by overexpressing PHD-finger transcription factor gene OsMsr16 . Genomics Appl. Biol. 35, 1820–1827. doi: 10.13417/j.gab.035.001820 [DOI] [Google Scholar]
- Zhang Z., Li J., Zhao X.-Q., Wang J., Wong G. K.-S., Yu J. (2006). KaKs_Calculator: calculating ka and ks through model selection and model averaging. Genom. Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.-X., Gong H.-J., Yin J.-L. (2019). Role of silicon in mediating salt tolerance in plants: a review. Plants 8, 147. doi: 10.3390/plants8060147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .