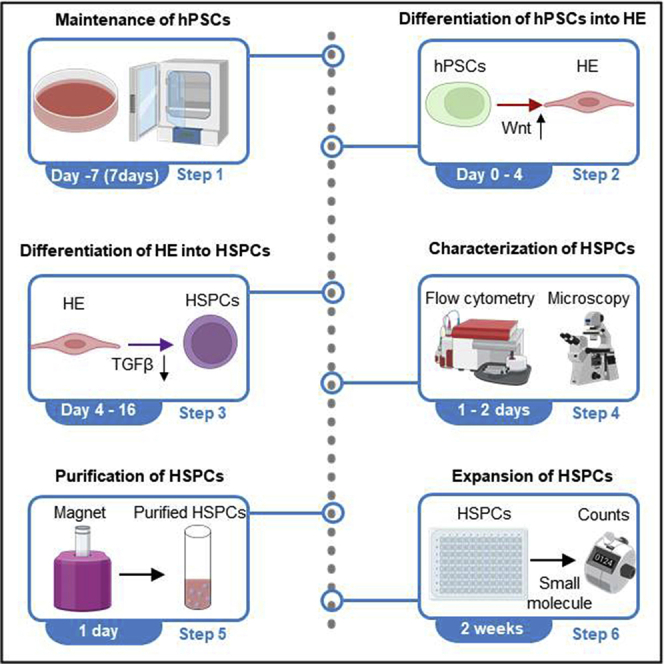
Summary
Here, we present a protocol to efficiently direct human pluripotent stem cells (hPSCs) into hematopoietic stem and progenitor cells (HSPCs) under a chemically defined, albumin-free system. We describe the induction of aorta-gonad-mesonephros-like hematopoiesis from hPSCs into SOX17+ hemogenic endothelium and then into CD34+CD45+ HSPCs via application of Wnt activator and TGFβ inhibitor, respectively. The generated HSPCs, characterized by flow cytometry and colony-forming unit assay, express definitive hematopoiesis markers and exhibit multilineage differentiation potential and the capacity to expand.
For complete details on the use and execution of this protocol, please refer to Chang et al. (2022a, 2022b).1,2
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Stem Cells, Cell Differentiation, Biotechnology and bioengineering
Graphical abstract
Highlights
-
•
A chemically defined protocol for generating homogenous definitive hematopoietic cells
-
•
In vitro hematopoietic differentiation mimics aorta-gonad-mesonephros hematopoiesis
-
•
Application of Wnt activator and TGFβ inhibitor to promote hPSC-HE-HSPC hematopoiesis
-
•
Resulting HSPCs exhibit multilineage differentiation potential and expansion capacity
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol to efficiently direct human pluripotent stem cells (hPSCs) into hematopoietic stem and progenitor cells (HSPCs) under a chemically defined, albumin-free system. We describe the induction of aorta-gonad-mesonephros-like hematopoiesis from hPSCs into SOX17+ hemogenic endothelium and then into CD34+CD45+ HSPCs via application of Wnt activator and TGFβ inhibitor, respectively. The generated HSPCs, characterized by flow cytometry and colony-forming unit assay, express definitive hematopoiesis markers and exhibit multilineage differentiation potential and the capacity to expand.
Before you begin
Institutional permissions
The human embryonic stem cell lines used in this study are approved for research use by Purdue University Institutional Biosafety Committee and are eligible for use in National Institutes of Health (NIH) funded research. Others who wish to replicate this protocol will need approval from their respective funding agencies and/or institution committees.
Preparations
Timing: 2 days to 3 weeks
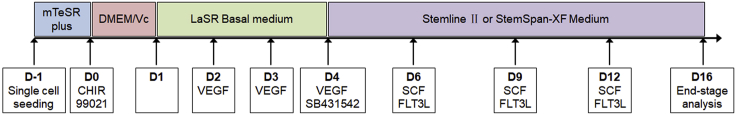
A detailed schematic of human pluripotent stem cell (hPSC) differentiation through hemogenic endothelium (HE) to hematopoietic stem and progenitor cells (HSPCs) is shown in Figure 1. For additional details on hPSC differentiation into hemogenic endothelium (Figures 2A and 2B) and HSPCs (Figures 3A–3C), please refer to Chang et al.1,2 We have successfully performed HSPC differentiation in 10 hPSC lines, including H1, H9, and H13 human embryonic stem cell (hESC) lines, as well as 6-9-9 and 19-9-11 human induced pluripotent stem cell (hiPSC) lines. We expect similar differentiation results in other hPSC lines. To perform the differentiation processes, please follow these steps:
-
1.
Obtain all key reagents mentioned in the key resources table.
-
2.
Prepare all stock solutions listed below and working aliquots.
-
3.
Order antibodies listed in the key resources table to assess differentiation efficiency.
-
4.
Prepare media following the steps below when needed. Media can be stored at 4°C for up to 2 weeks.
-
5.
Obtain/thaw hPSCs and maintain as undifferentiated stem cell cultures in mTeSR1, mTeSR Plus or other stem cell culture media following the protocol below. hPSCs need to be passaged at least twice (more than 1 week in culture) before starting the differentiation.
Figure 1.
Schematic illustration of definitive hematopoietic stem and progenitor cell differentiation protocol from human pluripotent stem cells
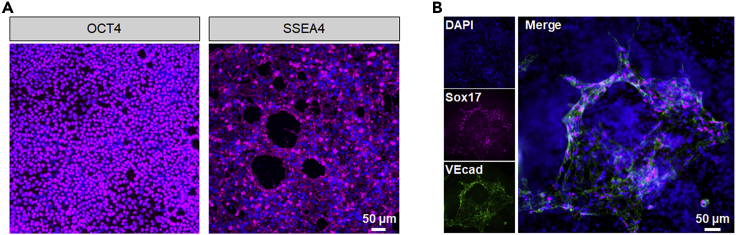
Figure 2.
Characterization of hPSC and hPSC-derived endothelial progenitor cells
(A) Immunostaining analysis for OTC4 and SSEA4 expression on human pluripotent stem cells.
(B) Immunostaining analysis for Sox17 and VE-cadherin (VEcad) expression on hPSC-derived endothelial progenitors (EPs) on day 6.
Scale bars, 50 μm.
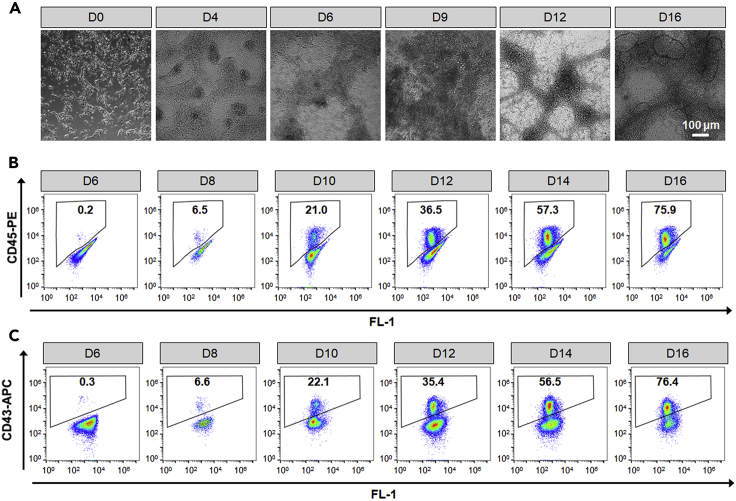
Figure 3.
Characterization of hPSC-derived hematopoietic stem and progenitor cells
(A) Brightfield images of differentiated hematopoietic cells at the indicated days. Scale bars, 100 μm.
(B and C) Representative flow cytometry plots of CD45 (B) and CD43 (C) expression at the indicated days.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45-PE (1: 50) | BD Biosciences | Cat#555483; RRID: AB_395875 |
| CD45-APC (1: 100) | BD Biosciences | Cat#555485; RRID: AB_398600 |
| CD34-APC (1: 100) | BD Biosciences | Cat#555824; RRID: AB_398614 |
| CD45-FITC (1: 100) | BioLegend | Cat#304006; RRID: AB_314394 |
| CD44-APC (1: 50) | BD Biosciences | Cat#559942; RRID: AB_398683 |
| SOX17-APC (1: 50) | R&D Systems | Cat#IC1924A; RRID: AB_1964715 |
| VE-cadherin (1: 100) | Santa Cruz | Cat#sc9989; RRID: AB_2077957 |
| SSEA-4 Ms IgG3 (1: 100) | Santa Cruz | Cat#sc-21704; RRID: AB_628289 |
| OCT-3/4 Ms IgG2b (1: 100) | Santa Cruz | Cat#sc-5279; RRID: AB_628051 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | Cat#354277 |
| iMatrix-511 | Iwai North America Inc | Cat#N-892021 |
| mTeSR™ plus | STEMCELL Technologies | Cat#100-0276 |
| MethoCult™ H4434 | STEMCELL Technologies | Cat#04434 |
| Stemline II medium | Sigma-Aldrich | Cat#S0192 |
| EDTA | Thermo Fisher Scientific | Cat#15575020 |
| Versene solution | Thermo Fisher Scientific | Cat#15040066 |
| Y-27632 | Cayman Chemical | Cat#10005583 |
| CHIR99021 | Cayman Chemical | Cat#13122 |
| DMEM | Thermo Fisher Scientific | Cat#11965118 |
| DMEM/F12 | Thermo Fisher Scientific | Cat#11320033 |
| Ascorbic acid (L-ascorbic acid 2- phosphate) | Sigma-Aldrich | Cat#A8960 |
| Bovine serum albumin | Sigma-Aldrich | Cat#A1923 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050079 |
| Human VEGF | PEPROTECH | Cat#100-20 |
| Human SCF | PEPROTECH | Cat#300-07 |
| Human Flt3-ligand | PEPROTECH | Cat#300-19 |
| SB-431542 | Cayman Chemical | Cat#13031 |
| 16% Formaldehyde | Thermo Fisher Scientific | Cat#28906 |
| Triton X-100 | Thermo Fisher Scientific | Cat#A16046.AE |
| Blotting-grade blocker (dry milk) | Bio-Rad Laboratories, Inc | Cat#1706404 |
| Hoechst 33342 | Invitrogen | Cat#H3570 |
| Accumax™ | Innovative Cell Technologies, Inc | Cat#AM-105 |
| Dulbecco’s phosphate-buffered saline | Sigma-Aldrich | Cat#D8537 |
| BIO | Cayman Chemical | Cat#13123 |
| UM171 | STEMCELL Technologies | Cat#72914 |
| SR-1 (StemRegenin 1) | Cayman Chemical | Cat#14268 |
| TEPA | Cayman Chemical | Cat#23745 |
| Accutase | Innovative Cell Technologies, Inc | Cat# AT104-500 |
| GW9962 | Cayman Chemical | Cat#70785 |
| Experimental models: Cell lines | ||
| Human embryonic stem cell: H9 | WiCell | RRID: CVCL_9773 |
| Human embryonic stem cell: H9 RUNX1c Reporter | Drs. Edouard Stanley & Andrew Elefanty at the Murdoch Children’s Research Institute | N/A |
| Human induced pluripotent stem cell: 19-9-11 | WiCell | RRID: CVCL_K054 |
| Software and algorithms | ||
| FlowJo | BD Biosciences | http://www.flowjo.com/ |
| ImageJ | Schindelin et al.3 | https://imagej.nih.gov/ij/ |
| Other | ||
| EasySep Human FITC Positive Selection Kit II | STEMCELL Technologies | Cat#17662 |
| EasySep Magnet | STEMCELL Technologies | Cat#18000 |
| 5 mL Round-bottom tube with a cell-strainer cap | Falcon | Cat#352235 |
| 15 mL Centrifuge tube | Falcon | Cat#14-959-53A |
| 50 mL Centrifuge tube | Falcon | Cat#14-432-22 |
| Corning tissue culture plates (6 well) | Corning | Cat#3516 |
| Corning tissue culture plates (12 well) | Corning | Cat#3513 |
| Corning tissue culture plates (24 well) | Corning | Cat#3526 |
| Corning tissue culture plates (96 well) | Corning | Cat#3596 |
| Sterile biosafety cabinets | Thermo Fisher Scientific | Cat#1323TS |
| Humidified tissue culture incubator (37°C, 5% CO2) | Thermo Fisher Scientific | Cat#51026282 |
| Serological pipettes 5 mL | Thermo Fisher Scientific | Cat#13-678-11D |
| Leica DMi8 microscope | Leica Microsystems | N/A |
| Serological pipettes 10 mL | Thermo Fisher Scientific | Cat#13-678-11E |
| BD Accuri C6 Plus Cytometer | BD Biosciences | N/A |
Materials and equipment
We recommend that cell culture performed under a sterile environment without the use of antibiotics. Aseptic technique should be sufficient to maintain sterile conditions, and STEMCELL Technologies, producer of mTeSR1 or mTesR plus stem cell maintenance medium, recommends antibiotic-free culture of undifferentiated human pluripotent stem cells. We suggest to purchase sterile cell culture materials such as sterile centrifuge tubes, microcentrifuge tubes, serological pipettes, and DNase/RNase free filter pipette tips. Routine spraying of gloved hands with 70% ethanol will effectively reduce the chance of contamination.
Media should be prepared under sterile and endotoxin-free conditions or can be sterilized using a 0.22 μm filter prior to use. We recommend that media should be warmed at room temperature or in a water bath at 37°C on the day of use for at least 30 min.
For dissociating cells, Accutase solution should be warmed in a 37°C water bath for 10–30 min prior to use, and Versene could be directly used.
Y-27632 ROCK inhibitor (5 mM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Y-27632 | 5 mM | 10 mg |
| DPBS | N/A | 6.24 mL |
| Total | N/A | 6.24 mL |
Aliquot and store at −20°C up to 1 year. Keep a single working aliquot at 4°C up to a month. Avoid freeze-thaw cycles.
CHIR99021 solution (36 mM)
| Reagent | Final concentration | Amount |
|---|---|---|
| CHIR99021 | 36 mM | 25 mg |
| DMSO | N/A | 1.49 mL |
| Total | N/A | 1.49 mL |
Aliquot and store at −20°C for up to 3 months. Keep a single working aliquot at 4°C for up to 2 weeks. Avoid freeze-thaw cycles.
VEGF solution (50 μg/mL)
| Reagent | Final concentration | Amount |
|---|---|---|
| VEGF | 50 μg/mL | 50 μg |
| DPBS | N/A | 1 mL |
| Total | N/A | 1 mL |
Aliquot and store at −20°C for up to 3 months. Keep a single aliquot at 4°C for up to 1 week. Avoid freeze-thaw cycles.
SB431542 (10 mM)
| Reagent | Final concentration | Amount |
|---|---|---|
| SB431542 | 10 mM | 5 mg |
| DMSO | N/A | 1.3 mL |
| Total | N/A | 1.3 mL |
Aliquot and store at −20°C for up to 3 months. Keep a single aliquot at 4°C for up to 1 month. Avoid freeze-thaw cycles.
SCF (50 μg/mL)
| Reagent | Final concentration | Amount |
|---|---|---|
| SCF | 50 μg/mL | 50 μg |
| DPBS | N/A | 1 mL |
| Total | N/A | 1 mL |
Aliquot and store at −20°C for up to 3 months. Keep a single aliquot at 4°C for up to 1 week. Avoid freeze-thaw cycles.
FLT3L (50 μg /mL)
| Reagent | Final concentration | Amount |
|---|---|---|
| FLT3L | 50 μg /mL | 50 μg |
| DPBS | N/A | 1 mL |
| Total | N/A | 1 mL |
Aliquot and store at −20°C for up to 3 months. Keep a single aliquot at 4°C for up to 1 week. Avoid freeze-thaw cycles.
Dry milk solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Non-fat dry milk | 5% (wt/vol) | 0.5 g |
| 1% Triton X-100 | 0.4% (vol/vol) | 4 mL |
| DPBS | N/A | 6 mL |
| Total | N/A | 10 mL |
We do not recommend storing this solution.
mTeSR plus Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Basal medium | N/A | 400 mL |
| 5× supplement provided from mTeSR plus complete kit | 1 × | 100 mL |
| Total | N/A | 500 mL |
Store this solution at 4°C for 2 weeks and −20°C for 3 months.
LaSR Basal Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Advanced DMEM/F-12 | N/A | 500 mL |
| GlutaMAX | 1.23% (vol/vol) | 6.25 mL |
| L-ascorbic acid 2-phosphate | 0.006% (wt/vol) | 0.03 g |
| Total | N/A | 506.25 mL |
Store this solution at 4°C for 1 month and −20°C for 3 months.
DMEM/Vc medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 500 mL |
| L-ascorbic acid 2-phosphate | 0.01% (wt/vol) | 50 mg |
| Total | N/A | 500 mL |
Store this solution at 4°C for 1 month and −20°C for 3 months.
Flow buffer-1
| Reagent | Final concentration | Amount |
|---|---|---|
| Bovine serum albumin | 0.5% (wt/vol) | 2.5 g |
| DPBS | N/A | 500 mL |
| Total | N/A | 500 mL |
Store at 4°C up to 3 months. Aliquot Flow buffer-1 solution could prevent the contamination from frequent use.
Flow buffer-2
| Reagent | Final concentration | Amount |
|---|---|---|
| Bovine serum albumin | 0.5% (wt/vol) | 2.5 g |
| DPBS | N/A | 500 mL |
| 1% Triton X-100 | 0.09% (vol/vol) | 50 mL |
| Total | N/A | 550 mL |
Store at 4°C up to 6 months. Aliquot Flow buffer-2 solution could prevent the contamination from frequent use.
Step-by-step method details
Preparing Matrigel-coated plates
Timing: 1.5 h
These steps describe preparation of tissue culture plates for cell culture.4
-
1.Preparing Matrigel-coated plates.
-
a.Transfer 32 mL of ice-cold DMEM/F-12 medium into a 50 mL conical tube.
-
b.Prepare cell culture plates for coating. One aliquot of Matrigel solution (2.5 mg, approximately 0.25 mL) is enough for coating 5 plates.
-
c.Place the Matrigel on the ice after removing from the −80°C freezer.
-
d.Matrigel was transferred into DMEM/F-12 medium using a P1000 pipette. Gently mix the Matrigel and DMEM/F-12 medium well using a 10 mL pipette.
-
e.Add 1 mL of Matrigel solution to each well of 6 well plate, 0.5 mL of Matrigel to each well of 12 well plate, and 0.25 mL of Matrigel solution to each well of 24 well plate using a P1000 pipette. Gently shake the plates (3 quick, short, side-to-side, and back-and-forth motions) to make sure the Matrigel solution fully cover the surface of the well.
-
f.Place the coated plates in a humidified incubator at 37°C for at least 30 min before cell culture.
-
g.Add extra DMEM/F-12 medium (0.5–1 mL) to prevent drying out 2–3 days after coating.
-
a.
Note: 1–2 mL of DMEM/F-12 medium is suggested to refill the well every 3–7 days. Other coating substrates, such as Synthemax II-SC and Cultrex, can also be used for routine hPSC maintenance.
CRITICAL: Matrigel-coated plates can be stored in the incubator (humidified, 5% CO2, 37°C), and these coated plates should be used within 12 days. For long-term storage, Matrigel-coated plates should be sealed by parafilm and stored at 4°C. If there are some dry spots in the coated plates due to DMEM/F-12 evaporation, they cannot be used for stem cell culture.
Thawing frozen human pluripotent stem cells
Timing: 1 h
These steps describe thawing of human pluripotent stem cell lines to recover healthy cells suitable for hematopoietic stem and progenitor cell differentiation.
-
2.Thawing of human pluripotent stem cells.
-
a.Warm mTeSR Plus medium at room temperature for approximately 30 min.
-
b.Transfer 5 mL of mTeSR Plus to a 15 mL conical tube using a 10 mL serological pipette.
-
c.Remove a frozen cell cryovial of human pluripotent stem cells (passage number: 10–20) from a liquid nitrogen tank.
-
i.Immerse the cryovial in a 37°C water bath without submerging the cap.
-
ii.Gently swirl the cryovial for 2–3 min until the contents are completely thawed.
-
i.
-
d.Spray the cryovial with 70% (vol/vol) ethanol before placing it into a biosafety tissue culture cabinet. Gently transfer cell mixture from the cryovial to a sterile 15 mL conical tube containing 5 mL of prewarmed mTeSR Plus medium using a P1000 pipette.Note: Over-heating the cryovial in the water bath would reduce cell viability.
-
e.Centrifuge the cell mixture at 200 × g for 5 min at room temperature.
-
f.Aspirate the supernatant from the 15-mL tube via vacuum aspiration and remove the Matrigel solution from the coated plate by a P1000 pipette or vacuum aspiration.
-
g.Gently resuspend the cell pellet in 2 mL of mTeSR Plus medium containing 5 μM Y27632 by a P1000 pipette, and transfer them into an empty Matrigel-coated well using the P1000 pipette.
-
h.Clearly label the plates with hPSC line, passage number, data frozen, data thawed, and initials.
-
i.Return the plate to the incubator (humidified, 5% CO2, 37°C) and gently shake the plate back and forth for a few times to evenly distribute the cells.
 CRITICAL: Including ROCK inhibitors (Y27632) is essential for efficient attachment of human pluripotent stem cells after thawing.
CRITICAL: Including ROCK inhibitors (Y27632) is essential for efficient attachment of human pluripotent stem cells after thawing.
-
a.
Maintenance of human pluripotent stem cells
Timing: 45 min
These steps describe daily maintenance and passage of human pluripotent stem cells. Human pluripotent stem cells split at a 1:6 ratio should be passaged at 70%–90% confluency approximately every 3–4 days. If split at a 1:12 ratio, these human pluripotent stem cells should be passaged approximately every 4–5 days.5
-
3.Daily maintenance of human pluripotent stem cells.
-
a.Warm mTeSR Plus medium at room temperature for approximately 30 min.
-
b.Aspirate the old medium from each well of hPSC-containing 6-well plate and replace with 2 mL of fresh prewarmed mTeSR Plus medium. Repeat this step daily until cells are ready for passaging (∼80%–90% confluency).
-
a.
-
4.Passaging human pluripotent stem cells using Versene.
-
a.Warm mTeSR Plus medium at room temperature for approximately 30 min.
-
b.Aspirate the old medium from each well of hPSC-containing 6-well plate and add 1 mL of room temperature Versene.
-
c.Incubate the plate at an incubator (humidified, 5% CO2, 37°C) for 5 min.Note: Versene incubation time may vary for different hPSC lines. If cells do not detach easily from the bottom after incubation for 5 min, leave the plates longer (e.g., 7–10 min) in the incubator. If cells detach easily from the bottom, incubate the plate for less than 5 min (e.g., 3 or 4 min).
-
d.During Versene incubation, add 2 mL of mTeSR Plus medium with 5 μM Y27632 into a new Matrigel-coated well after aspirating Matrigel solution.
-
e.Aspirate the Versene from the hPSC-containing plate.
-
f.Using a P1000 pipette, collect 1 mL of medium in step 4d and dispense this medium over the surface of the hPSC-containing well treated with Versene until all the colonies are detached (∼3 or 4 times) under a microscope with a 10× objective.
-
i.After most cells (over 90% cells) are detached, gently pipette to mix the cell mixture using a P1000 pipette.
-
ii.Transfer ∼160 μL (at a ratio of 1:6) to the new Matrigel-coated well in step 4d.
 CRITICAL: Human pluripotent stem cells should be gently passaged to reduce and avoid spontaneous differentiation. The split ratio is variable, and generally a ratio between 1:6 and 1:12 (or 20,000–40,000 cells per cm2) is appropriate when using Versene for passaging.
CRITICAL: Human pluripotent stem cells should be gently passaged to reduce and avoid spontaneous differentiation. The split ratio is variable, and generally a ratio between 1:6 and 1:12 (or 20,000–40,000 cells per cm2) is appropriate when using Versene for passaging.
-
i.
-
g.Return the plate to the incubator after plating the cells. Gently shake the plate in 3 quick, short, back- and front- and side-to-side motions to evenly disperse the cell across the surface of the wells.
-
a.
Directed differentiation of human pluripotent stem cells to definitive hematopoietic stem and progenitor cells
Timing: 18 days
These steps describe the directed differentiation of human pluripotent stem cells to definitive hematopoietic stem and progenitor cells. For the background on the development of this protocol and differentiation to hematopoietic progenitor cells, please see.1,2 In this part of the protocol, we will describe differentiation of human pluripotent stem cells into hematopoietic cells in a full 24-well plate, which requires approximately 2.4 million human pluripotent stem cells or one well of 6-well plate with confluent cells. We denote Day 0 as the day when we treat cells with CHIR99021 to initiate the differentiation.
-
5.Day -1.
-
a.Warm mTeSR Plus medium at room temperature for approximately 30 min.
-
b.Aspirate the old medium from stem cell culture plate via vacuum aspiration, and add 1 mL of Accutase to the same well using a P1000 pipette.
-
c.Incubate the plate at the incubator (37°C, 5% CO2, humidified) for 5 min.
-
d.Gently pipette to dissociate all the cells from surface with a P1000 pipette, manually count the cells in a cell hemocytometer, and use a P1000 pipette to transfer 5 × 107 cells (about 1/3 of cell suspension) to a 15-mL conical tube containing 2 mL of mTeSR Plus as a quenching medium.
 CRITICAL: Initial cell seeding density is critical for efficient hematopoietic stem and progenitor cell differentiation. We suggest a starting cell split ratio of 1:12 or 5× 105 cells/cm2. The cells may need to expand for 1 or 2 more days in mTeSR plus before CHIR99021 treatment if attached cell density (below 1 × 105 cells/cm2) on Day 0 was too low to initiate the differentiation.
CRITICAL: Initial cell seeding density is critical for efficient hematopoietic stem and progenitor cell differentiation. We suggest a starting cell split ratio of 1:12 or 5× 105 cells/cm2. The cells may need to expand for 1 or 2 more days in mTeSR plus before CHIR99021 treatment if attached cell density (below 1 × 105 cells/cm2) on Day 0 was too low to initiate the differentiation. -
e.Centrifuge the cells at 200 × g for 5 min at room temperature.
-
f.After centrifugation, the cells were resuspended in 12 mL of mTeSR Plus medium containing 5 μM Y27632 (ROCK inhibitor) and 0.5 μg/mL iMatrix-511 using a 10 mL serological pipette.Note: The concentration of iMatrix-511 is critical for cell attachment. We recommend using 0.5–1.5 μg/mL iMatrix-511.
-
g.Gently mix the cell mixture and transfer 0.5 mL into each well of a 24-well plate using a P1000 pipette or 5 mL serological pipette.
-
h.Place the plate with cells to an incubator (37°C, 5% CO2, humidified) and gently shake the plate back and forth for 2–3 times to evenly distribute the cells.
-
a.
-
6.Day 0.
-
a.Warm DMEM/Vc medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of DMEM/Vc medium containing 6 μM CHIR99021.
-
c.Aspirate the old mTeSR plus medium via vacuum aspiration, transfer 0.5 mL of DMEM/Vc with CHIR99021 in each well of the 24-well plate using a 5 mL serological pipette, and record the time.
 CRITICAL: Recording the time of adding CHIR99021 is important as CHIR99021 treatment should be around 24 h for the cell lines we tested. 2-day CHIR99021 treatment in LaSR basal medium also works effectively.5 Optimization of CHIR99021 treatment may be needed for a different cell line.
CRITICAL: Recording the time of adding CHIR99021 is important as CHIR99021 treatment should be around 24 h for the cell lines we tested. 2-day CHIR99021 treatment in LaSR basal medium also works effectively.5 Optimization of CHIR99021 treatment may be needed for a different cell line. -
d.Return the plate to the incubator (37°C, 5% CO2, humidified).
-
a.
-
7.Day 1.
-
a.Warm LaSR Basal medium in a 37°C water bath for approximately 30 min.
-
b.Aspirate the old DMEM/Vc medium via vacuum aspiration, and transfer 0.5 mL LaSR Basal medium in each well of the plate using a 5 mL serological pipette.
-
c.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
8.Day 2.
-
a.Warm LaSR Basal medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of LaSR Basal medium containing 50 ng/mL VEGF.Note: While VEGF is not required for male hPSC lines,6 the addition of exogenous VEGF may reduce differentiation variability resulting from different initial cell seeding densities.
-
c.Aspirate the old medium via vacuum aspiration, and transfer 0.5 mL of the prepared medium in each well of the plate using a 5 mL serological pipette.
-
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
9.Day 3.
-
a.Warm LaSR Basal medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of LaSR Basal medium containing 50 ng/mL VEGF.
-
c.Aspirate the old medium via vacuum aspiration, and transfer 0.5 mL of the prepared medium in each well of the plate using a 5 mL serological pipette.
-
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
10.Day 4.
-
a.Warm Stemline II medium in a 37°C water bath for approximately 30 min.Note: While we identified Stemline II as the optimal medium (about 80% HSPCs at day 15), StemSpan-XF also works effectively (about 70% HSPCs at day 15).
-
b.Prepare a conical tube with 12 mL of Stemline II medium containing 50 ng/mL VEGF and 10 μM SB431542.
-
c.Aspirate the old medium via vacuum aspiration, and transfer 0.5 mL of the prepared medium in each well of the plate using a 5 mL serological pipette.
-
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
11.Day 6.
-
a.Warm Stemline II medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of Stemline II medium containing 50 ng/mL SCF and 50 ng/mL FLT3L.
-
c.Aspirate the old medium via vacuum aspiration, and transfer 0.5 mL of the prepared medium in each well of the plate using a 5 mL serological pipette.
-
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
12.Day 9.
-
a.Warm Stemline II medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of Stemline II medium containing 50 ng/mL SCF and 50 ng/mL FLT3L.
-
c.Aspirate top half of the old medium using a P1000 pipette, and transfer 0.5 mL of the prepared medium in each well of the plate using a P1000 pipette.
 CRITICAL: Floating hematopoietic cells start to produce from day 9, and be careful when aspirating top half of the old medium. If many floating cells present in top half medium due to unexpected plate shaking during medium change, 0.5–1 mL fresh medium could be directly added to the differentiated wells without removing old medium.
CRITICAL: Floating hematopoietic cells start to produce from day 9, and be careful when aspirating top half of the old medium. If many floating cells present in top half medium due to unexpected plate shaking during medium change, 0.5–1 mL fresh medium could be directly added to the differentiated wells without removing old medium. -
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
a.
-
13.Day 12.
-
a.Warm Stemline II medium in a 37°C water bath for approximately 30 min.
-
b.Prepare a conical tube with 12 mL of Stemline II medium containing 50 ng/mL SCF and 50 ng/mL FLT3L.
-
c.Aspirate top half of the old medium using a P1000 pipette, and transfer 0.5 mL of the prepared medium in each well of the plate using a 5 mL serological pipette.
-
d.Return the plate to the cell culture incubator (37°C, 5% CO2, humidified).
-
e.Perform half-medium change every three days until day 18 or functional analysis (Figure 3A).
-
a.
Characterization of differentiated hematopoietic stem and progenitor cells
Timing: 2 days (hand-on time: 1 h on first day and 30 min on second day, and additional time (1 h/plate) is needed for imaging) (for step 14)
Timing: 1.5 h (for step 15)
These steps describe immunostaining and flow cytometry analysis on the differentiated cells.
-
14.Immunostaining analysis.Note: We recommend staining 3 wells of a 24-well plate for each condition.
-
a.Aspirate the old medium from the differentiated well using a P1000 pipette. Gently wash the cells once with PBS solution and fix the cells with 0.2 mL of 4% PFA solution at room temperature for 15 min.
-
b.Aspirate PFA solution from the wells with a P1000 pipette and use 0.5 mL PBS to gently wash fixed cells twice. Fixed cells could be stored in PBS at 4°C for up to 2 weeks.
-
c.Add 200 μL blocking solution into each well using a P1000 pipette and incubated for 1 h at room temperature or 8–24 h at 4°C. PBS solution with 0.1% (vol/vol) Triton X-100 and 0.5% (wt/vol) dry milk or BSA could be used as blocking solution.
-
d.Dilute primary antibody at a ratio 1:50 to 1:100 in blocking solution.
-
e.Remove the blocking solution from the wells using a P1000 pipette and add 200 μL diluted antibody solution to each well. Incubate the cell plate at room temperature for 1 h or 4°C for 8–24 h in dark.Note: The use of conjugated antibodies can shorten staining time compared with the 2-step primary and secondary antibody staining.
-
f.After incubation, aspirate the primary antibody solution using a P1000 pipette and gently wash the cells with PBS solution twice.
-
g.If non-conjugated antibody is used, dilute appropriate secondary antibody in blocking solution at a ratio of 1:500 to 1:1,000 according the manufacturer’s instruction.
-
i.Add 200 μL diluted secondary antibody solution into the wells using a P1000 pipette.
-
ii.Incubate at room temperature in dark for 0.5 h or 4°C for 8–24 h.
-
i.
-
h.Aspirate the secondary antibody solution with a P1000 pipette, and use PBS to gently wash the cells twice.
-
i.Add 200 μL of diluted 1 μg/mL Hoechst or DAPI solution to each well for nuclear staining and incubate the plate in the dark at room temperature for 5 min.Note: The incubation time or concentration of Hoechst or DAPI can be slightly increased for a stronger signal.
-
j.Aspirate the nuclear staining solution, and gently use PBS to wash twice. Use a P1000 pipette to add 500 μL of PBS solution to each well to avoid drying out.
-
k.Image stained cells in a fluorescence microscope by selecting desired emission filter (e.g., FITC, DAPI, and CY7) and objective lens (e.g., 10× and 20×). Take and save the images after focusing, locating and centering the specimen.Note: We suggest imaging at a 10× or 20× magnification in a Leica DMi8 microscope according to the manufacturer’s instruction.
-
l.Process the fluorescent images using Fiji/ImageJ or a similar software according to the manufacturer’s instruction.
-
i.Brightness/Contrast adjustment is usually applied to the fluorescence images to reduce signal noise and improve intensity visualization.
-
ii.Images from different channels are processed individually and also merged together (Figures 2A and 2B).
-
i.
-
a.
-
15.Flow cytometry analysis.Note: These steps describe the use of flow cytometry to quantify hPSC differentiation efficiency by analyzing hematopoietic markers, including CD34, CD45, CD43, Runx1c and CD44. For intracellular protein detection, cells will be first fixed in 1% (wt/vol) PFA for 20 min and permeabilized in ice cold 90% (vol/vol) methanol for half an hour.7 We also suggest at least 3 samples for each condition and 0.5 to 1 million cells for each sample.
-
a.For attached cells, e.g., day 6 endothelial cells, incubate them with 0.5 mL/well Accutase for 5–10 min in 37°C.
-
i.Use a P1000 pipette to detach and singularize differentiated cells from the bottom.
-
ii.For floating hematopoietic cells after day 6, use a P1000 pipette to collect and filter through a 70 μm strainer sitting on a 15 or 50-mL conical tube.
-
i.
-
b.For Accutase-dissociated cells, equal volume of DMEM medium with 10% FBS is added for neutralizing.
-
c.Centrifuge the cells at 200 × g for 5 min at room temperature.
-
d.Aspirate the supernatant via vacuum aspiration and wash pelleted cells with Flow buffer-1 solution twice to remove dead cells.
-
e.Prepare 100 μL/sample of conjugated antibody solution at a ratio of 1:50 to 1:100 in Flow buffer-1 according to the key resources table. Incubate the cell and antibody mixture at room temperature in dark for half an hour or 4°C for 4–24 h.
-
f.Add 2 mL Flow buffer-1 solution to each tube using a 10 mL serological pipette, and centrifuge at 200 × g for 5 min at room temperature.
-
g.Aspirate the supernatant via vacuum aspiration and wash the cell pellet once with 2 mL Flow buffer-1.
-
h.Aspirate the supernatant via vacuum aspiration, use a P1000 pipette to resuspend the cell pellet with 300 μL of Flow buffer-1, and transfer the cell mixture into a 5 mL FACS round bottom tube via the cap filter for flow cytometry analysis.Note: The FACS round bottom tube with a filter lid is recommended and the cells should be filtered right before flow analysis.
-
i.Keep the prepared cells on ice until analysis. Run prepared samples in a BD Accuri C6 Plus flow cytometer according to the manufacturer’s instruction.
-
i.Once the flow machine is ready, select desired sample well, run settings (e.g., run unlimited or run with limits in events, volume or time) and fluidics (e.g., slow, medium or fast) in the instrumental panel under the Collect tab.
-
ii.By default, a forward (FSC) vs side scatter (SSC) dot plot is created for data visualization, and other plot types (e.g., density plot and histogram) may be selected with x-axis and y-axis changed to desired filter channels (e.g., FITC, PE and APC). After setting is done, load the filtered sample, start and stop data acquisition.
-
i.
-
j.Save the collected data and exported as a .fcs file for further analysis.
-
i.Open the .fcs file in the FlowJo or a similar software (e.g., Accuri C6 itself can be used to visualize and analyze the data) to visualize and analyze collected flow data.
-
ii.Under the FSC vs SSC plot, gate the live cell population and apply to desired channels (e.g., PE and APC in Figures 3B and 3C) for quantitative analysis of targeted cells.
-
i.
-
a.
Purification of differentiated hematopoietic stem and progenitor cells
Timing: 2.5 h
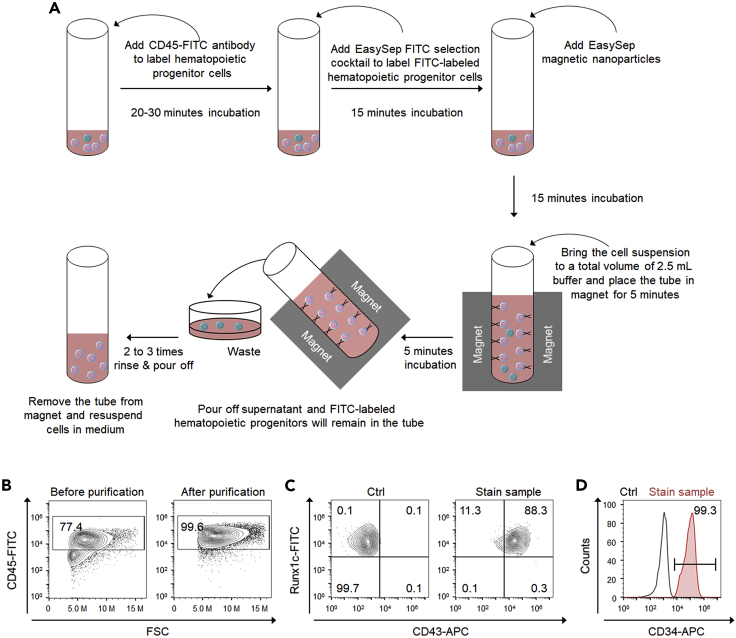
These steps describe the purification of differentiated hematopoietic stem and progenitor cells using magnetic activated cell sorting (MACS) (Figures 4A–4D).
-
16.Purification of differentiated hematopoietic stem and progenitor cells.
-
a.Pipette differentiated day 12 to day 16 hematopoietic stem and progenitor cells about 10 times using a P1000 pipette, and filter the cells through a 40 μm cell strainer into a 50 mL conical tube containing 5 mL of DMEM medium with 10% FBS.
-
b.Count the total cell number with a hemocytometer or automated cell counter, and centrifuge at 200 × g for 5 min at room temperature.
-
c.Aspirate the supernatant via vacuum aspiration and use 10 mL Flow buffer-1 solution to wash the cells.
-
d.Aspirate the supernatant after centrifuge, and resuspend the cells at a concentration of 107 cells/100 μL in Flow buffer-1 solution.
-
e.Add 2 μL CD45-FITC antibody per 100 μL cell mixture and incubate in dark at room temperature for 30 min.Note: Other FITC-conjugated antibodies, such as CD43-FITC and CD44-FITC, can also be used.
-
f.Add 2 mL Flow buffer-1 solution using a 5 mL serological pipette, centrifuge at 200 × g for 5 min at room temperature, and aspirate the supernatant via vacuum aspiration.
-
g.Resuspend the cell pellet with Flow buffer-1 at a concentration of 107 cells/100 μL and add EasySep™ FITC Positive Selection Cocktail at 10 μL per 100 μL cell solution.
-
h.Gently mix the cell and selection cocktail solution using a P1000 pipette, and incubate at room temperature for 15 min.
-
i.Add well-mixed magnetic nanoparticles at 5 μL per 100 μL cell solution, mix well and incubate at room temperature for another 10 min.
-
j.Bring the cell suspension to a total volume of 2.5 mL Flow buffer-1 solution using a 5 mL serological pipette, mix well and transfer into a 5-mL round-bottom flow tube.
-
k.Place the flow tube (without cap) into the magnet and set aside for 5 min.
-
l.Pick up the magnet, and in one continuous motion invert the magnet and flow tube, pouring off the supernatant fraction. Leave the magnet and tube inverted for 2–3 s, and then return to the upright position.
-
m.Remove the flow tube from the magnet, add 2.5 mL Flow buffer-1 solution using a 5 mL serological pipette, pipette up and down for 2–3 times to mix the cell suspension. Place the flow tube (without cap) back in the magnet and set aside for 5 min.
-
n.Repeat step l and m two to three times, and then step l once more. Remove the flow tube from the magnet and resuspend cells in appropriate amount of desired medium for subsequent culture or other applications.
-
a.
Figure 4.
Purification and analysis of hPSC-derived hematopoietic stem and progenitor cells
(A) Schematic of the protocol for the purification of hPSC-derived hematopoietic stem and progenitor cells.
(B) Representative flow cytometry plots of CD45 expression before and after purification.
(C) Representative flow cytometry plots of CD43 and RUNX1c expression on the purified definitive hematopoietic cells derived from H9 Sox17-mCherry RUNX1c-GFP dual reporter.
(D) Representative histogram plot of CD34 expression on the purified definitive hematopoietic cells.
Colony-forming unit (CFU) assay to analyze hematopoietic stem and progenitor cells
Timing: 17 days
These steps describe colony-forming unit assay for culturing and analyzing differentiated hematopoietic stem and progenitor cells (Figure 5).
-
17.Colony-forming unit (CFU) assay on the differentiated hematopoietic stem and progenitor cells.
-
a.Thaw the complete MethoCult™ medium at room temperature (15°C–25°C) for half an hour or overnight (16–24 h) at 2°C–8°C.
-
b.Prepare a 12-well plate for CFU assay by marking sample wells in the center and fill all neighboring wells with 2 mL sterile water to keep assayed wells under an appropriate moisture environment.Note: Only wells in the center of a 12-well plate are appropriate for CFU assay and sterile water should be used to fill the empty wells around the assayed wells.
-
c.Prepare the cell solution for CFU assay by diluting the cells from step 16n with Stemline II medium at a concentration of 2 × 104 cells/mL.Note: The final cell concentration suitable for CFU assay is around 2,000 cells per well of a 12-well plate.
-
d.Add 50 μL of prepared cell solution into 2 mL of thawed MethoCult™ medium.
-
e.Vortex the tube vigorously for at least 4 s to mix the content thoroughly.
-
f.Let stand for at least for 5 min to allow the bubbles to rise to the top.
-
g.Transfer cell-containing MethoCult™ medium into the prepared 12 well plate.Note: Pipette tips can be used to break the bubbles generated during medium transfer.
-
h.Distribute the medium evenly across the surface of the well and rotate the plate to allow MethoCult™ medium to attach to the wall of the well on all sides by gently shaking and rotating the plates.
-
i.Return the plate to a tissue-culture incubator (37°C, 5% CO2, humidified).
-
j.After a 16-day incubation, image and count the CFU colonies at a magnification of 10× or 20× under a microscope (Figure 5).
-
a.
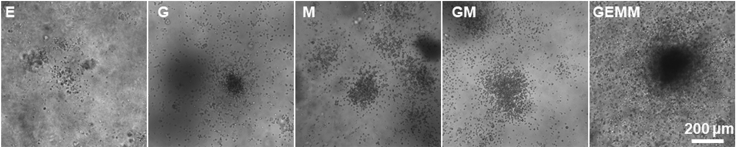
Figure 5.
hPSC-derived hematopoietic cells were assessed for myeloid potential via the colony-forming unit (CFU) assay in methylcellulose medium
After 2 weeks, the resulting hematopoietic colonies were scored for CFUs according to their morphologies: erythroid (E), granulocyte/macrophage (GM), macrophage (M), and multilineage progenitor (GEMM) colonies. Scale bars, 200 μm.
Expansion of hematopoietic stem and progenitor cells with small molecules
Timing: up to 16 days
These steps describe the expansion of differentiated hematopoietic stem and progenitor cells under a feeder-free system with various small molecules (Figures 6A and 6B).
-
18.Expansion of differentiated hematopoietic stem and progenitor cells under feeder-free conditions.
-
a.Prepare fresh culture media with Stemline II, 50 ng/mL SCF, 50 ng/mL FLT3L, and different small molecules (750 nM SR-1, 250 nM UM171, 0.5 μM BIO, 5 μM TEPA, and 1 μM GW9962).
-
b.Seed 2 × 105 hematopoietic stem and progenitor cells from step 16n per well of 24-well plate in 1 mL Stemline II medium with different small molecules.Note: For the 12-well plate, cells from step 16n should be resuspended in 2 mL culture medium at a density of 4 × 105 cells/well. StemSpan-XF medium could also be used to replace Stemline II medium.
-
c.Perform half-medium change with small molecule-containing Stemline II medium every 3 days.
-
d.After a 2-week expansion, gently pipette the cells up and down, collect all the cells and run flow cytometry analysis according to step 15 with CD34 and CD44 or other conjugated antibodies to count the total number of CD34+ HSPCs.
-
a.
Figure 6.
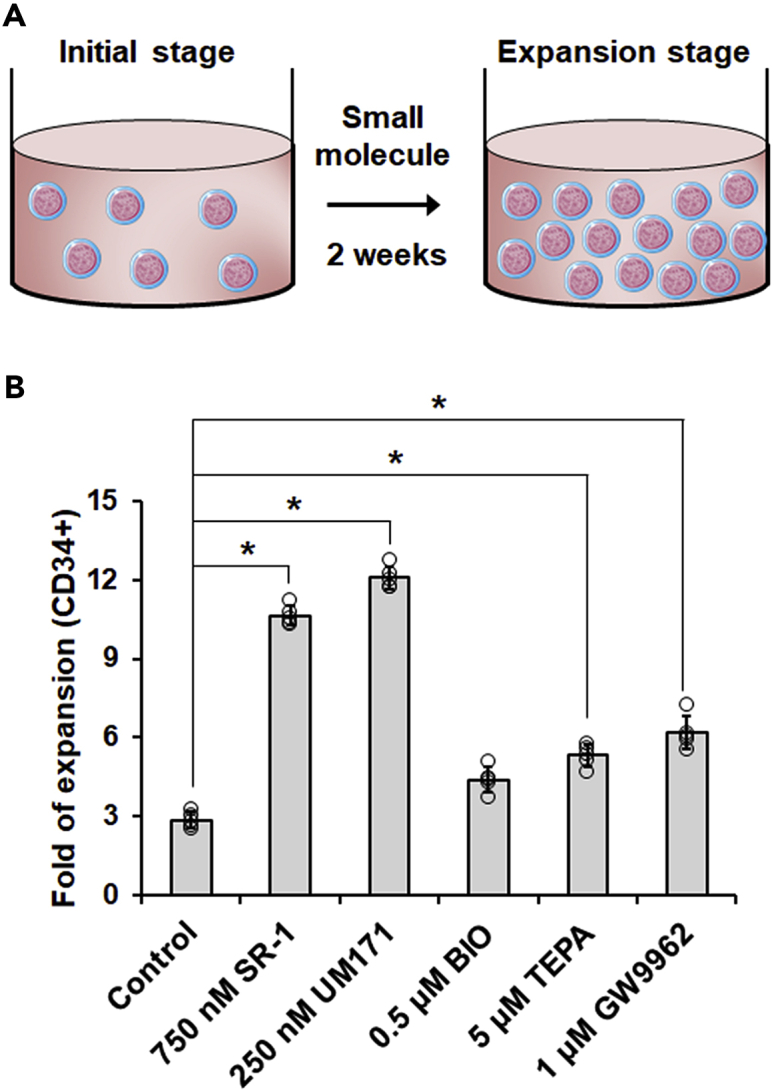
Expansion for hPSC-derived hematopoietic stem and progenitor cells
(A) Schematic of the protocol for the expansion of hPSC-derived hematopoietic cells.
(B) The fold expansion of hematopoietic cells after small molecule treatment was measured and quantified. ∗p < 0.05.
Expected outcomes
This protocol describes a method to differentiate human pluripotent stem cells into definitive hematopoietic stem and progenitor cells. Before differentiation, human pluripotent stem cells should present a morphology of compact colonies with no evidence of spontaneous differentiation and should exhibit high expression (>90%) of pluripotency markers including OCT4 and SSEA4. CHIR99021 treatment induced human pluripotent stem cells into mesoendoderm progenitors (>90%). At day 6 of differentiation, these cells should acquire a typical phenotype of endothelial progenitor cells (∼15%–40% CD34+VEcad+ cells). Subsequently, successful differentiation of hematopoietic stem and progenitor cells (>75% HSPCs within the floating cells) is characterized by the expression of typical hematopoietic markers including CD34, CD43, CD44, CD45, and RUNX1c by fluorescence microscope and flow cytometry analysis. The multipotent potential of hPSC-derived hematopoietic stem and progenitor cells is characterized by the CFU assay. In the presence of different small molecules, hPSC-derived hematopoietic stem and progenitor cells could expand up to 12 fold in 2 weeks. These hematopoietic progenitor cells can also be further differentiated into T, natural killer (NK) cells, neutrophils or other cells as described in our earlier publications.1,2
Quantification and statistical analysis
For applications of human pluripotent stem cells and differentiated hematopoietic progenitor cells, we suggest using at least 3 replicates and repeating the experiment at least 3 times independently. Additionally, we recommend that these 3 independent differentiations are in at least 2 different human pluripotent stem cell lines. Finally, we recommend performing statistical analysis on the data across all 3 differentiations as a blinded variable.
Limitations
We have successfully performed this hematopoietic differentiation protocol in 10 hPSC lines. However, the differentiation efficiency of this protocol may vary in other human pluripotent stem cell lines. Thus, it’s recommended to at least optimize the initial seeding density of undifferentiated hPSCs on day -1 and the concentration of CHIR99021 on day 0 for different hPSC lines. Our protocol described above only allowed 2-week expansion of differentiated hematopoietic stem and progenitor cells. Further optimization of expansion medium for longer-term culture with other cytokines will be needed to for a broader application of our hPSC-derived hematopoietic stem and progenitor cells.
Troubleshooting
Problem 1
At steps 2g and 5f, poor attachment of human pluripotent stem cells to Matrigel-coated plates may occur if ROCK inhibitor is not in the medium when replating singularized cells. Alternatively, unqualified Matrigel may not support cell adhesion.
Potential solution
Include ROCK inhibitor in replating medium (steps 2g and 5f); use a hPSC qualified ECM substrate.
Problem 2
At step 6d, cell detachment or death on day 1 after the treatment of CHIR99021 may be due to the non-optimal initial cell seeding density or concentration of CHIR99021.
Potential solution
Optimize initial seeding density (step 5f) and day 0 CHIR99021 concentration.
Problem 3
The cells detach from the bottom at day 4 (step 10e).
Potential solution
Pre-warm culture medium before medium change. Gently add the medium back to the well during the medium change as the mechanical force generated during medium change may reduce cell attachment.
Problem 4
The number of differentiated hematopoietic stem and progenitor cells (<1 × 105/well of 24-well-plate) at day 12 (step 13d) was low.
Potential solution
Change only top half of the old medium during differentiation process after day 6.
Problem 5
The purification efficiency (<90%) of differentiated hematopoietic stem and progenitor cells was low (step 16n).
Potential solution
Repeat the step 16m for one or two more times. Alternatively, fluorescence-activated cell sorting (FACS) may be used.
Problem 6
CFU counts are low (below 40 colonies) (step 17j).
Potential solution
Increase initial cell seeding density (step 17c). The kinetics of differentiation may vary among different batches.
Problem 7
The expansion or proliferation ability of differentiated hematopoietic stem and progenitor cells was decreased over time (step 18d).
Potential solution
Do not expand differentiated hematopoietic stem and progenitor cells with a too high or too low seeding density. If differentiated hematopoietic stem and progenitor cells are seeded at a very low density, they may not proliferate. High cell seeding density may induce the differentiation of these progenitor cells.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, [Xiaoping Bao] (bao61@purdue.edu).
Materials availability
This study did not generate any new reagents. Human pluripotent stem cells can be purchased from WiCell (https://www.wicell.org) upon completion of a material transfer agreement. Other materials can be obtained by contacting Xiaoping Bao upon completion of a material transfer agreement with Purdue University.
Acknowledgments
This study was supported by startup funding from the Davidson School of Chemical Engineering and the College of Engineering at Purdue (X.B.), Showalter Research Trust (Young Investigator Award to X.B.), NSF CBET (grant no. 2143064 to X.B.), NSF CBET (grant no. 1943696 to X.L.L.), and NIH NCI (grant no. R37CA265926 to X.B.).
Author contributions
Conceptualization, J.J., Y.C., Q.D., X.L., X.B.; Methodology, Y.C., X.B.; Investigation, Y.C., J.J.; Validation, Y.C., J.J.; Writing - original draft, Y.C., J.J.; Writing - review and editing, Y.C., X.L.L., X.B.; Visualization, Y.C., J.J.; Supervision, X.B.; Funding acquisition, X.L.L., X.B.
Declaration of interests
Y.C., Q.D., and X.B. are inventors on provisional US patent applications related to differentiation of human pluripotent stem cells into human hematopoietic progenitor cells.
Contributor Information
Yun Chang, Email: chang692@purdue.edu.
Xiaoping Bao, Email: bao61@purdue.edu.
Data and code availability
This study did not generate/analyze any datasets or codes that were submitted to any repositories.
References
- 1.Chang Y., Syahirah R., Oprescu S.N., Wang X., Jung J., Cooper S.H., Torregrosa-Allen S., Elzey B.D., Hsu A.Y., Randolph L.N., et al. Chemically-defined generation of human hemogenic endothelium and definitive hematopoietic progenitor cells. Biomaterials. 2022;285:121569. doi: 10.1016/J.BIOMATERIALS.2022.121569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y., Syahirah R., Wang X., Jin G., Torregrosa-Allen S., Elzey B.D., Hummel S.N., Wang T., Li C., Lian X., et al. Engineering chimeric antigen receptor neutrophils from human pluripotent stem cells for targeted cancer immunotherapy. Cell Rep. 2022;40:111128. doi: 10.1016/j.celrep.2022.111128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao X., Lian X., Dunn K.K., Shi M., Han T., Qian T., Bhute V.J., Canfield S.G., Palecek S.P. Chemically-defined albumin-free differentiation of human pluripotent stem cells to endothelial progenitor cells. Stem Cell Res. 2015;15:122–129. doi: 10.1016/j.scr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian X., Bao X., Al-Ahmad A., Liu J., Wu Y., Dong W., Dunn K.K., Shusta E.V., Palecek S.P. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Rep. 2014;3:804–816. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randolph L.N., Bao X., Oddo M., Lian X.L. Sex-dependent VEGF expression underlies variations in human pluripotent stem cell to endothelial progenitor differentiation. Sci. Rep. 2019;9:16696. doi: 10.1038/s41598-019-53054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X., Lian X., Qian T., Bhute V.J., Han T., Palecek S.P. Directed differentiation and long-term maintenance of epicardial cells derived from human pluripotent stem cells under fully defined conditions. Nat. Protoc. 2017;12:1890–1900. doi: 10.1038/nprot.2017.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze any datasets or codes that were submitted to any repositories.