Abstract
This article reviews the molecular genetic data pertaining to the major surface glycoprotein (MSG) gene family of Pneumocystis carinii and its role in surface variation and compares this fungal system to antigenic variation systems in the protozoan Trypanosoma brucei and the bacteria Borrelia spp.
PNEUMOCYSTIS CARINII
Pneumocystis carinii is a fungus that can cause pneumonia in immunocompromised mammals (100, 174). While P. carinii was recognized nearly a century ago and has been a significant human pathogen since the beginning of the AIDS epidemic, this organism is not well understood. Much of the blame for the lack of basic information about this organism and its natural history can be placed on its failure to grow well in culture. Populations of the organism can be maintained in broth culture, but growth in culture has not been good enough to allow either production of large numbers or derivation of clonally derived stocks (95). Therefore, in vitro culture has been of limited utility for studying the genetics and biochemistry of P. carinii.
By contrast, large numbers of organisms can be routinely obtained from laboratory animals. Most of what is known about P. carinii has been learned from studies with animals, and most of what is known about the surface antigen genes was determined from studies on organisms from laboratory rats. Reliance on rats as the source of P. carinii has had its advantages and disadvantages, which have shaped the course of research on antigenic variation. It is important, therefore, to preface this review with a brief description of the general features of rat models of P. carinii infection.
The rat model of P. carinii pneumonia (Pcp) has its roots in studies that showed that P. carinii infections can be provoked by treating rats with chemical immunosuppressants (18, 24, 38, 53). These infections appeared to be caused by growth of P. carinii that was present in the rats at the time of immunosuppression (reactivation of latent infections). Most, if not all, commercial rat colonies harbor P. carinii in a latent form, where it can persist for up to a year (103, 162, 178). It seems most plausible that P. carinii maintains itself in rat colonies via airborne transmission from older rats to neonates, and evidence for airborne transmission has been obtained in laboratory settings (53, 55, 176). However, other mechanisms of transmission from adults to neonates have not been excluded, and observations suggestive of transplacental transfer have been reported (17).
Even though latent infections are common in laboratory rats, simply immunosuppressing rats does not always lead to the production of large numbers of organisms per animal. This observation led to the implementation of a protocol known as the natural transmission method. In this method, rats are bred in colonies known to harbor latent infections. Young animals in such colonies are immunosuppressed and housed in open-air cages. Some or all of the immunosuppressed animals develop heavy infections. When new young animals from another source (such as a commercial vendor) are introduced into the colony and immunosuppressed, the P. carinii they produce in their lungs can be from either or both of two sources: (i) the airborne organisms from other infected rats in the immunosuppressed colony and (ii) the latent organisms that entered the animals while they were in the commercial vendor colony.
The natural transmission method has provided a reliable source of large amounts of P. carinii organisms, which is its chief advantage. Consequently, this rat model has been a mainstay of P. carinii research. However, the method suffers from the disadvantage that the experimenter has little control over the kind of organisms in the animals, the time of infection, or the number of organisms acquired. An alternative method to propagate P. carinii in rats is to introduce P. carinii into the rat lung via inoculation (15). The inoculation method was developed to aid in testing drugs for activity against P. carinii. Consequently, millions of P. carinii organisms were introduced into each animal to ensure infection. However, recently it has been shown that as few as 10 P. carinii organisms can cause infection (25). Hence, the low-dose inoculation method holds promise as a means to produce the clonal stocks that will be needed in order to fully understand regulation of surface antigens. Indeed, preliminary indications are that P. carinii from rats inoculated with a low dose of P. carinii show far less surface variation than P. carinii from rats infected via natural transmission (see below).
DIVERSITY WITHIN THE GENUS PNEUMOCYSTIS
Several other laboratory animal models of P. carinii infection have been developed. The most popular are mouse, ferret, and rabbit (reviewed in reference 26). It is important at this point to make clear that no animal model produces the organism that causes Pneumocystis pneumonia in humans. The human form of the Pneumocystis organism has been found only in humans. Similarly, each animal species appears to have its own form of Pneumocystis organism. The evidence for such diversity and host specificity is summarized below. Suffice it to say here that the genus Pneumocystis is sufficiently diverse to predict that the details derived from the study of one member of the genus will not necessarily pertain to all members of the genus. Nevertheless, so far, data from animal-derived Pneumocystis organisms has been roughly indicative of what occurs in the human organism, as far as this is understood.
The data supporting the view that there is a great deal of diversity in the genus Pneumocystis are as follows. Histological surveys of the lungs of animals from laboratories farms, ranches, zoos, and the wild have suggested that members of the genus Pneumocystis are widespread in mammals; animals typically exhibit low numbers of organisms (between 103 and 104 organisms) in the lungs (56, 68, 92). Infection of non mammals has not been reported (133). Prior to 1990, the Pneumocystis organisms from different host species were widely regarded to be essentially the same, even though morphological differences could be discerned (37) and attempts to transfer P. carinii from one host species into another (cross-species inoculation studies) were nearly always unsuccessful. Despite these data, the idea that the genus contained multiple members was met with skepticism because the morphological differences between P. carinii from different hosts could have been caused by residence in a particular host and some cross-species inoculation experiments studies had been reported to have resulted in growth in a host species other than the one from which the P. carinii were obtained. Soon after 1990, the diversity within the genus became objectively clear from analysis of DNA sequences, which established that each host species harbors at least one distinct genetic variety of the microbe (30, 114, 127, 129, 134, 139, 140, 141). The genetic divergence among these varieties of P. carinii is high enough to suggest that the genus Pneumocystis contains multiple species (58, 139). However, proof that the genus Pneumocystis contains multiple biological species (i.e., that the different varieties cannot mate productively) is lacking because mating experiments cannot be performed. As a result, the terms special form and formae specialis are used to designate the varieties of P. carinii (4, 32, 91, 142, 172). The special form of P. carinii that has been most studied is P. carinii f. sp. carinii, which is the most common of the two special forms found in laboratory rats.
After the methods to distinguish between special forms were established, it was possible to perform controlled cross-species inoculation experiments, which showed that P. carinii from one host species did not cause disease in another host species, even when millions of organisms were introduced (42). By contrast, inoculation within a host species routinely leads to heavy infections (42). These data suggest that the P. carinii special forms from one host species are specific for that species and cannot reproduce in a different host species. Earlier reports to the contrary were from experiments performed prior to the development of the molecular genetic markers needed to distinguish one special form from another. Hence, the organisms observed in the test animals may not have been the ones that were introduced during the experiment, but rather could have emerged from a latent stage of the P. carinii special form normally found in that species.
MAJOR SURFACE ANTIGEN OF P. CARINII
The principal protein antigen on the surface of P. carinii has been assigned a variety of names, but lately it has primarily come to be known either as the MSG (65, 79, 87, 112, 117, 150, 173) or as gpA (40, 41). Here, we will refer to it as MSG.
Large and very abundant, MSG is conspicuous after separation of total P. carinii proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Initially, the study of MSG was via biochemical and immunological approaches, which had significant limitations because P. carinii are difficult to grow, making it difficult to obtain MSG in amounts large enough to allow thorough purification. Despite these difficulties, biochemical studies established that the 120-kDa band contained a glycosylated protein that was on the organism's surface (40, 41, 65, 76, 79, 80, 86, 87, 101, 106, 112, 117, 150, 173, 177). Material in the 120-kDa band was shown to be recognized by serum antibodies and T cells from exposed hosts (35, 36, 39, 43, 45, 48, 66, 76, 88, 101, 111, 136, 151, 152, 154), and to bind to several host proteins, including fibronectin, vitronectin, surfactant protein A, and surfactant protein D (34, 74, 75, 93, 104, 115, 116, 183; A. H. Limper, Editorial, J. Lab. Clin. Med. 125:12–13, 1995; S. T. Pottratz, Editorial, J. Lab. Clin. Med. 126:414–415, 1995). Hence, MSG came to be regarded as a crucial player in host-pathogen interactions.
MSG GENES: NUMBER AND ORGANIZATION
The composition of the 120-kDa SDS-PAGE band known as MSG became much clearer after MSG genes became accessible. An MSG cDNA was first cloned from ferret P. carinii by screening a cDNA library with an antibody that reacted with the 120-kDa band on a Western blot (50). The same approach yielded several MSG cDNA clones from rat P. carinii (67, 166). These cDNAs each encoded a different, but related, protein. The presence of multiple MSG cDNAs seemed to indicate the presence of a gene family. Southern blot experiments supported this view: a single cDNA hybridized to all of the chromosomes (67, 144). Subsequent sequencing of segments of the genome cloned by virtue of their ability to hybridize to a cloned MSG cDNA confirmed that the genome contains multiple open reading frames, each encoding a different isoform of a protein with the properties expected of a member of an MSG protein family (molecular mass of approximately 120 kDa, rich in cysteine residues that were located at the same positions in different isoforms) (39, 143, 148, 166). These predicted isoforms of MSG varied in sequence by as much as 35%. While the DNA sequencing results were consistent with the presence of multiple MSG genes, the functionality of the open reading frames encoding MSG isoforms remains unproven because none of the predicted individual protein isoforms has been characterized biochemically in P. carinii organisms. Nevertheless, in the genomics era it is standard practice to assume that long open reading frames are functional genes, and, lacking evidence to the contrary, it seems reasonable to assume that all MSG open reading frames are able to produce an isoform of the MSG protein in P. carinii.
The number of MSG genes in the family is not completely clear but is probably between 50 and 100. Early DNA hybridization experiments produced an estimate of approximately 100 MSG genes per haploid genome in P. carinii f. sp. carinii (148). Sequencing data have shown the presence and expression of at least 26 different MSG genes in a single organism population (147).
MSG genes appear to be on all of the chromosomes in the genome of P. carinii f. sp carinii because all of the 13 to 15 chromosome-sized bands resolved by pulsed-field gel electrophoresis (PFGE) hybridized to an MSG DNA probe (63, 67, 148, 166, 170). (The number of PFGE bands varies among isolates of rat P. carinii. The number of chromosomes appears to be 15 to 18 because a few of the PFGE bands stain more intensely with DNA stains than others do [24, 86, 180].) As far as is known, all members of the MSG family reside at the ends of chromosomes. MSG genes were mapped to the ends of P. carinii f. sp carinii chromosomes by showing that they are sensitive to digestion with an exonuclease (146). Supporting evidence for telomeric MSG genes came from cloning of P. carinii chromosome ends. One such cloned DNA fragment began with the 3′ end of an MSG gene and ended with a sequence known to be at subtelomeric regions of the P. carinii f. sp. carinii genome (158).
MSG genes are organized as clusters of 2 to 4 genes (39, 144, 148, 166, 167, 179). If each of the 30 to 36 chromosome ends in the genome has 2 to 4 MSG genes, then the number of genes per genome is between 60 and 144. These numbers bracket the initial estimate of 100 genes per genome obtained by DNA hybridization. More than the foregoing is known about the structure MSG gene clusters, which have recently been discovered to contain members of other gene families along with MSG genes, but these features are best left to be described in the context of gene expression (see below).
MSG GENE TRANSCRIPTS
The initial cDNA cloning experiments suggested that multiple members of the MSG gene family can be transcribed; otherwise, cDNAs encoding different MSG isoforms would not have been present in the library. However, interpretation of the cDNA cloning data was complicated by the fact that the organisms used to prepare the library were from rats infected by an unknown number of strains. In addition, organisms from different rats were sometimes pooled in order to increase the yield of P. carinii RNA available for cDNA library construction. Hence, the multiplicity of MSG cDNAs in the library could have been due to expression of one or a few MSG genes in each of several different P. carinii strains. Subsequent analysis showed that pooling P. carinii from different rats was not required to obtain multiple different MSG cDNAs. In that analysis, a cDNA library made from P. carinii f. sp. carinii from a single infected rat was hybridized to a single cloned MSG cDNA. Seven positive clones were isolated and sequenced. All seven clones encoded an MSG, but none of the clones had the same sequence (77). Other studies had indicated that this rat was infected by a single strain of P. carinii f. sp. carinii (23).
The number of MSG genes expressed at any one time in an individual organism remained unclear. In theory, expression of multiple MSG genes within a population of P. carinii might come about in either of two ways. Each organism might express multiple genes. Alternatively, each organism might express only one MSG gene, in which case the multiplicity of MSG cDNAs would be due to different genes being expressed in different organisms within the population. The latter possibility seemed more probable because it was more consistent with a role for MSG in surface variation, which is a feature of many pathogenic protozoa and bacteria.
Determining the number of MSG genes expressed in an individual P. carinii organism was hampered by the technical problems attendant to not having access to clonally derived P. carinii. This problem precluded simple experiments to determine the number and kinds of MSGs expressed by a population produced from a single, founder P. carinii organism (i.e., a clonally derived population). Fortunately, basic studies on the structure of messenger RNAs encoding MSGs revealed a feature at the 5′ end of these messages that came to be called the upstream conserved sequence (UCS). The existence of the UCS ultimately made it possible to make progress in the study of MSG gene expression by studying populations of P. carinii from rats infected by the natural transmission method.
The UCS was discovered by sequencing the 5′ ends of mRNAs encoding MSG. The first experiments of this kind analyzed 21 mRNAs, all of which were identical in sequence for the first 429 bp (the UCS), after which they diverged (170). The divergent sequences downstream of the UCS encoded different isoforms of MSG. These findings were confirmed by another group using a similar approach (31).
Given the seemingly uniform presence of the UCS at the beginning of MSG mRNAs, it seemed odd that the UCS was not present in the several MSG gene sequences that were known at the time. The strangeness of the UCS became more evident when it was then shown to be absent from the majority of MSG genes in the genome because the UCS hybridized to only one of the 15 chromosomes resolved by PFGE, whereas MSG gene probes hybridized to all 15 chromosomes (31, 170). The restriction of the UCS to a single PFGE band suggested that it might be unique in the genome. Quantitative hybridization experiments supported this hypothesis (146).
Further progress in understanding the structure and copy number of the DNA encoding the UCS found on mRNAs came from analysis of genomic clones. Lambda clones carrying the UCS were obtained by screening genomic libraries from P. carinii f. sp. carinii with a UCS DNA probe (146, 170). The number of UCS clones in the library was low enough to suggest that the UCS is present only once in the genome. Multiple UCS-MSG genomic clones were isolated, and all of these contained an MSG gene attached directly to the UCS. Therefore, it appears that the UCS locus is always adjoined to an MSG gene. Each of the cloned UCS loci contained a different MSG gene attached to the UCS. The UCS-linked MSG gene was often followed by other MSG copies lacking the UCS, showing that the clustering of MSG genes previously observed was also occurring at the UCS locus. Since the UCS is linked to MSG genes, and MSG genes are telomeric, it was expected that the UCS would also be telomeric. Experiments with exonuclease-digested chromosomes showed this to be the case (146).
The fact that each cloned UCS was upstream of a different cluster of MSG genes predicted that the UCS locus in a population of P. carinii f. sp. carinii would be heterogeneous in structure at points downstream of the UCS and that these downstream regions might differ between populations. Mapping of restriction enzyme cleavage sites downstream of the UCS produced results consistent with this prediction (31, 146). The downstream heterogeneity of the UCS locus was consistent with the previously observed presence of mRNAs encoding different MSG isoforms, yet each starting with a copy of the UCS.
In contrast with the restriction site polymorphism of the region downstream of the UCS locus, the region upstream of this locus (10 kb upstream of the UCS) appeared to have a single map for restriction enzyme cleavage sites (146). These data indicated either that there is only one UCS locus or that there are two UCS loci that are identical in structure for at least 10 kb upstream. The former alternative seems more probable, but this issue will not be resolved until both ends of the chromosome carrying the UCS are characterized. This goal should be reached soon because the P. carinii f. sp. carinii genome is being sequenced.
MODEL FOR CONTROL OF MSG GENE TRANSCRIPTION
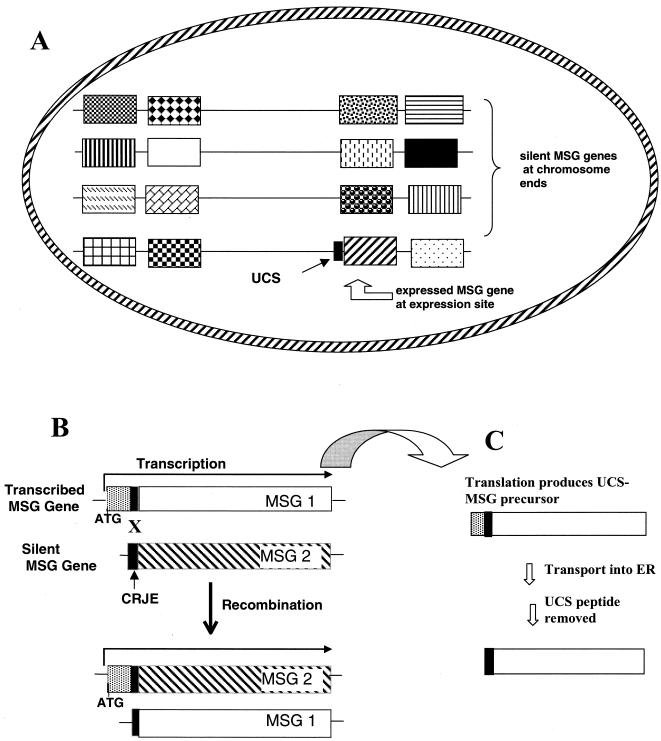
The structural features of MSG mRNAs, of the UCS locus, of MSG genes at the UCS locus, and of MSG genes elsewhere in the genome can be combined to produce a model for control of expression of the MSG gene family (Fig. 1). In the model, the vast majority of MSG genes are organized in transcriptionally silent clusters that are located at the ends of each of the chromosomes (Fig. 1A). Only the MSG gene that is attached to the UCS locus is transcribed, and therefore, only one isoform of MSG is expressed on the surface (Fig. 1A and B). The surface MSG can be changed by changing the MSG gene that is at the UCS locus (Fig. 1B). These changes generate diversity at the UCS locus within populations, thereby causing production of mRNAs encoding diverse MSGs within a single population.
FIG. 1.
Expression site model. (A) MSG genes shown as boxes at chromosome ends. Only the gene at the UCS locus is transcribed. The surface of the organism has the MSG isoform encoded by this gene. (B) Recombination switches the gene at the expression site, possibly via crossovers between copies of the CRJE. X, a site of crossover. (C) Translation starts in the UCS; the preprotein is made and sent into the ER. The UCS-encoded portion is removed.
While this model explained all of the features of MSG gene structure and expression known at the time, direct evidence linking expression of MSG mRNAs to the structure of the UCS locus was lacking and other possible mechanisms of MSG mRNA synthesis had not been excluded. The principal rival to the model in Fig. 1 suggested itself from the similarity between MSG mRNAs and mRNAs from kinetoplastid protozoans such as Trypanosoma brucei. In the kinetoplastida, all mRNAs begin with the same sequence, but not because transcribed genes all reside next to a locus encoding this sequence. Instead, the 5′ leader is transcribed from a separate locus and then added to pre-mRNAs by trans-splicing (160). To determine if trans-splicing was involved in putting the UCS on the 5′ end of each MSG mRNA, the structure of the UCS-MSG junctions in mRNAs was compared to the structure of the UCS-MSG junctions in the genome. Six populations of P. carinii f. sp. carinii, each from a single rat, were studied. Each of the six populations expressed a different subset of the MSG gene family. All of the populations studied had the same UCS-MSG junctions in mRNAs as in the genome (147). These data are most consistent with the model shown in Fig. 1, whereby only the MSG gene that is directly attached to the UCS locus is transcribed. If this were not the case, and the UCS were attached posttranscriptionally by RNA splicing, then one would expect to find mRNAs for which there is no corresponding UCS-linked MSG gene.
Additional data supporting the hypothesis that transcription of MSG genes is restricted to only UCS-linked MSG genes came from analysis of MSG proteins. These studies indicated that the UCS is probably required for both translation of mRNAs and transport of the nascent MSG peptide into the endoplasmic reticulum (ER). The UCS has a potential translational start codon near its 5′ end (indicated by the ATG in Fig. 1B) (31, 170). This start codon is followed by a sequence encoding a putative signal peptide that could serve to direct the MSG into the ER from whence it would presumably be sent to the cell surface via the Golgi apparatus. Analysis of total proteins from P. carinii f. sp. carinii with an antibody raised against the UCS-encoded peptide provided evidence that the peptide encoded by the UCS is part of an MSG protein precursor. Total protein from P. carinii contained a large protein (apparent molecular mass, 160 kDa) containing both UCS and MSG epitopes, which suggested that it is a UCS-containing MSG precursor (Fig. 1C) (145). The UCS epitopes were not present on the mature MSG found on the cell surface, suggesting that the UCS peptide is removed from the UCS-MSG precursor by a protease (Fig. 1C). The fate of the UCS after cleavage is not known. If it is removed after the precursor reaches the surface, which is suggested by the presence of the PRT protease on the surface (see below), then it may survive as a free unit. While there is no evidence either for or against the persistence of free UCS peptides, such a situation would explain the high degree of polymorphism in the UCS genomic copy (146).
There is experimental evidence for the functionality of the UCS peptide in sending the MSG precursor into the ER. A gene encoding a UCS-MSG protein produced a glycosylated protein in insect cells, which presumably required transit of the nascent polypeptide into the ER and Golgi apparatus. By contrast, a gene that encoded only an MSG open reading frame expressed a protein that was not glycosylated (146). These data suggest that the UCS is necessary and sufficient to direct the nascent polypeptide into the compartments known to be traversed by proteins bound for the cell surface. If the behavior of MSG transcripts and precursor peptides is the same in P. carinii f. sp. carinii as it is in insect cells, then MSG genes that are not linked to the UCS would not be expressed as surface proteins, even if such genes were to be transcribed to produce RNAs lacking a 5′ UCS. Hence, the UCS appears to exert control over MSG transcription, translation, processing, and transport.
The UCS and MSG portions of the predicted precursor protein are divided by an invariant amino acid string of eight residues that contains a site that would be cut by a subtilisin-like protease. Molecular genetic analysis has identified a family of genes in P. carinii f. sp. carinii (the PRT family) that encodes proteases that might serve this purpose (83, 84, 126, 169). At least some PRT family members appear to be surface proteins, which would explain why they vary in sequence (84, 126, 169). It is intriguing that PRT and MSG genes tend to be linked in the same orientation. This linkage presents the possibility that the organism coordinates expression of the two gene families, thereby achieving surface variation with respect to both proteins. Such coordinate expression can be imagined to occur through transcriptional read-through followed by processing of the resulting bicistronic transcript. Alternatively, residence at the UCS locus might relieve repression at a promoter adjacent to the PRT gene. At this point, however, little is known about PRT expression and function. It is not yet known if the PRT gene family is regulated at all, let alone in concert with MSG. Nevertheless, the genetic linkage of these genes evokes speculation about the possibility of coordinate regulation and the benefits of same. As already mentioned, coregulation would serve to vary both surface proteins, which would seem to be important even if the protease does not cleave the MSG protein. Alternatively, the protease may serve to cut MSG on the organism's surface. Such proteolysis could serve any of several ends, including removal of the UCS peptide, further modification of surface MSG by clipping off other small bits, and clipping off entire MSG molecules in order to shed an old coat. Here it is necessary to point out that shedding of MSG may not require proteolysis at all. MSG proteins lack a sequence of amino acids hydrophobic enough to be predicted to traverse the cell membrane; hence, MSGs are presumably attached to the cell in another way. One possibility is attachment via a glycosyl-phosphatidyl-inositol linkage (GPI). While GPI has not been shown to be on the MSG obtained from P. carinii organisms, neither has it been shown to be absent. In addition, MSG has been shown to able to accept a GPI moiety in mammalian cells (47). In these studies, a cDNA encoding the carboxyl-terminal part of an MSG from P. carinii f. sp. mustelae (ferret) was expressed in a mammalian cell line in such a way as to direct it to the cell surface. The protein made in the mammalian cells was found to be on the cell surface and to have GPI. Thus, if P. carinii organisms process MSG in the same way as mammalian cells do, MSG would be expected to acquire a GPI linker.
The PRT family is also notable for reasons separate from consideration of MSG. Other fungal genomes encode related proteases (such as the kexins of Saccharomyces cerevisiae [57]), but P. carinii f. sp. carinii appears to encode many more different kexin-like proteases than other fungi (83, 84, 126, 169). The reason for the large number of proteases in P. carinii f. sp. carinii is not known, but it may be related to surface variation. However, it should be noted that mouse P. carinii (P. carinii f. sp. muris) has been reported to contain a single protease gene, which was called kex1 (71). It is possible that the mouse and rat special forms of P. carinii differ with respect to the number of protease genes, in which case this difference underscores how different the special forms can be. Alternatively, both special forms may have multiple protease genes, in which case the kex1 gene would appear to not be a member of the mouse PRT gene family.
The model shown in Fig. 1 posits that only one MSG isoform is expressed per organism. Indirect immunofluorescence studies were consistent with this view; not all organisms within a population could be labeled with an antibody directed against a subset of MSG isoforms (3, 146). In addition, the fraction of organisms labeled by such an antibody varied among populations. Additional phenotypic evidence of limited MSG expression was provided by Western blotting, which showed that some P. carinii f. sp. carinii populations contained a particular MSG epitope in abundance and other populations did not (165).
The most direct evidence for the UCS locus exerting complete control over the MSG that is on the cell surface came from recent work with a monoclonal antibody called RA-C11, which recognizes a very small subset of the MSG isoforms encoded in the genome (78). In that study, the C11 epitope and the nucleotide sequence encoding it (C11 epitope-encoding sequence [EES]) were mapped. Then, three populations of P. carinii f. sp. carinii were identified that varied over a range of tenfold with respect to the fraction of cells with the C11 EES at the UCS locus. The same populations were analyzed by immunofluorescence to determine the fraction of cells with the C11 epitope on their surface. There was a strong correlation between the proportion of C11-reactive organisms and the proportion of organisms with the C11 EES attached to the expression site (128).
The model in Fig. 1 proposes that the MSG protein on the surface can be switched by changing the MSG gene at the expression site. How this is accomplished is not known. Figure 1B depicts one possible recombination mechanism, whereby a crossover occurs between the expressed gene and an unexpressed gene. Such crossovers could involve a site-specific recombinase because all MSG genes possess a common 23-bp sequence called the conserved recombination junction element (CRJE) (170), which could serve as the target of such a recombinase. However, recent findings have shown that the CRJE differs among special forms, so if a recombinase is at work, it would appear to have different targets in different special forms (49, 127, 179).
Once recombination is initiated, crossing over is not necessarily needed to generate diversity at the UCS locus. Instead, an MSG gene at another locus might donate some or all of its sequence to the UCS-linked MSG gene in a nonreciprocal manner (not shown in Fig. 1). If such gene conversion events were to occur, the number of different MSG isoforms that could be formed at the UCS locus would be virtually unlimited. In addition, gene conversion might tap a second source of sequence diversity, the MSR gene family, also known as variant MSG and type II MSG (54, 130, 168). This family was named MSR, which stands for MSG related, because MSR gene family members share sequence homology with MSG genes but are distinct from MSG genes by virtue of lacking a CRJE and linkage to the UCS (130). While whole MSR genes are not associated with the UCS locus, parts of MSR genes could be used to generate antigenic variation in UCS-linked MSG genes via gene conversion.
In closing this section, it is important to emphasize that the model shown in Fig. 1 was constructed based exclusively on data from P. carinii f. sp. carinii. It would not be surprising if regulation of MSG expression in other special forms differs in some ways from regulation in P. carinii f. sp. carinii. Indeed, studies on the other special form of P. carinii commonly found in laboratory rats, P. carinii f. sp. ratti, have revealed dramatic differences in structure between both the expression site and some MSG genes in this special form and those in P. carinii f. sp. carinii (127, 129). In both special forms, the UCS locus contains two exons. In P. carinii f. sp. carinii, both exons and the intron between them are located at a single locus, the MSG expression site. In P. carinii f. sp. ratti, exon I and the intron are at a single locus but copies of exon II are present upstream of MSG genes located at multiple loci, which are located on multiple chromosomes (127, 129). Thus, the uniqueness of the MSG expression site is conserved in the two special forms, but only part of the UCS (exon I and the intron) is encoded uniquely at the expression site in P. carinii f. sp. ratti, while in P. carinii f. sp. carinii, all of the UCS is encoded uniquely at the expression site. The presence of a unique expression site locus in both special forms suggests that this site controls transcription of MSG genes in both, but the differences in MSG gene and expression site structure suggest that switching the MSG gene at the expression site may occur by different mechanisms in different special forms. P. carinii f. sp. ratti has available relatively large tracts of sequence identity (copies of UCS exon II) that could support homologous recombination between expressed and silent MSG genes, while P. carinii f. sp. carinii lacks such tracts, the only region of sequence identity being the 23 bp provided by the CRJE, which is too short to support homologous recombination. Consequently, a site-specific recombination enzyme targeted to the CRJE remains the leading candidate recombination mechanism in P. carinii f. sp. carinii but may not be involved in the other special form. Whatever the functional consequences may be, this dramatic difference in these two special forms underscores the genetic differences between special forms of P. carinii and bolsters the notion that special forms are, in fact, different species.
FREQUENCY OF MSG SWITCHING
While the natural transmission model (i.e., infection via activation of latent organisms combined with inhalation of P. carinii-laden air) made it easy to observe variation at the UCS locus, studying switching was not possible in this system because this model does not permit the investigator to control three variables of infection, as follows: (i) the number of P. carinii that enter the rat, (ii) the time at which these organisms enter, and (iii) the MSG genes that are being expressed in the invading microbes. Without control of these variables, it is not possible to trace the pathway by which variation at the UCS locus arises. Recently, however, it was reported that intratracheal inoculation with a low dose of P. carinii can cause infections in rats with high efficiency (25). This finding encourages the hope that this approach will allow switching at the UCS locus to be studied. Indeed, the results of the initial low-dose inoculation studies suggest that switching might occur at a high rate (60). In these experiments, the complexity of the UCS-MSG junction was found to be a function of dose. Populations formed from 107 or 104 input organisms exhibited the same complex set of junctions as the input population, suggesting that these doses generated a duplicate of the input population. By contrast with the results obtained with rats inoculated with 104 or more organisms, the eight rats inoculated with 10 organisms produced P. carinii populations with far fewer different UCS-MSG junctions than the input population. In some rats in this group, 90% of the organisms recovered had the same DNA sequence at the UCS locus. The UCS-MSG junctions in the other 10% of the organisms were different from each other and from the majority UCS-MSG sequence. These data suggest that one of two things happened in these low-dose rats. Either (i) more than one organism propagated, but one expanded its numbers much more than the others, or (ii) only one of the 10 organisms introduced propagated, but some organisms changed the UCS-linked MSG as the population expanded. If the latter occurred, then switching can be calculated to have occurred at a frequency as high as 1 × 10−2 events per cell per generation (60). Further work is required to determine the cause of the UCS locus variation observed in the P. carinii recovered from animals inoculated with a low dose. It should be possible to use the quasiclonal organisms obtained from the low-dose-inoculated rats to establish infections in new animals, again via low-dose inoculation. Because the quasiclonal population is 90% homogeneous with respect to MSG, such a procedure would essentially eliminate the chance of introducing into a given rat organisms expressing different MSGs. Any heterogeneity at the UCS would then be unambiguously ascribable to switching.
COMPARISON OF P. CARINII TO OTHER MICROBIAL ANTIGEN VARIATION SYSTEMS
Surface variation is widespread in nature. In addition to P. carinii, eukaryotes that are known to exhibit this feature include another fungus, Candida albicans (2), and at least three protozoa, Giardia lamblia (149), Plasmodium falciparum (13, 120), and Trypanosoma brucei (121, 123). Special mechanisms that create phenotypic variation are also present in bacterial species such as Neisseria spp. (97, 102), Haemophilus influenzae (89), and Borrelia spp. (5, 118). Space limitations prohibit discussion of all of these systems. Besides, the mechanisms of most of them appear to have little in common with the variation mechanism employed by P. carinii. By contrast, the remarkable similarities between P. carinii and T. brucei and Borrelia spp. (29) warrant special consideration.
Both T. brucei and Borrelia hermsii enter the blood stream of a mammal and multiply there until they begin to be killed by an immune response directed against the major antigen that coats the pathogen's surface. Despite being exposed to the full force of the immune response, the microbe is not eliminated because variants with a coat that is not recognized by the host's defenses arise in the pathogen population. In T. brucei, the major variable surface antigen is called VSG. In Borrelia hermsii, VMP is the major variable antigen. In other Borrelia species, molecules similar to VMP are variable. The antigen coats of T. brucei and Borrelia spp. are dense and thick and apparently shield other membrane molecules from immune attack. Functions other than shielding are probable, but not much is known in this regard. VSGs are thought to inhibit phagocytosis by host defense cells (157). VMPs may confer the ability to adapt to different microenvironments within the host (16).
In T. brucei, switching the structure of the VSG coat is made possible by the VSG gene family, which has on the order of 1,000 members per trypanosome nucleus. In a given T. brucei organism at a given time, only one of the VSG genes is transcribed. Switching the surface VSG is accomplished by switching off the expressed VSG gene and switching on a previously silent one (14, 20, 98, 123). This switch can occur in either of the following two ways. (i) A DNA recombination event can put a previously silent VSG gene (or part of one) into an active expression site (10, 11, 27, 70, 81, 107–110, 122, 124, 159). (The genome contains about 20 loci, all telomeric, that are each capable of transcribing a linked VSG gene in the bloodstream form of the parasite, but only one such locus is active in a given parasite cell.) (ii) The expression of the active VSG gene can be curtailed, with a formerly silent VSG gene being turned on in situ (9, 20, 69, 98). How expression sites are activated and inactivated is not known, but recent evidence suggests that allowing elongation of transcripts may be key to activation (161; reviewed in reference 125). A third possible source of variation is mutation of VSG genes in situ (82, 119).
Importation of DNA from silent VSG genes appears to be mediated by DNA sequence homology and recombination events that can occur either in the VSG genes themselves or in a region occupied by 70-bp repeats (94, 155, 159). A site-specific nuclease targeted to the repeats may serve to initiate recombination (14). Recombination can be either reciprocal or by gene conversion (124). Reliance on homologous recombination may explain the nonrandomness of VSG expression that has been observed in many experiments. Certain VSGs tend to be expressed early in infection, and other VSGs are seen only at later times. These expression patterns have been proposed to be due to the higher probability that two closely related genes will undergo homologous exchange (155). Another possible influence on observed switching patterns is the dimerization of VSG. When a switch occurs, it may be necessary for the new VSG to be compatible with the old one during the transition period while both are present. An alternative hypothesis is that the nonrandom expression patterns observed are artifacts imposed by studying parasite lines that have been adapted to growth in the laboratory (see below) (121).
While several switching mechanisms (activation of an expression site, gene conversion of a gene already in an active expression site, and reciprocal exchange between active and inactive expression sites) have been observed in T. brucei, the relative contributions of these various mechanisms to pathogen survival in the wild is not clear (7, 121). A related area of uncertainty is the rate of switching, which has been estimated to be as low as 10−7 and as high as 10−2 events per cell per generation. The lower rates came from studies on laboratory lines of T. brucei, which were derived through repeated passaging by injection of large numbers of parasites from one laboratory animal into another. These lines are monomorphic, generating only long, slender parasites typical of the bloodstream stage and not short, stumpy parasites (7). By contrast, trypanosome lines that have not been passaged in this way remain pleomorphic. These pleomorphic lines have been reported to switch at a frequency of approximately 10−2 switches per cell per generation (156). It seems, then, that switching is at least 10,000-fold less frequent in monomorphic lines. More importantly, in pleomorphic lines, the switching mechanism that predominates involves recombination in the 70-bp repeats, while this type of switching is less of a factor in monomorphic lines (121). From these data, it has been suggested that the high-frequency recombination switching mechanism that is important in the wild is defective in monomorphic laboratory stocks (90, 121, 124).
The P. carinii MSG system resembles the trypanosome VSG system in several ways. Both organisms utilize a telomeric expression site to regulate expression of a gene family, and both are able to change the gene at the expression site via recombination. However, P. carinii has only one expression site, while trypanosomes have up to 20. In T. brucei, an expression site supports transcription of the battery of genes downstream of it (12). P. carinii may also be able to switch expression of more than one gene family (such as the protease and MSG families) in a concerted manner by transferring whole arrays of genes to the expression site. Both systems may initiate the DNA recombination events used for switching via a special mechanism. In both organisms, circumstantial evidence suggests that the initiator might be a site-specific nuclease. The CRJE of P. carinii and the 70-bp repeats of T. brucei may be targets for such nucleases. A major difference between P. carinii and trypanosomes is in the number of roles played by the expression site. The P. carinii expression site not only provides the RNA polymerase promoter needed to transcribe an MSG mRNA, it also furnishes the RNA sequence (UCS) needed to translate MSG mRNAs into proteins. Moreover, because translation begins in the UCS, it produces a protein that starts with a UCS-derived peptide sequence that seems to be required to get the MSG to the cell surface. By contrast, the trypanosome expression site appears to serve only to accomplish transcription of downstream genes, while not providing sequences needed for translation, peptide trafficking, and peptide processing.
In Borrelia spp., different VMPs are encoded by genes located on linear plasmids that have hairpin telomeres (6). In B. hermsii, the expressed gene is located at a telomeric expression site. Switches in VMP gene expression can occur either via gene conversion that changes the VMP gene at an expression site or via a deletion event involving an active VMP gene and an upstream inactive or pseudogene sequence (64, 118, 181). Conversion events seem to be initiated and terminated at 17-bp repeats that flank donor and recipient sequences. New VMP phenotypes arise readily during infections but not in vitro (181, 182). Phenotypic typing studies of organisms from infections imply that switching occurs frequently, at a rate as high as 10−4 to 10−3 switches per cell per generation (137). The regulation of switching (it does not take place in vitro), its high rate, and the involvement of the 17-bp repeats all suggest that a site-specific endonuclease may break the recipient DNA, thereby initiating double-strand break repair utilizing DNA from a donor gene. The CRJE of P. carinii may be used in a similar manner. Activation of VMP genes is often accompanied by the introduction of multiple point mutations in the coding region of the activated gene (118). The same thing may happen at the P. carinii UCS locus, which is much more polymorphic than other unique loci in this organism (129, 146).
It is striking that the fungus P. carinii, the protozoan T. brucei, and the bacterium B. hermsii all have a telomeric variable surface protein gene family, one member of which is transcribed from a telomeric expression site. The occurrence of a variation system constructed along the same general lines in such diverse taxa implies that the telomeric location of silent and expressed members of a gene family encoding variants of a surface protein arose early in the history of life. Alternatively, deposition of such genes at telomeres evolved more than once. Either way, a telomeric location for genes encoding variable surface antigens would appear to be very advantageous. The possible advantages of a telomeric location include allowing a silent gene to be moved to the expression site via a reciprocal exchange event without translocating major segments of the two participating chromosomes. At the same time, genes downstream of the locus that becomes joined to the expression site locus are moved downstream of a transcriptionally active locus, providing a means to switch on expression of a battery of linked genes. Another possible advantage of the telomeric system would be that new genes can be generated via unequal crossing over between existing genes, again with little effect on overall chromosome structure.
IMPORTANCE OF MSG SYSTEM
While the function of the MSG system is not known, its similarities to antigenic variation systems of other pathogenic microbes argue that its purpose is to favor P. carinii in its interactions with a mammalian host. These interactions may have two modes. One mode, conspicuous disease, appears in immunoincompetent hosts. The second mode, covert colonization, may occur in an immunocompetent host. Covert colonization is known to occur in laboratory animals and in wild animals (1, 26, 38, 68, 92, 162, 175, 176). Although P. carinii is rarely if ever detectable in healthy people (33, 96, 113, 131, 132) and recurrent Pcp in some AIDS patients appears to be due to exogenous infection (59, 61, 62), it seems likely that P. carinii can establish itself transiently in humans whose immune systems are functional but suboptimal. Some of the earliest reports of P. carinii were of people who were housed in crowded dwellings, eating poorly, and otherwise under stress, e.g., people in orphanages (22). More recently, there have been several reports of P. carinii in the lungs of patients who have been medically immunosuppressed after receiving an organ transplant (46, 52, 72; G. Nevez, C. Raccurt, V. Jounieaux, E. Dei-Cas, and E. Mazars, Letter, AIDS 13:535–536, 1999). PCR-detectable P. carinii DNA was also reported in nonimmunosuppressed patients with primary lung disease that was not P. carinii pneumonia (21, 135), and in healthy health-care workers (163), but the clinical significance of these low-level infections is controversial (E. Visconti, S. Marinaci, M. Zolfo, P. Mencarini, E. Tamburrini, G. Pagliari, E. Ortona, and A. Siracusano, Letter, J. Clin. Microbiol. 38:1307–1308, 2000). Nonetheless, its seems quite probable that P. carinii infects immunonaive humans, where it may cause serious disease (105, 164). It is clear that immunonaive animals are susceptible to infection and disease (26; B. Soulez, E. Dei-Cas, P. Charet, G. Mougeit, M. Callaux, and D. Camus, Letter, J. Infect. Dis. 160:355–356, 1989).
P. carinii may use antigenic variation to prolong its survival in a host that can raise an effective immune response. It has been clear for years that the cellular immune response is very important in protection from P. carinii infections (51). Perhaps the MSG system helps P. carinii to dull the cellular immune response. How this might happen is suggested by recent data on the interactions of T cells with a variable antigen from Trypanosoma cruzi. When T cells that were reactive with one version of the antigen were exposed to a mixture containing that version of the antigen plus other closely related peptides, normal activation did not occur (99). The antagonistic effect of peptides that are slightly different from a peptide agonist is thought to be an aspect of the system that normally regulates T-cell development and responsiveness (73). Microbes like P. carinii may produce mixtures of related antigens during their latent phase in order to exploit this system.
While the above mentioned considerations are intriguing, testing these ideas at the functional level is a very difficult problem. Presently, little is known about the possible influence of MSG variation on the immune response and on the fate of P. carinii at the hands of this response. Immunization studies have not been very informative. Attempts to use vaccination to elicit a protective immune state in laboratory animals have met with variable degrees of success, depending on the animal species and special form of P. carinii studied and the type of antigen used as the immunizing agent. In rats, immunization with either whole, dead P. carinii f. sp. carinii or partially purified MSG significantly reduced the number of P. carinii f. sp. carinii in the lungs of animals challenged with live P. carinii (153). In mice, only whole P. carinii f. sp. muris was protective (44). Purified MSG was not effective in this system. The reason for the different outcomes with rats and mice is not clear. Even if the data from mice and rats were concordant, there would still be a problem because either result, protection or lack of protection by MSG, fits with MSG variation. Protection can develop despite variation if a strong immune response is directed against a nonvariable part of MSG. On the other hand, lack of protection might be due to a failure of the immunization procedure to elicit an immune response to attack nonvariable MSG determinants. Progress in this area will be possible only when the MSG family is fully characterized and variable and nonvariable determinants are identified. Other advances in understanding the interplay between the MSG family and the immune system are sure to come from improvements in animal model systems. In the meantime, it seems reasonable to hold the view that the P. carinii MSG gene family is used to prolong survival in the face of immune attack.
Of course, the utility of the MSG family need not be limited to evasion of the immune system. One other function for this diverse family of surface proteins is the capacity to adhere to a variety of host structures (reviewed in reference 8). While MSG binding has been well studied, nearly all such experiments have used MSG prepared from P. carinii populations that must be presumed to be heterogeneous with respect to MSG. Therefore, the properties of individual MSG isoforms are largely unknown. Clearly, understanding this aspect of MSG function will require more details about the expression of MSG isoforms.
P. carinii may also use its collection of different MSG isoforms to allow it to live in more than one environment. P. carinii DNA has been detected in air traps set in unpopulated areas, suggesting that P. carinii may propagate itself outside of a host (171). Perhaps MSG complexity reflects occupation of multiple environmental niches. In addition, there are two morphological forms of P. carinii, called the cyst form and the trophic form, which tend to coexist in infected animals. These forms may be stages of a life cycle or may serve some other purpose. Little is known about MSG expression in these different morphological forms, but MSG is present in both (3, 28). Evidence for form-specific antigens has been presented, and it would not be surprising if some MSGs are form specific (19).
ACKNOWLEDGMENT
This work was supported by grant RO1 AI 36701 from the National Institutes of Health.
REFERENCES
- 1.Aliouat E M, Martinez A, Jimenez E, Dei-Cas E, Mullet C, Delcourt P, Gargallo-Viola D. Development of pneumocystosis animal models: corticosteroid-treated Wistar rat, SCID mouse and nude rat. J Eukaryot Microbiol. 1997;44:41S–42S. doi: 10.1111/j.1550-7408.1997.tb05765.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J M, Soll D R. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus C W, Tu A, Vogel P, Qin M, Kovacs J A. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J Exp Med. 1996;183:1229–1234. doi: 10.1084/jem.183.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. Revised nomenclature for Pneumocystis carinii. The Pneumocystis Workshop. J Eukaryot Microbiol. 1994;41:121S–122S. [PubMed] [Google Scholar]
- 5.Barbour A G, Burman N, Carter C J, Kitten T, Bergstrom S. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 7.Barry J D. The relative significance of mechanisms of antigen variation in African Trypanosomes. Parasitol Today. 1997;13:212–218. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- 8.Benfield T L, Lundgren J D. The Pneumocystis carinii major surface glycoprotein (MSG): its potential involvement in the pathophysiology of pneumocystosis. FEMS Immunol Med Microbiol. 1998;22:129–134. doi: 10.1111/j.1574-695X.1998.tb01197.x. [DOI] [PubMed] [Google Scholar]
- 9.Bernards A, de Lange T, Michels P A, Liu A Y, Huisman M J, Borst P. Two modes of activation of a single surface antigen gene of Trypanosoma brucei. Cell. 1984;36:163–170. doi: 10.1016/0092-8674(84)90085-0. [DOI] [PubMed] [Google Scholar]
- 10.Bernards A, Kooter J M, Borst P. Structure and transcription of a telomeric surface antigen gene of Trypanosoma brucei. Mol Cell Biol. 1985;5:545–553. doi: 10.1128/mcb.5.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernards A, Van der Ploeg L H, Frasch A C, Borst P, Boothroyd J C, Coleman S, Cross G A. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]
- 12.Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 13.Borst P, Bitter W, McCulloch R, Van Leeuwen F, Rudenko G. Antigenic variation in malaria. Cell. 1995;82:1–4. doi: 10.1016/0092-8674(95)90044-6. [DOI] [PubMed] [Google Scholar]
- 14.Borst P, Rudenko G, Taylor M C, Blundell P A, Van Leeuwen F, Bitter W, Cross M, McCulloch R. Antigenic variation in trypanosomes. Arch Med Res. 1996;27:379–388. [PubMed] [Google Scholar]
- 15.Boylan C J, Current W L. Improved rat model of Pneumocystis carinii pneumonia: induced laboratory infections in Pneumocystis-free animals. Infect Immun. 1992;60:1589–1597. doi: 10.1128/iai.60.4.1589-1597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadavid D, Thomas D D, Crawley R, Barbour A G. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J Exp Med. 1994;179:631–642. doi: 10.1084/jem.179.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cere N, Drouet-Viard F, Dei-Cas E, Chanteloup N, Coudert P. In utero transmission of Pneumocystis carinii sp. f. oryctolagi. Parasite. 1997;4:325–330. doi: 10.1051/parasite/1997044325. [DOI] [PubMed] [Google Scholar]
- 18.Chandler F W, Jr, Frenkel J K, Campbell W G., Jr Pneumocystis pneumonia. Animal model: Pneumocystis carinii pneumonia in the immunosuppressed rat. Am J Pathol. 1979;95:571–574. [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterton J M, Joss A W, Pennington T H, Ho-Yen D O. Differences exist in the immunoblotting profiles of cyst and trophozoite antigens of Pneumocystis carinii. J Med Microbiol. 1995;42:120–126. doi: 10.1099/00222615-42-2-120. [DOI] [PubMed] [Google Scholar]
- 20.Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Contini C, Villa M P, Romani R, Merolla R, Delia S, Ronchetti R. Detection of Pneumocystis carinii among children with chronic respiratory disorders in the absence of HIV infection and immunodeficiency. J Med Microbiol. 1998;47:329–333. doi: 10.1099/00222615-47-4-329. [DOI] [PubMed] [Google Scholar]
- 22.Cushion M T. Transmission and epidemiology. In: Walzer P D, editor. Pneumocystis carinii pneumonia. New York, N.Y: Marcel Dekker; 1994. pp. 123–140. [Google Scholar]
- 23.Cushion M T. Genetic heterogeneity of rat-derived Pneumocystis. FEMS Immunol Med Microbiol. 1998;22:51–58. doi: 10.1111/j.1574-695X.1998.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 24.Cushion M T, Kaselis M, Stringer S L, Stringer J R. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cushion M T, Linke M J, Collins M, Keely S P, Stringer J R. The minimum number of Pneumocystis carinii f. sp. carinii organisms required to establish infections is very low. J Eukaryot Microbiol. 1999;46:111S. [PubMed] [Google Scholar]
- 26.Dei-Cas E, Brun-Pascaud M, Bille-Hansen V, Allaert A, Aliouat E M. Animal models of pneumocystosis. FEMS Immunol Med Microbiol. 1998;22:163–168. doi: 10.1111/j.1574-695X.1998.tb01201.x. [DOI] [PubMed] [Google Scholar]
- 27.de Lange T, Kooter J M, Michels P A, Borst P. Telomere conversion in trypanosomes. Nucleic Acids Res. 1983;11:8149–8165. doi: 10.1093/nar/11.23.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Stefano J A, Myers J D, Du Pont D, Foy J M, Theus S A, Walzer P D. Cell wall antigens of Pneumocystis carinii trophozoites and cysts: purification and carbohydrate analysis of these glycoproteins. J Eukaryot Microbiol. 1998;45:334–343. doi: 10.1111/j.1550-7408.1998.tb04545.x. [DOI] [PubMed] [Google Scholar]
- 29.Donelson J E. Mechanisms of antigenic variation in Borrelia hermsii and African trypanosomes. J Biol Chem. 1995;270:7783–7786. doi: 10.1074/jbc.270.14.7783. [DOI] [PubMed] [Google Scholar]
- 30.Edlind T D, Bartlett M S, Weinberg G A, Prah G N, Smith J W. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992;6:3365–3373. doi: 10.1111/j.1365-2958.1992.tb02204.x. [DOI] [PubMed] [Google Scholar]
- 31.Edman J C, Hatton T W, Nam M, Turner R, Mei Q, Angus C W, Kovacs J A. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 1996;15:989–999. doi: 10.1089/dna.1996.15.989. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson O E. Pneumocystis carinii, a parasite in lungs of mammals, referred to a new family and order (Pneumocystidaceae, Pneumocystidales, Ascomycota) Syst Ascomycetum. 1994;13:165–180. [Google Scholar]
- 33.Esterly J A. Pneumocystis carinii in lungs of adults at autopsy. Am Rev Respir Dis. 1968;97:935–937. doi: 10.1164/arrd.1968.97.5.935. [DOI] [PubMed] [Google Scholar]
- 34.Ezekowitz R A, Williams D J, Koziel H, Armstrong M Y, Warner A, Richards F F, Rose R M. Uptake of Pneumocystis carinii mediated by the macrophage mannose receptor. Nature. 1991;351:155–158. doi: 10.1038/351155a0. [DOI] [PubMed] [Google Scholar]
- 35.Fisher D J, Gigliotti F, Zauderer M, Harmsen A G. Specific T-cell response to a Pneumocystis carinii surface glycoprotein (gp120) J Protozool. 1991;38:22S–23S. [PubMed] [Google Scholar]
- 36.Fisher D J, Gigliotti F, Zauderer M, Harmsen A G. Specific T-cell response to a Pneumocystis carinii surface glycoprotein (gp120) after immunization and natural infection. Infect Immun. 1991;59:3372–3376. doi: 10.1128/iai.59.10.3372-3376.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frenkel J K. Pneumocystis jiroveci n. sp. from man: morphology, physiology, and immunology in relation to pathology. Natl Cancer Inst Monogr. 1976;43:13–30. [PubMed] [Google Scholar]
- 38.Frenkel J K, Good J T, Shultz J A. Latent Pneumocystis infection of rats, relapse, and chemotherapy. Lab Investig. 1966;15:1559–1577. [PubMed] [Google Scholar]
- 39.Garbe T R, Stringer J R. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994;62:3092–3101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gigliotti F. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;165:329–336. doi: 10.1093/infdis/165.2.329. [DOI] [PubMed] [Google Scholar]
- 41.Gigliotti F, Ballou L R, Hughes W T, Mosley B D. Purification and initial characterization of a ferret Pneumocystis carinii surface antigen. J Infect Dis. 1988;158:848–854. doi: 10.1093/infdis/158.4.848. [DOI] [PubMed] [Google Scholar]
- 42.Gigliotti F, Harmsen A G, Haidaris C G, Haidaris P J. Pneumocystis carinii is not universally transmissible between mammalian species. Infect Immun. 1993;61:2886–2890. doi: 10.1128/iai.61.7.2886-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gigliotti F, McCool T. Glycoprotein A is the immunodominant antigen of Pneumocystis carinii in mice following immunization. Parasitol Res. 1996;82:90–91. doi: 10.1007/s004360050075. [DOI] [PubMed] [Google Scholar]
- 44.Gigliotti F, Wiley J A, Harmsen A G. Immunization with Pneumocystis carinii gpA is immunogenic but not protective in a mouse model of P. carinii pneumonia. Infect Immun. 1998;66:3179–3182. doi: 10.1128/iai.66.7.3179-3182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graves D C, McNabb S J, Worley M A, Downs T D, Ivey M II. Analyses of rat Pneumocystis carinii antigens recognized by human and rat antibodies by using Western immunoblotting. Infect Immun. 1986;54:96–103. doi: 10.1128/iai.54.1.96-103.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gryzan S, Paradis I L, Zeevi A, Duquesnoy R J, Dummer J S, Griffith B P, Hardesty R L, Trento A, Nalesnik M A, Dauber J H. Unexpectedly high incidence of Pneumocystis carinii infection after lung-heart transplantation. Implications for lung defense and allograft survival. Am Rev Respir Dis. 1988;137:1268–1274. doi: 10.1164/ajrccm/137.6.1268. [DOI] [PubMed] [Google Scholar]
- 47.Guadiz G, Haidaris C G, Maine G N, Simpson-Haidaris P J. The carboxyl terminus of Pneumocystis carinii glycoprotein A encodes a functional glycosylphosphatidylinositol signal sequence. J Biol Chem. 1998;273:26202–26209. doi: 10.1074/jbc.273.40.26202. [DOI] [PubMed] [Google Scholar]
- 48.Haidaris C G, Fisher D J, Gigliotti F, Simpson-Haidaris P J. Antigenic properties of recombinant glycosylated and nonglycosylated Pneumocystis carinii glycoprotein A polypeptides expressed in baculovirus-infected insect cells. Mol Biotechnol. 1998;9:91–97. doi: 10.1007/BF02760811. [DOI] [PubMed] [Google Scholar]
- 49.Haidaris C G, Medzihradsky O F, Gigliotti F, Simpson-Haidaris P J. Molecular characterization of mouse Pneumocystis carinii surface glycoprotein A. DNA Res. 1998;5:77–85. doi: 10.1093/dnares/5.2.77. [DOI] [PubMed] [Google Scholar]
- 50.Haidaris P J, Wright T W, Gigliotti F, Haidaris C G. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;166:1113–1123. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 51.Harmsen A G, Stankiewicz M. Requirement for CD4+ cells in resistance to Pneumocystis carinii pneumonia in mice. J Exp Med. 1990;172:937–945. doi: 10.1084/jem.172.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauser P M, Blanc D S, Bille J, Nahimana A, Francioli P. Carriage of Pneumocystis carinii by immunosuppressed patients and molecular typing of the organisms. AIDS. 2000;14:461–463. doi: 10.1097/00002030-200003100-00022. [DOI] [PubMed] [Google Scholar]
- 53.Hendley J O, Weller T H. Activation and transmission in rats of infection with Pneumocystis. Proc Soc Exp Biol Med. 1971;137:1401–1404. doi: 10.3181/00379727-137-35798. [DOI] [PubMed] [Google Scholar]
- 54.Huang S N, Angus C W, Turner R E, Sorial V, Kovacs J A. Identification and characterization of novel variant major surface glycoprotein gene families in rat Pneumocystis carinii. J Infect Dis. 1999;179:192–200. doi: 10.1086/314558. [DOI] [PubMed] [Google Scholar]
- 55.Hughes W T. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 56.Hughes W, editor. Pneumocystis carinii pneumonitis. Boca Raton, Fla: CRC Press; 1987. pp. 57–70. [Google Scholar]
- 57.Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 58.Keely S, Pai H J, Baughman R, Sidman C, Sunkin S M, Stringer J R, Stringer S L. Pneumocystis species inferred from analysis of multiple genes. J Eukaryot Microbiol. 1994;41:94S. [PubMed] [Google Scholar]
- 59.Keely S P, Baughman R P, Smulian A G, Dohn M N, Stringer J R. Source of Pneumocystis carinii in recurrent episodes of pneumonia in AIDS patients. AIDS. 1996;10:881–888. doi: 10.1097/00002030-199607000-00011. [DOI] [PubMed] [Google Scholar]
- 60.Keely S P, Cushion M T, Stringer J R. Determination of the maximum frequency of genetic rearrangements associated with Pneumocystis carinii surface antigen variation. J Eukaryot Microbiol. 1999;46:128S. [PubMed] [Google Scholar]
- 61.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keely S P, Stringer J R, Baughman R P, Linke M J, Walzer P D, Smulian A G. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172:595–598. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 63.Kitada K, Wada M, Nakamura Y. Multi-gene family of major surface glycoproteins of Pneumocystis carinii: full-size cDNA cloning and expression. DNA Res. 1994;1:57–66. doi: 10.1093/dnares/1.2.57. [DOI] [PubMed] [Google Scholar]
- 64.Kitten T, Barbour A G. Juxtaposition of expressed variable antigen genes with a conserved telomere in the bacterium Borrelia hermsii. Proc Natl Acad Sci USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kovacs J A, Halpern J L, Swan J C, Moss J, Parrillo J E, Masur H. Identification of antigens and antibodies specific for Pneumocystis carinii. J Immunol. 1988;140:2023–2031. [PubMed] [Google Scholar]
- 66.Kovacs J A, Lundgren B, Masur H. Identification of antigens specific for Pneumocystis carinii. J Protozool. 1989;36:67S–69S. doi: 10.1111/j.1550-7408.1989.tb02706.x. [DOI] [PubMed] [Google Scholar]
- 67.Kovacs J A, Powell F, Edman J C, Lundgren B, Martinez A, Drew B, Angus C W. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J Biol Chem. 1993;268:6034–6040. [PubMed] [Google Scholar]
- 68.Laakkonen J. Pneumocystis carinii in wildlife. Int J Parasitol. 1998;28:241–252. doi: 10.1016/s0020-7519(97)00155-0. [DOI] [PubMed] [Google Scholar]
- 69.Laurent M, Pays E, Delinte K, Magnus E, Van Meirvenne N, Steinert M. Evolution of a trypanosome surface antigen gene repertoire linked to non-duplicative gene activation. Nature. 1984;308:370–373. doi: 10.1038/308370a0. [DOI] [PubMed] [Google Scholar]
- 70.Laurent M, Pays E, Magnus E, Van Meirvenne N, Matthyssens G, Williams R O, Steinert M. DNA rearrangements linked to expression of a predominant surface antigen gene of trypanosomes. Nature. 1983;302:263–266. doi: 10.1038/302263a0. [DOI] [PubMed] [Google Scholar]
- 71.Lee L H, Gigliotti F, Wright T W, Simpson-Haidaris P J, Weinberg G A, Haidaris C G. Molecular characterization of KEX1, a kexin-like protease in mouse Pneumocystis carinii. Gene. 2000;242:141–150. doi: 10.1016/s0378-1119(99)00533-8. [DOI] [PubMed] [Google Scholar]
- 72.Leigh T R, Wakefield A E, Peters S E, Hopkin J M, Collins J V. Comparison of DNA amplification and immunofluorescence for detecting Pneumocystis carinii in patients receiving immunosuppressive therapy. Transplantation. 1992;54:468–470. doi: 10.1097/00007890-199209000-00016. [DOI] [PubMed] [Google Scholar]
- 73.Levelt C N, Mizoguchi E, Huang X, Zacks R, Bhan A K, Tonegawa S. Inhibition of intrathymic T cell development by expression of a transgenic antagonist peptide. Proc Natl Acad Sci USA. 1998;95:14349–14354. doi: 10.1073/pnas.95.24.14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Limper A H, Pottratz S T, Martin W J. Modulation of Pneumocystis carinii adherence to cultured lung cells by a mannose-dependent mechanism. J Lab Clin Med. 1991;118:492–499. [PubMed] [Google Scholar]
- 75.Limper A H, Standing J E, Hoffman O A, Castro M, Neese L W. Vitronectin binds to Pneumocystis carinii and mediates organism attachment to cultured lung epithelial cells. Infect Immun. 1993;61:4302–4309. doi: 10.1128/iai.61.10.4302-4309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Linke M J, Cushion M T, Walzer P D. Properties of the major antigens of rat and human Pneumocystis carinii. Infect Immun. 1989;57:1547–1555. doi: 10.1128/iai.57.5.1547-1555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Linke M J, Smulian A G, Stringer J R, Walzer P D. Characterization of multiple unique cDNAs encoding the major surface glycoprotein of rat-derived Pneumocystis carinii. Parasitol Res. 1994;80:478–486. doi: 10.1007/BF00932694. [DOI] [PubMed] [Google Scholar]
- 78.Linke M J, Smulian A G, Yoshihara P, Walzer P D. Production and characterization of monoclonal antibodies specific for the major surface glycoprotein of Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:99S–100S. [PubMed] [Google Scholar]
- 79.Linke M J, Walzer P D. Analysis of a surface antigen of Pneumocystis carinii. J Protozool. 1989;36:60S–61S. doi: 10.1111/j.1550-7408.1989.tb02701.x. [DOI] [PubMed] [Google Scholar]
- 80.Linke M J, Walzer P D. Identification and purification of a soluble species of gp120 released by zymolyase treatment of Pneumocystis carinii. J Protozool. 1991;38:176S–178S. [PubMed] [Google Scholar]
- 81.Liu A Y, Van der Ploeg L H, Rijsewijk F A, Borst P. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences. J Mol Biol. 1983;167:57–75. doi: 10.1016/s0022-2836(83)80034-5. [DOI] [PubMed] [Google Scholar]
- 82.Lu Y, Alarcon C M, Hall T, Reddy L V, Donelson J E. A strand bias occurs in point mutations associated with variant surface glycoprotein gene conversion in Trypanosoma rhodesiense. Mol Cell Biol. 1994;14:3971–3980. doi: 10.1128/mcb.14.6.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lugli E B, Allen A G, Wakefield A E. A Pneumocystis carinii multi-gene family with homology to subtilisin-like serine proteases. Microbiology. 1997;143:2223–2236. doi: 10.1099/00221287-143-7-2223. [DOI] [PubMed] [Google Scholar]
- 84.Lugli E B, Bampton E T, Ferguson D J, Wakefield A E. Cell surface protease PRT1 identified in the fungal pathogen Pneumocystis carinii. Mol Microbiol. 1999;31:1723–1733. doi: 10.1046/j.1365-2958.1999.01306.x. [DOI] [PubMed] [Google Scholar]
- 85.Lundgren B, Cotton R, Lundgren J D, Edman J C, Kovacs J A. Identification of Pneumocystis carinii chromosomes and mapping of five genes. Infect Immun. 1990;58:1705–1710. doi: 10.1128/iai.58.6.1705-1710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lundgren B, Koch C, Mathiesen L, Nielsen J O, Hansen J E. Glycosylation of the major human Pneumocystis carinii surface antigen. APMIS. 1993;101:194–200. [PubMed] [Google Scholar]
- 87.Lundgren B, Lipschik G Y, Kovacs J A. Purification and characterization of a major human Pneumocystis carinii surface antigen. J Clin Investig. 1991;87:163–170. doi: 10.1172/JCI114966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lundgren B, Lundgren J D, Nielsen T, Mathiesen L, Nielsen J O, Kovacs J A. Antibody responses to a major Pneumocystis carinii antigen in human immunodeficiency virus-infected patients with and without P. carinii pneumonia. J Infect Dis. 1992;165:1151–1155. doi: 10.1093/infdis/165.6.1151. [DOI] [PubMed] [Google Scholar]
- 89.Maskell D J, Szabo M J, Butler P D, Williams A E, Moxon E R. Molecular biology of phase-variable lipopolysaccharide biosynthesis by Haemophilus influenzae. J Infect Dis. 1992;165(Suppl. 1):S90–S92. doi: 10.1093/infdis/165-supplement_1-s90. [DOI] [PubMed] [Google Scholar]
- 90.Matthews K R, Shiels P G, Graham S V, Cowan C, Barry J D. Duplicative activation mechanisms of two trypanosome telomeric VSG genes with structurally simple 5′ flanks. Nucleic Acids Res. 1990;18:7219–7227. doi: 10.1093/nar/18.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mazars E, Dei-Cas E. Epidemiological and taxonomic impact of Pneumocystis biodiversity. FEMS Immunol Med Microbiol. 1998;22:75–80. doi: 10.1111/j.1574-695X.1998.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 92.Mazars E, Guyot K, Fourmaintraux S, Renaud F, Petavy F, Camus D, Dei-Cas E. Detection of Pneumocystis in European wild animals. J Eukaryot Microbiol. 1997;44:39S. doi: 10.1111/j.1550-7408.1997.tb05763.x. [DOI] [PubMed] [Google Scholar]
- 93.McCormack F X, Festa A L, Andrews R P, Linke M, Walzer P D. The carbohydrate recognition domain of surfactant protein A mediates binding to the major surface glycoprotein of Pneumocystis carinii. Biochemistry. 1997;36:8092–8099. doi: 10.1021/bi970313f. [DOI] [PubMed] [Google Scholar]
- 94.McCulloch R, Barry J D. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merali S, Frevert U, Williams J H, Chin K, Bryan R, Clarkson A B., Jr Continuous axenic cultivation of Pneumocystis carinii. Proc Natl Acad Sci USA. 1999;96:2402–2407. doi: 10.1073/pnas.96.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meuwissen J H. Infections with Pneumocystis carinii. Natl Cancer Inst Monogr. 1976;43:133–136. [PubMed] [Google Scholar]
- 97.Meyer T F, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 98.Michels P A, Van der Ploeg L H, Liu A Y, Borst P. The inactivation and reactivation of an expression-linked gene copy for a variant surface glycoprotein in Trypanosoma brucei. EMBO J. 1984;3:1345–1351. doi: 10.1002/j.1460-2075.1984.tb01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Millar A E, Wleklinski-Lee M, Kahn S J. The surface protein superfamily of Trypanosoma cruzi stimulates a polarized Th1 response that becomes anergic. J Immunol. 1999;162:6092–6099. [PubMed] [Google Scholar]
- 100.Mills J. Pneumocystis carinii and Toxoplasma gondii infections in patients with AIDS. Rev Infect Dis. 1986;8:1001–1011. doi: 10.1093/clinids/8.6.1001. [DOI] [PubMed] [Google Scholar]
- 101.Nakamura Y, Tanabe K, Egawa K. Structure of major surface determinants and DNA diagnosis of Pneumocystis carinii. J Protozool. 1989;36:58S–60S. doi: 10.1111/j.1550-7408.1989.tb02699.x. [DOI] [PubMed] [Google Scholar]
- 102.Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol. 1993;8:719–725. doi: 10.1111/j.1365-2958.1993.tb01615.x. [DOI] [PubMed] [Google Scholar]
- 103.O'Leary T J, Tsai M M, Wright C F, Cushion M T. Use of semiquantitative PCR to assess onset and treatment of Pneumocystis carinii infection in rat model. J Clin Microbiol. 1995;33:718–724. doi: 10.1128/jcm.33.3.718-724.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O'Riordan D M, Standing J E, Limper A H. Pneumocystis carinii glycoprotein A binds macrophage mannose receptors. Infect Immun. 1995;63:779–784. doi: 10.1128/iai.63.3.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Panero A, Roggini M, Papoff P, Moretti C, Contini C, Bucci G. Pneumocystis carinii pneumonia in preterm infants: report of two cases successfully diagnosed by non-bronchoscopic bronchoalveolar lavage. Acta Paediatr. 1995;84:1309–1311. doi: 10.1111/j.1651-2227.1995.tb13555.x. [DOI] [PubMed] [Google Scholar]
- 106.Paulsrud J R, Queener S F, Bartlett M S, Smith J W. Isolation and characterization of rat lung Pneumocystis carinii gp120. J Protozool. 1991;38:10S–11S. [PubMed] [Google Scholar]
- 107.Pays E, Guyaux M, Aerts D, Van Meirvenne N, Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- 108.Pays E, Lheureux M, Steinert M. The expression-linked copy of a surface antigen gene in Trypanosoma is probably the one transcribed. Nature. 1981;292:265–267. doi: 10.1038/292265a0. [DOI] [PubMed] [Google Scholar]
- 109.Pays E, Van Assel S, Laurent M, Darville M, Vervoort T, Van Meirvenne N, Steinert M. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell. 1983;34:371–381. doi: 10.1016/0092-8674(83)90371-9. [DOI] [PubMed] [Google Scholar]
- 110.Pays E, Van Meirvenne N, Le Ray D, Steinert M. Gene duplication and transposition linked to antigenic variation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1981;78:2673–2677. doi: 10.1073/pnas.78.5.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peglow S L, Smulian A G, Linke M J, Pogue C L, Nurre S, Crisler J, Phair J, Gold J W, Armstrong D, Walzer P D. Serologic responses to Pneumocystis carinii antigens in health and disease. J Infect Dis. 1990;161:296–306. doi: 10.1093/infdis/161.2.296. [DOI] [PubMed] [Google Scholar]
- 112.Pesanti E L, Shanley J D. Glycoproteins of Pneumocystis carinii: characterization by electrophoresis and microscopy. J Infect Dis. 1988;158:1353–1359. doi: 10.1093/infdis/158.6.1353. [DOI] [PubMed] [Google Scholar]
- 113.Peters S E, Wakefield A E, Sinclair K, Millard P R, Hopkin J M. A search for Pneumocystis carinii in post-mortem lungs by DNA amplification. J Pathol. 1992;166:195–198. doi: 10.1002/path.1711660217. [DOI] [PubMed] [Google Scholar]
- 114.Peters S E, Wakefield A E, Whitwell K E, Hopkin J M. Pneumocystis carinii pneumonia in thoroughbred foals: identification of a genetically distinct organism by DNA amplification. J Clin Microbiol. 1994;32:213–216. doi: 10.1128/jcm.32.1.213-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pottratz S T, Martin W J. Role of fibronectin in Pneumocystis carinii attachment to cultured lung cells. J Clin Investig. 1990;85:351–356. doi: 10.1172/JCI114445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pottratz S T, Paulsrud J, Smith J S, Martin W J. Pneumocystis carinii attachment to cultured lung cells by pneumocystis gp 120, a fibronectin binding protein. J Clin Investig. 1991;88:403–407. doi: 10.1172/JCI115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Radding J A, Armstrong M Y, Ullu E, Richards F F. Identification and isolation of a major cell surface glycoprotein of Pneumocystis carinii. Infect Immun. 1989;57:2149–2157. doi: 10.1128/iai.57.7.2149-2157.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Restrepo B I, Barbour A G. Antigen diversity in the bacterium B. hermsii through “somatic” mutations in rearranged vmp genes. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 119.Rice-Ficht A C, Chen K K, Donelson J E. Point mutations during generation of expression-linked extra copy of trypanosome surface glycoprotein gene. Nature. 1982;298:676–679. doi: 10.1038/298676a0. [DOI] [PubMed] [Google Scholar]
- 120.Roberts D J, Craig A G, Berendt A R, Pinches R, Nash G, Marsh K, Newbold C I. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Robinson N P, Burman N, Melville S E, Barry J D. Predominance of duplicative VSG gene conversion in antigen variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roth C, Bringaud F, Layden R E, Baltz T, Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc Natl Acad Sci USA. 1989;86:9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rudenko G, Cross M, Borst P. Changing the end: antigenic variation orchestrated at the telomeres of African trypanosomes. Trends Microbiol. 1998;6:113–116. doi: 10.1016/s0966-842x(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 124.Rudenko G, McCulloch R, Dirks-Mulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- 125.Rudenko G. Genes involved in phenotypic and antigenic variation in African trypanosomes and malaria. Curr Opin Microbiol. 1999;2:651–656. doi: 10.1016/s1369-5274(99)00039-9. [DOI] [PubMed] [Google Scholar]
- 126.Russian D A, Andrawis-Sorial V, Goheen M P, Edman J C, Vogel P, Turner R E, Klivington D L, Angus C W, Kovacs J A. Characterization of a multicopy family of genes encoding a surface-expressed serine endoprotease in rat Pneumocystis carinii. Proc Assoc Am Physicians. 1999;111:347–356. doi: 10.1046/j.1525-1381.1999.99118.x. [DOI] [PubMed] [Google Scholar]
- 127.Schaffzin J K, Garbe T R, Stringer J R. Major surface glycoprotein genes from Pneumocystis carinii f. sp. ratti. Fungal Genet Biol. 1999;28:214–226. doi: 10.1006/fgbi.1999.1171. [DOI] [PubMed] [Google Scholar]
- 128.Schaffzin J K, Stringer J R. Direct correlation of genomic localization and surface expression of the major surface glycoprotein of Pneumocystis carinii. J Eukaryot Microbiol. 1999;46:127S. [PubMed] [Google Scholar]
- 129.Schaffzin J K, Stringer J R. The major surface glycoprotein expression sites of two special forms of rat Pneumocystis carinii differ in structure. J Infect Dis. 2000;181:1729–1739. doi: 10.1086/315438. [DOI] [PubMed] [Google Scholar]
- 130.Schaffzin J K, Sunkin S M, Stringer J R. A new family of Pneumocystis carinii genes related to those encoding the major surface glycoprotein. Curr Genet. 1999;35:134–143. doi: 10.1007/s002940050442. [DOI] [PubMed] [Google Scholar]
- 131.Sedaghatian M R, Singer D B. Pneumocystis carinii in children with malignant disease. Cancer. 1972;29:772–777. doi: 10.1002/1097-0142(197203)29:3<772::aid-cncr2820290333>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 132.Settnes O P, Genner J. Pneumocystis carinii in human lungs at autopsy. Scand J Infect Dis. 1986;18:489–496. doi: 10.3109/00365548609021652. [DOI] [PubMed] [Google Scholar]
- 133.Settnes O P, Nielsen P B, Bucala R, Linke M J, Cushion M T. A survey of birds in Denmark for the presence of Pneumocystis carinii. Avian Dis. 1994;38:1–10. [PubMed] [Google Scholar]
- 134.Sinclair K, Wakefield A E, Banerji S, Hopkin J M. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 135.Sing A, Roggenkamp A, Autenrieth I B, Heesemann J. Pneumocystis carinii carriage in immunocompetent patients with primary pulmonary disorders as detected by single or nested PCR. J Clin Microbiol. 1999;37:3409–3410. doi: 10.1128/jcm.37.10.3409-3410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smulian A, Walzer P. Serological studies of Pneumocystis carinii infection. In: Walzer P, editor. Pneumocystis carinii pneumonia. New York, N.Y: Marcel Dekker; 1994. pp. 141–149. [Google Scholar]
- 137.Stoenner H G, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stringer J R. The identity of Pneumocystis carinii: not a single protozoan, but a diverse group of exotic fungi. Infect Agents Dis. 1993;2:109–117. [PubMed] [Google Scholar]
- 139.Stringer J R. Pneumocystis carinii: what is it, exactly? Clin Microbiol Rev. 1996;9:489–498. doi: 10.1128/cmr.9.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stringer J R, Cushion M T. The genome of Pneumocystis carinii. FEMS Immunol Med Microbiol. 1998;22:15–26. doi: 10.1111/j.1574-695X.1998.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 141.Stringer J R, Stringer S L, Zhang J, Baughman R, Smulian A G, Cushion M T. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 142.Stringer J R, Wakefield A E, Cushion M T, Dei-Cas E. Pneumocystis taxonomy and nomenclature: an update. J Eukaryot Microbiol. 1997;44:5S–6S. doi: 10.1111/j.1550-7408.1997.tb05736.x. [DOI] [PubMed] [Google Scholar]
- 143.Stringer S L, Garbe T, Sunkin S M, Stringer J R. Genes encoding antigenic surface glycoproteins in Pneumocystis from humans. J Eukaryot Microbiol. 1993;40:821–826. doi: 10.1111/j.1550-7408.1993.tb04481.x. [DOI] [PubMed] [Google Scholar]
- 144.Stringer S L, Hong S T, Giuntoli D, Stringer J R. Repeated DNA in Pneumocystis carinii. J Clin Microbiol. 1991;29:1194–1201. doi: 10.1128/jcm.29.6.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sunkin S M, Linke M J, McCormack F X, Walzer P D, Stringer J R. Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii. Infect Immun. 1998;66:741–746. doi: 10.1128/iai.66.2.741-746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sunkin S M, Stringer J R. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol Microbiol. 1996;19:283–295. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- 147.Sunkin S M, Stringer J R. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol Microbiol. 1997;25:147–160. doi: 10.1046/j.1365-2958.1997.4461806.x. [DOI] [PubMed] [Google Scholar]
- 148.Sunkin S M, Stringer S L, Stringer J R. A tandem repeat of rat-derived Pneumocystis carinii genes encoding the major surface glycoprotein. J Eukaryot Microbiol. 1994;41:292–300. doi: 10.1111/j.1550-7408.1994.tb01509.x. [DOI] [PubMed] [Google Scholar]
- 149.Svard S G, Meng T C, Hetsko M L, McCaffery J M, Gillin F D. Differentiation-associated surface antigen variation in the ancient eukaryote Giardia lamblia. Mol Microbiol. 1998;30:979–989. doi: 10.1046/j.1365-2958.1998.01125.x. [DOI] [PubMed] [Google Scholar]
- 150.Tanabe K, Takasaki S, Watanabe J, Kobata A, Egawa K, Nakamura Y. Glycoproteins composed of major surface immunodeterminants of Pneumocystis carinii. Infect Immun. 1989;57:1363–1368. doi: 10.1128/iai.57.5.1363-1368.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Theus S A, Andrews R P, Linke M J, Walzer P D. Characterization of rat CD4 T cell clones specific for the major surface glycoprotein of Pneumocystis carinii. J Eukaryot Microbiol. 1997;44:96–100. doi: 10.1111/j.1550-7408.1997.tb05944.x. [DOI] [PubMed] [Google Scholar]
- 152.Theus S A, Andrews R P, Steele P, Walzer P D. Adoptive transfer of lymphocytes sensitized to the major surface glycoprotein of Pneumocystis carinii confers protection in the rat. J Clin Investig. 1995;95:2587–2593. doi: 10.1172/JCI117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Theus S A, Smulian A G, Steele P, Linke M J, Walzer P D. Immunization with the major surface glycoprotein of Pneumocystis carinii elicits a protective response. Vaccine. 1998;16:1149–1157. doi: 10.1016/s0264-410x(98)80113-8. [DOI] [PubMed] [Google Scholar]
- 154.Theus S A, Walzer P D. Adoptive transfer of specific lymphocyte populations sensitized to the major surface glycoprotein of Pneumocystis carinii decreases organism burden while increasing survival rate in the rat. J Eukaryot Microbiol. 1997;44:23S–24S. doi: 10.1111/j.1550-7408.1997.tb05750.x. [DOI] [PubMed] [Google Scholar]
- 155.Thon G, Baltz T, Giroud C, Eisen H. Trypanosome variable surface glycoproteins: composite genes and order of expression. Genes Dev. 1990;4:1374–1383. doi: 10.1101/gad.4.8.1374. [DOI] [PubMed] [Google Scholar]
- 156.Turner C M, Barry J D. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- 157.Turner M J. Trypanosome variant suface glycoprotein. In: Englund P E, Sher A, editors. The biology of parasitism. New York, N.Y: Alan R. Liss; 1988. pp. 349–369. [Google Scholar]
- 158.Underwood A P, Louis E J, Borts R H, Stringer J R, Wakefield A E. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol Microbiol. 1996;19:273–281. doi: 10.1046/j.1365-2958.1996.374904.x. [DOI] [PubMed] [Google Scholar]
- 159.Van der Ploeg L H, Bernards A, Rijsewijk F A, Borst P. Characterization of the DNA duplication-transposition that controls the expression of two genes for variant surface glycoproteins in Trypanosoma brucei. Nucleio Acids Res. 1982;10:593–609. doi: 10.1093/nar/10.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Van Xong H, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 162.Vargas S L, Hughes W T, Wakefield A E, Oz H S. Limited persistence in and subsequent elimination of Pneumocystis carinii from the lungs after P. carinii pneumonia. J Infect Dis. 1995;172:506–510. doi: 10.1093/infdis/172.2.506. [DOI] [PubMed] [Google Scholar]
- 163.Vargas S L, Ponce C A, Gigliotti F, Ulloa A V, Prieto S, Munoz M P, Hughes W T. Transmission of Pneumocystis carinii DNA from a patient with P. carinii pneumonia to immunocompetent contact health care workers. J Clin Microbiol. 2000;38:1536–1538. doi: 10.1128/jcm.38.4.1536-1538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vargas S L, Ponce C A, Hughes W T, Wakefield A E, Weitz J C, Donoso S, Ulloa A V, Madrid P, Gould S, Latorre J J, Avila R, Benveniste S, Gallo M, Belletti J, Lopez R. Association of primary Pneumocystis carinii infection and sudden infant death syndrome. Clin Infect Dis. 1999;29:1489–1493. doi: 10.1086/313521. [DOI] [PubMed] [Google Scholar]
- 165.Vasquez J, Smulian A G, Linke M J, Cushion M T. Antigenic differences associated with genetically distinct Pneumocystis carinii from rats. Infect Immun. 1996;64:290–297. doi: 10.1128/iai.64.1.290-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wada M, Kitada K, Saito M, Egawa K, Nakamura Y. cDNA sequence diversity and genomic clusters of major surface glycoprotein genes of Pneumocystis carinii. J Infect Dis. 1993;168:979–985. doi: 10.1093/infdis/168.4.979. [DOI] [PubMed] [Google Scholar]
- 167.Wada M, Nakamura Y. MSG gene cluster encoding major cell surface glycoproteins of rat Pneumocystis carinii. DNA Res. 1994;1:163–168. doi: 10.1093/dnares/1.4.163. [DOI] [PubMed] [Google Scholar]
- 168.Wada M, Nakamura Y. Type-II major-surface-glycoprotein family of Pneumocystis carinii under the control of novel expression elements. DNA Res. 1999;6:211–217. doi: 10.1093/dnares/6.4.211. [DOI] [PubMed] [Google Scholar]
- 169.Wada M, Nakamura Y. Immunological characterization of surface subtilisin-like protease (SSP) of Pneumocystis carinii. J Eukaryot Microbiol. 1999;46:151S–152S. [PubMed] [Google Scholar]
- 170.Wada M, Sunkin S M, Stringer J R, Nakamura Y. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J Infect Dis. 1995;171:1563–1568. doi: 10.1093/infdis/171.6.1563. [DOI] [PubMed] [Google Scholar]
- 171.Wakefield A E. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wakefield A E. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol Med Microbiol. 1998;22:5–13. doi: 10.1111/j.1574-695X.1998.tb01182.x. [DOI] [PubMed] [Google Scholar]
- 173.Walzer P D, Linke M J. A comparison of the antigenic characteristics of rat and human Pneumocystis carinii by immunoblotting. J Immunol. 1987;138:2257–2265. [PubMed] [Google Scholar]
- 174.Walzer P D, Perl D P, Krogstad D J, Rawson P G, Schultz M G. Pneumocystis carinii penumonia in the United States. Epidemiologic, diagnostic, and clinical features. Ann Intern Med. 1974;80:83–93. doi: 10.7326/0003-4819-80-1-83. [DOI] [PubMed] [Google Scholar]
- 175.Walzer P D, Powell R D, Jr, Yoneda K. Experimental Pneumocystis carinii pneumonia in different strains of cortisonized mice. Infect Immun. 1979;24:939–947. doi: 10.1128/iai.24.3.939-947.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Walzer P D, Schnelle V, Armstrong D, Rosen P P. Nude mouse: a new experimental model for Pneumocystis carinii infection. Science. 1977;197:177–179. doi: 10.1126/science.301657. [DOI] [PubMed] [Google Scholar]
- 177.Walzer P D, Stanforth D, Linke M J, Cushion M T. Pneumocystis carinii: immunoblotting and immunofluorescent analyses of serum antibodies during experimental rat infection and recovery. Exp Parasitol. 1987;63:319–328. doi: 10.1016/0014-4894(87)90179-2. [DOI] [PubMed] [Google Scholar]
- 178.Weisbroth S H, Geistfeld J, Weisbroth S P, Williams B, Feldman S H, Linke M J, Orr S, Cushion M T. Latent Pneumocystis carinii infection in commercial rat colonies: comparison of inductive immunosuppressants plus histopathology, PCR, and serology as detection methods. J Clin Microbiol. 1999;37:1441–1446. doi: 10.1128/jcm.37.5.1441-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wright T W, Bissoondial T Y, Haidaris C G, Gigliotti F, Haidaris P J. Isoform diversity and tandem duplication of the glycoprotein A gene in ferret Pneumocystis carinii. DNA Res. 1995;2:77–88. doi: 10.1093/dnares/2.2.77. [DOI] [PubMed] [Google Scholar]
- 180.Yoganathan T, Lin H, Buck G A. An electrophoretic karyotype and assignment of ribosomal genes to resolved chromosomes of Pneumocystis carinii. Mol Microbiol. 1989;3:1473–1480. doi: 10.1111/j.1365-2958.1989.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 181.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 182.Zhang J R, Norris S J. Kinetics and in vivo induction of genetic variation of v1sE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. . (Erratum 67:468, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zimmerman P E, Voelker D R, McCormack F X, Paulsrud J R, Martin W J. 120-kD surface glycoprotein of Pneumocystis carinii is a ligand for surfactant protein A. J Clin Investig. 1992;89:143–149. doi: 10.1172/JCI115554. [DOI] [PMC free article] [PubMed] [Google Scholar]