Summary
Spermatogenesis carries the task of precise intergenerational transmission of genetic information from the paternal genome and involves complex developmental processes regulated by the testicular microenvironment. Studies performed mainly in mouse models have established the theoretical basis for spermatogenesis, yet the wide interspecies differences preclude direct translation of the findings, and farm animal studies are progressing slowly. More than 32,000 cells from prepubertal (3-month-old) and pubertal (24-month-old) buffalo testes were analyzed by using single-cell RNA sequencing (scRNA-seq), and dynamic gene expression roadmaps of germ and somatic cell development were generated. In addition to identifying the dynamic processes of sequential cell fate transitions, the global cell-cell communication essential to maintain regular spermatogenesis in the buffalo testicular microenvironment was uncovered. The findings provide the theoretical basis for establishing buffalo germline stem cells in vitro or culturing organoids and facilitating the expansion of superior livestock breeding.
Subject areas: Cell biology, Cell, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Single-cell RNA sequencing uncovers heterogeneity of cell types in buffalo testes
-
•
Trajectory analysis reveals sequential differentiation events in germ cells
-
•
Pseudotime analysis uncovers dynamic roadmap of germ cell development
-
•
Prepubertal and pubertal buffalo testes: Comparative global cell-cell communication
Cell biology; Cell; Omics; Transcriptomics
Introduction
The water buffalo (Bubalus bubalis) is a valuable animal resource that significantly contributes to the agricultural economy of many developing Asian countries by providing milk, meat, and draft power.1,2 Nevertheless, buffalo has a poor reproductive performance, and the major cause is abnormal spermatogenesis in males.3,4 Spermatogenesis begins at puberty and depends on the maturity of the somatic microenvironment.5 Testicular spermatogenic cell division begins at about 12 months, but the ejaculate contains a high percentage of viable spermatozoa only after 24–30 months.6 In the testicular seminiferous tubules, some spermatogonial stem cell (SSC) populations maintain stem cell pools by self-renewal, whereas the other SSCs differentiate to form highly proliferative spermatogonia (SPG).5,7 SPG proliferate into primary spermatocytes by mitosis, which then undergo two meiotic divisions in response to hormones such as follicle-stimulating hormone and testosterone to form haploid spermatids,6 and subsequently spermiogenesis into spermatozoa.8 Testicular somatic cells regulate spermatogenesis by secreting cytokines and regulating transduction of specific signals, thus controlling the balance between SSC self-renewal and differentiation, as well as secreting hormones/immunomodulatory factors, leading to an immune-privileged environment, and physical barriers.9,10,11,12,13,14 Obviously, mammalian male pubertal development involves an increase in testicular volume, complex hormonal and molecular regulation, and cellular interactions, which ensure the success of cell proliferation, differentiation, and spermatogenesis.5,6,15,16,17,18,19
Spermatogenesis carries out the precise intergenerational transmission of paternal genetic information, representing a critical guarantee for the reproduction and continuation of the species.20,21,22 Traditional bulk RNA sequencing (RNA-seq) requires total RNA collected from hundreds or thousands of cells, and the resulting expression matrix fails to distinguish among the distinct gene expression patterns from every spermatogenic cell type.23 Purification of transient cells remains exceedingly challenging, whereas single-cell RNA-seq (scRNA-seq) technology makes it possible to visualize the status of these cells without purification and provides numerous distinctive cell stage-specific markers.24 It helps in detecting the cell lineage and heterogeneity of all cell types in the original physiological state, revealing the different molecular events of spermatogenic cell sequential occurrence and gene expression regulatory mechanisms during spermatogenesis.25
Recent studies have obtained stage-specific gene markers in human testicular germ cells by scRNA-seq, such as HMGA1, PIWIL4, TEX29, SCML1, and CCDC112.26 At the single-cell level, several studies elucidated adult testicular germ cell–somatic cell interactions and human pubertal testicular development.27,28 Besides, the developmental landscape of the human male germline and its somatic cells was illustrated in another single-cell study, which contributed to studies of SSC therapy.29 In human, macaque, and mouse testes, scRNA-seq has been used to identify transcriptional signatures of major germ cells and somatic cell types, revealing spermatogonial stem/progenitor cell pools, differentiation markers, potential meiosis regulators, and germ cell–somatic cell communication mechanisms.30 Furthermore, single-cell transcript levels of various cell types during spermatogenesis in sheep and pigs have been demonstrated.31,32 Hence, the development patterns and gene expression of spermatogenic cells during spermatogenesis have both similarities and differences among species.33
Nevertheless, mammalian spermatogenesis is mostly understood from studies on rodents, monkeys, and humans.13,25,27,28,34,35,36,37,38,39 Conversely, significant differences exist among species in the seminiferous epithelial cycle, spermatogenesis cycle, the developmental pattern of SPG, specific molecular markers, etc.17,40,41,42,43 These differences between species make it difficult to establish germline stem cells or perform organoid culture in vitro, which prevents us from understanding the biological functions of germline stem cells, the complexity of spermatogenesis, and the regulation of the male germline.44 Similarly, it also hinders investigation into the expansion of superior livestock breeding and the establishment of livestock models.45,46,47 In the present study, single-cell RNA-seq data was obtained from prepubertal (PRE, 3-month-old) and pubertal (PERI, 24-month-old) buffalo testes. Spermatogenic and somatic cell clusters were identified; a high precision single-cell transcriptional atlas of spermatogenesis and somatic cell development was generated; and the similarities and differences in cell-cell communication networks between prepubertal and pubertal testes and stage-specific markers were elucidated. Our work reveals single-cell gene expression patterns in prepubertal and pubertal buffalo testes, providing crucial insights into the molecular mechanisms that underpin spermatogenesis and germ cell development. Furthermore, it presents a molecular theoretical foundation for investigating mechanisms in stem cell maintenance and organoid culture.
Results
Single-cell transcriptome of buffalo prepubertal and pubertal testes
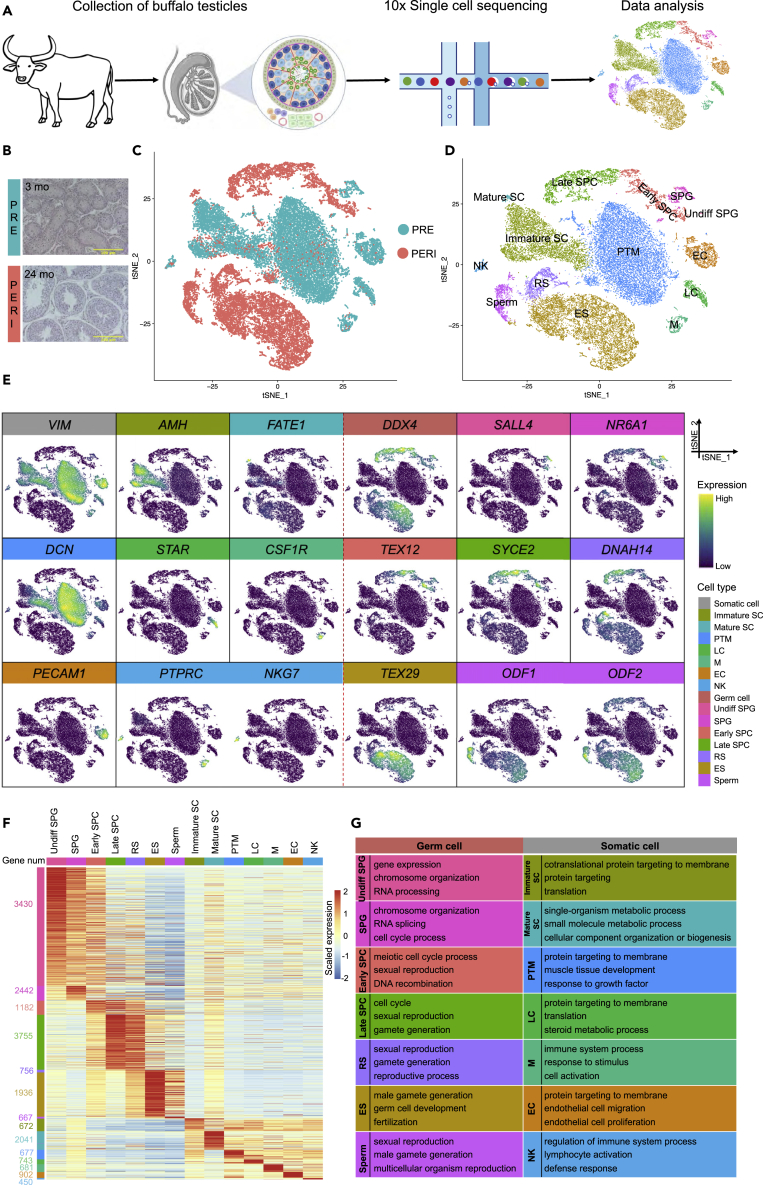
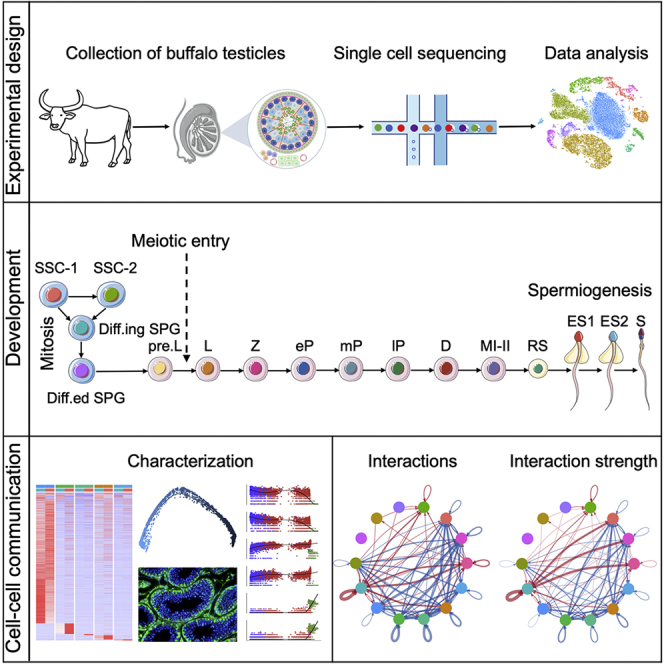
To characterize the cell types of buffalo testes, cells were isolated from 3-month-old (prepuberty) and 24-month-old (puberty) healthy buffalo testicular samples and obtained a total of 32,847 cells (after quality control filtration) were obtained by 10x single-cell sequencing (Figures 1A, S1A, and S1B). Furthermore, after morphological analysis (hematoxylin and eosin staining) of buffalo prepubertal and pubertal testes, it was observed that the prepubertal testis lacked a tubular lumen, whereas the pubertal testis already developed a complete tubular lumen with germ and somatic cells at all stages, and spermatogenesis was complete (Figure 1B). Subsequently, the t-distributed stochastic neighbor embedding (tSNE) analysis was performed on all sample sequencing data using the Seurat package in R, resulting in a total of 14 major clusters, which were subsequently annotated using identified genetic markers (Figures 1C and 1D).
Figure 1.
Single-cell transcriptome analysis of buffalo prepubertal and pubertal testes
(A) Schematic diagram of the experimental workflow.
(B) Histomorphometric analysis (H&E staining) of prepubertal and pubertal buffalo testes. Scale bar, 100 μm.
(C) Dimension reduction presentation (by tSNE) of combined single-cell transcriptome data from prepubertal and pubertal testes (a total of 32,847 cells). Each point represents a single cell and is colored by sample.
(D) tSNE plot of prepubertal and pubertal testicular cell clusters defined by scRNA-seq analysis, colored according to cell type.
(E) Expression pattern of selected markers on the tSNE plot.
(F) Heatmap of normalized expression in DEGs (Wilcoxon rank sum test, p-value < 0.05) for each cell type. Labels indicate cell type (columns) and the number of genes (rows).
(G) GO terms (biological process) for identified cell types (p-value <0.05).
The cell types and markers are as follows: Immature Sertoli cell (Immature SC, AMH+/INHA+/CLU+), mature Sertoli cell (Mature SC, FATE1+/HMGN5+/CLDN11+), peritubular myoid cell (PTM, DCN+/PDGFRA+/PTCH1+), Leydig cell (LC, STAR+/HSD3B1+/INSL3+), macrophage (M, CSF1R+/CD74+/MAFB+), endothelial cell (EC, PECAM1+/CLDN5+/ECSCR+), natural killer cell (NK, PTPRC+/NKG7+/KLRF1+/CD94+), undifferentiated spermatogonia (Undiff SPG, SALL4+/ZBTB16+/ELAVL2+), spermatogonia (SPG, NR6A1+/FGFR3+/FMR1+), early-stage primary spermatocyte (from preleptotene to zygotene) (Early SPC, TEX12+/SMCHD1+/PRSS50+), late-stage spermatocyte (from pachytene to secondary spermatocyte) (Late SPC, SYCE2+/BOLL+/MLH1+), round spermatid (RS, DNAH14+/IZUMO4+/NKAPL+), elongating/elongated spermatid (ES, TEX29+/FBXO24+/ACRV1+), and spermatozoa (Sperm, ODF1+/ODF2+/OAZ3+/AKAP4+) (Figures 1E, S1C, and S1D).
In the germ cell cluster (DDX4+), 3430, 2442, 1182, 3755, 756, 1936, and 667 differentially expressed genes (DEGs) were obtained for Undiff SPG, SPG, Early SPC, Late SPC, RS, ES, and Sperm, respectively; whereas in the somatic cell cluster (VIM+), Immature SC, Mature SC, PTM, LC, M, EC, and NK cells had 672, 2041, 677, 743, 681, 902, and 450 DEGs, respectively (Figure 1F and Table S1). Gene ontology (GO) terms for each cell type supported the definition. For instance, clusters in germ cell development contained “chromosome organization”, “sexual reproduction”, and “male gamete generation”. Also, somatic cells reflected the GO terms for functional characteristics in the physiological state (Figure 1G and Table S2).
Trajectory analysis of spermatogonial cells
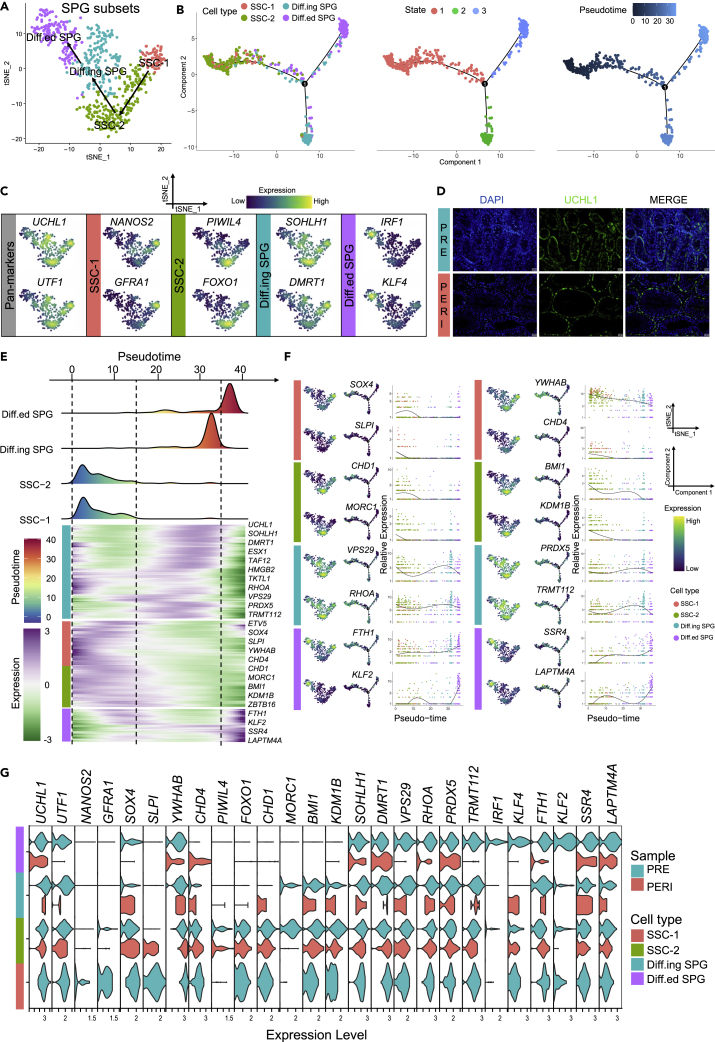
Spermatogonial mitosis is the first step to mammalian spermatogenesis.48 The original clustering analysis (Figure 1D) showed that spermatogonia consisted of two cell clusters (Undiff SPG and SPG, 626 cells). To further characterize the cellular subtypes and transitional relationships of spermatogonia from prepubertal and pubertal buffalo testes, Undiff SPG and SPG clusters were re-clustered to obtained SSC-1, SSC-2, Diff.ing SPG (differentiating) and Diff.ed SPG (differentiated) clusters (Figures 2A and S2A). A total of 1516, 1553, 510, and 310 DEGs with elevated expression at different developmental stages were determined by the Seurat package (Figure S2B and Table S3). Subsequently, cell trajectory analysis of the four cell types was performed using the Monocle package, and the pseudotime value confirmed the differentiation relationship of the re-clustered cells (Figure 2B).
Figure 2.
Identification of SPG subsets in prepubertal and pubertal buffalo
(A) Re-clustering tSNE plot of SPG subsets in buffalo testes.
(B) Pseudotime trajectories of buffalo SPG subsets, colored by cell type, state, and pseudotime value.
(C) Feature plots of marker genes for SSC-1, SSC-2, Diff.ing SPG, and Diff.ed SPG.
(D) IF analysis of antibody staining against UCHL1 on paraffin sections of prepubertal and pubertal buffalo testes. Scale bar, 20 μm.
(E) Heatmap of DEGs (q-value < 0.01) expression (rows) along the pseudotime (columns). The heatmap shows cell types (left) and the representative genes (right) for each cell type.
(F) Feature plots, trajectory plots, and pseudotime trajectory expression patterns of selected marker genes in SSC-1, SSC-2, Diff.ing SPG, and Diff.ed SPG.
(G) Violin plot of the selected gene expression split by sample.
The SPG subsets all broadly expressed UCHL1 and UTF1 (SPG pan-marker).28,49 Many previously identified and new SSC marker genes were expressed by SSC-1 and SSC-2, including DAZL,50 male germline stem cell asymmetric division-related genes (e.g., ZBTB16, ELAVL2, and HUWE1). ITGA6, ETV4, ETV5, and ST3GAL2 showed high expression levels in SSC-1 (NANOS2+/GFRA1+). In SSC-2 (PIWIL4+/FOXO1+), RBM47, PRC1, HELLS, and CXADR were exceedingly expressed. Furthermore, Diff.ing SPG (SOHLH1+/DMRT1+) strikingly expressed genes promoting spermatogonial differentiation (ESX1, CCDC124, TMEM147, and NDUFA4). Finally, Diff.ed SPG specifically expressed the spermatogonial promitogenic factor (IRF1),51 pluripotency transcription factors (TFs) (KLF4),52,53CLEC3B, DBI, PPIB, and IER2 (Figures 2C and S2C). As expected, immunofluorescence (IF) staining found that UCHL1 was located on the basal membrane of testicular seminiferous tubules in both PRE and PERI, consistent with the location of spermatogonial distribution, demonstrating that it could function as a pan-marker for every stage of SPG in buffalo (Figure 2D).
Monocle-derived pseudotime analysis showed dynamic expression patterns of multiple genes along the SPG differentiation timeline (Figure 2E). In addition, SOX4 was highly expressed in SSC-1, whereas SLPI, YWHAB, CHD4, HMGA1, RRBP1, ZNF518A, and RAC1 showed expression trends in the pseudotime axis consistent with the tSNE re-clustering results. CHD1 was specifically expressed in SSC-2, and we also found that MORC1, BMI1, KDM1B, ITGB1, LPIN2, GTF2A1, and RBBP8 shared the same expression pattern.
Besides, VPS29, RHOA, PRDX5, TRMT112, ATP5F1E, EIF1AX, NDUFV2, and PSMG4 showed a similar pattern to markers SOHLH1, ESX1, and DMRT1, which are known to be present in differentiating spermatogonia. Several pluripotency markers (KLF2 and KLF4), FTH1, SSR4, LAPTM4A, HSPB1, ANXA1, SELENOM, and FAU showed similar pseudotime expression patterns and were highly expressed in Diff.ed SPG (Figures 2F and S2D). Thus, the data revealed the dynamic expression pattern of SPG subsets along the developmental pseudotime trajectory, and these stage-specific markers could be used to construct regulatory networks of male germ cells in breeding livestock.
To further elucidate the heterogeneity of prepubertal and pubertal buffalo spermatogonial differentiation, the expression of spermatogonial stage markers was split by sample (PERI vs. PRE). Importantly, NANOS2, GFRA1, ST3GAL2, and MORC1 were only found to be specifically expressed in PRE-derived SSC, whereas IRF1, KLF2, and KLF4 were only found in PRE-derived Diff.ed SPG. All phases of PRE- and PERI-derived SPG subsets were identified by the remaining candidate markers (Figures 2G and S2E).
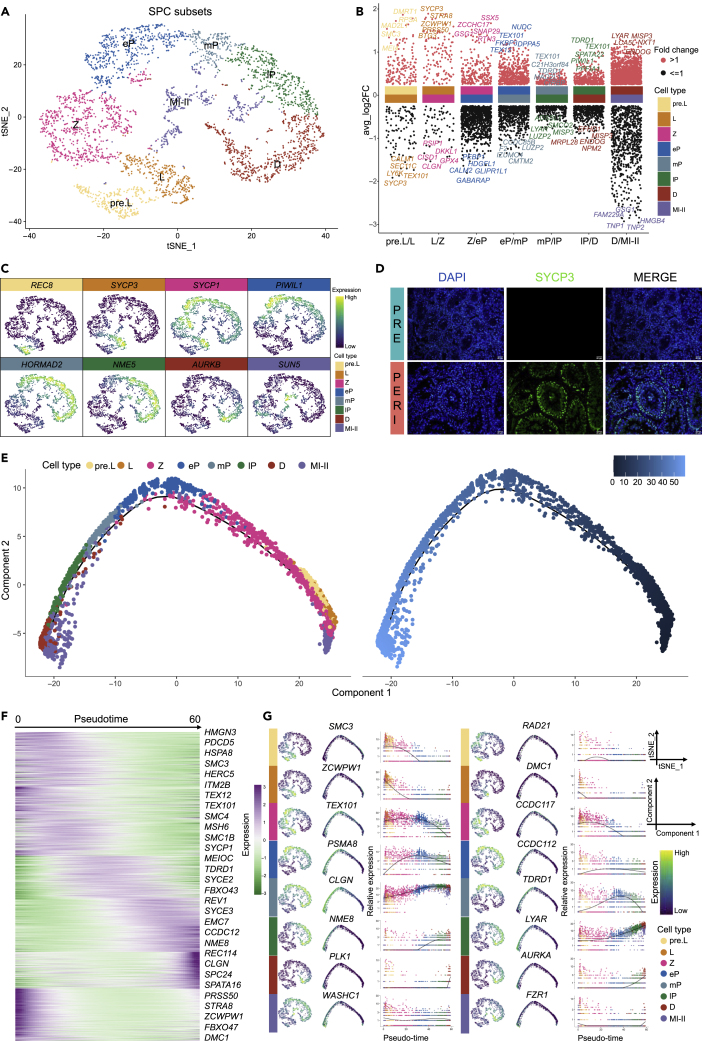
Identification of spermatocytes in buffalo
The second step of mammalian spermatogenesis is spermatocyte meiosis.54 SPC subsets (Early SPC and Late SPC, 3,782 cells) were re-clustered into cell populations, yielding eight clusters of spermatocytes at distinct developmental phases, namely, pre.L (preleptotene), L (leptotene), Z (zygotene), eP (early pachytene), mP (middle pachytene), lP (late pachytene), D (diplotene), and MI-II (mixed cells of primary and secondary spermatocytes) (Figures 3A and S3A, and Table S3). Next, meiosis-associated genes were calculated by the FindMarkers function of Seurat package for actively high-variable genes across sequential spermatocyte stages during meiosis (Figure 3B and Table S4). Meiosis-associated genes were linked to the following spermatocyte stages: DNA synthesis, sister chromatid cohesion, axial elements, homologous chromosome pairing, DNA double-strand break (DSB) formation, synaptonemal complex formation, recombination, nuclear envelope breakdown, and a series of molecular events during spermatogenesis from pre-meiosis to meiosis.55,56,57,58,59,60
Figure 3.
Identification of SPC subsets in buffalo
(A) Re-clustering tSNE plot of SPC subsets in buffalo testes.
(B) Multi-volcano plot showing meiotic genes (test.use = “wilcox”, min.pct = 0.65) in pairwise comparisons in all cell types. The graph displayed the top five annotated genes with the most fold change for each group.
(C) Feature plots of representative marker genes for pre.L, L, Z, eP, mP, lP, D, and MI-II.
(D) IF analysis of antibody staining against SYCP3 on paraffin sections of prepubertal and pubertal buffalo testes. Scale bar, 20 μm.
(E) Pseudotime trajectories of buffalo SPC subsets, colored by cell type (left) and pseudotime value (right).
(F) Heatmap showing DEGs (q-value < 0.01) expression (rows) along the pseudotime (columns) axis.
(G) Feature plots, trajectory plots, and pseudotime trajectory expression patterns of selected genes in pre.L, L, Z, eP, mP, lP, D, and MI-II.
The meiosis-associated genes REC8, SMC3, and DMRTB135,57,61 were found to be specifically expressed in pre.L spermatocytes. Further, the meiosis initiation marker STRA8, synaptonemal complex protein 3 (SYCP3), homologous chromosome pairing-related genes at meiosis (ZCWPW1), and DNA meiotic recombinase 1 (DMC1)62,63,64 tended to be highly expressed in L spermatocytes. In Z spermatocytes, the meiosis-specific genes SYCP1, TEX101, and MEIOB65 were acutely expressed. Moreover, the piwi family members PIWIL1 and CCDC11226,37 are biomarkers for the beginning of expression in eP spermatocytes. In mP spermatocytes, the central element deposition-related marker HORMAD2 and the pseudoautosomal region-related marker REC11466,67,68 were substantially expressed. In lP spermatocytes, the tubulin-binding and microtubule motor activity-related genes NME5 and NME826,32,65 were expressed. The D spermatocytes express aurora kinase A/B (AURKA and AURKB).25,26 Finally, meiotic nuclear division-related genes SUN5, SUN3, and FZR126,38,64,65 were expressed in MI-II spermatocytes (Figures 3C, S3B, and S3C). Actually, IF staining showed that the distribution of leptotene spermatocytes corresponded to the localization of SYCP3 in buffalo testes from PRE and PERI (Figure 3D).
The classification of cells at each stage of SPC subsets was confirmed by pseudotime trajectory analysis of buffalo spermatocytes (Figure 3E). To better understand the sequential gene expression patterns during meiosis, spermatocyte-specific marker gene expression at every stage was matched to cell developmental trajectories (Figure 3F). In pre-meiotic spermatocytes (pre. L), RAD21 (nuclear meiotic cohesin complex), SMC1A (meiotic cell cycle gene), SMC3 (cohesin complex), DMRTB1(coordinate the developmental transition from spermatogonial differentiation to meiotic entry), HMGN3 (the chromatin-binding protein), and MIF (glycosylation-inhibiting factor) were specifically expressed, which decreases as pseudotime development increases, and essentially no expression occurs after Z spermatocytes. In L spermatocytes, ZCWPW1, DMC1, STRA8, PRSS50, RAD21L1, and BTG3 were strongly expressed, whereas following Z spermatocytes, their expression was almost absent. TEX101, CCDC117, SETX, TOP2B, WDR82, and C11H14orf39 were highly expressed in Z and underexpressed in eP spermatocytes.
Synaptonemal complex protein 2 (SYCP2), meiotic spindle-related genes (HSPA2, SELENOW, and PEBP1), and meiotic cell cycle-related gene (PSMA8) showed the highest expression in eP spermatocytes and gradually decreased with the increase in pseudotime. Meiotic cell cycle-related genes (CLGN and TDRD1), meiotic recombination protein (REC114), GPATCH4, SAMD15, and MRPS26 were continually expressed with the pseudo-development axis, whereas CLGN and TDRD1 expression in the pre.L–mP spermatocyte stage progressively increased and the expression of mP–D spermatocyte stage gradually decreased.
Furthermore, NME8 and cell growth-regulating nucleolar protein (LYAR, ADAM2, MLLT10, CCDC38, and CCDC85B) were continuously expressed in the lP–D spermatocyte stage, with the maximum expression in lP. Then, PLK1 (meiotic cytokinesis), AURKA (meiotic spindle), MISP3, NPM2 (positive regulation of meiotic nuclear division), INCENP (meiotic spindle midzone assembly), and CCNA1 (meiotic cell cycle) began to be expressed in lP and peaked in the D spermatocyte stage. Ultimately, WASHC1 (meiotic spindle assembly), FZR1 (regulation of meiotic nuclear division), CCER1, CLDND2, FBXO39 (F-box protein family member), and TMEM89 were specifically expressed only in MI-II spermatocytes (Figures 3G and S3D). In summary, the data presented candidate-specific gene markers for the above stages of spermatocytes.
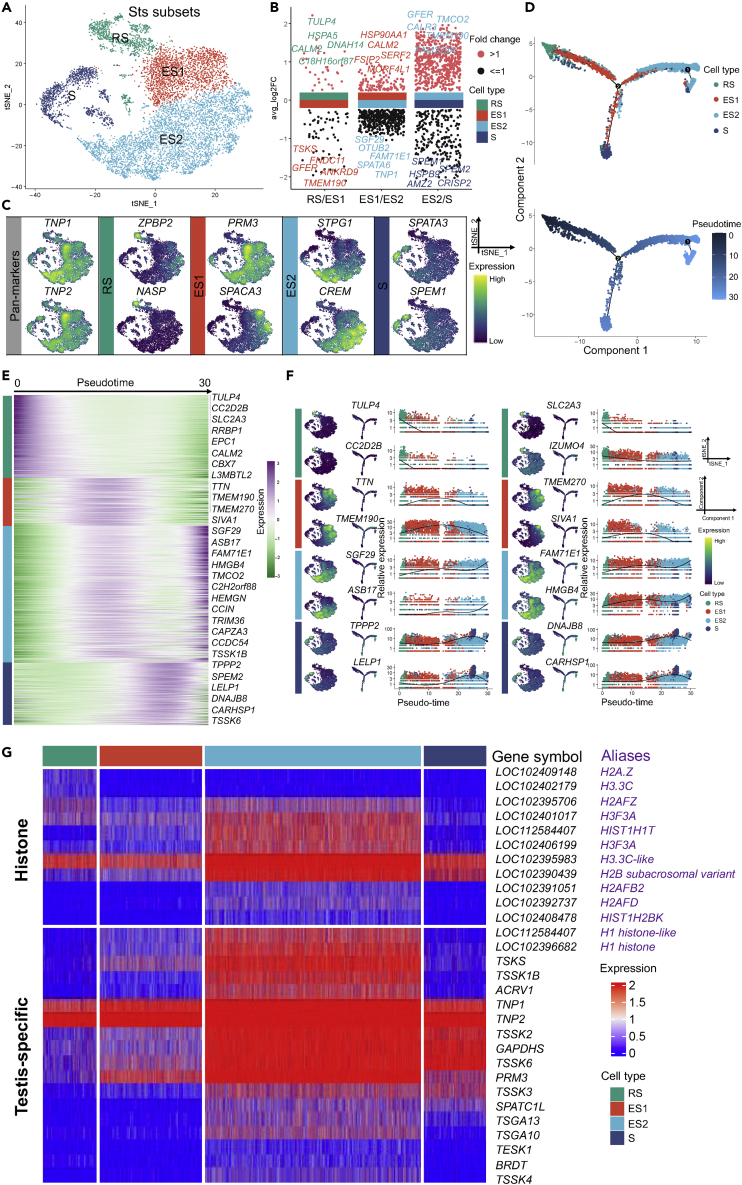
Dynamic gene expression patterns during spermiogenesis
Spermiogenesis is the final stage of spermatogenesis.8,69 Original RS, ES, and Sperm clusters (Sts subsets, 9,629 cells) were re-clustered into four haploid spermatids—RS (round spermatid), ES1 (elongating spermatid), ES2 (elongated spermatid), and S (spermatozoa) (Figures 4A, S4A, and S4B, and Table S3). To obtain highly variable genes that are active during spermiogenesis, spermiogenesis-related genes were calculated by the FindMarkers function of the Seurat package (Figure 4B and Table S4). The spermatid pan-marker transition protein (TNP1 and TNP2)70 is expressed in RS, ES1, ES2, and S. Initially, RS was identified by the zona pellucida binding protein 2 (ZPBP2) and nuclear autoantigenic sperm protein (NASP).27,71 For ES1, protamine 3 (PRM3) and sperm acrosome-associated 3 (SPACA3)27,72 were marked. Next, ES2 was marked by sperm tail PG-rich repeat containing 1 (STPG1) and cAMP response element modulator (CREM).73 Finally, S was characterized by spermatogenesis-associated 3 (SPATA3) and spermatid maturation 1 (SPEM1)74,75,76 (Figure 4C).
Figure 4.
Identification of Sts subsets in buffalo
(A) Re-clustering tSNE plot of Sts subsets in buffalo testes.
(B) Multi-volcano plot showing spermiogenesis-related genes (test.use = “wilcox”, min.pct = 0.65) in pairwise comparisons in all cell types (RS, ES1, ES2, and S).
(C) Feature plots of representative marker genes for RS, ES1, ES2, and S.
(D) Pseudotime trajectories of buffalo Sts subsets, colored by cell type (up) and pseudotime value (down), respectively.
(E) Heatmap showing DEGs (q-value < 0.01) expression (rows) along the pseudotime (columns) axis.
(F) Feature plots, trajectory plots, and pseudotime trajectory expression patterns of selected genes in RS, ES1, ES2, and S.
(G) Heatmap of histone and testis-specific gene expression in RS, ES1, ES2, and S.
The analysis of pseudotime trajectories provided evidence to support the categorization of spermatid development (Figure 4D), and the identification of expression patterns of multiple genes related to spermatid differentiation, resulting in distinct genetic markers for individual spermatid stages (Figure 4E). For instance, the expression of TULP4, CC2D2B, SLC2A3, IZUMO4, SPATA22, and STRBP was specifically detected at the RS stage, with decreasing expression levels as pseudotime increased. Specifically, TTN, TMEM190, TMEM270, SIVA1, SPACA1, and TEX29 were considered candidate genes for ES1-specific expression. Genes related to spermatid development (SGF29, ASB17, FAM71E1, and HMGB4) and sperm tail development-related genes (MEIG1 and KLC3)77 were categorized into candidate genes for ES2. Finally, TPPP2, LELP1, DNAJB8, CARHSP1, AKAP4, and SPACA9 were recognized as candidate markers for the S cluster (Figures 4F, S4C, and S4D).
Histone-to-protamine transition is a central molecular event during spermiogenesis.78 Consequently, the patterns of histone variants and testis-specific genes during spermiogenesis were examined, and these expressions were the strongest in ES2, demonstrating that transcriptional activity peaking during ES2 was associated with a histone-to-protamine transition (Figures 4G and S4E).
Spermatogenic cell lineage development in buffalo
To further characterize the dynamic transcriptome on the developmental trajectory of buffalo male germ cells, the expression patterns of 16 germ cell clusters (14,037 cells) with elevated DEGs (15,097 genes) were analyzed (Figure S5A). GO analysis demonstrated that individual germ cells undergo different biological processes (Figure 5A and Table S5). These activities were consistent with the sequential molecular events involved in mammalian spermatogenic cells.8,18,20,79
Figure 5.
Transcriptome functional characteristics and sequential expression patterns of buffalo male germ cells
(A) Top five most significant GO terms (p-value < 0.05) of spermatogenic cells (by DAVID), colored by cell type.
(B) Development stage-specific gene expression patterns in buffalo spermatogenic cells, with each color palette, and nucleus color-coordinated with cell type.
(C) Cell-surface specific marker gene expression levels of spermatogenic cells.
During spermatogonial differentiation, SSC-1 and SSC-2 were mainly enriched in chromatin organization and transcription-related activities. Diff.ing SPG was coupled with activities such as mitochondrial energy, mRNA splicing, and translation, whereas Diff.ed SPG was enriched in translation and the transforming growth factor beta receptor signaling pathway.
The processes of sister chromatid cohesion and spermatogenesis were particularly enriched in early primary spermatocytes (pre.L–Z). Late primary spermatocytes (eP–D) were involved in translation, ATP synthesis-related, cell division, and cilium assembly processes, revealing that transition from meiosis to spermiogenesis was a sequential step, with genes involved in sperm structural assembly already being expressed in the lP–D spermatocytes stage. Consistent with the results of the pseudotime analysis of spermatocytes, MI-II was enriched in spermatogenesis, spermatid development, flagellated sperm motility, fusion of sperm to egg plasma membrane and fertilization, showing evidence that MI-II not only significantly expressed genes related to the breakdown of the nuclear envelope but also participated in the molecular events of early and late meiosis.
During spermiogenesis, in addition to mRNA and protein activities, RS was responsible for microtubule-based movement and other biological processes closely related to spermatid development. ES1 and ES2 were predominantly involved in spermatogenesis, spermatid development, and fertilization. Except for enrichment to spermatogenesis, S was also broadly associated with protein catabolic process and sperm motility. The GO enrichment analysis showed that several biological processes were active or cell-type-specific in the buffalo male germline cell.
Moreover, the markers with stage-specific/pseudotime-sequential expression of individual germ cell clusters were summarized in the present study (Figure 5B). To investigate cell-surface specific marker genes contributing to the cultivation of buffalo primary spermatogenic cells in vitro, the linear expression relationship of markers along the differentiation timeline was observed by prediction of membrane protein (Figures 5C and S5B, and Table S6).
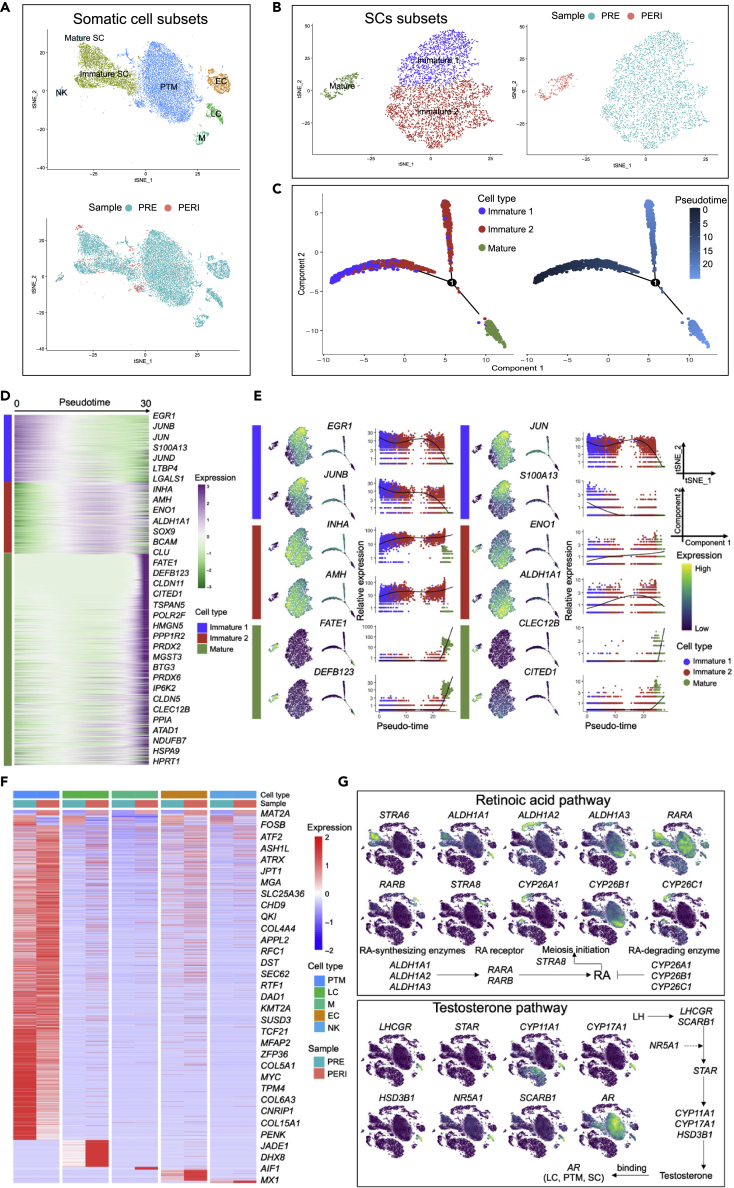
Characterization of the somatic lineages in buffalo testes
Testicular cell-cell interactions are required for complete spermatogenesis.16,80 Somatic cell subsets (Figures 1D, 18,810 cells) were investigated individually to characterize the gene expression patterns that establish the spermatogenic microenvironment during prepuberty and puberty (Figure 6A).
Figure 6.
Characterization of buffalo somatic lineages
(A) tSNE plot of somatic cell subsets in prepubertal and pubertal buffalo testes, colored by cell type (up) and sample (down).
(B) Re-clustering tSNE plot of SC lineages in buffalo testes, colored by cell type (left) and sample (right).
(C) Pseudotime trajectories of buffalo SCs subsets, coloring by cell type (left) and pseudotime value (right), respectively.
(D) Heatmap showing DEGs (q-value < 0.01) expression (rows) along the pseudotime (columns) axis.
(E) Feature plots, transcriptional trajectory plots, and pseudotime trajectory expression patterns of selected genes in Immature 1, Immature 2, and Mature clusters.
(F) Heatmap of differential genes in buffalo somatic cells (PTM, LC, M, EC, and NK) split by samples (PRE and PERI).
(G) tSNE plots showing gene relative expression levels of retinoic acid (up) and testosterone pathways (down).
SCs are the only somatic cells in seminiferous tubules that directly interact with germ cells to regulate spermatogenesis.9 To reveal SC differentiation, the SCs subsets (Immature and Mature SC, 5,219 cells) were re-clustered into two Immature SC clusters (Immature 1 and Immature 2) and one Mature SC cluster (Mature) (Figures 6B and S6A and Table S3). Immature 1 (AMH+/SOX9−), Immature 2 (AMH+/SOX9+), and Mature SC (SOX9+/AMH−) were marked by LGALS1 and MMP2,81,82GATA4 and TIMP2,39,83WT1 and CLDN11,84,85,86 respectively (Figure S6B). Strikingly, Immature 2 and Mature clusters were positive for SOX9, and IF staining showed that SOX9 was partially expressed in cells near the basement membrane of prepubertal and pubertal testes (Figure S6C).
Likewise, the pseudotime trajectory analysis proved that Immature 1 developed before Immature 2, which gradually differentiated into Mature SC (Figure 6C). In the dynamic gene expression pattern of SCs on the pseudotime developmental axis, genes such as EGR1, JUNB, JUN, and S100A13 showed elevated expression in the Immature 1 cluster and decreased expression with SC differentiation. Also, INHA and AMH broadly marked Immature SC, whereas expression was significantly higher in Immature 2 but negative for the Mature cluster, and genes with similar patterns included ENO1, ALDH1A1, CLU, and BCAM. Mature cluster specifically expressed FATE1, DEFB123, CLEC12B, CITED1, and TSPAN5 (Figures 6D and 6E). Notably, mitochondrial function was enhanced along with the pseudotime developmental differentiation of SCs (Figures S6D and S6E and Table S5).
Heatmap showing the expression patterns of prepubertal and pubertal somatic cell marker genes. (Figure 6F and Table S7). Afterward, the focus was only on the expression of several paracrine factors, cytokines, growth factors, and ligand-receptor genes which have been associated with spermatogenesis. For instance, some immune-related factors such as interleukin (IL) 1 (IL1A and IL1B) and CSF1 were specifically secreted by M cells in buffalo. AMH, inhibin (INHA and INHBB), and desert hedgehog signaling molecule (DHH) are produced by SCs; moreover, DHH and AMH are the essential downstream gene of SRY (sex determining region Y)-box9 (SOX9).87 As expected, DHH and INHBB were expressed specifically in Immature SC and Mature SC; AMH and INHA were only expressed in Immature SC. Remarkably, many cytokines and hormones require to bind with their receptors to function, and we still found expression patterns of some ligand-receptor signaling pathways, such as CLCL12-CXCR4, KITLG-KIT, and CGA-FSHR (Figure S6F). These findings further suggested that the somatic cells also form strong interactions to create a microenvironment for spermatogenesis.
Retinoic acid (RA) is essential for male spermatogonial development and spermatocyte meiosis in both humans and mice.54,88 Importantly, the membrane receptor STRA6, which is responsible for the intake of retinol, was specifically expressed in SCs (Immature 2) of buffalo testes. Similarly, ALDH1A2 was expressed from pachytene to diplotene spermatocytes in postnatal mouse testes.89In situ hybridization showed that the RA-degrading enzyme CYP26B1 was only expressed in PTM cells in mice,89 but its transcript was detected in both buffalo SCs (Immature 1) and PTM cells ALDH1A1 transcripts were found in immature SC (Immature 1 and Immature 2) and LC, and ALDH1A3 was found in SCs (Immature 1 and Mature), PTM, and LC, suggesting that buffalo LC and PTM cells participated in RA synthesis. As for RA receptors, RARA was predominantly expressed in SSC-1 (NANOS2+/GFRA1+), SCs (Immature 1, Immature 2, and Mature), and PTM cells, but was only weakly expressed in LC, EC, and M. RARB was more abundant in SPG subsets (SSC-1, SSC-2, and Diff.ing SPG) and pre.L primary spermatocytes. These findings suggested that SCs, PTM, LC, EC, and M are involved in the RA regulation of spermatogonial differentiation (NANOS2+/GFRA1+) and meiotic initiation in buffalo (Figures 6G and S6G).
Testicular endocrine function is accomplished by androgen biosynthesis by LCs.90 Testosterone is one of the major representative androgens regulating spermatogenesis in mammals by binding to the androgen receptor (AR).91,92 Here, steroidogenesis-related genes were specifically expressed in LC. AR was expressed in LC, PTM, and SC in buffalo testes but negative for germline cells, suggesting that testosterone promotes spermatogenesis by the interactions among LC, PTM, and SC (Figure 6G).
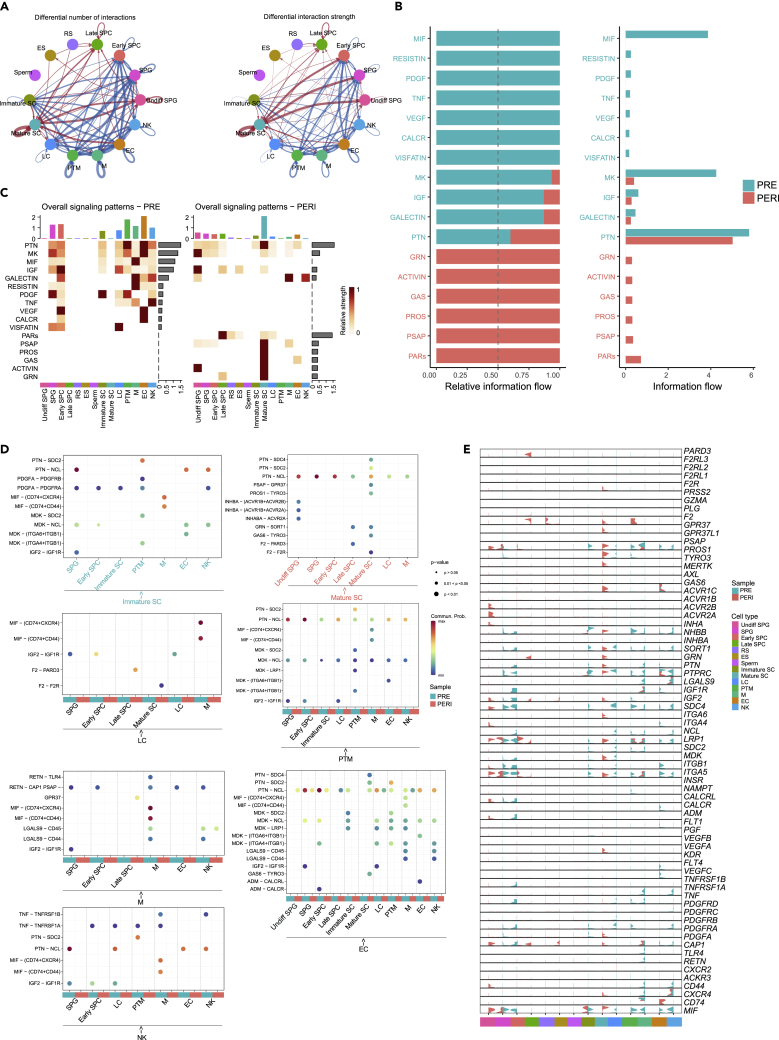
Testicular cell-cell communication
Significant differences were observed in the number and strength of interactions (secreted signaling) among different testicular cell populations (Figure 7A and Table S8), with prepubertal cells showing a higher overall strength of interactions than pubertal cells, as evidenced by interactions between somatic cells (Figures S7A and S7B). Comparing the overall information flow of each signaling pathway revealed 17 signaling pathways, with PRE- and PERI-derived cells sharing four signaling pathways (PTN, MK, IGF, and GALECTIN) (Figure 7B). Notably, Mature SC was placed at the center of testicular intercellular communication, occupying the strongest signaling sender and receiver of five signaling pathways (GRN, GAS, PROS, PSAP, and PTN). Meanwhile, in the ACTIVIN pathway that promotes spermatogenesis,93 cell-cell communication was only from Mature SC to Undiff SPG (Figures 7C, S7C, and S7D).
Figure 7.
Global testicular cell-cell communication network in buffalo
(A) Number of interactions and interaction strength among all testicular cell types.
(B) Overall information flow of each signaling pathway from PRE and PERI.
(C) Heatmap comparison of overall signaling in PRE- and PERI-derived testicular cells.
(D) Overview of ligand-receptor interactions (p-value < 0.05) of buffalo testicular cells. The size of the circle represents the p-value, with red representing the communication probability, whereas the color of the square represents the sample resource, with turquoise representing PRE and red representing PERI.
(E) Violin plots showing the expression of ligand-receptor pairs split by sample.
Furthermore, ligand-receptor pairs that function in the signaling pathway were examined, and somatic cells were found to interact with somatic/germ cells by paracrine/autocrine signaling (Figure 7D and Table S9). Especially, Undiff SPG majorly expressed receptors of the ACTIVIN (ACVR1B and ACVR2A) and PTN (NCL) pathway, whereas prepubertal spermatogonia were primarily represented by receptors of the IGF, MK, PDGF, PTN, and RESISTIN pathways—IGF1R, NCL, PDGFRA, NCL, and CAP1, respectively. Therefore, somatic cells might communicate with SSC by secreted signaling to maintain self-renewal or differentiation.
Simultaneously, prepubertal testicular somatic cell communication was predominantly clustered in the IGF, MIF, MK, PTN, GALECTIN, RESISTIN, CALCR, and TNF pathways, whereas pubertal cells were concentrated in PARs, GALECTIN, PSAP, GAS, and PTN pathways. PTPRC (CD45) was increased in abundance in M and NK cells of PERI, demonstrating that the GALECTIN pathway is involved in PTPRC+ cell proliferation, differentiation, angiogenesis (formed by EC expressing ligand LGALS9), and immune response during buffalo testis development94,95 (Figures 7E and S7E). Briefly, our data showed similarities and dissimilarities of microenvironments in prepubertal and pubertal spermatogenic cells, provided evidence for understanding cellular interactions during spermatogenesis, and highlighted the mechanisms of stem cell maintenance.
Discussion
In mammals, male fertility requires the coordination of endocrine and exocrine testicular functions (androgen secretion and spermatogenesis), both of which are directly controlled by the hypothalamic-pituitary-gonadal (HPG) axis.17,26,28,96 This study presents a comprehensive high-precision single-cell transcriptional cell atlas during spermatogenesis in buffalo, elucidating a gene expression roadmap and critical molecular markers for testicular cell development. In summary, scRNA-seq analysis was used to reveal sequentially distinct molecular events in spermatogenic cells and cell-cell communication mechanisms in the buffalo spermatogenic microenvironment, which can establish molecular foundations for the cultivation of germline stem cells in vitro, organoid culture, and agricultural superior livestock breeding.
Categorization of testicular germ and somatic cells
Spermatogenic cells were distinguished from microenvironment somatic cells according to DDX4- and VIM-positive expression32,97 (Figures 1E, S1C, and S1D). Adult SSCs in mice were reported to compose only 0.03% of total germ cells.98,99 Similarly, the undifferentiated SPG (buffalo) consisted of PERI only (42 cells), which expressed many identified markers such as pluripotency TFs (SALL4 and ETV5), ELAVL2, and PIWIL4.28,33,97,100,101 A recent spermatogenesis scRNA-seq study that failed to mark adult SSCs in sheep highlights the challenge of identifying SSCs in domestic animals.31 Notably, GO enrichment revealed that DEGs were primarily involved in gene expression, chromosome organization, and RNA processing, and these biological processes were enriched in SSC-related TFs (TCF3 and ZBTB16),29 ARID domain-containing proteins (KDM family) and epigenetic modifiers (DNMT3A),102 and RNA processing factors (LSM, RBM, and SNRP families). It showed that the buffalo undifferentiated SPG most likely contained a population of SSCs responsible for maintenance and self-renewal, which was consistent with previous findings in humans and mice.39,102
SPG (PRE: 558 cells; PERI: 26 cells) were labeled by genetic markers in spermatogonial differentiation such as NR6A1, FGFR3, and FMR1, which were also commonly highly expressed in spermatogonial cells of humans and mice.27,29,39 SPG was shown to be predominantly involved in chromosome organization, RNA splicing, and cell cycle processes, with genes promoting SSC self-renewal (UCHL1, UTF1, CHD1, FOXO1, and BMI1) and spermatogonial proliferation expansion signals (DMRT1, SOHLH1, KIT, TKTL1, and ESX1).36,103,104,105 These observations suggested that SPG was a mixed cluster of spermatogonia with multiple differentiation states and required further re-clustering for identification.
For the spermatocytes, Early SPC was marked by TEX12, SMCHD1, and PRSS50.27,65,106 Late SPC was labeled by synaptonemal complex central element protein 2 (SYCE2), DAZ gene family member (BOLL), and mutL homolog 1 (MLH1).55,107 In cynomolgus macaque testes, the single-cell analysis showed that IZUMO4 is expressed in round spermatids,97 and TEX29 is expressed in the early spermatid stage.26 Moreover, studies have shown that ODF1, ODF2, ODF3, OAZ3, and AKAP4 are sperm tail-related genes, supporting the initial classification of haploid spermatids (RS, ES, Sperm).77,108 In summary, buffalo prepubertal and pubertal testicular germ cells were defined at the single-cell level (multiple differentiation states existed within a cluster in the initial classification). Nonetheless, genetic markers and expression profiles specifically for seven germ cell stages were found to characterize spermatogenesis (Table S1).
Microenvironment somatic cells (VIM+) secrete multiple cytokines, growth factors, hormones, and paracrine factors participating in the regional regulatory mechanism of spermatogenesis, and spermatogenesis depends on the maturity of somatic cells.5,96,109 The differential expression of AMH and CLDN11 in the original classification categorized SCs into two distinct cell clusters, despite the lack of a suitable marker to represent the overall status of SCs (Immature SC and Mature SC).109 The prepubertal male mammalian SC is predominantly characterized by anti-Müllerian hormone (AMH), which lacks androgen receptor expression, and lacks the blood-testis barrier (BTB) as well.110 The appearance of CLDN11 transcripts signals that SCs entered the mature stage, which is characterized by loss of proliferation activity, BTB formation, and providing nutritional and structural support to germ cells at all developmental stages of spermatogenesis.11,111KITLG (KIT ligand, stem cell factor) plays a distinct role in regulating the fate of mouse spermatogonia.112,113 The maturation roadmap of SCs was demonstrated by pseudotime trajectory analysis, and the time of KITLG expression initiation in buffalo SCs was found to be the same as SOX9, indicating a potential regulatory relationship that requires further verification. Specifically, SHBG, rather than SOX9, WT1, or GATA4, was a more comprehensive molecular marker for prepubertal and pubertal SCs in buffalo. The buffalo PTM expressed DHH receptor (PTCH1), PDGF receptor (PDGFRA), and AR, indicating that it interacts with other somatic cells to mediate spermatogenesis by participating in hedgehog, PDGF, and androgen signaling pathways. These results corroborate the findings of a previous study on humans.67
In the interstitial compartment of buffalo testis, LC specifically expressed testosterone biosynthesis-related genes (STAR, HSD3B1, CYP17A1, CYP11A1, and NR5A1), which match those observed in earlier studies.114,115 High expression levels of AR in buffalo SC, PTM, and LC indicated that androgen signaling promotes sex differentiation, gonadal development, and spermatogenesis by the cooperative work of SC, PTM, and LC, with androgen signaling being subsequently transmitted to germ cells by SC.116 Furthermore, weak AR expression was observed in buffalo EC (PECAM+/CLDN5+/ECSCR+), which was consistent with previous observations that testosterone interacts with EC.117 In addition, two classes of PTPRC+ cells—M (CSF1R+/CD74+/MAFB+) and NK (PTPRC+/NKG7+/KLRF1+/CD94+)—were identified. M is a “niche” in the interstitial compartment of prepubertal and adult mice SSCs, interacting with LCs to promote steroid synthesis, expressing the SPG proliferation and differentiation-inducing molecule CSF1, and still being involved in the RA biosynthetic pathway.118 IF staining of adult mouse testis revealed that CSF1 was expressed in LC, PTM, and M, whereas CSF1R was expressed in spermatogonial cells.118,119 Conversely, we found that CSF1 and CSF1R were specifically expressed in M in the present study. Therefore, CSF1 is unlikely to influence the proliferation or self-renewal of SSC by binding to CSF1R in buffalo.
Spermatogenic cell development
Re-clustering analysis was performed on the original cluster to identify highly variable genes for cell fate transition between neighboring spermatogenic cells. SSC-1 (NANOS2+/GFRA1+) differentiated the earliest in the cell trajectory analysis of SPG subsets, expressing receptors (GFRA1 and RET), and transcription factors (ETV5) associated with the GDNF signaling pathway. Thus, the GDNF signaling pathway is required to maintain SSC self-renewal in buffalo, similar to mice, rats, and humans.33,101,120 SSC-2 (PIWIL4+/FOXO1+) specifically expresses PRC1, BMI1, and TAF4B. In particular, ZBTB16 and TAF4B were found to be under differential signaling control in prepubertal human spermatogonia and mouse SSC, which might prevent differentiation without being directly involved in self-renewal.33 Moreover, human SSEA4+ SSCs had high transcript levels of PRC1 and BMI1.39DMRT1 is expressed in mouse SCs and spermatogonia (all stages) and promotes spermatogonial differentiation by activating SOHLH1 and inhibits meiotic initiation by specific interruption of STRA8 transcription (RA pathway).121 Similarly, the scRNA-seq profile revealed that DMRT1 and SOHLH1 simultaneously marked Diff.ing SPG, and STRA8 initiated weak expression in Diff.ed SPG (IRF1+/KLF4+), peaking in L primary spermatocytes.
Compared with other mammals, buffalo shared some conserved SSCs, such as UCHL1, UTF1, ZBTB16, DAZL, SOX4, NANOS2, GFRA1, PAX7, SALL4, ITGA6, RET, PIWIL4, FOXO1, MORC1, TAF4B, NR6A1, and FGFR3.27,97 Nevertheless, few identified SSC markers were not detected in buffalo, including NGN3, PLPPR3, TSPAN33, GPR125, SSEA4, MAGEA4, DPPA4, BCL6B, and FOXO3.26,29,122,123 Of interest, RBM47 and SLPI were shown to be specifically expressed in mouse primordial germ cells (PGCs).124 In contrast, NANOS2, which is similar to SLPI, was particularly expressed in SSC-1 in the present study, whereas RBM47 was specifically expressed in SSC-2. The earlier systematic investigation of spermatogonial cell subtypes in the domestic animal field came from a single-cell examination of a 150-day-old Guanzhong pig.32 In short, this detailed characterization of the single-cell transcriptional atlas of buffalo spermatogonia can contribute to theoretical models of spermatogonial development in livestock.
Spermatogonial mitotic division and differentiation into pre.L spermatocytes all occur in the basal compartment, whereas other spermatocytes and post-meiotic spermatids are separated in the adluminal compartment.125 Increasing advancements in scRNA-seq allowed for the classification of spermatocytes into more specialized stages, and a recent study reported 12 phases of spermatocytes during mouse spermatogenesis and listed the following stages: G1, ePL, mPL, and lPL (preleptotene stage), L, Z, eP, mP, lP, D, MI, and MII stage.38 Likewise, our data showed the following spermatocyte subtypes: pre.L (REC8+/SMC3+), L (SYCP3+/ZCWPW1+), Z (SYCP1+/TEX101+), eP (PIWIL1+/CCDC112+), mP (HORMAD2+/REC114+), lP (NME5+/NME8+), D (AURKA+/AURKB+) and MI-II (SUN5+/FZR1+) (Figures 3 and S3). Multiple interspecies conserved spermatocyte phase-specific markers were identified using scRNA-seq, including ZCWPW1, TEX101, PIWIL1/CCDC112, and AURKA/SPATA16, which broadly marked L, Z, P, and D primary spermatocytes, respectively, in human, mouse, monkey, pig, sheep, and buffalo.64 Several unconserved genetic markers were observed in buffalo, presumably because the cycle of seminiferous epithelium differs among animals. Human, cynomolgus macaque, and mouse showed DMRTB1 expression in differentiated spermatogonia, whereas DMRTB1 expression was detected in pre.L spermatocytes in buffalo.97HERC5 was highly expressed in human late primary spermatocytes,27 whereas it was detected in the buffalo pre.L cluster. Similarly, SYCP3 was expressed in mouse Z spermatocytes,64 but was expressed in the buffalo L stage. To form species-specific maturing spermatozoa, round spermatids undergo a series of morphological transformations and chromatin remodeling.77 Dynamic transcriptional signatures of buffalo spermiogenesis were revealed in the present study, and the major stage of histone-to-protamine chromatin remodeling was found to occur at the ES2 stage.
To improve understanding of male germ cells, detailed information on TMHMM-predicted proteins is provided in Table S6. The surface marker ST3GAL2 has been identified as specific to prepubertal SSC in buffalo. Several other surface genes were selected as candidate markers for the buffalo spermatogenic lineage, such as ITGA6, KNTC1, RHOA, and ITGB5 (Figure 5C). Furthermore, it would be interesting to explore the process of transforming germ cell fate during puberty based on cell surface markers isolating single-cell-defined testicular germ cells in vitro. Hence, single-cell transcriptome analysis was used to define the dynamic transformation of spermatogenesis in buffalo, thereby contributing to deepening our understanding of the regulatory basis of male germ cell development.
Global testicular cell-cell communication network in buffalo
SSCs undergo spermatogenesis in a microenvironment created by testicular somatic cells, a complex process that involves self-renewal and differentiation into spermatozoa.21 Nevertheless, the mechanism of mammalian testicular cell communication in the physiological state is poorly defined. In addition to examining the expression distribution of paracrine factors (e.g., DHH, NRG1, POMC, and INSL3), cytokines (e.g., IL1A, IL1B, CSF1, and INHBB), growth factors (e.g., IGF2, TGFβs, PDGFs, HGF, FGF1, VEGFA, and NGF), CXCL12/CXCR4 signaling, KITLG/KIT signaling, RA biosynthetic pathway, and testosterone pathway (Figures 6G, S6F, and S6G), global paracrine/autocrine signaling interactions were observed between prepubertal and pubertal buffalo testicular cells by CellChat analysis (Figures 7 and S7). Of note, the limitation of CellChat is that high expression of ligands and receptors at the RNA level does not necessarily correlate with actual cell-cell signaling because cell signaling occurs at the protein level.126 Nevertheless, CellChat analysis is still a good indicator for detecting potential signaling pathways in intercellular communication.
During the information flow from niche cells to sender cells, the IGF pathway was detected in the interaction pattern of prepubertal somatic cells (Immature SC, LC, PTM, M, EC, and NK). Of interest, germ cells (Undiff SPG, SPG, and Early SPC) also expressed IGF2, and the IGF pathway was evident in the flow of information from germ cells to niche cells. Thus, the data further supported that IGF is the most critical positive regulator of immature SC proliferation, promoting the proliferation, differentiation, and development of PTM, LC, EC, M, and NK cells.127 Meanwhile, the IGF pathway communicates bidirectionally between germ and niche cells. The PDGF pathway enhances steroid production in LC, promotes PTM contractions, and stimulates germ cell differentiation and proliferation.128,129 Similarly, PDGFA showed specific expression in buffalo SCs, and PDGF receptors (PDGFRA and PDGFRB) were predominantly expressed in Immature SC and PTM, and slightly expressed in LC. The buffalo PDGF signaling pathway was regulated by PDGF/PDGFR during testis development by the interaction of SC, PTM, and LC. Furthermore, although ACTIVIN receptors (ACVR2A and ACVR1B) were expressed in both prepubertal and pubertal-derived SPGs in buffalo, the ACTIVIN communication signal was only detected in adult buffalo when the sender cell is adult SC and the receiver cell is mature SC. Hence, the data further confirmed that the ACTIVIN pathway promotes the expansion of spermatogonia in adult mammals (including humans and mice).130
M-specific secreted immune-related cytokines, IL1 (IL1A and IL1B) in prepubertal and pubertal buffalo, whereas IL1A is secreted by SC in humans and rats, and has a direct effect on germ cell differentiation and testicular steroid hormone production.131,132 The GALECTIN pathway was found in buffalo EC and PTPRC+ immune cells (M and NK), confirming a crucial role in angiogenesis and the maintenance of an immunologically privileged environment in the testis.94 Importantly, TNF is a pro-inflammatory cytokine produced by SC and LC, which increases the secretion of transferrin in SC.133 However, only TNF and its receptor TNFRSF1B were detected in buffalo M and NK cells, whereas TNFRSF1A was present mainly in PTM, LC, M, immature SC, and EC. The results indicate that TNF was mainly expressed in immune cells of buffalo testis and functioned as a signaling cascade by cellular communication.
Several ligands share the same receptor, demonstrating that paracrine factors most likely crosstalk between multiple signals. For instance, MDK (MK pathway) and PTN (PTN pathway) share three receptors (NCL, SDC2, and SDC4); F2 and PLG (PARs pathway) share two receptors F2R and PARD3; MIF (MIF pathway) and LGALS9 (GALECTIN pathway) share receptor CD44; and GAS6 (GAS pathway) and PROS1 (PROS pathway) share receptor TYRO3. Our data further supported that the MK/PTN family promotes angiogenesis134 and found that NCL was also expressed in germ cells (Undiff SPG, SPG, Early SPC, and Late SPC), indicating that the MK/PTN family was also involved in constructing the “microenvironment” of buffalo spermatogenesis. The role of angiogenic signaling in maintaining self-renewal in SSCs is yet to be evaluated functionally, which is worthy of further investigation. In short, the paracrine/autocrine signaling interactions in prepubertal and pubertal buffalo testicular cells that create distinct spermatogenic microenvironments were elucidated, yielding a potential ligand-receptor list for establishing germline stem cells and organoid culture in vitro.
This study establishes a single-cell transcriptional atlas of prepubertal and pubertal healthy buffalo testes, uncovering dynamic and complex processes in the sequential cell fate transition from SSCs to spermatozoa and developmental trajectory of microenvironment somatic cells during spermatogenesis. Also, this study presents a global buffalo testicular cell-cell communication network that maintains regular spermatogenesis under physiological conditions. Overall, our dataset provides comprehensive, distinctive, and valuable information on buffalo spermatogenesis and contributes to the molecular basis for studies of the male germline from livestock models in vitro.
Limitations of the study
The present study focused on scRNA-seq analysis and IF staining of key markers in the testis. However, further experimental validation is required for the expression and localization of novel markers, because commercial antibodies do not easily function in buffalo. Only the single-cell data of prepubertal and pubertal buffalo testes was compared, which does not entirely reveal the gradual molecular process of the testis from prepuberty to puberty. Further studies using sequencing data from multiple continuous development samples may reveal additional details. Besides, integrating multi-species single-cell data to investigate spermatogenesis contributes to enhancing our understanding of male fertility and spermatogenesis. Although two SSC clusters were identified in the present study without performing functional assays, further studies involving the isolation of SSC xenografts by cell surface markers can improve the current findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal [EPR4118] to PGP9.5; dilution: 1:1000 | Abcam | Cat# ab108986; RRID:AB_10891773 |
| Rabbit polyclonal to SCP3; dilution: 1:1000 | Abcam | Cat# ab15093; RRID:AB_301639 |
| Rabbit monoclonal [EPR3776] to Vimentin; dilution: 1:1000 | Abcam | Cat# ab92547; RRID:AB_10562134 |
| Rabbit monoclonal [EPR14335-78] to SOX9; dilution: 1:1000 | Abcam | Cat# ab185966; RRID:AB_2728660 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488); dilution: 1:1000 | Abcam | Cat# ab150077; RRID:AB_2630356 |
| Biological samples | ||
| Prepubertal (PRE) buffalo testicular tissue | 3 months old | N/A |
| Pubertal (PERI) buffalo testicular tissue | 24 months old | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| type IV collagenase | Worthington Biochemical Corporation | Cat# LS004188 |
| 0.25% Trypsin-EDTA | Sangon Biotech | Cat# E607002-0100 |
| Deoxyribonuclease I (DNase I) | Sigma-Aldrich | Cat# D4527 |
| DMEM | ThermoFisher | Cat# C11995500BT |
| Fetal Bovine Serum (FBS) | ThermoFisher | Cat# 16140071 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A9647 |
| 0.4% Trypan Blue staining solution | Sigma-Aldrich | Cat# T8154 |
| Critical commercial assays | ||
| Metenyi Dead Cell Removal Kit | Metenyi Biotec | Cat# 130-090-101 |
| 10x Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 | 10x Genomics | Cat# 1000147 |
| Deposited data | ||
| Single-cell RNA-seq data | This paper | GEO: GSE190477 |
| Software and algorithms | ||
| Cell Ranger v 3.1.0 | 10x Genomics | RRID:SCR_017344 |
| Seurat R package v 3.1.1 | 135 | RRID:SCR_016341 |
| Harmony R package v 0.1.0 | 136 | RRID:SCR_022206 |
| Monocle R package v 2.22.0 | 137 | RRID:SCR_016339 |
| CellChat R package v 1.1.3 | 126 | RRID:SCR_021946 |
| TMHMM Server v 2.0 | 138 | RRID:SCR_014935 |
| ggplot2 R package v 3.3.6 | https://cran.r-project.org/web/packages/ggplot2/index.html | RRID:SCR_014601 |
| GO (DAVID) | 139,140 | RRID:SCR_001881 |
| R version 4.1.3 | https://cran.r-project.org/ | RRID:SCR_001905 |
| Other | ||
| 70 μm cell strainer | NEST | Cat# 258368 |
| 40 μm cell strainer | NEST | Cat# 258369 |
| Illumina NovaSeq 6000 | Illumina | RRID:SCR_016387 |
| UOA_WB_1 buffalo genome assembly | NCBI | RefSeq assembly accession: GCA_003121395.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Qiang Fu (gxfuq@gxu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All strategies related to testicular experiments were conducted under CIOMS protocols approved by the Animal Experimentation Ethics Committee (AEEC) of Guangxi University, Nanning, China. The testes of a prepubertal (3-month-old) and a pubertal (24-month-old) normal healthy male buffalo were obtained from the Guangxi Key Laboratory of Buffalo Genetics, Reproduction and Breeding, and from a local slaughterhouse, respectively.
Method details
Buffalo testes sample preparation
Testes from a 3-month-old (prepuberty) and a 24-month-old (puberty) healthy buffalo were washed three times with pre-cooled DPBS to remove the testicular white membrane and epithelium, then instrumented to disrupt testicular tissue and cut into fractions. Single testicular cells were isolated with a two-step enzymatic digestion protocol described previously.29 Briefly, the cells were incubated with 1 mg/mL type IV collagenase for 8 minat 37°C with a shaker (100 rpm) and washed twice with pre-cooled DPBS. Subsequently, 0.25% Trypsin-EDTA and 0.25 mg/mL DNase I were added and incubated in an incubator at 37°C for 10 min with 15 gentle pulses every 3 min with a blunt tip. Afterward, the digestion was stopped by adding an equal volume of DMEM medium containing 10% FBS. Single testicular cells were obtained by filtering through 70 μm and 40 μm cell strainer, centrifuged at 800 g for 5 min, and washed with pre-cooled DPBS. Then, cells were counted with a hemocytometer and resuspended in phosphate-buffered saline adding 0.4% bovine serum albumin at an adjusted concentration of ∼1000 cells/μL for scRNA-seq loading. Cell viability was 92% for prepubertal samples and 86% for pubertal samples using the Metenyi Dead Cell Removal Kit to remove dead cells and cell debris.
10x genome library preparation and sequencing
Single-cell suspensions were obtained in tiny volumes and an equal volume of 0.4% trypan blue staining solution was added for cell quality control, and the available cell concentration was adjusted to the desired concentration (1000–2000 cells/μL). Afterward, the cell suspension was loaded onto a 10x Genomics GemCode Single-cell instrument, and cell capture and library preparation were performed according to the Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 instructions. In addition, high-throughput sequencing of libraries was performed using the PE150 sequencing mode of the Illumina sequencing platform (NovaSeq 6000) at Guangzhou Genedenovo Biotechnology Co., Ltd.
Genome alignment and gene expression quantification
10x Genomics Cell Ranger software (v3.1.0) was used to convert the raw BCL files to FASTQ files, as well as for alignment, and counts quantification. Later, reads with low-quality barcodes and unique molecular identifiers (UMIs) were filtered out and then mapped to the reference genome (UOA_WB_1). In particular, reads uniquely mapped to the transcriptome and intersecting an exon at least 50% were considered for UMI counting. Before quantification, the UMI sequences were corrected for sequencing errors, and valid barcodes were identified based on the EmptyDrops method.141 The cell-by-gene matrices were produced by UMI counting and cell barcode calling.
Cell clustering and annotation
The cell-by-gene matrices for each sample were individually imported to Seurat (v3.1.1)135 for downstream analysis. Primarily, the filtering conditions were as follows: the percentage of the mitochondrial gene was less than 10%, the number of genes that could be detected per cell was 200, and each gene was expressed in a minimum of three cells. Moreover, data was normalized using the LogNormalize method, and the FindVariableFeatures function was used to identify highly variable genes. Samples were integrating using Harmony136 to remove batch effects and select the first 30 dimensions for analysis. Eventually, tSNE dimensionality reduction was performed using the Seurat function RunTSNE. The Seurat function FindAllMarkers (test used “wilcox” and “min.pct” set to 0.25) was used to calculate marker genes for each cell cluster (Tables S1 and S3). Meiosis-associated and spermiogenesis-related genes were calculated by the Seurat function FindMarkers (min.pct = 0.65, only test genes detected in a minimum fraction of 65% cells in either of the two populations), and the remaining parameters use default settings (Figures 3B, 4B and Table S4). GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using DAVID (https://david.ncifcrf.gov/home.jsp).139,140
Single-cell trajectory analysis
The single-cell trajectory was analyzed using the matrix of cells and gene expression by the Monocle package in R software (v2.22.0)137 by the documentation provided. After annotating cell types in Seurat and constructing cell developmental trajectories according to unsupervised analysis methods (DEGs: mean_expression ≥0.1 and num_cells_expressed >10), the reverse graph embedding “DDRTree” algorithm was used to dimensionalize the data after downscaling and aligning cells in pseudotime. The cell trajectory plots and heatmap visualization were performed based on the feature values of pseudotime, cell type, and specified genes. All line plots were constructed according to the “loess model” fitted curve in the ggplot2 package, with the horizontal coordinates plotted according to the cell type ordered by the pseudotime value.
Prediction of transmembrane (TM) helices in proteins
TMHMM is an HMM-based (Hidden Markov Model) TM protein prediction tool.138 The protein sequence was extracted in the FASTA format and calculated by default. The output is presented as following: The length of the amino acid sequence of the protein encoded by the gene is represented by length. ExpAA shows the predicted number of amino acids in the TM helix, and if this score is higher than 18, the protein is the most likely TM or contains a signal peptide structure. First60 refers to the expectation value of amino acids predicted to be TM helices in the first 60 amino acids in the N-terminal protein, predicting the likelihood of the TM helix structure at the N-terminal to be the signal peptide. PredHel predicts the number of TM helices by N-best. The N-best topology determines the final TM result, with “i” indicating that the protein is associated with the inner membrane and “o” indicating that the protein is associated with the extra membrane.
Comparison analysis of PRE- and PERI-derived testicular cells using CellChat
After homologous transformation of buffalo genes to human genes and subsequent analysis, the testicular cell-cell communication network was calculated using the CellChat package in R software (v1.1.3).126 The expression matrices after standardization of PRE- and PERI-derived cells were extracted from the Seurat objects and then merged by upgrading the CellChat objects after their creation, followed by comparative analysis.
Hematoxylin and eosin staining
The prepared paraffin slices of prepubertal and pubertal testes were deparaffinized, transferred to distilled water for 5 min, stained in hematoxylin for 15 min, and then flushed under flowing water for 3 s in 1% hydrochloric acid solution. Subsequently, the slices were rinsed for 20 min under flowing water, followed by sequential immersion in an alcohol gradient of 70%, 80%, and 90% for 5 min per concentration, and then immersed into 1% eosin staining solution for 10 min. The slices were sequentially dehydrated and made transparent in 95% alcohol (5 min), 100% alcohol I (5 min), 100% alcohol II (5 min), dimethylbenzene : alcohol = 1 : 1 (10 min), dimethylbenzene I (10 min), dimethylbenzene II (10 min), air-dried, and sealed with coverslips until microscopic examination and image capture.
Immunostaining of buffalo testicular tissue
To improve the permeability of the cell membrane, paraffin sections were deparaffinized, and antigen retrieved (pH 6.0 sodium citrate buffer) and incubated in a 1% Triton X-100-PBST solution for more than 30 min. Thereafter, a 5% BSA blocking solution was poured dropwise onto the sections and sealed at room temperature for 1 h. After incubating each slide overnight at 4°C with the appropriate proportion of diluted primary antibody poured dropwise, the appropriate dilution of fluorescent secondary antibody was added and incubated for 1 hat room temperature, avoiding light, DAPI stained (nuclei), blocked, and observed under a fluorescence microscope.
Quantification and statistical analysis
The statistical details of the scRNA-seq data can be found in the figure legends. In Figures 1F, 3B, 4B, S2B–S4B, and S6A and Tables S1, S3, and S4, Wilcoxon rank sum test was performed. The CellChat function netVisual_bubble was used to visualize ligand-receptor interactions (Figure 7D and Table S9). Detailed information on functional enrichment was included in Figures 1G, 5A, and S6E and Tables S2 and S5. The significant level described above was defined as p-value <0.05. In Figures 2E, 3F, 4E, 6D, and S4E, the pseudotime-dependent genes (q-value <0.01) were calculated by trajectory analysis and were for visualization. R (v4.1.3) was used for data processing and visualization.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 32160789).
Author contributions
Conceptualization: L.F.H. and J.J.Z.
Methodology: L.F.H., J.J.Z., and X.C.H.
Investigation: L.F.H., P.F.Z., W.H.Y., R.F.L., and Q.Q.S.
Visualization: L.F.H., J.J.Z., and P.F.Z.
Funding acquisition: M.Z.
Project administration: M.Z.
Supervision: M.Z., Q.F., and Y.Q.L.
Writing – original draft: L.F.H.
Writing – review and editing: M.Z. and Q.F.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105733.
Contributor Information
Yangqing Lu, Email: lyq@gxu.edu.cn.
Ming Zhang, Email: mingzhang@gxu.edu.cn.
Qiang Fu, Email: gxfuq@gxu.edu.cn.
Supplemental information
Data and code availability
-
•
All sequencing data in this paper have been deposited at GEO (GSE190477) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Perera B.M.A.O. Reproductive cycles of buffalo. Anim. Reprod. Sci. 2011;124:194–199. doi: 10.1016/j.anireprosci.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Michelizzi V.N., Dodson M.V., Pan Z., Amaral M.E.J., Michal J.J., McLean D.J., Womack J.E., Jiang Z. Water buffalo genome science comes of age. Int. J. Biol. Sci. 2010;6:333–349. doi: 10.7150/ijbs.6.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Occhio M.J., Ghuman S.S., Neglia G., Della Valle G., Baruselli P.S., Zicarelli L., Visintin J.A., Sarkar M., Campanile G. Exogenous and endogenous factors in seasonality of reproduction in buffalo: a review. Theriogenology. 2020;150:186–192. doi: 10.1016/j.theriogenology.2020.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Khatun M., Kaur S., Kanchan, Mukhopadhyay C.S. Subfertility problems leading to disposal of breeding bulls. Asian-Australas. J. Anim. Sci. 2013;26:303–308. doi: 10.5713/ajas.2012.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oatley J.M., Brinster R.L. The germline stem cell niche unit in mammalian testes. Physiol. Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGowan M., Holland M.K., Boe-Hansen G. Review: ontology and endocrinology of the reproductive system of bulls from fetus to maturity. Animal. 2018;12:s19–s26. doi: 10.1017/S1751731118000460. [DOI] [PubMed] [Google Scholar]
- 7.Law N.C., Oatley M.J., Oatley J.M. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat. Commun. 2019;10:2787. doi: 10.1038/s41467-019-10596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staub C., Johnson L. Review: spermatogenesis in the bull. Animal. 2018;12:s27–s35. doi: 10.1017/S1751731118000435. [DOI] [PubMed] [Google Scholar]
- 9.Griswold M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 10.Bhang D.H., Kim B.-J., Kim B.G., Schadler K., Baek K.-H., Kim Y.H., Hsiao W., Ding B.-S., Rafii S., Weiss M.J., et al. Testicular endothelial cells are a critical population in the germline stem cell niche. Nat. Commun. 2018;9:4379. doi: 10.1038/s41467-018-06881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarulli G.A., Stanton P.G., Meachem S.J. Is the adult sertoli cell terminally differentiated? Biol. Reprod. 2012;87:13. doi: 10.1095/biolreprod.111.095091. 1-11. [DOI] [PubMed] [Google Scholar]
- 12.Chen S.R., Liu Y.X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction. 2015;149:R159–R167. doi: 10.1530/REP-14-0481. [DOI] [PubMed] [Google Scholar]
- 13.Tan K., Song H.-W., Wilkinson M.F. Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development. 2020;147:dev183251. doi: 10.1242/dev.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhushan S., Meinhardt A. The macrophages in testis function. J. Reprod. Immunol. 2017;119:107–112. doi: 10.1016/j.jri.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Barrio R., de Luis D., Alonso M., Lamas A., Moreno J.C. Induction of puberty with human chorionic gonadotropin and follicle-stimulating hormone in adolescent males with hypogonadotropic hypogonadism. Fertil. Steril. 1999;71:244–248. doi: 10.1016/s0015-0282(98)00450-6. [DOI] [PubMed] [Google Scholar]
- 16.Skinner M.K. Cell-cell interactions in the testis. Endocr. Rev. 1991;12:45–77. doi: 10.1210/edrv-12-1-45. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings N., Evans A.C.O., Chandolia R.K., Bagu E.T. Sexual maturation in the. Bull. Reprod. Domest. Anim. 2008;43:295–301. doi: 10.1111/j.1439-0531.2008.01177.x. [DOI] [PubMed] [Google Scholar]
- 18.Neto F.T.L., Bach P.V., Najari B.B., Li P.S., Goldstein M. Spermatogenesis in humans and its affecting factors. Semin. Cell Dev. Biol. 2016;59:10–26. doi: 10.1016/j.semcdb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 19.De Rooij D.G. The spermatogonial stem cell niche. Microsc. Res. Tech. 2009;72:580–585. doi: 10.1002/jemt.20699. [DOI] [PubMed] [Google Scholar]
- 20.Krausz C., Riera-Escamilla A. Genetics of male infertility. Nat. Rev. Urol. 2018;15:369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S., Wistuba J., Pock T., Schlatt S., Neuhaus N. Spermatogonial stem cells: updates from specification to clinical relevance. Hum. Reprod. Update. 2019;25:275–297. doi: 10.1093/humupd/dmz006. [DOI] [PubMed] [Google Scholar]
- 22.de Kretser D.M., Loveland K.L., Meinhardt A., Simorangkir D., Wreford N. Hum. Reprod. 1998;13:1–8. doi: 10.1093/humrep/13.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Navin N.E. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., He Y., Zhang Q., Ren X., Zhang Z. Direct comparative analyses of 10X Genomics Chromium and smart-seq2. Dev. Reprod. Biol. 2021;19:253–266. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J., Sosa E., Chitiashvili T., Nie X., Rojas E.J., Oliver E., DonorConnect. Plath K., Hotaling J.M., Stukenborg J.B., et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell. 2021;28:764–778.e4. doi: 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Liu X., Chang G., Chen Y., An G., Yan L., Gao S., Xu Y., Cui Y., Dong J., et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23:599–614.e4. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Guo J., Grow E.J., Mlcochova H., Maher G.J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L., et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J., Nie X., Giebler M., Mlcochova H., Wang Y., Grow E.J., DonorConnect, Kim R., Tharmalingam M., Matilionyte G., et al. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell. 2020;26:262–276.e4. doi: 10.1016/j.stem.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sohni A., Tan K., Song H.-W., Burow D., de Rooij D.G., Laurent L., Hsieh T.-C., Rabah R., Hammoud S.S., Vicini E., Wilkinson M.F. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 2019;26:1501–1517.e4. doi: 10.1016/j.celrep.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shami A.N., Zheng X., Munyoki S.K., Ma Q., Manske G.L., Green C.D., Sukhwani M., Orwig K.E., Li J.Z., Hammoud S.S. Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Dev. Cell. 2020;54:529–547.e12. doi: 10.1016/j.devcel.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Ma J., Wan Z., Wang Q., Wang Z., Zhao J., Wang F., Zhang Y. Characterization of sheep spermatogenesis through single-cell RNA sequencing. Faseb. J. 2021;35 doi: 10.1096/fj.202001035RRR. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Li F., Lei P., Guo M., Liu R., Wang L., Yu T., Lv Y., Zhang T., Zeng W., et al. Single-cell RNA-sequencing reveals the dynamic process and novel markers in porcine spermatogenesis. J. Anim. Sci. Biotechnol. 2021;12:122. doi: 10.1186/s40104-021-00638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X., Schmidt J.A., Avarbock M.R., Tobias J.W., Carlson C.A., Kolon T.F., Ginsberg J.P., Brinster R.L. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc. Natl. Acad. Sci. USA. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukassen S., Bosch E., Ekici A.B., Winterpacht A. Single-cell RNA sequencing of adult mouse testes. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grive K.J., Hu Y., Shu E., Grimson A., Elemento O., Grenier J.K., Cohen P.E. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L., Yao C., Xing X., Jing T., Li P., Zhu Z., Yang C., Zhai J., Tian R., Chen H., et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 2020;11:5683. doi: 10.1038/s41467-020-19414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green C.D., Ma Q., Manske G.L., Shami A.N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S.J., Moore B.B., et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell. 2018;46:651–667.e10. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., Wang Q., Xie N., Hua R., Liu M., et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28:879–896. doi: 10.1038/s41422-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J., Grow E.J., Yi C., Mlcochova H., Maher G.J., Lindskog C., Murphy P.J., Wike C.L., Carrell D.T., Goriely A., et al. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell. 2017;21:533–546.e6. doi: 10.1016/j.stem.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrobel K.H. Prespermatogenesis and spermatogoniogenesis in the bovine testis. Anat. Embryol. 2000;202:209–222. doi: 10.1007/s004290000111. [DOI] [PubMed] [Google Scholar]
- 41.Mahla R.S., Reddy N., Goel S. Spermatogonial stem cells (SSCs) in buffalo (Bubalus bubalis) testis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida S. From cyst to tubule: innovations in vertebrate spermatogenesis. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:119–131. doi: 10.1002/wdev.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boitani C., Di Persio S., Esposito V., Vicini E. Spermatogonial cells: mouse, monkey and man comparison. Semin. Cell Dev. Biol. 2016;59:79–88. doi: 10.1016/j.semcdb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Lord T., Oatley J.M. Functional assessment of spermatogonial stem cell purity in experimental cell populations. Stem Cell Res. 2018;29:129–133. doi: 10.1016/j.scr.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giassetti M.I., Ciccarelli M., Oatley J.M. Spermatogonial stem cell transplantation: insights and outlook for domestic animals. Annu. Rev. Anim. Biosci. 2019;7:385–401. doi: 10.1146/annurev-animal-020518-115239. [DOI] [PubMed] [Google Scholar]
- 46.Fahrenkrug S.C., Blake A., Carlson D.F., Doran T., Van Eenennaam A., Faber D., Galli C., Gao Q., Hackett P.B., Li N., et al. Precision genetics for complex objectives in animal agriculture. J. Anim. Sci. 2010;88:2530–2539. doi: 10.2527/jas.2010-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.González R., Dobrinski I. Beyond the mouse monopoly: studying the male germ line in domestic animal models. ILAR J. 2015;56:83–98. doi: 10.1093/ilar/ilv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun R.E. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 49.Goel S., Reddy N., Mandal S., Fujihara M., Kim S.-M., Imai H. Spermatogonia-specific proteins expressed in prepubertal buffalo (Bubalus bubalis) testis and their utilization for isolation and in vitro cultivation of spermatogonia. Theriogenology. 2010;74:1221–1232. doi: 10.1016/j.theriogenology.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 50.Mikedis M.M., Fan Y., Nicholls P.K., Endo T., Jackson E.K., Cobb S.A., de Rooij D.G., Page D.C. DAZL mediates a broad translational program regulating expansion and differentiation of spermatogonial progenitors. Elife. 2020;9 doi: 10.7554/eLife.56523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian H., Cao Y.-X., Zhang X.-S., Liao W.-P., Yi Y.-H., Lian J., Liu L., Huang H.-L., Liu W.-J., Yin M.-M., et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis. 2013;4:e617. doi: 10.1038/cddis.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanatsu-Shinohara M., Lee J., Inoue K., Ogonuki N., Miki H., Toyokuni S., Ikawa M., Nakamura T., Ogura A., Shinohara T. Pluripotency of a single spermatogonial stem cell in mice. Biol. Reprod. 2008;78:681–687. doi: 10.1095/biolreprod.107.066068. [DOI] [PubMed] [Google Scholar]
- 53.Jeong H.-S., Bhin J., Joon Kim H., Hwang D., Ryul Lee D., Kim K.-S. Transcriptional regulatory networks underlying the reprogramming of spermatogonial stem cells to multipotent stem cells. Exp. Mol. Med. 2017;49:e315. doi: 10.1038/emm.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griswold M.D. Spermatogenesis: the commitment to meiosis. Physiol. Rev. 2016;96:1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cahoon C.K., Hawley R.S. Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 2016;23:369–377. doi: 10.1038/nsmb.3208. [DOI] [PubMed] [Google Scholar]
- 56.Bhalla N., Dernburg A.F. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Page S.L., Hawley R.S. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 58.Bolcun-Filas E., Handel M.A. Meiosis: the chromosomal foundation of reproduction. Biol. Reprod. 2018;99:112–126. doi: 10.1093/biolre/ioy021. [DOI] [PubMed] [Google Scholar]
- 59.Loidl J. Conservation and variability of meiosis across the eukaryotes. Annu. Rev. Genet. 2016;50:293–316. doi: 10.1146/annurev-genet-120215-035100. [DOI] [PubMed] [Google Scholar]
- 60.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.-C., Gildersleeve H., Lehle J.D., Mayo M., Westernströer B., et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667.e8. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koubova J., Menke D.B., Zhou Q., Capel B., Griswold M.D., Page D.C. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson E.L., Baltus A.E., Roepers-Gajadien H.L., Hassold T.J., de Rooij D.G., van Pelt A.M.M., Page D.C. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng Y., Qiao H. The application of single-cell RNA sequencing in mammalian meiosis studies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.673642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ernst C., Eling N., Martinez-Jimenez C.P., Marioni J.C., Odom D.T. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 2019;10:1251. doi: 10.1038/s41467-019-09182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baudat F., Imai Y., de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 67.Chen S., An G., Wang H., Wu X., Ping P., Hu L., Chen Y., Fan J., Cheng C.Y., Sun F. Human obstructive (postvasectomy) and nonobstructive azoospermia – insights from scRNA-Seq and transcriptome analysis. Genes Dis. 2022;9:766–776. doi: 10.1016/j.gendis.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acquaviva L., Boekhout M., Karasu M.E., Brick K., Pratto F., Li T., van Overbeek M., Kauppi L., Camerini-Otero R.D., Jasin M., Keeney S. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature. 2020;582:426–431. doi: 10.1038/s41586-020-2327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou R., Wu J., Liu B., Jiang Y., Chen W., Li J., He Q., He Z. The roles and mechanisms of Leydig cells and myoid cells in regulating spermatogenesis. Cell. Mol. Life Sci. 2019;76:2681–2695. doi: 10.1007/s00018-019-03101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Y.E., Zhang Y., Unni E., Shirley C.R., Deng J.M., Russell L.D., Weil M.M., Behringer R.R., Meistrich M.L. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc. Natl. Acad. Sci. USA. 2000;97:4683–4688. doi: 10.1073/pnas.97.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Rand M.G., Richardson R.T., Zimmerman L.J., Widgren E.E. Sequence and localization of human NASP: conservation of a Xenopus histone-binding protein. Dev. Biol. 1992;154:37–44. doi: 10.1016/0012-1606(92)90045-i. [DOI] [PubMed] [Google Scholar]
- 72.Kierszenbaum A.L., Tres L.L. Structural and transcriptional features of the mouse spermatid genome. J. Cell Biol. 1975;65:258–270. doi: 10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walker W.H., Sanborn B.M., Habener J.F. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc. Natl. Acad. Sci. USA. 1994;91:12423–12427. doi: 10.1073/pnas.91.26.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Girault M.-S., Dupuis S., Ialy-Radio C., Stouvenel L., Viollet C., Pierre R., Favier M., Ziyyat A., Barbaux S. Deletion of the Spata3 gene induces sperm alterations and in vitro hypofertility in mice. Int. J. Mol. Sci. 2021;22:1959. doi: 10.3390/ijms22041959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng H., Stratton C.J., Morozumi K., Jin J., Yanagimachi R., Yan W. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc. Natl. Acad. Sci. USA. 2007;104:6852–6857. doi: 10.1073/pnas.0701669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Donnell L., Nicholls P.K., O’Bryan M.K., McLachlan R.I., Stanton P.G. Spermiation: the process of sperm release. Spermatogenesis. 2011;1:14–35. doi: 10.4161/spmg.1.1.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lehti M.S., Sironen A. Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 2017;97:522–536. doi: 10.1093/biolre/iox096. [DOI] [PubMed] [Google Scholar]
- 78.Bao J., Bedford M.T. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction. 2016;151:R55–R70. doi: 10.1530/REP-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohkura H. Meiosis: an overview of key differences from mitosis. Cold Spring Harbor Perspect. Biol. 2015;7:a015859. doi: 10.1101/cshperspect.a015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skinner M.K. Cell-cell interactions in the testis. Ann. N. Y. Acad. Sci. 1987;513:158–171. doi: 10.1111/j.1749-6632.1987.tb25006.x. [DOI] [PubMed] [Google Scholar]
- 81.Dettin L., Rubinstein N., Aoki A., Rabinovich G.A., Maldonado C.A. Regulated expression and ultrastructural localization of galectin-1, a proapoptotic beta-galactoside-binding lectin, during spermatogenesis in rat testis. Biol. Reprod. 2003;68:51–59. doi: 10.1095/biolreprod.102.006361. [DOI] [PubMed] [Google Scholar]
- 82.Yao P.-L., Lin Y.-C., Richburg J.H. TNF alpha-mediated disruption of spermatogenesis in response to Sertoli cell injury in rodents is partially regulated by MMP2. Biol. Reprod. 2009;80:581–589. doi: 10.1095/biolreprod.108.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ulisse S., Farina A.R., Piersanti D., Tiberio A., Cappabianca L., D’Orazi G., Jannini E.A., Malykh O., Stetler-Stevenson W.G., D’Armiento M. Follicle-stimulating hormone increases the expression of tissue inhibitors of metalloproteinases TIMP-1 and TIMP-2 and induces TIMP-1 AP-1 site binding complex(es) in prepubertal rat Sertoli cells. Endocrinology. 1994;135:2479–2487. doi: 10.1210/endo.135.6.7988435. [DOI] [PubMed] [Google Scholar]
- 84.Wang H., Wen L., Yuan Q., Sun M., Niu M., He Z. Establishment and applications of male germ cell and Sertoli cell lines. Reproduction. 2016;152:R31–R40. doi: 10.1530/REP-15-0546. [DOI] [PubMed] [Google Scholar]
- 85.Kanatsu-Shinohara M., Ogonuki N., Matoba S., Ogura A., Shinohara T. Autologous transplantation of spermatogonial stem cells restores fertility in congenitally infertile mice. Proc. Natl. Acad. Sci. USA. 2020;117:7837–7844. doi: 10.1073/pnas.1914963117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park C.J., Ha C.M., Lee J.E., Gye M.C. Claudin 11 inter-sertoli tight junctions in the testis of the Korean soft-shelled turtle (Pelodiscus maackii) Biol. Reprod. 2015;92:96. doi: 10.1095/biolreprod.114.117804. [DOI] [PubMed] [Google Scholar]
- 87.Mäkelä J.A., Koskenniemi J.J., Virtanen H.E., Toppari J. Testis development. Endocr. Rev. 2019;40:857–905. doi: 10.1210/er.2018-00140. [DOI] [PubMed] [Google Scholar]
- 88.Busada J.T., Chappell V.A., Niedenberger B.A., Kaye E.P., Keiper B.D., Hogarth C.A., Geyer C.B. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev. Biol. 2015;397:140–149. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vernet N., Dennefeld C., Rochette-Egly C., Oulad-Abdelghani M., Chambon P., Ghyselinck N.B., Mark M. Retinoic acid metabolism and signaling pathways in the adult and developing mouse testis. Endocrinology. 2006;147:96–110. doi: 10.1210/en.2005-0953. [DOI] [PubMed] [Google Scholar]
- 90.Tsai M.Y., Yeh S.D., Wang R.S., Yeh S., Zhang C., Lin H.Y., Tzeng C.R., Chang C. Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc. Natl. Acad. Sci. USA. 2006;103:18975–18980. doi: 10.1073/pnas.0608565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heinrich A., DeFalco T. Essential roles of interstitial cells in testicular development and function. Andrology. 2020;8:903–914. doi: 10.1111/andr.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Hara L., Smith L.B. Androgen receptor roles in spermatogenesis and infertility. Best Pract. Res. Clin. Endocrinol. Metabol. 2015;29:595–605. doi: 10.1016/j.beem.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 93.Sofikitis N., Giotitsas N., Tsounapi P., Baltogiannis D., Giannakis D., Pardalidis N. Hormonal regulation of spermatogenesis and spermiogenesis. J. Steroid Biochem. Mol. Biol. 2008;109:323–330. doi: 10.1016/j.jsbmb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 94.Meggyes M., Lajko A., Fulop B.D., Reglodi D., Szereday L. Phenotypic characterization of testicular immune cells expressing immune checkpoint molecules in wild-type and pituitary adenylate cyclase-activating polypeptide-deficient mice. Am. J. Reprod. Immunol. 2020;83 doi: 10.1111/aji.13212. [DOI] [PubMed] [Google Scholar]
- 95.Miko E., Meggyes M., Doba K., Barakonyi A., Szereday L. Immune checkpoint molecules in reproductive immunology. Front. Immunol. 2019;10:846. doi: 10.3389/fimmu.2019.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jarow J.P., Zirkin B.R. The androgen microenvironment of the human testis and hormonal control of spermatogenesis. Ann. N. Y. Acad. Sci. 2005;1061:208–220. doi: 10.1196/annals.1336.023. [DOI] [PubMed] [Google Scholar]
- 97.Lau X., Munusamy P., Ng M.J., Sangrithi M. Single-cell RNA sequencing of the cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev. Cell. 2020;54:548–566.e7. doi: 10.1016/j.devcel.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 98.Tegelenbosch R.A., de Rooij D.G. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 99.Fayomi A.P., Orwig K.E. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lovelace D.L., Gao Z., Mutoji K., Song Y.C., Ruan J., Hermann B.P. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143:1893–1906. doi: 10.1242/dev.132761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schmidt J.A., Avarbock M.R., Tobias J.W., Brinster R.L. Identification of glial cell line-derived neurotrophic factor-regulated genes important for spermatogonial stem cell self-renewal in the rat. Biol. Reprod. 2009;81:56–66. doi: 10.1095/biolreprod.108.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oatley J.M., Avarbock M.R., Telaranta A.I., Fearon D.T., Brinster R.L. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z., Xu X., Li J.L., Palmer C., Maric D., Dean J. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat. Commun. 2019;10:5196. doi: 10.1038/s41467-019-13193-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki H., Ahn H.W., Chu T., Bowden W., Gassei K., Orwig K., Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 2012;361:301–312. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen W., Zhang Z., Chang C., Yang Z., Wang P., Fu H., Wei X., Chen E., Tan S., Huang W., et al. A bioenergetic shift is required for spermatogonial differentiation. Cell Discov. 2020;6:56. doi: 10.1038/s41421-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davies O.R., Maman J.D., Pellegrini L. Structural analysis of the human SYCE2-TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol. 2012;2 doi: 10.1098/rsob.120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vereecke G., Defreyne J., Van Saen D., Collet S., Van Dorpe J., T’Sjoen G., Goossens E. Characterisation of testicular function and spermatogenesis in transgender women. Hum. Reprod. 2021;36:5–15. doi: 10.1093/humrep/deaa254. [DOI] [PubMed] [Google Scholar]
- 108.Kumar N., Singh A.K. The anatomy, movement, and functions of human sperm tail: an evolving mystery. Biol. Reprod. 2021;104:508–520. doi: 10.1093/biolre/ioaa213. [DOI] [PubMed] [Google Scholar]
- 109.Hai Y., Hou J., Liu Y., Liu Y., Yang H., Li Z., He Z. The roles and regulation of Sertoli cells in fate determinations of spermatogonial stem cells and spermatogenesis. Semin. Cell Dev. Biol. 2014;29:66–75. doi: 10.1016/j.semcdb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 110.Al-Attar L., Noël K., Dutertre M., Belville C., Forest M.G., Burgoyne P.S., Josso N., Rey R. Hormonal and cellular regulation of Sertoli cell anti-Müllerian hormone production in the postnatal mouse. J. Clin. Invest. 1997;100:1335–1343. doi: 10.1172/JCI119653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nicholls P.K., Stanton P.G., Chen J.L., Olcorn J.S., Haverfield J.T., Qian H., Walton K.L., Gregorevic P., Harrison C.A. Activin signaling regulates sertoli cell differentiation and function. Endocrinology. 2012;153:6065–6077. doi: 10.1210/en.2012-1821. [DOI] [PubMed] [Google Scholar]
- 112.Rossi P., Sette C., Dolci S., Geremia R. Role of c-kit in mammalian spermatogenesis. J. Endocrinol. Invest. 2000;23:609–615. doi: 10.1007/BF03343784. [DOI] [PubMed] [Google Scholar]
- 113.Liu S., Chen X., Wang Y., Li L., Wang G., Li X., Chen H., Guo J., Lin H., Lian Q.-Q., Ge R.S. A role of KIT receptor signaling for proliferation and differentiation of rat stem Leydig cells in vitro. Mol. Cell. Endocrinol. 2017;444:1–8. doi: 10.1016/j.mce.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 114.Ge R.S., Dong Q., Sottas C.M., Chen H., Zirkin B.R., Hardy M.P. Gene expression in rat Leydig cells during development from the progenitor to adult stage. Biol. Reprod. 2005;72:1405–1415. doi: 10.1095/biolreprod.104.037499. [DOI] [PubMed] [Google Scholar]
- 115.Huang L., Xiao K., Zhang J., Zhang P., He W., Tang Y., Yang W., Huang X., Liu R., Liang X., et al. Comparative transcriptome analysis reveals potential testosterone function-related regulatory genes/pathways of Leydig cells in immature and mature buffalo (Bubalus bubalis) testes. Gene. 2021;802 doi: 10.1016/j.gene.2021.145870. [DOI] [PubMed] [Google Scholar]
- 116.Cooke P.S., Walker W.H. Nonclassical androgen and estrogen signaling is essential for normal spermatogenesis. Semin. Cell Dev. Biol. 2022;121:71–81. doi: 10.1016/j.semcdb.2021.05.032. [DOI] [PubMed] [Google Scholar]
- 117.Smith L.B., Walker W.H. The regulation of spermatogenesis by androgens. Semin. Cell Dev. Biol. 2014;30:2–13. doi: 10.1016/j.semcdb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.DeFalco T., Potter S.J., Williams A.V., Waller B., Kan M.J., Capel B. Macrophages contribute to the spermatogonial niche in the adult testis. Cell Rep. 2015;12:1107–1119. doi: 10.1016/j.celrep.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oatley J.M., Oatley M.J., Avarbock M.R., Tobias J.W., Brinster R.L. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development. 2009;136:1191–1199. doi: 10.1242/dev.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 121.Matson C.K., Murphy M.W., Griswold M.D., Yoshida S., Bardwell V.J., Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell. 2010;19:612–624. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoshida S., Sukeno M., Nakagawa T., Ohbo K., Nagamatsu G., Suda T., Nabeshima Y.I. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- 123.Seandel M., James D., Shmelkov S.V., Falciatori I., Kim J., Chavala S., Scherr D.S., Zhang F., Torres R., Gale N.W., et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao J., Lu P., Wan C., Huang Y., Cui M., Yang X., Hu Y., Zheng Y., Dong J., Wang M., et al. Cell-fate transition and determination analysis of mouse male germ cells throughout development. Nat. Commun. 2021;12:6839. doi: 10.1038/s41467-021-27172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao Y., Xiao X., Lui W.-Y., Lee W.M., Mruk D., Cheng C.Y. Cell polarity proteins and spermatogenesis. Semin. Cell Dev. Biol. 2016;59:62–70. doi: 10.1016/j.semcdb.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Neirijnck Y., Kühne F., Mayère C., Pavlova E., Sararols P., Foti M., Atanassova N., Nef S. Tumor suppressor PTEN regulates negatively sertoli cell proliferation, testis size, and sperm production in vivo. Endocrinology. 2019;160:387–398. doi: 10.1210/en.2018-00892. [DOI] [PubMed] [Google Scholar]
- 128.Gnessi L., Basciani S., Mariani S., Arizzi M., Spera G., Wang C., Bondjers C., Karlsson L., Betsholtz C. Leydig cell loss and spermatogenic arrest in platelet-derived growth factor (PDGF)-A-deficient mice. J. Cell Biol. 2000;149:1019–1026. doi: 10.1083/jcb.149.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Basciani S., Mariani S., Spera G., Gnessi L. Role of platelet-derived growth factors in the testis. Endocr. Rev. 2010;31:916–939. doi: 10.1210/er.2010-0004. [DOI] [PubMed] [Google Scholar]
- 130.Wijayarathna R., de Kretser D.M. Activins in reproductive biology and beyond. Hum. Reprod. Update. 2016;22:342–357. doi: 10.1093/humupd/dmv058. [DOI] [PubMed] [Google Scholar]
- 131.Hedger M.P., Meinhardt A. Cytokines and the immune-testicular axis. J. Reprod. Immunol. 2003;58:1–26. doi: 10.1016/S0165-0378(02)00060-8. [DOI] [PubMed] [Google Scholar]
- 132.Sarkar O., Mathur P.P., Cheng C.Y., Mruk D.D. Interleukin 1 alpha (IL1A) is a novel regulator of the blood-testis barrier in the rat. Biol. Reprod. 2008;78:445–454. doi: 10.1095/biolreprod.107.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huleihel M., Lunenfeld E. Regulation of spermatogenesis by paracrine/autocrine testicular factors. Asian J. Androl. 2004;6:259–268. [PubMed] [Google Scholar]
- 134.Sorrelle N., Dominguez A.T.A., Brekken R.A. From top to bottom: midkine and pleiotrophin as emerging players in immune regulation. J. Leukoc. Biol. 2017;102:277–286. doi: 10.1189/jlb.3MR1116-475R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 139.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 140.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., participants in the 1st Human Cell Atlas Jamboree. Marioni J.C. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All sequencing data in this paper have been deposited at GEO (GSE190477) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.