Abstract
Random amplified polymorphic DNA typing was used to study the genetic diversity of Pseudomonas aeruginosa strains from (i) ventilated patients with nosocomial pneumonia who were hospitalized in intensive care units, (ii) cases of bacteremia in cancer patients with severe neutropenia, and (iii) rivers and swimming pools. Genetic diversity was determined by three phylogenetic methods and by statistical analysis of population genetics. The population studied undergoes epidemic clonality with a high rate of genetic recombination. P. aeruginosa bacteremia and pneumonia are not caused by specific clones within this species.
Nosocomial bacterial pneumonia often occurs in ventilated patients hospitalized in intensive care units (ICU) (7, 22) and is the second most frequent nosocomial infection (1, 3), although it has the highest mortality rate, at about 30% (17). Pseudomonas aeruginosa is the gram-negative bacillus most frequently isolated from patients with nosocomial pneumonia (3, 5). Diagnosis of bacterial pneumonia in ventilated patients is difficult because its clinical, radiologic, and biologic signs are nonspecific (8). Quantitative cultures of distal bronchial specimens collected with a protected specimen brush and/or by bronchoalveolar lavage (2, 6, 9) has been proposed to establish the diagnosis of nosocomial pneumonia.
Basically, two situations can lead to opportunistic infections caused by P. aeruginosa. (i) Immunosuppression of a patient can allow strains with low pathogenicity to invade tissues. For example, in cancer patients, when counts of polymorphonuclear cells are below 100/mm3 of peripheral blood (26), P. aeruginosa bacteremia occurs more frequently. (ii) Specific strains within a basically nonpathogenic species acquire characteristics that confer increased virulence, as shown for Escherichia coli strains causing bacteremia (13, 19, 20). The population structure of P. aeruginosa strains is still a matter of debate. Nineteen percent of a collection of 573 P. aeruginosa strains isolated from various clinical cases and from the environment were found to belong to the same clonal group (23). Similar homogeneity was found among P. aeruginosa strains isolated either from cases of clinical infection and from gasoline sources (12). Other results, however, suggested that the population structure of a collection of 257 clinical isolates of P. aeruginosa was panmictic (4, 21). The problem is one of relevance, since in a panmictic microorganism, individual multilocus genotypes are ephemeral due to high rates of genetic recombination. They cannot then be used as stable markers for epidemiological tracking.
In the present study, we first assessed the genetic diversity of P. aeruginosa strains isolated first from three sources: cases of nosocomial pneumonia in ventilated patients hospitalized in ICU, cases of bacteremia in cancer patients with neutropenia, and water from swimming pools and rivers. In the light of these results, we then considered the question of P. aeruginosa population structure.
We compared three groups of P. aeruginosa strains. The first included 36 strains, numbered 1 to 36, isolated from cases of nosocomial pneumonia. Inclusion criteria were isolation of the strain from a ventilated adult patient hospitalized for more than 48 h between January 1994 and September 1997 in one of eight ICU, three of them in hospital A and one each in hospitals B, C, D, E, and F, and suspected to have developed nosocomial pneumonia for two reasons, (i) the presence of a recent pulmonary infiltrate and (ii) the presence of a monomicrobial P. aeruginosa culture from a bronchoalveolar lavage sample in counts of >104 CFU/ml or from a protected specimen brush sample in counts of >103 CFU/ml, as previously described (2, 6). The second group included 20 strains, numbered 37 to 56, isolated from blood cultures drawn from neutropenic cancer patients with less than 100 polymorphonuclear neutrophils/mm3 of peripheral blood at the onset of bacteremia who were hospitalized in the oncology departments of four hospitals (B, G, H, and I). The nine hospitals (A to I) that provided samples were located in seven different cities (I to VII). The third group comprised seven strains, numbered 57 to 63, isolated from swimming pools in eastern France (V) and 11 strains, numbered 64 to 74, from four rivers on Guadeloupe Island (rivers 1 to 4). Details of these clinical and environmental strains are presented at the website http://www.chez.com /rruimy. Seventeen reference strains of P. aeruginosa, one of each known serotype (CIP 5933 to CIP 5941, CIP 5943 to 5945, CIP 6092, CIP 7212, CIP 7213, and ATCC 27853), were also included as standards, and type strains Pseudomonas stutzeri CIP 103022T and Pseudomonas putida CIP 52191T were included as outgroups for the phylogenetic study. Equivalence of strains numbers of the Collection Institut Pasteur (Paris, France) and the American Type Culture Collection is reported at website http://www.chez.com/rruimy.
Strains were identified as P. aeruginosa on the basis of typical morphology by gram-negative staining, a positive oxidase reaction, and conventional biochemical tests using the API 20NE system (API-System, BioMérieux, Marcy l'Etoile, France). Serotyping was performed by slide agglutination using commercially available O antisera (Sanofi Diagnostics Pasteur, Marne La Coquette, France) as recommended by the manufacturer. All strains were stored at −80°C. Strains were grown in Mueller-Hinton broth for 18 h with shaking. The volume of culture harvested corresponded to about 4 × 109 bacteria as measured by optical density at 600 nm. DNA was extracted from the bacterial pellet using an AXG 20 nucleobond column (Macherey-Nagel, Hoerdt, France) as recommended by the manufacturer, dried, and suspended in 200 μl of TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA). All DNA preparations were diluted to 10 ng/μl before storage at −20°C.
To select primers that generated reproducible polymorphic random amplified polymorphic DNA (RAPD) patterns, we performed 22 RAPD assays in duplicate, using 22 primers on DNA extracted from one randomly chosen strain in each group of our collection (strain 32, 37, or 64) and on the DNA of P. aeruginosa ATCC 27853. Eleven of these primers had been previously used for RAPD assays on P. aeruginosa strains (18), and 11 others were selected for their various GC percentages. Nine primers were selected from the results of this preliminary study because they generated reproducible RAPD patterns with one to five major bands comprising between 500 and 1,500 bp. Six of the primers, 208, 241, 272, 275, 277, and 287, were previously described (18). Three of them, Pyo 15 (5′-GCA GGG TGT T-3′), B10 (5′-CTG CTG GGA C-3′), and B7 (5′-GGT GAC GAC G-3′), were designed. These nine primers were further used to perform duplicate RAPD assays on each strain of the various test and control groups described above. The RAPD mixture contained 30 ng of bacterial DNA, one primer at 400 nM, 250 μM each deoxynucleoside triphosphate (Boehringer GmbH, Mannheim, Germany), 1× reaction buffer supplied by the manufacturer with 1.5 mM MgCl2 (Boehringer), and 1 U of Taq DNA polymerase (Boehringer) in a final volume of 50 μl. DNA was amplified in a PE 9600 thermocycler (Perkin-Elmer Applied Biosystems, Langen, Germany) using 45 cycles, each consisting of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C, with a final elongation step of 10 min at 72°C. The amplified fragments were separated by horizontal gel electrophoresis (1.5% [wt/vol] agarose gel, 1× Tris-borate-EDTA buffer, and 4 V/cm) for 4 h and stained with ethidium bromide.
In the patterns resulting from RAPD assays, only the major reproducible bands which were detected by UV fluorescence and comprised 500 to 1,500 bp were taken into account in the analysis, as previously described (29). RAPD patterns were analyzed by phylogenetic methods and by a population genetic approach based on the analysis of linkage disequilibrium, i.e., the nonrandom association of the genotypes observed at different loci (29).
The unweighted pair-group method with arithmetic averages (25), neighbor joining (NJ), and the Wagner phylogenetic method, all computed with the PHYLIP software package (11), were used to assess the phylogenetic relationships between strains. P. putida and P. stutzeri were chosen as outgroups to root trees. Only branch topologies found to be similar by all three methods were considered significant, because agreement among the results of several phylogenetic methods has been shown to be a strong indication that phylogeny has been correctly determined (14, 15). Lastly, the robustness of the branches was estimated by the bootstrap method (10). The bootstrap method used in the phylogenies is a statistical method of resampling characters with replacement, to create a series of bootstrap samples of the same size as the original data. Each of these is analyzed, and the variation among the resulting estimates is taken to indicate the size of the error involved in making estimates from the original data (10).
To assess the population structure of the sample under study, we used the previously described f test (28), which is based on the analysis of linkage disequilibrium (i.e., the nonrandom association of the genotypes occurring at different loci) by Monte Carlo simulations with 104 iterations. The f test takes panmixia, a situation in which genetic exchange occurs randomly, as the null hypothesis. It evaluates the probability of observing, by chance, a linkage disequilibrium as high as, or higher than, the one actually observed in the sample (28). A panmixia situation implies that recombination is free and that the expected probability of a given multilocus genotype is the product of the observed probabilities of the individual genotypes of which it is composed. Inhibition of recombination leads to linkage disequilibrium or to nonrandom association among loci (when the predictions of expected probabilities for multilocus genotypes are no longer satisfied) (for a review, see reference 27).
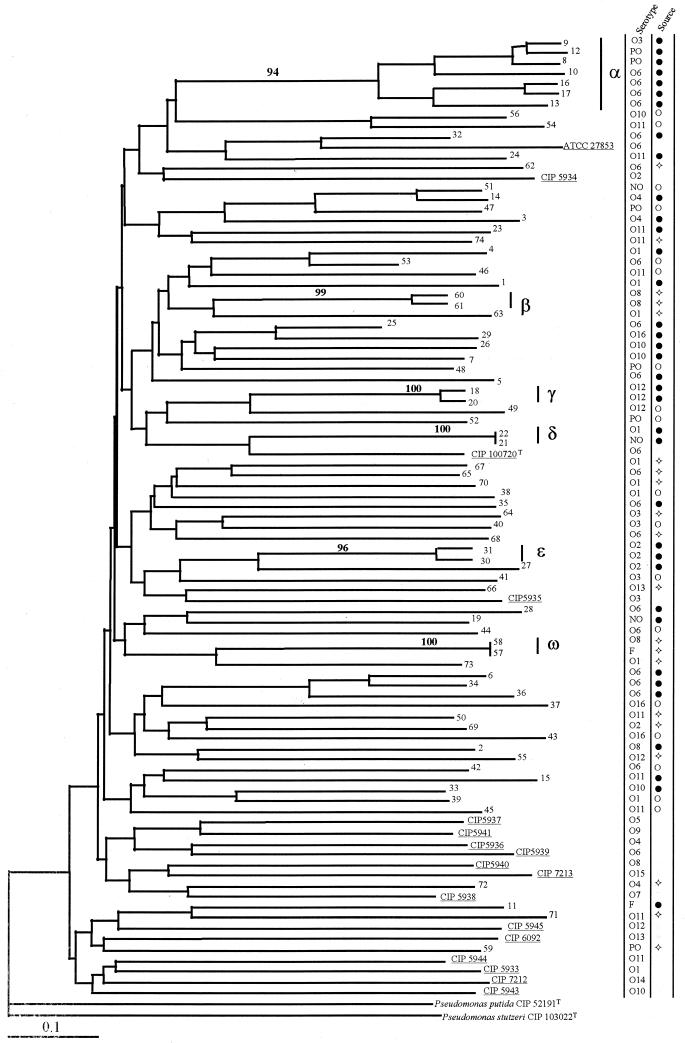
The results in Fig. 1 show that 63 of the 74 strains analyzed agglutinated with only one serotype. Only two strains agglutinated with the poly-F (O7, O8, O11, and O12) serum. Three strains were nonagglutinable, and six were polyagglutinable. The serotypes of the 17 reference strains are also given in Fig. 1.
FIG. 1.
The topology shown is a tree obtained by the neighbor-joining method. Values above the lines are bootstrap values expressed as percentages (only values of greater than 50% are shown). Clusters α to ω were also found by the Wagner phylogenetic method and the unweighted pair-group method with arithmetic averages. The underlined strains are reference strains. Abbreviations: NO, nonagglutinable; PO, polyagglutinable; F, agglutinable with O-serogroup F-containing antisera O7, O8, O11, and O12. Symbols: ●, ventilated patient with pneumonia; ○, cancer patients with bacteremia; ◊, rivers and swimming pools.
The results of the phylogenetic analysis are shown in Fig. 1. Although the general morphology of the tree suggested great diversity, we identified one major cluster consisting of seven strains (cluster α) and five secondary clusters consisting of two strains each (clusters β through ω). All of these clusters were identified by the three phylogenetic methods used and by significance of bootstrap analysis values of greater than 90% (Fig. 1). For all of the remaining strains, the results of the three phylogenetic methods of analysis did not identify any other branch with significant bootstrap values.
Taken together, these results indicate that there was no association between a specific cluster and the fact that the isolates originated from patients with nosocomial pneumonia, except for major cluster α and the pairs of strains constituting clusters β, δ, and ɛ. We also found no specific cluster group among the strains isolated from cases of bacteremia. We observed no association between a common environmental origin and a given cluster, except for the strains constituting clusters β and δ. Also, strains with the same geographic origin were not clustered together. Lastly, no clustering association was found between strains of the same serotype.
The linkage disequilibrium f test was significant for all of the stocks considered together (f = 0.007). The f-test result also remained significant (f = 0.006) when the strains from the outgroup and one strain from each pair which exhibited identical genotypes and composed clusters δ and ω were excluded from the analysis. However, the f test result was no longer significant when we excluded from this analysis either six of the seven strains in cluster α (f = 0.045) or one strain of each pair with closely related genotypes constituting clusters β, γ, and ɛ (f = 0.011), respectively. These results indicate that the linkage disequilibrium originated from the concomitant presence of both cluster α and the pairs of strains of clusters β, γ, and ω.
We found that the linkage disequilibrium f test was only significant because of clusters α, β, γ, and ɛ and was not altered by the pairs of repeat genotypes constituting clusters δ and ɛ. Only for these clusters were the branches of the trees convergent and the bootstrap values significant. The pairs of strains constituting clusters γ, δ, and ɛ were probably repeat isolates of the same strain because there were obvious epidemiological links (e.g., hospitalization in the same ICU) between the pairs of ventilated patients from whom each pair of strains was isolated. The same hypothesis can be formulated for the two pairs of strains isolated from environmental water that constituted clusters β and ω. The results of serotyping also argue in favor of the repeat isolate hypothesis. The O serotype from the two strains in each cluster was either the same, as in clusters β, γ, and ɛ, or only slightly different, as in clusters δ and ω. Such minor serotype differences were previously observed between strains from a single clone (16). As for cluster α, an epidemiological link was easily found among five of the isolates which originated from patients hospitalized in the same ICU (ICU 3 in hospital A) and between two other strains isolated from patients hospitalized in another ICU (hospital B). However, we could not find any epidemiological link between the patients hospitalized in these two hospitals. We also cannot exclude the possibility of an epidemiological link between these two groups. One can also postulate that the same clone was present in the environments of both units, even though they were about 400 km apart, as previously reported (23). Serotyping of strains from cluster α showed that they did not belong to the same serotype. However, it has been shown both in vivo and in vitro that serotypes are subject to changes within a single clonal group after antibiotic treatments (16). Antibiotic treatments of ventilated patients are frequent.
Among the three main models of structure previously proposed for bacterial populations (24), the present results favor that of epidemic clonality, i.e., occasional bouts of clonal propagation in a basically sexual species, as postulated for Neisseria meningitidis (24). In fact, if the P. aeruginosa population were really panmictic, as previously proposed (4, 21), we would not have found any significant linkage disequilibrium values. The lack of linkage disequilibrium when only one strain of cluster α or only one strain of cluster β, γ, and ɛ was included in the analysis does not argue in favor of a clonal structure such as that proposed for E. coli (24). Moreover, if the population structure of P. aeruginosa was indeed the result of long-term clonal evolution, as is the case for Trypanosoma cruzi (28, 29), the linkage disequilibrium would not vanish simply because repeat genotypes or genotypes belonging to the same cluster were removed. The fact that the linkage disequilibrium was easily eliminated from our sample when this was done, and the scarcity of repeat genotypes, suggested that the impact of genetic recombination was much stronger in the sample under examination than in the E. coli population (13), in which the linkage disequilibrium is more stable.
In conclusion, the genetic relationship among the three groups of P. aeruginosa strains tested here showed that neither nosocomial pneumonia nor bacteremia was caused by specific genotypes or groups of genotypes within this species. The evolution of P. aeruginosa strains has not allowed the establishment of a clear phylogenetic separation between pathogenic and nonpathogenic strains. The existence of a population which seems to undergo epidemic clonality, probably with a high rate of genetic recombination, suggests that individual genotypes of P. aeruginosa are unstable and should be used as epidemiological markers with great caution, on small scales of space and time only, and with highly discriminant markers.
REFERENCES
- 1.Anonymous. National Nosocomial Infections Surveillance (NNIS) report, data summary from October 1986–April 1996, issued May 1996. A report from the National Nosocomial Infections Surveillance (NNIS) System. Am J Infect Control. 1996;24:380–388. [PubMed] [Google Scholar]
- 2.Chastre J, Fagon J Y, Bornet-Lecso M, Calvat S, Dombret M C, al Khani R, Basset F, Gibert C. Evaluation of bronchoscopic techniques for the diagnosis of nosocomial pneumonia. Am J Respir Crit Care Med. 1995;152:231–240. doi: 10.1164/ajrccm.152.1.7599829. [DOI] [PubMed] [Google Scholar]
- 3.Comité Technique des Infections Nosocomiales, Cellule Infections Nosocomiales, CCLIN Est, CCLIN Ouest, CCLIN Paris-Nord, and C. Sud-Ouest. Enquête nationale de prévalence des infections nosocomiales, 1996. Bull Epidemiol Hebd. 1997;36:161–163. [Google Scholar]
- 4.Denamur E, Picard B, Decoux G, Denis J B, Elion J. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol Lett. 1993;110:275–280. doi: 10.1111/j.1574-6968.1993.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 5.Emori T G, Gaynes R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fagon J Y, Chastre J, Domart Y, Trouillet J L, Gibert C. Mortality due to ventilator-associated pneumonia or colonization with Pseudomonas or Acinetobacter species: assessment by quantitative culture of samples obtained by a protected specimen brush. Clin Infect Dis. 1996;23:538–542. doi: 10.1093/clinids/23.3.538. [DOI] [PubMed] [Google Scholar]
- 7.Fagon J Y, Chastre J, Domart Y, Trouillet J L, Pierre J, Darne C, Gibert C. Nosocomial pneumonia in patients receiving continuous mechanical ventilation. Prospective analysis of 52 episodes with use of a protected specimen brush and quantitative culture techniques. Am Rev Respir Dis. 1989;139:877–884. doi: 10.1164/ajrccm/139.4.877. [DOI] [PubMed] [Google Scholar]
- 8.Fagon J Y, Chastre J, Hance A J, Domart Y, Trouillet J L, Gibert C. Evaluation of clinical judgment in the identification and treatment of nosocomial pneumonia in ventilated patients. Chest. 1993;103:547–553. doi: 10.1378/chest.103.2.547. [DOI] [PubMed] [Google Scholar]
- 9.Fagon J Y, Chastre J, Hance A J, Guiguet M, Trouillet J L, Domart Y, Pierre J, Gibert C. Detection of nosocomial lung infection in ventilated patients. Use of a protected specimen brush and quantitative culture techniques in 147 patients. Am Rev Respir Dis. 1988;138:110–116. doi: 10.1164/ajrccm/138.1.110. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP: phylogenetic inference package, version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 12.Foght J M, Westlake D W, Johnson W M, Ridgway H F. Environmental gasoline-utilizing isolates and clinical isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology. 1996;142:2333–2340. doi: 10.1099/00221287-142-9-2333. [DOI] [PubMed] [Google Scholar]
- 13.Hilali F, Ruimy R, Saulnier P, Barnabe C, Lebouguenec C, Tibayrenc M, Andremont A. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect Immun. 2000;68:3983–3989. doi: 10.1128/iai.68.7.3983-3989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huelsenbeck J P, Hillis D M. Success of phylogenetic methods in the four-taxon case. Syst Biol. 1992;42:247–264. [Google Scholar]
- 15.Kim J. Improving the accuracy of phylogenetic estimation by combining different methods. Syst Biol. 1993;43:331–340. [Google Scholar]
- 16.Kobayashi I, Hasegawa M, Miyazaki S, Nishida M, Goto S. In vitro and in vivo changes of serotype in Pseudomonas aeruginosa isolates by anti-pseudomonal drugs. J Antibiot (Tokyo) 1994;47:72–79. doi: 10.7164/antibiotics.47.72. [DOI] [PubMed] [Google Scholar]
- 17.Leu H S, Kaiser D L, Mori M, Woolson R F, Wenzel R P. Hospital-acquired pneumonia. Attributable mortality and morbidity. Am J Epidemiol. 1989;129:1258–1267. doi: 10.1093/oxfordjournals.aje.a115245. [DOI] [PubMed] [Google Scholar]
- 18.Mahenthiralingam E, Campbell M E, Foster J, Lam J S, Speert D P. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opal S M, Cross A S, Gemski P, Lyhte L W. Aerobactin and alpha-hemolysin as virulence determinants in Escherichia coli isolated from human blood, urine, and stool. J Infect Dis. 1990;161:794–796. doi: 10.1093/infdis/161.4.794. [DOI] [PubMed] [Google Scholar]
- 21.Picard B, Denamur E, Barakat A, Elion J, Goullet P. Genetic heterogeneity of Pseudomonas aeruginosa clinical isolates revealed by esterase electrophoretic polymorphism and restriction fragment length polymorphism of the ribosomal RNA gene region. J Med Microbiol. 1994;40:313–322. doi: 10.1099/00222615-40-5-313. [DOI] [PubMed] [Google Scholar]
- 22.Richards M J, Edwards J R, Culver D H, Gaynes R P. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 23.Romling U, Wingender J, Muller H, Tummler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J M, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sneath P H A, Sokal R R. Numerical taxonomy. The principle and practice of numerical classification. San Francisco, Calif: W. H. Freeman & Co.; 1973. [Google Scholar]
- 26.Tancrede C H, Andremont A O. Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J Infect Dis. 1985;152:99–103. doi: 10.1093/infdis/152.1.99. [DOI] [PubMed] [Google Scholar]
- 27.Tibayrenc M. Towards a unified evolutionary genetics of microorganisms. Annu Rev Microbiol. 1996;50:401–429. doi: 10.1146/annurev.micro.50.1.401. [DOI] [PubMed] [Google Scholar]
- 28.Tibayrenc M, Kjellberg F, Ayala F J. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. . (Erratum, 87:8185.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibayrenc M, Neubauer K, Barnabe C, Guerrini F, Skarecky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]