Abstract
Myocardial infarctions affect approximately 735,000 people annually in the United States and have a substantial impact on quality of life. Neonates have an enhanced capability of repairing cardiovascular damage, while adults do not. The mechanistic basis for this age-dependent difference in regenerative capacity remains unknown. Recent studies have shown that microRNAs (miRNAs) play a significant role in regulating the regenerative ability of cardiovascular cells. This report defines the alterations in miRNA expression within the cardiovascular repair zone of infarcted sheep hearts following intracardiac injection of neonatal islet-1+ cardiovascular progenitor cells. Sheep were infarcted via left anterior descending coronary artery ligation. After 3 to 4 weeks of infarction, sheep neonatal islet-1+ cardiovascular progenitor cells were injected into the infarcted area for repair. Cell-treated sheep were euthanized 2 months following cell injection, and their hearts were harvested for the analysis of miRNA and gene expression within the cardiovascular repair zone. Ten miRNAs were differentially regulated in vivo, including miR-99, miR-100, miR-302a, miR-208a, miR-665, miR-1, miR-499a, miR-34a, miR-133a, and miR-199a. These miRNAs promote stemness, cell division, and survival. Several signaling pathways are regulated by these miRNAs, including Hippo, Wnt, and Erythroblastic Leukemia Viral Oncogene B (ERBB). Transcripts encoding Wnt, ERBB, and Neuregulin 1 (NRG1) were elevated in vivo in the infarct repair zone. Wnt5a signaling and ERBB/NRG1 transcripts contribute to activation of Yes-Associated Protein 1. MiRNAs that impact proliferation, cell survival, and signaling pathways that promote regeneration were induced during cardiovascular repair in the sheep model. This information can be used to design new approaches for the optimization of miRNA-based treatments for the heart.
Keywords: cardiac, miRNAs, myocardial infarction, cardiac progenitor cells
Introduction
The human heart loses its regenerative ability shortly after the neonatal period1. Stem cell–based therapies aimed at cardiovascular repair must be capable of activating the cell cycle2, promoting paracrine effects3, recruiting immune mediators to the site of repair, and ultimately, activating Yes-Associated Protein 1 (YAP1) to effectively restore cardiac function4. MicroRNAs (miRNAs) have recently been identified as key modulators of these processes. There are more than 2,000 recognized miRNAs, many of which have a regulatory role in the development and function of the cardiovascular system5. Zebrafish and neonatal mouse models, which are capable of cardiovascular repair, have provided insight into miRNA expression patterns that contribute to effective regeneration6.
The feasibility of miRNA administration in a therapeutic setting has recently gained considerable attention due, in large part, to studies supporting the concept of single miRNA delivery for sufficient induction of cardiovascular repair7. In swine, administration of miRNA-199a, for example, reduces infarct size, activates cardiomyocyte proliferation, and improves overall cardiac function8. This concept has also been demonstrated using miR-5909. Despite these encouraging data, careful follow-up has identified the potential for adverse effects resulting from long-term overexpression of specific miRNAs. Additional knowledge defining the miRNA profile in the cardiovascular repair zone would benefit the design of alternative approaches. Understanding which miRNAs facilitate repair as a group may contribute to the development of enhanced regenerative therapies in which specific miRNAs are applied. Currently, information defining these miRNAs in large animal models is limited. Here, we identify miRNAs that are induced in the cardiovascular repair zone following administration of neonatal islet-1+ cardiovascular progenitor cells (CPCs) post–myocardial infarction in sheep. Our objective is to provide insight regarding the molecular mechanisms that contribute to cardiovascular repair in this model. In the present study, we identify the miRNAs that play a role in promoting a proregenerative environment through the regulation of YAP1, Erythroblastic Leukemia Viral Oncogene B (ERBB), and Wnt signaling.
Materials and Methods
Sheep Model of Cardiovascular Repair
A left anterior descending coronary artery ligation procedure was performed on four sheep (less than 1 year old) which were purchased from Nebeker Ranch (Lancaster, CA, USA). The coronary artery ligation model is a well-documented and well-accepted experimental model for myocardial infarction10,11. Three to four weeks after infarction, 10 million cloned, fluorescently labeled sheep neonatal CPCs were directly injected into the myocardium in the peri-infarct region. Ten aliquots of one million cells each were prepared for administration into the left ventricle immediately surrounding the infarction. The border region located between the infarcted and non-infarcted tissue is the cardiovascular repair zone and is the region adjacent to the edges of the infarction where the newly introduced neonatal islet-1+ cells were administered. The retention of transplanted cells was confirmed via carboxyfluorescein succinimidyl ester (CFSE) labeling12. Neonatal islet-1+ sheep CPC clones used for cell therapy were previously harvested, characterized, and expanded in our laboratory13. Ten million early-stage allogeneic CPC cell clones expressing high levels of islet-1 and low levels of c-kit were used for transplantation. Two months after cell injection, the sheep were sacrificed, and their hearts were collected for RNA purification and analysis. Infarcted and non-infarcted tissue samples were collected from the left ventricle of cell-treated sheep and were classified according to their proximity to the infarcted cell injection site. Additional controls included tissue isolated from the left ventricle of three normal, non-infarcted sheep and tissue isolated from the infarcted region of the left ventricle in three sheep which received an infarction but did not receive any post-infarct cell-based treatment. Our study design was approved by the Institutional Animal Care and Use Committee (IACUC) of Loma Linda University and performed within the regulations of the Animal Welfare Act.
Islet-1+ Cardiac Progenitor Cell Isolation and Clonogenic Culture
Sheep islet-1+ cardiac progenitor cell clones were used for the cell transplantation experiments and human islet-1+ cardiac progenitor cell clones were used for the Wnt5a activation experiments and the miRNA inhibition experiments that were done in vitro. CPCs that express the transcription factor islet-1 have been well documented in the literature to have the ability to differentiate into cardiomyocytes, endothelial cells, and smooth muscle cells14–16. This work has been validated by numerous laboratories using both in vivo and in vitro approaches. The clonal progenitors that we used in this study were isolated by selection of cells that express islet-1 and low levels of c-kit. These cells can differentiate into all cells of the cardiovascular lineage17. In addition, we chose to use these cells because they can be expanded as clonal populations, they are well-characterized which provides reproducibility, and because neonatal CPCs have an enhanced capacity for cardiac repair. The cells were cultured and grown on 0.1% gelatin-coated six-well plates in medium 199 (Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (Thermo Scientific, Waltham, MA, USA), 100 µg/ml penicillin-streptomycin (Life Technologies), and 1% minimum essential medium non-essential amino acids (Life Technologies, Carlsbad, CA, USA). The Institutional Review Board at Loma Linda University approved the protocol for using discarded human cardiovascular tissue obtained after cardiac surgery without the use of identifiable patient information, following a waiver of informed consent. The use of sheep for islet-1+ progenitor cell isolation was approved by the Institutional Animal Care and Use Committee of Loma Linda University and was performed within the regulations of the Animal Welfare Act.
Analysis and Selection of miRNAs
MiRNAs used in this study were selected on the basis of predicted function and were included in custom-designed assay plates (Qiagen, Valencia, CA, USA). The databases TargetScan (http://www.targetscan.org/), miRTarBase, DIANA-mirPath (http://www.microrna.gr/miRPathv3), Kyoto Encyclopedia of Genes and Genomes (KEGG), and miRBase (http://www.mirbase.org) were used to determine the predicted targets of the miRNAs. These databases use a context score model that takes approximately 14 features of each individual miRNA and predicts their targets18. The miRNAs examined in this study are listed in Table 1.
Table 1.
Mature MicroRNA (miRNA) Sequences for Primers Used in RT-PCR.
| miRNA | Sequence (5′ → 3′) |
|---|---|
| hsa-miR-34a-5p | UGG CAG UGU CUU AGC UGG UUG U |
| hsa-miR-302a-3p | UAA GUG CUU CCA UGU UUU GGU GA |
| hsa-miR-302b-3p | UAA GUG CUU CCA UGU UUU AGU AG |
| hsa-miR-302c-3p | UAA GUG CUU CCA UGU UUC AGU GG |
| hsa-miR-302d-3p | UAA GUG CUU CCA UGU UUG AGU GU |
| hsa-miR-367-3p | AAU UGC ACU UUA GCA AUG GUG A |
| hsa-miR-371a-5p | ACU CAA ACU GUG GGG GCA CU |
| hsa-miR-373-3p | GAA GUG CUU CGA UUU UGG GGU GU |
| hsa-miR-135a-5p | UAU GGC UUU UUA UUC CUA UGU GA |
| hsa-miR-141-3p | UAA CAC UGU CUG GUA AAG AUG G |
| hsa-miR-200a-3p | UAA CAC UGU CUG GUA ACG AUG U |
| hsa-miR-200b-3p | UAA UAC UGC CUG GUA AUG AUG A |
| hsa-miR-200c-3p | UAA UAC UGC CGG GUA AUG AUG GA |
| hsa-miR-181b-5p | AAC AUU CAU UGC UGU CGG UGG GU |
| hsa-miR-218-5p | UUG UGC UUG AUC UAA CCA UGU |
| hsa-miR-487b-3p | AAU CGU ACA GGG UCA UCC ACU U |
| hsa-miR-539-3p | AUC AUA CAA GGA CAA UUU CUU U |
| hsa-miR-665 | ACC AGG AGG CUG AGG CCC CU |
| hsa-let-7i-5p | UGA GGU AGU AGU UUG UGC UGU U |
| hsa-miR-199a-3p | ACA GUA GUC UGC ACA UUG GUU A |
| hsa-miR-15a-5p | UAG CAG CAC AUA AUG GUU UGU G |
| hsa-miR-16-5p | UAG CAG CAC GUA AAU AUU GGC G |
| hsa-miR-195-5p | UAG CAG CAC AGA AAU AUU GGC |
| hsa-miR-133a-3p | UUU GGU CCC CUU CAA CCA GCU G |
| hsa-miR-499a-5p | UUA AGA CUU GCA GUG AUG UUU |
| hsa-miR-208a-3p | AUA AGA CGA GCA AAA AGC UUG U |
| hsa-miR-1-3p | UGG AAU GUA AAG AAG UAU GUA U |
| hsa-miR-17-5p | CAA AGU GCU UAC AGU GCA GGU AG |
| hsa-miR-18a-5p | UAA GGU GCA UCU AGU GCA GAU AG |
| hsa-miR-20a-5p | UAA AGU GCU UAU AGU GCA GGU AG |
| hsa-miR-92a-3p | UAU UGC ACU UGU CCC GGC CUG U |
| hsa-miR-99a-5p | AAC CCG UAG AUC CGA UCU UGU G |
| hsa-miR-100-5p | AAC CCG UAG AUC CGA ACU UGU G |
RT-PCR: reverse transcription polymerase chain reaction.
RNA and cDNA Preparation From Sheep Cardiac Tissue
The left ventricle of infarcted sheep hearts was sectioned and labeled according to distance from the infarct. Total RNA was purified from 30 mg or ~1 cm3 of ovine cardiac tissue using the miRNeasy Mini Kit (Qiagen), per the manufacturer’s instructions. Tissue samples were homogenized in QIAzol prior to isolation as recommended by the manufacturer. In all, 200 ng of total RNA was reverse-transcribed into complementary DNA (cDNA) using the miRCURY Locked Nucleic Acid (LNA) Reverse Transcriptase (RT) Kit (Qiagen). For experiments that were done using individual miRNA primers, 500 ng of total RNA was reverse-transcribed into cDNA using miScript RT II Kit (Qiagen). Primers for genes of interest are listed in Table 2. cDNA was synthesized with SuperScript III (Invitrogen, Carlsbad, CA, USA).
Table 2.
Primer Sequences Used for RT-qPCR.
| Primer | Sequence (5′ → 3′) |
|---|---|
| KI67-Fwd | TGG CAC AAA ATA CCA TTT CCG T |
| KI67-Rev | AGC CAA AAG TGT ACA CAG GTC A |
| NRG1-Fwd | TGG TGA TCG CTG CCA AAA CT |
| NRG1-Rev | CAG CTG TGA CTG GGA GTC TG |
| ERBB3-Fwd | TGA GAT TGT GCT CAC GGG AC |
| ERBB3-Rev | ATC TCG GTC CCT CAC GAT GT |
| Wnt5a-Fwd | CTT CGC CCA GGT TGT AAT TGA AGC |
| Wnt5a-Rev | CTG CCA AAA ACA GAG GTG TTA TCC |
| Wnt9a-Fwd | GTA CCA GTT CCG CTT TGA GC |
| Wnt9a-Rev | CTG CCC ACT GGG TAA GTC A |
| Wnt11-Fwd | GAG GAA GAA AGT CCA GTC CCG |
| Wnt11-Rev | CCG CGA GTC TTG CTA GAT GT |
| YAP1-Fwd | TCC CAG ATG AAC GTC ACA GC |
| YAP1-Rev | TCA TGG CAA AAC GAG GGT CA |
| CTGF-Fwd | CAC CCG GGT TAC CAA TGA CA |
| CTGF-Rev | TCC GGG ACA GTT GTA ATG GC |
| TEAD1-Fwd | AAC TCA GGA CAG GCA AGA CG |
| TEAD1-Rev | GGC TTG ACG TCT TGT GAG GA |
| SOX2-Fwd | AAC CAG CGC ATG GAC AGT TA |
| SOX2-Rev | GAC TTG ACC ACC GAA CCC AT |
| sheep NRG1-Fwd | TGG TGA TCG CTG CCA AAA CT |
| sheep NRG1-Rev | CAG CTG TGA CTG GGA GTC TG |
| sheep ERBB1-Fwd | GAC TTT ACT GGG GCC TGA CC |
| sheep ERBB1-Rev | ACG TGT TAC CTG GAA GGC TG |
| sheep ERBB2-Fwd | AGA TCC TCA AGG GAG GGG TC |
| sheep ERBB2-Rev | GAA GGT ATA ACG CCC CTC GG |
| sheep ERBB3-Fwd | AGT GCC TAT CTT GCC GGA AC |
| sheep ERBB3-Rev | CTT GTA GAT GGG GCC CTT GG |
| sheep ERBB4-Fwd | AGT CAC AGG CTA CGT GTT GG |
| sheep ERBB4-Rev | CAG GTT GGA AGG CCA TGG AT |
| sheep PIK3C2B-Fwd | CGC AGG TGC CCA GAC A |
| sheep PIK3C2B-Rev | GTA GAG TGG TTG GAC AGC CC |
RT-qPCR: quantitative reverse transcription polymerase chain reaction.
Quantitative RT-PCR
Quantitative real-time polymerase chain reaction (RT-qPCR) was performed using the miRCURY LNA SYBR Green PCR kit and the miRCURY LNA miRNA Custom PCR-Panels (Qiagen). Custom PCR-Panels were designed with miRNA primers listed in Table 1, four miRNA reference genes (U6 snRNA, SNORD48, SNORD68, cel-miR-39-3p), two UniSp3 Inter-plate Calibrators (IPC), and UniSp6 cDNA synthesis positive controls (Qiagen). The IPC controls were required for comparison between multiple plates. The UniSp6 control was required to confirm the success of cDNA synthesis. This protocol requires an initial heat activation of 2 min at 95°C, 40 cycles of denaturation for 10 s at 95°C, and combined annealing/extension for 60 s at 56°C. The Bio-Rad CFX96 PCR machine was used for all quantitative reverse transcription polymerase chain reaction (RT-PCR) experiments (Bio-Rad, Hercules, CA, USA). Single miScript primer assays (Qiagen) for the quantification of mature miRNAs miR-99a, miR-100, and miR-199a using RT-qPCR required an initial activation of 15 min at 95°C, followed by 40 cycles of a three-step cycling series: denaturation for 15 s at 94°C, annealing for 30 s at 55°C, and an extension of 30 s at 70°C.
Treatment of Human Neonatal Islet-1+ Cardiac Progenitor Cells With Wnt5a
Human neonatal islet-1+ CPC clones grown to 60%–70% confluency were treated with 100 ng/ml of recombinant human/mouse Wnt5a (R&D Systems, Minneapolis, MN, USA) for 72 h and then examined for YAP1 expression by RT-qPCR and Western blot. Untreated CPCs were grown in normal CPC growth media for use as a control. Protein was isolated from treated and untreated islet-1+ CPCs for Western blot analysis at the 96- and 144-h timepoints.
RNA Purification From Cells and RT-qPCR
Purification of total RNA from Wnt5a-treated human neonatal islet-1+ CPC samples was performed using an RNeasy mini kit (Qiagen) following the manufacturer’s instructions. The integrity of the RNA was validated using gel electrophoresis prior to preparing cDNA for each sample. Two micrograms of RNA was used to prepare cDNA using SuperScript III (Invitrogen). RT-qPCR was performed with a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad). PCR plates were run under the following conditions: 94°C for 10 min, 94°C for 15 s, 58°C for 1 min, 72°C for 30 s, repeated for 40 cycles. The primers for our genes of interest were developed by NCBI Primer-BLAST (Table 2). RT-qPCR products were visualized using 1%–2% agarose gel electrophoresis and low-mass DNA ladder (Invitrogen).
Protein Purification and Western Blot
Human neonatal islet-1+ CPCs were either not treated and served as controls or were treated with Wnt5a for 72 h before protein was isolated at the 96 and 144-h time periods. Cells were washed with cold phosphate-buffered saline and treated with cold trypsin. Trypsinized cells were placed on ice at room temperature until all cells rose. Samples were collected and agitated for 1 h at 4°C in protein lysis buffer consisting of RIPA (radioimmunoprecipitation assay) buffer, 0.5 M EDTA, protease inhibitor cocktail, sodium orthovanadate, and sodium fluoride, before being centrifuged at 14,000 × g, and aliquoted for use. Protein was quantified using the Micro BCA Protein Assay Kit (ThermoFisher, Waltham, MA, USA). Protein Simple Wes (Protein Simple, San Jose, CA, USA) was used to analyze YAP1 protein levels. Rabbit anti-YAP antibodies were purchased from Cell Signaling (Danvers, MA, USA) and were used at a 1:200 dilution. Antibodies to actin were used as controls.
miRNA Inhibition
Human neonatal islet-1+ CPC clones (n = 3) were individually plated at a confluency of 40% in six-well plates using media without antibiotics. The cells were transfected with 200 ng of mirVana miRNA 302a inhibitor or mirVana negative control inhibitor using lipofectamine 2000 (Invitrogen). The mirVana miRNA inhibitors are single-stranded oligonucleotides that are specifically designed to inhibit endogenous miRNA in cells. The RNAi and lipofectamine mixture was prepared according to the protocol provided by the manufacturer. After a 20-min incubation at room temperature, the solution was added dropwise to each well containing cells and media. The treated cells were maintained at 37°C in a CO2 incubator for 7 h prior to removing the transfection reagent by changing the media. RNA was isolated from the treated cells after 48 h. The viability of the cells at the time of RNA isolation was 96%–98%. cDNA was prepared from the RNA for RT-qPCR.
Statistical Analysis
The PCR data for the miRCURY LNA Custom PCR-Panels were sent to Qiagen’s GeneGlobe Analysis software. The data were normalized by the geNorm method for a more accurate and reliable normalization of miRNA expression19. Other PCR data not using this system were analyzed via Prism v8. The miRNA data presented in this study represent a comparison of miRNA expression in tissue collected in the cardiovascular repair zone near the infarct site relative to tissue that was collected from a distal portion of the same heart of each respective, cell-treated sheep. Data are reported as mean with standard error of the mean.
Results
MiRNA Expression in the Regenerating Myocardium of Sheep
The retention of CFSE+ CPC in the cardiovascular repair zone was quantified in frozen sections of cardiovascular tissue using Image Pro Plus Version 5 software. An average of,2400 DAPI+ cells were counted in the cardiovascular repair zone of four cell-treated, infarcted sheep. The blue fluorescent DNA stain DAPI binds to AT-rich regions of double-stranded DNA and was used to count the number of nuclei in the stained sections. The proportion of CFSE+ CPC was 19% on when the counts from all sheep in the study were averaged, reflecting retention of a proportion of newly introduced islet-1+ cardiovascular progenitors at 2 months following cell injection. CFSE+ cells that were retained in the cardiovascular repair zone of one of the representative sheep are shown in Fig. 1A. To identify which miRNAs were differentially regulated within the cardiovascular repair zone, DIANA-mirPath v3 and TargetScan databases were used to select miRNAs with potential involvement in cell-based repair. The design was based on predicted targets associated with cardiovascular stem cell proliferation, survival, and differentiation (Fig. 1B). These features are necessary for improving cardiac function and regeneration20,21. The changes in miRNA expression that were identified within the cardiovascular repair zone following islet-1+ neonatal CPC transplantation are shown in Fig. 1C. Ten differentially expressed miRNAs were identified as significant: miRNAs 99a, 100, 302a, 208a, 665, 1, 499a, 34a, 133a, and 199a.
Figure 1.
The cardiovascular repair zone of infarcted, islet-1+ cell–injected sheep hearts demonstrates ongoing presence of stem cells at the site of infarction and activation of select microRNA (miRNA) transcripts. Labeled CFSE+ cells (green) were quantified within the cardiovascular repair zone. The black arrows in the Masson’s trichrome–stained tissue section identify the cardiovascular repair zone where viable tissue (red) borders nonviable tissue (blue) in the infarcted heart (A). MiRNAs involved in various aspects of cardiovascular repair, including proliferation, differentiation, and survival, were identified using TargetScan and existing literature (B). Quantitative real-time polymerase chain reaction was done to identify the relative expression of miRNAs involved in cardiovascular repair. The heatmap represents the changes in miRNA expression in the cardiovascular repair zone following neonatal islet-1+ cardiovascular progenitor cell treatment in infarcted sheep relative to the non-infarcted area (C). CFSE: carboxyfluorescein succinimidyl ester.
Predicted Pathways Targeted by Significantly Altered MiRNAs
Using DIANA-mirPath v.3 software, the top predicted pathways targeted by the 10 miRNAs of interest were identified (Fig. 2). These pathways, and in particular the Hippo22, Wnt23, and ERBB24 signaling pathways, are well documented as critical for cardiovascular repair. The inhibition of the Hippo pathway increases active YAP1 which is a known promoter of proliferation and cell cycle reentry. Interestingly, the most significant pathway to be impacted by the predicted target genes is cell cycle, which, in the context of repair, is crucial for adequate regeneration of lost tissue.
Figure 2.
Predicted pathways targeted by miR-302a, miR-99a, miR-100, miR-199a, miR-92a, miR-208a, miR-499a, miR-1, miR-133a, and miR-665. Prediction is based on the results from DIANA-mirPath v.3 software.
MiRNAs Involved in Cardiovascular Repair Displayed Expression Patterns Consistent With Regeneration
A significant reduction in miR-99a and miR-100 transcripts (0.38 ± 0.03-fold change; 0.34 ± 0.07-fold change, respectively) occurred in the islet-1+ CPC-treated region of infarcted sheep hearts compared with the control/non-infarct regions (Fig. 3A, B). Interestingly, a similar trend has been reported in regenerating zebrafish and neonatal mouse hearts where miR-99a and miR-100 are downregulated during cardiovascular repair25. miR-99a and miR-100 transcript levels were significantly lower in infarcted plus cell-treated hearts when compared with transcript levels identified after myocardial infarction alone (Fig. 3). MiRNA-199a was found to be significantly induced (7.61 ± 0.36-fold change) in the infarct repair region in the sheep model compared with control/non-infarct zones or when compared with the infarcted region of sheep which did not receive cell treatment (Fig. 3C). The expression of miR-199a declines with age, coinciding with the loss of regenerative capacity in adults26. MiRNA-199a also enhances ERBB signaling, which was elevated in cell-treated, infarcted sheep (Fig. 3D). Neuregulin 1 (NRG1) induction in vivo further elevates ERBB signaling to maintain the expression of this proproliferative pathway (Fig. 3E). NRG1 was not found to be elevated in infarcted sheep without cell treatment.
Figure 3.
Differentially expressed miRNAs in the cardiovascular repair zone and activation of ERBB signaling and NRG1 in the CPC-treated infarcted region. A decrease in miR-99a and miR-100 transcripts (A, B) and an increase in miRNA-199a compared with controls that included infarct only or the control/non-infarcted area of infarcted, cell-treated sheep support cell cycle re-entry and cardiovascular regeneration (C). ERBB signaling pathway transcripts are elevated as predicted based on the miRNA profile (D) and NRG1 transcripts are activated in the cardiovascular repair zone (E). Fold changes are displayed as mean ± SEM. ERBB: Erythroblastic Leukemia Viral Oncogene B; NRG1: Neuregulin 1; CPC: cardiovascular progenitor cell. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
MiRNAs Associated With Proliferation Are Elevated During Cardiovascular Repair
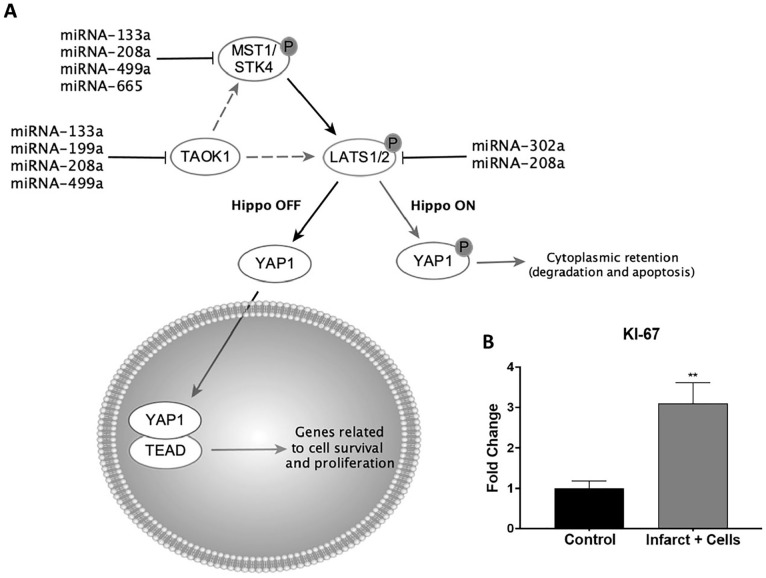
Following treatment with neonatal islet-1+ CPCs, the infarcted area of the heart displayed an increase in the expression of miRNAs associated with proliferation. These miRNAs included miR-133a (1.965 ± 0.2926-fold change), miR-208a (2.86 ± 0.52-fold change), miR-499a (2.97 ± 0.57-fold change), miR-665 (16.96 ± 1.89-fold change), and miR-302a (2.7 ± 0.49-fold change) as shown in Fig. 4A–E, respectively, compared with control/non-infarct zones. None of these miRNAs were elevated in the infarcted region of the heart in sheep that received an infarct without cell treatment (Fig. 4). Induction of these miRNAs is associated with Hippo signaling regulation (Fig. 5A). To validate these changes in our model, real-time PCR was used to investigate the level of KI-67 transcripts as a marker of proliferation in the cardiovascular repair zone. As shown in Fig. 5B, KI-67 transcripts were elevated (3.1 ± 0.3-fold) at the site of repair, relative to the non-infarcted region of each respective sheep. Similar trends of KI-67 expression were observed when comparing infarcted cardiovascular tissue from cell-treated sheep with normal non-infarcted sheep that did not receive cell transplantation (data not shown). Furthermore, validation of the role of the select miRNAs that were differentially regulated and predicted to activate the Hippo and Wnt signaling pathways was provided by PCR and Western blotting12. As the Hippo pathway consists of a kinase cascade resulting in YAP1 phosphorylation and decreased proliferation27, miRNAs that inhibit the Hippo pathway elevate active YAP1 levels and maintain cell proliferation (Fig. 5A). In the sheep model of cardiovascular repair, Hippo regulators miR-133a28, miR-208a (TargetScan)18, miR-499a (TargetScan)18, miR-199a29, miR-302a, and miR-66530 were significantly induced (Figs 1, 3, and 4). A previous publication from our laboratory showed that YAP1 transcripts were elevated in the infarct repair zone of sheep transplanted with islet-1+ CPCs, validating the effects of the listed miRNAs12. The activation of YAP1 is associated with an increase in cell survival and proliferation. miRNA 302a, which targets LATS2 and enhances nuclear localization of YAP 1, was elevated in the cardiovascular repair zone of cell-treated, infarcted sheep hearts (Figs. 1 and 4). miRNA inhibitors such as 302ai reduce the expression of CTGF, TEAD and SOX2, which are elevated when YAP1 is translocated into the nucleus, supporting the role of miRNA-mediated regulation of this process (Supplementary Fig. 1).
Figure 4.
MicroRNAs (miRNAs) associated with proliferation are upregulated in the infarct repair zone when compared with controls that included infarct only or the control/non-infarcted area of infarcted, cell-treated sheep. MiRNA-133a (A), miR-208a (B), miR-499a (C), miR-665 (D), and miR-302a (E) expression is positively correlated with an induction of proliferation. Fold changes are displayed as mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Figure 5.
Signaling pathways responsible for driving proliferation are regulated by miRNAs. An induction of miRNAs that promote YAP1 activation was observed (A). Elevated levels of KI-67 are shown by real-time polymerase chain reaction in the islet-1+ CPC-treated infarcted tissue region compared with the control/non-infarcted normal tissue (B). Fold changes are displayed as mean ± SEM. CPC: cardiovascular progenitor cell. **P ≤ 0.01.
Wnt Signaling Is Induced by MiRNAs in the Cardiovascular Repair Zone
The expression of noncanonical Wnt (Wnt5a, Wnt11) and canonical Wnt transcripts (Wnt3a, Wnt9a) was elevated in the cardiovascular repair zone after transplantation of islet-1+ neonatal CPCs as predicted by the miRNA profile. Noncanonical Wnt transcripts, Wnt5a, and Wnt11 were significantly increased (2.3 ± 0.2-fold change; 6.9 ± 0.2-fold change, respectively) (Fig. 6A). Canonical Wnt transcripts, Wnt3a, and Wnt9a exhibited a 2.9 ± 0.01-fold change and 5.3 ± 0.2-fold change, respectively (Fig. 6B). Transcripts for canonical and noncanonical Wnt were similarly upregulated in infarcted cardiovascular tissue from cell-treated sheep relative to normal non-infarcted sheep without cell intervention (data not shown), indicating that either control led to the same conclusion. Both noncanonical and canonical Wnt-signaling pathways are necessary but insufficient on their own for cardiac differentiation31. MiRNA-499a is a positive regulator of the Wnt signaling pathway32. The increase in miR-499a expression in the sheep model, along with the activation of canonical and noncanonical Wnts, provides mechanistic context for the efficacy of transplanted neonatal CPCs for cardiovascular repair.
Figure 6.
Wnt signaling is induced after islet-1+ neonatal cardiovascular progenitor cell transplantation as shown by RT-qPCR. Noncanonical Wnt ligands Wnt5a and Wnt11 are significantly elevated in the islet-1+ CPC-treated infarcted region by 2.3-fold and 6.9-fold, respectively, compared with the control/non-infarct normal tissue (A). Canonical Wnt ligands Wnt3a and Wnt9a are also significantly increased in the islet-1+ CPC-treated infarcted region compared with the control/non-infarcted area (B); fold changes are displayed as mean ± SEM. RT-qPCR: quantitative reverse transcription polymerase chain reaction; CPC: cardiovascular progenitor cell. **P ≤ 0.01; ****P ≤ 0.0001.
The Interaction of Wnt5a, ERBB, and YAP1 Forms a Proliferative, Prosurvival Signaling Loop Induced by MiRNAs in the Cardiovascular Repair Zone
Treatment of neonatal CPC with Wnt5a elevates YAP1 transcript levels and YAP1 protein (Fig. 7A–C). Intranuclear translocation of YAP1 is followed by activation of NRG-1 which is downstream of YAP1. NRG1 participates in an autocrine loop effect by further activating the ERBB receptor on islet-1+ CPCs (Fig. 7E, F). Activated ERBB signals through PI3K to further elevate the expression of intranuclear YAP1. This interactive signaling pathway identified in vitro demonstrates a potential mechanism by which the miRNA environment induced by neonatal CPC-mediated repair contributes to cardiomyocyte proliferation, dedifferentiation, growth, and survival. Understanding the interaction of signaling pathways activated by cell-based treatment and the mechanistic basis by which they function provides new opportunities for the design of potential treatment options for patients suffering from heart failure.
Figure 7.
ERBB/YAP1/NRG1 signaling forms an autocrine loop. NRG1 binding to the ERBB3/ERBB2 receptor stimulates downstream signaling to the PI3K-AKT pathway to promote cell survival. Activation of PDK1 indirectly inhibits the Hippo kinase cascade which activates YAP1. An autocrine loop interaction may occur when the expression of intranuclear YAP1 is elevated, resulting in the upregulation of NRG1 downstream of YAP1. NRG1 is transported through the cell membrane to rebind with ERBB3/ERBB2 receptors on the cell surface (A). If YAP1 protein levels are induced as shown by Western blot following exposure to Wnt5a (B, C), transcripts encoding NRG1 and ERBB3 are also induced as shown by RT-qPCR (D–F). Fold changes are shown as mean ± SEM. PCR samples were run in triplicates and were normalized to actin. ERBB: Erythroblastic Leukemia Viral Oncogene B; NRG1: Neuregulin 1; CPC: cardiovascular progenitor cell; YAP1: Yes-Associated Protein 1; RT-qPCR: quantitative reverse transcription polymerase chain reaction.
Discussion
The ability of neonatal CPCs to enhance cardiovascular repair has been well documented; however, the mechanistic basis for this finding is still under investigation. MiRNAs actively participate in cardiovascular regeneration and play a role in modulating multiple signaling pathways that are relevant for repair. Understanding the miRNAs that participate in neonatal CPC-based cardiovascular repair provides new insight into the regenerative process. We previously reported that the miRNA profile of human neonatal CPCs differs significantly from that of comparable adult CPCs17; however, the miRNAs identified in the cardiovascular repair zone following neonatal CPC administration in vivo have not been characterized to our knowledge. Here, we identified 10 miRNAs whose expression is significantly altered in the cardiovascular repair zone following transplantation of neonatal islet-1+ CPCs in a sheep model of myocardial infarction: miR-99, miR-100, miR-199a, miR-302a, miR-208a, miR-665, miR-1, miR-499a, miR-34a, and miR-133a. These miRNAs enhance proliferation, differentiation, and stemness; activate YAP1, ERBB, and Wnt signaling; and inhibit both apoptosis and fibrosis33–35. Collectively, the miRNAs defined here contribute to the stimulation of cardiac repair observed in this model.
In the current study, 7 of the 10 differentially regulated miRNAs activate proliferation. Low levels of miR-34a post-infarction induce proliferation, improve cardiac function, and reduce fibrosis36. Other miRNAs that impact proliferation include miR-99a and miR-100, whose Gene Ontology (GO) processes are predicted to include proliferation pathways, chromatin remodeling, and morphogenesis25. The decline in miR-99a and miR-100 expression is consistent with the expression profile of zebrafish and neonatal mice, both of which are capable of efficient cardiovascular regeneration6,37. Activation of miRNAs 199a38, 302a39, 133a28, and 499a40 improves proliferative and migratory capacity. These miRNAs collectively modulate the observed behavior of transplanted CPCs in our model. Proliferation of transplanted CPCs in the infarcted area of the heart is required for the formation of new tissue to repair the damaged myocardium. Meanwhile, differentiation of cells toward cardiac lineages as well as dedifferentiation of existing mature cells is necessary to enhance regenerative capacity.
Dedifferentiation of resident cardiovascular cells and reprogramming events provide a necessary contribution to optimal cardiovascular repair given the limited retention of newly introduced progenitor cells. Dedifferentiation and reprogramming are known to be regulated by miRNAs41,42. MiRNA-133a enhances the reprogramming of cardiovascular cells toward a cardiomyocyte-like state while reducing fibrosis and apoptosis43. Additional miRNAs involved in modulating dedifferentiation/reprogramming include miR-99/miR-100 when expressed at low levels25 and activated miR-302a44, miR-1, miR-499a, and miR-208a. Dedifferentiation restores the proliferative capacity of terminally differentiated cells, enabling them to re-enter the cell cycle and potentially replenish damaged cardiomyocytes45. In the zebrafish model of cardiovascular regeneration, mature cardiomyocytes undergo a dedifferentiation step to proliferate within the damaged myocardium and redifferentiate to replace lost cells46. It has only recently been found that dedifferentiation also contributes to cardiovascular repair in large animal models. Collectively, the miRNAs that facilitate neonatal CPC-mediated repair in sheep are capable of initiating these steps. The regulatory role of miRNAs can be leveraged to develop novel miRNA therapeutics for regeneration.
Direct administration of miRNAs at the site of infarction has been addressed in several laboratories as a potential approach to achieving functional cardiovascular recovery. Studies done in multiple animal models have provided data supporting the concept that cell-free therapies mediated by miRNAs can be effective9,47. For example, miR-199a administration can reintroduce regenerative capacity in adult mice and can significantly improve cardiac function in swine8. Upregulation of miR-199a expression activates cardiomyocyte proliferation by interaction with the Hippo pathway to activate YAP148,49. While the activation of YAP1 via miR-199a administration is a crucial step in cardiovascular regeneration, whether or not it is sufficient as a standalone treatment remains to be determined. In our model, the induction of miR-199a along with other miRNAs that regulate dedifferentiation and proliferation has the potential to improve outcomes when applied in the context of CPC-based repair. The 10 miRNAs described in this study jointly contribute to cardiovascular repair through multiple modes of action.
The beneficial effects of miRNA treatment are mediated predominantly via activation of signaling pathways that facilitate repair. In our study, miR-302a was significantly elevated in the infarct repair region and regulates both the Wnt and Hippo signaling pathways39,50. Wnt signaling is of significance during cardiogenesis for its role in differentiation, proliferation, and self-renewal post-infarction51. KI-67 transcripts were elevated in neonatal CPC-treated sheep, which infers that proliferation is transpiring within this repair model. Noncanonical (Wnt5a, Wnt11) and canonical (Wnt3a, Wnt9a) Wnt signaling transcripts were also elevated, suggesting the presence of both differentiating and proliferating CPCs in the cardiovascular repair zone. Canonical Wnt signaling is activated in the early developmental stages, while noncanonical Wnt-signaling pathways are activated later when driving the production of new cardiomyocytes31. Wnt5a can also induce the expression of YAP152, resulting in elevated NRG1 and ERBB signaling, which functions as an autocrine loop to maintain YAP1 levels. Activation of the NRG1-induced ERBB3 pathway contributes to cardiomyocyte proliferation, dedifferentiation, growth, and survival.
Hippo signaling is regulated by miR-302a-mediated inhibition of LATS2, which increases YAP1 and creates a proproliferative cell state12. In addition to miR-302a, it is known that miR-199a, miR-665, miR-499a, miR-208a, and miR-133a all interact with the Hippo pathway. These miRNAs were regulated in a manner that induces YAP1 following neonatal islet-1+ cell transplantation in sheep. The mechanism of regulation differs among these miRNAs. For example, while miR-302a and miR-208a directly inhibit the immediate upstream kinases/inhibitors of YAP1, Large Tumor Suppressor Kinase 1 (LATS1)/Large Tumor Suppressor Kinase 2 (LATS2), miR-133a, miR-499a, and miR-665 act to prevent any kinase cascade within the Hippo pathway. In the context of cardiovascular repair, emerging evidence suggests that the Hippo pathway is a critical regulator of regeneration4. Interestingly, the miRNAs that are suggested to be sufficient for cardiogenesis simultaneously target the Hippo pathway to induce YAP1 expression.
In conclusion, our study shows that neonatal islet-1+ CPC transplantation regulates multiple miRNA transcripts that target Hippo at various levels, promoting intranuclear expression of YAP1 and cardiovascular repair. It is interesting to speculate that multiple levels of YAP1 activation are preferable, as the evidence surrounding this pathway continues to strengthen. The new information provided here characterizes the miRNA expression profile following neonatal islet-1+ CPC treatment in the sheep myocardial infarction model. This work provides insight into the mechanistic basis for the proregenerative alterations observed in neonatal cell-treated sheep. The identification of induced miRNAs that contribute to cardiovascular repair may lead to the development of novel miRNA-based therapies designed to promote regeneration.
Supplemental Material
Supplemental material, sj-jpg-1-cll-10.1177_09636897221136787 for MicroRNA Expression in the Infarcted Heart Following Neonatal Cardiovascular Progenitor Cell Transplantation in a Sheep Model of Stem Cell–Based Repair by Larry V. Lopez, Victor Camberos, Leonard L. Bailey, Nahidh Hasaniya, Christopher Ramos, Lorelei Hughes, Cole Knox and Mary K. Kearns-Jonker in Cell Transplantation
Footnotes
Author Contributions: LVL participated in research design, performance of the research, data analysis, formulation of figures, and writing of the paper. VC participated in performance of the research, data analysis, and writing of the paper. LVL and VC contributed equally to this paper. LLB contributed reagents and participated in the performance of the research. NH contributed reagents and participated in the performance of the research. CR participated in the performance of the research. CK contributed resources and participated in critical review of the paper. LH participated in data analysis and formulation of figures, and MKK-J participated in conceptualization, performance of the research, writing of the paper, and funding acquisition and contributed reagents. All authors have approved this article.
Ethical Approval: This study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Institutional Review Board of Loma Linda University (IRB# 5110115).
Statement of Human and Animal Rights: All animal studies were approved by the Institutional Animal Care and Use Committee of Loma Linda University and performed within the regulations of the Animal Welfare Act (IACUC approval no.: 8160018, May 27, 2016).
Statement of Informed Consent: The Institutional Review Board at Loma Linda University approved the protocol for using discarded human cardiovascular tissue obtained after cardiac surgery without the use of identifiable patient information, following a waiver of informed consent.
Data Availability Statement: Data will be shared upon reasonable request.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the California Institute for Regenerative Medicine (CIRM) Bridges to Stem Cell Research Training (grant number EDUC2-08418) and a Loma Linda University School of Medicine GCAT award (grant number 681130-2934).
ORCID iD: Mary K. Kearns-Jonker  https://orcid.org/0000-0002-8989-3698
https://orcid.org/0000-0002-8989-3698
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Haubner BJ, Schneider J, Schweigmann U, Schuetz T, Dichtl W, Velik-Salchner C, Stein J, Penninger JM. Functional recovery of a human neonatal heart after severe myocardial infarction. Circ Res. 2016;118(2): 216–21. [DOI] [PubMed] [Google Scholar]
- 2. Hesse M, Welz A, Fleischmann BK. Heart regeneration and the cardiomyocyte cell cycle. Pflugers Arch. 2018;470(2): 241–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tachibana A, Santoso MR, Mahmoudi M, Shukla P, Wang L, Bennett M, Goldstone AB, Wang M, Fukushi M, Ebert AD, Woo YJ, et al. Paracrine effects of the pluripotent stem cell-derived cardiac myocytes salvage the injured myocardium. Circ Res. 2017;121(6): e22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X, Li Y, Luo J, Hou N. Molecular mechanism of hippo–YAP1/TAZ pathway in heart development, disease, and regeneration. Front Physiol. 2020;11:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, Hart M, Abu-Halima M, Grässer FA, Lenhof H, Keller A, et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47(7): 3353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hodgkinson CP, Kang MH, Dal-Pra S, Mirotsou M, Dzau VJ. MicroRNAs and cardiac regeneration. Circ Res. 2015;116(10): 1700–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lock MC, Tellam RL, Darby JRT, Soo JY, Brooks DA, Seed M, Selvanayagam JB, Morrison JL. Identification of novel miRNAs involved in cardiac repair following infarction in fetal and adolescent sheep hearts. Front Physiol. 2020;11:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, Ali H, Braga L, Gorgodze N, Bernini F, Burchielli S, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569(7756): 418–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ Res. 2017;120(8): 1298–1304. [DOI] [PubMed] [Google Scholar]
- 10. Charles CJ, Elliott JM, Nicholls MG, Rademaker MT, Richards M. Myocardial infarction with and without reperfusion in sheep: early cardiac and neurohumoral changes. Clin Sci (Lond). 2000;98(6): 703–11. [PubMed] [Google Scholar]
- 11. De Villiers C, Riley PR. Mouse models of myocardial infarction: comparing permanent ligation and ischaemia-reperfusion. Dis Model Mech. 2020;13(11): dmm046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Camberos V, Baio J, Bailey L, Hasaniya N, Lopez LV, Kearns- Jonker M. Effects of spaceflight and simulated microgravity on YAP1 expression in cardiovascular progenitors: implications for cell-based repair. Int J Mol Sci. 2019;20(11):2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou X, Appleby N, Fuentes T, Longo LD, Bailey LL, Hasaniya N, Kearns-Jonker M. Isolation, characterization, and spatial distribution of cardiac progenitor cells in the sheep heart. J Clin Exp Cardiol. 2012; S6:004. [PMC free article] [PubMed] [Google Scholar]
- 14. Jia G, Preussner J, Chen X, Guenther S, Yuan X, Yekelchyk M, Kuenne C, Looso M, Zhou Y, Teichmann S, Braun T. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9(1): 4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6): 877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao R, Liang X, Cheedipudi S, Cordero J, Jiang X, Zhang Q, Caputo L, Günther S, Kuenne C, Ren Y, Bhattacharya S, et al. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. 2019;29(6): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuentes TI, Appleby N, Tsay E, Martinez JJ, Bailey L, Hasaniya N, Kearns-Jonker M. Human neonatal cardiovascular progenitors: unlocking the secret to regenerative ability. Plos One. 2013;8(10):e77464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Selvarajah GT, Bonestroo FAS, Timmermans Sprang EPM, Kirpensteijn J, Mol JA. Reference gene validation for gene expression normalization in canine osteosarcoma: a geNorm algorithm approach. BMC Vet Res. 2017;13:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S, Srivastava D. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173(1): 104–116.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Foo KS, Lehtinen ML, Leung CY, Lian X, Xu J, Keung W, Geng L, Kolstad TRS, Thams S, Wong AOT, Wong N, et al. Human ISL1+ ventricular progenitors self-assemble into an in vivo functional heart patch and preserve cardiac function post infarction. Mol Ther. 2018;26(7): 1644–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng M, Jacob J, Hung SH, Wang J. The hippo pathway in cardiac regeneration and homeostasis: new perspectives for cell-free therapy in the injured heart. Biomolecules. 2020;10(7): e77464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu WB, Wang WE, Zeng CY. Wnt signaling pathways in myocardial infarction and the therapeutic effects of Wnt pathway inhibitors. Acta Pharmacol Sin. 2019;40(1): 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang X, Ding Y, Zhang Y, Chai Y-H, He J, Chiu S-M, Gao F, Tse H-F, Lian Q. Activation of NRG1-ERBB4 signaling potentiates mesenchymal stem cell-mediated myocardial repairs following myocardial infarction. Cell Death Dis. 2015;6(5): e1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez- Ferrer E, Rodriguez-Esteban C, Kumar S, et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15(5): 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morrison JL, Zhang S, Tellam RL, Brooks DA, McMillen C, Porrello ER, Botting KJ. Regulation of microRNA during cardiomyocyte maturation in sheep. BMC Genomics. 2015;16(1): 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng J, Wang S, Dong Y, Yuan Z. The role and regulatory mechanism of hippo signaling components in the neuronal system. Front Immunol. 2020;11:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izarra A, Moscoso I, Levent E, Cañón S, Cerrada I, Díez-Juan A, Blanca V, Núñez-Gil I-J, Valiente I, Ruíz-Sauri A, Sepúlveda P, et al. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Rep. 2014;3(6): 1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Torrini C, Cubero RJ, Dirkx E, Braga L, Ali H, Prosdocimo G, Gutierrez MI, Collesi C, Licastro D, Zentilin L, Mano M, et al. Common regulatory pathways mediate activity of MicroRNAs inducing cardiomyocyte proliferation. Cell Rep. 2019;27(9): 2759–71.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9(10): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Mazzotta S, Neves C, Bonner RJ, Bernardo AS, Docherty K, Hoppler S. Distinctive roles of canonical and noncanonical Wnt signaling in human embryonic cardiomyocyte development. Stem Cell Rep. 2016;7(4): 764–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song JL, Nigam P, Tektas SS, Selva E. microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015;27(7): 1380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen C, Ponnusamy M, Liu C, Gao J, Wang K, Li P. MicroRNA as a therapeutic target in cardiac remodeling. Biomed Res Int. 2017;2017:1278436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Diez-Cuñado M, Wei K, Bushway PJ, Maurya MR, Perera R, Subramaniam S, Ruiz-Lozano P, Mercola M. miRNAs that induce human cardiomyocyte proliferation converge on the hippo pathway. Cell Rep. 2018;23(7): 2168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li N, Zhou H, Tang Q. miR-133: a suppressor of cardiac remodeling? Front Pharmacol. 2018;9:903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iannolo G, Sciuto MR, Raffa GM, Pilato M, Conaldi PG. MiR34 inhibition induces human heart progenitor proliferation. Cell Death Dis. 2018;9(3): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hodgkinson CP, Dzau VJ. Conserved MicroRNA program as key to mammalian cardiac regeneration. Circ Res. 2015;116(7): 1109–11. [DOI] [PubMed] [Google Scholar]
- 38. Tao Y, Zhang H, Huang S, Pei L, Feng M, Zhao X, Ouyang Z, Yao S, Jiang R, Wei K. miR-199a-3p promotes cardiomyocyte proliferation by inhibiting Cd151 expression. Biochem Biophys Res Commun. 2019;516(1): 28–36. [DOI] [PubMed] [Google Scholar]
- 39. Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, Snitow M, Morley M, Li D, Petrenko N, Zhou S, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7(279): 279ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Z, Han Y, Liu J, Jiang F, Hu H, Wang Y, Liu Q, Gong Y, Li X. MiR-135b-5p and MiR-499a-3p promote cell proliferation and migration in atherosclerosis by directly targeting MEF2C. Sci Rep. 2015;5(1): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee YJ, Ramakrishna S, Chauhan H, Park WS, Hong SH, Kim KS. Dissecting microRNA-mediated regulation of stemness, reprogramming, and pluripotency. Cell Regen. 2016;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yao S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol Proced Online. 2016; 18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao Y, Zhao J, Tuazon JP, Borlongan CV, Yu G. MicroRNA-133a and myocardial infarction. Cell Transplant. 2019;28(7): 831–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shim J, Nam JW. The expression and functional roles of microRNAs in stem cell differentiation. BMB Rep. 2016;49(1): 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yao Y, Wang C. Dedifferentiation: inspiration for devising engineering strategies for regenerative medicine. NPJ Regen Med. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang WE, Li L, Xia X, Fu W, Liao Q, Lan C, Yang D, Chen H, Yue R, Zeng C, Zhou L, et al. Dedifferentiation, proliferation, and redifferentiation of adult mammalian cardiomyocytes after ischemic injury. Circulation. 2017;136(9): 834–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song Y, Zhang C, Zhang J, Jiao Z, Dong N, Wang G, Wang Z, Wang L. Localized injection of miRNA-21-enriched extracellular vesicles effectively restores cardiac function after myocardial infarction. Theranostics. 2019;9(8): 2346–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abbas N, Perbellini F, Thum T. Non-coding RNAs: emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res Cardiol. 2020;115(5): 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Q, Ye B, Wang P, Yao F, Zhang C, Yu G. Overview of microRNA-199a regulation in cancer. Cancer Manag Res. 2019;11:10327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sebastiani G, Grieco GE, Brusco N, Ventriglia G, Formichi C, Marselli L, Marchetti P, Dotta F. MicroRNA expression analysis of in vitro dedifferentiated human pancreatic islet cells reveals the activation of the pluripotency-related microRNA cluster miR-302s. Int J Mol Sci. 2018;19(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pahnke A, Conant G, Huyer LD, Zhao Y, Feric N, Radisic M. The role of Wnt regulation in heart development, cardiac repair and disease: a tissue engineering perspective. Biochem Biophys Res Commun. 2016;473(3): 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Del Re DP. Beyond the cardiomyocyte: consideration of HIPPO pathway cell type specificity. Circ Res. 2018;123(1): 30–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-jpg-1-cll-10.1177_09636897221136787 for MicroRNA Expression in the Infarcted Heart Following Neonatal Cardiovascular Progenitor Cell Transplantation in a Sheep Model of Stem Cell–Based Repair by Larry V. Lopez, Victor Camberos, Leonard L. Bailey, Nahidh Hasaniya, Christopher Ramos, Lorelei Hughes, Cole Knox and Mary K. Kearns-Jonker in Cell Transplantation