Summary
Here, we have developed an automated image processing algorithm for segmenting lungs and individual lung tumors in in vivo micro-computed tomography (micro-CT) scans of mouse models of non-small cell lung cancer and lung fibrosis. Over 3000 scans acquired across multiple studies were used to train/validate a 3D U-net lung segmentation model and a Support Vector Machine (SVM) classifier to segment individual lung tumors. The U-net lung segmentation algorithm can be used to estimate changes in soft tissue volume within lungs (primarily tumors and blood vessels), whereas the trained SVM is able to discriminate between tumors and blood vessels and identify individual tumors. The trained segmentation algorithms (1) significantly reduce time required for lung and tumor segmentation, (2) reduce bias and error associated with manual image segmentation, and (3) facilitate identification of individual lung tumors and objective assessment of changes in lung and individual tumor volumes under different experimental conditions.
Subject areas: Cancer, Artificial intelligence, Machine learning
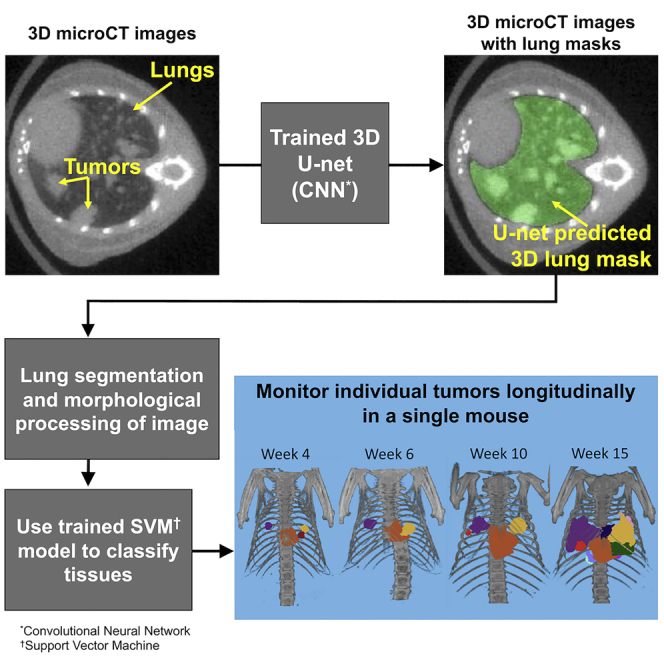
Graphical abstract
Highlights
-
•
Manually segmenting lungs/tumors in murine CT images is subjective and time consuming
-
•
Automated algorithm segments lungs and identifies individual lung tumors
-
•
Automated algorithm reduces bias and image processing time
-
•
Facilitates translational investigation of intra-subject tumor heterogeneity
Cancer; Artificial intelligence; Machine learning
Introduction
Lung cancer is the leading cause of cancer death, accounting for an estimated 18% of all cancer deaths globally in 2020.1 Expression of oncogenic mutant Kras and p53 genes in lung tissues of the KrasLsl.G12D; p53frt/frt; adenoCre.FLP genetically engineered mouse model (GEMM) drives development of lung adenocarcinomas resembling human lung cancers, and enables translational preclinical studies focused on treatment of non-small cell lung cancer.2 GEMMs of lung cancer have been shown to correlate with human tumor growth and response to treatment, providing insight into the development of new therapies.2 To effectively investigate these GEMMs, in vivo micro-CT scans of mouse lungs are used for quantification of lung tumor burden and detection of individual lung tumors. They can also be used to study longitudinal response of total tumor burden and individual tumors in response to treatment with anti-VEGF antibodies, chemotherapy and small molecule kinase inhibitors.2,3,4 Although manual tissue segmentation has the benefits of simplicity and increased control over the task, both preclinically and clinically, segmentation of the lungs and lung tumors by a human reader is time-consuming and impacted by inter- and intra-reader bias and variability. Automated image segmentation is a broad area of research that aims to replace or augment manual segmentation and reduce bias and variability.5 Although numerous machine learning and deep learning methods have been proposed for segmentation of lungs and lung tumors in clinical CT images, there are far fewer published preclinical studies that have utilized these techniques. Published preclinical lung and lung tumor segmentation methods utilize a wide range of techniques, including rules-based segmentation, a mix of rules-based and machine learning methods, machine learning methods with manual intervention and fully automated deep learning approaches.
For preclinical lung segmentation, multiple semi-automated methods have been proposed.6,7,8 However, they rely on a combination of manual input and rules-based segmentation, which can be time-consuming and potentially biased. More recently, several fully automated methods have been reported, which employ a mix of machine learning techniques with morphological and volumetric methods.3,9,10 Wang et al.11 adapted a clinical lung segmentation method from Dou et al.12 that involved a two-stage 3D Convolutional Neural Network (CNN) to segment multiple organs, including lungs. Schoppe et al.13 employed a 2D U-Net-like architecture to automatically segment the lungs in healthy mice and Sforazzini et al.14 trained a 2-D U-net architecture to automatically segment the lungs in mice with varying degrees of fibrosis. Most recently, Malimban et al15 compared 2D- and 3D-Unet approaches for automatically segmenting the left and right lungs, heart and spinal cord, where the models were trained and tested on 175 native and contrast enhanced micro-CT images from 30 healthy mice using the nnU-net method.16
For preclinical segmentation of the entire tumor burden as a single object, both Haines et al. and Ren et al. estimated total tumor and vasculature volume by calculating functional lung volume and subtracting it from the total lung volume, defined as the functional lung region plus tumor and blood vessels.6,7 Rodt et al. and Lalwani et al. employed region growing methods to measure total tumor burden starting with random seed points.17,18 Blocker et al.19 used tools from the 3D slicer toolbox20 for semi-automated whole-tumor segmentation. Barck et al. described automated estimation of lung soft tissue volume based on intensity thresholding, morphological operations and removal of the heart volume, providing an estimate of total tumor burden that correlated well with manual metrics.3 In 2021, Montgomery et al.21 adapted the framework from Barck et al. to segment whole tumor burden.
For preclinical segmentation of individual lesions, Li et al. proposed a semi-automated nodule segmentation method that involved manual detection of nodules to extract a region of interest.22 Rudyanto et al. devised a semi-automated approach to segment and track nodules during longitudinal studies by combining manual nodule identification with segmentation by the fast-marching growth method if the nodule was well circumscribed.8 Recently, Holbrook et al. proposed the first deep learning architecture to automatically detect and segment lung tumor nodules that employs a V-net convolutional neural network trained on a small number of micro-CT scans of tumor-bearing mice supplemented by inclusion of augmented versions of the scans.23 This deep learning approach improves on previous methods of segmentation but generates bounding boxes around each nodule instead of true segmentation of nodules.
Although progression of total tumor burden can be monitored by evaluating longitudinal changes in soft tissue content, automatic detection and segmentation of individual nodules facilitates tracking of change in individual lesion characteristics over time. Detecting these changes is valuable when researching the efficacy of new targeted therapies, especially in the preclinical setting where the growth responses of individual tumors to a given treatment could be related to the genetic profile of each tumor. To this aim, we are proposing a hierarchical, two-step method for fully automated segmentation of healthy, tumor-bearing and fibrotic lungs and lung tumors in micro-CT images using (1) a convolutional neural network (CNN) to segment the lungs and (2) a support vector machine (SVM)-based classifier to distinguish tumors from other tissues within the lungs.
Results
Automated lung segmentation
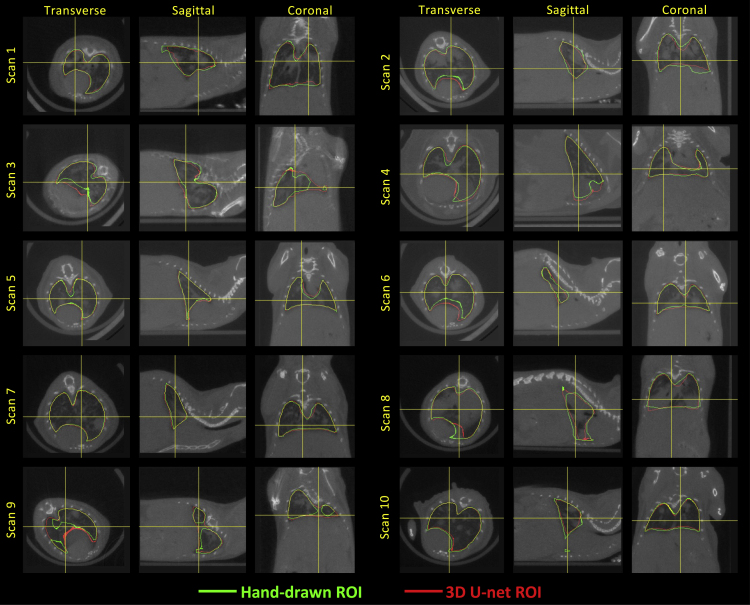
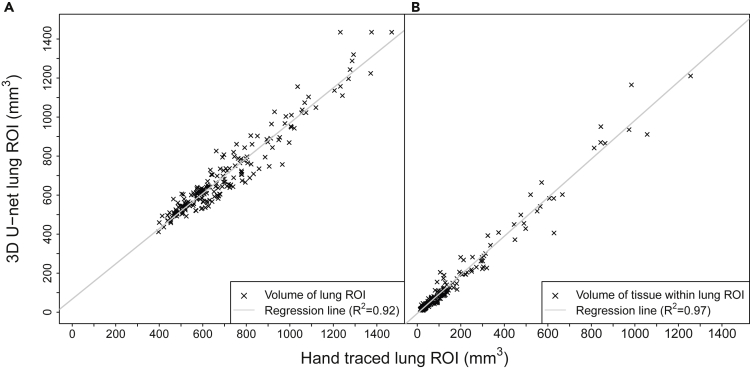
The 3D U-net was trained for 15 epochs (Figure S1), with final dice coefficients of 0.928, 0.926 and 0.920 for the training , cross-validation , and hold-out images, respectively. Figures 1 and Figure 1, Figure 2, Figure 3, Figure 4 show representative hand-drawn (green) and predicted (red) lung ROIs from 10 scans in the hold-out test data set; regions where the hand-drawn and predicted ROIs overlap are shown in yellow. Qualitatively, 3D U-net predicted ROIs closely match the hand-drawn ROIs or outperform the hand-drawn ROIs in cases where there are interpolation or other errors in the ground-truth ROIs, as seen in the transverse slices of scans 3 and 9 (Figure 1). Figure 2A compares the 3D U-net lung volumes to hand-traced, ground truth lung volumes, where a correlation of was observed between the two, whereas Figure 2B compares the soft tissue volumes (calculated based on the binarized image) within the 3D U-net lung ROI to soft tissue volumes within the hand-traced, ground truth lung ROIs, where a correlation of was observed between the two. Note that in this case is the square of the Pearson correlation coefficient. Figure S3 plots the dice coefficient versus lung soft tissue volume for GEMM (open circles) and fibrosis (closed circles) mouse models in the ct120 hold-out test set. The dice coefficient remains relatively constant as total tissue volume in the lung increases, decreasing slightly for larger tissue volumes (approximately greater than 300 mm3). Note that soft tissue volumes are primarily composed of blood vessels and tumors (GEMM) or fibrotic tissue (fibrosis model).
Figure 1.
micro-CT images with lung ROIs
Comparison of hand-drawn and 3D U-net-predicted lung ROIs for images from the hold-out test image set, where the green ROIs were manually drawn by a human reader and the red ROIs were predicted by the trained 3D U-net.
See also Figures S1, S2, and S8.
Figure 2.
Performance of trained 3D U-net for lung segmentation
3D U-net versus manually drawn lung ROIs for CT120 hold-out test scans used to calculate A) volumes of 3D U-net predicted lung ROIs vs. volumes of manually-drawn ground truth lung ROIs and B) volumes of soft tissue within the 3D U-net predicted lung ROIs vs. volumes of soft tissue within the manually-drawn ground truth lung ROIs.
See also Figure S3.
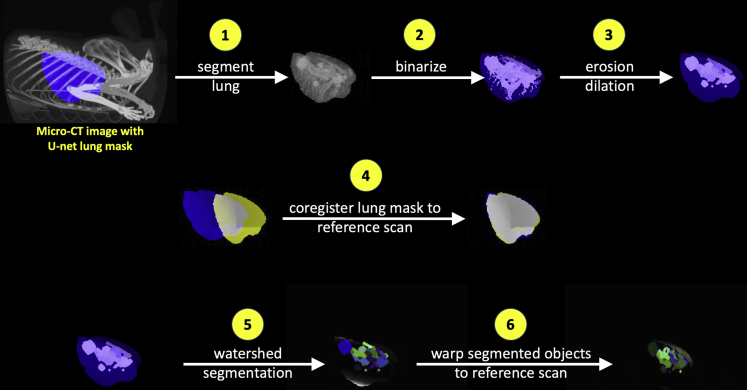
Figure 3.
3D image preprocessing steps for lung tumor segmentation
The key image processing steps for the lung tumor segmentation algorithm are summarized here, where (1) the lungs are segmented using the region of interest estimated by the trained 3D U-net, the resulting lung CT image is (2) binarized and (3) small objects are removed via erosion/dilation operations before (4) coregistering the lung ROI to a reference scan lung ROI; (5) watershed segmentation is then applied to the image generated by step 3, and (6) the resulting objects are warped to the reference scan coordinates using the affine transformation parameters calculated in step 4.
See also Figure S10.
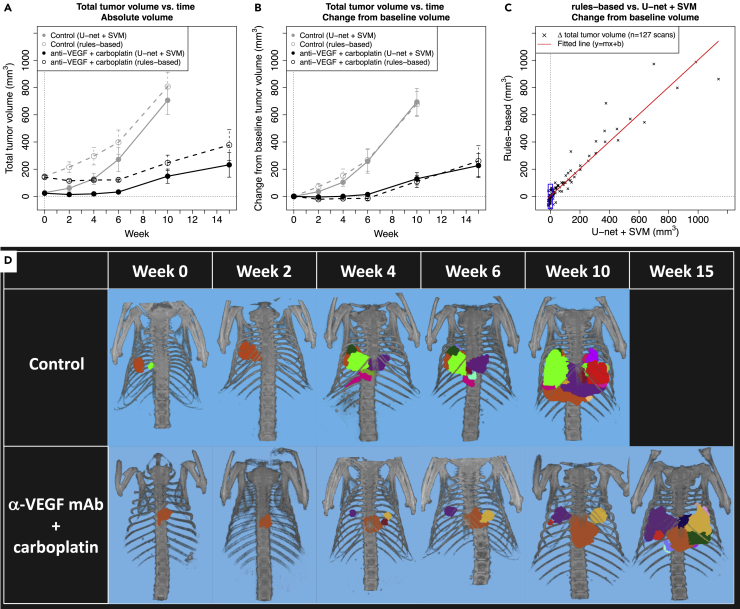
Figure 4.
Segmentation of total tumor burden and individual tumors
(A and B) Comparison of A) average change in absolute total tumor volume and B) change in total tumor volume from baseline volume in control vs. treated GEMM mice as calculated by the U-net + SVM approach (solid lines) and the rules-based approach by Barck et al. (dotted lines). Gray circles and lines represent the untreated control group and black circles and lines represent the treated group, where the untreated control group contained , 14, 14, 13, 5 and 0 mice and the treated group contained , 13, 13, 12, 10 and 4 mice at , 2, 4, 6, 10 and 15 weeks, respectively. Mean +/- S.E.M. (standard error of the mean) of calculated tumor volumes are shown at each time point.
(C) Per-scan comparison of change in total tumor volume from baseline volume as calculated by the rules-based (y-axis) and U-net + SVM (x-axis) approaches. The red line was fitted to the per-scan data using linear regression, where slope , y-intercept and . Comparison of tumor growth in control vs. anti-VEGF + carboplatin treated GEMM mice, where treatment began at week 0 of the study.
(D) Visualization of individual tumor growth in representative mice where individual lesions identified by the U-net + SVM algorithm have been manually color coded.
See also Figures S5 and S9.
Figure S4 shows the log file for all steps in the lung segmentation workflow for the 12 MILabs scans in the hold-out test set, where prediction of a single lung mask took approximately 2 seconds per scan.
Automated tumor segmentation
The 2305 tissue objects generated by the image preprocessing steps (Figure 3) were manually labeled using the labeling tool (Figure S5), yielding 781 objects labeled as “tumor”, 1020 objects labeled as “vessel”, and 504 objects labeled as “other”. Objects labeled as “other” were typically very small (mean object volume approximately 0.5 mm diameter) fragments of tissue that could not be identified by visual inspection and likely were composed of small blood vessel fragments generated during the watershed segmentation step. Lymph nodes are not typically visible in the lungs on mouse micro-CT scans; however, we cannot rule out the possibility that some very small tissue fragments in the “other” category are lymph nodes. A Support Vector Machine classifier with a Gaussian kernel was determined to be the best model with respect to F1 score for tumor object classification, where Table 1 summarizes the hyperparameter search space and final hyperparameter values. A priori normalization of the feature array was performed before hyperparameter optimization. The F1 score for the final SVM model was 0.93 for the training set and 0.85 for the hold-out test set. Performance of other tissue classification algorithms considered here are summarized in Table S1 and Figure S6, where SVM and Classification Ensemble methods yielded the highest F1 score of 0.85 on the hold-out test set. However, the Classification Ensemble method appeared to over-fit the training data set with an F1 score of 1 (Table S1), so the SVM model was selected as the final model.
Table 1.
Optimal hyperparameters for final classification model
| Model | Hyperparameter | Type | Search space | Optimal Hyperparameter |
F1 score |
|
|---|---|---|---|---|---|---|
| training | hold-out | |||||
| Support vector machine | Coding | categorical | onevsall, onevsone | onevsone | 0.93 | 0.85 |
| Box Constraint | real | [0.001, 1000] | 4.7927 | |||
| Kernel Scale | real | [0.001, 1000] | 3.4563 | |||
| Kernel Function | categorical | gaussian, linear, polynomial | gaussian | |||
| Polynomial Order | integer | [2, 4] | N/A | |||
| Standardize | categorical | true, false | false | |||
Figure S7 shows the log file for all steps in the lung tumor segmentation workflow for the 12 MILabs scans in the hold-out test set, where prediction of lesions for a single scan took approximately 2 minutes. Image coregistration was the most time-consuming step and accounted for approximately 80% of the total processing time.
Segmentation of total tumor burden
The trained 3D U-net/SVM model was used to assess changes in total and individual tumor volumes in 27 GEMM mice that were not a part of the training or hold-out test sets for the U-net lung or SVM tumor segmentation algorithms. Figures 4A and 4B plot tumor growth in the control and treatment groups, both as absolute tumor volume (Figure 4A) and change from tumor volume at baseline (Figure 4B). The U-net + SVM based estimates of total tumor volume (solid circles and lines) show that the rate of tumor growth is attenuated in the VEGF + carboplatin treated group such that average total tumor burden at week 10 is approximately 150 mm3 versus 700 mm3 in the untreated control group (Figure 4A). The rules-based method (open circles and dashed lines) describes a similar trend, but with a bias of approximately 120 +/- 60 mm3 on a per-scan basis across all time points and treatment groups because of incomplete removal of non-tumor tissue from the lung volume. When plotting change in total tumor volume from baseline (Figure 4B), the tumor growth profiles calculated using the U-net + SVM and rules-based methods are similar.
Figure 4C compares change in total tumor volume from baseline on a per-scan basis as estimated by the 3D U-net/SVM and rules-based methods. When normalizing to baseline tumor volume, the correlation between the two methods is high, with an R2 of 0.92 and a fitted line close to the identity line, with a slope of approximately 1 and y-intercept of 3.32 mm3. However, there is a significant amount of variability associated with estimated changes in tumor volume when using the rules-based approach, such that in cases where the 3D U-net/SVM approach estimates change in tumor volume to be close to zero (−15 to 15 mm3), the rules based approach returns estimated changes in tumor volume ranging from approximately −90 to 90 mm3 because of presence of residual heart and other tissues within the lung ROI (as indicated by bounding box in Figure 4C).
Segmentation of individual tumors
Individual tumor segmentation allows visualization of individual lesion response to treatment, as shown in Figure 4D for two representative mice, where the mouse in the control group had 5 imaging timepoints and the animal in the treatment group survived until week 15 after treatment and thus had 6 imaging time points. Lesions were manually color coded to facilitate tracking of individual lesion response to treatment, where formation of tumors can be observed and then tracked across time, as seen at week 4 in the treated (purple and yellow lesions) and control (purples and green lesions) animals. In both animals, the growth of the primary lesion observed at week 0 can be tracked across all imaging time points.
Discussion
In this study we utilized a large, high-quality data set of approximately 3000 micro-CT images and hand-drawn lung masks to train a 3D U-net to automatically segment the lungs in scans of healthy, tumor-bearing and fibrotic mice. Performance of the trained 3D U-net was qualitatively excellent, with a dice coefficient of 0.92 on a hold-out test set of 200 scans and based on linear regression of the 3D U-net versus hand-drawn lung ROI volumes (Figure 2A). By comparison, the extended rules-based framework recently described by Montgomery et al., based on Barck et al., obtained a dice score of approximately 0.85 when comparing model-predicted lung ROIs to manually drawn ROIs (Montgomery 2021). The nnU-net-based lung segmentation approach described by Malimban et al15 is similar to what we describe here and accurately (dice score = 0.97) segments the right and left lung in native micro-CT scans of healthy mice, where there is less variability in lung morphology compared to mice with lung tumors or lung fibrosis.
In practice, performance of the trained 3D U-net appears to be equivalent to a human reader because the hand-drawn lung masks include errors where the 3D lung ROI is constructed by interpolating the manually-drawn 2D ROIs, whereas the 3D U-net generates an ROI on every slice of the image. Importantly, the U-net trained on images acquired by the CT120 scanner is able to accurately segment micro-CT images acquired on other scanners, as demonstrated on the respiratory-gated micro-CT images acquired from the MILabs scanner for training the lung tumor segmentation model which required only resampling and normalization of the HU distribution to achieve compatibility with the trained U-net (Figure S8).
The morphological features included in the tumor segmentation algorithm (Figure 3 and Table 2) and can be loosely grouped into four categories: (1) Tissue object size (volume, surface area, eq. diameter, principal axis length), (2) shape (extent, convex volume, solidity, frac. anisotropy), (3) orientation and (4) location (centroid). The mean and maximum intensities (HU) of each tissue object were also used as features. HU intensity of each object and features in each of the four main categories are correlated to one another to varying degrees, but a priori analysis of the feature array suggests that metrics of tissue object size and location are strongly correlated with tissue class. Figure S9 describes the results of a priori analysis of the full feature array using neighborhood component analysis.24
Table 2.
Radiomic features used for object classification
| Feature | Description |
|---|---|
| Volume | Number of voxels in the object |
| Surface area | Distance around the boundary of the object |
| Equivalent diameter | Diameter of a sphere with the same volume as the object |
| Extent | Ratio of voxels in the object to voxels in the total bounding box |
| Convex volume | Number of voxels in smallest convex polygon that can contain the object |
| Solidity | Proportion of the voxels in the convex hull that are also in the object |
| Centroid | Center of mass of the object |
| Fractional anisotropy | Scalar between 0 and 1 which trends toward 0 for increasingly spherical objects |
| Principal axis length | Length of the major axes of the ellipsoid that have the same normalized second central moment as the object |
| Orientation | Euler angles of the object |
| Mean intensity | Mean of all the intensity values in the object |
| Maximum intensity | Highest single-voxel intensity in the object |
The features used here are primarily a function of each tissue object’s size, shape and location (volume, surface area, equivalent diameter, extent, convex volume, solidity, centroid, fractional anisotropy, principal axis lengths). The orientation of each object within the 3-dimensional coordinate system and micro-CT intensities corresponding to each object are also included as features. The bounding box and convex hull are the smallest cuboid and convex polygon, respectively, that can contain the tissue object.
Micro-CT images from the anti-VEGF mAb + carboplatin study versus untreated control mice were acquired using the CT120 scanner and analyzed using 1) the 3D U-net/SVM model trained on respiratory-gated MILabs images and 2) the rules-based segmentation model by Barck et al.3 The two approaches performed similarly overall when calculating change in total tumor burden from baseline (Figure 4B). However, when estimating absolute changes in tumor volume over time the rules-based approach introduced a bias because of incomplete removal of the heart and other tissues during the lung segmentation process (Figure 4A). This bias appears to be relatively constant across scans when evaluating mean tumor growth dynamics for each group, but when considering individual scans (Figure 4C) the rules-based approach demonstrates reduced sensitivity to changes in total tumor burden at lower volumes, on the order of approximately 100 mm3.
Qualitative evaluation of individual tumor segmentation on individual scans shows that the 3D U-net/SVM approach is a useful tool with respect to segmenting and tracking individual lesions over a time course (Figure 4D). Although the SVM classification model for tumor segmentation was trained on the higher-quality, respiratory-gated MILabs scans, it appears to perform well on scans acquired using the non-respiratory-gated CT120 scanner because the SVM classifier was trained on MILabs that were pre-processed to have a similar spatial resolution and HU distribution as compared to the CT120 scanner training set.
Current efforts are focused on automating the temporal linking of individual tumors across longitudinal micro-CT scans. The current approach automatically segments and quantifies individual tumors in a single scan, but for longitudinal scans of a single animal the tracking of individual lesions across time required manual intervention. Ideally, we aim to fully automate segmentation and quantification of individual tumors across time points to investigate variability among tumors, which may have heterogeneous responses to treatment because of factors such as genetic, metabolic, and location differences. In addition, we are investigating use of the SVM-generated tumor masks as a training set for a CNN-based tumor segmentation algorithm.
In summary, our method (1) reduced hands-on time required to perform lung segmentation from approximately 10 min to a few seconds per scan, and reduced hands-on time required to perform lung tumor segmentation from approximately 15–20 min to 2 min per scan and (2) qualitatively reduced bias and error associated with manual segmentation of images. We have also shown that (3) our automated lung tumor segmentation approach performs consistently with previous methods that we have developed for assessment of changes in total tumor burden in GEMM models under various treatment conditions (Figures 4A–4C), and furthermore provides qualitatively reasonable segmentation of individual lung tumors (Figure 4D). Additional studies are required to quantify the amount of bias/error associated with the manually versus automatically segmented lungs and lung tumors.
Conclusions
Here, we have developed an automated tool (3D U-net) for segmentation of lungs in healthy mice, a genetically-engineered mouse model of lung cancer and the bleomycin mouse model of lung fibrosis. Performance is qualitatively on par with a human reader (dice score = 0.92), enabling us to replace time-consuming and potentially error and bias-prone manual segmentation of the lungs with an accurate, automated method that can be applied to micro-CT scans acquired from different scanners. The U-net for lung segmentation also serves as the first step in an automated lung tumor segmentation tool (support vector machine), which automates segmentation of individual lesions and facilitates tracking of individual tumor response to treatment over time.
Limitations of the study
Limitations of this study include (1) the subjective nature of the training set for lung tumor segmentation, where a human reader annotated all tissue objects generated by watershed segmentation as tumor or non-tumor; (2) the trained tumor segmentation model can perform only as well as a human reader and is unlikely to detect tumors or tumor fragments smaller then approximately 0.5 mm diameter; (3) although the lung tumor segmentation algorithm predicts lung tissue objects most likely to belong to the tumor class, longitudinal tracking of individual lesions in a single mouse is not automated and requires manual image processing.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Selected CT images | This paper |
https://github.com/gzferl/LungTumorSegmentationCode https://doi.org/10.5281/zenodo.7366902 |
| Software and algorithms | ||
| Matlab | Mathworks | https://www.mathworks.com |
| Analyze | Analyze Direct | https://analyzedirect.com |
| Python | Python Software Foundation | https://www.python.org |
| Tensorflow | Abadi et al. (2016) | https://www.tensorflow.org |
| Keras | Chollet et al. (2015) | https://keras.io |
| Code for lung and tumor segmentation | This paper |
https://github.com/gzferl/LungTumorSegmentationCode https://doi.org/10.5281/zenodo.7366902 |
Resource availability
Lead contact
Further information and requests for data should be directed to and will be fulfilled by the lead contact, Gregory Z. Ferl (ferlg@gene.com).
Materials availability
This study did not generate new unique reagents.
Methods details
Genetically-engineered mouse model (GEMM) of lung cancer
KrasLsl.G12D;p53frt/frt positive animals were generated2,3 and maintained on a C57Bl/6J-Tyr strain background. Tumors were induced in the KrasLsl.G12D;p53frt/frt mice by infection with infectious units of Adeno-FLPe/IRES/CRE at 7 to 9 weeks of age. All animals were dosed and monitored according to the guidelines from the Institutional Animal Care and Use Committee at Genentech, Inc (South San Francisco, CA).
Mouse model of lung fibrosis
Lung fibrosis was induced with bleomycin, which causes inflammation and alveolar tissue damage leading to fibrosis. Bleomycin at various doses was delivered by either intratracheal or oropharyngeal instillation, or systemic delivery via subcutaneously implanted osmotic pumps.
Data set for training 3D U-net lung segmentation model
3145 lung scans were acquired from a variety of GEMM lung cancer (approx. 2925 scans) and bleomycin induced lung fibrosis (approx. 220 scans) studies. Individual animals were scanned up to five times, with at least two weeks between scans. This resulted in a variety of disease states represented in the data sets, ranging from healthy mice to severe lung tumor burden or lung fibrosis.
Mice were scanned on an in vivo micro-CT system (eXplore CT120, Trifoil Imaging, Chatsworth, CA, USA) using an in-house built animal holder that allows four mice to be scanned simultaneously. The scanning parameters were: 900 projections per full rotation, 16 ms integration time, 75 keV photon energy, 40 mA tube current and 4 × 4 detector binning. The images from the four mice were reconstructed within individual regions of 275 x 275 x 300 dimensions and 100 m isotropic voxel size. The animals were anesthetized with 2% isoflurane in medical air, and the scan time was approximately five minutes per four mice resulting in an average scan time of 1.25 minutes per mouse.
Data set for training tumor classification model
71 lung scans were acquired from a GEMM lung cancer study where individual animals were scanned at one or two time points, yielding a range of imaged tumor burdens from no visible tumor to severe tumor burden. An MILabs micro-CT system (U-CTUHR, MILabs, The Netherlands) was used to scan one mouse at a time25 to enable acquisition of higher-quality respiratory gated micro-CT images and generation of a correspondingly higher-quality ground truth data set for training the tumor segmentation algorithm. The scanning parameters were: 360 projections per full rotation, 20 ms integration time, 65 keV photon energy, 0.13 mA tube current and 2 × 2 detector binning. Retrospective respiratory gating was applied to use only expiration phase projection images for reconstruction. For all animals, a scan was acquired approximately 16 weeks after administration of adenoCRE.FLP, when tumor burden was very low. For animals with two imaging time points, scans were performed either 24 ( animals) or 75 days ( animals) after the first scan.
Treatment study
The study design and animal dosing protocol was similar to that described by Singh et al.,2 where anti-VEGF (B20–4.1.1, mouse IgG2a) and control (anti-ragweed, mouse IgG2a) antibodies prepared and purified at Genentech, Inc. were dosed through intraperitoneal injection at 5 mg/kg twice weekly until the end of study and chemotherapy (carboplatin: Paraplatin from Bristol-Myers Squibb, New York, NY, or generic from Pliva d.d., Zagreb, Croatia.) was dosed intraperitoneally at 25 mg/kg for the first 5 days of the study. The treatment groups were control ( mice) and anti-VEGF + carboplatin ( mice) where each animal was imaged prior to treatment and then at 2 to 5 additional time points selected from 14, 29, 42, 70 and 105 days post-treatment for a total of 127 scans (61 control, 66 treatment). Mice were scanned on the eXplore CT120 micro-CT system as described here.
Lung ROI tracing for training image set
The total intra-thoracic space within the rib cage, excluding heart, mediastinum, liver and diaphragm, was segmented by eight human readers with Analyze software (AnalyzeDirect, Lenexa, KS) by manually drawing regions in the transverse plane on approximately eight evenly spaced slices and propagating (shape based interpolation) between the regions to include all slices in the region of interest (ROI). All manually drawn lung regions of interest were visually inspected for accuracy prior to inclusion in this study, where 174 of 3319 scans were excluded due to qualitatively significant errors in ROI placement. Time required to manually draw lung ROIs was approximately 10 minutes per image.
Convolutional neural network: Training
A 3D U-net was trained on the micro-CT images with manually-drawn ground truth lung ROIs to perform automated lung segmentation, where a “U-net” is a type of convolutional neural network commonly used to process and segment images.26 Briefly, we used a 3D U-Net CNN, which consists of 4 downsampling blocks and 4 upsampling blocks with two convolutional layers of 16 features per block and a ReLU activation. One minus the dice similarity coefficient (DSC) was used as a loss function, where a total of 2900 images were used during training, with an 80/20 split between training and cross-validation sets. An additional 200 images of mice that did not appear in the training or cross-validation sets were excluded from the training process as a hold-out set, for a total of 3100 images. The CNN was trained on 4 GPU compute nodes, with a batch size of 4 images per step and 600 steps per epoch. The CNN was trained using an RMSProp optimizer and a learning rate of 0.0001 were used for training after hyperparameter tuning and the approximately 120,000 model parameters were initialized to random values. All images were downsampled from the original image dimensions of approximately 275 x 275 x 300 by a factor of 0.8 and padded to dimensions of 256 x 256 x 256, which is the largest image size the U-net can accommodate.
Convolutional neural network: Forward prediction
When predicting lung ROIs for the 200 image hold-out test using the trained 3D U-net, the following image processing steps are performed: 1) as in the training set, downsample each image by a factor of 0.8 and pad to dimensions of 256 x 256 x 256, 2) predict the lung mask using the trained U-net, 3) retain only the largest connected object in the predicted lung mask, discarding all others, 4) perform a 3D hole-filling operation on the remaining object, i.e., the lung mask and 5) upsample the predicted lung mask to the original image dimensions.
Image reconstruction and scaling
The 3D U-net for lung segmentation was trained on images acquired from the high-throughput eXplore CT120 system, however, the lung tumor segmentation algorithm was trained using the higher-quality respiratory-gated images acquired from the MILabs system. As such, preprocessing for the MILabs images was required to achieve compatibility with the trained U-net lung segmentation model. Briefly, the MILabs images were reconstructed to the same field of view and dimensions as the CT120 images (275 x 275 x 300 voxels, 100 m voxel size) and the image intensities were scaled to match the CT120 images based on Hounsfield units (HU) associated with air and muscle tissue.
Image preprocessing and watershed segmentation
To prepare images for watershed segmentation, each micro-CT image undergoes 4 preprocessing steps: 1) segment the lungs using the trained U-net model, 2) extract and binarize soft tissues in the resulting image by setting all voxels with intensities between −300 and 200 HU to 1 and all other voxels to zero, 3) apply erosion and dilation operations to remove small objects from the binarized image, and 4) coregister the lung ROI to a reference scan lung ROI using a linear, affine transformation with 12 degrees of freedom. Segmentation of the preprocessed images is split into 2 steps, where 1) a watershed segmentation algorithm is applied to the image, segmenting out discrete objects in the form of a 3D label map, and 2) the 3D label map is warped to the reference scan using the affine transformation array generated in step 4. Matlab R2018b (Natick, MA) was used for steps 1–4 and 6; Analyze software (AnalyzeDirect, Lenexa, KS) was used for step 5. Representative images from each step are shown in Figures 3 and S10.
Briefly, the watershed segmentation algorithm used here accepts a binary image as input, which is eroded repeatedly until all voxels are removed and the connected components of the voxels removed in the final erosion are assigned to unique objects. The algorithm then works backwards, where the voxels removed at the previous erosion level are considered and are assigned to the closest existing object. Voxels which are not connected to an existing object form new objects. This is repeated until all original voxels are assigned.
Feature array for training data set
A total of 2305 segmented objects were generated across the 71 scans (40 mice), where 12 scans (6 mice) were set aside as a hold-out test set such that the training set contained 1941 objects and the test set contained 364 objects. A custom object labeling tool (Figure S5) was developed using MATLAB’s image processing toolbox, facilitating manual assignment of tissue class (tumor, blood vessel or unknown) to each object generated by the watershed segmentation step. Time required to manually label segmented objects was approximately 15–20 minutes per image. Subsequent to object labeling, 12 features describing each object’s shape, size, location and intensity are calculated (Table 2) and saved in the feature array used for classifier training.
Classifier training for lung tumor segmentation
k-nearest neighbors clustering,27support vector machine28 and ensemble methods29,30,31 were evaluated with respect to their ability to discriminate between watershed objects that were manually labeled as ‘tumor’ versus objects labeled as ‘blood vessel’ or ‘other’. The hyperparameter space for k-nearest neighbors (KNN), support vector machine (SVM) and classification/regression ensemble (cEnsemble/rEnsemble) machine learning algorithms (Table S1) was searched using a Bayesian optimization algorithm and the final model and hyperparameter values were determined using the F1 score calculated on the hold-out test set as a goodness-of-fit metric. The classification models were evaluated with and without a priori normalization of the feature array.
Quantification and statistical analysis
Unless otherwise stated in the figure legends, data is shown as mean SEM. The associated with each plot is found in the figure legends, where represents the number of mouse micro-CT scans. Performance of the tissue classification algorithms is assessed by F1 score and accuracy, as defined in the legend for Table S1. Neighborhood component analysis was used for a priori tissue object feature analysis as described in the legend for Figure S9. The 3D U-net was implemented using Tensorflow,32 Keras33 and Python. Watershed segmentation was performed using Analyze (AnalyzeDirect, Lenexa, KS) and Matlab R2020a (Mathworks, Natick, MA) was used to train and optimize tissue classifiers, perform a priori feature array analysis and perform all other image pre- and post-processing tasks. See key resources table for additional details.
Acknowledgments
Not applicable.
Author contributions
Conceptualization: R.A.D.C., K.H.B., M.R.J., G.Z.F.; Methodology: R.A.D.C., G.Z.F., K.H.B., and S.J.; Software and Formal Analysis: G.Z.F., S.J., E.J.M, and J.P.; Investigation and Data Curation: K.H.B., J.H.C., A.L., and J.E.L.; Resources: R.A.D.C. and M.R.J.; Writing – Original Draft, Review and Editing: G.Z.F., K.H.B., R.A.D.C., S.J., E.J.M, and J.P.
Declaration of interests
All authors are current or former employees of Genentech. G.Z.F, K.H.B., S.J., A.L., J.E.L., J.H.C., M.R.J., and R.A.D.C. are current or past stockholders in Genentech, a Roche company.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105712.
Contributor Information
Gregory Z. Ferl, Email: ferlg@gene.com.
Kai H. Barck, Email: kbarck@gene.com.
Supplemental information
Data and code availability
-
•
micro-CT image files corresponding to Figures 1 and 4D and S8 have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
The complete training and hold-out test data sets are available for non-commercial use from the authors upon reasonable request and with necessary data access agreements in place.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. from the lead contact upon request.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Parkin D.M., Piñeros M., Znaor A., Bray F. Cancer statistics for the year 2020: an overview. Int. J. Cancer. 2021;149:778–789. doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 2.Singh M., Lima A., Molina R., Hamilton P., Clermont A.C., Devasthali V., Thompson J.D., Cheng J.H., Bou Reslan H., Ho C.C.K., et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat. Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 3.Barck K.H., Bou-Reslan H., Rastogi U., Sakhuja T., Long J.E., Molina R., Lima A., Hamilton P., Junttila M.R., Johnson L., Carano R.A.D. Quantification of tumor burden in a genetically engineered mouse model of lung cancer by micro-CT and automated analysis. Transl. Oncol. 2015;8:126–135. doi: 10.1016/j.tranon.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merchant M., Moffat J., Schaefer G., Chan J., Wang X., Orr C., Cheng J., Hunsaker T., Shao L., Wang S.J., et al. Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marten K., Auer F., Schmidt S., Kohl G., Rummeny E.J., Engelke C. Inadequacy of manual measurements compared to automated CT volumetry in assessment of treatment response of pulmonary metastases using RECIST criteria. Eur. Radiol. 2006;16:781–790. doi: 10.1007/s00330-005-0036-x. [DOI] [PubMed] [Google Scholar]
- 6.Haines B.B., Bettano K.A., Chenard M., Sevilla R.S., Ware C., Angagaw M.H., Winkelmann C.T., Tong C., Reilly J.F., Sur C., Zhang W. A quantitative volumetric micro-computed tomography method to analyze lung tumors in genetically engineered mouse models. Neoplasia. 2009;11:39–47. doi: 10.1593/neo.81030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren R., Somayajula S., Sevilla R., Vanko A., Wiener M.C., Dogdas B., Zhang W. Medical Imaging 2013: Biomedical Applications in Molecular, Structural, and Functional Imaging SPIE. 2013. Automated 3D mouse lung segmentation from CT images for extracting quantitative tumor progression biomarkers. [DOI] [Google Scholar]
- 8.Rudyanto R.D., Bastarrika G., de Biurrun G., Agorreta J., Montuenga L.M., Ortiz-de Solorzano C., Muñoz-Barrutia A. Individual nodule tracking in micro-CT images of a longitudinal lung cancer mouse model. Med. Image Anal. 2013;17:1095–1105. doi: 10.1016/j.media.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Xu Z., Bagci U., Mansoor A., Kramer-Marek G., Luna B., Kubler A., Dey B., Foster B., Papadakis G.Z., Camp J.V., et al. Computer-aided pulmonary image analysis in small animal models. Med. Phys. 2015;42:3896–3910. doi: 10.1118/1.4921618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan D., Zhang Z., Luo Q., Yang X. A novel mouse segmentation method based on dynamic contrast enhanced micro-CT images. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Han Y., Chen Z., Hu R., Chatziioannou A.F., Zhang B. Prediction of major torso organs in low-contrast micro-CT images of mice using a two-stage deeply supervised fully convolutional network. Phys. Med. Biol. 2019;64 doi: 10.1088/1361-6560/ab59a4. [DOI] [PubMed] [Google Scholar]
- 12.Dou Q., Chen H., Jin Y., Lin H., Qin J., Heng P.A. Medical Image Computing and Computer Assisted Intervention – MICCAI 2017. Springer; 2017. Automated pulmonary nodule detection via 3D ConvNets with online sample filtering and hybrid-loss residual learning. [DOI] [Google Scholar]
- 13.Schoppe O., Pan C., Coronel J., Mai H., Rong Z., Todorov M.I., Müskes A., Navarro F., Li H., Ertürk A., Menze B.H. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nat. Commun. 2020;11:5626–5714. doi: 10.1038/s41467-020-19449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sforazzini F., Salome P., Moustafa M., Zhou C., Schwager C., Rein K., Bougatf N., Kudak A., Woodruff H., Dubois L., et al. Deep learning–based automatic lung segmentation on multiresolution CT from healthy and fibrotic lungs in mice. Radiol. Artif. Intell. 2022;4:e210095. doi: 10.1148/ryai.210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malimban J., Lathouwers D., Qian H., Verhaegen F., Wiedemann J., Brandenburg S., Staring M. Deep learning-based segmentation of the thorax in mouse micro-CT scans. Sci. Rep. 2022;12:1822–1912. doi: 10.1038/s41598-022-05868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 17.Rodt T., von Falck C., Halter R., Ringe K., Shin H.O., Galanski M., Borlak J. In vivo microCT quantification of lung tumor growth in SPC-raf transgenic mice. Front. Biosci. 2009;14:1939–1944. doi: 10.2741/3353. [DOI] [PubMed] [Google Scholar]
- 18.Lalwani K., Giddabasappa A., Li D., Olson P., Simmons B., Shojaei F., Van Arsdale T., Christensen J., Jackson-Fisher A., Wong A., et al. Contrast agents for quantitative microCT of lung tumors in mice. Comp. Med. 2013;63:482–490. doi: 10.2741/3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blocker S.J., Mowery Y.M., Holbrook M.D., Qi Y., Kirsch D.G., Johnson G.A., Badea C.T. Bridging the translational gap: implementation of multimodal small animal imaging strategies for tumor burden assessment in a co-clinical trial. PLoS One. 2019;14 doi: 10.1371/journal.pone.0207555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery M.K., David J., Zhang H., Ram S., Deng S., Premkumar V., Manzuk L., Jiang Z.K., Giddabasappa A. Mouse lung automated segmentation tool for quantifying lung tumors after micro-computed tomography. PLoS One. 2021;16 doi: 10.1371/journal.pone.0252950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M., Jirapatnakul A., Biancardi A., Riccio M.L., Weiss R.S., Reeves A.P. Growth pattern analysis of murine lung neoplasms by advanced semi-automated quantification of micro-CT images. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holbrook M.D., Clark D.P., Patel R., Qi Y., Bassil A.M., Mowery Y.M., Badea C.T. Detection of lung nodules in micro-CT imaging using deep learning. Tomography. 2021;7:358–372. doi: 10.3390/tomography7030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W., Wang K., Zuo W. Neighborhood component feature selection for high-dimensional data. J. Comput. 2012;7:161–168. [Google Scholar]
- 25.Sengupta-Ghosh A., Dominguez S.L., Xie L., Barck K.H., Jiang Z., Earr T., Imperio J., Phu L., Budayeva H.G., Kirkpatrick D.S., et al. Muscle specific kinase (MuSK) activation preserves neuromuscular junctions in the diaphragm but is not sufficient to provide a functional benefit in the SOD1G93A mouse model of ALS. Neurobiol. Dis. 2019;124:340–352. doi: 10.1016/j.nbd.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Ronneberger O., Fischer P., Brox T. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer; 2015. U-Net: convolutional networks for biomedical image segmentation. [DOI] [Google Scholar]
- 27.Cunningham P., Delany S.J. k-Nearest neighbour classifiers - a Tutorial. ACM Comput. Surv. 2022;54:1–25. doi: 10.1145/3459665. [DOI] [Google Scholar]
- 28.Cristianini N., Shawe-Taylor J. Cambridge University Press; 2000. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. [Google Scholar]
- 29.Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. doi: 10.1007/BF00058655. [DOI] [Google Scholar]
- 30.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 31.Freund Y., Schapire R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997;55:119–139. doi: 10.1006/jcss.1997.1504. [DOI] [Google Scholar]
- 32.Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G.S., Davis A., Dean J., Devin M., et al. TensorFlow: large-scale machine learning on heterogeneous systems. arXiv. 2016 doi: 10.48550/arXiv.1603.04467. Preprint at. [DOI] [Google Scholar]
- 33.Chollet F., et al. Keras. 2015. https://keras.io
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
micro-CT image files corresponding to Figures 1 and 4D and S8 have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
The complete training and hold-out test data sets are available for non-commercial use from the authors upon reasonable request and with necessary data access agreements in place.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. from the lead contact upon request.