Abstract
Leber congenital amaurosis (LCA), an inherited retinal degeneration, causes severe visual dysfunction in children and adolescents. In patients with LCA, pathogenic variants, such as RPE65, are evident in specific genes, related to the functions of retinal pigment epithelium and photoreceptors. In contrast to the original Cas9, base editing tools can correct pathogenic substitutions without generation of DNA double-stranded breaks (DSBs). In this study, dual adeno-associated virus (AAV) vectors containing split adenine base editors (ABEs) with trans-splicing intein were prepared for in vivo base editing in retinal degeneration of 12 (rd12) mice, an animal model of LCA, possessing a nonsense mutation of C to T transition in the Rpe65 gene (p.R44X). Subretinal injection of AAV-ABE in retinal pigment epithelial cells of rd12 mice resulted in an A to G transition. The on-target editing was sufficient for recovery of wild-type mRNA, RPE65 protein, and light-induced electrical responses from the retina. Compared with our previous therapeutic editing strategies using Cas9 and prime editing, or with the gene transfer strategy shown in the current study, our results suggest that, considering the editing efficacy and functional recovery, ABEs could be a strong, reliable method for correction of pathogenic variants in the treatment of LCA.
Keywords: MT: RNA/DNA editing, base editing, CRISPR, Leber congenital amaurosis, RPE65, adeno-associated virus, retinal pigment epithelium, retina
Graphical abstract
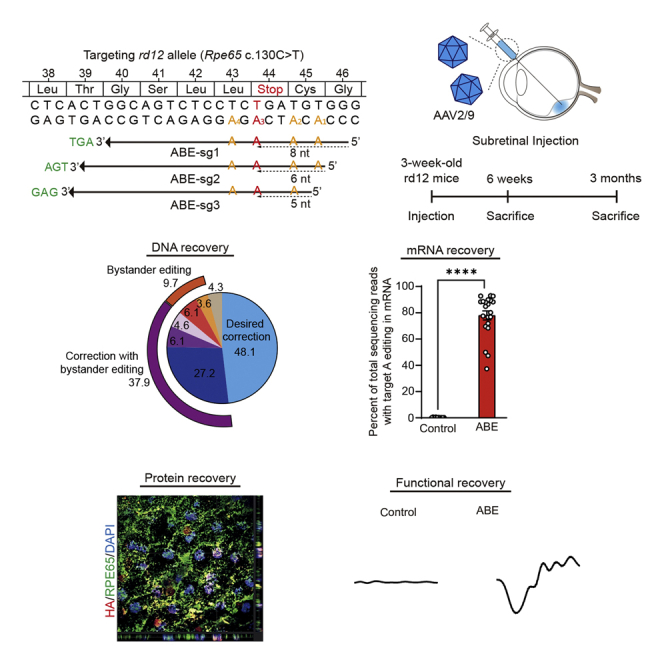
Subretinally administered adenine base editors in dual adeno-associated viral vectors corrected a nonsense mutation of C to T transition in the Rpe65 gene in rd12 mice, an animal model of Leber congenital amaurosis. The on-target editing was sufficient to recover wild-type mRNA, RPE65 protein, and light-induced electroretinographic responses.
Introduction
Leber congenital amaurosis (LCA), an inherited retinal degeneration, is caused by biallelic mutations in the RPE65 gene and/or other genes associated with the functions of retinal pigment epithelium (RPE) and photoreceptors.1,2 Severe early-onset visual dysfunction is typical in patients with LCA.3 There was no cure for LCA until approval of first-in-class gene therapy using adeno-associated virus (AAV) (voretigene neparvovec; Luxturna, Spark Therapeutics). In addition to the AAV-based gene therapy, with the aim of delivering the normal RPE65 gene into the subretinal region,4 direct modifications of pathogenic variants in the genome using CRISPR-associated genome editing tools can provide alternative modalities for the treatment of patients with LCA.5 It is noteworthy that permanent correction of pathogenic mutations by genome editing tools occurs at the endogenous locus.
As previously demonstrated by Maeder and colleagues, small insertions and deletions (indels) generated by CRISPR-initiated DNA double-stranded breaks (DSBs) can result in the correction of a frameshift intron variant of the CEP290 gene (IVS26; c.2991+1655 A>G), the most frequent one in patients with LCA type 10.6 In addition, the therapeutic potential of a CRISPR-initiated homology-directed repair (HDR) strategy was demonstrated by our group in a previous study.7 Use of dual AAVs carrying CRISPR-Cas9 and donor DNA in each vector resulted in ∼1.2% of HDR and ∼1.6% of in-frame 1-codon deletion in retinal degeneration of 12 (rd12) mice, with restoration of visual responses.7 Although the use of CRISPR-initiated indels or HDR was a plausible method for gene recovery,8 it appeared that the correction efficiencies were insufficient to guarantee clinical efficacy.7 There are also critical concerns that CRISPR-mediated DSBs frequently induce unexpected large deletion, chromosomal depletion, and complex rearrangement.9,10,11
As alternatives, base editing or prime editing tools can result in the correction and recovery of disease phenotypes without the generation of acute DSBs.12 DNA base editors (BEs), including cytosine base editors (CBEs) and adenine base editors (ABEs), consist of a cytidine/adenosine deaminase fused to nuclease-deficient Cas9 nickase (nCas9), with the conversion of C to T and A to G, respectively. In contrast, prime editors (PEs) are composed of an engineered reverse transcriptase and nCas9, with the introduction of all types of base conversions and small indels. PEs are more versatile and expansible than BEs; however, considering that BEs generally show higher activity than PEs, BEs might be preferred over PEs for targets that lack bystander nucleotides or when bystander edits are acceptable.13 In addition, according to the ClinVar database, approximately 75% of likely pathogenic/pathogenic variants of the RPE65 gene, one of the most frequently mutated genes in patients with LCA,14,15 are substitutions. Therefore, although only a few studies have been reported to date, the base editing approach can be a reliable method for the treatment of patients with LCA. As a proof-of-concept study, Suh et al. reported that the use of lentiviral vector-delivered ABE enables effective restoration of visual function in adult rd12 mice.16 Our group recently demonstrated that purified ABE ribonucleoprotein (RNP) complex could induce in vivo gene correction of the disease-causing mutation in rd12 mice with negligible off-target DNA and RNA editing.17 However, to date, comprehensive study comparing ABE-mediated gene editing and AAV-based normal gene transfer that can be readily applied in clinics has not yet been reported.
Here, we demonstrated the therapeutic potential of adenine base conversion of a nonsense mutation in the Rpe65 gene of rd12 mice. Subretinal delivery of ABEs via a dual-AAV serotype 9 vector effectively induced an A to G transition in the RPE in vivo. The use of therapeutic base editing of the Rpe65 gene resulted in an increase of light-induced electrical responses from the retina after dark adaptation in young rd12 mice that show no responses without treatment, similar to gene-transferred rd12 mice. In addition, the proportion of desired gene correction was sustained even at 3 months after the injection in the corrected rd12 mice, suggesting that base editing can provide a therapeutic opportunity in the treatment of patients with LCA.
Results
In vitro selection of target sites for adenine base editing of a nonsense mutation in rd12 mice
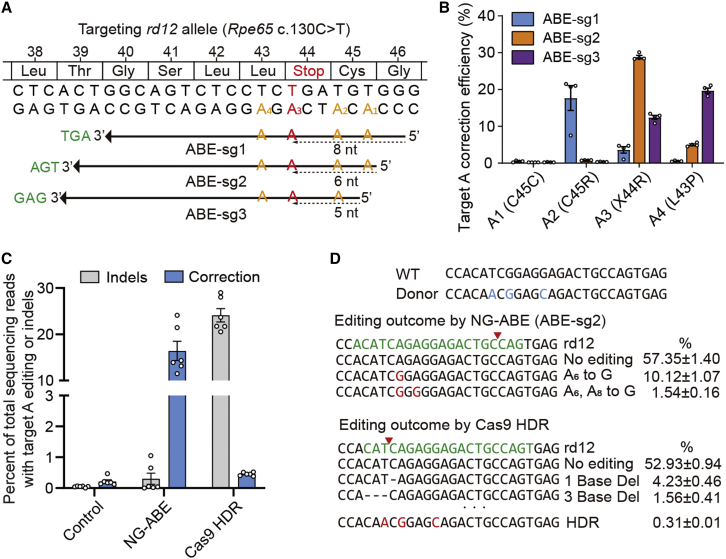
Similar to patients with LCA type 2, rd12 mice possess a nonsense mutation of C to T transition at the 130th position in exon 3 of the Rpe65 gene (c.130C>T, p.R44X) (Figure 1A).18 Of note, it was reported that this R44X mutation is also included in humans (http://www.ncbi.nlm.nih.gov/clinvar/variation/374497).15,19,20 An optimized version of ABE, ABEmax, was utilized to correct the nonsense mutation of the Rpe65 gene in rd12 mice,21 and further engineering was performed in order to employ the protospacer-adjacent motif (PAM) of NG sequences instead of the original NGG,22 named NG-ABE.23 Proper base editing with NG-ABE requires positioning of target adenosines within the editing activity window, from approximately three to eight positions counting from the 5′ end of the protospacer.21 To this end, three possible single-guide RNAs (sgRNAs) were designed with different protospacers and PAMs; the target adenine was placed at positions 8, 6, and 5 with corresponding AGT, TGA, and GAG PAMs, respectively, named ABE-sg1, ABE-sg2, and ABE-sg3 (Figure 1A). Testing of each sgRNA in vitro with mouse embryonic fibroblasts from rd12 mice was performed in order to determine an optimal sgRNA for correction of the nonsense mutation. For ABE-sg3, an original ABEmax was also used because it can employ NAG PAM with less content, in addition to canonical NGG PAM. Because ABEs convert target adenines as well as bystander adenines within the editing activity window, we performed targeted deep sequencing in order to examine the editing activity of all possible adenines. According to our results, ABE-sg2 was selected for use in further experiments because, compared with other sgRNAs, it induced higher on-target editing (∼28%) with fewer bystander edits (Figure 1B). Using this approach with ABE-sg2, the disease-associated point mutation (c.130C>T) was placed at the sixth position (A6) (Figure 1A).
Figure 1.
Adenine base editing to correct a nonsense mutation in rd12 mice
(A) The C>T nonsense mutation (red) at position 130 (c.130C>T; p.R44X). Using a PAM (TGA, green) for NG-ABEmax (ABE-sg2), the targeted adenosine (A3) is located at protospacer position 6, counting the PAM as positions 21–23. The other adenosine within the editing window of ABEmax is indicated in yellow-orange letters. The other two sgRNAs employing GAG and AGT PAM are indicated by the top (ABE-sg1) and bottom (ABE-sg3) arrows, respectively. (B) Percentage of sequencing reads with target A (A1, A2, A3, and A4) editing in rd12 mouse embryonic fibroblast cells after transfection of ABE and sgRNA-encoding plasmids. (C) Percentage of sequencing reads with A to G editing or indels in rd12 mouse embryonic fibroblasts according to treatments (n = 6). Control, Rpe65, and GFP-encoding plasmid; NG-ABE, NG-ABEmax-encoding plasmid, and sgRNA-encoding plasmid; Cas9 HDR, Cas9-encoding plasmid, sgRNA-encoding plasmid, and single-stranded oligodeoxynucleotide donor. (D) Representative results of deep sequencing analysis of mouse embryonic fibroblasts. The sequences of wild-type (WT) and donor sequences are shown at the top (3′ to 5′) for reference. Red letters highlight edited sequences. Green letters highlight the sgRNA-targeting region. Horizontal dashed lines indicate deleted sequences. The red inverted triangles indicate the predicted cleavage site.
A CRISPR-mediated HDR strategy for the correction of this pathogenic variant in rd12 mice for the treatment of LCA was suggested by our group in the previous publication.7 Although 1.9% correction efficiency including deletion of a pathogenic stop codon and functional recovery of rd12 mice was achieved using the previous strategy, a large amount of unwanted indels (∼20%) was regarded as a limitation. To evaluate the advantages of ABE-mediated gene correction, the HDR experiments were repeated using similar experimental conditions and the editing outcomes were compared between ABE and HDR methods. According to the results, significantly higher editing efficiency was observed with lower indels in ABEs compared with the HDR method (Figures 1C and 1D), suggesting that base editing is a more efficient and relevant method for the treatment of disease, as demonstrated in other studies.24,25
Optimization of in vivo adenine base editing based on a dual-AAV vector system in rd12 mice
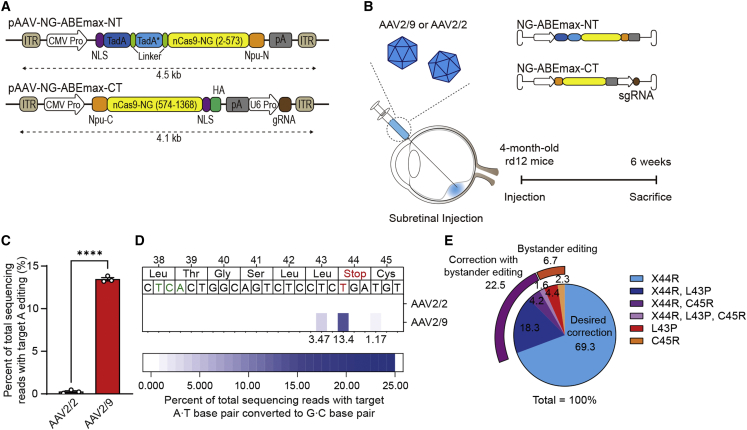
Currently, it is generally thought that AAV is a reliable, effective method for gene therapy including in vivo adenine base editing.26 It is also noteworthy that AAV-based gene therapy is a clinical approach approved by the Food and Drug Administration of the United States for the treatment of patients with LCA.4 The NG-ABE was divided into halves using a trans-splicing intein in order to bypass the size limit of the cargo of AAVs (∼4.8 kb) (Figure 2A).27,28 Prior to performing in vivo delivery of the dual-AAV system, testing was performed in human HEK293T cells to determine whether dual plasmids encoding each half of NG-ABE had sufficient editing activities. As a result, editing efficiency comparable with intact NG-ABE was observed at 10 different endogenous targets, demonstrating the validity of our split NG-ABE system (Figure S1A). In vitro and in vivo reconstitution of NG-ABE after delivery of split NG-ABE components was confirmed by detecting the full size of NG-ABE protein using western blot analysis (Figures S1B and S1C).
Figure 2.
In vivo adenine base editing corrects a nonsense mutation in the RPE of rd12 adult mice
(A) A schematic drawing of the dual-AAV vectors for ABE delivery. CMV Pro, CMV promoter; gRNA, guide RNA; ITR, inverted terminal repeat; nCas9-NG, NG PAM ABEmax; NLS, nuclear localization signal; Npu-C, C-intein from N. punctiforme; Npu-N, N-intein from N. punctiforme; pA, polyA; TadA, tRNA-specific adenosine deaminase; TadA∗, the evolved version of TadA; U6 Pro, U6 promoter. (B) A schematic diagram depicting the design of animal experiments. The dual-AAV vectors were packaged into AAV serotype 2 or 9. Three-week-old or 4-month-old rd12 mice received subretinal injection of AAV-ABE. Six weeks later, the eyes were enucleated for further analyses. (C) Quantitation of the intended A to G correction at position 6 (A6) in RPE cells at 6 weeks after subretinal injection of AAV serotype 2 (AAV2/2) or serotype 9 (AAV2/9) containing NG-ABEmax in 4-month-old rd12 mice (n = 3). (D) The heatmap of DNA reads according to the edited nucleotide positions within the Rpe65. (E) The editing outcomes including the intended (X44R) and bystander edits in RPE cells at 6 weeks after subretinal injection of AAV serotype 9 containing NG-ABEmax in 4-month-old rd12 mice (n = 3). ∗∗∗∗p < 0.0001, two-tailed Student’s t test.
Prompted by these data, we decided to perform subretinal injection of the dual-AAV vectors in order to test the therapeutic efficacy of ABEs (Figure 2B). Because it is not clear which serotypes are most effective for adenine base editing of RPE cells, especially in the delivery of dual-AAV vectors, we first attempted to determine optimal AAV serotypes. AAV serotype 2 (i.e., AAV2/2) and serotype 9 (i.e., AAV2/9) were selected because these serotypes have been utilized in a gene therapy drug (voretigene neparvovec) and in our previous study for Cas9-mediated HDR for treatment of LCA, respectively7; the effects of adenine base editing were then examined in 4-month-old rd12 mice. Six weeks after subretinal injection of each dual-AAV vector, bulk RPE cells were isolated and subjected to targeted deep sequencing. According to the results, higher on-target editing (A6 to G) efficiency was observed with AAV2/9 (13.5% ± 0.2%) compared with AAV2/2 (0.3% ± 0.1%) (Figures 2C and 2D). Therefore, dual-AAV2/9 vectors were used in further experiments.
As mentioned above, when ABE-sg2 was used, neighboring adenosines (A1, A3, A8, and A11) could also be converted, in addition to the desired adenosine (A6). Theoretically, the anticipated products of bystander editing at A1, 3, 8, or 11 could induce C45C (synonymous), C45R, L43P, or L42P (non-synonymous), respectively. Among all base-edited reads at any sites, the top five products were X44R (the desired edit, 69.3%), X44R and L43P (18.3%), L43P (4.4%), X44R and C45R (4.2%), and C45R (2.3%) (Figure 2E). It is worthy of note that L43P and C45R variants have not been reported in patients with inherited retinal degeneration. In addition, use of the L43P variant, the more prominent bystander product, results in non-functional RPE65 without definite toxicity, as demonstrated in HEK293 cells transfected with L43P variant plasmids.16
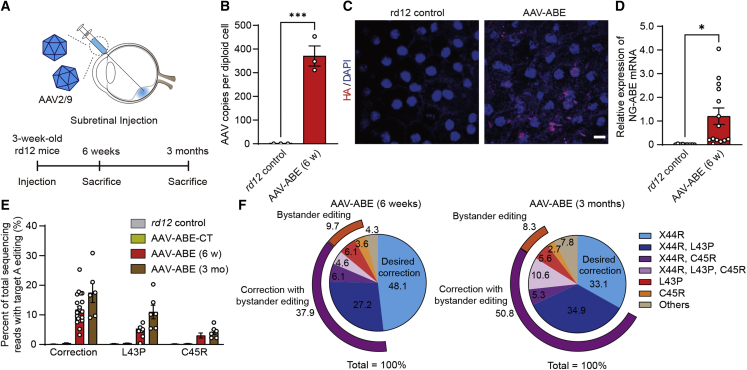
In vivo adenine base editing with a dual-AAV vector system in 3-week-old rd12 mice
Base editing for young mice is required in order to examine the therapeutic effect and functional recovery of rd12 mice. To this end, rd12 mice were prepared at the age of 3 weeks and subretinal injection of dual-AAV2/9 vectors containing NG-ABE was performed (i.e., AAV-ABE) (Figure 3A). Six weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice, multiple AAV copies were observed in RPE cells of rd12 mice (Figure 3B). In addition, AAV-mediated ABE delivery was supported by the prominent nuclear and cytoplasmic expression of a hemagglutinin (HA) tag (Figure 3C) encoded by the sequence in the AAV vectors flanked by the C-term parts of the NG-ABE and nuclear localization sequence (Figure 2A). These data were also supported by results of further analyses using quantitative real-time polymerase chain reactioin (qRT-PCR) with a probe targeting ABEmax mRNA (Figure 3D). Targeted deep sequencing with bulk RPE cells was also performed. According to results, AAV-ABE effectively induced on-target (i.e., A6) editing of 11.8% ± 1.1% (range, 3.2%–25.3%) (Figure 3E), similar to the results described above with 4-month-old mice. Among base-edited reads, the top five products were X44R (48.1%), X44R and L43P (27.2%), X44R and C45R (6.1%), L43P (6.1%), followed by X44R, C45R, and L43P (4.6%) (Figure 3F).
Figure 3.
Comprehensive analysis of in vivo adenine base editing to correct a nonsense mutation in the RPE of rd12 juvenile mice
(A) A schematic diagram depicting the design of animal experiments. (B) Quantitation of AAV copies per diploid cell of the RPE at 6 weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 3). (C) Representative photographs of confocal microscopy of the RPE of rd12 mice at 6 weeks after subretinal injection of AAV-ABE. Scale bar, 10 μm. (D) Quantitation of relative NG-ABEmax mRNA expression in the RPE at 6 weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 9 and 14 for each group). (E) Quantitation of the intended A to G correction at position 6 (A6) in the RPE at 6 weeks (AAV-ABE (6 w)) and 3 months (AAV-ABE (3 mo)) after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 11, 8, 21, and 6 for each group). AAV-ABE-CT, rd12 mice receiving one AAV vector containing the C-term part of NG-ABEmax. (F) The editing outcomes including the intended (X44R) and bystander edits in RPE cells at 6 weeks and 3 months after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 21 and 6 for each group). ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, two-tailed Student’s t test.
To determine whether the gene correction is maintained for a long time, AAV-ABE was injected into the subretinal space of 3-week-old rd12 mice and analyses were performed 3 months after subretinal injection of AAV-ABE (Figure 3A). Results of targeted deep sequencing with bulk RPE cells showed slightly increased editing efficiency of each base, whereas the proportion of the desired correction without bystander edits was sustained at 3 months after the subretinal injection (6.0% ± 0.6% at 6 weeks versus 6.1% ± 0.9% at 3 months after the subretinal injection) (Figures 3E and 3F; Table S1).
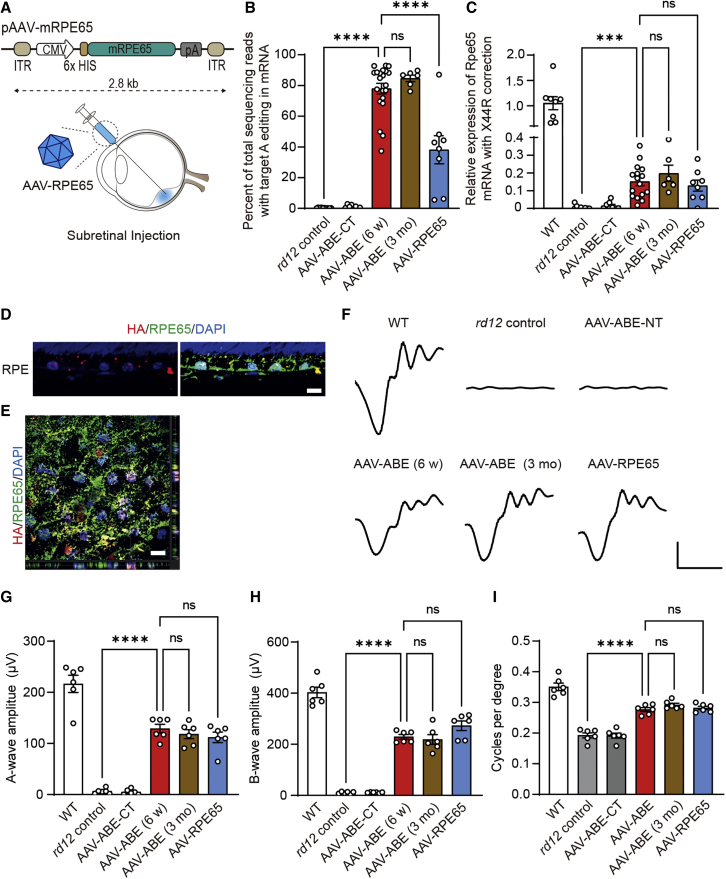
In vivo adenine base editing recovers RPE65 protein in the RPE and improves the visual function of rd12 mice
In the treatment of patients with LCA who possess the pathogenic variants in the RPE65 gene, restoration of the functional RPE65 protein, which is essential in the visual cycle, is the purpose of both gene therapy and genome editing.1,29 To examine the functional consequences of AAV-ABE, AAV2/9 vectors encoding the cytomegalovirus (CMV) promoter-driven mouse Rpe65 gene, named AAV-RPE65, were prepared as a control for gene transfer (Figure 4A). mRNA as well as genomic DNA were isolated from retinal and RPE tissues of AAV-treated mice and analyzed (Figure S2A). Results of mRNA sequencing analyses showed that most (80.0% ± 3.4% and 84.6% ± 1.9%) of the reads of the Rpe65 mRNA from RPE tissues of AAV-ABE-treated mice at 6 weeks and 3 months after treatment were the products of A6 correction (Figure 4B), which were higher than the levels of DNA correction efficiency (Figure 3E). These discrepancies between DNA and mRNA correction have been repeatedly reported by several groups.25,30,31 In AAV-ABE-treated rd12 mice, surveillance mechanisms, such as nonsense-mediated decay, might decrease the mutant transcripts and increase the proportion of the corrected transcripts.32 As expected from the targeted deep sequencing, the top two reads of the Rpe65 mRNA by adenine base editing corresponded to X44R (40.9% and 36.9% at 6 weeks and 3 months after the administration of AAV-ABE, respectively) and X44R and L43P (22.9% and 27.2% at 6 weeks and 3 months after the administration of AAV-ABE, respectively) (Figure S2B). On the other hand, 38.2% ± 8.5% of Rpe65 mRNA of RPE tissue from rd12 mice treated with gene transfer (AAV-RPE65) were A6-corrected mRNA, which resulted from exogenous Rpe65 gene delivery.
Figure 4.
In vivo adenine base editing recovers RPE65 protein in the RPE and improves visual function of rd12 mice
(A) A schematic drawing of the AAV vectors for RPE65 gene delivery. CMV Pro, CMV promoter; mRPE65, mouse RPE65 protein coding sequence; 6X HIS, 6X HIS epitope tag; ITR, inverted terminal repeat; pA, polyA. (B) Percentage of sequencing reads with the A6-correction in Rpe65 mRNA of the RPE from rd12 mice according to the treatment (n = 10, 8, 21, 6, and 8 for each group). (C) Quantitation of relative expression of A6-corrected Rpe65 mRNA of the RPE from WT C57BL/6 and rd12 mice according to the treatment (n = 8, 9, 8, 16, 6, and 8 for each group). (D and E) Representative photographs of confocal microscopy of the RPE in the paraffin section (D) and wholemount preparation of an RPE-choroid-scleral complex (E) at 6 weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice. Scale bar, 10 μm. (F) Representative waveforms of ERG of WT C57BL/6 and rd12 mice according to the treatment. x axis, 30 ms; y axis, 50 mV. (G) Quantitative analyses of amplitudes of a-waves of ERG of C57BL/6 and rd12 mice according to the treatment (n = 6). (H) Quantitative analyses of amplitudes of b-waves of ERG of C57BL/6 and rd12 mice according to the treatment (n = 6). (I) Quantitative analyses of the visual acuity of C57BL/6 and rd12 mice according to the treatment (n = 6). ns, p > 0.05; ∗∗∗∗p < 0.0001, one-way ANOVA with post hoc Tukey’s multiple comparison tests. WT, wild-type C57BL/6; AAV-ABE-CT, rd12 mice receiving one AAV vector containing the C-term part of NG-ABEmax; AAV-ABE (6 weeks) and AAV-ABE (3 months), rd12 mice receiving AAV-ABE at the age of 3 weeks and prepared at 6 weeks and 3 months post injection, respectively.
Wild-type Rpe65-specific probes were designed for quantitative analysis of functional Rpe65 mRNA expression (Figure S2A). Results of qRT-PCR analyses showed significant increases in expression of functional Rpe65 from AAV-ABE-treated mice (15.2% ± 2.3% and 19.8% ± 4.2% of that of wild-type C57BL/6 mice, at 6 weeks and 3 months after the administration of AAV-ABE, respectively) compared with rd12 control mice, which was comparable with treatment using gene transfer (12.9% ± 2.9%) (Figure 4C). On the other hand, treatment with AAV-ABE did not alter the mRNA expression of the Best1 gene (Figure S2C), whose function is related to RPE.33 rd12 mice lack the RPE65 protein due to a nonsense mutation of R44X in the Rpe65 gene.16,18 In contrast, RPE cells in AAV-ABE-treated rd12 mice expressed the RPE65 protein with a prominent expression of an HA tag in the nucleus (Figures 4D and 4E).
To determine whether restoration of the RPE65 protein by AAV-ABE resulted in functional recovery, electroretinography (ERG) was performed using wild-type C57BL/6 and rd12 mice after dark adaptation. As in the previous study employing lentiviral vectors for delivery of ABE,16 AAV-ABE increased light-induced electrical responses of retinal tissues in rd12 mice (Figures 4F and S3). The amplitudes of a- and b-waves of dark-adapted ERG responses of AAV-ABE-treated rd12 mice at 0 dB light stimuli were 59.6% ± 9.5% and 56.9% ± 5.5% of those of wild-type mice, respectively (Figure 4F). In addition, results of linear regression analyses showed that the on-target DNA editing efficiencies were related to both a- and b-waves of ERG responses (Figure S4). It is also noteworthy that the amplitudes of ERG were maintained at 3 months after administration and were comparable with those in rd12 mice after the Rpe65 gene transfer (Figures 4G and 4H). The optomotor responses were measured further to respond to the rotating stimuli in a virtual cylinder. In this experiment estimating the visual function, significant recovery of the visual thresholds was observed for AAV-ABE-treated rd12 mice compared with untreated mice (Figure 4I). No significant differences were observed among rd12 mice at 6 weeks and 3 months after treatment with AAV-ABE and at 6 weeks after Rpe65 gene transfer (AAV-RPE65) (Figure 4I).
Analysis of potential off-target effects of in vivo adenine base editing
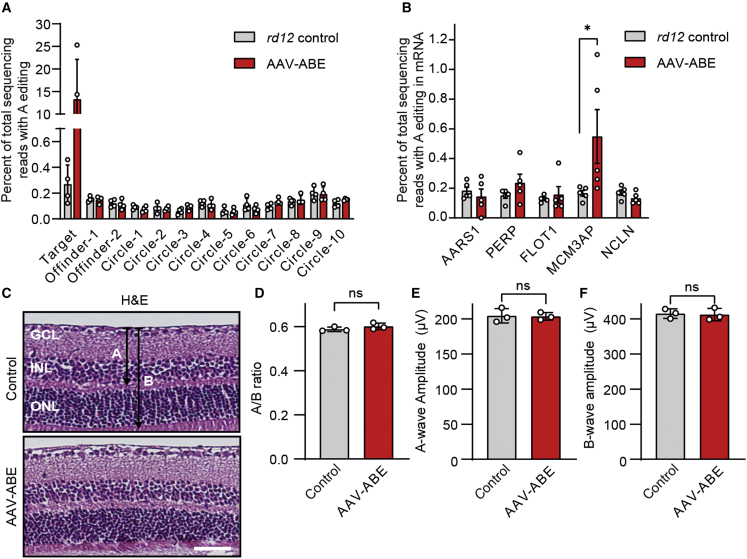
Although ABEs rarely induce DNA DSBs, there are still concerns regarding sgRNA-dependent off-targeting of DNA (A to G conversion at other sites in the genome)34 and sgRNA-independent RNA deamination35,36,37 by excess adenosine deaminases (TadA) included in ABE. Despite the effective on-target editing, negligible off-target editing of DNA was observed at the two predicted sites from the Cas-OFFinder38 and the 10 predicted sites from the Circle-seq16,39 in the RPE samples of AAV-ABE-treated rd12 mice (Figure 5A and Table S2). Among the five tested RNA off-target sites,35,36,37 only one differential RNA deamination pattern was observed at the MCM3AP transcript, which was predicted on the sequence similarity to the TadA substrate (GCUCGGCUACGAACCGAG) (Figure 5B and Table S3).40 Further transcriptomic analyses upon in vivo adenine base editing might be beneficial to guarantee the safety for clinical application.
Figure 5.
Analysis of potential off-target effects of in vivo adenine base editing
(A) On- and off-target editing rates of specific sites of the genomic DNA of RPE tissues at 6 weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 4). Off-target sites were predicted from Cas-OFFinder (two sites) and CIRCLE-seq (six sites). (B) Off-target editing rates of RNA of RPE tissues at 6 weeks after subretinal injection of AAV-ABE in 3-week-old rd12 mice (n = 5). ∗p < 0.05; Mann-Whitney U test. Three off-target sites were selected based on the sequence similarity to the TadA substrate. (C) Representative photographs of H&E-stained retinal tissues at 6 weeks after subretinal injection of AAV-ABE in C57BL/6 mice (n = 3). A and B indicate the thickness from the internal limiting membrane to the inner nuclear layer and that from the internal limiting membrane to the outer nuclear layer, respectively. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar, 50 μm. (D) Quantitation of the A/B ratios at 6 weeks after subretinal injection of AAV-ABE in C57BL/6 mice (n = 3). (E) Quantitative analyses of amplitudes of b-waves of ERG at 6 weeks after subretinal injection of AAV-ABE in C57BL/6 mice (n = 3). (F) Quantitative analyses of amplitudes of b-waves of ERG at 6 weeks after subretinal injection of AAV-ABE in C57BL/6 mice (n = 3). ns, p > 0.05; two-tailed Student’s t test.
Regarding clinical application, minimizing toxicity to adjacent tissues is essential. In the present study, the CMV promoter was utilized for the delivery of ABEmax to RPE tissues via subretinal injection of AAV-ABE (Figure 1C). As expected, we observed expression of HA in retinal layers (Figure S5A) and on-target DNA editing of 8.5% ± 1.6% from retinal tissues (Figure S5B). However, gross changes of retinal layers (Figures 5C and 5D) and ERG responses in wild-type C57BL/6 were not induced by AAV-ABE (Figures 5E and 5F). These data implied that significant nonspecific tissue damage was not induced by the local administration of AAV-ABE. Nonetheless, target-cell-specific promoters might eliminate toxicity concerns.
Discussion
The findings of this study demonstrated that the use of adenine base editing could result in the correction of a nonsense mutation and restoration of visual function in rd12 mice, an animal model of LCA. Notably, approximately 6% of editing efficiencies were achieved in the intended correction with clinically applicable AAV vectors, leaving aside the additional approximately 5% of readthrough edits.41 AAV vectors were injected into the subretinal space of less than half of the murine retina. Accordingly, we speculated that the functional outcomes obtained by adenine base editing could be improved in humans compared with mice.7 In patients with LCA, AAV vectors were injected in the selected area near the macula by retinal surgeons to enable the delivery of high-dose AAV-ABE and enhance the potential for functional recovery of the central vision.4 In contrast, in the experiments using mice, editing rates were measured in the whole RPE, although less than half of RPE cells near the subretinal bleb were exposed to viral vectors. The editing efficiencies might be higher when normalized to the transduction efficiency.
It is also remarkable that a dual-AAV vector was utilized for in vivo adenine base editing. Similar to our results, Suh et al. previously reported that lentiviral vector-delivered ABE targeting NAG PAM enables effective restoration of visual function in adult rd12 mice. As mentioned by the authors in the paper, lentiviral vectors in their current form are for proof-of-concept studies.16 Use of AAV-ABE resulted in on-target editing comparable with the lentivirus-based approach, which might be due to a different ABE construct and sgRNA sequence. AAV-ABE also enabled higher editing efficiencies than ABE RNP complex, which only induced 1.8% DNA editing in rd12 mice.17 ABE RNP recovered RPE65 protein expression in RPE cells but failed to recover ERG responses.17 Still, novel delivery methods are required to enhance the editing efficiency of ABE RNP, which makes AAV-ABE a more plausible method of clinical application in the current status.42
In addition, our group recently demonstrated that the use of AAV-delivered PEs resulted in the exact correction of the disease-causing mutation in rd12 mice with an editing ratio of 6.4% in bulk RPE cells.43 Although PE rarely showed bystander edits or unwanted indels in rd12 mice, considering the higher gene correction efficiency (∼10%), ABE can still be regarded as a strong therapeutic strategy.
As in our previous study employing HDR and in-frame deletion using CRISPR-Cas9,7 we observed higher mRNA (Figures 4B and 4C) and functional recovery (Figures 4F–4I) than expected from DNA editing (∼10%). Approximately 80% of the total reads of the Rpe65 mRNA from RPE tissues of AAV-ABE-treated rd12 mice were the products of the corrected DNA sequence. In addition, the amplitudes of ERG waves in the treated rd12 mice were ∼60% compared with those in wild-type C57BL/6 mice. We speculated that these phenomena might be associated with nonsense-mediated decay of mutant mRNA from the DNA sequence with a nonsense mutation.32 Augmented clinical responses at the levels of mRNA, protein, and functions can result from a permanent correction in the DNA.
Taken together, we suggest AAV-based adenine base editing as a permanent and direct therapeutic approach for the treatment of patients with LCA-associated substitutions. Base editing is a better method than HDR for correcting substitutions, due in part to the higher efficiency and lower indels. In terms of specificity and efficacy, the impact of base editing might be greater with the improved versions of BEs, such as ABE8e, ABE8eW,44 ABE8eWQ,45 and ABE8s with V106W.46 However, ABE8e, which is known to show enhanced on-target editing, also induced substantial levels of bystander editing compared with ABEmax.16,47 In this context, it is still notable that the use of AAV-ABE in its current form resulted in the effective correction of a pathogenic variant and improved visual function in rd12 mice. We expect that this AAV-based adenine base editing will be applied to patients with LCA who only have limited treatment options.
Materials and methods
Molecular cloning and virus production
All plasmids were constructed using Gibson-cloning with NEBuilder HiFi DNA Assembly master mix (NEB). For the construction of the N-terminal part of the split NG-ABEmax-encoding vector (AAV-NT-ABEmax), the N-terminal coding sequence of ABEmax along with the CMV promoter (pCMV-ABEmax from our previous study17) and N-intein from Nostoc punctiforme was PCR amplified with matching overlaps and cloned into an AAV2-ITR backbone (NotI and XbaI restriction digest of pAAV-nEF-Cas9 plasmid; Plasmid #87115, addgene). For the construction of the C-terminal part of the split NG-ABEmax-encoding vector (AAV-CT-ABEmax), C-Intein from N. punctiforme along with the CMV promoter, the C-terminal coding sequence of ABEmax and gRNA along with the U6 promoter were PCR-amplified with matching overlaps and cloned into an AAV2-ITR backbone. For the construction of the pAAV-mRPE65 plasmid, mRPE constructs (Plasmid #41019, addgene) were PCR amplified with matching overlaps and cloned into an AAV2-ITR backbone. Recombinant AAV packaging (AAV2/2 and AAV2/9) was performed by Vigene Biosciences.
Cell culture and transfection
For correction of the rd12 mutation in vitro, mouse embryonic fibroblasts from rd12 mice were maintained in DMEM supplemented with 10% FBS, 1% penicillin-streptomycin (WELGENE), and 4 mM glutamine (Glutamax-I, Gibco). For ABE-mediated gene correction, mouse embryonic fibroblasts (1.0 × 105) were electroporated with ABEmax- or NG-ABEmax-encoding plasmids (500 ng) and a guide RNA-encoding plasmid (167 ng) using the Neon Transfection System (Thermo).
Animals
C57BL/6 mice and mating pairs of rd12 mice (stock no. 005379, The Jackson Laboratory) were purchased from Central Laboratory Animal and maintained under a 12-h dark-light cycle. Mating of rd12 mice was performed for the production of offspring. Subretinal injection was administered in 3-week-old and 4-month-old rd12 mice. All animal experiments were performed according to the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research. The protocols were approved by the Institutional Animal Care and Use Committee of Seoul National University.
Subretinal injection of AAV
As previously described, after administration of deep anesthesia, injection of AAV-NT-ABEmax and AAV-CT-ABEmax (5.4 × 1010 viral genomes for AAV2/2 and 7.3 × 1010 viral genomes for AAV2/9 each in 3 μL of PBS) into the subretinal space of mice was performed using a customized Nanofil syringe with a 33G blunt needle (World Precision Instrument) under an operating microscope (Leica).48
Isolation of retinal and RPE tissues
From the enucleated eyes, the entire neural retina was removed after the removal of the cornea, iris, and lens. The RPE-choroid-scleral complexes were then incubated in RNAprotect Tissue Reagent ( catalog no. 76104, Qiagen) on ice for 30 min, followed by gentle vortexing for isolation of RPE tissues. Following the removal of the remaining choroid-scleral complexes, a buffer containing RPE cells was centrifuged to obtain RPE cells for use in further analyses.
Targeted deep sequencing
Extraction of genomic DNAs and total RNAs from retinal or RPE tissues was performed at 6 weeks and 3 months after subretinal injection for sequencing of DNA or RNA at on-target or off-target sites. Each tissue sample was sonicated for a few seconds with the buffer LBP1 from NucleoSpin RNA Plus kits (MACHEREY-NAGEL) and divided into two tubes. The lysates were then purified to genomic DNA and RNA using NucleoSpin Tissue Kits and NucleoSpin RNA Plus kits (MACHEREY-NAGEL), respectively, according to the manufacturer’s instructions. Reverse transcription of the purified RNA (total 30 μg) was performed using PrimeScript RT Master Mix (Takara). Amplification of on-target or off-target sites of genomic DNA or cDNA was performed using KOD Multi and Epi PCR kits (Toyobo) for the generation of sequencing libraries (see the Table S4 for the primer sequences). Sequencing of these libraries was performed using MiniSeq with a TruSeq HT Dual Index system (Illumina) as previously described.49 Briefly, equal amounts of the PCR amplicons were subjected to paired-end sequencing using the Illumina MiniSeq platform. After MiniSeq, analysis of paired-end reads was performed by comparing wild-type and mutant sequences using a BE-analyzer.50
Analyses of editing outcomes
An analysis of sequencing reads from AAV-ABE-treated mice was performed for the evaluation of bystander editing. The top 10 frequent patterns of reads and patterns of reads with C conversion in the editing window were calculated among total reads with substitution in the sgRNA-targeting region (see Table S3). The proportions of each pattern of reads were averaged and visualized. Patterns of reads with frequency <0.3% were excluded.
qRT-PCR
As previously described, quantitative estimation of AAV copies in RPE tissues was performed using the AAVpro Titration Kit (Takara).7 qRT-PCR was performed according to the manufacturer’s instructions on AAVs extracted from RPE tissues. Calculation of the number of AAV genome copies was performed using the standard curve with the positive control included in the kit. A conversion factor of 1.6 × 104 cells per 100 μg of genomic DNA was used for the estimation of the number of diploid mouse cells. Quantitative estimation of the Rpe65 mRNA with correction of disease genotype (A6 correction), Best1 mRNA, or NG-ABE mRNA in the RPE tissues was performed using qRT-PCR using TaqMan probes (Thermo) or laboratory-made primers. Information regarding primers or probes is shown in Table S4.
Western blot analyses
Western blot was performed for analysis of the detection of NG-ABEmax protein in HEK 293T cells after transfection of dual plasmids encoding split NG-ABEmax and RPE tissues from rd12 mice. Cell lysates were prepared from HEK293T cells 1 day after transfection and RPE tissues at 6 weeks after subretinal injection of AAV-ABE using radioimmunoprecipitation assay buffer (Sigma) supplemented with Protease inhibitor cocktails (Sigma). A bicinchoninic acid assay kit (Thermo) was used for the measurement of protein concentrations. Equal amounts of proteins were loaded onto Mini Protean TGX Protein Gels (Bio-Rad) and run at 80 V for 20 min and 120 V for 40 min. After the transfer of the proteins to a nitrocellulose membrane, the blots were incubated with anti-Cas9 (#844301, BioLegend), anti-HA (#3724, Cell Signaling), and anti-GAPDH (#2118, Cell Signaling) antibodies, followed by incubation with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (#7076, Cell Signaling). Chemiluminescence from the HRP reaction was detected using a Fusion SL gel chemiluminescence documentation system (Vilber Lourmat) or iBright CL750 Imaging System (Thermo).
Immunofluorescence
Paraffin sections and RPE-choroid-scleral complexes were prepared from enucleated eyes 6 weeks after subretinal injection. Immunostaining of the tissues was performed using anti-HA antibody (1:100; catalog no. 26183-D680, Thermo) and anti-RPE65 antibody (1:100; catalog no. NB100-355AF488, Novus). Nuclear staining was performed using 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma). The slides were then observed under a confocal microscope (Leica).
ERG
Mice were dark adapted overnight. After administration of deep anesthesia, pupils were dilated by topical administration of phenylephrine hydrochloride (5 mg/mL) and tropicamide (5 mg/mL). Full-field electroretinography was performed using the universal testing and electrophysiologic system 2000 (UTAS E-2000, LKC). The responses were recorded at a gain of 2 k utilizing a notch filter at 60 Hz and were bandpass filtered between 0.1 and 1,500 Hz. In this study, 0 dB flash corresponds to 2.5 cd⋅s/m2. Visualization of graphs and estimation of amplitudes was performed using the Prism 9 (GraphPad). The amplitudes of the a-wave were measured from the baseline to the lowest negative-going voltage, whereas peak b-wave amplitudes were calculated from the trough of the a-wave to the highest peak of the positive b-wave. Based on our previous experience,7 we speculated that n = 6 was sufficient to produce significant differences in the treatment groups. Data from six mice in each group were randomly selected using the random allocation tool.
Optomotor response
The virtual optokinetic system (OptoMotry HD, CerebralMechanics) was used for measurement of the grating acuity as visual thresholds to drive head tracking of mice to a rotating virtual cylinder, according to the manufacturer’s instructions and original publications on the system.51,52 Briefly, mice were placed on a platform where they were exposed to a view of the rotating cylinder on the monitors. A staircase procedure was used for the determination of visual thresholds for the detection of the maximum spatial frequency (cycles/degrees) above which the mice did not respond to the rotating stimuli.
Histologic evaluation
Paraffin blocks were prepared from enucleated eyes 6 weeks after subretinal injection. After H&E staining, thin sections were evaluated for histologic toxicity. To examine the histologic toxicity of adenine base editing, the assessment of H&E-stained slides was performed for measurement of the ratio of the thickness of retinal layers (the thickness from the internal limiting membrane to the inner nuclear layer to that from the internal limiting membrane to the outer nuclear layer).53
Statistics
All group results are expressed as mean ± SEM, if not stated otherwise. One-way ANOVA and Tukey post hoc multiple comparison tests were performed for comparisons between groups. Statistical analyses were performed using Prism 9 (GraphPad). p < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the New Faculty Startup Fund from Seoul National University; the Research Grant from Seoul National University Hospital (no 0320213010 to J.H.K.); the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (no. KGM5362111 to J.H.K.); the Korea Drug Development Fund funded by the Ministry of Science and ICT; the Ministry of Trade, Industry and Energy; and the Ministry of Health and Welfare (no. HN21C0917 to J.H.K.); Kun-hee Lee Child Cancer & Rare Disease Project, Republic of Korea (no. 202200004004 to J.H.K.); and by grants from the National Research Foundation of Korea (NRF) (no. 2022M3A9E4017127 and no. 2018M3D1A1058826 to J.H.K.; no. 2020M3A9I4036074 to D.H.J.; and no. 2021R1A2C3012908 and no. 2021M3A9H3015389 to S.B.).
Author contributions
D.H.J., H.K.J., C.S.C., J.H.H., G.R., and Y.J. performed the experiments. D.H.J. and H.K.J. analyzed the data and wrote the manuscript. S.B. and J.H.K. designed the experiments and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.11.021.
Contributor Information
Sangsu Bae, Email: sbae7@snu.ac.kr.
Jeong Hun Kim, Email: steph25@snu.ac.kr.
Supplemental information
Data availability statement
All data generated during this study are included in this published article and its supplemental information files or are available upon request.
References
- 1.Cideciyan A.V. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog. Retin. Eye Res. 2010;29:398–427. doi: 10.1016/j.preteyeres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahel J.A. Spotlight on childhood blindness. J. Clin. Invest. 2011;121:2145–2149. doi: 10.1172/JCI58300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cideciyan A.V., Jacobson S.G. Leber congenital amaurosis (LCA): potential for improvement of vision. Invest. Ophthalmol. Vis. Sci. 2019;60:1680–1695. doi: 10.1167/iovs.19-26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiCarlo J.E., Mahajan V.B., Tsang S.H. Gene therapy and genome surgery in the retina. J. Clin. Invest. 2018;128:2177–2188. doi: 10.1172/jci120429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A., et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 7.Jo D.H., Song D.W., Cho C.S., Kim U.G., Lee K.J., Lee K., Park S.W., Kim D., Kim J.H., Kim J.S., et al. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci. Adv. 2019;5:eaax1210. doi: 10.1126/sciadv.aax1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang H.K., Song B., Hwang G.H., Bae S. Current trends in gene recovery mediated by the CRISPR-Cas system. Exp. Mol. Med. 2020;52:1016–1027. doi: 10.1038/s12276-020-0466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q. Large deletions induced by Cas9 cleavage. Nature. 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 11.Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V., et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019;10:1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habib O., Habib G., Hwang G.-H., Bae S. Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res. 2022;50:1187–1197. doi: 10.1093/nar/gkab1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Hollander A.I., Roepman R., Koenekoop R.K., Cremers F.P.M. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Astuti G.D.N., Bertelsen M., Preising M.N., Ajmal M., Lorenz B., Faradz S.M.H., Qamar R., Collin R.W.J., Rosenberg T., Cremers F.P.M. Comprehensive genotyping reveals RPE65 as the most frequently mutated gene in Leber congenital amaurosis in Denmark. Eur. J. Hum. Genet. 2016;24:1071–1079. doi: 10.1038/ejhg.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh S., Choi E.H., Leinonen H., Foik A.T., Newby G.A., Yeh W.H., Dong Z., Kiser P.D., Lyon D.C., Liu D.R., Palczewski K. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 2021;5:169–178. doi: 10.1038/s41551-020-00632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang H.K., Jo D.H., Lee S.N., Cho C.S., Jeong Y.K., Jung Y., Yu J., Kim J.H., Woo J.S., Bae S. High-purity production and precise editing of DNA base editing ribonucleoproteins. Sci. Adv. 2021;7:abg2661. doi: 10.1126/sciadv.abg2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang J.J., Chang B., Hawes N.L., Hurd R.E., Davisson M.T., Li J., Noorwez S.M., Malhotra R., McDowell J.H., Kaushal S., et al. Retinal degeneration 12 (rd12): a new, spontaneously arising mouse model for human Leber congenital amaurosis (LCA) Mol. Vis. 2005;11:152–162. [PubMed] [Google Scholar]
- 19.Jespersgaard C., Fang M., Bertelsen M., Dang X., Jensen H., Chen Y., Bech N., Dai L., Rosenberg T., Zhang J., et al. Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 2019;9:1219. doi: 10.1038/s41598-018-38007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong Z., Rong F., Dai Y., Yibulayin A., Zeng L., Liao J., Wang L., Huang Z., Zhou Z., Chen J. Seven novel variants expand the spectrum of RPE65-related Leber congenital amaurosis in the Chinese population. Mol. Vis. 2019;25:204–214. [PMC free article] [PubMed] [Google Scholar]
- 21.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H., et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong Y.K., Yu J., Bae S. Construction of non-canonical PAM-targeting adenosine base editors by restriction enzyme-free DNA cloning using CRISPR-Cas9. Sci. Rep. 2019;9:4939. doi: 10.1038/s41598-019-41356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong Y.K., Song B., Bae S. Current status and challenges of DNA base editing tools. Mol. Ther. 2020;28:1938–1952. doi: 10.1016/j.ymthe.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborn M.J., Newby G.A., McElroy A.N., Knipping F., Nielsen S.C., Riddle M.J., Xia L., Chen W., Eide C.R., Webber B.R., et al. Base editor correction of COL7A1 in recessive dystrophic epidermolysis bullosa patient-derived fibroblasts and iPSCs. J. Invest. Dermatol. 2020;140:338–347.e5. doi: 10.1016/j.jid.2019.07.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy J.M., Yeh W.H., Pendse N., Davis J.R., Hennessey E., Butcher R., Koblan L.W., Comander J., Liu Q., Liu D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens A.J., Sekar G., Shah N.H., Mostafavi A.Z., Cowburn D., Muir T.W. A promiscuous split intein with expanded protein engineering applications. Proc. Natl. Acad. Sci. USA. 2017;114:8538–8543. doi: 10.1073/pnas.1701083114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong D.J.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redmond T.M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J.X., Crouch R.K., Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 30.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D., et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 31.Yang L., Wang L., Huo Y., Chen X., Yin S., Hu Y., Zhang X., Zheng R., Geng H., Han H., et al. Amelioration of an inherited metabolic liver disease through creation of a de novo start codon by cytidine base editing. Mol. Ther. 2020;28:1673–1683. doi: 10.1016/j.ymthe.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esumi N., Kachi S., Hackler L., Jr., Masuda T., Yang Z., Campochiaro P.A., Zack D.J. BEST1 expression in the retinal pigment epithelium is modulated by OTX family members. Hum. Mol. Genet. 2009;18:128–141. doi: 10.1093/hmg/ddn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang P., Xie X., Zhi S., Sun H., Zhang X., Chen Y., Chen Y., Xiong Y., Ma W., Liu D., et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 2019;10:67. doi: 10.1038/s41467-018-07988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rees H.A., Wilson C., Doman J.L., Liu D.R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 2019;5:eaax5717. doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R., et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 38.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J., Malashkevich V., Roday S., Lisbin M., Schramm V.L., Almo S.C. Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry. 2006;45:6407–6416. doi: 10.1021/bi0522394. [DOI] [PubMed] [Google Scholar]
- 41.Lee C., Hyun Jo D., Hwang G.H., Yu J., Kim J.H., Park S.E., Kim J.S., Kim J.H., Bae S. CRISPR-pass: gene rescue of nonsense mutations using adenine base. Mol. Ther. 2019;27:1364–1371. doi: 10.1016/j.ymthe.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo D.H., Bae S., Kim H.H., Kim J.S., Kim J.H. In vivo application of base and prime editing to treat inherited retinal diseases. Prog. Retin. Eye Res. 2022;101132:101132. doi: 10.1016/j.preteyeres.2022.101132. [DOI] [PubMed] [Google Scholar]
- 43.Jang H., Jo D.H., Cho C.S., Shin J.H., Seo J.H., Yu G., Gopalappa R., Kim D., Cho S.R., Kim J.H., Kim H.H. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2022;6:181–194. doi: 10.1038/s41551-021-00788-9. [DOI] [PubMed] [Google Scholar]
- 44.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong Y.K., Lee S., Hwang G.H., Hong S.A., Park S.E., Kim J.S., Woo J.S., Bae S. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 2021;39:1426–1433. doi: 10.1038/s41587-021-00943-2. [DOI] [PubMed] [Google Scholar]
- 46.Gaudelli N.M., Lam D.K., Rees H.A., Solá-Esteves N.M., Barrera L.A., Born D.A., Edwards A., Gehrke J.M., Lee S.-J., Liquori A.J., et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020;38:892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 47.Banskota S., Raguram A., Suh S., Du S.W., Davis J.R., Choi E.H., Wang X., Nielsen S.C., Newby G.A., Randolph P.B., et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell. 2022;185:250–265.e16. doi: 10.1016/j.cell.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S.W., Kim J.H., Park W.J., Kim J.H. Limbal approach-subretinal injection of viral vectors for gene therapy in mice retinal pigment epithelium. J. Vis. Exp. 2015:e53030. doi: 10.3791/53030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae S., Kweon J., Kim H.S., Kim J.S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 50.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Douglas R.M., Alam N.M., Silver B.D., McGill T.J., Tschetter W.W., Prusky G.T. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis. Neurosci. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- 52.Prusky G.T., Alam N.M., Beekman S., Douglas R.M. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest. Ophthalmol. Vis. Sci. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- 53.Kim J.H., Kim C., Kim J.H., Lee B.J., Yu Y.S., Park K.H., Kim K.W. Absence of intravitreal bevacizumab-induced neuronal toxicity in the retina. Neurotoxicology. 2008;29:1131–1135. doi: 10.1016/j.neuro.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article and its supplemental information files or are available upon request.