Abstract
Protective monoclonal antibodies (MAbs) to the major outer membrane protein (MOMP) of species of the family Chlamydiaceae, which is the primary vaccine candidate antigen, recognize nonlinear epitopes conferred by the oligomeric conformation of the molecule. Protective MAbs failed to recognize oligomeric MOMP of the variant strain LLG, which bears amino acid substitutions in variable segments (VSs) 1, 2, and 4, and competed with monomer-specific MAbs mapping to these VSs in reference strain 577. The results suggest that multiple sites located in the three VSs contribute to the epitope of protective MAbs.
Chlamydophila abortus, also known as Chlamydia psittaci serotype 1, is an important pathogen of ruminants causing abortion late in gestation. The organism can also be transmitted to humans, and pregnant women coming in contact with the bacterium are also at risk of abortion (3). Antibiotic treatment as a prophylactic measure is commonly used in some countries. Other countries practice vaccination using commercially available vaccines consisting of inactivated chlamydial elementary bodies (EB) or a mutant strain (13). The development of an alternative subunit vaccine that is cheap to produce, safe, and effective is being sought both to prevent chlamydial abortions and to diminish the spread and the risk of zoonosis. Currently the best subunit vaccine candidate antigen is the 40-kDa major outer membrane protein (MOMP). The MOMP is the most prevalent protein on the surface of the EB (60% of outer membrane protein content) and constituted the major antigen of a subcellular vaccine that conferred protection in pregnant ewes (15). Sequence comparisons of the MOMPs from several chlamydial species indicated the existence of four variable sequences (VS1 to -4) flanked by five domains of conserved sequences (10). In Chlamydia trachomatis, epitopes located within the VS regions that had species, subspecies, and serovar specificity were protective (20). In contrast to C. trachomatis infections, immunity to C. abortus infection has been shown to be conferred by a 110-kDa oligomeric, probably trimeric (7, 11, 18), form of the MOMP. Monoclonal antibodies (MAbs) A11 and F3 that neutralized chlamydial infectivity in vitro and protected pregnant mice from abortion after passive transfer reacted only with the oligomer (1, 4, 7, 11). The epitopes that are recognized by these neutralizing MAbs are unknown as yet.
We have previously reported the existence of naturally occurring variant isolates of C. abortus in Greece. Strains LLG and POS were not recognized by several MAbs, including the anti-MOMP MAb 188 (17). Furthermore, we observed that the protective MAbs A11 and F3 failed to react with inclusions of the two variant strains. Sequencing of variant antigens from escape mutants is a useful approach to obtain important information concerning epitopes that inhibit infectivity. In this report we have determined the amino acid changes in the MOMP variant by DNA sequencing and defined the localization of protective epitopes against C. abortus.
Strains LLG and POS were isolated in Greece from the lungs of aborted caprine and ovine fetuses (17). Strain 577 was from the American Type Culture Collection (Rockville, Md.). C. abortus isolates were grown and purified as described previously (11, 17). Genomic DNA of strain LLG was obtained as described by McClenaghan et al. (12), and the MOMP gene (omp1) was amplified using the Expand Long Template PCR system (Roche Molecular Biochemicals, Lewes, East Sussex, United Kingdom) and primers flanking the MOMP coding region (forward primer, 5′-CAGGAYATCTTGTCTGGCTTTAA-3′; reverse primer, 5′-GGGCGAATTCTTATGCGAATGGAT-3′). The 1.4-kb PCR product was ligated into T-vector pGEM-T (Promega, Southampton, United Kingdom) and used to transform, by heat shock, Epicurian Coli XL-1 Blue ultracompetent cells (Stratagene, La Jolla, Calif.). Recombinant plasmid DNA was purified using a QIAprep Spin Plasmid Miniprep kit (Qiagen Ltd., Crawley, United Kingdom) and sequenced on an ABI 377 automated DNA sequencer (Advanced Biotechnology Centre, Charing Cross Hospital, London, United Kingdom). The characterization of the antimonomer-specific MAbs 188 and 4/11 has been previously reported (11, 14, 17). MAb 202, a genus-specific immunoglobulin G2a, was raised against a Greek abortion strain and was further characterized in this study. The properties of oligomer-specific MAbs A11 and F3 have been described elsewhere (1, 4, 11). MAb 1B8, a serotype 1-specific immunoglobulin G2b, was raised against abortion strain S26/3 and was provided by H. Kennedy (Queens University, Belfast, United Kingdom). The specificity of the novel MAbs 202 and 1B8 was tested using an immunoperoxidase assay on chlamydial inclusions as described previously (17). Further characterization of the MAbs included Western blotting, peptide scanning, and neutralization, competition, and blocking assays. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting have been described in detail elsewhere (17), as have the synthesis of decapeptides spanning the four VSs of the MOMP and the mapping of epitopes (14). The neutralization assays were performed as previously recommended (5), in hamster kidney cells (HaK, ATCC no. CCL15) by using purified MAbs and by following a complement-assisted protocol (2). Inclusions were stained in an immunoperoxidase assay with a genus-specific MAb and counted using an inverted microscope at 200× magnification (Zeiss Axiovert). Neutralization was considered efficient when the infectivity was reduced by 50%. The blocking assay was initially developed for the measurement of anti-MOMP antibodies in sera, and the technique has been described in detail elsewhere (14). For the competition assay, the competitor antibody was purified using a protein A-agarose column (Pharmacia, Uppsala, Sweden) and biotinylated with biotin succinimide as described previously (16). The biotinylated MAbs were first titrated on enzyme-linked immunosorbent assay plates coated with periodate-treated EB to determine the saturation point. Competition was assessed with serial dilutions of the biotinylated competitor MAb and constant amounts of the unlabeled inhibitor MAb. In the blocking assays the competition was between a constant amount of the biotinylated competitor and serial dilutions of the nonlabeled inhibitor.
Reactivity of the MOMP-specific MAbs.
The reactivity of MAbs against the MOMP monomer and oligomer is summarized in Table 1. The monomer-specific MAbs 188, 4/11, and 202 did not cause an efficient neutralization of strain 577 in the presence of complement (reduction of infectivity by 30, 30, and 45%, respectively), whereas the oligomer-specific MAb 1B8 reduced the infectivity by 85%. The neutralizing capabilities of A11 and F3 were similar to those originally reported (1), with 80 and 62%, respectively. The antioligomer MAbs did not react with any of 38 previously (14) synthesized overlapping peptides spanning the four variable segments of the MOMP. This result was in accordance with the previous findings concerning the conformational nature of the corresponding epitopes. MAb 202 mapped to the hydrophobic domain in VS4, which is conserved among species of the family Chlamydiaceae (Table 1).
TABLE 1.
Reactivity of MOMP-specific MAbs
| MAb | Amino acid sequence from epitope scanning (location within MOMP) | Western blotting results | Reactivity to variant strain LLG inclusionsb | Neutralizationc (%) |
|---|---|---|---|---|
| 188 | PTGTAAANYKTP (VS1)a | MOMP monomerd | No | 30 |
| 4/11 | GSSIAADQLP (VS2)a | MOMP monomer/oligomere | Yes | 30 |
| 202 | AAVLNLTTWNPTLL (VS4) | MOMP monomerf | Yes | 45 |
| A11 | No reactivity | MOMP oligomere | No | 80g |
| F3 | No reactivity | MOMP oligomere | No | 62g |
| 1B8 | No reactivity | MOMP oligomerf | No | 85 |
As reported in reference 14.
Reactivity was tested by immunoperoxidase staining as described elsewhere (17), and comparable findings were reported for MAbs 188 and 4/11 (17).
Neutralization studies were performed with strain 577 in the presence of 5% complement. Values shown were obtained at 1.5 μg of MAb/well.
Probed with heated EB of strain 577; comparable findings were reported in reference 17.
Probed with heated and unheated EB of strain S26/3; comparable findings were reported in reference 11.
Probed with heated and unheated EB of strain 577.
Comparable findings were reported in reference 1.
Failure of the neutralizing MAbs to react with the MOMP oligomer of the variant strain.
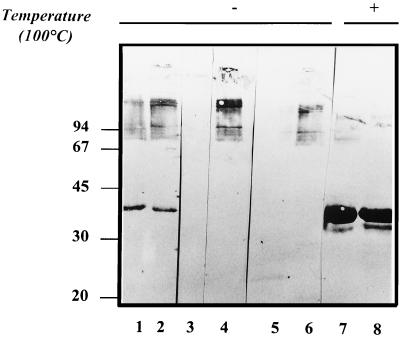
To determine whether any of the neutralizing MAbs would recognize the MOMP oligomer of the variant strain, we performed Western blotting with either unheated or boiled EB of the reference strain 577 and the variant strain LLG (Fig. 1). The results with strain 577 show that the neutralizing MAbs A11 (lane 4) and 1B8 (lane 6) recognized two major regions above the marker at 94 kDa, corresponding to the 110-kDa MOMP oligomer previously identified with A11 (11). MAb F3 had an identical pattern to A11 (data not shown). The corresponding regions of the MOMP oligomer of the variant strain LLG were not recognized by the neutralizing MAbs (lanes 3 and 5), in agreement with the results obtained upon immunoperoxidase staining of inclusions (Table 1). The anti-VS2 MAb 4/11 reacted with the MOMP monomers of both strains 577 and LLG (lanes 7 and 8, respectively). MAb 4/11 also weakly recognized the MOMP oligomers of both the reference and the variant strains (lanes 1 and 2) and so has mixed monomer and oligomer specificity, as previously indicated (11).
FIG. 1.
Comparison of immunoblots after sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis of purified EB from the reference strain 577 and the variant strain LLG with monomer- and oligomer-specific MAbs. EB were either loaded unboiled (lanes 1 to 6) or heat inactivated for 5 min at 100°C prior to loading (lanes 7 and 8). Strain LLG EB are in lanes 1, 3, 5 and 8; strain 577 EB are in lanes 2, 4, 6 and 7. Blots were probed with MAbs 4/11 (lanes 1, 2, 7, and 8), A11 (lanes 3 and 4), and 1B8 (lanes 5 and 6). Molecular masses in kilodaltons are indicated on the left.
Sequence variations in the omp1 gene of strain LLG.
To determine the sequence difference(s) between abortifacient strains LLG and 577, the entire omp1 of strain LLG was sequenced and compared with published sequence data (accession number M73036) (10). The comparison showed seven nucleotide changes at positions 286, 493, 612, 856, 966, 976, and 989 in the coding sequence. The variations at nucleotide (nt) 286 and 493 were within VS1 and VS2 and resulted in amino acid changes at positions 96 (Asn to Asp) and 165 (Ile to Val). The substitutions at nt 612 and 966 were silent. The variations at nt 856, 976, and 989 resulted in three amino acid replacements, one before and two within the VS4 domain. 286Ser was replaced by Gly, 326Ala was replaced by Thr, and 330Ser was replaced by Asn.
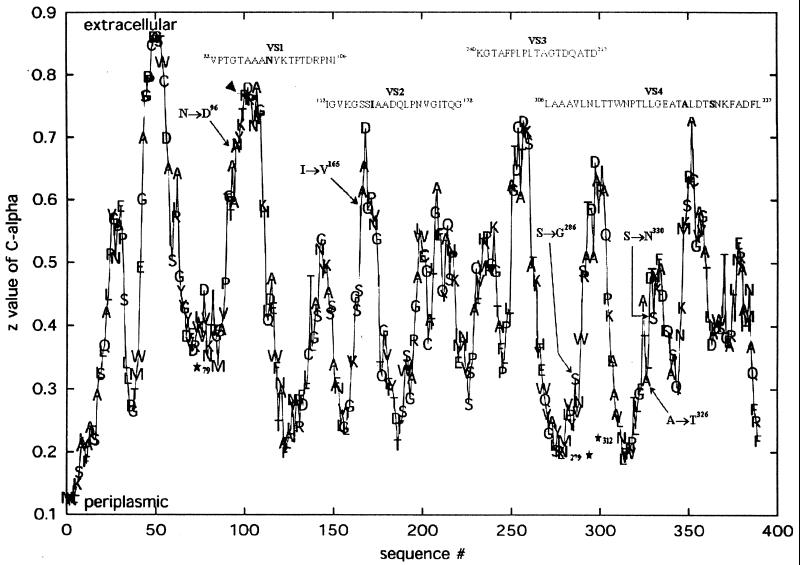
Topology prediction of the variant positions in the MOMP.
To locate the positions of the substituted amino acids in the MOMP molecule, we used a neural network (NN), which is accessible online (http://strucbio.biologie.uni-konstanz.de), to predict the topology of outer membrane (OM) proteins based on their sequence. The NN predicts the z coordinate of Cα atoms in a coordinate frame with the OM in the xy plane. Low z values (below 0.4) indicate periplasmic turns, medium z values indicate transmembrane β-strands, and high z values (>0.6) indicate extracellular loops (8). The output of the NN for the MOMP of reference is shown in Fig. 2. The five amino acid substitutions in LLG-MOMP are indicated (Fig. 2). The mutations in VS1 and VS2 were predicted to be located in extracellular loops, which correlates well with their localization within a surface-exposed variable segment. The z values of the two conservative replacements in VS4 were <0.6. The topology prediction of the nonconservative exchange of 286Ser to Gly was periplasmic. This is in good agreement with the fact that a Gly residue is conserved in most MOMP alleles, with the exception of the reference abortion strains 577 (10) and S26/3 (9). 286Gly is replaced by Ser in these two alleles.
FIG. 2.
Prediction for the topology of the abortifacient MOMP strain S26/3 (9) (identical to reference strain 577) as calculated by an artificial NN trained for bacterial β-strand OM proteins (porins). The four VSs (VS1 to 4) are located above the corresponding peaks. The five amino acid substitutions in LLG are indicated in bold letters and are also marked in the figure by single arrows. The conserved 103Arg at the tip of the VS1 loop that may be involved in the ion selectivity of the channel (see text) is marked by an arrowhead. Asterisks indicate three putative N-glycosylation sites.
Competition assays between antimonomer and antioligomer MAbs.
The replacement of 96Asn by Asp in VS1 was located within the epitope of MAb 188 and probably affected the binding of this MAb to strain LLG (Table 1) (17). This suggestion was reconfirmed by using synthetic peptides. Two decapeptides corresponding to the epitope of MAb 188, but with the substitution found in strain LLG, were synthesized and tested. Binding of MAb 188 to decapeptide 89PTGTAAANYK98 decreased by 20%, and binding to peptide 91GTAAANYKPT100 decreased by 75%, both after substitution of 96Asn by Asp.
The DNA sequencing results also implied that amino acids 96 in VS1, 165 in VS2, and 326 and 330 in VS4 were directly or indirectly involved in the epitope(s) of the neutralizing MAbs A11, F3, and 1B8. While conclusions relevant to linear epitopes can be reconfirmed by the use of synthetic peptides, those concerning conformational epitopes are more difficult to support by direct evidence. We used the competition approach. Antimonomer MAbs 188, 4/11, and 202 (against VS1, VS2, and VS4, respectively) (Table 1) were labeled with biotin and were allowed to compete with the antioligomer, neutralizing MAbs F3, A11, and 1B8 for binding to strain 577 EB-coated enzyme-linked immunosorbent assay plates. As shown in the double-reciprocal plots in Fig. 3A and B, the binding of MAbs 188 and 4/11 to EB was inhibited in a competitive manner by the presence of MAbs 1B8, A11, and F3. Interestingly, MAb 188 inhibited the binding of MAb 4/11 and vice versa (data not shown), indicating that VS1 and VS2 are in close proximity. The data obtained with the anti-VS4 MAb 202 differentiated the reactivity of the neutralizing MAbs A11, F3, and 1B8. The results (Fig. 3C) showed that MAbs A11 and F3 at 10 μg/ml competitively inhibited the binding of MAb 202, whereas MAb 1B8 stimulated the binding of MAb 202 at the same concentration and noncompetitively inhibited it at 0.01 μg/ml. The presence of the nonprotective MAb 188 did not affect the binding of MAb 202 (Fig. 3C). Blocking assays were also performed with a fixed concentration of labeled MAb 188 that was determined by titration to 75% of maximum binding. The concentrations of the blocking antibody that reduced the binding of the labeled MAb by 50% were determined to be 7.5, 9.0, 0.1, and 1.5 μg/ml for MAbs A11, F3, 1B8, and 4/11, respectively. Correspondingly, the binding of labeled MAb 4/11 was reduced by 50% with blocking antibody concentrations of 5.4, 7.5, 0.05, and 10 μg/ml for MAbs A11, F3, 1B8, and 188, respectively. The results suggest that multiple sites located in the three VSs (VS1, VS2, and VS4) are involved in the formation of the epitopes of the neutralizing anti-MOMP MAbs.
FIG. 3.
(A and B) Double-reciprocal plots illustrating competition of biotin-labeled MAbs 188 (A) and 4/11 (B) with unlabeled A11, F3, and 1B8 in binding to strain 577 EB-coated plates. Ten micrograms of the inhibitor MAb was used for each point. (C) Competition of biotinylated MAb 202 with unlabeled MAbs A11, F3, 188, 1B8b (10 μg of each), and 1B8 (0.01 μg). The binding of MAb 202 decreased after labeling with biotin, and saturation was achieved at a low dilution of this antibody. Bound labeled MAb was visualized with streptavidin-conjugated peroxidase and tetramethyl benzidine (TMB) substrate. The experiments were repeated three times. A, absorbance.
The chlamydial MOMP has been recently characterized as a porin (18). Membrane-extracted MOMP or recombinant MOMP formed ion channels when incorporated into planar lipid bilayers (18, 19). However, although the function and secondary structure of MOMP have been documented, structural studies are still incomplete. The NN used here for the topology prediction of the MOMP has been trained with porins of known structure, and the evaluation showed in general a good correlation between the predicted and experimental results (8). According to the topology prediction of this NN, the sequence variations of the MOMP of strain LLG in VS1 and VS2 are within highly exposed regions of extracellular loops. The semiconservative variations in VS4 are less exposed. The introduction of an Asp residue in VS1 and of Val in VS2 of strain LLG changed the z value of the loops by 4.6 and 2.8%, respectively. The importance of these changes on the infectivity of the variant strain is not known. However, it should be kept in mind that LLG was repeatedly found to be 10 times more infectious than other C. abortus strains (17).
Variations in surface-exposed amino acids, while not necessarily having any effect on the function of the protein (porin), can affect the binding of antibodies. The results presented here show that this is the case for MAb 188 and the neutralizing MAbs A11, F3, and 1B8, which do not bind the MOMP of strain LLG. It has been recently shown that the presence of MAb A11 reduced the single-channel conductance of the porin, most probably by occluding the channel (18, 19). The presented data suggest that the protective MAbs A11 and F3 (in contrast to nonprotective MAb 188) make contact with residues in VS4 in addition to their binding sites in VS1 and VS2 and in so doing possibly span the barrel of the porin. This might result in a partial obstruction of the channel and the reduction in single-channel conductance. The same seems to apply also to MAb 1B8, which sterically or allosterically affects the binding of MAb 202. It is conceivable that a highly exposed loop such as the VS1 loop can fold into the barrel, thus reducing the bridging distance between the multiple sites of the epitope. Indeed, such a pore-confined external loop has been identified in other bacterial porins whose crystal structures have been determined (6). This L3 loop, known as the “eyelet” loop, contributes to a constriction of the channel where charge distribution affects the ion selectivity. Residues near the tip of the L3 loop influence the ion selectivity of the channel. It is interesting therefore that weakly cation-selective porins such as OmpF have a conserved negatively charged residue at the tip of L3 (117Glu in OmpF) (6). The MOMP of C. abortus, which is weakly anion selective (18), has in contrast a conserved, positively charged residue (103Arg) at the tip of the predicted VS1 loop (Fig. 2).
In conclusion, the identification of epitopes that induce protective immunity as presented in this study will prove useful for the development of a chlamydial antiabortion vaccine.
Nucleotide sequence accession number.
The complete coding sequence of the LLG strain MOMP has been deposited in the GenBank DNA database and assigned accession number AF272945.
Acknowledgments
We thank A. A. Andersen (Agricultural Research Service, Ames, Iowa) for the gift of MAbs A11 and F3 and H. Kennedy (Queens University, Belfast, United Kingdom) for MAb 1B8.
This work was supported by EU contract CT93-0957 (AIR-3).
REFERENCES
- 1.Andersen A A, Van Deusen R A. Production and partial characterization of monoclonal antibodies to four Chlamydia psittaci isolates. Infect Immun. 1988;56:2075–2079. doi: 10.1128/iai.56.8.2075-2079.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando S, Takashima I, Hashimoto N. Neutralization of Chlamydia psittaci with monoclonal antibodies. Microbiol Immunol. 1993;37:753–758. doi: 10.1111/j.1348-0421.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 3.Buxton D. Potential danger to pregnant women of Chlamydia psittaci from sheep. Vet Rec. 1986;118:510–511. doi: 10.1136/vr.118.18.510. [DOI] [PubMed] [Google Scholar]
- 4.Buzoni-Gatel D, Bernard F, Andersen A A, Rodolakis A. Protective effect of polyclonal and monoclonal antibodies against abortion in mice infected by Chlamydia psittaci. Vaccine. 1990;8:342–346. doi: 10.1016/0264-410x(90)90092-z. [DOI] [PubMed] [Google Scholar]
- 5.Byrne G I, Stephens R S, Ada G, Caldwell H D, Su H, Morrison R P, Van der Pol B, Bavoil P M, Bobo L, Everson S, Ho Y, Hsia R-C, Kennedy K, Kuo C-C, Montgomery P C, Peterson E, Swanson A, Whitaker C, Whittum-Hudson J, Yang C L, Zhang Y-X, Zhong G M. Workshop on in vitro neutralization of Chlamydia trachomatis: summary of proceedings. J Infect Dis. 1993;168:415–420. doi: 10.1093/infdis/168.2.415. [DOI] [PubMed] [Google Scholar]
- 6.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 7.De Sa C, Souriau A, Bernard F, Salinas J, Rodolakis A. An oligomer of the major outer membrane protein of Chlamydia psittaci is recognized by monoclonal antibodies which protect mice from abortion. Infect Immun. 1995;63:4912–4916. doi: 10.1128/iai.63.12.4912-4916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieterichs K, Freigang J, Umhau S, Breed J. Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herring A J, Tan T W, Baxter S, Inglis N F, Dunbar S. Sequence analysis of the major outer membrane protein gene of an ovine abortion strain of Chlamydia psittaci. FEMS Microbiol Lett. 1989;65:153–158. doi: 10.1016/0378-1097(89)90383-2. [DOI] [PubMed] [Google Scholar]
- 10.Kaltenboeck B, Kousoulas K G, Storz J. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J Bacteriol. 1993;175:487–502. doi: 10.1128/jb.175.2.487-502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCafferty M C, Herring A J, Andersen A A, Jones G E. Electrophoretic analysis of the major outer membrane protein of Chlamydia psittaci reveals multimers which are recognized by protective monoclonal antibodies. Infect Immun. 1995;63:2387–2389. doi: 10.1128/iai.63.6.2387-2389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClenaghan M, Herring A J, Aitken L D. Comparison of Chlamydia psittaci isolates by DNA restriction endonuclease analysis. Infect Immun. 1984;45:384–389. doi: 10.1128/iai.45.2.384-389.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodolakis A, Salinas J, Papp J R. Recent advances on ovine chlamydial abortion. Vet Res. 1998;29:275–288. [PubMed] [Google Scholar]
- 14.Salti-Montesanto V, Tsoli E, Papavassiliou P, Psarrou E, Markey B, Jones G E, Vretou E. Diagnosis of ovine enzootic abortion, using a competitive ELISA based on monoclonal antibodies against variable segments 1 and 2 of the major outer membrane protein of Chlamydia psittaci serotype 1. Am J Vet Res. 1997;58:228–235. [PubMed] [Google Scholar]
- 15.Tan T W, Herring A J, Anderson I E, Jones G E. Protection of sheep against Chlamydia psittaci infection with a subcellular vaccine containing the major outer membrane protein. Infect Immun. 1990;58:3101–3108. doi: 10.1128/iai.58.9.3101-3108.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vretou E, Goswami P C, Bose S K. Adherence of multiple serovars of Chlamydia trachomatis is mediated by thermolabile protein(s) to a common receptor on HeLa and McCoy cells. J Gen Microbiol. 1989;135:3229–3237. doi: 10.1099/00221287-135-12-3229. [DOI] [PubMed] [Google Scholar]
- 17.Vretou E, Loutrari E, Mariani L, Costelidou K, Eliades P, Conidou G, Karamanou S, Mangana O, Siarkou V, Papadopoulos O. Diversity among abortion strains of Chlamydia psittaci demonstrated by inclusion morphology, polypeptide profiles and monoclonal antibodies. Vet Microbiol. 1996;51:275–289. doi: 10.1016/0378-1135(96)00048-x. [DOI] [PubMed] [Google Scholar]
- 18.Wyllie S, Ashley R H, Longbottom D, Herring A J. The major outer membrane protein of Chlamydia psittaci functions as a porin-like ion channel. Infect Immun. 1998;66:5202–5207. doi: 10.1128/iai.66.11.5202-5207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie S, Longbottom D, Herring A J, Ashley R H. Single channel analysis of recombinant major outer membrane protein porins from Chlamydia psittaci and Chlamydia pneumoniae. FEBS Lett. 1999;445:192–196. doi: 10.1016/s0014-5793(99)00121-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y-X, Stewart S, Joseph T, Taylor H R, Caldwell H D. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J Immunol. 1987;138:575–581. [PubMed] [Google Scholar]