Abstract
Monoclonal antibodies (MAbs) were prepared against the putative binding domain of botulinum neurotoxin A (BoNT/A), a nontoxic 50-kDa fragment. Initially, all fusion products were screened against the holotoxin BoNT/A and against the binding fragment, BoNT/A HC. Eleven neutralizing hybridomas were cloned, and their specific binding to BoNT/A HC was demonstrated by surface plasmon resonance, with dissociation constants ranging from 0.9 to <0.06 nM. Epitope mapping by real-time surface plasmon resonance showed that the antibodies bound to at least two distinct regions of the BoNT/A HC fragment. These MAbs will be useful tools for studying BoNT/A interactions with its receptor, and they have potential diagnostic and therapeutic applications.
The anaerobic bacterium Clostridium botulinum produces seven immunologically distinct but structurally similar neurotoxins designated BoNT/A to BoNT/G that are associated with food-borne, infant, and wound botulism (12, 15, 16). Due to their unique properties, BoNTs have been used to treat a variety of human muscle disorders (13). After synthesis, highly active neurotoxin is generated by proteolytic cleavage of the clostridial neurotoxins. The active neurotoxin contains two polypeptide chains connected via a disulfide linkage. The location of the enzymatic subunit of the CNTs has been mapped to the light chain (∼50 kDa), which has Zn endopeptidase activity (3, 12, 17). On the other hand, the binding and translocation motifs are located within the heavy (H) chain (∼100 kDa).
Probably due to the unusually high toxicity of BoNTs, previous attempts to produce large numbers of high-affinity neutralizing monoclonal antibodies (MAbs) against these neurotoxins have been unsuccessful. Since vaccination with the nontoxic binding fragment (the 50-kDa carboxy-terminal fragment of the heavy chain [HC]) of BoNT/A can induce protective immunity in mice (5), we reasoned that it should be possible to generate neutralizing antibodies by using this fragment. We report herein that vaccination with BoNT/A HC elicited neutralizing MAbs. We have characterized these antibodies in detail, demonstrated their biochemical detection of BoNT/A and its binding fragment, determined their ability to neutralize the neurotoxin, measured their affinity, and mapped their epitope binding sites.
Antigens.
BoNT/A was purchased from the University of Wisconsin Food Research Institute (Madison, Wis.), and BoNT/A HC, BoNT/B HC, and BoNT/E HC were made and purified to homogeneity at our institute (U.S. Army Medical Research Institute of Infectious Diseases, Frederick, Md.). The BoNT/A HC preparation was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide) under reducing conditions and was at least 95% pure.
Laboratory animals.
Pathogen-free BALB/c (H-2d) mice, 10 to 12 weeks old, were obtained from the Frederick Cancer Research and Development Center (Frederick, Md.). The mice were maintained under pathogen-free conditions and fed laboratory chow and water ad libitum.
Vaccination and hybridoma production.
The mice were vaccinated intraperitoneally five times at 4-week intervals with 0.1 to 2 μg of BoNT/A HC in 100 μl of phosphate-buffered saline. Splenic mononuclear cells from the mice with the highest titers were collected and fused with myeloma cells, and after the HAT (hypoxanthine, aminopterin, and thymine) selection, the supernatants of the hybridoma cells were tested by enzyme-linked immunosorbent assay (ELISA) for the presence of antibodies to BoNT/A and BoNT/A HC. Some of the positive hybridoma supernatants were tested for their ability to neutralize BoNT/A, as described below. Limiting dilution was used to clone the hybrids that produced neutralizing antibodies.
Screening of hybridoma supernatants, antibody purification, and quantification of MAbs.
The binding of the cell-free hybridoma supernatants to BoNT/A or BoNT/A HC were measured by ELISA. The bound antibody was detected with horseradish peroxidase-labeled goat anti-mouse immunoglobulin G (IgG) (Cappel/Organon Teknika Corp., West Chester, Pa.), and the mean of duplicate optical density (OD) measurements of each sample at 620 nm was obtained after incubation with 3,3′,5,5′-tetramethylbenzidine peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Wells were scored as positive if the OD was greater than twice that of the negative controls in which antigen or serum was omitted.
MAbs were purified by using protein G conjugated to Sepharose (Pharmacia Biotech, Uppsala, Sweden), and the IgG was quantitated as specified by the manufacturer. The isotype and subclass of each MAb were determined by standard direct ELISA as specified by the manufacturer (Bio-Rad Laboratories, Inc., Melville, N.Y.).
Neutralization assays.
Serial dilutions of MAbs were incubated with various lethal doses (5 to 20 mouse 50% lethal doses [LD50]) of BoNT/A for 1 h at room temperature. The toxin-antibody mixture was administered intraperitoneally at a dose of 0.2 ml per mouse. Five days after challenge, the mice were scored for survivors. In initial neutralization assays, mice were observed for up to 20 days.
Measurement of binding kinetics using SPR.
The affinities of the MAbs were determined with an optical biosensor using real-time surface plasmon resonance technology (SPR) (BIAcore 1000 with upgrade; Pharmacia Biacore, Piscataway, N.J.). Briefly, anti-mouse Fc antibody was coupled to the chip as specified by the manufacturer and MAb was captured and immobilized by the chip. Kinetic analyses were carried out at a flow rate of 25 μl/min with 5 to 200 nM BoNT/A HC in HEPES-buffered saline containing 3 mM EDTA and 0.005% Tween (HBS). Values for the apparent equilibrium dissociation constant (KD) were calculated from the ratio of the dissociation (koff) and association (kon) rate constants obtained with the BIAevaluation 2.1 software package supplied by the vendor. Two 30-s pulses of 10 mM glycine (pH 1.8) were used to remove bound MAbs and regenerate the biosensor chips with anti-IgG Fc antibody.
Epitope mapping with biosensor technology.
Epitope mapping of the MAbs was carried out by SPR at a flow rate of 5 μl/min. Affinity-purified antibody to mouse IgG Fc (8,045 response units [RU]) was immobilized onto the chip, and a series of reagents were each passed over the chip for 3 min with a 2-min wash with HBS between injections. First, purified BoNT/A HC MAb was captured by the Fc-specific antibody. Second, nonspecific sites were blocked by passing a saturating concentration (0.10 mg/ml) of an unrelated MAb over the matrix surface. Third, the antigen (200 nM BoNT/A HC in HBS) was allowed to bind to the captured MAb. Finally the second MAb was injected, and its binding was determined. The biosensor chip was regenerated as described above, and the process was repeated to test the ability of all MAbs to bind as a second MAb by using each as the first MAb. Thus, all antibody pairs were tested in both directions. The use of controls demonstrated that binding of BoNT/A HC required the presence of bound first MAb and that binding of the second MAb required the presence of bound BoNT/A HC.
Generation of neutralizing MAbs to BoNT/A.
We hypothesized that neutralizing antibodies against BoNT/A might best be generated if we vaccinated animals with BoNT/A HC, the protective, nontoxic receptor binding fragment of BoNT/A. To test this hypothesis, spleen cells from the highly seropositive (titers, >105) mice were fused with myeloma cells. To avoid isolation of antibodies with little or no reactivity to the whole toxin, all of the fusion product supernatants were screened by a direct ELISA on 96-well plates containing intact BoNT/A or BoNT/A HC. This vaccination approach produced 660 hybridoma fusions that recognized BoNT/A HC, and 488 of these 660 hybridoma fusions recognized the native toxin in an ELISA-based assay.
To increase the possibility of finding neutralizing antibodies, we examined the protective ability of some of the fusion products that recognized the toxin in an in vivo mouse lethality model before cloning and subsequently subcloning the hybrids. The cells corresponding to the positive wells were transferred to six-well plates and cultured for 7 to 10 days. The culture supernatants were tested for their ability to protect mice against 10 LD50 of BoNT/A. We obtained 33 hybrids that produced neutralizing antibody, and these fusion products were cloned. Candidate hybridomas were subjected to a second round of expansion and screening and were subcloned. Eleven hybridomas were cloned, and antibodies were immunopurified on protein G columns. The names and isotypes of all MAbs were determined and are listed in Table 1. The isotypes of MAbs 6B2-2 and 6C2-4 were IgG1, and those of the remaining nine MAbs were IgG2a. When MAbs were tested against 5 to 10 LD50 of BoNT/A, all the mice were protected (Table 1).
TABLE 1.
Specificity of BoNT/A neutralizing MAbs
| MAb | Isotypea | BoNT serotype | ELISA resultb | LD50c | No. live/ no. deadd |
|---|---|---|---|---|---|
| 4A2-2 | IgG2a | A | +++ | 10 | 5/0 |
| B | ND | 5 | 0/5 | ||
| E | ND | 5 | 0/5 | ||
| 4A2-4 | IgG2a | A | +++ | 10 | 0/5 |
| 6B2-2 | IgG1 | A | +++ | 10 | 5/0 |
| B | ND | 5 | 0/5 | ||
| E | ND | 5 | 0/5 | ||
| 6C2-4 | IgG1 | A | +++ | 10 | 5/0 |
| B | ND | 5 | 0/5 | ||
| E | ND | 5 | 0/5 | ||
| 6E9-1 | IgG2a | A | +++ | 10 | 5/0 |
| B | ND | 5 | 0/5 | ||
| E | ND | 5 | 0/5 | ||
| 6E9-3 | IgG2a | A | +++ | 10 | 5/0 |
| 6E9-4 | IgG2a | A | +++ | 10 | 5/0 |
| 6E10-4 | IgG2a | A | +++ | 10 | 5/0 |
| 6E10-5 | IgG2a | A | +++ | 10 | 5/0 |
| 6E10-8 | IgG2a | A | +++ | 10 | 5/0 |
| 6E10-10 | IgG2a | A | +++ | 10 | 5/0 |
The isotypes were determined as described in the text.
ELISA wells were coated with BoNT/A, BoNT/B, or BoNT/E. The MAbs were added to the wells at 100 ng/well, and bound antibodies were detected with horseradish peroxidase-labeled antibody. OD readings above 1.2 are presented as +++, while OD readings below 0.05 are shown as ND (not detected).
The antibody was incubated for 1 h with 10 LD50 of BoNT/A or 5 LD50 of BoNT/B or BoNT/E before being administered to the mice. Lethality was recorded 7 days after the challenge dose.
Final lethality was statistically scored using Fisher exact tests. P < 0.001 indicates protection against BoNT/A versus BoNT/B or BoNT/E.
Next, we analyzed the specificity of the MAbs by ELISA. After preliminary characterization of all 11 MAbs against BoNT/A, five of them (4A2-2, 4A2-4, 6B2-2, 6C2-4, and 6E9-1) that represented both isotypes were selected for further evaluation against two heterologous BoNTs. All antibodies recognized BoNT/A HC and did not bind to the HC fragment of the other BoNTs, nor did they protect against either BoNT/B or BoNT/E (Table 1). These results indicate that vaccination with the protective fragment of BoNT/A elicited the production of substantial numbers of homologous neutralizing MAbs.
Characterization of the neutralizing MAbs.
To explore the binding characteristics of the MAbs, we examined the interaction of the neutralizing MAbs with intact BoNT/A and BoNT/A HC under a variety of conditions, including Western immunoblotting and immunoprecipitation. All of the MAbs recognized a 50-kDa protein corresponding to BoNT/A HC after the protein was Western immunoblotted, and the antibodies did not recognize other portion of BoNT/A (data not shown).
We also evaluated the abilities of the MAbs to detect BoNT/A HC under more physiological conditions. BoNT/A HC was incubated with the neutralizing MAbs, and the immune complexes were captured by protein A-Sepharose. After being washed, they were eluted, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and detected with rabbit anti-BoNT/A HC. All MAbs immunoprecipitated BoNT/A HC with similar relative intensities, suggesting that their affinities may be similar (data not shown).
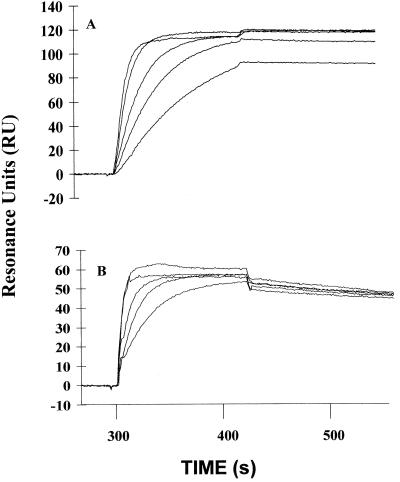
To characterize the kinetic interactions between the neutralizing MAbs and BoNT/A HC, we used the SPR biosensor technology. Each antibody was captured by anti-mouse IgG Fc on a biosensor chip, various concentrations of BoNT/A HC were passed through the flow cell, and the binding kinetics were recorded. The kinetic rate constants kon and koff were determined from the ascending rate of the BIAcore signal during binding and the descending rate during the wash-off interval, respectively. Examples of the binding kinetics are shown in Fig. 1A for MAb 6E9-4 and Fig. 1B for MAb 6B2-2. We calculated the apparent KDs of each MAb for BoNT/A HC from the ratio koff/kon. As seen in Table 2, all of the MAbs had similar on rates, and their overall values for KD ranged from about 1 to 0.1 nM. The KD for one MAb, 6B2-2, was significantly lower (<0.06 nM) but was difficult to resolve accurately due to its very low rate of dissociation (Fig. 1B) compared with the other MAbs. Although we observed differences between the MAbs in overall affinity, it is noteworthy that a feature common to all of the neutralizing MAbs is their very high affinity.
FIG. 1.
Kinetic analyses of MAb binding to BoNT/A HC by SPR. Biosensor chips with immobilized anti-mouse Fc were used to capture 104 RU of MAb 6E9-4 (A) or 190 RU of MAb 6B2-2 (B). After equilibration, a series of concentrations of the antigen BoNT/A HC (30, 60, 100, 200, and 300 nM) was passed over the chip for 2 min, and this was followed by a washout phase with HBS buffer. Controls for a small amount of binding of BoNT/A HC when no MAb was bound were run, and the results have been subtracted for each concentration of antigen. Kinetic constants were calculated using BiaEvaluation software version 2.1.
TABLE 2.
Kinetic constants of BoNT/A MAbs binding to BoNT/A HCa
| MAb | RUb of MAb | koff (M−1 s−1) | kon (s−1) | KD (nM) |
|---|---|---|---|---|
| 4A2-2 | 150 | 3.5 × 10−4 | 6.9 × 105 | 0.51 |
| 4A2-4 | 128 | 4.2 × 10−4 | 7.8 × 105 | 0.54 |
| 6B2-2 | 263 | <0.2 × 10−4 | 3.1 × 105 | <0.06 |
| 6C2-4 | 148 | 1.9 × 10−4 | 17 × 105 | 0.11 |
| 6E9-1 | 170 | 2.5 × 10−4 | 11 × 105 | 0.22 |
| 6E9-3 | 121 | 2.8 × 10−4 | 11 × 105 | 0.25 |
| 6E9-4 | 104 | 6.7 × 10−4 | 7.8 × 105 | 0.86 |
| 6E10-4 | 80 | 5.3 × 10−4 | 11 × 105 | 0.48 |
| 6E10-5 | 128 | 3.9 × 10−4 | 4.8 × 105 | 0.81 |
| 6E10-8 | 124 | 2.1 × 10−4 | 4.9 × 105 | 0.42 |
| 6E10-10 | 90 | 3.1 × 10−4 | 14 × 105 | 0.22 |
The kinetic constants were measured by SPR as described in the text.
The amount of MAb captured by the chip-immobilized anti-mouse Fc is shown as RU (response units).
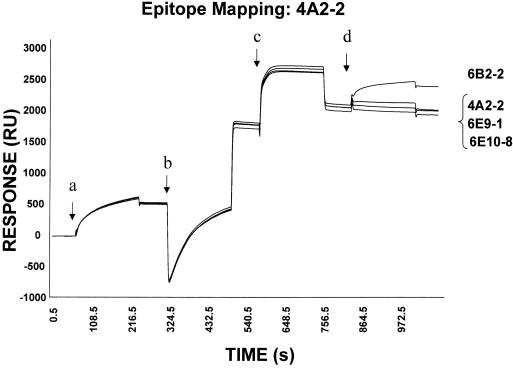
Next, we used SPR to characterize the binding sites of the MAbs. The BoNT/A HC MAb was captured by anti-mouse Fc, and any remaining anti-mouse binding sites were blocked with an unrelated MAb. After binding the antigen BoNT/A HC, a second MAb was injected and its binding was determined. This experiment was repeated to examine the ability of all MAbs to bind as the second MAb by using each as the first MAb, thus testing all pairs of antibodies in both directions. As seen in an example in Fig. 2, MAb 4A2-2 was captured by anti-mouse Fc on the sensor chip and then allowed to bind its antigen, BoNT/A HC. As expected, when the same antibody 4A2-2 was injected, no additional binding was observed, since the BoNT/A HC epitope for this MAb was already occupied by interaction with chip-immobilized MAb 4A2-2. Other heterologous antibodies, i.e., 6B2-2, 6E9-1, or 6E10-8, were tested similarly, and only MAb 6B2-2 bound, showing that its epitope on the antigen was distinct from that of MAb 4A2-2. We observed no binding of MAbs 6E9-1 and 6E 10-8, which indicated that their binding epitopes were the same or similar to that of MAb 4A2-2. All combinations of antibodies were tested likewise, and the data obtained are summarized in Fig. 3.
FIG. 2.
Epitope-mapping analyses of neutralizing MAbs by SPR. A biosensor chip with immobilized anti-mouse Fc was equilibrated with HBS buffer, and the following solutions were passed sequentially over the chip with a 2-min injection of HBS buffer between each reagent: MAb 4A2-2 for 3 min (a), unrelated mouse MAb for 3 min (to block all remaining anti-mouse Fc sites) (b), 10 μg of antigen BoNT/A HC per ml for 3 min (c), and the second MAb for 3 min, either 4A2-2, 6B2-2, 6E9-1, or 6E10-10 as indicated (d). There is a response, sometimes large, at each injection due to the change in the refractive index of the medium.
FIG. 3.
Summary of epitope-mapping studies of neutralizing MAbs by SPR. We propose at least two distinct neutralizing epitopes from the MAb-mapping studies. One site is defined by MAb 6B2-2. Another site is defined by MAb 6E9-1 and is shared by all other MAbs tested except 6B2-2 and 6C2-4. The epitope of MAb 6C2-4 overlaps that defined by MAbs 4A2-2 and 4A2-4, which must therefore be overlapping rather than identical to the 6E9-1 binding site.
The epitope-mapping studies defined three groups of MAbs, corresponding to two distinct and one overlapping protective epitope regions on BoNT/A HC. One particular region of the antigen was defined by MAb 6B2-2, while MAbs 4A2-2, 4A2-4, 6E9-1, 6E9-3, 6E9-4, 6E10-4, 6E10-5, 6E10-8, and 6E10-10 bound to a distinct site, clearly defining two protective epitopes. However, the results with MAb 6C2-4 were more complex. When captured as first antibody, 6C2-4 bound and presented BoNT/A HC in a way that it bound all other MAbs except 4A2-2 and 4A2-4. When 6C2-4 was tested as second MAb, it bound well (a level typical of all other interactions) only to antigen presented by 6B2-2. It bound to a lower extent to antigens presented by 6E9-1, 6E9-3, 6E9-4, 6E10-4, 6E10-5, 6E10-8, and 6E10-10, and it did not bind significantly to antigens presented by 4A2-2 and 4A2-4. Thus, the MAb 6C2-4 epitope was distinct from the binding epitope of 6B2-2 as well as from that of 6E9-1 and all other 6E9 and 6E10 MAbs, but it overlapped with that of MAbs 4A2-2 and 4A2-4.
Interpretation of the results.
A major obstacle in obtaining protective antibodies against BoNTs is the extreme toxicity of these toxins. To circumvent this, various approaches have been employed (4, 7, 8). Here we used BoNT/A HC as the antigen and obtained numerous neutralizing MAbs against BoNT/A. Amersdorfer et al. (1) have also vaccinated mice with BoNT/A HC to generate a large number of MAbs. Spleens were obtained from the vaccinated mice, and phage libraries were constructed from their Ig VH and VK genes. After using phage display to create and screen millions of antibodies for binding, they characterized 28 single-chain MAb fragments. The single-chain antibodies were found by SPR to bind to four distinct epitopes with dissociation constants ranging from 1 to 70 nM. Some of the constructs showed partial antagonism of toxin-induced neuroparalysis, indicating that two of the four epitopes may be somewhat protective in vitro. However, these small antibody fragments are rapidly cleared from the bloodstream and could not be meaningfully tested in vivo.
The MAbs reported here recognized neither BoNT/B HC nor BoNT/E HC and are specific for BoNT/A HC and BoNT/A holotoxin. Although the various BoNT serotypes have significant amino acid sequence homology, there is little to no cross-protection among heterologous neurotoxins. This may be explained by the lack of sequence conservation within the last ∼150 C-terminal residues of the HC region (9–11, 18).
Previous attempts to identify linear, protective epitopes of BoNT/A suggest that these epitopes are easily denatured and may be discontinuous or conformational (1, 2, 4, 6). Results using a display library to identify epitopes to the MAbs described here were also consistent with their binding to discontinuous epitopes (M. Segal and S. Bavari, unpublished results). We have used the MAbs to define at least two (possibly three) distinct, protective epitopes on BoNT/A HC. MAbs 6C2-4 and 6E family bound to distinct epitopes, but both interfered with the binding of MAb 4A2-2. Thus, these three MAbs may have bound to sites clustered in one region of the toxin that appeared to contain two distinct epitopes and may be part of one large protective epitope. MAb 6B2-2 bound to a fully independent site. For each of the three epitopes, at least one MAb bound with very high affinity, ranging from 0.2 to 0.06 nM. Further characterization of these epitopes and identification of their locations within the BoNT/A HC sequence may help to create a more effective peptide-based vaccine.
A second benefit of defining the protective epitopes on BoNT/A HC antigen will be to improve in vitro diagnostic assays. The current ELISA is not sufficiently reliable to predict protective immunity because it measures the antibody response to the whole antigen, which can include both protective and nonprotective antibodies (14). The MAbs to defined, protective epitopes on the BoNT/A HC antigen described in this study could enable the development of competitive ELISAs to determine the magnitude and duration of a protective immune response in mice. The goal is to develop an in vitro correlate of immunity in mice and ultimately an in vitro correlate of immunity in humans. In addition, one or more of the high-affinity MAbs described here may be useful in monitoring food for contamination with BoNT/A. For example, a rapid, sensitive assay specific for BoNT/B was recently reported that uses a serospecific MAb to capture the toxin from food samples (19).
To summarize, we obtained neutralizing MAbs by vaccination with the nontoxic binding fragment BoNT/A HC. These MAbs have very high affinity and recognized at least two neutralizing epitopes. They will be important tools in attempts to monitor immunity developed in vaccinations, to transfer passive immunity, and to further characterize the protective epitopes on the binding domain of BoNT/A.
REFERENCES
- 1.Amersdorfer P, Wong C, Chen S, Smith T, Deshpande S, Sheridan R, Finnern R, Marks J D. Molecular characterization of murine humoral immune response to botulinum neurotoxin type A binding domain as assessed by using phage antibody libraries. Infect Immun. 1997;65:3743–3752. doi: 10.1128/iai.65.9.3743-3752.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavari S, Pless D D, Torres E R, Lebeda F J, Olson M A. Identifying the principal protective antigenic determinants of type A botulinum neurotoxin. Vaccine. 1998;16:1850–1856. doi: 10.1016/s0264-410x(98)00175-3. [DOI] [PubMed] [Google Scholar]
- 3.Blasi J, Chapman E R, Binz L E, T, Yamasaki S, De Camilli P, Sudhof T C, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP25. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown D R, Lloyd J P, Schmidt J J. Identification and characterization of a neutralizing monoclonal antibody against botulinum neurotoxin, serotype F, following vaccination with active toxin. Hybridoma. 1997;16:447–456. doi: 10.1089/hyb.1997.16.447. [DOI] [PubMed] [Google Scholar]
- 5.Clayton M A, Clayton J M, Brown D R, Middlebrook J L. Protective vaccination with a recombinant fragment of Clostridium botulinum neurotoxin serotype A expressed from a synthetic gene in Escherichia coli. Infect Immun. 1995;63:2738–2742. doi: 10.1128/iai.63.7.2738-2742.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dertzbaugh M T, West M W. Mapping of protective and cross-reactive domains of the type A neurotoxin of Clostridium botulinum. Vaccine. 1996;14:1538–1544. doi: 10.1016/s0264-410x(96)00094-1. [DOI] [PubMed] [Google Scholar]
- 7.Kiyatkin N, Maksymowych A B, Simpson L L. Induction of an immune response by oral administration of recombinant botulinum toxin. Infect Immun. 1997;65:4586–4591. doi: 10.1128/iai.65.11.4586-4591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubota T, Watanabe T, Yokosawa N, Tsuzuki K, Indoh T, Moriishi K, Sanda K, Maki Y, Inoue K, Fujii N. Epitope regions in the heavy chain of Clostridium botulinum type E neurotoxin recognized by monoclonal antibodies. Appl Environ Microbiol. 1997;63:1214–1218. doi: 10.1128/aem.63.4.1214-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacy D B, Stevens R C. Sequence homology and structural analysis of the clostridial neurotoxins. J Mol Biol. 1999;291:1091–1104. doi: 10.1006/jmbi.1999.2945. [DOI] [PubMed] [Google Scholar]
- 10.Lacy D B, Stevens R C. Unraveling the structures and modes of action of bacterial toxins. Curr Opin Struct Biol. 1998;8:778–784. doi: 10.1016/s0959-440x(98)80098-5. [DOI] [PubMed] [Google Scholar]
- 11.Lacy D B, Tepp W, Cohen A C, DasGupta B R, Stevens R C. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 12.Montecucco C, Schiavo G. Mechanism of action of tetanus and botulinum neurotoxins. Mol Microbiol. 1994;13:1–8. doi: 10.1111/j.1365-2958.1994.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 13.Schantz E J, Johnson E A. Botulinum toxin: the story of its development for the treatment of human disease. Perspect Biol Med. 1997;40:317–327. doi: 10.1353/pbm.1997.0032. [DOI] [PubMed] [Google Scholar]
- 14.Siegel L S. Human immune response to botulinum pentavalent (ABCDE) toxoid determined by a neutralization test and by an enzyme-linked immunosorbent assay. J Clin Microbiol. 1988;26:2351–2356. doi: 10.1128/jcm.26.11.2351-2356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson L L. The origin, structural and pharmacological activity of botulinum toxin. Pharmacol Rev. 1981;33:155–188. [PubMed] [Google Scholar]
- 16.Simpson L L. The study of clostridial and related toxins. The search for unique mechanisms and common denominators. J Physiol. 1990;84:143–151. [PubMed] [Google Scholar]
- 17.Tonello F, Morante S, Rossetto O, Schiavo G, Montecucco C. Tetanus and botulism neurotoxins: a novel group of zinc-endopeptidases. Adv Exp Med Biol. 1996;389:251–260. [PubMed] [Google Scholar]
- 18.Umland T C, Wingert L M, Swaminathan S, Furey W F, Schmidt J J, Sax M. Structure of the receptor binding fragment HC of tetanus neurotoxin. Nat Struct Biol. 1997;4:788–792. doi: 10.1038/nsb1097-788. [DOI] [PubMed] [Google Scholar]
- 19.Wictome J, Newton K, Jameson K, Hallis B, Dunnigan P, Mackay E, Clarke S, Taylor R, Gaze J, Foster K, Shone C. Development of an in vitro bioassay for Clostridium botulinum type B neurotoxin in foods that is more sensitive than the mouse bioassay. Appl Environ Microbiol. 1999;65:3787–7923. doi: 10.1128/aem.65.9.3787-3792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]