Highlights
-
•
circ-ZEB1 is highly expressed in colorectal cancer tissues.
-
•
circ-ZEB1 knocking down inhibits CRC cells proliferation and metastasis in vitro and in vivo.
-
•
circ-ZEB1 regulates EMT and chemotherapeutic resistance in CRC cells.
-
•
circ-ZEB1 exerts its regulatory effect through sponging miR-200c.
-
•
Our research provides new targets for colorectal cancer treatment.
Keywords: circ-ZEB1, miR-200c, EMT, Chemoresistance, Colorectal cancer
Abbreviations: CRC, Colorectal cancer; EMT, Epithelial-mesenchymal Transition
Abstract
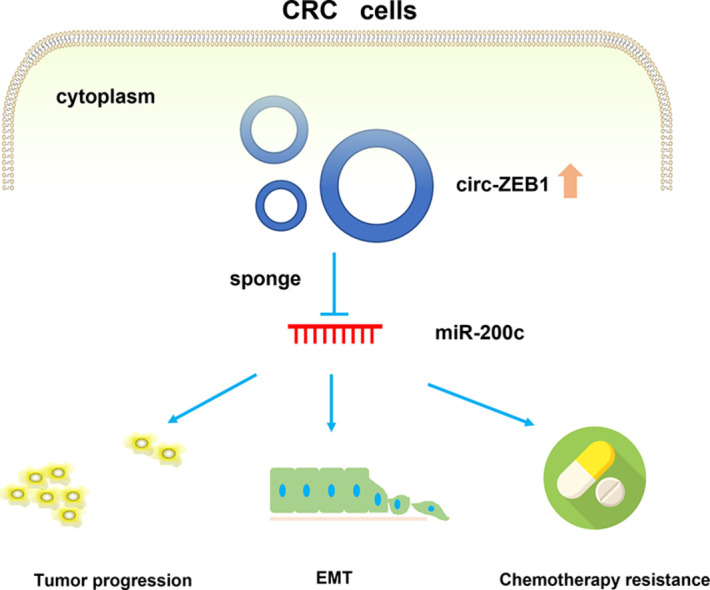
Circular RNAs (circRNAs) have been demonstrated to be important regulators in human malignant tumors, including colorectal cancer (CRC). While the role circ-ZEB1 played in CRC remains unclear. In this study, we aim to explore the biological function and the underlying mechanism of circ-ZEB1 in CRC. RNAscope was used to analyze the expression and localization of circ-ZEB1 in CRC tissues. Loss of function experiments were conducted, including CCK-8, transwell assays, flow cytometry analysis, and murine xenograft models, so as to detect the effect of circ-ZEB1 on CRC cells. IC50 assay was used to evaluate the influence of circ-ZEB1 on the chemoresistance of CRC cells. Epithelial-mesenchymal transition (EMT) related markers were detected. The relationship between circ-ZEB1 and miR-200c-5p was investigated by FISH, dual-luciferase reporter assay, and RIP assay. We found in our study that circ-ZEB1 was significantly upregulated in CRC tissues. Downregulation of circ-ZEB1 inhibited cell proliferation, colony formation, as well as cell migration and invasion abilities of CRC cell lines. In vivo experiments indicated that knockdown of circ-ZEB1 suppressed tumorigenesis and distant metastasis of CRC cells in nude mice. What's more, EMT and chemoresistance of CRC cells were also attenuated following circ-ZEB1 knockdown. Mechanistically, we proved that circ-ZEB1 could directly bind with miR-200c and functioned as miR-200c sponge to exert its biological functions in CRC cells. In conclusion, circ-ZEB1 could promote CRC cells progression, EMT, and chemoresistance via acting on miR-200c, elucidating a potential therapeutic target to inhibit CRC progression.
Graphical abstract
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors. Its incidence ranks third among all tumors (10% of total), and the mortality of CRC ranks second, with approximately 935,000 deaths in 2020 [1]. Approximately 25% of patients develop distant metastasis at presentation, which is the main cause of poor prognosis and death in patients with CRC. Despite the improvements in treatments, the survival rate of patients with CRC remains unsatisfactory. So, exploring the pathophysiology mechanisms underlying the occurrence and metastasis of CRC will help us to improve the treatment strategy and prognosis of the patients.
Oxaliplatin and 5-fluorouracil (5-FU) are the most prevalently used chemotherapeutics in clinical practice for patients with advanced colorectal cancer [2]. However, de novo or acquired chemoresistance during chemotherapy remains challenging [3]. So, there is an urgent need to identify those key molecular determinants of sensitivity to chemotherapy drugs, which may constitute novel predictive biomarkers of drug resistance. Epithelial-mesenchymal transition (EMT) plays a central role in tumor development. It converts epithelial cells to a mesenchymal-like phenotype and is associated with the loss of cell contacts, resulting in the increased motility and invasion ability of cancer cells [4,5]. EMT is related to cancer progression, recurrence, metastasis, and chemotherapy resistance. Numerous studies have proved that non-coding RNAs are involved in the regulation of EMT process of cancer cells [6,7].
Circular RNAs (circRNAs) are a newly discovered class of endogenous noncoding RNAs that differ from traditional linear RNAs. They are characterized by a stably closed-loop lacking the 5’ cap and the 3’ end of the poly (A) tail, forming a ring structure with covalent bonds, thus obtaining a high degree of stability and sequence conservation [8,9]. They exert regulatory functions at both transcriptional and post-transcriptional levels [10]. Circular RNAs are found to play prominent roles in tumorigenesis and tumor progression, which have recently become the focus of molecular oncology research. As widely reported, circRNAs always act as competing endogenous RNAs (ceRNAs) by sponging miRNAs and blocking the inhibition of their target gene expression [11]. In CRC, many circRNAs have been proven to play regulatory roles, such as circSPARC [12], circ_0124554 [13], and CircMYH9 [14], etc., which affect different malignant phenotypes of CRC cancer cells, including proliferation, metastasis, apoptosis, and resistance to chemotherapy or radiotherapy.
Previously, we explored the expression profiles of circRNAs in CRC tissues through high-throughput RNA sequencing and identified circ-ZEB1 as a differentially expressed circRNA. It derives from ZEB1 gene with a higher expression in cancerous tissues than normal colorectal mucosa. Bioinformatic prediction showed that circ-ZEB1 might bind with miR-200c-5p. While both ZEB1 and miR-200c are reported to be involved in the regulation of EMT, as well as chemotherapy resistance. So, we presume that circ-ZEB1 may also regulate EMT and drug resistance of CRC cells. To our knowledge, there is still no study about the role circ-ZEB1 played in CRC. In this study, we aim to explore the expression and biological functions of circ-ZEB1 in CRC. we hope our study may contribute to revealing the pathophysiologic mechanism of CRC and may provide a new target for CRC treatment.
Materials and methods
Patients and specimens
A total of 44 pairs of colorectal cancer tissue samples and adjacent normal mucosal tissue samples were collected by our research group from Beijing Chao-Yang Hospital, Capital Medical University between October 1, 2020 and August 31, 2021. The samples were further used to construct tissue microarray (TMA). All specimens were confirmed by the results of pathological diagnosis. None of the patients received chemoradiotherapy or targeted therapy before surgery. This study was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University, and was conducted in accordance with the ethical standards stated in the Declaration of Helsinki. All patients were aware and signed the relevant informed consent.
Cell lines
The human colorectal cancer cell lines were purchased from the American Type Culture Collection. Among them, SW480 and RKO were cultured in RPMI 1640. All cells were cultured in complete medium consisting of 90% basal medium, 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin (penicillin 100 U/ml, streptomycin 100 ug/ml). The cells were cultured at 37°C with 5% CO2.
Oligonucleotides transfection
Small interfering RNA (siRNA) against circ-ZEB1 (sense: 5′- GGAUCAUUGGAGUUUUCAGTT -3′, antisense: 5′- CUGAAAACUCCAAUGAUCCTT -3′), miR-200c mimics, miR-200c inhibitor and respective negative control oligonucleotides used in this experiment were purchased from GenePharma (GenePharma Corporation, Shanghai, China). When the cells were in good condition as long as about 50% confluence, we transfected oligonucleotides according to the instructions of lipofectamine 3000 (Invitrogen, USA).
Lentivirus construction and infection
To stably knock down circ-ZEB1, lentiviruses expressing short-hairpin RNA (shRNA) directly targeting circ-ZEB1 or negative control (NC) were synthesized by Hanheng Biotechnology Corporation (Shanghai, China). For cloning the shRNA and negative control, pHBLVCMV-MCS-EF1-puro lentiviral vector (Hanheng Biotechnology Corporation, Shanghai, China) was used. RKO cells were further infected with the lentiviruses and puromycin was used for selection for at least 14 days.
Total RNA extraction, reverse transcription, and qRT-PCR
Extraction of total RNA was conducted using TRIzol ® Reagent (Invitrogen, USA). The concentration and quality of the total RNA were measured by a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The corresponding cDNA was further obtained by reverse transcription using the PrimeScript ™ RT reagent Kit (TAKARA, Japan) and miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN BIOTECH, Beijing). The quantitative real-time polymerase chain reaction (qRT-PCR) assays were performed on ABI 7500 real-time PCR machine (Applied Biosystems, USA) using the TB Green® Premix Ex Taq™ II (TAKARA, Japan) and miRcute Plus miRNA qPCR Kit (TIANGEN BIOTECH, Beijing). The primers of miR-200c (CD201-0544) and U6 (CD201-0145) were purchased from TIANGEN BIOTECH. The primer sequences of other targets are shown in Supplementary Table 1.
RNAscope
The expression of circ-ZEB1 in tissues from CRC patients was detected by RNAscope technology. A RNAScope 2.5 High-Definition Assay kit (Advanced Cell Diagnostics) was applied, and circ-ZEB1 target probe was provided by Advanced Cell Diagnostics (ACD). TMA was used for ISH analysis based on the manufacturer's instructions. The data were subsequently analyzed using Aperio ImageScope software (Leica Biosystems). The signals were visually detected, and those cells with cytoplasm or nucleus stained with red dots were identified as positive results. Each sample was scored according to the percentage of positive cells in the section.
Western blotting
The cells were lysed with RIPA buffer (Solarbio, Beijing, China), After the precipitate was discarded, the concentration was measured using BCA Protein assay kit (Beyotime, China). Proteins were separated on SDS–PAGE and transferred on polyvinylidene fluoride (PVDF) membranes (Immobilon-P Millipore, Bedford, MA). Subsequently, the membranes were blocked in 5% nonfat dry milk (NFDM) in TBST buffer for 2h and incubated overnight at 4°C with the diluted antibodies shown in Supplementary Table 2. After four washes with the Tris-Buffered-Saline-Tween (TBST), the membranes were incubated with IRDye® 680RD Goat anti-Mouse IgG Secondary Antibody or IRDye® 680RD Goat anti-Rabbit IgG Secondary Antibody (1:10000, LI-COR Biosciences, U.S) for 1h. Finally, the blots were visualized using Odyssey® Imaging System (LI-COR Biosciences, U.S).
Nuclear and cytoplasmic RNA fractionation
To determine the subcellular distribution of circ-ZEB1, a PARIS Kit (AM1921, Thermo Fisher Scientific) was used following the manufacturer's protocol. The expression level of circ-ZEB1 in the nuclear and cytoplasmic fractions was determined using qRT-PCR. The expression level of GAPDH and U6 was served as controls.
Cell viability assay
We monitored cell viability using the Cell Counting Kit-8 (Hanheng Biotechnology Corporation, Shanghai, China). Cells transfected with siRNA against circ-ZEB1 or other oligonucleotides as indicated in figure legends were evenly seeded into 96-well plates, and the values of OD450 were measured. The 50% inhibitory concentration (IC50) values of oxaliplatin and 5-FU were calculated using GraphPad Prism.
Colony formation assay
Approximately 1000 cells were plated into each well of six-well plates. When cell colonies were visible, the plates were washed twice using cold phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 20min, and stained with crystal violet until the colonies could be observed. Then the colonies were photographed and analyzed.
Transwell assay
We used transwell inserts to conduct transwell migration and invasion assays. In the migration assay, CRC cells were resuspended in serum-free medium. The upper layer of each transwell insert was added with 200ul cell suspension and the lower layer of each transwell insert was added with 500ul complete medium. After 24 hours, the inserts were collected, fixed using 4% paraformaldehyde, and stained with crystal violet. We further observed the inserts using a Leica DM4000B microscope (Leica), photographed randomly selected areas, and calculated cell numbers. For the invasion assay, 50ul of matrix gel diluted at a ratio of 1:8 was added to the upper layer of the chamber, and the chamber was collected after 48 hours. The remaining steps were the same as in the migration assay.
Cell cycle analysis
Cell cycle assay was measured by flow cytometry. After transfection, SW480 or RKO cells were digested, washed with PBS, and fixed in 75% ethanol at 4°C overnight. Before detection, the cells were incubated with PI/RNase Staining Buffer (KeyGen Biotech, Nanjing, China) for 30 min. The data of the cell cycle were analyzed using FlowJo software.
RNA immunoprecipitation (RIP) assay
To conduct the RIP assay, the EZ-Magna RIP Kit (Millipore) was used according to the manufacturer's instructions. Twenty million SW480 cells or RKO cells were harvested and then lysed with an equal volume of the RIP lysis buffer. After being incubated on ice for 5 min, the lysate was incubated with magnetic beads conjugated with anti-Ago2 antibody (Abcam, ab186733) or IgG (PP64B) with rotating for 3 hours at 4°C. The protein in the immunoprecipitated was digested with the proteinase K buffer at 55°C for 30 minutes with shaking. Subsequently, the RNA was extracted with phenol: chloroform: isoamyl alcohol. Finally, the RT-qPCR was conducted to analyze the immunoprecipitated RNA.
RNA fluorescence in situ hybridization (FISH)
The probes of circ-ZEB1 and miR-200c were synthesized by GenePharma (Shanghai, China). The probe of circ-ZEB1 was Cy3-labeled. The probe of miR-200c was FAM-labeled. The assay was conducted according to the manufacturer's instructions. The cell nuclei were stained with DAPI. The subcellular distribution of circ-ZEB1 and miR-200c was observed by a confocal microscope (Leica SP5).
Xenograft tumor formation
For the tumor formation experiments, 1 × 106 RKO cells (circ-ZEB1 knocking down or negative control) were injected subcutaneously into each female BALB/c nude mouse (6-7 weeks old). Three weeks later, mice were sacrificed and the tumors were harvested. The volume of tumors was calculated as the following method: 0.52 × length × width2.
For the liver metastasis experiments, the 2 × 106 RKO cells were injected under the capsule of the spleen. About six weeks later, the mice were sacrificed and the livers and spleens were excised. All the specimens were afterwards confirmed through histological analysis.
The animal studies were approved by the Animal Experiments and Experimental Animal Welfare Committee of Capital Medical University and were performed in compliance with the Animal Protection Law of the People's Republic of China-2009 for experimental animals.
Dual-Luciferase Reporter Assay
The circ-ZEB1 sequences containing mutant or wild-type miR-200c binding sites were synthesized and cloned into psiCHECK-2 plasmid (Promega, Madison, WI, USA). We used the Lipofectamine 3000 reagent (Invitrogen) to transfect the plasmids into SW480 cells together with miR-200c mimics or the negative control. Then the transfected SW480 cells were cultured for 48h before we measured the luciferase activity using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA).
Statistical analysis
All the analyses were performed using GraphPad Prism version 8.0. The data were presented as the mean ± standard deviation (SD) from at least three independent experiments. Student's t-test or Wilcoxon rank test was used to compare between two groups. One-way analysis of variance was used to compare among multiple groups. The statistically significant threshold was set as p-value < 0.05.
Results
circ-ZEB1 is highly expressed in colorectal cancer
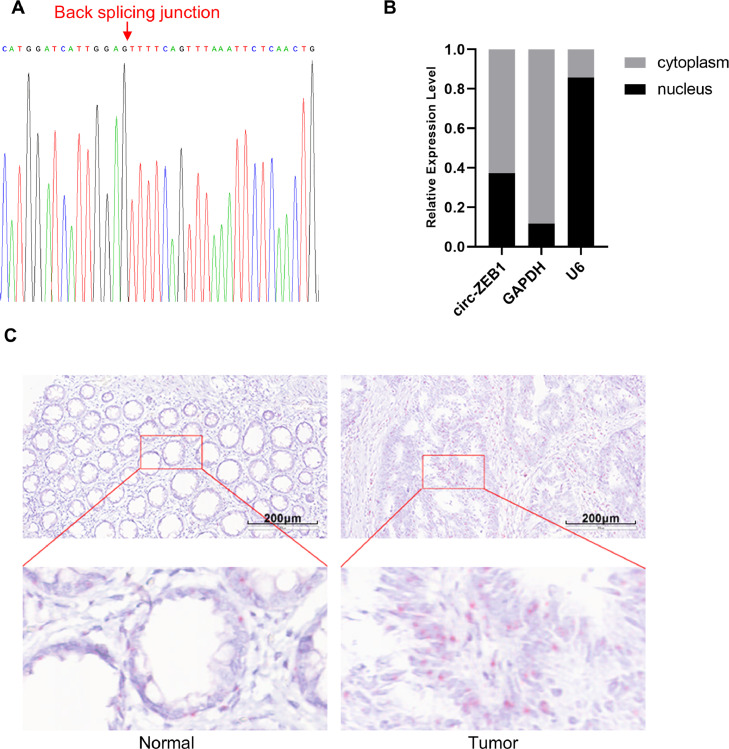
To clarify the role of circRNAs in CRC, we screened the differentially expressed circRNAs between cancerous tissues and adjacent normal tissues of CRC patients through high-throughput sequencing in our previous work [15] and found that circ-ZEB1 (hsa_circ_0000229) was highly expressed in CRC tumor tissues (logFC=1.85, P=0.0076). According to circBase (http://www.circbase.org/), circ-ZEB1 was generated from ZEB1 through back-splicing. The back-splicing junction site of circ-ZEB1 was validated by Sanger sequencing (Fig. 1A). To determine the subcellular localization of circ-ZEB1, we detected the expression of circ-ZEB1 in SW480 cell fractions by qRT-PCR and found that circ-ZEB1 mainly located in the cytoplasm (Fig. 1B). Besides, we further examined the expression and localization of circ-ZEB1 in CRC tissues by the RNAscope method (Fig. 1C). We found that circ-ZEB1 was expressed in both normal and cancerous tissues from CRC patients, and it was expressed in both the cytoplasm and nucleus of cells. Notably, in addition to parenchymal cells, circ-ZEB1 was also abundantly expressed in stromal cells, indicating that it may contribute to the tumor microenvironment of CRC. Statistical analysis of the 44 pairs of tissues included in this study suggested that the expression of circ-ZEB1 was significantly higher in CRC tumor tissues than in adjacent normal tissues (Z=-4.8, P < 0.001), which validated the sequencing results and suggested that it might play an oncogenic role in CRC disease progression.
Fig. 1.
Characterization and expression of circ-ZEB1 in CRC. (A) The back-splicing junction of circ-ZEB1 was confirmed by Sanger sequencing. (B) The subcellular distribution of circ-ZEB1 in SW480 cells. (C) The expression level of circ-ZEB1 was higher in CRC tissues (right) than adjacent normal tissues (left) as detected by RNAscope. Circ-ZEB1 was localized both in parenchymal cells and stromal cells.
circ-ZEB1 knocking down inhibits malignant phenotypes of CRC cells in vitro
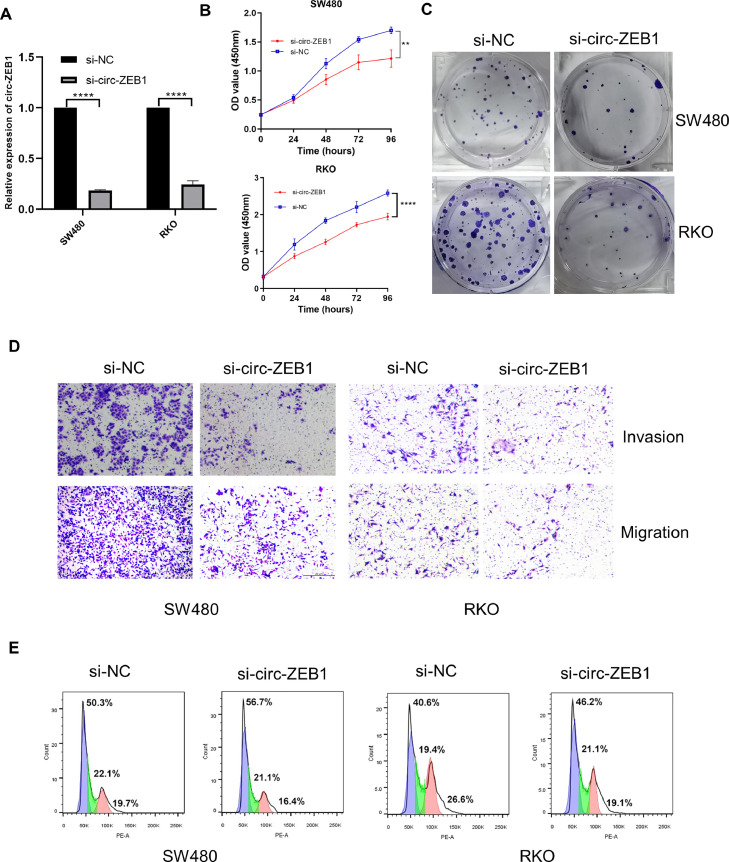
To explore the functions of circ-ZEB1 in CRC cells, a siRNA against circ-ZEB1 was designed, and its interference efficiency was verified by qPCR in SW480 and RKO cells, with a significantly reduced expression of circ-ZEB1 in the si-circ-ZEB1 group (Fig. 2A). Then, CCK-8 and colony formation assay were employed to test the cell proliferation ability. Results revealed a significant reduction in cell viability (Fig. 2B) and colony-forming ability (Fig. 2C) following si-circ-ZEB1 treatment relative to the si-NC group from 48h after transfection. Next, we investigated the effect of circ-ZEB1 on the invasion and migration of CRC cells by transwell assay and found that the invasion and migration capabilities of CRC cells were significantly inhibited after interfering circ-ZEB1(Fig. 2D). Finally, flow cytometry results indicated that the cells underwent G0/G1 arrest following interfering circ-ZEB1 expression (Fig. 2E).
Fig. 2.
Silencing circ-ZEB1 inhibits malignant phenotypes of CRC cells in vitro. (A) The expression level of circ-ZEB1 was detected by qRT-PCR in SW480 and RKO cells transfected with si-NC or si-circ-ZEB1. (B&C) The effect of knocking down circ-ZEB1 on proliferation in SW480 and RKO cells was detected by CCK-8 assay and colony formation assay. (D) Transwell assay was performed to detect the invasion and migration abilities of SW480 and RKO cells after knocking down circ-ZEB1. (E) The cell cycle distributions of SW480 and RKO cells after transfection with si-NC or si-circ-ZEB1. **p < 0.01, ****p < 0.0001.
circ-ZEB1 knocking down suppresses CRC cells tumorigenicity and metastasis in vivo
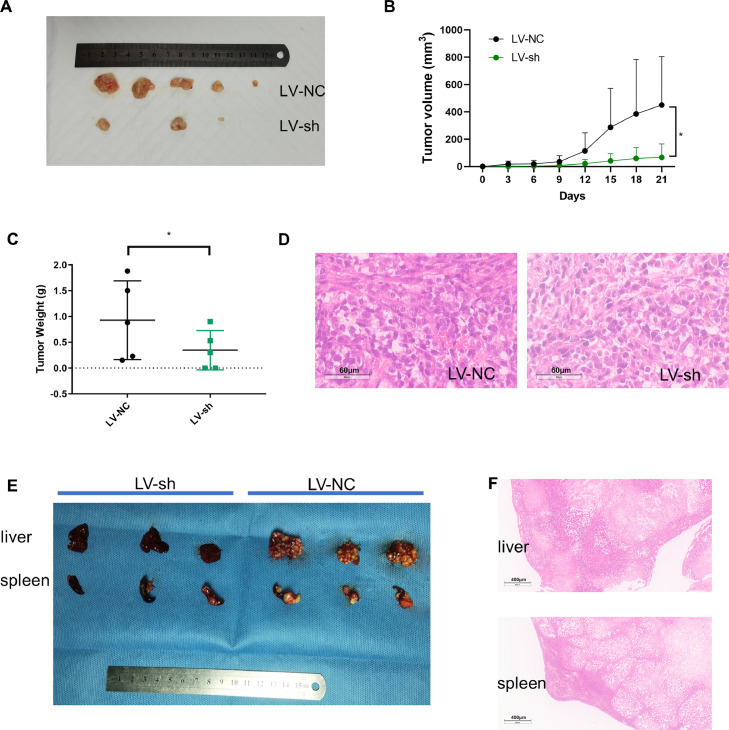
To investigate how circ-ZEB1 affects CRC tumor growth and metastasis in vivo, we developed RKO cells with circ-ZEB1 stably knocking down using pHBLVCMV-MCS-EF1-puro lentiviral vectors. Subsequently, a nude mice xenograft model was applied to determine the effect of circ-ZEB1 on tumorigenesis in vivo. RKO cells stably transfected with LV-NC or LV-sh-circ-ZEB1 plasmid were subcutaneously injected into each mouse (n=5 in each group). Results showed that circ-ZEB1 knockdown reduced tumor volume and weight significantly when compared with the NC group (Fig. 3A-C). For metastasis study, RKO cells stablely transfected with LV-NC or LV-sh-circ-ZEB1 plasmid were injected into spleens of mice through operation. For all mice in the NC group, there was in situ tumorigenesis in the spleen and prominent metastatic tumors in the liver; while in the circ-ZEB1 knocking down group, only two mice had in situ tumor tumorigenesis in the spleen, of which only one had few metastatic tumors in liver (Fig. 3E). All formed tumors, including subcutaneous and spleen tumor tissues, along with liver metastatic tumors were pathologically confirmed by HE staining (Fig. 3D&F). On the basis of these results, we concluded that knocking down circ-ZEB1 can inhibit tumorigenesis and metastasis of CRC cells in vivo.
Fig. 3.
Silencing circ-ZEB1 suppresses CRC cells tumorigenicity and metastasis in vivo. (A) Photographs of subcutaneous xenograft tumors in nude mice (n = 5). (B) The subcutaneous tumors were measured every 3 days to monitor the growth of tumors. (C) The weight of tumors was recorded. (D) Images of HE-stained subcutaneous xenograft tumors. (E) Photographs of livers and spleens excised from nude mice. (F) Images of HE-stained liver metastasis tumor tissues and spleen tumor tissues. *p < 0.05.
circ-ZEB1 regulates EMT and chemotherapy resistance in CRC cells
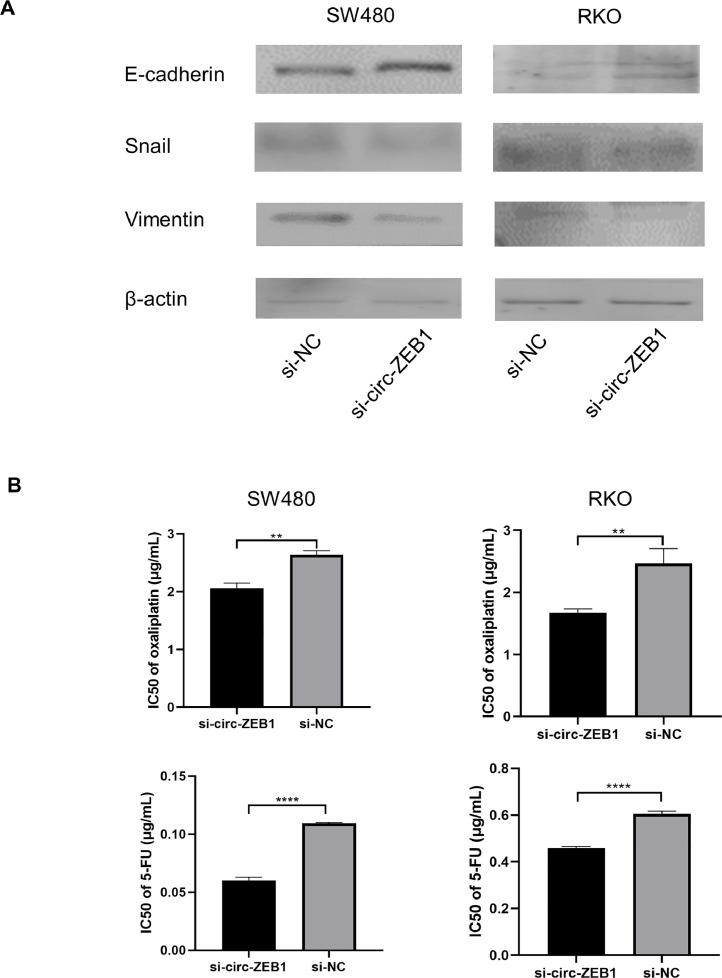
EMT is one of the most important features during tumor progression. We detected the expression of EMT-related proteins in SW480 and RKO cells, and results showed that the expression level of Vimentin and Snail were downregulated following knockdown of circ-ZEB1, while E-cadherin was upregulated (Fig. 4A). These results suggested that knockdown of circ-ZEB1 inhibited the EMT process in colorectal cancer cells.
Fig. 4.
circ-ZEB1 regulates EMT and chemotherapy resistance in CRC cells. (A) Western blotting analysis of EMT markers expression in SW480 and RKO cells following treatment of si-circ-ZEB1 or control. (B) The IC50 values was calculated after treatment with oxaliplatin or 5-FU in SW480 and RKO cells. **p < 0.01, ****p < 0.0001.
To assess the influence of circ-ZEB1 on cellular sensitivity to oxaliplatin and 5-FU, we next transfected si-circ-ZEB1 or si-NC into SW480 and RKO cells. Cell viability assay showed that inhibition of circ-ZEB1 significantly attenuates oxaliplatin or 5-FU resistance when compared with the si-NC groups, with the IC50 values decreased in both cell lines (Fig. 4B).
These results indicated that knocking down of circ-ZEB1 was sufficient to reduce the chemoresistance of SW480 and RKO cells to oxaliplatin and 5-FU.
circ-ZEB1 sponges miR-200c
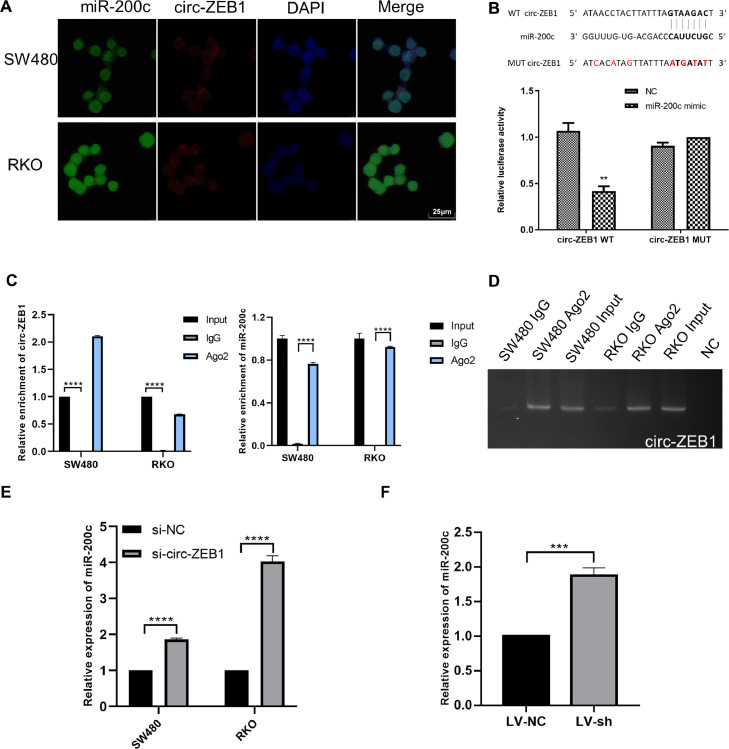
As circRNAs might act as sponges for miRNAs and further regulate downstream targets, we further predicted the miRNAs that could interact with circ-ZEB1 through miRanda. Among the predicted miRNAs, we chose miR-200c for a further study. Then, RNA FISH was performed in CRC cells to observe the subcellular localization of circ-ZEB1 and miR-200c, results exhibited both cytoplasmatic and nucleus distribution of them, which indicated that circ-ZEB1 and miR-200c are co-localized (Fig. 5A). Based on these data, we further verify the interaction through dual-luciferase reporter assay and RIP assay. As shown in Fig. 5B, the miR-200c mimics significantly reduced the luciferase activity of the cells transfected with WT-circ-ZEB1 plasmids, while barely no effect in the cells with Mut-circ-ZEB1 plasmids. RIP assay shows (Fig. 5C-D) that both miR-200c and circ-ZEB1 are remarkably enriched in the anti-Ago2 group compared with that in the anti-IgG group as negative control.
Fig. 5.
circ-ZEB1 sponges miR-200c. (A) Colocalization of circ-ZEB1 and miR-200c in SW480 and RKO cells. Green, miR-200c; red, circ-ZEB1; blue, DAPI. (B) Dual-luciferase reporter assays demonstrated that miR-200c-5p was a direct target of circ-ZEB1. (C) qPCR analysis of circ-ZEB1 and miR-200c in the RIP products of SW480 and RKO cells. (D) Nucleic acid electrophoresis analysis was used to analyze the qPCR products. (E) Relative expression of miR-200c in SW480 and RKO cells after transfected with si-circ-ZEB1/negative control. (F) Relative expression of miR-200c in LV-sh or LV-NC xenograft tumor tissues. **p < 0.01, ***p < 0.001, ****p < 0.0001.
Furthermore, the expression of miR-200c was prominently elevated after transfection with siRNA-circ-ZEB1 (Fig. 5E). We also detected the expression of miR-200c in xenograft tumor tissues obtained from the above in vivo experiments, results showed a significantly decreased expression in the si-circ-ZEB1 group than the si-NC group (Fig. 5F).
circ-ZEB1 exerts its regulatory effect through acting on miR-200c
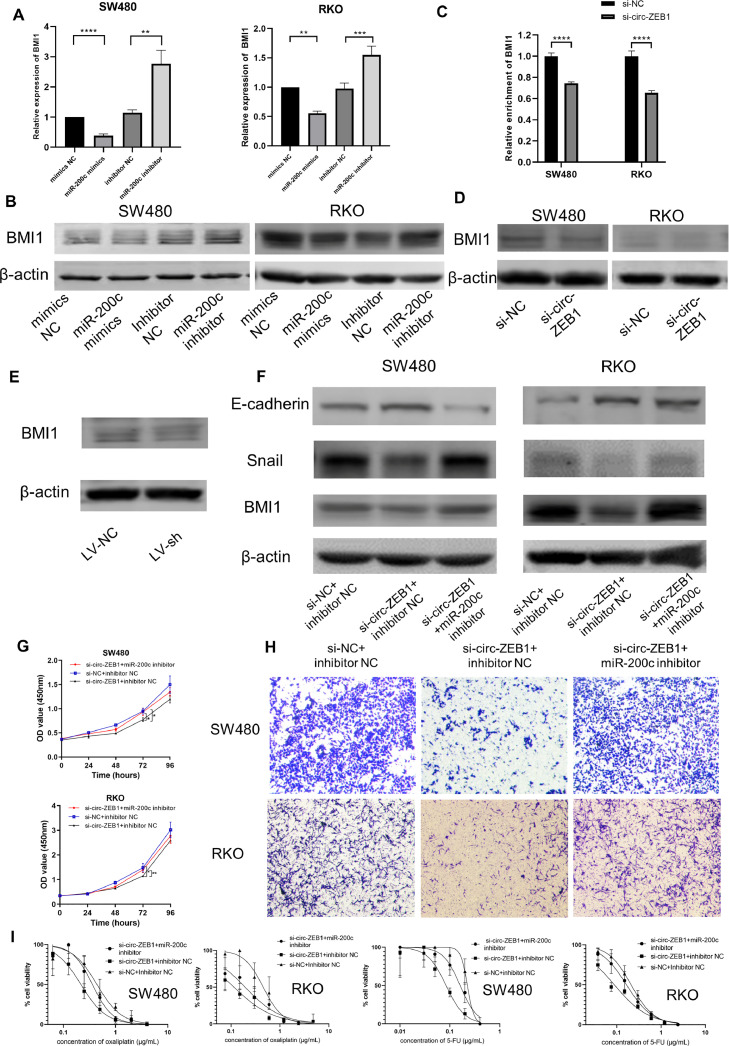
Previous studies have shown that BMI1 was a target of miR-200c, which has been verified in different types of cancers, including CRC [16]. We showed in SW480 and RKO cells that miR-200c mimics could significantly reduce the expression level of BMI1, while miR-200c inhibitor played the opposite role (Fig. 6A-B). Furthermore, inhibition of circ-ZEB1 could suppress the expression of BMI1 both in CRC cells (Fig. 6C-D) and in xenograft tumor tissues (Fig. 6E), which could be rescued by miR-200c inhibitors (Fig. 6F).
Fig. 6.
circ-ZEB1 exerts its regulatory effect through acting on miR-200c. (A&B) Expression of BMI1 in SW480 and RKO cells after transfected with miR-200c mimics or miR-200c inhibitors and their negative controls was detected by qRT-PCR (A) and western blotting (B). (C&D) Expression of BMI1 in SW480 and RKO cells after transfected with si-circ-ZEB1 or negative control was detected by qRT-PCR (C) and western blotting (D). (E) Expression of BMI1 in xenograft tumor tissues of LV-sh-circ-ZEB1 and LV-NC group. (F) Expression of E-cadherin, Snail, and BMI1 in SW480 and RKO cells transfected with indicated oligonucleotides. (G) The cell viability was detected by CCK-8 in SW480 and RKO cells transfected with indicated oligonucleotides. (H) The cell migration ability was detected by transwell assay in SW480 and RKO cells transfected with indicated oligonucleotides. (I) The cell viability was detected after treatment with oxaliplatin or 5-FU in SW480 and RKO cells transfected with indicated oligonucleotides. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To determine whether circ-ZEB1 promotes CRC progression by acting on miR-200c, rescue experiments were performed by co-transfecting si-circ-ZEB1 and miR-200c inhibitors into SW480 and RKO cells. The cell proliferation assay showed that miR-200c inhibitors effectively reversed the si-circ-ZEB1-induced inhibition of proliferation in SW480 and RKO cells (Fig. 6G). Likewise, transwell assay indicated that miR-200c inhibitors attenuated the inhibitory effect of si-circ-ZEB1 on cell migration (Fig. 6H). Besides, the regulatory effect of circ-ZEB1 on EMT-related markers, such as E-cadherin and Snail, could also be rescued by miR-200c inhibitors (Fig. 6F). Moreover, the regulatory effect of inhibition of circ-ZEB1 on chemoresistance to oxaliplatin and 5-FU in CRC cell lines could also be rescued by miR-200c inhibitors (Fig. 6I). Taken together, these results indicated that circ-ZEB1 exerts its regulatory effect through acting on miR-200c in CRC cell lines.
Discussion
Accumulating studies have shown that circRNAs play a crucial role in various cancers, and they hold the potential to regulate tumorigenesis and metastasis [9,17,18]. A variety of circRNAs have been proven to be involved in the development of CRC, such as hsa_circ_0006401 [19], hsa_circ_0000231 [20], etc circ-ZEB1 is dysregulated in different types of tumors, and have recently been studied. In hepatocellular carcinoma, Gong et al. [21] found that circ-ZEB1.33 promotes the proliferation of cancer cells by sponging miR-200a-3p and upregulating CDK6, while Liu et al. [22] found that circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma; Pei et al. [23] found that circ-ZEB1 was highly expressed in triple negative breast cancer (TNBC) tumor tissues and tumor cell lines, it acted as an oncogene by affecting cell proliferation and apoptosis via sponging miR-448. However, whether circ-ZEB1 participates in the tumorigenesis and development of CRC remains unknown. In this research, we found that circ-ZEB1 was upregulated in CRC tissues, and it was located both in tumor cells and the stromal cells; knocking down of circ-ZEB1 can inhibit malignant phenotypes of CRC cells in vitro and in vivo, which indicate that circ-ZEB1 might be a potential therapeutic target for CRC patients.
As have been mentioned above, previous reports have shown that circ-ZEB1 promotes the progression of tumors through sponging miRNAs [21], [22], [23]. Our results in this study showed that circ-ZEB1 could bind with miR-200c, and inhibition of miR-200c abolished the effect of interfering with circ-ZEB1 in regulating EMT and chemotherapeutic sensitivity. miR-200c has well been studied and is considered as a tumor suppressor in human malignant tumors. It is a highly studied miRNA in terms of oncogenesis, EMT, and chemotherapy resistance [24]. In CRC, Dermani et al. found that miR-200c can regulate EMT by regulating the expression of ZEB1 [25]; XU et al. found that sodium butyrate inhibited colorectal cancer cell migration by down-regulating BMI-1 through enhancing miR-200c expression [16]. In our research, we found that knocking down of circ-ZEB1 downregulated the expression level of the miR-200c target gene BMI1, which could be reversed by inhibiting miR-200c. Through the above results, we illustrated that circ-ZEB1 exerts its regulatory function through sponging miR-200c and regulating its downstream target genes.
EMT, as a characteristic behavior of malignant tumors, is one of the core mechanisms in tumor progression, recurrence, metastasis, and chemotherapeutic resistance [5]. It was revealed that circRNAs are involved in regulating EMT of malignancies. Yi et al. found that circular RNA PLCE1 promotes EMT of CRC cells [26]; circRNA_0005075 suppresses carcinogenesis via regulating miR-431/p53/EMT axis in gastric cancer [27], and circular RNA circ_PVT1 induces EMT to promote metastasis of cervical cancer cells [28]. However, whether circ-ZEB1 is involved in the EMT process of cancer cells remains unknown. In our research, inhibition of circ-ZEB1 suppressed the expression of Vimentin and Snail, while the expression of E-cadherin was increased. The results indicated that circ-ZEB1 was involved in the EMT of CRC. However, when co-transfecting si-ZEB1 and miR-200c inhibitor, the regulatory effect of circ-ZEB1 on EMT of CRC cells was rescued, which disclosed that circ-ZEB1 silencing suppressed EMT of CRC cells partly through sponging miR-200c in vitro. Actually, it has already been demonstrated that miR-200c plays a vital role in EMT, it could suppress the EMT process in a series of malignant tumors, including lung cancer [29], breast cancer [30], colorectal cancer [31], et al., which have been well documented [24]. Our study provided a novel aspect that miR-200c involved in EMT.
Chemotherapy is recommended for the treatment of CRC patients with advanced stages. 5-fluorouracil and oxaliplatin are the main components of different regimens available for CRC patients. However, it is estimated that a large portion of CRC patients would develop resistance to chemotherapy during the treatment process [32], resulting in high risk of cancer recurrence and poor prognosis. Unfortunately, there is still a lacking of reliable biomarkers for early prediction of chemoresistance in CRC patients. Uncovering the detailed mechanisms underlying drug resistance and finding predictive biomarkers would be of crucial importance for the patients. Previous reports have demonstrated that circRNAs can serve as crucial regulators of tumor chemoresistance [33,34] in different malignancies, including CRC. Cheng performed the circRNA expression profile in two CRC cell lines and their homologous 5-FU resistant cells by high-throughput sequencing, and they identified circ_0002813 and circ_0000236 could be involved in the formation of 5-FU resistance [35]; Wang et al. [36] found that exosome-delivered circRNA, ciRS-122, promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in CRC; Jiang et al. found in their work that circPTK2 knockdown constrained cancer progression and chemoresistance to 5-FU and oxaliplatin [37]. In this study, we found that circ-ZEB1 regulates chemoresistance of CRC cell lines to 5-FU and oxaliplatin, which have not been reported previously. Mechanism study showed that the regulatory effect of si-circ-ZEB1 on IC50 values of CRC cells to 5-FU and oxaliplatin could be rescued by miR-200c inhibitor, indicating that circ-ZEB1 regulates chemoresistance of CRC cells through acting on miR-200c. On one hand, previous studies have already proved that miR-200c is widely involved in the chemoresistance to 5-FU and cisplatin in different tumors [38], [39], [40], on the other hand, rare studies have been conducted in CRC before. Herein, we demonstrated a circ-ZEB1/miR-200c axis participates in the regulation of chemoresistance of CRC cells.
There are limitations of this study. Firstly, only loss of function experiments were performed, while there is an absence of experiments of overexpression of circ-ZEB1; Secondly, although we found that knocking down of circ-ZEB1 can improve chemotherapeutic sensitivity in CRC cells, in vivo studies are needed to verify the potential therapeutic effect of targeting circ-ZEB1; In addition, the molecular targets downstream of the circ-ZEB1/miR-200c axis were not fully elucidated.
Conclusions
In conclusion, the present study demonstrated that circ-ZEB1 acts as an oncogene in CRC, it regulates progression, EMT and chemoresistance of CRC cells through sponging miR-200c. Our study provided a new potential therapeutic target for CRC patients.
Funding
This work was supported by National Natural Science Foundation of China (81802353, 82002471).
CRediT authorship contribution statement
Hongyu Chen: Data curation, Software, Writing – original draft. Jianwei Zhang: Data curation, Methodology, Writing – original draft. Lei Yang: Methodology, Investigation. Yansen Li: Methodology, Investigation. Zhenjun Wang: Writing – review & editing, Supervision. Chunxiang Ye: Validation, Methodology, Resources, Formal analysis, Writing – review & editing.
Declarations of Interest
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101604.
Contributor Information
Zhenjun Wang, Email: drzhenjun@163.com.
Chunxiang Ye, Email: chunxiang-ye@163.com.
Appendix. Supplementary materials
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Yoshino T, Arnold D, Taniguchi H, et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann. Oncol. 2018;29(1):44–70. doi: 10.1093/annonc/mdx738. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 4.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat. Rev. Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 5.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 6.Heery R, Finn SP, Cuffe S, Gray SG. Long Non-coding RNAs: key regulators of epithelial-mesenchymal transition, Tumour drug resistance and cancer stem cells. Cancers (Basel) 2017;9(4) doi: 10.3390/cancers9040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Jiang Z, Li Y, Zhang Y, Han Y, Gao L. Non-coding RNAs regulating epithelial-mesenchymal transition: Research progress in liver disease. Biomed. Pharmacother. 2022;150 doi: 10.1016/j.biopha.2022.112972. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 10.Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Zhang Y, Song H, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol. Cancer. 2021;20(1):81. doi: 10.1186/s12943-021-01375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang J, Zhang C, Huang Y, et al. CircRNA circ_0124554 blocked the ubiquitination of AKT promoting the skip lymphovascular invasion on hepatic metastasis in colorectal cancer. Cell Death. Dis. 2021;12(3):270. doi: 10.1038/s41419-021-03565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Liu Y, Liu Z, et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol. Cancer. 2021;20(1):114. doi: 10.1186/s12943-021-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018;246(2):166–179. doi: 10.1002/path.5125. [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Tao J, Chen P, et al. Sodium butyrate inhibits colorectal cancer cell migration by downregulating Bmi-1 through enhanced miR-200c expression. Mol. Nutr. Food Res. 2018;62(6) doi: 10.1002/mnfr.201700844. [DOI] [PubMed] [Google Scholar]
- 17.Yu YZ, Lv DJ, Wang C, et al. Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol. Cancer. 2022;21(1):12. doi: 10.1186/s12943-021-01480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Hu ZQ, Yu SY, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022;82(6):1055–1069. doi: 10.1158/0008-5472.CAN-21-1259. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Zhou X, Geng X, et al. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death. Dis. 2021;12(5):443. doi: 10.1038/s41419-021-03714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Wang B, Lin Y, et al. hsa_circ_0000231 Promotes colorectal cancer cell growth through upregulation of CCND2 by IGF2BP3/miR-375 dual pathway. Cancer Cell Int. 2022;22(1):27. doi: 10.1186/s12935-022-02455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18:116. doi: 10.1186/s12935-018-0602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Zheng L, Zhang R, et al. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol. Cancer. 2022;21(1):72. doi: 10.1186/s12943-022-01529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei X, Zhang Y, Wang X, Xue B, Sun M, Li H. Circular RNA circ-ZEB1 acts as an oncogene in triple negative breast cancer via sponging miR-448. Int. J. Biochem. Cell Biol. 2020;126 doi: 10.1016/j.biocel.2020.105798. [DOI] [PubMed] [Google Scholar]
- 24.Mutlu M, Raza U, Saatci Ö, Eyüpoğlu E, Yurdusev E, Şahin Ö. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J. Mol. Med. 2016;94(6):629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 25.Karimi Dermani F, Amini R, Saidijam M, Najafi R. miR-200c, a tumor suppressor that modulate the expression of cancer stem cells markers and epithelial-mesenchymal transition in colorectal cancer. J. Cell. Biochem. 2018;119(7):6288–6295. doi: 10.1002/jcb.26880. [DOI] [PubMed] [Google Scholar]
- 26.Yi B, Dai K, Yan Z, Yin Z. Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered. 2022;13(3):6243–6256. doi: 10.1080/21655979.2021.2003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Chen Z, Song Y, et al. CircRNA_0005075 suppresses carcinogenesis via regulating miR-431/p53/epithelial-mesenchymal transition axis in gastric cancer. Cell Biochem. Funct. 2020;38(7):932–942. doi: 10.1002/cbf.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Wei M, Kang Y, Xing J, Zhao Y. Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to promote metastasis of cervical cancer. Aging. 2020;12(20):20139–20151. doi: 10.18632/aging.103679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommerova L, Ondrouskova E, Martisova A, Zoumpourlis V, Galtsidis S, Hrstka R. ZEB1/miR-200c/AGR2: a new regulatory loop modulating the epithelial-mesenchymal transition in lung adenocarcinomas. Cancers. 2020;12(6) doi: 10.3390/cancers12061614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi D, Guo L, Sun X, et al. UTMD inhibit EMT of breast cancer through the ROS/miR-200c/ZEB1 axis. Sci. Rep. 2020;10(1):6657. doi: 10.1038/s41598-020-63653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Cai X, Ma L. Curcumin modifies epithelial-mesenchymal transition in colorectal cancer through regulation of miR-200c/EPM5. Cancer Manag. Res. 2020;12:9405–9415. doi: 10.2147/CMAR.S260129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol. Ther. 2020;206 doi: 10.1016/j.pharmthera.2019.107447. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Zhang J, Cao G, Hua J, Shan G, Lin W. Emerging roles of circular RNAs in gastric cancer metastasis and drug resistance. J. Exp. Clin. Cancer Res. 2022;41(1):218. doi: 10.1186/s13046-022-02432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sang C, Rao D, Wu C, Xia Y, Si M, Tang Z. Role of circular RNAs in the diagnosis, regulation of drug resistance and prognosis of lung cancer. Oncol. Lett. 2022;24(3):302. doi: 10.3892/ol.2022.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng PQ, Liu YJ, Zhang SA, et al. RNA-Seq profiling of circular RNAs in human colorectal cancer 5-fluorouracil resistance and potential biomarkers. World. J. Gastrointest. Oncol. 2022;14(3):678–689. doi: 10.4251/wjgo.v14.i3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Z, Hou Z, Liu W, Yu Z, Liang Z, Chen S. Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered. 2022;13(1):810–823. doi: 10.1080/21655979.2021.2012952. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Posch F, Prinz F, Balihodzic A, et al. MiR-200c-3p modulates cisplatin resistance in biliary tract cancer by ZEB1-independent mechanisms. Cancers. 2021;13(16) doi: 10.3390/cancers13163996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Zhou Y, Cheng H, Tian J, Yang S. Roles of a TMPO-AS1/microRNA-200c/TMEFF2 ceRNA network in the malignant behaviors and 5-FU resistance of ovarian cancer cells. Exp. Mol. Pathol. 2020;115 doi: 10.1016/j.yexmp.2020.104481. [DOI] [PubMed] [Google Scholar]
- 40.Jiang T, Dong P, Li L, et al. MicroRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. Oncol. Rep. 2017;38(1):151–158. doi: 10.3892/or.2017.5659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.